ABSTRACT
Nuclear pore complexes (NPCs) are indispensable for cell function and are at the center of several human diseases. NPCs provide access to the nucleus and regulate the transport of proteins and RNA across the nuclear envelope. They are aqueous channels generated from a complex network of evolutionarily conserved proteins known as nucleporins. In this Cell Science at a Glance article and the accompanying poster, we discuss how transport between the nucleoplasm and the cytoplasm is regulated, what we currently know about the structure of individual nucleoporins and the assembled NPC, and how the cell regulates assembly and disassembly of such a massive structure. Our aim is to provide a general overview on what we currently know about the nuclear pore and point out directions of research this area is heading to.
KEY WORDS: Nuclear pore complex, Nucleus, Transport
Introduction
A defining difference between eukaryotic and prokaryotic cells is the evolution of an endomembrane system and the presence of a nucleus, resulting in the separation of the genetic material from the rest of the cell. By restricting access to the nucleus and separating gene transcription from protein translation, eukaryotic cells have evolved to control gene expression in a highly regulated manner. Although separated by the double-membrane nuclear envelope, the interior of the nucleus is not completely isolated. Embedded throughout the nuclear envelope are large protein complexes known as nuclear pore complexes (NPCs) that sit in circular openings where the outer nuclear membrane is fused with the inner nuclear membrane. Proteins and RNA can cross the nuclear envelope in a tightly regulated process by traveling through these aqueous protein channels. NPCs are considered to be the gatekeepers of the nucleus and facilitate almost all transport between the nucleoplasm and cytoplasm.
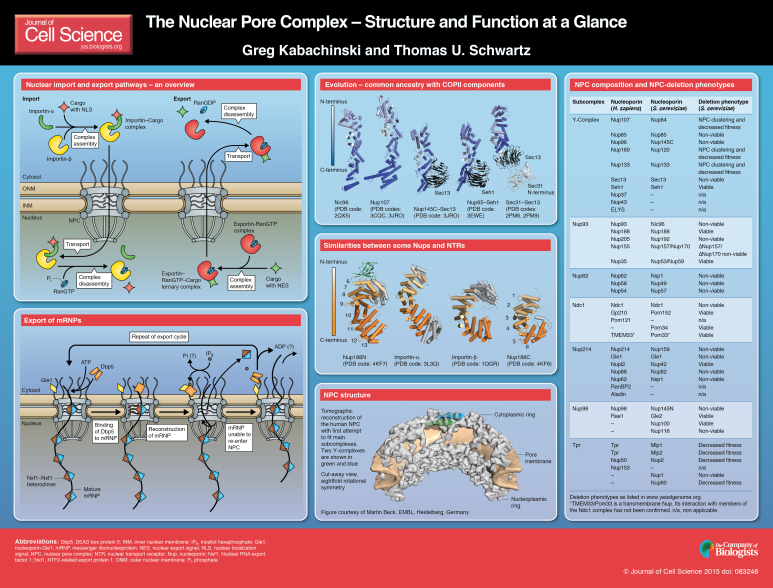
The nuclei of HeLa cells – the commonly used human tissue culture cell line – contain on average 3000 NPCs (Dultz and Ellenberg, 2010; Maul et al., 1972). A single NPC comprises ∼500, mainly evolutionarily conserved, individual protein molecules that are collectively known as nucleoporins (Nups) (Alber et al., 2007). A fully assembled human NPC has an estimated molecular mass of ∼125 MDa (Reichelt et al., 1990), making it one of the largest protein complexes in the cell. Despite its size, the NPC is generated from a limited number of Nups (∼30) that appear in multiple copies (see poster, ‘NPC composition and NPC-deletion phenotypes'). Since the discovery of the NPC more than 60 years ago (Callan and Tomlin, 1950), much has been learned about its composition, structure and function.
In this Cell Science at a Glance article and accompanying poster, we examine how nuclear transport is regulated and highlight the key differences between the transport requirements of proteins and different types of RNA. We discuss some common protein folds found in many Nups and how they contribute to the overall NPC structure and barrier function, before discussing recent discoveries that show evolutionary links between Nups and vesicle coats, and Nups and nuclear transport receptors (NTRs) (see Box 1). Finally, we will review how the cell orchestrates and regulates the assembly of the NPC at different times in the cell cycle. For the purpose of readability, we predominantly use the human nucleoporin nomenclature to address those evolutionarily conserved properties of the NPC that are likely to apply to NPCs of all eukaryotes; we use the fungal nomenclature only when addressing specific aspects that are different when comparing eukaryotic lineages. A brief overview of NPC function, structure and assembly is provided, and more-detailed reviews regarding specific aspects of NPC biology are referenced throughout the text.
Box 1. Evolution of the NPC.
In addition to the architectural information gained from high-resolution structural studies of the NPC, these experiments have also shed light on the evolution of Nups and the NPC as a whole. It has been hypothesized that the NPC shares common ancestry with vesicle coat complexes, including COPI, COPII and clathrin (Devos et al., 2004). This hypothesis was initially supported by predictions that similar elements of protein structures are found in both vesicle coats and NPCs – mainly β-propellers and α-helical solenoids – and the fact that Sec13 is a bona fide member of both the NPC and a vesicle coat complex (COPII). The proposed evolution from a common ancestor gained further support from high-resolution structural studies, which found that – despite having less sequence similarity – several Nups (Nup85, Nup96, Nup93 and Nup107) as well as vesicle coat proteins (Sec31 and Sec16) contain a conserved tripartite element, the so-called ancestral coatomer element 1 (ACE1) (Brohawn et al., 2008).
Recently, an additional evolutionary link between the NPC and NTRs has been uncovered. The defining characteristic of the karyopherin protein family, which includes importin-α, importin-β and the exportins, is that they all contain superhelical stacked α-helical units, which allows them to interact with FG-repeats and shuttle cargo through the NPC. Structural analysis of Nup188 (M. thermophile) and its mammalian homolog Nup205 (Nup192 in S. cerevisiae and Chaetomium thermophilium), members of the Nup93 subcomplex, showed they also adopt a structure of stacked α-helices (Andersen et al., 2013; Flemming et al., 2012; Sampathkumar et al., 2013; Stuwe et al., 2014). Such superhelical structures are abundant in eukaryotic cells and are used in many functional contexts. However, similar to karyopherins, Nup188 and Nup205/192 were both shown to specifically bind to FG-repeats and to be able to transverse the NPC, characteristics typically associated with NTRs (Andersen et al., 2013).
The evolutionary links between the NPC, coat-proteins and transport receptors that have been provided by high-resolution structural studies might help to better understand how the NPC is formed, how Nups interact with each other and, ultimately, how each Nup functions in nuclear transport.
The nuclear import–export cycle
Whereas metabolites, ions and molecules smaller than ∼40 kDa can pass freely across the nuclear envelope, larger macromolecules (e.g. proteins, mRNA, tRNA, ribosome subunits and viruses) typically cannot diffuse, but must be actively transported through the NPC (Görlich and Kutay, 1999; Weis, 2003). Each class of macromolecule has a specific way in which it is transported across the nuclear envelope.
