Abstract
The transcription factor nuclear factor erythroid 2-related factor 2 (Nrf2) regulates various cellular activities, including redox balance, detoxification, metabolism, autophagy, proliferation, and apoptosis. Several studies have demonstrated that Nrf2 regulates hepatocyte proliferation during liver regeneration. The aim of this study was to investigate how Nrf2 modulates the cell cycle of replicating hepatocytes in regenerating livers. Wild-type and Nrf2 null mice were subjected to 2/3 partial hepatectomy (PH) and killed at multiple time points for various analyses. Nrf2 null mice exhibited delayed liver regrowth, although the lost liver mass was eventually restored 7 days after PH. Nrf2 deficiency did not affect the number of hepatocytes entering the cell cycle but did delay hepatocyte mitosis. Mechanistically, the lack of Nrf2 resulted in increased mRNA and protein levels of hepatic cyclin A2 when the remaining hepatocytes were replicating in response to PH. Moreover, Nrf2 deficiency in regenerating livers caused dysregulation of Wee1, Cdc2, and cyclin B1 mRNA and protein expression, leading to decreased Cdc2 activity. Thus, Nrf2 is required for timely M phase entry of replicating hepatocytes by ensuring proper regulation of cyclin A2 and the Wee1/Cdc2/cyclin B1 pathway during liver regeneration.
Keywords: nuclear factor erythroid 2-related factor 2, hepatocyte mitosis, hepatocyte proliferation
nuclear factor erythroid 2-related factor 2 (Nrf2) is a basic leucine zipper region-containing transcription factor that is abundantly expressed in many tissues such as the liver (6). The activity of Nrf2 is regulated by multiple mechanisms, including gene transcription, kelch-like ECH-associated protein 1-mediated proteasome degradation, and kinase-mediated phosphorylation. The sequential events leading to Nrf2 activation include nuclear translocation, heterodimer formation with small Maf portions, binding to an antioxidant response element (ARE), and transactivation of Nrf2 target genes. There are both direct and indirect Nrf2 target genes involved in various cellular functions, including detoxification, metabolism, autophagy, proliferation, differentiation, and apoptosis (3, 13, 17, 26). Previous studies have demonstrated that Nrf2 regulates the hepatocyte proliferative response to liver mass loss (2, 33). The aim of the present study was to investigate how Nrf2 modulates the cell cycle progression of replicating hepatocytes during liver repair.
Partial hepatectomy (PH) induces highly synchronized entry and progression of the cell cycle in residual hepatocytes. Thus, PH is widely used as an in vivo model to study the regulation of cell proliferation (23). We performed PH on wild-type and Nrf2 null mice and compared the cell cycle progression of replicating hepatocytes. Our data indicate that Nrf2 is a regulator of hepatocyte mitosis.
MATERIALS AND METHODS
Mice and PH.
The animals used for this study were wild-type and Nrf2-deficient male mice (3 mo old) with a C57BL6/129SV mixed background (6). The mice were housed in plastic cages at 22 ± 1°C on a 12:12-h light-dark cycle with lights on from 6:00 A.M. to 6:00 P.M. Standard rodent chow and water were provided ad libitum throughout the entire feeding period. A standard 70% PH was performed between 10:00 A.M. and 12:00 P.M. according to the procedure described previously (7, 12). The gall bladders were kept intact. All of the animal experiments were conducted in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals. The protocols for the care and use of animals were approved by the Indiana University-Purdue University Indianapolis Animal Care and Use Committee.
Immunohistochemistry.
Formalin-fixed and paraffin-embedded liver sections were subjected to standard immunohistochemistry. Ki-67 immunostaining was performed to visualize and count the proliferating hepatocytes. Hematoxylin- and eosin-stained liver sections were used to quantify mitotic figures in hepatocytes. The Ki-67-positive hepatocytes and mitotic figures were counted in five randomly chosen microscope fields per section at ×200 and ×100 magnifications, respectively.
Western blot analysis.
Liver homogenates (10 or 30 μg) were separated by polyacrylamide gel electrophoresis under reducing conditions. The proteins were then electrophoretically transferred to polyvinylidene difluoride membranes. The following antibodies were used as probes: cyclin D1 (no. 2922) and cyclin B1 (no. 4138) (Cell Signaling Technology, Danvers, MA); cyclin A2 (1540-1) and NQO-1 (2618-1) (Epitomics, Burlingame, CA); cyclin E1 (SC-481), Wee1 (SC-9037), p-Cdc2 p34 (Tyr15) (SC-7898), Cdc2 p34 (SC-54), and glyceraldehyde 3-phosphate dehydrogenase (SC-25778) (Santa Cruz Biotechnology, Santa Cruz, CA). The immune complexes were detected using an enhanced chemiluminescence system (Pierce, Rockford, IL).
Quantitative real-time polymerase chain reaction.
TRIzol reagent was used to prepare total RNA from frozen liver tissue according to the manufacturer's protocol (Invitrogen, Carlsbad, CA). The cDNAs were synthesized from total RNA (1 μg) of each sample using a Verso cDNA Kit (Thermo Scientific). The cDNAs were diluted four times with water and subjected to quantitative real-time polymerase chain reaction to quantify mRNA levels. TaqMan Universal PCR Master Mix, primers, and TaqMan MGB probes of mouse cyclin A2 (Mm00438063_m1), cyclin B1 (Mm03053893_gh), Cdc2 (Mm00772472_m1), Wee1 (Mm00494175_ml), and β-actin (Mm00607939_s1) were purchased from Applied Biosystems (Foster City, CA). The amplification reactions were performed with the ABI Prism 7900 sequence detection system (Applied Biosystems) with initial hold steps (50°C for 2 min followed by 95°C for 10 min) and 40 cycles of a two-step PCR (92°C for 15 s and 60°C for 1 min). The comparative CT method was used for the relative quantification of the amount of mRNA in each sample. The data were normalized to the β-actin transcript levels.
