Abstract
The cell-biological program termed the epithelial-mesenchymal transition (EMT) confers on cancer cells mesenchymal traits and an ability to enter the cancer stem cell (CSC) state. However, the interactions between CSCs and their surrounding microenvironment are poorly understood. Here we show that tumor-associated monocytes and macrophages (TAMs) create a CSC-niche via juxtacrine signaling with CSCs. We performed quantitative proteomic profiling and found that the EMT program upregulates the expression of CD90/Thy1 and EphA4, which mediate the physical interactions of CSCs with TAMs by directly binding with their respective counter-receptors on these cells. In response, the EphA4 receptor on the carcinoma cells activates Src and NF-κB, the latter results in the secretion of a variety of cytokines by the CSCs; these cytokines serve to sustain the stem-cell state. Indeed, admixed macrophages enhance the CSC activities of carcinoma cells. These findings underscore the significance of TAMs as important components of the CSC niche.
Despite improvements in diagnosis and treatment, breast cancer-associated mortality remains high due to clinical relapse associated with metastasis to distant organs. During primary tumor progression, breast carcinoma cells may pass through an EMT, thereby acquiring traits associated with high-grade malignancy, including motility, invasiveness, and an increased resistance to apoptosis1, 2. Furthermore, passage of both normal and neoplastic mammary epithelial cells through an EMT confers on the cells many of the properties associated with normal mammary stem cells (MaSCs) and cancer stem cells (CSCs) respectively3–5. Importantly, carcinoma cells that have passed through an EMT exhibit heightened resistance to conventional chemotherapeutic agents and hence may regenerate tumor growth after initial drug treatment is halted6–8.
Various types of stromal cells have been found to influence the CSC state through paracrine signaling. As an example, mesenchymal stem cells contribute to the formation of CSCs by secreting prostaglandin E2 (PGE2), IL-6, IL-8 and Gro-α, which help to trigger activation of the previously latent EMT program in nearby carcinoma cells9. Periostin (POSTN) released by lung stromal fibroblasts fosters creation of a metastatic niche for breast cancer CSCs10. Some have speculated that a physical CSC-niche maintained by both paracrine and juxtacrine signaling exists in colon cancers similar to the normal colon stem cell niche supported by myofibroblasts11. Nevertheless, a juxtacrine-mediated CSC niche that maintains these cells in their stem-cell state has not been described.
Tumor-associated monocytes and derived macrophages (TAMs) have been shown to be involved in many aspects of tumor initiation and progression12, 13. These observations have not, however, shed light on how CSCs interact with TAMs in a manner different from non-CSCs, and whether heterotypic interactions contribute to the formation and maintenance of the stem-cell niche.
Some have reported that normal MaSC function requires the continuing presence of macrophage-derived factors14, but the nature of these factors has not been explored. Moreover, it remains unclear whether the interactions between macrophages and normal MaSCs are relevant to carcinoma pathogenesis. In the present study, we describe a key mechanism by which already-formed mammary CSCs interact with niche-forming TAMs in a contact-dependent manner in order to maintain their residence in the mesenchymal/stem-like state.
RESULTS
Quantitative proteomic profiling of cell-surface proteins
We and others have found extensive similarities between the stem-cell program of normal MaSCs and that of mammary CSCs3–5. To identify cell-surface proteins that enable normal and neoplastic mammary stem cells (SCs) to interact with nearby stromal cells, we performed quantitative proteomic profiling of membrane-associated proteins before and after immortalized human mammary epithelial cells (HMLE)15 were forced experimentally to undergo an EMT (and thereby acquire mesenchymal and SC traits). We employed stable isotope labeling by amino acids in cell culture (SILAC)16 followed by membrane-associated protein fractionation and mass spectrometry analysis (MS; Supplementary Fig. 1).
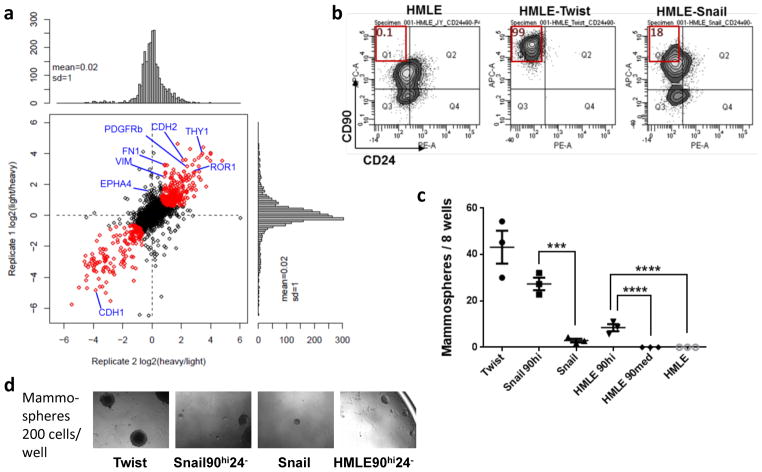
Plasma membrane- and extracellular matrix- associated proteins were represented by the most abundant peptides detected by MS (Supplementary Table 1). 2,607 proteins were reproducibly identified with at least two peptide SILAC ratios measured in each replicate, with 460 proteins found to be either upregulated (277) or downregulated (183) significantly (P<0.05) in the EMT/stem-like cells after applying a moderated T-test17. Several known EMT marker proteins E-cadherin (CDH1), N-cadherin (CDH2), vimentin (VIM) and fibronectin (FN1) showed significant expression level changes in the expected directions, confirming the specificity of the proteomic profiling (Fig. 1a).
Figure 1. Quantitative proteomic profiling identified EMT-induced membrane protein changes in HMECs.
(a) Expression ratios of proteins detected in the mesenchymal HMLE-Twist cells over the epithelial HMLE-vector cells plotted for two biological replicate experiments. Ratios highlighted in red were found to be significantly differentially expressed (p<0.05) after standardizing and applying a moderated T-test corrected by the Benjamini Hochberg method17. Gene names were labeled for known EMT marker proteins and candidates that were followed up in this study. (b) The HMLE, HMLE-Twist and HMLE-Snail cells were stained with anti-CD90 and anti-CD24 antibodies and analyzed by flow cytometry. The percentage of the CD90hiCD24− population was shown on each plot. (c,d) The cells were FACS sorted based on CD90 and CD24 levels and seeded into 3D mammosphere medium. The numbers of mammospheres were plotted (c), and representative pictures of mammospheres formed by the indicated cell populations are shown (d). n= 3 independent experiments. *** p<0.001; **** p<0.0001 (Student’s t-test). Source data provided in Supplementary Table 7.
We focused subsequent analyses on the CD90/Thy1 protein, which was among the most strongly upregulated plasma membrane proteins (12.4 fold) following passage through an EMT. CD90 is a GPI-anchored glycoprotein and has been shown to interact with integrins displayed by adjacent cells18, 19. Its mRNA was also substantially upregulated during the EMT programs induced in HMLE cells by expression of the Twist, Snail, Slug or Zeb1 EMT-TFs (Supplementary Fig. 2a).
Presence of stem-like cells in the CD90hi cell population
Because human mammary epithelial cells that have undergone an EMT can acquire stem-like properties4, we postulated that CD90 expression was upregulated in MaSCs and breast CSCs. We analyzed the gene-expression data from two published reports and found a significant enrichment of CD90 expression in freshly isolated human mammary SC and CSC fractions in primary patient tissues across multiple specimens20, 21 (Supplementary Fig. 2b,c).
We then examined the cell-surface antigen profile of HMLE-derived cells using anti-CD90 and -CD24 antibodies. CD24 has been found to be mostly expressed in differentiated mammary epithelial cells21. Flow cytometry analysis revealed that 99% of HMLE-Twist cells and 18% of HMLE-Snail cells were CD90hiCD24−, whereas the HMLE-vector control cells only contained 0.1% cells in the same compartment (Fig. 1b), indicating that forced expression of either the Twist or Snail EMT-TFs led to substantial increases in CD90hiCD24− cells. The CD90hiCD24− cells from the three cell lines formed mammospheres22 with far higher efficiency than cell populations with lower CD90 expression (Fig. 1c,d), providing further support for the association of CD90 expression with the MaSC state.
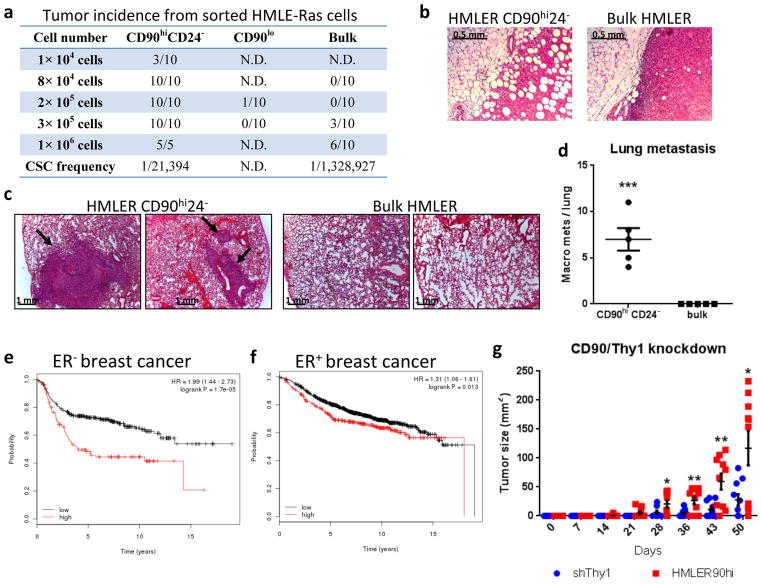
We subsequently undertook to measure the association of tumor-initiating ability with the CD90hiCD24− state in HMLE cells that had been transformed with the H-RasV12 oncogene (HMLER); this ability represents the operational definition of CSCs. Sorted HMLER cells were implanted orthotopically at limiting dilutions into NOD-SCID mice. The CD90hiCD24− HMLER cells initiated tumors at a 60-fold higher efficiently than the bulk HMLER cells and the tumors were more invasive and metastatic; the CD90lo populations rarely formed tumors (Fig. 2a–d). Similar results were observed when the sorted cell populations were implanted orthotopically into Nude mice (Supplementary Fig. 2d). CD90hi cells from the MB-MDA-435 human breast cancer cell line also formed larger and more aggressive tumors when compared to CD90lo cell populations (Supplementary Fig. 2e–g). Consistently, the expression level of CD90 is negatively correlated with patient survival, especially in ER− breast cancer (Fig. 2e,f). Taken together, these data demonstrated that the CD90 marker served to enrich for CSCs in carcinoma cells of mammary origin.