The import and export of proteins across the nuclear membrane is regulated by a cycle of interactions between protein cargo, NTRs (e.g. importins, exportins, transportins and karyopherins) and the small GTPase Ran, which regulates the ability of both importins and exportins to transport their cargo (see poster ‘Nuclear import and export pathways – an overview’). For a protein to actively pass through the NPC, it must contain a nuclear localization signal (NLS) sequence. Although the NLS can be complex, the classic NLS is a stretch of basic residues (i.e. KKKRK) (Kalderon et al., 1984). Simply adding an NLS to a non-nuclear protein is often sufficient to localize that protein into the nucleus (Goldfarb et al., 1986). Recognition of the NLS of a cargo is the first step in assembling an import-complex. Canonical nuclear import involves the recognition of the NLS by the adaptor protein importin-α, followed by binding of the karyopherin importin-β, thereby forming a trimeric import complex (Cook et al., 2007; Fried and Kutay, 2003; Stewart, 2007b). Importin-β acts as the transport factor and carries the cargo through the NPC. In other cases, such as when the cargo contains an atypical NLS, importin-β binds to the cargo directly (Cingolani et al., 2002; Lee et al., 2006; Lee et al., 2003). Through either direct interaction with the NLS or through an adaptor protein, the NTRs ultimately determine which cargo is permitted to pass through the nuclear pore.
Fluorescent microscopy studies have shown that translocation through the pore is a rapid process with first-order kinetics, which occurs at a rate of ∼1000 translocations every second (Ribbeck and Görlich, 2001; Yang et al., 2004). Interestingly, the pore itself does not determine the directionality of the import complex. In fact, the movement of the import complex within the pore appears to be random. The directionality of cargo import is determined by a gradient of nuclear RanGTP. Once an import complex enters the nucleus, RanGTP binds importin-β, which releases the cargo. The importin-β–RanGTP complex itself is transported back to the cytosol, disassembled by GTP hydrolysis and ready for the next round of import. Because a dynamic equilibrium of the import complex is maintained in the pore, disassembly of the complex in the nucleus results in a net flow of cargo towards the nucleoplasm. This directional flow is dependent on a RanGTP gradient, whereby the concentration of RanGTP is greater in the nucleus than in the cytosol. This RanGTP gradient is maintained by a Ran guanine nucleotide exchange factor (GEF) that is preferentially located in the nucleus and by RanGTPase-activating proteins (GAPs) in the cytosol (Stewart, 2007b).
Conceptually, the export of proteins from the nucleus occurs in an process that is analogous to nuclear import but reversed (see poster, ‘Nuclear import and export pathways – an overview’). An export complex forms inside the nucleus between cargo displaying a nuclear export signal (NES) – typically a sequence that is leucine rich – a cognate export karyopherin and RanGTP (Ossareh-Nazari et al., 2001). This ternary export complex enters the NPC and, upon exiting the nuclear pore, encounters RanGAP – the Ran-specific GTPase-activating protein – that catalyzes GTP hydrolysis, resulting in disassembly and the release of the cargo (Cook et al., 2007). Also here, the established RanGTP gradient is the driving force for the directionality and nuclear export of proteins.
RNA export
The export of some classes of RNA is similar to the export of proteins. For example, tRNAs and small nuclear RNAs (snRNAs) use the RanGTP gradient and are transported through the NPC by their specific karyopherins exportin-t and Crm1, respectively (Cook and Conti, 2010; Rodriguez et al., 2004). The export of mature ribosomal subunits is also dependent on the RanGTP gradient and uses karyopherin-like transport receptors. However, because of the size of the ribosome, the precise process is still under intense investigation and, overall, remains poorly understood (Panse and Johnson, 2010; Tschochner and Hurt, 2003; Zemp and Kutay, 2007).
The export of mRNA is considerably different from that of proteins and other RNAs. mRNA is not exported alone but, instead, as a large messenger ribonucleoprotein (mRNP) complex, in which a single mRNA molecule is surrounded by hundreds of proteins that have a function in processing, capping, splicing and polyadenylation. The export of mRNPs, thus, presents the cell with a new set of challenges because (1) the diameter of the mRNP cargo is extremely large, (2) the cell must be able to distinguish between correctly (mature) and incorrectly (immature) packaged RNPs, and (3) the mRNA within the RNP may adopt topologies that need to be remodeled before translation can occur (Grünwald et al., 2011).
To overcome these unique challenges, cells have developed a separate mode for transport of mRNPs through the NPC. Before an mRNP particle can enter the NPC, a quality control step is performed by members of the TRAMP (Trf4–Air2–Mtr4p polyadenylation) and exosome protein complexes, which survey mRNA, and identify and degrade any defective mRNPs (Chlebowski et al., 2013; Makino et al., 2013). A mature mRNP is then recruited to the nucleoplasmic side of the NPC or – more specifically – the nuclear basket (see below) by the TREX2 (transcription-export complex 2) and THO complexes. This occurs co-transcriptionally and TREX2 and THO complexes are, therefore, essential in linking active gene transcription to mRNA export (Köhler and Hurt, 2007).
Once a mature mRNP is assembled and targeted to the NPC, it is transported through the channel by a non-karyopherin transport receptor, the Nxf1–Nxt1 heterodimer (Mex67-Mtr2 in yeast) (Grüter et al., 1998; Segref et al., 1997; Stewart, 2010). Although the exact stoichiometry is unknown, several Nxf1–Nxt1 heterodimers bind to the mRNP. Nxf1 and Mex67 neither bind to Ran nor do they use the RanGTP gradient that is crucial for protein transport. Instead, mRNA export (see poster, ‘Export of mRNPs’) is driven by ATP rather than GTP hydrolysis. The energy is required to establish transport directionality by remodeling the mRNP once it reaches the cytoplasm (Stewart, 2007a). Inside the central NPC channel, the mRNP can move forward and backward. However, once part of the mRNP reaches the cytoplasmic face of the NPC, the DEAD-box RNA helicase Dbp5 (also known as SON in humans), whose activity is regulated by the nucleoporin Gle1 and inositol hexaphosphate (IP6), binds to the mRNA and alters the structure of the mRNP, thereby removing the transport receptor Nxf1–Nxt1 heterodimer in an ATP-dependent manner (Montpetit et al., 2011; Tran et al., 2007; Weirich et al., 2006). Removal of Nxf1–Nxt1 prevents the respective part of the mRNP from returning to the central channel. By repeating this process, the mRNP is fully extracted out of the pore into the cytoplasm. For a more in-depth review of the process, see (Bonnet and Palancade, 2014; Oeffinger and Zenklusen, 2012).