Transient transfection and luciferase reporter activity assay.
Hepa-1 cells were cultured in Dulbecco's modified Eagle's medium (Mediatech) supplemented with 10% fetal bovine serum (Biomeda) at 37°C in a 5% CO2 atmosphere. The cells were plated in 24-well plates at ∼1 × 105 cells/well. After overnight culture, FuGENE6 (Promega, Madison, WI) was used to transfect a mcyclin A2-Luc construct (300 ng/well) harboring a firefly luciferase reporter driven by a 700-bp 5′-flanking fragment of mouse cyclin A2 gene (kindly provided by Dr. Jean Marine Blanchard, University of Montpellier, Montpellier, France) and a mouse Nrf2 expression plasmid (50 ng/well) or a balance plasmid (pGL-3 basic vector, 50 ng/well). A Renilla luciferase expression vector (10 ng/well) was cotransfected as an internal control for normalization of transfection efficiency. A hHO1ARE-Luc reporter construct (300 ng/well) containing six copies of an antioxidant response element (ARE) identified in the human heme oxygenase 1 gene was used as a positive control reporter. The cells were harvested 48 h after treatment, and then firefly and Renilla luciferase activities were measured using the Dual Luciferase Reporter Assay System (Promega).
Cdc2 kinase activity assay.
Cdc2 protein was immunoprecipitated from each liver lysate, and the kinase activity was determined by a kinase assay kit. Briefly, each liver sample was homogenized in a lysis buffer (T-PER Tissue Protein Extraction Reagent; Thermo Scientific) containing a protease and phosphatase inhibitor cocktail (Halt Protease & Phosphatase Inhibitor Single-Use Cocktail; Pierce) and incubated on ice for 30 min. The homogenate was centrifuged at 10,000 g for 10 min at 4°C. The protein concentration of the supernatant was measured with Pierce 660 nm Protein Assay Reagent (Thermo Scientific). A portion of the supernatant containing 100 μg of liver protein was adjusted to 300 μl with PBS and incubated with 20 μl of agarose-conjugated mouse monoclonal anti-Cdc2 antibody (SC-54 AC; Santa Cruz Biotechnology) with gentle shaking overnight at 4°C. The immunoprecipitate was collected by centrifugation at 1,000 g and washed with 1 ml of PBS four times. Cdc2 kinase activity was evaluated with the Cdc2-cyclin B kinase assay kit according to the manufacturer's instructions (CycLex, Nagano, Japan). The average value for wild-type mice at 24 h after PH was set to one.
Statistical analysis.
The data are shown as means ± SD. All statistical analyses were performed using a one-way analysis of variance. The comparisons of means were determined by post hoc analysis. Significant differences were defined when P < 0.05.
RESULTS
Nrf2 deficiency causes the impairment of liver regrowth.
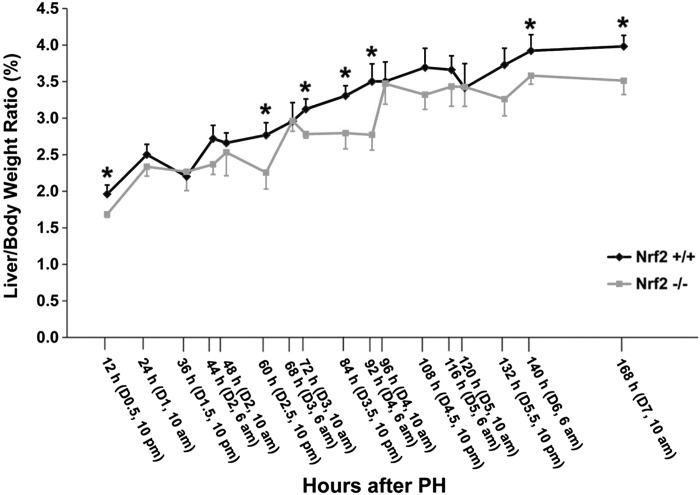
Mice lacking Nrf2 have a reduced liver size and a lower liver-to-body weight ratio than wild-type mice (4.52 ± 0.09 vs. 4.91 ± 0.10; P < 0.05). This observation is consistent with previous reports (2, 15). After 70% PH, the remaining livers in each genotype of mice regenerated to the original liver mass (Fig. 1).1 In wild-type mice, each wave of hepatocyte replication drove a further increase in the liver mass. Thus, the regenerating livers exhibited a gradual and consistent increase from 44 h (the first peak of hepatocyte mitosis) to 116 h (the last peak of hepatocyte mitosis) after PH. However, Nrf2 knockout mice showed a sluggish liver regrowth pattern compared with wild-type mice. The liver regrowth in Nrf2 mutants was reduced at 60 h but normal at 68 h. Additionally, the mutants had reduced liver growth again at 92 h but normal liver size at 96 h after PH. Thus, the Nrf2-deficient mice exhibited an impaired regenerative response to liver mass loss. The results indicate that Nrf2 is required for the normal progression of liver regeneration.
Fig. 1.
Liver regrowth following partial hepatectomy (PH) in nuclear factor erythroid 2-related factor 2 (Nrf2)-sufficient (+/+) and -deficient (Nrf2−/−) mice. Mice were subjected to PH and killed at the indicated time points. Liver-to-body weight ratios were used as a liver regrowth index. D, day. Results are presented as the mean liver-to-body weight ratios ± SD (n = 3–5 mice/time point for each genotype; *P < 0.05).
The lack of Nrf2 results in a delay in hepatocyte mitosis during the first wave of hepatocyte replication following PH.