Figure 2. Enrichment of CD90 protein on the cell surface of EMT/stem-like cells.
(a) HMLE-Ras cells were FACS-separated and then injected orthotopically into NOD-SCID mice in a limiting dilution assay. The tumor initiation rates are shown, with the estimated CSC-frequency displayed for the bulk and the CD90hiCD24− populations. N.D., not determined. (b,c) H&E staining comparing xenograft tumors (b) and lungs (c) of mice receiving injections from the bulk and CD90hiCD24− HMLER cells. Black arrows (c) indicate macro-metastasis from the CD90hiCD24− tumors. The numbers of macro-metastases per lung are shown in (d; n=5 mice). (e,f) Correlation of CD90 level in patient tumors with relapse-free survival. High CD90 expression (red line) was significantly correlated with poor prognosis in ER− (H; n=457, p=1.7×10−5) and ER+ (I; n=1413, p=0.013) breast cancer patients. The plots were generated using http://kmplot.com (probe: 213869_x_at).58 (g) HMLER90hi cells with or without shCD90 were injected orthotopically into mice and the tumor size were measured weekly during a course of 50 days. n=10 mice. Knockdown of CD90 by the shRNA was confirmed by flow cytometry analysis (Supplementary Fig. 2h). All error bars shown indicate S.E.M. * p<0.05; ** p<0.01; *** p<0.001 (Student’s t-test).
Physical interaction between CD90hi CSC and TAMs
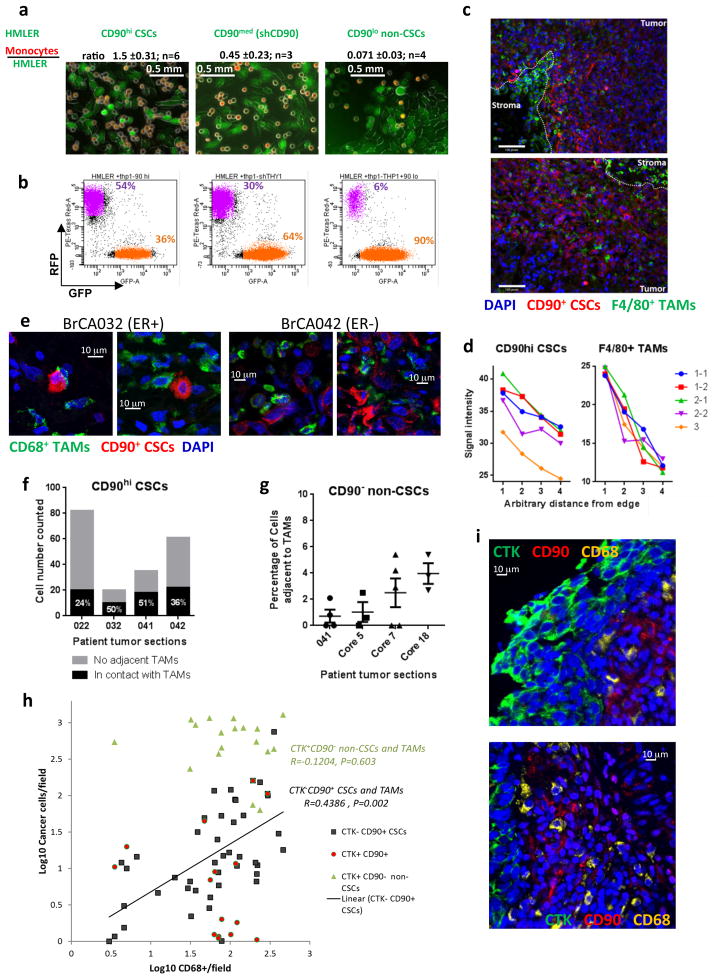
Reduced CD90 expression by shRNA knockdown resulted in delayed tumor onset and decreased tumor size (Fig. 2g) by the HMLER90hi cells, suggesting that CD90 has a functional role in tumor growth. CD90 has been shown to be expressed on activated endothelial cells, where it mediates the adhesion of monocytes via its counter-receptor MAC1 (integrin alphaM/betaV)19. We speculated that CD90 also serves as a physical anchor used by carcinoma stem cells to tether monocytes and derived macrophages. To test this notion in cell culture, we employed a human monocytic cell line THP123. GFP-expressing HMLE-Ras cells were sorted into CD90hiCD24− (HMLER90hi) and CD90loCD24+ (HMLER90lo) populations (Supplementary Fig. 2h) and expanded in monolayer culture. RFP-expressing THP1 monocytes, normally propagated in suspension, were then added above the cell monolayers. We found that the RFP+ monocytes adhered in large numbers to the GFP+ monolayer of HMLER90hi cells, but rarely stay attached to the HMLER90lo cells (a 22-fold difference; Fig. 3a,b). ShRNA-mediated knockdown of CD90 in HMLER90hi cells reduced the adherent monocytes by 3-fold, confirming the requirement of CD90 expression on the surface of the stem-like cells for efficient physical interaction with the monocytes (Fig. 3a, b middle panels).
Figure 3. CD90 serves as an anchor for the adherence of monocytes and macrophages.
(a) THP1-RFP monocytes were cocultured with monolayer HMLER-GFP cells. Monocytes that did not adhere to the HMLER monolayer were washed away after 1 hr and the adherent cells were imaged and then quantified by flow cytometry based on GFP and RFP expression. Representative images (a) and flow cytometry profiles (b) are shown. The ratios of the adhered monocytes to the HMLER cells were calculated from the flow-cytometry quantitation in panel (b); data presented as mean ± s.e.m. (c) Immunofluorescence (IF) staining of CD90 (red) and F4/80 (green) in HMLER90hi xenograft tumors. The dotted lines delineate the border between the tumor cells and stroma. Scale bar: 0.1mm. (d) Quantitation of CD90 signal and F4/80 signal intensity from tumor edge to center. n=5 tumors. (e) 60x confocal images of primary patient breast tumor sections co-stained for CD90+ CSCs (red) and CD68+ TAMs (green) showed juxtaposed CSC-TAM pairs. (f,g) Numbers of CD90high CSCs (f) or CD90−CTK+ non-CSCs with or without adjacent TAMs were counted from 3–6 representative fields of 20x or 25x images (n=4 patients). The percentages shown indicate the proportion of cancer cells with adjacent TAMs. Error bars indicate S.E.M. Source data for g provided in Supplementary Table 7. (h) The numbers of CD90+CTK+, CD90+CTK−, CD90−CTK+ cancer cells and CD68+ macrophages in the same microscopic field (0.6 mm2) were quantified from 25 core biopsy sections. The number of CD90+CTK− CSCs correlates with the number of CD68+ TAMs. The CD90+ endothelial cells and fibroblasts were excluded from the cancer cell counts based on morphology. (i) Typical images showing infiltration of CD68+ TAMs into the CD90+ CSC-rich areas but not the CD90−CTK+ cancer cell islands.
Association of TAMs with CD90hi carcinoma cells in living tissues
In the xenograft tumors formed by HMLER90hi cells, CD90 expression was most prominent at the outer edge of tumor sections, where a high density of infiltrating tumor-associated macrophages (TAMs) was observed intermingled with CD90hi cancer cells (Fig. 3c,d), suggesting that CD90-mediated adhesion of TAMs to CSCs also occurs in vivo.
CD90 expression has been detected in primary human breast carcinomas on invading single cells or cell clusters24; in metastases the CD44+CD90+ subpopulation represents a major phenotype25. While TAM density in primary breast cancer has been correlated with poor prognosis26–28, a CSC-niche involving TAMs has not been demonstrated microscopically in patient tumor tissues. We undertook to verify direct contact between TAMs and CD90hi CSCs in patient breast cancer tissue by immunofluorescence analyses with anti-CD90 and -CD68 (a cell-surface protein expressed largely by macrophages) antibodies. Juxtaposition of CD68+ TAMs with single-invading CD90hi CSCs was readily detected at the tumor-stroma interface (Fig. 3e; Supplementary Fig. 3a). Indeed, 24–51% of observed CD90hi cancer cells showed the presence of TAMs in close contact; in contrast very few (0 to 5%) CD90− cytokeratin+ (CTK+) cells were localized adjacent to TAMs (Fig. 3f,g; Supplementary Fig. 3b). This direct visualization of the CSC-TAM juxtaposition in patient samples provided further support for the notion that CSCs and TAMs engage in direct contact in human breast tumors, where the latter cells help to form a niche for the CSCs.