The FG barrier
The NPC is an extremely versatile protein complex as it has to enable the selective transport of both proteins and mature RNA with sizes that range from 40 kDa to entire ribosomal subunits, while preventing other molecules of similar sizes from passing. How does the NPC create a barrier while regulating nuclear transport? This question still remains an intensely debated topic in the field. One issue that is agreed upon is that the regulatory function of the NPC is achieved by a subset of specific Nups, collectively known as FG-Nups. FG-Nups typically contain a structured domain that serves as an NPC anchor point, from which a largely unstructured, filamentous and hydrophilic extension emanates, which is studded with multiple (5–50) hydrophopic phenylalanine–glycine (FG)-repeats (Terry and Wente, 2009).
Over the past decade, a number of models have been proposed on how FG-Nups form the transport barrier (Lim et al., 2007; Patel et al., 2007; Peters, 2005; Ribbeck et al., 2002; Rout et al., 2003). Recent work has provided strong evidence for the so-called ‘selective-phase model’ (Hülsmann et al., 2012; Labokha et al., 2013). In this model, FG-Nups line the central channel and extend their FG-repeat regions into the middle of the channel. The high local concentration of FG-repeats due to the many FG-Nups that localize to the channel (∼200 FG-Nups per channel) generates a hydrogel, in which the FG-repeats bind cohesively to form a ‘sieve’ with mesh size of ∼5 nm. A macromolecule larger than ∼5 nm (∼40 kDa) passes through this barrier by means of a transport receptor that has the ability to bind the FG-repeats, thereby locally ‘melting’ the FG-mediated sieve. The selective-phase model has shortcomings because it does not yet explain the interplay between the different FG nucleoporins; moreover, it is largely based on in vitro data using one FG-protein at the time. Also, many FG-Nups are – for so-far-unknown reasons – heavily glycosylated. Finally, the influence of NTRs as possible constitutive elements of the FG barrier is still mostly unexplored. Thus, the FG barrier remains a central research topic that is passionately and controversially discussed (Atkinson et al., 2013; Kapinos et al., 2014; Peters, 2009; Yamada et al., 2010; Zilman et al., 2010).
NPC structure
The NPC is one of the largest protein complexes in the cell and easily recognizable using scanning electron microscopy (EM). Early EM studies revealed an eightfold rotational ring symmetry for the entire structure. In addition, the main NPC structure contains rings that are situated on its cytoplasmic and nucleoplasmic sides, giving the NPC an apparent twofold symmetry across the nuclear membrane (Beck et al., 2004; Stoffler et al., 2003). A recent electron tomographic study has delineated the structure of the core NPC scaffold at a resolution of 3.2 nm (Bui et al., 2013). This work, together with other studies, shows that the NPC has a thickness of ∼50 nm, an outer diameter of ∼80–120 nm and an inner diameter of ∼40 nm (Bui et al., 2013; Grossman et al., 2012; Maimon et al., 2012). In addition, the obtained images reveal that the NPC contains a structure, resembling a basket that extends into the nucleoplasm (the nuclear basket) and filaments that extend into the cytoplasm (referred to as cytoplasmic filaments).
Despite its enormous size, the NPC is made up of only ∼30 Nups that are largely conserved throughout eukaryotic evolution (Cronshaw et al., 2002; DeGrasse et al., 2009; Rout et al., 2000). Nups are organized into subcomplexes that are biochemically defined by their affinity to each other. In humans, the Nup62 complex comprises Nup62, Nup58 (Uniprot ID: Q9BVL2) and Nup54, all of which contain FG-repeats and are found in the central pore (Finlay et al., 1991; Grandi et al., 1993; Hu et al., 1996) (see poster, ‘NPC composition and NPC-deletion phenotypes’, for corresponding Nup nomenclature in budding yeast Saccharomyces cerevisiae). Nup62 is also a member of the human Nup214 subcomplex, where it interacts with Nup88 and the FG-containing Nup214, and is located on the cytoplasmic side of the NPC (Fornerod et al., 1997; Macaulay et al., 1995). Two other essential subcomplexes provide the structural scaffold for the entire NPC and serve as adaptor proteins that link the FG-Nups to the nuclear membrane. The first is the Y-complex that, in several EM studies, was shown to be elongated and branched, resembling the letter Y – hence its name (Bui et al., 2013; Kampmann and Blobel, 2009; Lutzmann et al., 2002). In humans, this subcomplex contains ten proteins, Nup107, Nup85, Nup96, Nup160, Nup133, Sec13, Seh1, Nup37, Nup43 and ELYS (Loïodice et al., 2004; Lutzmann et al., 2002; Rasala et al., 2006; Siniossoglou et al., 1996). Another NPC subcomplex is the Nup93 complex, which comprises Nup93, Nup188, Nup205, Nup155 and Nup35 in humans (Vollmer and Antonin, 2014). Although the structure of the entire Nup93 subcomplex is still unknown, the individual structures of all its components have been published. Surprisingly, even though the NPC spans two membranes (the outer- and inner-nuclear membrane), only four of the human Nups contain transmembrane (TM) domains, namely Ndc1, Gp210, TMEM33 and Pom121. Biochemical data indicate that these TM proteins connect to the NPC scaffold through the Nup93 complex (Eisenhardt et al., 2014; Mitchell et al., 2010; Yavuz et al., 2010).
A high-resolution structure of the NPC, in which the position of each individual protein is resolved, is a formidable goal for structural biologists. Efforts over the past decade have shown that a hybrid approach using different experimental and computational methods is likely to be needed to fulfill this goal (Alber et al., 2007; Bui et al., 2013).
Assembly and disassembly of the NPC
In addition to being one of the largest protein complexes in the cell, the structural scaffold of the NPC is also one of the longest lived (Savas et al., 2012). Whereas most mammalian proteins have an average half-life of a few days (Cambridge et al., 2011; Price et al., 2010), recent whole animal pulse-chase experiments demonstrated that, in postmitotic tissues, the structural scaffold of the NPC, or at least components thereof, persists over the entire lifetime of a cell (Toyama et al., 2013). In dividing cells, NPCs undergo a cycle of assembly and disassembly that is in concert with the cell cycle. NPC assembly occurs during two stages of the cell cycle, in interphase and immediately after mitosis.
During interphase, the nuclear envelope increases its surface area, and the number of NPCs doubles in preparation for mitosis and to allow the cell to handle the concomitant increase in transcription that necessitates additional mRNA export as well as synthesis and import of histones. These new NPCs form de novo and assemble from both sides of the intact nuclear envelope (D'Angelo et al., 2006).
A model for NPC assembly in interphase has been proposed on the basis of several studies, in which the members of the Nup93 subcomplex are first recruited to the TM-Nups (Flemming et al., 2009; Makio et al., 2009; Onischenko et al., 2009). This association, facilitated by membrane-deforming proteins (e.g. reticulons) then bends the outer- and inner-nuclear membrane toward each other until they eventually fuse (Dawson et al., 2009). Live-imaging kinetics studies have shown that members of the Y-complex also appear around this time (Dultz and Ellenberg, 2010). The localization of the Y-complex during assembly, and NPC assembly itself, is dependent on the components of the Ran cycle and importin-β (D'Angelo et al., 2006; Ryan and Wente, 2002; Ryan et al., 2003; Ryan et al., 2007). Finally, after all the scaffolding Nups have been assembled, the FG-Nups are localized to the NPC, generating the transport barrier.