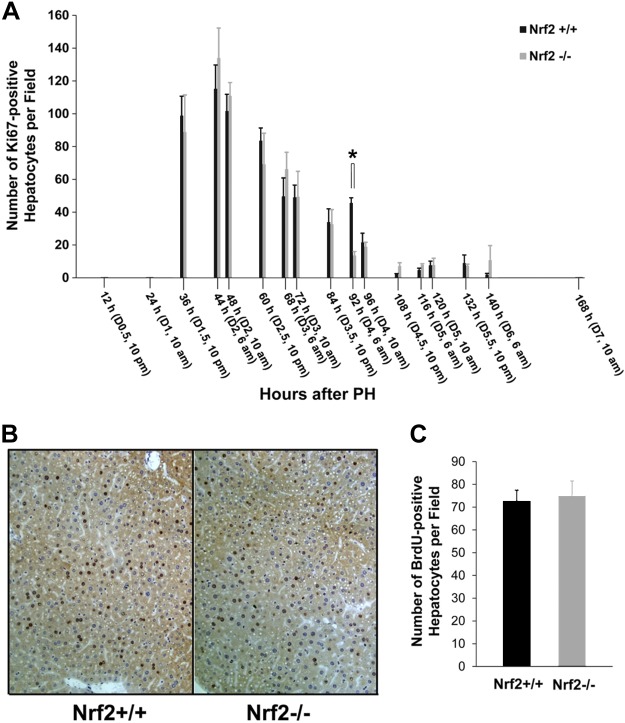
PH-induced liver regeneration consists of four consecutive waves of hepatocyte replication that are indicated by four rhythmic mitosis peaks (36). We first evaluated the numbers of Ki-67-positive hepatocytes at various time points after PH in the two genotypes of mice. This assessment allowed us to determine whether the absence of Nrf2 affects the total number of hepatocytes undergoing each wave of the cell cycle because Ki-67 is expressed in each active phase of the cell cycle. We observed that there were significantly fewer proliferative hepatocytes at 92 h after PH in Nrf2 null mice than in their wild-type littermates (Fig. 2A). This result indicates that the Nrf2 null mutation inhibits hepatocyte replication during the third wave of the hepatic proliferative cycle (84–108 h post-PH).
Fig. 2.
Nos. of proliferating hepatocytes after PH in Nrf2+/+ and Nrf2−/− mice. Mice were subjected to PH and killed at the indicated time points. A: assessment of total proliferating hepatocytes. Ki-67 immunostaining was performed with liver sections. Ki-67-positive hepatocytes were counted at ×200 magnification in 5 randomly chosen fields/section. Results are shown as the means per field ± SD (n = 3–5 mice/time point for each genotype; *P < 0.05). B and C: no. of hepatocytes undergoing DNA synthesis at 36 h after PH. Mice were killed at 36 h post-PH. One hour before death, bromodeoxyuridine (BrdU) was injected (100 mg/kg ip). Liver sections were subjected to BrdU immunostaining. B: representative liver sections show BrdU-positive hepatocytes. C: BrdU-positive hepatocytes were counted at ×100 magnification in 5 randomly chosen fields/section. Data are shown as the means per field ± SD (n = 3–5 mice/time point for each genotype).
To investigate cell cycle progression, we counted bromodeoxyuridine (BrdU)-positive (S phase) hepatocytes at 36 h following PH. At this time point, DNA synthesis peaks during the first and major wave of hepatocyte proliferation in PH-induced liver regeneration (7, 10, 21, 23, 36). Thus, a significant difference in the numbers of hepatocytes undergoing DNA synthesis was not observed when the two strains of mice were compared (Fig. 2, B and C). These data demonstrate that Nrf2 deficiency does not significantly affect S phase entry and progression in replicating hepatocytes during the first round of the cell cycle after PH.
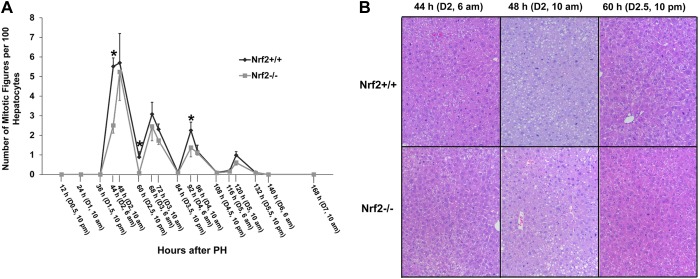
We then evaluated the numbers of hepatocytes undergoing mitosis (M phase hepatocytes) throughout the course of liver regrowth by quantifying hepatocyte mitotic figures. As shown in Fig. 3, during the first wave of hepatocyte replication (24–60 h after PH), the number of mitotic hepatocytes in Nrf2 null mutants was approximately half of that in wild types at 44 h. The number of mitotic hepatocytes subsequently became comparable with the wild type at 48 h. These data indicate that the lack of Nrf2 causes a delay in hepatocyte mitosis during the initial and strongest wave of hepatocyte replication. As the liver regeneration progressed through the remaining three hepatocyte proliferative cycles, the M phase hepatocytes were significantly reduced in Nrf2 nulls compared with wild types at 92 h following PH. This finding is consistent with the Ki-67 assessment at the same time point (Fig. 2A). Thus, without Nrf2, the third wave of hepatocyte proliferation (84–108 h after PH) was suppressed.
Fig. 3.
Nos. of hepatocytes undergoing mitosis after PH in Nrf2+/+ and Nrf2−/− mice. Mice were subjected to PH and killed at the indicated time points. Liver sections were stained with hematoxylin and eosin. A: hepatocytes and hepatic mitotic figures were counted at ×100 magnification in 5 randomly chosen fields/liver section. Data are shown as the means of the nos. of mitotic hepatocytes/100 hepatocytes ± SD (n = 3–5 mice/time point for each genotype; *P < 0.05). B: representative liver sections showing mitotic figures in hepatocytes at the indicated time points following PH.