We then expanded our analysis to include 25 core biopsy samples from patient tissues with invasive ductal carcinoma obtained during initial diagnostic core biopsies. The majority of CD90+ carcinoma cells showed reduced levels of cytokeratin expression, consistent with their passage through an EMT (Supplementary Fig. 3c). The number of CD68+ TAMs per microscopic field correlated with number of CD90+CTK− CSCs (r=0.44, P=0.002) but not with CD90−CTK+ non-CSCs (r=−0.12, P=0.60; Fig. 3h). This indicated that TAMs often infiltrate CD90+ CSC-rich areas, allowing the juxtacrine signaling to occur. Accordingly, we could observe intermingling of CD68+ TAMs with CD90+ CSCs, but the macrophages rarely infiltrated into islands of CD90−CTK+ non-CSCs (Fig. 3i). Finally, consistent with this, analysis of the TCGA invasive breast carcinoma dataset revealed that CD90 expression in breast tumors positively correlated with CD68 expression (Supplementary Fig. 3e).
Monocytes/macrophages facilitate tumor outgrowth by the CSCs
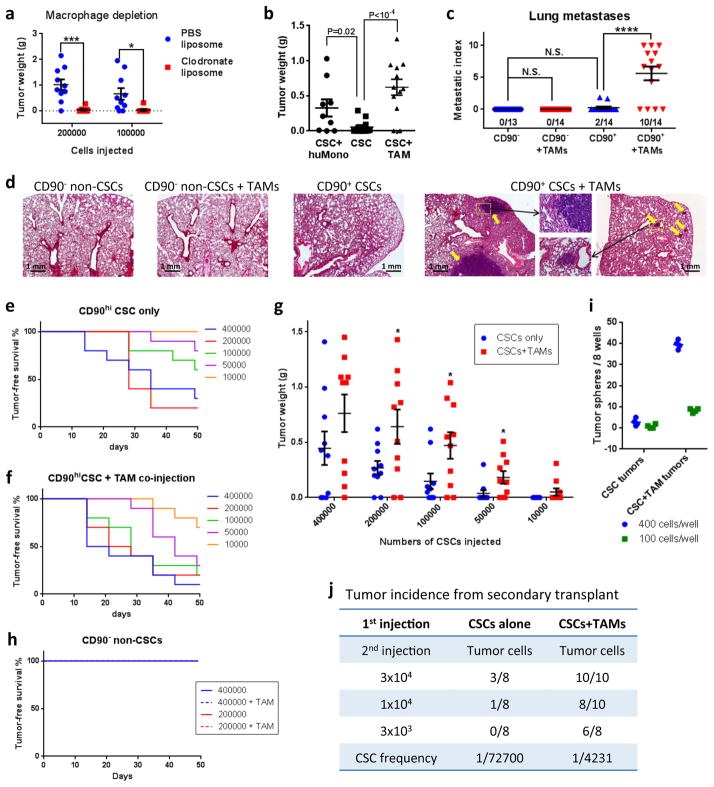
Monocytes and macrophages in tumors have great functional plasticity12, 29. To test whether the CSC-TAM interaction that we observed is important to tumor formation, we depleted endogenous macrophages in the mammary fat pads at the time of implantation by co-injecting the HMLER90hi cells with clodronate liposomes14. Nude mice were used in this and subsequent experiments instead of NOD-SCID hosts because of the documented defect in macrophage activation in the latter. Strikingly, tumor initiation was almost completely blocked by macrophage depletion, suggesting that endogenous macrophages in the tumor microenvironment are critical for the survival and proliferation of implanted CSCs (Fig. 4a). To examine the opposite effect, we co-injected HMLER90hi cells with either primary human monocytes or mouse TAMs isolated from HMLER90hi xenografts (Supplementary Fig. 4a). The admixture of either human monocytes or mouse TAMs resulted in higher tumor incidence and size, indicating that they promoted tumor outgrowth by CSCs (Fig. 4b).
Figure 4. Macrophages enhance tumor initiation by the CD90hi CSC-like cells.
(a) CD90hi CSCs were mixed with PBS- or clodronate- liposomes and orthotopically implanted into Nude mice (n=10 per group). Data were pooled from 2 independent experiments, each assessing 5 mice per group. The tumors were harvested 50 days later and weighed. (b) 105 HMLER90hi cells were injected either alone or co-mixed with 5×104 human monocytes (huMono) or mice TAMs, into the mammary fat pads of female Nude mice. n=9,12,12 mice (left to right). Reduced amount of Matrigel was used for the injection mix. The tumor weight was measured after 5 weeks. Data from 3 independent experiments were pooled and plotted. (c,d) Naturally arising MMTV-PyMT tumors were dissociated and tumor cells were sorted based on CD90 expression, whereas TAMs were isolated by F4/80 expression. 3x105 CD90+ or CD90− tumor cells were orthotopically transplanted into syngeneic wildtype Fvb mice with or without comixed 1.5x105 TAMs. (c) The numbers of lung macro-metastases from tumor-bearing mice were quantified and normalized to primary tumor weight (per gram), shown as metastatic index. Data were pooled from 3 independent experiments. n=13,14,14,14 mice from left to right (15 mice were injected for each group, but not all mice developed tumors; see Supplementary Fig. 4d). (d) H&E staining of the lungs of tumor-bearing mice. (e–g) CD90hi CSCs were injected alone (e) or with admixed TAMs (f) at limiting dilution orthotopically (n=10 mice per group). Data were pooled from 2 independent experiments, each assessing 5 mice per group. Tumor incidence was determined by weekly palpation, and tumors were harvested and weighed at 50 days post injection (g). (h) CD90lo non-CSCs were injected alone or with admixed TAMs orthotopically but no tumor formed. Data were pooled from 2 independent experiments, each assessing 5 mice per group. (i,j) HMLER90hi cells injected alone or together with TAMs were allowed to grow in the mammary fat pads of NOD-SCID mice for 3 weeks; the tumor cells were then harvested and seeded for tumorsphere forming assay (i; presents data from 4 technical replicates) or injected as secondary tumor transplants into recipient NOD-SCID mice (j) to measure tumor-initiating capability. Data shown are from one experiment out of two biological replicates; source data provided in Supplementary Table 7 (i,j). Error bars indicate S.E.M. * p<0.05; *** p<0.001; *** p<0.0001 (Student’s t-test for a,b,g and one way ANOVA for c).
The TAMs within tumors arising from CSCs showed low expression of Nos2 (M1 polarization), and high expression of Arg1, MRC1, FIZZ1 and VEGF (M2 polarization30), which resembled the expression patterns of tumor-promoting TAMs31 isolated from mammary tumors in the MMTV-PyMT mice (Supplementary Fig. 4b). Moreover, when analyzing the CD68+ TAMs from the patient samples described above, we found that these cells were predominantly CD163+HLADR−/dim (Supplementary Fig. 4c), indicative of an M2, tumor-promoting phenotype32.
Various types of tumor-infiltrating immune cells, including lymphocytes, have been shown to modulate the function of TAMs33. Therefore we examined the effect of TAMs on CSC-activities in immunocompetent mice. The MMTV-PyMT mice are known to develop mammary tumors that harbor CD90+ CSCs, which efficiently metastasize to the lungs when injected intravenously10. We sorted freshly-isolated mouse mammary carcinoma cells based on CD90 expression from the MMTV-PyMT-driven metastatic tumors and then injected the resulting CD90+ and CD90− populations, with or without TAMs, orthotopically into wild-type syngeneic hosts. All groups developed tumors of similar weight (Supplementary Fig. 4d); lymphocyte infiltration was readily detected within the tumors, resembling spontaneously arising MMTV-PyMT tumors (Supplementary Fig. 4e,f). However, tumors that arose from CD90− non-CSCs (with or without co-injected TAMs) were largely necrotic and contained cysts filled with fluid (Supplementary Fig. 4g,h), with low numbers of proliferating cells (Supplementary Fig. 4i,j) and no metastasis in the lungs (Fig. 4c,d). CD90+ CSCs injected on their own resulted in tumors with less necrosis and more proliferating cells but yielded very few lung metastases. In contrast, the CD90+ CSCs with co-injected TAMs resulted in tumors that contained little necrosis, were more proliferative, and metastasized to lung with a 22-fold higher efficiency than CD90+ CSCs injected alone. These data supported the notion that the co-injected macrophages promoted more robust primary tumor growth and enhanced the metastatic CSC-activity of the CD90+ carcinoma cells in the context of an intact immune system.
We noted that both the CD90+ and CD90− MMTV-PyMT carcinoma cells initiated primary tumors efficiently. This is likely due to the fact that the tumor cells were isolated from spontaneously arising MMTV-PyMT tumors at the stage of metastatic dissemination, when most of the carcinoma cells had evolved to be rapidly proliferating following weeks of robust tumor growth.
Effects of admixed macrophages on tumor initiation
We reasoned that the direct contact of macrophages with the CSCs might allow the TAMs to provide signals important for the maintenance of their CSC state. To determine whether TAMs enhance tumor initiation rates and thus enrich for CSCs we performed tumor xenograft experiments by implanting the carcinoma cells in limiting dilutions. Co-injecting macrophages with the CD90hi CSCs resulted in earlier tumor onset (Fig. 4e,f), higher tumor incidence and burden compared to CSCs injected alone (Fig. 4g); in contrast, CD90lo non-CSCs did not initiate tumors either with or without admixed TAMs (Fig. 4h).
To measure more precisely the frequency of CSCs from the tumors with admixed TAMs, we injected CSCs in the presence or absence of TAMs into mouse mammary fat pads and harvested the tumors three weeks later when they were no larger than 0.2 gram (Supplementary Fig. 4k). The cancer cells from the TAMs+CSCs group formed more tumorspheres and initiated tumors with a 17-fold higher efficiency in a secondary transplantation experiment than those from the CSCs-only injected tumors (Fig. 4i,j). Taken together, these data confirmed directly the ability of TAMs to boost the representation of CSCs in these carcinoma cell populations, ostensibly by enabling CSCs to maintain their residence in the stem cell state.