For post-mitotic NPCs assembly it is important to consider the vast differences between the fate of the nuclear envelope in different organisms. In S. cerevisiae, the nuclear envelope remains closed throughout mitosis, as the microtubule organizing center (MTOC) is embedded in it. In many other organisms, MTOCs are cytoplasmic, which necessitates the nuclear envelope to break down for correct chromosome segregation to occur. Between these extremes of ‘open’ and ‘closed’ mitosis, there are a number of cell types that undergo variations of a ‘semi-closed’ mitosis (De Souza and Osmani, 2007; Güttinger et al., 2009). To what extend these variations affect NPC dissembly and/or reassembly, and how this might have generated fundamentally different post-mitotic versus interphase assembly pathways is an ongoing discussion (D'Angelo and Hetzer, 2008; Doucet et al., 2010). However, it is clear that NPCs are fully disassembled during open mitosis (Laurell et al., 2011) and partially disassembled during ‘semi-closed’ mitosis (De Souza et al., 2004).
Post-mitotic assembly of NPCs begins at the same time as the nuclear envelope starts to reform (Schooley et al., 2012). This assembly process is proposed to be initiated by the targeting of the Y-complex to chromatin, which is facilitated by small-DNA-binding motifs (AT hooks) that are predicted to be present in ELYS (Rasala et al., 2008). As with NPC assembly during interphase, Ran – as well as its effectors – and importin-β regulate Nup recruitment to chromatin (Harel et al., 2003; Walther et al., 2003). However, once ELYS is localized to chromatin, assembly continues with the remainder of the Nup107 subcomplex, which interacts with the TM-Nups Ndc1 and Pom121 (Dultz et al., 2008; Rasala et al., 2008). Members of the Nup93 subcomplex are then recruited soon after (Dultz et al., 2008) and thought to assist in membrane fusion and pore stabilization (Eisenhardt et al., 2014; Ródenas et al., 2009). Once all the scaffolding Nups have been incorporated into the pore, the FG-Nups are localized and nuclear pore activity is restored (Dultz et al., 2008).
Despite the overall stability of the NPC in non-dividing cells, NPCs must nevertheless disassemble in order for mitosis to proceed in dividing cells. NPC disassembly is fast and occurs in a stepwise-regulated manner. Although the exact steps and order in which they occur are currently unknown, disassembly does not appear to be a simple reversal of the assembly steps described above (Dultz et al., 2008). The trigger to initiate the disassembly process is the phosphorylation of several Nups by mitotic kinases (e.g. Cdk1) (Glavy et al., 2007; Laurell et al., 2011; Onischenko et al., 2005). Following their phosphorylation and release from the NPC, some nuclear pore subcomplexes also have additional roles in mitosis. For example, the Y subcomplex is localized to kinetochores, and regulates mitotic spindle assembly and chromosome congression (Orjalo et al., 2006; Zuccolo et al., 2007).
Perspectives
The NPC is a multifaceted and intricate protein complex that is essential to all eukaryotic life. In addition to the main function of the Nups in nucleocytoplasmic transport, new roles – especially in the regulation of gene expression – have recently been identified (Texari et al., 2013; Van de Vosse et al., 2013). Therefore, it is not surprising that aberrant functions of Nups and NPCs have been implicated in many and diverse human pathologies, including autoimmune diseases, viral infections, cardiomyopathies and various cancers (Capelson and Hetzer, 2009; Chow et al., 2012; Hatch and Hetzer, 2014; Simon and Rout, 2014). To better understand large, complicated protein complexes such as the NPC, a multipronged approach using hybrid methods is needed. Already, single-particle EM, EM tomography, super-resolution microscopy, crystallography, immunoprecipitation, crosslinking, mass spectrometry and numerous other methods are being used and combined to help to generate a highly detailed model of the NPC and to identify so-far-unknown Nup functions. Given the intensity of research in the field, it seems realistic that we might know the structure of the NPC, including the positions of all scaffold nucleoporins in the foreseeable future. This, in turn, will give researchers the opportunity to specifically address the many aspects of NPC biology, which will no doubt disclose exciting secrets of cell biology.
Supplementary Material
Acknowledgments
We apologize to the authors whose work we were unable to cite due to space limitations. We thank Allyson Anding for her help in generating some of the figures, Kasper Anderson for critically reading the manuscript and Martin Beck for providing the tomographic figure.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Funding
This work of our laboratory was supported by the National Institutes of Health [grant number GM077537] to T.U.S. Deposited in PMC for release after 12 months.
Cell science at a glance
A high-resolution version of the poster is available for downloading in the online version of this article at jcs.biologists.org. Individual poster panels are available as JPEG files at http://jcs.biologists.org/lookup/suppl/doi:10.1242/jcs.083246/-/DC2.
References
- Alber F., Dokudovskaya S., Veenhoff L. M., Zhang W., Kipper J., Devos D., Suprapto A., Karni-Schmidt O., Williams R., Chait B. T. et al. (2007). The molecular architecture of the nuclear pore complex. Nature 450, 695–701 10.1038/nature06405 [DOI] [PubMed] [Google Scholar]
- Andersen K. R., Onischenko E., Tang J. H., Kumar P., Chen J. Z., Ulrich A., Liphardt J. T., Weis K., Schwartz T. U. (2013). Scaffold nucleoporins Nup188 and Nup192 share structural and functional properties with nuclear transport receptors. eLife 2, e00745 10.7554/eLife.00745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkinson C. E., Mattheyses A. L., Kampmann M., Simon S. M. (2013). Conserved spatial organization of FG domains in the nuclear pore complex. Biophys. J. 104, 37–50 10.1016/j.bpj.2012.11.3823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck M., Förster F., Ecke M., Plitzko J. M., Melchior F., Gerisch G., Baumeister W., Medalia O. (2004). Nuclear pore complex structure and dynamics revealed by cryoelectron tomography. Science 306, 1387–1390 10.1126/science.1104808 [DOI] [PubMed] [Google Scholar]