Nrf2 activity progressively increases during liver regeneration.
NAD(P)H-quinone oxidoreductase 1 (NQO-1) is a typical target gene of Nrf2 (5). During liver injury, both the mRNA and protein expression of the gene is solely regulated by Nrf2 (1). Thus, we evaluated hepatic NQO-1 protein expression to estimate the activity of Nrf2 in regenerating livers (Fig. 4A). The result showed that, in wild-type mice, the protein level of hepatic NQO-1 gradually increased as liver regrowth progressed. In contrast, the protein was only weakly detected in both resting and regenerating livers in Nrf2 null mice. This observation indicates that Nrf2 activity is gradually upregulated, suggesting that progressively increased Nrf2 activity may be required for sustained liver regeneration. In addition, our data further prove that NQO-1 is a reliable marker for Nrf2 activity because the loss of Nrf2 results in almost diminished NQO-1 expression when the liver undergoes a variety of biological changes in response to PH.
Fig. 4.
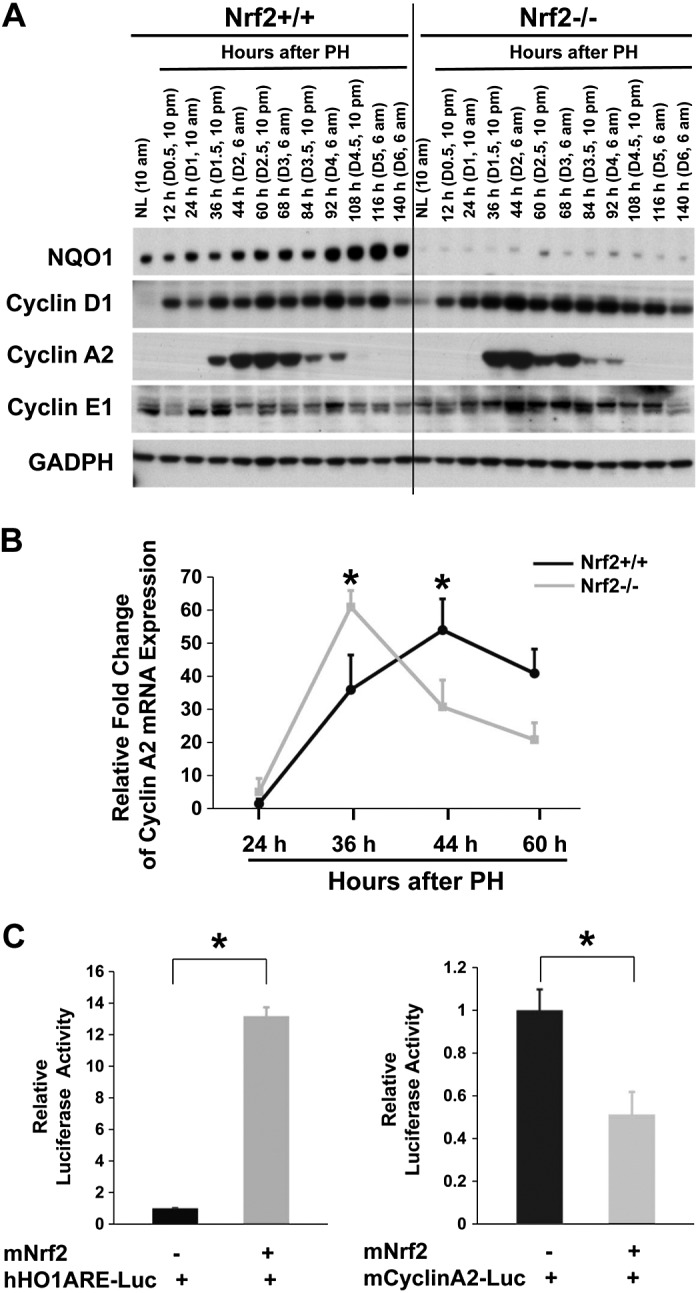
A: protein expression of a subset of cell cycle components in regenerating livers of Nrf2+/+ and Nrf2−/− mice. Livers were collected from normal mice and mice subjected to PH at the indicated time points after surgery. Liver lysates prepared from 3 mice/time point for each genotype were pooled with equal amounts of proteins from each preparation. Western blotting was performed with antibodies against the proteins indicated. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used as a loading control. NL, normal liver. B: mRNA expression of cyclin A2 in regenerating livers. Total RNA was prepared from the livers at the indicated time points after PH. Hepatic mRNA levels of cyclin A2 were measured by quantitative real-time polymerase chain reaction (qRT-PCR) and are expressed as the mean fold change relative to the mRNA level at 24 h post-PH in Nrf2+/+ mice ± SD (n = 3 mice/time point for each genotype; *P < 0.05 between Nrf2+/+ and Nrf2−/− mice). C: Nrf2 negatively regulates cyclin A2 transcription. A Nrf2 expression vector (mNRF2) and a mouse cyclin A2 promoter reporter construct (mcyclinA2-Luc) or a construct containing 6 copies of an antioxidant response element (ARE) of human heme oxygenase 1 gene (hHO1ARE-Luc, positive control) were cotransfected into Hepa-1 cells as described in materials and methods. Note that Nrf2 overexpression increased the activity of the ARE in hHO1ARE-Luc but reduced the activity of the mouse cyclin A2 promoter. Data are representative results from 3 independent experiments. *P < 0.05.
Nrf2 absence leads to the dysregulation of a subset of cell cycle components in regenerating livers.