Monocytes/macrophages-stimulated cytokine production in the stem-like cells
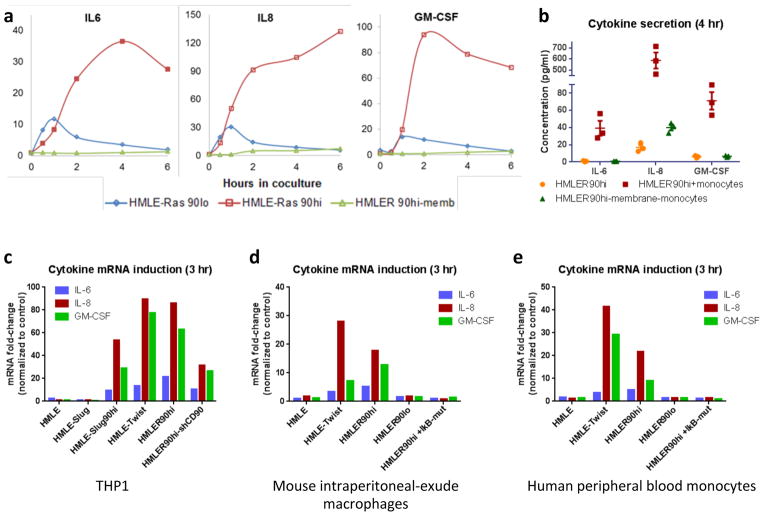
We then sought to dissect the molecular mechanism for the interaction between CSCs and monocytes/macrophages. Because cytokines are known to mediate the interactions between cancer cells and TAMs29, we measured the expression of various cytokine mRNAs in both HMLER90hi cells and THP1 monocytes that had been separated by FACS (fluorescence-activated cell sorting) after 3 hours of coculture (Supplementary Fig. 5a). Widespread changes in cytokine expression were already apparent in both cell populations within this time period (Supplementary Tables 2,3). Strikingly, we observed rapid and robust induction of mRNAs encoding IL-6, IL-8 and GM-CSF in the HMLER90hi cells, whose levels far exceeded the cytokines produced by the cocultured monocytes.
The mRNA levels of IL-6, IL-8 and GM-CSF rose abruptly upon monocyte coculture and persisted for at least 6 hrs (Fig. 5a). HMLER90lo cells, used as controls, only showed a modest and transient increase in these cytokines in response to monocyte coculture. Importantly, the induction of these cytokines in the HMLER90hi cells was completely blocked when they were separated from the monocytes by a cell-impermeable insert, indicating that the observed signaling between the two cell types required direct cell-cell contact, i.e., juxtracrine signaling. Virtually no cytokines were detected in the conditioned medium by ELISA from mock-treated HMLER90hi cells in contrast with the robust increases of cytokine levels after 4 hrs of coculture (Fig. 5b); membrane-separated indirect cocultures yielded only marginal if any increases in the three cytokines. Knockdown of CD90 by shRNA resulted in 2- to 3-fold reduction in cytokine mRNA induction in the HMLER90hi cells (Fig. 5c), likely due to the reduced number of monocytes adhering to these cells.
Figure 5. Monocytes induce cytokine production in the CSCs upon contact.
(a) Induction of cytokine mRNA in the HMLER90hi or HMLER90lo cells when cocultured with the THP1 monocytes for indicated time. HMLER90hi-memb, an indirect coculture where the two cell types were separated by a cell-impermeable insert. Data from one experiment was shown as averages of three technical replicates. Two biological replicates were performed. (b) ELISA measurement of the secreted cytokines from the coculture. n=3 independent experiments. Error bars indicate S.E.M. Source data shown in Supplementary Table 7. (c) Cytokine mRNA fold-change stimulated by THP1 monocytes (relative to corresponding no monocytes control) in the epithelial HMLE, mesenchymal HMLE-Twist, HMLE-Slug CD90hiCD24−, HMLER90hi cells and HMLER90hi with shCD90. (d,e) Cytokine mRNA fold-changes in the HMLE-derived cells stimulated by mouse primary macrophages (d) or human blood monocytes (e). The HMLER90hi-IκBmut cells lacking NF-κB activation showed no cytokine production. Typical data shown from one experiment out of 3 (a,b) and 2 (c–e) biological replicates, only technical replicate data are plotted. Source data provided in Supplementary Table 7.
We also noted that the induction of IL-6, IL-8 and GM-CSF in the stem-like cells did not require the actions of H-RasV12, because the CD90hiCD24− cells from the non-transformed HMLE-Twist and HMLE-Slug stem-like cells also responded to monocyte coculture by producing these cytokines, whereas the parental epithelial HMLE cells, used here as controls, showed only a marginal upregulation of the cytokines (Fig. 5c). In addition, cytokine induction in the CD90hi cells was also observed when primary mouse macrophages or primary human monocytes were used instead of THP1 monocytes (Fig. 5d,e), which indicated that the above observations applied to a variety of monocytes/macrophages, and that the signaling between mouse macrophages and the human HMLE-derived CD90hiCD24− cells was not compromised by inter-species signaling incompatibilities.
IL-6 and IL-8 have been shown to induce and maintain the CSC-state in autocrine and paracrine fashions9, 34, 35. In HMLER90hi cells, individual knockdown of IL-6 or IL-8 led to cessation of cell proliferation in culture, whereas the proliferation of epithelial HMLE-Ras cells was unaffected by the knockdowns (Supplementary Fig. 5b,c). In addition, IL-8 overexpression or treatment with recombinant IL-6 and/or IL-8 resulted in more efficient tumorsphere formation by the HMLER90hi cells, indicating that IL-6 and IL-8 can indeed act on the HMLER90hi cells to promote the expansion of CSCs (Supplementary Fig. 5d–f). GM-CSF is known to promote the differentiation of TAMs into tumor-supporting phenotypes12, 33, and may act in a paracrine fashion to perpetuate the CSC-TAM interaction. The three cytokines therefore may further reinforce and sustain the stem cell-state of the HMLER90hi CSCs.
Role of EphA4 in mediating signaling activation in the stem-like cells
We undertook to determine the molecular mechanisms that were responsible for the marked induction of cytokines following exposure of normal and neoplastic MaSCs to monocytes. CD90 lacks an intracellular domain and hence is poorly equipped to transduce signals across the plasma membrane18. As an alternative, we considered the well-documented CSF1-EGF paracrine loop between TAMs and breast cancer cells31, 36; however, the expression of EGFR is strongly downregulated during passage through an EMT6, 37, 38, rendering it unlikely to mediate the response of CSCs to TAMs.
We therefore searched for other cell-surface receptor candidates that could mediate the juxtacrine signaling from our proteomics analysis. Three receptor tyrosine kinases (RTKs) – ROR1 (a pseudokinase39), PDGFRβ and EphA4 – were strongly upregulated in the stem-like cells (Fig. 1a, Supplementary Table 1, Supplementary Fig. 6a); among these, only EphA4 is known to mediate contact-dependent signaling following binding with plasma membrane-associated Ephrin ligands displayed on closely apposed cells40. We noted that while peptides shared by several Eph RTKs showed little change in levels, MS analysis of the peptides unique to EphA4 revealed its upregulation by over 20-fold following passage through an EMT (see Methods).
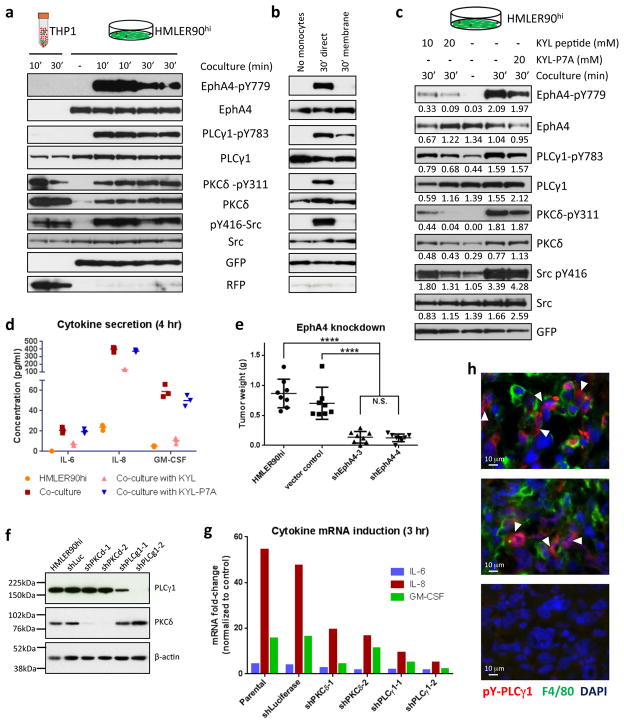
We measured the activation of the RTKs upon contact with monocytes by immunoblotting after removal of monocytes following coculture (Supplementary Fig. 6b). Only EphA4 showed a robust increase in activation in the stem-like HMLER90hi cells and HMLE-Twist cells upon direct monocyte coculture (Fig. 6a,b; Supplementary Fig. 6c).
Figure 6. EphA4 mediates signal transduction in the CSCs upon stimulation by the monocytes.