- Bonnet A., Palancade B. (2014). Regulation of mRNA trafficking by nuclear pore complexes. Genes (Basel) 5, 767–791 10.3390/genes5030767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brohawn S. G., Leksa N. C., Spear E. D., Rajashankar K. R., Schwartz T. U. (2008). Structural evidence for common ancestry of the nuclear pore complex and vesicle coats. Science 322, 1369–1373 10.1126/science.1165886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bui K. H., von Appen A., DiGuilio A. L., Ori A., Sparks L., Mackmull M. T., Bock T., Hagen W., Andrés-Pons A., Glavy J. S. et al. (2013). Integrated structural analysis of the human nuclear pore complex scaffold. Cell 155, 1233–1243. [DOI] [PubMed] [Google Scholar]
- Callan H. G., Tomlin S. G. (1950). Experimental studies on amphibian oocyte nuclei. I. Investigation of the structure of the nuclear membrane by means of the electron microscope. Proc. R. Soc. B 137, 367–378 10.1098/rspb.1950.0047 [DOI] [PubMed] [Google Scholar]
- Cambridge S. B., Gnad F., Nguyen C., Bermejo J. L., Krüger M., Mann M. (2011). Systems-wide proteomic analysis in mammalian cells reveals conserved, functional protein turnover. J. Proteome Res. 10, 5275–5284 10.1021/pr101183k [DOI] [PubMed] [Google Scholar]
- Capelson M., Hetzer M. W. (2009). The role of nuclear pores in gene regulation, development and disease. EMBO Rep. 10, 697–705 10.1038/embor.2009.147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chlebowski A., Lubas M., Jensen T. H., Dziembowski A. (2013). RNA decay machines: the exosome. Biochim. Biophys. Acta 1829, 552–560. [DOI] [PubMed] [Google Scholar]
- Chow K-H., Factor R. E., Ullman K. S. (2012). The nuclear envelope environment and its cancer connections. Nat. Rev. Cancer 12, 196–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cingolani G., Bednenko J., Gillespie M. T., Gerace L. (2002). Molecular basis for the recognition of a nonclassical nuclear localization signal by importin beta. Mol. Cell 10, 1345–1353 10.1016/S1097-2765(02)00727-X [DOI] [PubMed] [Google Scholar]
- Cook A. G., Conti E. (2010). Nuclear export complexes in the frame. Curr. Opin. Struct. Biol. 20, 247–252 10.1016/j.sbi.2010.01.012 [DOI] [PubMed] [Google Scholar]
- Cook A., Bono F., Jinek M., Conti E. (2007). Structural biology of nucleocytoplasmic transport. Annu. Rev. Biochem. 76, 647–671 10.1146/annurev.biochem.76.052705.161529 [DOI] [PubMed] [Google Scholar]
- Cronshaw J. M., Krutchinsky A. N., Zhang W., Chait B. T., Matunis M. J. (2002). Proteomic analysis of the mammalian nuclear pore complex. J. Cell Biol. 158, 915–927 10.1083/jcb.200206106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Angelo M. A., Hetzer M. W. (2008). Structure, dynamics and function of nuclear pore complexes. Trends Cell Biol. 18, 456–466 10.1016/j.tcb.2008.07.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Angelo M. A., Anderson D. J., Richard E., Hetzer M. W. (2006). Nuclear pores form de novo from both sides of the nuclear envelope. Science 312, 440–443 10.1126/science.1124196 [DOI] [PubMed] [Google Scholar]
- Dawson T. R., Lazarus M. D., Hetzer M. W., Wente S. R. (2009). ER membrane-bending proteins are necessary for de novo nuclear pore formation. J. Cell Biol. 184, 659–675 10.1083/jcb.200806174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Souza C. P. C., Osmani S. A. (2007). Mitosis, not just open or closed. Eukaryot. Cell 6, 1521–1527 10.1128/EC.00178-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Souza C. P. C., Osmani A. H., Hashmi S. B., Osmani S. A. (2004). Partial nuclear pore complex disassembly during closed mitosis in Aspergillus nidulans. Curr. Biol. 14, 1973–1984 10.1016/j.cub.2004.10.050 [DOI] [PubMed] [Google Scholar]
- DeGrasse J. A., DuBois K. N., Devos D., Siegel T. N., Sali A., Field M. C., Rout M. P., Chait B. T. (2009). Evidence for a shared nuclear pore complex architecture that is conserved from the last common eukaryotic ancestor. Mol. Cell. Proteomics 8, 2119–2130 10.1074/mcp.M900038-MCP200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devos D., Dokudovskaya S., Alber F., Williams R., Chait B. T., Sali A., Rout M. P. (2004). Components of coated vesicles and nuclear pore complexes share a common molecular architecture. PLoS Biol. 2, e380 10.1371/journal.pbio.0020380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doucet C. M., Talamas J. A., Hetzer M. W. (2010). Cell cycle-dependent differences in nuclear pore complex assembly in metazoa. Cell 141, 1030–1041 10.1016/j.cell.2010.04.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dultz E., Ellenberg J. (2010). Live imaging of single nuclear pores reveals unique assembly kinetics and mechanism in interphase. J. Cell Biol. 191, 15–22 10.1083/jcb.201007076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dultz E., Zanin E., Wurzenberger C., Braun M., Rabut G., Sironi L., Ellenberg J. (2008). Systematic kinetic analysis of mitotic dis- and reassembly of the nuclear pore in living cells. J. Cell Biol. 180, 857–865 10.1083/jcb.200707026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenhardt N., Redolfi J., Antonin W. (2014). Interaction of Nup53 with Ndc1 and Nup155 is required for nuclear pore complex assembly. J. Cell Sci. 127, 908–921 10.1242/jcs.141739 [DOI] [PubMed] [Google Scholar]
- Finlay D. R., Meier E., Bradley P., Horecka J., Forbes D. J. (1991). A complex of nuclear pore proteins required for pore function. J. Cell Biol. 114, 169–183 10.1083/jcb.114.1.169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flemming D., Sarges P., Stelter P., Hellwig A., Böttcher B., Hurt E. (2009). Two structurally distinct domains of the nucleoporin Nup170 cooperate to tether a subset of nucleoporins to nuclear pores. J. Cell Biol. 185, 387–395 10.1083/jcb.200810016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flemming D., Devos D. P., Schwarz J., Amlacher S., Lutzmann M., Hurt E. (2012). Analysis of the yeast nucleoporin Nup188 reveals a conserved S-like structure with similarity to karyopherins. J. Struct. Biol. 177, 99–105 10.1016/j.jsb.2011.11.008 [DOI] [PubMed] [Google Scholar]