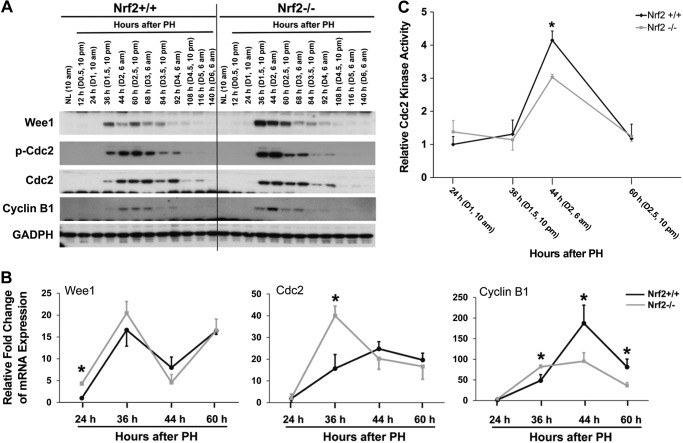
We analyzed the protein expression of major cell cycle components in regenerating livers in wild-type and Nrf2 null mice to evaluate the impact of Nrf2 ablation on hepatocyte proliferation at the molecular level. As observed in Fig. 4A, the lack of Nrf2 affected the levels but not the protein expression kinetics of hepatic cyclins D1, A2, and E1. Most notably, Nrf2 null mutants accumulated more cyclin A2 protein at 36 h following PH than wild types. This observation suggests that Nrf2 plays an inhibitory role in the initiation of cyclin A2 protein expression and thus that the absence of Nrf2 leads to increased cyclin A2 protein synthesis in regenerating livers.
We next quantified the mRNA expression of hepatic cyclin A2 during the first wave of hepatocyte replication to determine whether the Nrf2-dependent dysregulation of cyclin A2 expression occurs at the mRNA level. As shown in Fig. 4B, the level of cyclin A2 mRNA peaked at 36 h after PH in Nrf2 null livers but at 44 h in wild-type regenerating livers. The data indicate a stronger initiation of cyclin A2 transcription in cells lacking Nrf2. This result is consistent with the observation at the protein level. Furthermore, a transient transfection assay showed that overexpression of Nrf2 reduced the activity of the mouse cyclin A2 promoter (Fig. 4C). Collectively, these data demonstrate that Nrf2 is a transcriptional suppressor of cyclin A2.
Nrf2 null mutation causes the dysregulation of the Wee1/Cdc2/cyclin B1 pathway during the first round of hepatocyte replication after PH.
The appropriate regulation of the Wee1/Cdc2/cyclin B1 pathway is crucial for a normal G2/M transition during the cell cycle. Increased phosphorylation of Cdc2 at Tyr15 by Wee1 kinase reduces the activity of Cdc2/cyclin B complex and leads to the inhibition of M phase entry (11). During liver regeneration, this pathway regulates the timing of mitosis in replicating hepatocytes (22). Therefore, we postulated that the dysregulation of this pathway may cause the delayed M phase entry of proliferating hepatocytes lacking Nrf2. To address this hypothesis, we first examined the mRNA and protein expression of the components in this pathway in the resting and regenerating livers of wild-type and Nrf2 null mice. We found that Wee1, Cdc2, and cyclin B1 were altered at both the mRNA and protein levels when Nrf2 was absent (Fig. 5, A and B). Moreover, the lack of Nrf2 led to the hyperphosphorylation of Cdc2 protein at 36 and 44 h after PH, which is consistent with the excessive accumulation of Wee1 caused by Nrf2 deficiency. The data demonstrate that Nrf2 deficiency results in the dysregulation of Wee1, Cdc2, and cyclin B1. These findings prompted us to analyze the activity of Cdc2 in the regenerating livers of both genotypes of mice. We found that Nrf2 absence caused a significant decrease in Cdc2 activity during hepatocyte mitosis (44 h following PH) (Fig. 5C). Thus, these data link the dysregulated Wee1/Cdc2/cyclin B1 pathway and decreased Cdc2 activity to delayed mitosis during liver regeneration in Nrf2 null mice.
Fig. 5.
A: protein expression of Wee1, Cdc2, and cyclin B1 in regenerating livers of Nrf2+/+ and Nrf2−/− mice. The same set of protein samples described in Fig. 4A was used for Western blotting with antibodies against the proteins indicated. GAPDH was used as a loading control. B: mRNA expression of Wee1, Cdc2, and cyclin B1 in regenerating livers of Nrf2+/+ and Nrf2−/− mice. The same set of total RNA samples described in Fig. 4B was used. Hepatic mRNA levels of Wee1, Cdc2, and cyclin B1 were measured by qRT-PCR and are expressed as the mean fold change relative to mRNA levels at 24 h post-PH in Nrf2+/+ mice ± SD (n = 3 mice/time point for each genotype; *P < 0.05 between Nrf2+/+ and Nrf2−/− mice). C: Cdc2 kinase activity in regenerating livers of Nrf2+/+ and Nrf2−/− mice. Liver lysates were prepared from both genotypes of mice at the indicated time points following PH. A Cdc2 kinase activity assay was performed as described in materials and methods. The average value for Nrf2+/+ mice at 24 h after PH was set to 1. Results are presented as the mean fold change relative to data at 24 h post-PH in Nrf2+/+ mice ± SD (n = 3 mice/time point for each genotype; *P < 0.05 between Nrf2+/+ and Nrf2−/− mice).