(a) Phospho- and total-protein levels of EphA4 as well as downstream proteins PLCγ1, PKCδ and Src in the two cocultured cell types were detected by western blot. GFP and RFP were only detected in the corresponding cell populations with minimum cross-contamination. Numbers above each lane indicate minutes of coculture. Phospho-EphA4 was detected with an antibody that non-discriminatively recognizes the phosphorylated forms of EphA3, 4 and 5. (b) Indirect coculture (membrane) with monocytes did not lead to activation of EphA4 and downstream proteins in the CSCs. (c) Treatment of HMLER90hi cells with an EphA4-specific inhibitor peptide KYL prior to and during coculture with monocytes resulted in decreased phosphorylation. A scrambled peptide (KYL-P7A) served as control. The largely diminished phosphorylation level with KYL treatment indicated that the signals detected by the phospho-EphA3/4/5 antibody were mostly from activated EphA4. The numbers below each panel are the intensities of the bands quantified using Fiji software and normalized to GFP (bottom panel) of the corresponding samples. (d) Cytokine secretion from coculture treated with KYL or KYL-P7A was measured by ELISA. Technical replicate data are plotted. Source data provided in Supplementary Table 7. (e) 1x105 HMLER90hi parental cells or HMLER90hi with sh-Luciferase or sh-EphA4 were orthotopically implanted into NOD-SCID mice. n=8 mice (Data were pooled from 2 independent experiments, each assessing 4 mice per group). The tumors were harvested 40 days after implantation and weighed. **** p<0.0001 (One way ANOVA). Error bars indicate S.D. (f) Western blots measuring levels of knockdown with 2 pairs of shRNA each for PLCγ1 and PKCδ in the HMLER90hi cells. (g) Cytokine mRNA induction in the HMLER90hi cells with PLCγ1 and PKCδ knockdown. Technical replicate data are presented as mRNA fold-changes in the HMLER90hi cells upon monocyte coculture. Source data provided in Supplementary Table 7. (h) IF staining of xenograft tumors with antibodies against phopho-PLCγ1 (Tyr783) and F4/80. White arrowheads point to the phospho-PLCγ1+ cells (red) surrounded by F4/80+ TAMs (green). Areas with no TAMs showed no phopho-PLCγ1 signals (bottom panel).
A KYL peptide has been shown to specifically inhibit EphA4 activation41. Treatment of cocultures with the KYL peptide, but not the KYL-P7A control peptide41, blocked 90% of EphA4 phosphorylation in the HMLER90hi cells and significantly reduced the secretion of the three cytokines (Fig. 6c,d). Consistently, shRNA knockdown of EphA4 in the HMLER90hi cells hindered cytokine mRNA induction by monocytes (Supplementary Fig. 6d,e). Taken together, these data demonstrated the involvement of EphA4 in monocyte-stimulated cytokine production in EMT-induced stem-like cells. This was further supported by our observation that knockdown of EphA4 in the CSCs led to a 4-fold reduction in the size of resulting tumors (Fig. 6e).
Because EphA4 activation requires direct engagement of its ligand displayed on the surface of monocytes, it is likely that CD90 physically anchors the monocytes to the HMLER90hi cells, thereby providing the physical association that is essential for the Ephrin ligand-receptor juxtacrine signaling. As expected, we observed that EphA4 phosphorylation in the CD90 knockdown cells was 40% lower than in the control cells when cocultured with monocytes (Supplementary Fig. 6f).
EphA4 receptor can promiscuously bind to both types of human Ephrin ligands, including five glycosylphosphatidylinositol (GPI)-linked ephrin-A ligands and three transmembrane ephrin-B ligands40. Pretreatment of the monocytes with recombinant PI-specific PLC prior to coculture resulted in decreased phosphorylation of the EphA4 receptor, indicating that the ephrin ligand(s) displayed by the monocytes belong to the GPI-anchored type A family (Supplementary Fig. 6g). Consistently, the EphA2 receptor, which only recognizes ephrin-A ligands was also activated in the CSCs upon coculture with monocytes. Given that EphA4 can bind all five ephrin-A ligands, we did not pursue the precise identity of its cognate ephrin-A ligand(s) displayed by the monocytes/macrophages.
Intracellular signaling activated by the EphA4 receptor
In subsequent work, we sought to characterize the downstream signaling pathway of EphA4 activation. EphA4 has been shown to bind and phosphorylate phospholipase Cγ1 (PLCγ1)42, which can activate a variety of protein kinase C (PKC) enzymes via its production of diacylglycerol (DAG)43. In addition, Src family kinases have also been shown to mediate downstream signaling of EphA444. Indeed, we observed strong phosphorylation of PLCγ1 (Y783), PKCδ (Y311), and Src (Y416) in the CSC-like HMLER90hi cells in response to cocultured monocytes (Fig. 6a,b). Activation of these proteins was greatly reduced when treated with the KYL peptide (Fig. 6c)
PKCδ is known to mediate degradation of IκB, allowing NF-κB to translocate into the nucleus45, which can then drive the transcription of various genes including IL-6, IL-8 and GM-CSF46–48. Indeed, knockdown of PLCγ1 and PKCδ in the HMLER90hi cells resulted in a 2–10 fold reduction in the induction of the three cytokine mRNAs (Fig. 6f,g). Consistently, phosphorylation of PLCγ1 could be readily detected in the tumor cells adjacent to TAMs in the HMLER90hi tumor xenografts (Fig. 6h).
NF-κB activation in the CSC-like HMLER90hi cells
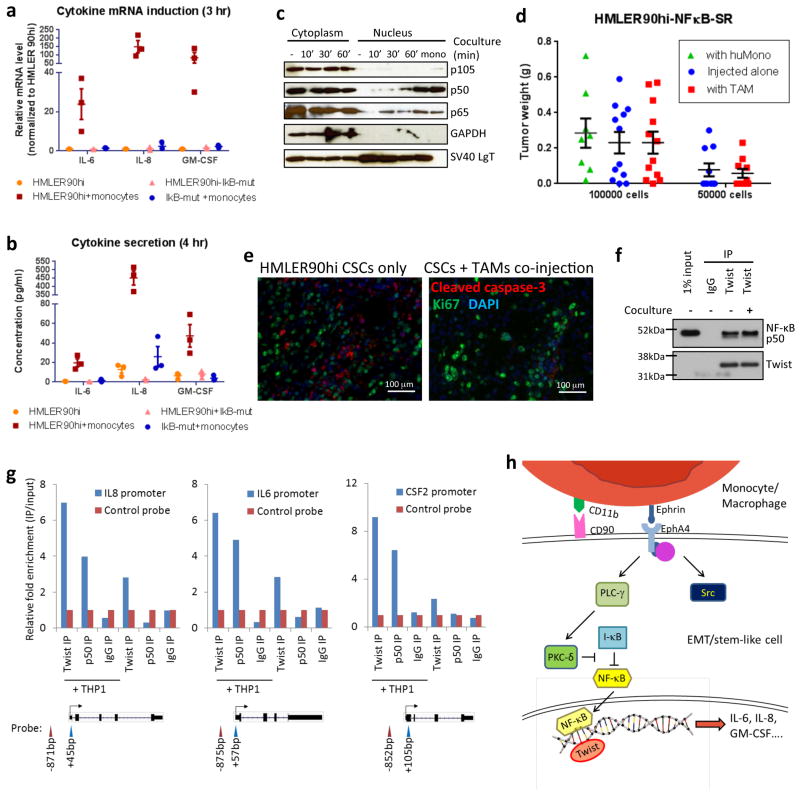
We also undertook to examine in more detail the involvement of the PKCδ-stimulated NF-κB signaling in the HMLER90hi cells. The expression of an NF-κB super-repressor (IκBα S32A/S36A)49 in the HMLER90hi cells blocked the cytokine mRNA induction in these cells following coculture with monocytes or macrophages (Fig. 7a,5d,5e). Secretion of the cytokines from the cocultured cells was also reduced by more than 10-fold when the NF-κB super-repressor was expressed in the HMLER90hi cells (Fig. 7b), which further demonstrated that the three cytokines were largely secreted by the HMLER90hi cells and not by the monocytes. Consistently, we observed a time-dependent nuclear accumulation of the NF-κB subunits p65 and, more strikingly, p50, in the CD90hi cells following coculture with the monocytes (Fig. 7c).
Figure 7. NF-κB activation is required for monocytes-stimulated cytokine production in the CSCs.
(a) The mRNA levels and (b) secreted cytokine concentrations of IL-6, IL-8 and GM-CSF in the HMLER90hi parental cells and the NF-κB super-repressor-expressing cells (HMLER90hi-IκB-mut) with or without monocyte coculture were measured by Q-PCR and ELISA. n=3 biological replicates (with 3 technical repeats each). Source data provided in Supplementary Table 7. Error bars indicate S.E.M. (c) The HMLER90hi cells, alone or in coculture with monocytes (the latter was removed prior to cell harvest), were lysed and fractionated to obtain the cytosolic and nucleus extracts. NF-κB subunits were immunoblotted to show the nuclear translocation. GAPDH and SV40-LgT served as cytosolic and nucleus protein controls. (d) The IκBα mutant-expressing HMLER90hi cells (CSC-Ikb-mut) were injected, either alone (□) or co-mixed with human monocytes (huMono, □) or with mice TAMs (□), into the mammary fat pads of female Nude mice. Tumor weight was measured after 7 weeks and data from 2 independent experiments were pooled. n=8,12,12,10,10 mice (left to right). Error bars indicate S.E.M. (e) IF staining of tumors from HMLER90hi CSCs alone or CSCs+TAMs with antibodies against cleaved caspase 3 (apoptosis marker) and Ki67 (proliferation marker). (f) The endogenous Twist proteins were immunoprecipitated from the HMLER90hi cells, and then immunoblotted for NF-κB subunit p50 to demonstrate complex formation. (g) Binding of both Twist and NF-κB at the promoters of IL6, IL8 and CSF2 (GM-CSF) in the HMLER90hi cells alone or in coculture was measured by ChIP-PCR. Data was represented as fold-enrichment of binding to the specific segments relative to background average. The positions of probes were illustrated below the graphs. Data plotted are technical replicates. Source data from two biological replicates are provided in Supplementary Table 7. (h) A depiction of the signaling transduction pathway in a CSC when stimulated by a monocyte/macrophage.