- Fornerod M., van Deursen J., van Baal S., Reynolds A., Davis D., Murti K. G., Fransen J., Grosveld G. (1997). The human homologue of yeast CRM1 is in a dynamic subcomplex with CAN/Nup214 and a novel nuclear pore component Nup88. EMBO J. 16, 807–816 10.1093/emboj/16.4.807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fried H., Kutay U. (2003). Nucleocytoplasmic transport: taking an inventory. Cell. Mol. Life Sci. 60, 1659–1688 10.1007/s00018-003-3070-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glavy J. S., Krutchinsky A. N., Cristea I. M., Berke I. C., Boehmer T., Blobel G., Chait B. T. (2007). Cell-cycle-dependent phosphorylation of the nuclear pore Nup107-160 subcomplex. Proc. Natl. Acad. Sci. USA 104, 3811–3816 10.1073/pnas.0700058104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldfarb D. S., Gariépy J. Schoolnik, G., and Kornberg, R. D(1986). Synthetic peptides as nuclear localization signals. Nature 322, 641–644 10.1038/322641a0 [DOI] [PubMed] [Google Scholar]
- Görlich D., Kutay U. (1999). Transport between the cell nucleus and the cytoplasm. Annu. Rev. Cell Dev. Biol. 15, 607–660 10.1146/annurev.cellbio.15.1.607 [DOI] [PubMed] [Google Scholar]
- Grandi P., Doye V., Hurt E. C. (1993). Purification of NSP1 reveals complex formation with ‘GLFG’ nucleoporins and a novel nuclear pore protein NIC96. EMBO J. 12, 3061–3071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossman E., Medalia O., Zwerger M. (2012). Functional architecture of the nuclear pore complex. Annu. Rev. Biophys. 41, 557–584 10.1146/annurev-biophys-050511-102328 [DOI] [PubMed] [Google Scholar]
- Grünwald D., Singer R. H., Rout M. (2011). Nuclear export dynamics of RNA-protein complexes. Nature 475, 333–341 10.1038/nature10318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grüter P., Tabernero C., von Kobbe C., Schmitt C., Saavedra C., Bachi A., Wilm M., Felber B. K., Izaurralde E. (1998). TAP, the human homolog of Mex67p, mediates CTE-dependent RNA export from the nucleus. Mol. Cell 1, 649–659 10.1016/S1097-2765(00)80065-9 [DOI] [PubMed] [Google Scholar]
- Güttinger S., Laurell E., Kutay U. (2009). Orchestrating nuclear envelope disassembly and reassembly during mitosis. Nat. Rev. Mol. Cell Biol. 10, 178–191 10.1038/nrm2641 [DOI] [PubMed] [Google Scholar]
- Harel A., Chan R. C., Lachish-Zalait A., Zimmerman E., Elbaum M., Forbes D. J. (2003). Importin beta negatively regulates nuclear membrane fusion and nuclear pore complex assembly. Mol. Biol. Cell 14, 4387–4396 10.1091/mbc.E03-05-0275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatch E., Hetzer M. (2014). Breaching the nuclear envelope in development and disease. J. Cell Biol. 205, 133–141 10.1083/jcb.201402003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu T., Guan T., Gerace L. (1996). Molecular and functional characterization of the p62 complex, an assembly of nuclear pore complex glycoproteins. J. Cell Biol. 134, 589–601 10.1083/jcb.134.3.589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hülsmann B. B., Labokha A. A., Görlich D. (2012). The permeability of reconstituted nuclear pores provides direct evidence for the selective phase model. Cell 150, 738–751 10.1016/j.cell.2012.07.019 [DOI] [PubMed] [Google Scholar]
- Kalderon D., Roberts B. L., Richardson W. D., Smith A. E. (1984). A short amino acid sequence able to specify nuclear location. 39, 499–509. [DOI] [PubMed] [Google Scholar]
- Kampmann M., Blobel G. (2009). Three-dimensional structure and flexibility of a membrane-coating module of the nuclear pore complex. Nat. Struct. Mol. Biol. 16, 782–788 10.1038/nsmb.1618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapinos L. E., Schoch R. L., Wagner R. S., Schleicher K. D., Lim R. Y. H. (2014). Karyopherin-centric control of nuclear pores based on molecular occupancy and kinetic analysis of multivalent binding with FG nucleoporins. Biophys. J. 106, 1751–1762 10.1016/j.bpj.2014.02.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köhler A., Hurt E. (2007). Exporting RNA from the nucleus to the cytoplasm. Nat. Rev. Mol. Cell Biol. 8, 761–773 10.1038/nrm2255 [DOI] [PubMed] [Google Scholar]
- Labokha A. A., Gradmann S., Frey S., Hülsmann B. B., Urlaub H., Baldus M., Görlich D. (2013). Systematic analysis of barrier-forming FG hydrogels from Xenopus nuclear pore complexes. EMBO J. 32, 204–218 10.1038/emboj.2012.302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurell E., Beck K., Krupina K., Theerthagiri G., Bodenmiller B., Horvath P., Aebersold R., Antonin W., Kutay U. (2011). Phosphorylation of Nup98 by multiple kinases is crucial for NPC disassembly during mitotic entry. Cell 144, 539–550 10.1016/j.cell.2011.01.012 [DOI] [PubMed] [Google Scholar]
- Lee S. J., Sekimoto T., Yamashita E., Nagoshi E., Nakagawa A., Imamoto N., Yoshimura M., Sakai H., Chong K. T., Tsukihara T. et al. (2003). The structure of importin-beta bound to SREBP-2: nuclear import of a transcription factor. Science 302, 1571–1575 10.1126/science.1088372 [DOI] [PubMed] [Google Scholar]
- Lee B. J., Cansizoglu A. E., Süel K. E., Louis T. H., Zhang Z., Chook Y. M. (2006). Rules for nuclear localization sequence recognition by karyopherin beta 2. Cell 126, 543–558 10.1016/j.cell.2006.05.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim R. Y. H., Fahrenkrog B., Köser J., Schwarz-Herion K., Deng J., Aebi U. (2007). Nanomechanical basis of selective gating by the nuclear pore complex. Science 318, 640–643 10.1126/science.1145980 [DOI] [PubMed] [Google Scholar]
- Loïodice I., Alves A., Rabut G., Van Overbeek M., Ellenberg J., Sibarita J-B., Doye V. (2004). The entire Nup107-160 complex, including three new members, is targeted as one entity to kinetochores in mitosis. Mol. Biol. Cell 15, 3333–3344 10.1091/mbc.E03-12-0878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutzmann M., Kunze R., Buerer A., Aebi U., Hurt E. (2002). Modular self-assembly of a Y-shaped multiprotein complex from seven nucleoporins. EMBO J. 21, 387–397 10.1093/emboj/21.3.387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macaulay C., Meier E., Forbes D. J. (1995). Differential mitotic phosphorylation of proteins of the nuclear pore complex. J. Biol. Chem. 270, 254–262 10.1074/jbc.270.1.254 [DOI] [PubMed] [Google Scholar]
- Maimon T., Elad N., Dahan I., Medalia O. (2012). The human nuclear pore complex as revealed by cryo-electron tomography. Structure 20, 998–1006 10.1016/j.str.2012.03.025 [DOI] [PubMed] [Google Scholar]
- Makino D. L., Halbach F., Conti E. (2013). The RNA exosome and proteasome: common principles of degradation control. Nat. Rev. Mol. Cell Biol. 14, 654–660 10.1038/nrm3657 [DOI] [PubMed] [Google Scholar]