DISCUSSION
The current study demonstrates that Nrf2 is a novel regulator of hepatocyte mitosis during liver regeneration. We found that the absence of Nrf2 does not affect S phase entry and progression but entry into M phase in regenerating hepatocytes. Mechanistically, we revealed that Nrf2 is indispensable for the proper regulation of cyclin A2 and the Wee1/Cdc2/cyclin B1 pathway during liver regeneration. As reported in our previous publication, the absence of Nrf2 leads to increased levels of cyclin A2 mRNA and protein in the maternal liver exhibiting hepatocyte proliferation during gestation, which demonstrates that Nrf2 is a cyclin A2 transcriptional suppressor (37). In this study, we found that Nrf2 deficiency results in elevated cyclin A2 mRNA and protein expression in the regenerating liver. Our previous and current findings collectively indicate that Nrf2 may function as a brake to prevent the overactivation of cyclin A2 transcription and translation when hepatocytes go through the cell cycle. It was previously demonstrated that timely degradation of cyclin A is a key event in M phase progression because nondegradable mutations in cyclin A result in cell arrest in metaphase (29, 32). Thus, the delayed mitosis in replicating hepatocytes caused by Nrf2 null mutation is at least partially due to the excessive accumulation of cyclin A2. Moreover, we found that Nrf2 genetic deletion dysregulates the components of the Wee1/Cdc2/cyclin B1 pathway. When Nrf2 is absent, each component in this pathway is abnormally regulated at the mRNA and/or protein level. These pathway alterations lead to an increased amount of Wee1 kinase, hyperphosphorylation of its substrate Cdc2 at Tyr15, and reduced Cdc2 activity. It has been well established that the activity of the Cdc2/cyclin B complex is essential for M phase entry (11). Thus, the dysregulation of this pathway also contributes to the delayed mitosis in hepatocytes lacking Nrf2. Collectively, these data demonstrate that Nrf2 modulates cell cycle progression in regenerating hepatocytes by ensuring the appropriate regulation of cyclin A2 and the Wee1/Cdc2/cyclin B1 pathway.
Nrf2 is highly activated in various tumor cells and promotes their proliferation through poorly understood mechanisms (31). Evidence shows that Nrf2 may increase K-Ras-induced proliferation and tumorigenesis (9). Moreover, Nrf2 regulates metabolic reprogramming by redirecting glucose and glutamine into anabolic pathways in cancer cells to support cancer cell proliferation (27). Future studies must determine whether these Nrf2-dependent mechanisms also operate during normal hepatocyte replication, or vice versa, whether Nrf2 regulates cyclin A2 and the Wee1/Cdc2/cyclin B pathway in tumor cells. Of note, Nrf2 null mice are highly susceptible to chemical-induced carcinogenesis (16, 19, 30, 34). The misregulation of mitotic entry is often oncogenic (35). Therefore, it is possible that the regenerative response to chemical-induced tissue injury in cells deficient in Nrf2 prevents timely M phase entry, which promotes tumor formation.
Previous reports and our findings indicate that Nrf2 plays multiple roles in a stage-dependent manner during liver regeneration. It has been shown that Nrf2 absence increases NF-kB activation, reduces insulin signaling, and enhances hepatocyte apoptosis in regenerating livers at 3 and/or 6 h after PH. The previous study has also reported aggravated steatosis at 48 h post-PH; aberrant numbers of S phase hepatocytes at 48, 72, and 120 h; and elevated oxidative stress at 72 h in Nrf2 null regenerating livers (2). Notch1 and the augmenter of liver regeneration have been identified as Nrf2 direct target genes in regenerating livers (8, 33). Additionally, the Sestrin2-mediated Nrf2, mTOR, and p53/p21 signaling network has been proposed to regulate hepatocyte proliferation in response to liver injury (4). We recently reported that Nrf2 is required to maintain newly regenerated hepatocytes in a fully differentiated state. We found that the lack of Nrf2 causes transient but massive impairment in hepatocyte identity 60 h following PH, which is indicated by marked reduction in hepatocyte volume and strong activation of progenitor marker gene expression (38). This observation may suggest a phenomenon of hepatocyte dedifferentiation caused by Nrf2 absence, which correlates with liver regrowth inhibition at the same time point after PH in Nrf2 null mice (Fig. 1). In this study, we found that Nrf2 deficiency results in a delay in hepatocyte mitosis due to dysregulation of cyclin A2 and the Wee1/Cdc2/cyclin B1 pathway during the first wave of hepatocyte replication following PH. Moreover, we also found that Nrf2 activity is gradually increased during liver regeneration (Fig. 4A). This may be induced by elevated oxidative stress at later stages of liver regeneration (2) and might be required to maintain hepatocytes in a differentiated state during the post-PH remodeling period (38). This hypothesis should be tested in future studies. Collectively, these studies demonstrate the requirement of Nrf2 for normal liver regeneration. However, Nrf2 activation exerts negative effects on liver regeneration. Weiner's group generated transgenic mice restricting the expression of an Nrf2 mutant lacking the Keap1 binding domain in hepatocytes (20). Using this mouse model, the authors demonstrated that constitutive activation of Nrf2 results in a delay in hepatocyte proliferation and an increase in hepatocyte apoptosis within 72 h after PH without affecting the final restoration of the lost liver mass. Mechanistically, this is due to temporary upregulation of the expression of the Nrf2 direct target genes p15 and Bcl2l11. Our recent report demonstrates that Nrf2 is not activated when hepatocytes undergo the first round of replication following PH. In addition, Keap1 knockdown-induced Nrf2 activation disrupts the redox cycle and hepatocyte cell cycle progression in the regenerating liver (14). Clearly, Nrf2 activity needs to be tightly controlled during liver regeneration because both overly activated and deficient Nrf2 activity impairs the hepatic regenerative response. The results from studies concerning the roles for Nrf2 in liver regeneration support the notion that Nrf2 functions as “a double-edge sword” in distinct pathological states (24, 31).