More importantly, human monocytes or TAMs failed to promote tumor-initiation or growth from the NF-κB super-repressor-expressing HMLER90hi cells, indicating that NF-κB activation and cytokine induction in the CSCs are functionally essential for the tumor-promoting effects of the TAMs (Fig. 7d). In addition, the tumors from co-injected CSCs and TAMs had substantially fewer apoptotic cells, consistent with the well-known roles of NF-κB pathway and Src in conferring resistance to apoptosis (Fig. 7e).
The Twist EMT-TF has been shown to mediate IL8 transcription in cooperation with RelA NF-κB subunit50. Indeed, we detected the association of the p50 NF-κB subunit with the endogenous Twist protein in the HMLER90hi cells by co-immunoprecipitation (Fig. 7f). Using chromatin immunoprecipitation (ChIP), we found that Twist was moderately enriched close to the transcription start-site of IL6, IL8 and CSF2 (encoding GM-SCF) in the absence of monocytes, whereas no binding of p50 was observed. Upon monocytes coculture, we detected significant increases in the binding of both Twist and p50 to the same promoter regions (Fig. 7g). Hence, it appears that when stimulated by monocytes/macrophages, the EphA4 protein on the HMLER90hi cells activates the NF-κB p50 subunit, which then enters the nucleus and cooperates with Twist to further increase its binding to the cytokine promoters, contributing to the robust cytokine induction in the stem-like cells (Fig. 7h).
DISCUSSION
The EMT program is an important component of carcinoma progression by conferring on the cancer cells invasiveness and by enriching for stem-cell properties2–4. Here, we have shown that EMT-induced stem-like HMECs (human mammary epithelial cells) interact with and respond to monocytes and macrophages far more robustly than do differentiated, non-stem like HMECs. In the context of cancer, these cell-cell contact-dependent interactions provide evidence of a physical CSC-niche supported by the TAMs. Such direct contact could be visualized in xenograft tumors as well as patient-derived breast tumor sections. Furthermore, we delineated the receptors and signaling pathways mediating such interactions in the CSCs, which were confirmed using tumor cell transplantation models. The observed responses of the CSCs appear to sustain their residence in the stem-cell state via self-reinforcing, positive-feedback loops (Supplementary Fig. 6h).
Intriguingly, co-injection of CD90+ MMTV-PyMT tumor cells with TAMs into the mammary fat pads of wildtype syngeneic mice resulted in more efficient lung metastases than CD90+ CSCs injected on their own. In addition to enhancing initial survival and proliferation of the CSCs, we suspect that the cohort of cytokines secreted in response to the TAM-CSC interaction facilitated the induction of tumor-supportive stroma, allowing tumor progression to metastatic carcinoma. In contrast, when CD90+ CSCs were injected alone into wildtype syngeneic hosts, in the absence of various tumor-promoting and immune-suppressing cytokines produced by the interacting CSCs and TAMs, the subsequently recruited immune cells may have adopted tumor-suppressive phenotypes33. The effects of the immune cells on the CSC activities await further investigation.
During embryonic development, macrophages derive from the yolk sac and fetal liver prior to hematopoiesis, arising as the earliest cells of the hematopoietic lineage12. Among other functions, these tissue macrophages likely participate in embryonic organ development by providing effectors necessary for the formation and function of various tissue stem-cell niches. It has been reported that mouse mammary stem cell (MaSC) function requires the continuing presence of macrophages in the developing mammary gland14. This suggests that components of the heterotypic interactions between normal MaSCs and monocytes/macrophages are recapitulated in the carcinoma SCs.
More broadly, the presently observed physical interactions between CSCs and TAMs may quite possibly operate in other carcinoma types. Increased expression of CD90 has been reported in CSCs of liver cancer, gastric cancer, colon cancer and glioma51–54. EphA4 overexpression has been observed in gastric cancer55 and liver cancer metastasis56. Additional Ephrin receptor family members EphA2 and EphB2 have been shown to be upregulated in glioblastoma tumor-propagating cells and in colon CSCs11, 57. Accordingly, CD90 and Eph receptor(s) may well mediate interactions of CSCs and TAMs in a variety of carcinomas in order to maintain and reinforce the CSC-state. Indeed, such interactions may one day suggest potential targets for therapeutic attack on CSCs.
METHODS
Cell culture
HMLE, HMLE-Twist, HMLE-Snail, HMLE-Slug and HMLE-Ras cells were grown in MEGM medium (Lonza). MDA-MB-435 cells (ATCC) were grown in DMEM containing 10% inactivated fetal bovine serum (IFS). THP1 cells (ATCC) were grown in RPMI1640 containing 10% IFS with 5 mM β-mercaptoethanol. Cells were routinely tested for mycoplasma contamination.
Coculture assays
The HMLE-derived cells were seeded into culture dishes at 30% confluency, and grown into a ~70% confluent monolayer 18 hr later. The THP1 monocytes were pelleted by centrifugation at 300×g, resuspended in the MEGM medium at 1 million cells/ml, and laid on top of the monolayer cells (cell number of monocytes: monolayer cells = 3:2). The non-adherent monocytes were gently rinsed away after 1 hr by two washes of PBS, and the cells in the culture are replenished with fresh MEGM medium. Cells were trypsinized at indicated time points and resuspended in 2% IFS/PBS for FACS sorting. For conditioned medium collection, fresh MEGM medium was added to the coculture (without removing existing medium) after 1 hr, and the conditioned medium was collected after filtering through a 0.2 micron Acrodisc syringe filter (Pall Life Science #PN4192). For the peptide inhibitor experiments, the monolayer HMLER90hi cells were treated with the indicated peptide or DMSO for 40 min before and during the monocytes coculture.
Fluorescence activated cell sorting (FACS) and flow cytometry
Cells were prepared according to standard protocols and suspended in 2% IFS/PBS. DAPI (Life Technologies) was used to exclude dead cells. Cells were sorted on BD FACSAria SORP and analyzed on BD LSRII, using BD FACSDiva Software (BD Biosciences). Antibodies used are anti-CD90-APC (BD Biosciences #559869), anti-CD24-PE (BD Biosciences #555428).
Mammosphere/tumorsphere culture
Mammosphere/tumorsphere culture was performed as previously described22. The number of cells seeded per well was labeled for each graph in figures. Sphere numbers were counted between days 8 to 12.
RNA preparation and qRT-PCR analysis
Total RNA was isolated using the RNeasy Plus Mini kit (Qiagen) and reverse transcription was performed with High Capacity RNA-to-cDNA Kit (Applied Biosystem), both according to the manufacturer’s protocol. A cDNA sample prepared from 1 μg total RNA was used for each PCR. The PCR reactions using SYBR Green Mix I (Roche Diagnostics), data collection, and data analysis were performed on the LightCycler® 480 System (Roche Diagnostics). The thermal cycling parameters for the PCR were as follows: 95°C for 5 min, followed by 45 cycles of 95°C for 10 sec, 49°C for 7 sec, and 72°C for 25 sec. The relative mRNA quantity was normalized against the relative quantity of GAPDH mRNA in the same sample. Primers used are indicated in Supplementary Table 4.
Enzyme-linked immune-sorbent assay (ELISA)
Concentrations of cytokines IL-6, IL-8 and GM-CSF in conditioned media were detected by ELISA kits from Ray Biotech (IL-6 and GM-CSF) and EBiosciences (IL-8) according to manufacturers’ protocols. The absorbance was measured on Multiskan EX (Thermo Electron Corporation).
Cell proliferation assay
Cells were seeded in 96-well assay plates (Corning #3903) at 1000 cells/well, and the culture medium was replenished every 2 days. Cell proliferation was measured at indicated days after seeding with WST1 reagent (Roche 11644807001) according to manufacturer’s protocol.
Isolation of primary mouse macrophages and human monocytes
To isolate mouse intraperitoneal-exude macrophages (IPMCs), mice were injected with 2% v/v Bio-Gel P-100 (Bio-Rad #150-4174) intraperitoneally (1 ml/mouse). 3 days later, the mice were euthanized; RPMI media was injected intraperitoneally and then retrieved. The cells were then treated with Red Blood Cell Lysing Buffer (Sigma-Aldrich) and washed with PBS. The purity of the IPMCs was analyzed by flow cytometry after staining with anti-CD11b-PE (BD Biosciences #557397), anti-Ly-6G-AlexaFlour700 (BD Biosciences #561236) and anti-F4/80-PE-Cy7 (EBiosciences 25-4801-82). Mouse tumor-associated macrophages (TAMs) were isolated from dissociated mouse tumors previously grown from the HMLE-Ras CD90hi cells. Human monocytes were isolated from human peripheral blood mononuclear cells from healthy donors. Mouse TAMs and human monocytes were purified by magnetic activated cell sorting (MACS) with anti-F4/80-PE-Cy7 followed by anti-PE microbeads (Miltenyi Biotec), and human CD14 microbeads (Miltenyi Biotec), according to manufacturer’s protocol. The purity of mouse TAMs were directly analyzed by flow cytometry after MACS. The purity of human monocytes was analyzed by flow cytometry after staining with anti-CD14-PE-Cy7 (25-0149-71), anti CD11b-APC (17-0118-71) antibodies (both from EBiosciences).