- Makio T., Stanton L. H., Lin C-C., Goldfarb D. S., Weis K., Wozniak R. W. (2009). The nucleoporins Nup170p and Nup157p are essential for nuclear pore complex assembly. J. Cell Biol. 185, 459–473 10.1083/jcb.200810029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maul G. G., Maul H. M., Scogna J. E., Lieberman M. W., Stein G. S., Hsu B. Y., Borun T. W. (1972). Time sequence of nuclear pore formation in phytohemagglutinin-stimulated lymphocytes and in HeLa cells during the cell cycle. J. Cell Biol. 55, 433–447 10.1083/jcb.55.2.433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell J. M., Mansfeld J., Capitanio J., Kutay U., Wozniak R. W. (2010). Pom121 links two essential subcomplexes of the nuclear pore complex core to the membrane. J. Cell Biol. 191, 505–521 10.1083/jcb.201007098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montpetit B., Thomsen N. D., Helmke K. J., Seeliger M. A., Berger J. M., Weis K. (2011). A conserved mechanism of DEAD-box ATPase activation by nucleoporins and InsP6 in mRNA export. Nature 472, 238–242 10.1038/nature09862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oeffinger M., Zenklusen D. (2012). To the pore and through the pore: a story of mRNA export kinetics. Biochim. Biophys. Acta 1819, 494–506 10.1016/j.bbagrm.2012.02.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onischenko E. A., Gubanova N. V., Kiseleva E. V., Hallberg E. (2005). Cdk1 and okadaic acid-sensitive phosphatases control assembly of nuclear pore complexes in Drosophila embryos. Mol. Biol. Cell 16, 5152–5162 10.1091/mbc.E05-07-0642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onischenko E., Stanton L. H., Madrid A. S., Kieselbach T., Weis K. (2009). Role of the Ndc1 interaction network in yeast nuclear pore complex assembly and maintenance. J. Cell Biol. 185, 475–491 10.1083/jcb.200810030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orjalo A. V., Arnaoutov A., Shen Z., Boyarchuk Y., Zeitlin S. G., Fontoura B., Briggs S., Dasso M., Forbes D. J. (2006). The Nup107-160 nucleoporin complex is required for correct bipolar spindle assembly. Mol. Biol. Cell 17, 3806–3818 10.1091/mbc.E05-11-1061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ossareh-Nazari B., Gwizdek C., Dargemont C. (2001). Protein export from the nucleus. Traffic 2, 684–689 10.1034/j.1600-0854.2001.21002.x [DOI] [PubMed] [Google Scholar]
- Panse V. G., Johnson A. W. (2010). Maturation of eukaryotic ribosomes: acquisition of functionality. Trends Biochem. Sci. 35, 260–266 10.1016/j.tibs.2010.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel S. S., Belmont B. J., Sante J. M., Rexach M. F. (2007). Natively unfolded nucleoporins gate protein diffusion across the nuclear pore complex. Cell 129, 83–96 10.1016/j.cell.2007.01.044 [DOI] [PubMed] [Google Scholar]
- Peters R. (2005). Translocation through the nuclear pore complex: selectivity and speed by reduction-of-dimensionality. Traffic 6, 421–427 10.1111/j.1600-0854.2005.00287.x [DOI] [PubMed] [Google Scholar]
- Peters R. (2009). Translocation through the nuclear pore: Kaps pave the way. BioEssays 31, 466–477 10.1002/bies.200800159 [DOI] [PubMed] [Google Scholar]
- Price J. C., Guan S., Burlingame A., Prusiner S. B., Ghaemmaghami S. (2010). Analysis of proteome dynamics in the mouse brain. Proc. Natl. Acad. Sci. USA 107, 14508–14513 10.1073/pnas.1006551107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasala B. A., Orjalo A. V., Shen Z., Briggs S., Forbes D. J. (2006). ELYS is a dual nucleoporin/kinetochore protein required for nuclear pore assembly and proper cell division. Proc. Natl. Acad. Sci. USA 103, 17801–17806 10.1073/pnas.0608484103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasala B. A., Ramos C., Harel A., Forbes D. J. (2008). Capture of AT-rich chromatin by ELYS recruits POM121 and NDC1 to initiate nuclear pore assembly. Mol. Biol. Cell 19, 3982–3996 10.1091/mbc.E08-01-0012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichelt R., Holzenburg A., Buhle E. L., Jr, Jarnik M., Engel A., Aebi U. (1990). Correlation between structure and mass distribution of the nuclear pore complex and of distinct pore complex components. J. Cell Biol. 110, 883–894 10.1083/jcb.110.4.883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribbeck K., Görlich D. (2001). Kinetic analysis of translocation through nuclear pore complexes. EMBO J. 20, 1320–1330 10.1093/emboj/20.6.1320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribbeck K., Görlich D. (2002). The permeability barrier of nuclear pore complexes appears to operate via hydrophobic exclusion. EMBO J. 21, 2664–2671 10.1093/emboj/21.11.2664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ródenas E., Klerkx E. P. F., Ayuso C., Audhya A., Askjaer P. (2009). Early embryonic requirement for nucleoporin Nup35/NPP-19 in nuclear assembly. Dev. Biol. 327, 399–409 10.1016/j.ydbio.2008.12.024 [DOI] [PubMed] [Google Scholar]
- Rodriguez M. S., Dargemont C., Stutz F. (2004). Nuclear export of RNA. Biol. Cell 96, 639–655 10.1016/j.biolcel.2004.04.014 [DOI] [PubMed] [Google Scholar]
- Rout M. P., Aitchison J. D., Suprapto A., Hjertaas K., Zhao Y., Chait B. T. (2000). The yeast nuclear pore complex: composition, architecture, and transport mechanism. J. Cell Biol. 148, 635–652 10.1083/jcb.148.4.635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rout M. P., Aitchison J. D., Magnasco M. O., Chait B. T. (2003). Virtual gating and nuclear transport: the hole picture. Trends Cell Biol. 13, 622–628 10.1016/j.tcb.2003.10.007 [DOI] [PubMed] [Google Scholar]
- Ryan K. J., Wente S. R. (2002). Isolation and characterization of new Saccharomyces cerevisiae mutants perturbed in nuclear pore complex assembly. BMC Genet. 3, 17 10.1186/1471-2156-3-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan K. J., McCaffery J. M., Wente S. R. (2003). The Ran GTPase cycle is required for yeast nuclear pore complex assembly. J. Cell Biol. 160, 1041–1053 10.1083/jcb.200209116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan K. J., Zhou Y., Wente S. R. (2007). The karyopherin Kap95 regulates nuclear pore complex assembly into intact nuclear envelopes in vivo. Mol. Biol. Cell 18, 886–898 10.1091/mbc.E06-06-0525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampathkumar P., Kim S. J., Upla P., Rice W. J., Phillips J., Timney B. L., Pieper U., Bonanno J. B., Fernandez-Martinez J., Hakhverdyan Z. et al. (2013). Structure, dynamics, evolution, and function of a major scaffold component in the nuclear pore complex. Structure 21, 560–571 10.1016/j.str.2013.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savas J. N., Toyama B. H., Xu T., Yates J. R., 3rd, Hetzer M. W. (2012). Extremely long-lived nuclear pore proteins in the rat brain. Science 335, 942 10.1126/science.1217421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schooley, A., Vollmer, B. and Antonin, W(2012). Building a nuclear envelope at the end of mitosis: coordinating membrane reorganization, nuclear pore complex assembly, and chromatin de-condensation. Chromosoma 121, 539–554 10.1007/s00412-012-0388-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segref A., Sharma K., Doye V., Hellwig A., Huber J., Lührmann R., Hurt E. (1997). Mex67p, a novel factor for nuclear mRNA export, binds to both poly(A)+ RNA and nuclear pores. EMBO J. 16, 3256–3271 10.1093/emboj/16.11.3256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon D. N., Rout M. P. (2014). Cancer and the nuclear pore complex. Adv. Exp. Med. Biol. 773, 285–307 10.1007/978-1-4899-8032-8_13 [DOI] [PubMed] [Google Scholar]