PH induces sequential proliferation of hepatic parenchyma and nonparenchyma cells, dynamic adjustments in hepatocyte size, liver progenitor cell activation, and hepatic tissue remodeling (18, 25, 28, 38). Liver-to-body weight ratio is widely used as a parameter for general assessment of liver size or regrowth following PH. We found that genetic deletion of Nrf2 caused inhibition of liver regrowth at multiple time points post-PH (Fig. 1). We recently reported that the lack of Nrf2 leads to reductions in hepatocyte size at 60 h (day 2.5) and 140 h (day 6) after PH (38). This observation can explain the decreases in liver-to-body weight ratio at the same time points following PH (Fig. 1). Furthermore, we observed that Nrf2 absence caused a decrease in hepatocyte proliferation at 92 h after PH (Figs. 2 and 3), which should contribute to concurrent suppression in liver regrowth (Fig. 1). However, Nrf2 null mice exhibited lower liver-to-body weight ratios at several additional time points (12, 72, 84, and 168 h) post-PH without significant differences in hepatocyte volume and proliferation compared with wild-type controls (Fig. 1) (38). The data suggest uncovered roles for Nrf2 during liver regeneration that need to be explored in future studies.
GRANTS
This work was supported by a grant from the National Institute of Diabetes and Digestive and Kidney Diseases (7RO1-DK-07596).
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the authors.
AUTHOR CONTRIBUTIONS
Author contributions: Y.Z., J.Y.C., and G.D. conception and design of research; Y.Z., M.H., J.L., S.M.N., V.G., and Q.B. performed experiments; Y.Z., M.H., J.L., S.M.N., V.G., Q.B., and G.D. analyzed data; Y.Z. and G.D. interpreted results of experiments; Y.Z. and G.D. prepared figures; Y.Z. and G.D. drafted manuscript; Y.Z. and G.D. edited and revised manuscript; Y.Z. and G.D. approved final version of manuscript.
Footnotes
REFERENCES
- 1.Aleksunes LM, Slitt AL, Maher JM, Dieter MZ, Knight TR, Goedken M, Cherrington NJ, Chan JY, Klaassen CD, Manautou JE. Nuclear factor-E2-related factor 2 expression in liver is critical for induction of NAD(P)H:quinone oxidoreductase 1 during cholestasis. Cell Stress Chaperones 11: 356–363, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beyer TA, Xu W, Teupser D, auf dem Keller U, Bugnon P, Hildt E, Thiery J, Kan YW, Werner S. Impaired liver regeneration in Nrf2 knockout mice: role of ROS-mediated insulin/IGF-1 resistance. EMBO J 27: 212–223, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bryan HK, Olayanju A, Goldring CE, Park BK. The Nrf2 cell defence pathway: Keap1-dependent and -independent mechanisms of regulation. Biochem Pharmacol 85: 705–717, 2013. [DOI] [PubMed] [Google Scholar]
- 4.Buitrago-Molina LE, Marhenke S, Longerich T, Sharma AD, Boukouris AE, Geffers R, Guigas B, Manns MP, Vogel A. The degree of liver injury determines the role of p21 in liver regeneration and hepatocarcinogenesis in mice. Hepatology 58: 1143–1152, 2013. [DOI] [PubMed] [Google Scholar]
- 5.Chan K, Han XD, Kan YW. An important function of Nrf2 in combating oxidative stress: detoxification of acetaminophen. Proc Natl Acad Sci USA 98: 4611–4616, 2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chan K, Lu R, Chang JC, Kan YW. NRF2, a member of the NFE2 family of transcription factors, is not essential for murine erythropoiesis, growth, and development. Proc Natl Acad Sci USA 93: 13943–13948, 1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dai G, He L, Bu P, Wan YJ. Pregnane X receptor is essential for normal progression of liver regeneration. Hepatology 47: 1277–1287, 2008. [DOI] [PubMed] [Google Scholar]
- 8.Dayoub R, Vogel A, Schuett J, Lupke M, Spieker SM, Kettern N, Hildt E, Melter M, Weiss TS. Nrf2 activates augmenter of liver regeneration (ALR) via antioxidant response element and links oxidative stress to liver regeneration. Mol Med 19: 237–244, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.DeNicola GM, Karreth FA, Humpton TJ, Gopinathan A, Wei C, Frese K, Mangal D, Yu KH, Yeo CJ, Calhoun ES, Scrimieri F, Winter JM, Hruban RH, Iacobuzio-Donahue C, Kern SE, Blair IA, Tuveson DA. Oncogene-induced Nrf2 transcription promotes ROS detoxification and tumorigenesis. Nature 475: 106–109, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fausto N, Campbell JS, Riehle KJ. Liver regeneration. Hepatology 43: S45–S53, 2006. [DOI] [PubMed] [Google Scholar]
- 11.Fisher D, Krasinska L, Coudreuse D, Novak B. Phosphorylation network dynamics in the control of cell cycle transitions. J Cell Sci 125: 4703–4711, 2012. [DOI] [PubMed] [Google Scholar]
- 12.Greene AK, Puder M. Partial hepatectomy in the mouse: technique and perioperative management. J Invest Surg 16: 99–102, 2003. [PubMed] [Google Scholar]
- 13.Hayes JD, Dinkova-Kostova AT. The Nrf2 regulatory network provides an interface between redox and intermediary metabolism. Trends Biochem Sci 39: 199–218, 2014. [DOI] [PubMed] [Google Scholar]