Creation of stable cell lines
GFP-labeled HMLE, HMLE-Twist and HMLE-Slug cell lines were made by retroviral transduction using the pWZL-blast-GFP expression vector59. HMLE-Ras cell line (HMLER) was made by retroviral transduction with the pMSCV-HRasV12-IRES-GFP expression vector60. The tdTomato fluorescent protein-labeled THP1 monocytes (THP1-RFP) were made by lentiviral transduction using the pLV-tdTomato expression vector61. NF-κB super-repressor expressing HMLER90hi cells were made by retroviral transduction using the pBabe-puro-IκBα-S32A/S36A expression vector (Addgene plasmid 12332)62. Short hairpin RNAs (shRNA) targeting the mRNAs encoding CD90 (Thy1), EphA4, PLCγ1, PKCδ, IL-6, IL-8 and GM-CSF were expressed from pLKO.1-puro (Open Biosystems); the target sequences of these shRNA hairpins are indicated in Supplementary Table 5. All stable cell lines were generated via retroviral infection using HEK293T cells, as previously described63, and followed by selection with puromycin (2 ug/ml) or by cell sorting.
Western blotting
Phospho- and total- protein lysates were prepared with urea lysis buffer (9 M urea, 20 mM HEPES, pH 8.0), with freshly added phosphatase inhibitors (1 mM sodium orthovanadate, 5 mM sodium pyrophosphate and 1 mM β-glycerol-phosphate). For the separation of monocytes from the monolayer HMLE-derived cells, phosphatase inhibitors (1 mM sodium orthovanadate, 5 mM sodium pyrophosphate) were added to the culture medium before harvesting and the monocytes were repeatedly flushed off with the medium; the monolayer cells were rinsed with cold PBS and immediately lysed on the culture dish. The monocytes were then pelleted by centrifugation at 300xg for 5 min and then lysed. For Fig. 7c, nuclear and cytoplasmic extracts were prepared using NE-PER kit (Pierce). Concentrations were determined using the Dc protein assay (Bio-Rad). For Supplementary Fig. 6a, total protein was extracted with RIPA lysis buffer. Protein lysates were resolved on a 4%–12% Bis-Tris Gel, transferred to PVDF membranes, probed with primary antibodies overnight at 4°C and then with HRP-linked secondary antibodies (Cell Signaling Technology, CST) and visualized with ECL reagents (Thermo Scientific). The dilution, clone and catalog numbers of antibodies used are listed in Supplementary Table 6.
Co-immunoprecipitation (coIP) and Chromatin Immunoprecipitation (ChIP) Assay
CoIP of Twist and NF-κB p50 was performed using the HMLER90hi cells with endogenous high level of Twist. The monolayer cells were rinsed with cold PBS and lysed in the IP buffer (20 mM Tris, pH 7.5, 200 mM NaCl, 5 mM MgCl2, 0.1% NP40-substitute, 10% Glycerol, with 1 mM DTT, 0.5 mM PMSF and 1x protease inhibitor (Roche) fresh added before lysis). Cell lysates were incubated at 4°C with the anti-Twist antibody (Abcam # ab50887) or control IgG (CST, #5415) overnight (Supplementary Table 6). The protein-antibody mixture was then precipitated with the protein G magnetic beads (Invitrogen) according to manufacturer’s protocols. NF-κB p50 was then detected with the anti-p105/p50 antibody (CST, #3035) by western blot. ChIP assay was performed as previously described with the same antibodies64. The dilution, clone and catalog numbers of antibodies used are listed in Supplementary Table 6. The PCR primers used for confirming the protein bindings were: Promoter probe for IL6: TTCCAATCAGCCCCACCCG/GCTGGCAGTTCCAGGGCTAAGG. Control probe for IL6: TCTGGAGACTGGAGGGACAACC/GGACGCAGGCACGGCTCTA. Promoter probe for IL8: CATCAGTTGCAAATCGTGGAAT/GAGTGCTCCGGTGGCTTTTTAT. Control probe for IL8: AAGAGCATGAAGCAACAGTGGC/AATAGGAGGGCTTCAATAGAGG. Promoter probe for CSF2: AAGTTCTCTGGAGGATGTGGCTGC/CCATCTCAGCAGCAGTGTCTCTAC. Control probe for CSF2: TGGGGGATGCTGGGGTTATGTA/TAGCCCCCAGAAGACACCAGGAG.
Phosphatidylinositol-Specific Phospholipase C Protein (PI-PLC) treatment
THP1 monocytes were resuspended in PBS at 2 million cells/ml, and incubated with 1u/ml PI-PLC (Invitrogen #P-6466) at 37°C for 1 hr. Monocytes were then washed with PBS once and resuspended in MEGM medium for coculture experiments.
Animal experiments
All research involving animals complied with protocols approved by the MIT Committee on Animal Care. The number of cells and the time for tumor growth are indicated in the figure legends. To measure tumor-initiating cell/CSC frequency (Fig. 2a,g, 4h, 6e; Supplementary Fig. 2d,f), serial dilutions of cancer cell suspensions in 25 μl of a 1:1 MEGM and Matrigel (BD Biosciences) mixture were injected into the 4th mammary fat pads of 2–3 months old female NOD-SCID mice. CSC frequencies of the samples were determined using the ELDA webtool65. In TAMs+CSCs co-injection experiments (including the CSCs only control groups; Fig. 4a–f & 7d), the amount of Matrigel was reduced by 6-fold to minimize any effects of growth factors from the Matrigel. Thus, the cells were suspended in 10 μl of a 4:1 MEGM and Matrigel mixture and injected into the 4th mammary fat pads of 2-month old female Nude mice. For the MMTV-PyMT tumor cell transplantation experiment (Fig. 4i,j; Supplementary Fig. 4 d–j), naturally arising MMTV-PyMT tumors from 10–14 week old females were harvested (with their lungs being checked for visible macrometastases to gauge the metastatic capabilities of the tumor cells). The tumor cells were dissociated into single-cell suspension, subjected to negative selection by CD45, CD31 and Ter-119 microbeads (Miltenyi) and then sorted for CD90 expression. Separate MMTV-PyMT tumors were dissociated for the isolation of TAMs. 3x105 tumor cells were injected with or without 1.5x105 TAMs in a volume of 10 μl including 2 μl Matrigel into the 4th mammary fat pads of 2-month old female wildtype Fvb mice. Tumors and lungs were harvested after two months. Tumor weight was measured after draining of fluid in the cysts-containing tumors. Investigator was not blinded to the group allocation for the assessment of outcome. Mice were randomly allocated to each group by an independent person; however, no particular method of randomization was used. No statistical method was used to predetermine sample size.
Patient samples
The tissues used in this study were collected under a University of Pittsburgh Internal Review Board (UPCI 04-162). All patients were consented prior to surgery or any other clinically indicated procedure. All specimens are de-identified by an honest broker system and distributed for research. Tissues used for immunohistology were processed within 2 hours of surgery.
Immunohistostaining
Mouse tumors
Formalin-fixed paraffin-embedded xenograft tumor tissue slides were stained with primary antibodies mouse anti-α-SMA (Sigma), rat anti-F4/80 (Invitrogen, MF48000), rabbit anti-PLCγ1 (pY783)(Abcam, #ab53125), rabbit anti-cleaved caspase 3 (CST #9664) and mouse anti-Ki67 (BD Biosciences #550609), and the secondary antibodies used are goat anti-mouse Alexa594, goat anti-rat Alexa488, and donkey anti-rabbit Alexa 555 (Supplementary Table 6). Nuclear staining was performed with DAPI (Invitrogen). Slides were mounted in Prolong Gold anti-fade reagent (Invitrogen). Immunofluorescent staining was observed and photographed using a Nikon Eclipse E800 upright fluorescence microscope.
Patient tumor tissues
All tissues BrCA022T, BrCA032T, BrCA041T (invasive ductal carcinomas, ER+/PR+) and BrCA042T (invasive ductal carcinoma, ER−/PR−) were formalin-fixed and paraffin-embedded. Antigen retrieval of tissue sections were performed at 125°C for 20 min in pH 6.0-EDTA buffer (Dako), washed and blocked with blocking solution (PBS, 5% goat serum, 0.05% Tween 20) to reduce nonspecific antibody binding. All wash steps were performed with DAKO wash buffer. Immunofluorescence staining was performed using the following primary antibodies overnight at 4°C: monoclonal rabbit anti-human CD90 (Abcam, Cat. No. ab133350), mouse anti-human CD68 (Dako Cat. No. M0876). Negative Control primary antibodies were purchased from Dako (N1698 and N1699). After washing, biotinylated secondary goat anti-rabbit biotin (Dako Cat. No.E0432), goat anti-mouse Alexa488 antibody (CD90/CD68 double staining, Invitrogen Cat. No. A11001), or goat anti-mouse Cy5 antibody (CD90/CD68/CTK triple staining; Abcam Cat. No. ab97037) were applied for 1 hour at room temperature, followed by incubation with streptavidin-Cy3 (Sigma, Cat. No. 6402) for 30 minutes at room temperature. For triple staining mouse anti-CTK-FITC [AE1/AE3] (eBioscience, Cat. No. 53-9003-80) was incubated for 1 hr at room temperature. For detailed information on antibodies used see Supplementary Table 6. Final nuclear staining was performed with DAPI (Invitrogen, Cat. No. D1306). Slides were mounted in Prolong Gold anti-fade reagent (Invitrogen, Cat. No. P36934). Immunofluorescent staining was observed and photographed using a Nikon 90i motorized upright epi-fluorescence microscope using a 20X objective. Whole sections were photographed as a composite of individual “tiles” captured and joined using the NIS-Elements software. Individual CD90+/CD68+ cellular doublets were identified from the 20X whole section tiled images. Confocal images of the doublets were obtained using a Olympus Fluoview 1000 microscope (version 2.16 software) and a Plan Apo N 60X 1.42 N.A. objective, 4x zoom, and 0.45 μm sections (number varied depending on cells imaged: 13–17). Stacks were processed using MetaMorph (version 7.7) and autocontrast was applied consistently to all wavelengths.