- Siniossoglou S., Wimmer C., Rieger M., Doye V., Tekotte H., Weise C., Emig S., Segref A., Hurt E. C. (1996). A novel complex of nucleoporins, which includes Sec13p and a Sec13p homolog, is essential for normal nuclear pores. Cell 84, 265–275 10.1016/S0092-8674(00)80981-2 [DOI] [PubMed] [Google Scholar]
- Stewart M. (2007a). Ratcheting mRNA out of the nucleus. Mol. Cell 25, 327–330 10.1016/j.molcel.2007.01.016 [DOI] [PubMed] [Google Scholar]
- Stewart M. (2007b). Molecular mechanism of the nuclear protein import cycle. Nat. Rev. Mol. Cell Biol. 8, 195–208 10.1038/nrm2114 [DOI] [PubMed] [Google Scholar]
- Stewart M. (2010). Nuclear export of mRNA. Trends Biochem. Sci. 35, 609–617 10.1016/j.tibs.2010.07.001 [DOI] [PubMed] [Google Scholar]
- Stoffler D., Feja B., Fahrenkrog B., Walz J., Typke D., Aebi U. (2003). Cryo-electron tomography provides novel insights into nuclear pore architecture: implications for nucleocytoplasmic transport. J. Mol. Biol. 328, 119–130 10.1016/S0022-2836(03)00266-3 [DOI] [PubMed] [Google Scholar]
- Stuwe T., Lin D. H., Collins L. N., Hurt E., Hoelz A. (2014). Evidence for an evolutionary relationship between the large adaptor nucleoporin Nup192 and karyopherins. Proc. Natl. Acad. Sci. USA 111, 2530–2535 10.1073/pnas.1311081111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terry L. J., Wente S. R. (2009). Flexible gates: dynamic topologies and functions for FG nucleoporins in nucleocytoplasmic transport. Eukaryot. Cell 8, 1814–1827 10.1128/EC.00225-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Texari L., Dieppois G., Vinciguerra P., Contreras M. P., Groner A., Letourneau A., Stutz F. (2013). The nuclear pore regulates GAL1 gene transcription by controlling the localization of the SUMO protease Ulp1. Mol. Cell 51, 807–818 10.1016/j.molcel.2013.08.047 [DOI] [PubMed] [Google Scholar]
- Toyama B. H., Savas J. N., Park S. K., Harris M. S., Ingolia N. T., Yates J. R., III and Hetzer M. W. (2013). Identification of long-lived proteins reveals exceptional stability of essential cellular structures. Cell 154, 971–982 10.1016/j.cell.2013.07.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran E. J., Zhou Y., Corbett A. H., Wente S. R. (2007). The DEAD-box protein Dbp5 controls mRNA export by triggering specific RNA:protein remodeling events. Mol. Cell 28, 850–859 10.1016/j.molcel.2007.09.019 [DOI] [PubMed] [Google Scholar]
- Tschochner H., Hurt E. (2003). Pre-ribosomes on the road from the nucleolus to the cytoplasm. Trends Cell Biol. 13, 255–263 10.1016/S0962-8924(03)00054-0 [DOI] [PubMed] [Google Scholar]
- Van de Vosse D. W., Wan Y., Lapetina D. L., Chen W-M., Chiang J-H., Aitchison J. D., Wozniak R. W. (2013). A role for the nucleoporin Nup170p in chromatin structure and gene silencing. Cell 152, 969–983 10.1016/j.cell.2013.01.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vollmer B., Antonin W. (2014). The diverse roles of the Nup93/Nic96 complex proteins – structural scaffolds of the nuclear pore complex with additional cellular functions. Biol. Chem. 395, 515–528 10.1515/hsz-2013-0285 [DOI] [PubMed] [Google Scholar]
- Walther T. C., Alves A., Pickersgill H., Loïodice I., Hetzer M., Galy V., Hülsmann B. B., Köcher T., Wilm M., Allen T. et al. (2003). The conserved Nup107-160 complex is critical for nuclear pore complex assembly. Cell 113, 195–206 10.1016/S0092-8674(03)00235-6 [DOI] [PubMed] [Google Scholar]
- Weirich C. S., Erzberger J. P., Flick J. S., Berger J. M., Thorner J., Weis K. (2006). Activation of the DExD/H-box protein Dbp5 by the nuclear-pore protein Gle1 and its coactivator InsP6 is required for mRNA export. Nat. Cell Biol. 8, 668–676 10.1038/ncb1424 [DOI] [PubMed] [Google Scholar]
- Weis K. (2003). Regulating access to the genome: nucleocytoplasmic transport throughout the cell cycle. Cell 112, 441–451 10.1016/S0092-8674(03)00082-5 [DOI] [PubMed] [Google Scholar]
- Yamada J., Phillips J. L., Patel S., Goldfien G., Calestagne-Morelli A., Huang H., Reza R., Acheson J., Krishnan V. V., Newsam S. et al. (2010). A bimodal distribution of two distinct categories of intrinsically disordered structures with separate functions in FG nucleoporins. Mol. Cell. Proteomics 9, 2205–2224 10.1074/mcp.M000035-MCP201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W., Gelles J., Musser S. M. (2004). Imaging of single-molecule translocation through nuclear pore complexes. Proc. Natl. Acad. Sci. USA 101, 12887–12892 10.1073/pnas.0403675101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yavuz S., Santarella-Mellwig R., Koch B., Jaedicke A., Mattaj I. W., Antonin W. (2010). NLS-mediated NPC functions of the nucleoporin Pom121. FEBS Lett. 584, 3292–3298 10.1016/j.febslet.2010.07.008 [DOI] [PubMed] [Google Scholar]
- Zemp I., Kutay U. (2007). Nuclear export and cytoplasmic maturation of ribosomal subunits. FEBS Lett. 581, 2783–2793 10.1016/j.febslet.2007.05.013 [DOI] [PubMed] [Google Scholar]
- Zilman A., Di Talia S., Jovanovic-Talisman T., Chait B. T., Rout M. P., Magnasco M. O. (2010). Enhancement of transport selectivity through nano-channels by non-specific competition. PLOS Comput. Biol. 6, e1000804 10.1371/journal.pcbi.1000804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuccolo M., Alves A., Galy V., Bolhy S., Formstecher E., Racine V., Sibarita J-B., Fukagawa T., Shiekhattar R., Yen T. et al. (2007). The human Nup107-160 nuclear pore subcomplex contributes to proper kinetochore functions. EMBO J. 26, 1853–1864 10.1038/sj.emboj.7601642 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.