- 14.Hu R, Saw CL, Yu R, Kong AN. Regulation of NF-E2-related factor 2 signaling for cancer chemoprevention: antioxidant coupled with antiinflammatory. Antioxid Redox Signal 13: 1679–1698, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang J, Tabbi-Anneni I, Gunda V, Wang L. Transcription factor Nrf2 regulates SHP and lipogenic gene expression in hepatic lipid metabolism. Am J Physiol Gastrointest Liver Physiol 299: G1211–G1221, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Iida K, Itoh K, Kumagai Y, Oyasu R, Hattori K, Kawai K, Shimazui T, Akaza H, Yamamoto M. Nrf2 is essential for the chemopreventive efficacy of oltipraz against urinary bladder carcinogenesis. Cancer Res 64: 6424–6431, 2004. [DOI] [PubMed] [Google Scholar]
- 17.Jaramillo MC, Zhang DD. The emerging role of the Nrf2-Keap1 signaling pathway in cancer. Genes Dev 27: 2179–2191, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Karaca G, Swiderska-Syn M, Xie G, Syn WK, Kruger L, Machado MV, Garman K, Choi SS, Michelotti GA, Burkly LC, Ochoa B, Diehl AM. TWEAK/Fn14 signaling is required for liver regeneration after partial hepatectomy in mice. PLoS ONE 9: e83987, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Khor TO, Huang MT, Prawan A, Liu Y, Hao X, Yu S, Cheung WK, Chan JY, Reddy BS, Yang CS, Kong AN. Increased susceptibility of Nrf2 knockout mice to colitis-associated colorectal cancer. Cancer Prev Res 1: 187–191, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kohler UA, Kurinna S, Schwitter D, Marti A, Schafer M, Hellerbrand C, Speicher T, Werner S. Activated Nrf2 impairs liver regeneration in mice by activation of genes involved in cell cycle control and apoptosis. Hepatology 60: 670–678, 2014. [DOI] [PubMed] [Google Scholar]
- 21.Liao Y, Shikapwashya ON, Shteyer E, Dieckgraefe BK, Hruz PW, Rudnick DA. Delayed hepatocellular mitotic progression and impaired liver regeneration in early growth response-1-deficient mice. J Biol Chem 279: 43107–43116, 2004. [DOI] [PubMed] [Google Scholar]
- 22.Matsuo T, Yamaguchi S, Mitsui S, Emi A, Shimoda F, Okamura H. Control mechanism of the circadian clock for timing of cell division in vivo. Science 302: 255–259, 2003. [DOI] [PubMed] [Google Scholar]
- 23.Michalopoulos GK. Liver regeneration after partial hepatectomy: critical analysis of mechanistic dilemmas. Am J Pathol 176: 2–13, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Michalopoulos GK. NRF2, not always friendly but perhaps misunderstood. Hepatology 60: 461–463, 2014. [DOI] [PubMed] [Google Scholar]
- 25.Michalopoulos GK, DeFrances M. Liver regeneration. Adv Biochem Eng Biotechnol 93: 101–134, 2005. [DOI] [PubMed] [Google Scholar]
- 26.Mitsuishi Y, Motohashi H, Yamamoto M. The Keap1-Nrf2 system in cancers: stress response and anabolic metabolism (Abstract). Frontier Oncol 2: 200, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mitsuishi Y, Taguchi K, Kawatani Y, Shibata T, Nukiwa T, Aburatani H, Yamamoto M, Motohashi H. Nrf2 redirects glucose and glutamine into anabolic pathways in metabolic reprogramming. Cancer Cell 22: 66–79, 2012. [DOI] [PubMed] [Google Scholar]
- 28.Miyaoka Y, Ebato K, Kato H, Arakawa S, Shimizu S, Miyajima A. Hypertrophy and unconventional cell division of hepatocytes underlie liver regeneration. Curr Biol 22: 1166–1175, 2012. [DOI] [PubMed] [Google Scholar]
- 29.Parry DH, O'Farrell PH. The schedule of destruction of three mitotic cyclins can dictate the timing of events during exit from mitosis. Curr Biol 11: 671–683, 2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ramos-Gomez M, Kwak MK, Dolan PM, Itoh K, Yamamoto M, Talalay P, Kensler TW. Sensitivity to carcinogenesis is increased and chemoprotective efficacy of enzyme inducers is lost in nrf2 transcription factor-deficient mice. Proc Natl Acad Sci USA 98: 3410–3415, 2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sporn MB, Liby KT. NRF2 and cancer: the good, the bad and the importance of context. Nat Rev 12: 564–571, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sullivan M, Morgan DO. Finishing mitosis, one step at a time. Nat Rev Mol Cell Biol 8: 894–903, 2007. [DOI] [PubMed] [Google Scholar]
- 33.Wakabayashi N, Shin S, Slocum SL, Agoston ES, Wakabayashi J, Kwak MK, Misra V, Biswal S, Yamamoto M, Kensler TW. Regulation of notch1 signaling by nrf2: implications for tissue regeneration (Abstract). Sci Signal 3: 52, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xu C, Huang MT, Shen G, Yuan X, Lin W, Khor TO, Conney AH, Kong AN. Inhibition of 7,12-dimethylbenz(a)anthracene-induced skin tumorigenesis in C57BL/6 mice by sulforaphane is mediated by nuclear factor E2-related factor 2. Cancer Res 66: 8293–8296, 2006. [DOI] [PubMed] [Google Scholar]
- 35.Yasutis KM, Kozminski KG. Cell cycle checkpoint regulators reach a zillion. Cell Cycle 12: 1501–1509, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zou Y, Bao Q, Kumar S, Hu M, Wang GY, Dai G. Four waves of hepatocyte proliferation linked with three waves of hepatic fat accumulation during partial hepatectomy-induced liver regeneration. PLoS ONE 7: e30675, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zou Y, Hu M, Bao Q, Chan JY, Dai G. Nrf2 participates in regulating maternal hepatic adaptations to pregnancy. J Cell Sci 126: 1618–1625, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zou Y, Lee J, Nambiar SM, Hu M, Rui W, Bao Q, Chan JY, Dai G. Nrf2 Is Involved in Maintaining Hepatocyte Identity during Liver Regeneration. PLoS ONE 9: e107423, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]