Statistical analysis
Data are presented as mean ± SEM. Student’s t test (unpaired) was used to compare two groups (p< 0.05 being considered significant). Holm-Sidak’s multiple comparisons test was performed when one-way ANOVA was used to compare between more than two groups. Sample sizes were indicated in each corresponding figure legends.
Proteomic analysis of membrane pellet samples
SILAC media for HMLE-vector and HMLE-Twist cells were made based on MCDB-17066. For the amino acid labeling, either normal L-lysine and L-arginine or heavy-labeled 13C6-15N2 lysine and 13C6-15N4 arginine were supplemented at concentrations of 40 mg/L and 120 mg/L, respectively. Labeled cells were harvested after 8 cell doublings.
Membrane protein fractionation
Membrane proteins isolation was described in Supplementary Fig. 1b,c. The buffers used are as follows. Lysis buffer: 10 mM Tris, pH 7.5, 5 mM MgCl2, 0.5 mM PMSF, add 0.5% PMSF and 1% protease inhibitors (Sigma) fresh before use. Sucrose buffer: 10 mM Tris, pH 7.5, 1.25 M Sucrose (MW 342.3). Membrane proteins were pelleted with ultraclear tube (Beckman #344057) in Beckman ultracentrifuge (model# L8-M) with rotor SW50.1.
Solubilization, digestion, and peptide fractionation by off-gel electrophoresis
1 mg membrane protein pellets were solubilized in urea and disulfide bonds were reduced and alkylated, then proteins were deglycosylated with PNGaseF and digested with Lys-C, and trypsin. Solutions that began cloudy upon initial reconstitution were clear after overnight digestion. ~50 μg samples of peptide digest were separated by off-gel electrophoresis (OGE) according to isoelectric point into 11 fractions using an Agilent 3100 OFFGEL Fractionator (Agilent Technologies, Wilmington, DE) and 13 cm Immobiline Drystrips pH 3–10 (GE Healthcare BioSciences AB, Uppsala, Sweden). Experimental details have been previously described67.
Mass Spectrometry
Peptide off-gel electrophoresis fractions were analyzed with an automated nano LC-MS/MS system, consisting of an Agilent 1100 nano-LC system (Agilent Technologies, Wilmington, DE) coupled to either an LTQ-Orbitrap or an LTQ Orbitrap XL Fourier transform mass spectrometer (Thermo Fisher Scientific, San Jose, CA) equipped with a nanoflow ionization source (James A. Hill Instrument Services, Arlington, MA). Peptides were eluted from a 10 cm column (Picofrit 75 um ID, New Objectives) packed in-house with ReproSil-Pur C18-AQ 3 μm reversed phase resin (Dr. Maisch, Ammerbuch Germany) using either a 120 or 133 min. gradient at a flow rate of 200 nl/min to yield ~20 s peak widths as previously described75. Data-dependent LC-MS/MS spectra were acquired in ~3 s cycles; each cycle was of the following form: one full Orbitrap MS scan at 60,000 resolution followed by 8 MS/MS scans in the ion trap on the most abundant precursor ions using an isolation width of 3 m/z. Dynamic exclusion was enabled with a mass width of +/− 25 ppm, a repeat count of 1 and an exclusion duration of 45 sec. Charge-state screening was enabled along with monoisotopic precursor selection and non-peptide monoisotopic recognition to prevent triggering of MS/MS on precursor ions with unassigned charge or a charge state of 1. Normalized collision energy was set to 30 with an activation Q of 0.25 and activation time of 30 ms. 2 biological replicates were performed.
Protein identification and quantitation
All MS data was interpreted using the Spectrum Mill software package v4.1 beta (co-developed by us with Agilent Technologies, Santa Clara, CA). Similar MS/MS spectra acquired on the same precursor m/z within +/− 45 sec were merged, MS/MS spectra with precursor charge >4 and poor quality MS/MS spectra, which failed the quality filter by not having a sequence tag length > 0 (i.e., minimum of two masses separated by the in-chain mass of an amino acid) were excluded from searching. MS/MS spectra were searched against a UniProt database containing 78,369 human sequences (including isoforms and excluding fragments) downloaded from the UniProt web site on June 30, 2010, along with 73 common laboratory contaminant proteins appended. Search parameters included: ESI linear ion-trap scoring parameters, trypsin enzyme specificity with cleavage at K-P or R-P allowed, a maximum of 4 missed cleavages, 35% minimum matched peak intensity, +/− 20 ppm precursor mass tolerance, +/−0.7 Da product mass tolerance, and fixed/mix modifications that included carbamidomethylation of cysteines and either unlabeled Arg, Lys or SILAC labeled Arg-10, Lys-8. Allowed variable modifications were oxidized methionine, deamidation of asparagine, pyro-glutamic acid modification at N-terminal glutamine, and pyro-carbamidomethyl modification at N-terminal cysteine with a precursor MH+ shift range of −18 to 64 Da.
Identities interpreted for individual spectra were automatically designated as confidently assigned using the Spectrum Mill autovalidation module to apply target-decoy based false-discovery rate (FDR) scoring threshold criteria via a two-step auto threshold strategy at the spectral and protein levels. First, peptide mode was set to allow automatic variable range precursor mass filtering with score thresholds optimized to yield at spectral level FDR of 1.6% for each precursor charge state in each LC-MS/MS run. Second, protein mode was applied to further filter all the peptide-level validated spectra combined from the 22 LC-MS/MS runs from both replicates of an experiment using a minimum protein score of 20 and a maximum protein-level FDR of zero. Since the maximum peptide score is 25, the protein level step filters the results so that each identified protein is comprised of multiple peptides unless a single excellent scoring peptide was the sole match. The above criteria yielded target-decoy based FDR estimates for each sample replicate of 0.9 to 1.0% at the peptide-spectrum match level and 1.3 to 1.5 % at the distinct peptide level.
For scores calculated at the protein level and reporting of identified proteins, redundancy is addressed as follows. The protein score is the sum of the scores of distinct peptides. A distinct peptide is the single highest scoring instance of a peptide identified by an MS/MS spectrum. MS/MS spectra for a particular peptide may have been recorded multiple times, (i.e. both heavy and light SILAC labels, different precursor charge states, isolated from adjacent OGE fractions, modified by deamidation at Asn or oxidation of Met) but are still counted as a single distinct peptide. When a peptide sequence >8 residues long is contained in multiple protein entries in the sequence database, the proteins are grouped together and the highest scoring one and its accession number are reported. In some cases when the protein sequences are grouped in this manner there are distinct peptides which uniquely represent a lower scoring member of the group (isoforms, family members). Each of these instances spawns a subgroup and multiple subgroups are reported and counted towards the total number of proteins with each given related protein.subgroup numbers (e.g., the ephrin receptor tyrosine kinase family shares sufficient sequence homology that the observed shared and distinct peptides resulted in one protein group with 4 subgroups corresponding to EPHA2, EPHA4, EPHB2, and EPHB4 and are listed as subgroup members 214.1 thru 214.4 in Supplementary Table 1). GO annotations were then used to categorize all of the identified protein subgroups as being plasma membrane associated, extra cellular matrix derived, and/or tyrosine kinases. EphA4 is complicated by the fact that there is over 50% sequence identity among the Ephrin family members with distinct peptides detected for EphA2, A4, B2, B4. In each replicate a single distinct A4 tryptic peptide was observed, different ones each time, and both showed > 20-fold expression increase (data not shown). The peptides that are shared amongst family members and those distinct to the other three isoforms do not show significant expression change. The marginal distinct A4 evidence resulted from the median ratio of all its constituent peptides being below the significant change threshold in each replicate (Fig. 1a, Supplementary Table 1).
SILAC relative abundance ratios of proteins were determined using extracted ion chromatograms (XIC’s) for each sequenced peptide’s light and heavy labeled precursor ion pair in the intervening high resolution FT-MS scans of the LC-MS/MS runs as previously described75. Each individual protein’s SILAC ratio was calculated as the median of all SILAC precursor ion ratios corresponding to MS/MS spectra confidently assigned to peptides from that protein. Proteins were considered quantifiable if they were observed in each of 2 replicate experiments with at least 2 SILAC ratios measured in each experiment. Ratios for common laboratory contaminant proteins were filtered out of each dataset. Because two biological replicate experiments were performed with a light/heavy label-swap between experiments the contaminants were easily recognizable not only by identity but also by extreme quantitation ratios with nearly all signals in the light channel. To assign regulated proteins (Fig. 1a, Supplementary Table 1). We used the Limma package in the R environment to calculate moderated T-test p-values corrected by the Benjamini Hochberg method, as described previously17. In order to perform the moderated T-test, the protein SILAC ratios were standardized (distributions centered at zero and replicate 1 vs 2 diagonalized). Each log 2 protein ratio in a replicate experiment was standardized by subtracting the median and dividing by the standard deviation of all log2 protein ratios in that replicate.
Repeatability of experiments
Representative images were shown in Fig. 1–4,6,7; Suppl. fig. 1–4, 6. The experiments have been repeated as follows:
Main Fig: (1b,d) 3 repeats, (2b,c) 5 repeats. (3a,b) see fig., (3c) 3 repeats, (3e) 5 patient tumors, (3i) 25 tumor biopsies. 4d, 3 repeats, (6a–c,f,7e) 3 repeats. (6h,7c,f) 2 repeats. Suppl. Fig: (S1c) 3 repeats. (S2e,h) 4 repeats. (S2g) 2 repeats. (S3a,c,d,4c) 5 patient tumors. (S3b) 25 tumor biopsies. (S4e–g,i,) 2 repeats, (S6a,f,g) 2 repeats. (S6c) 3 repeats.
Data Deposition
The original mass spectra may be downloaded from MassIVE (http://massive.ucsd.edu) using the identifier: MSV000078799. The data is accessible at ftp://MSV000078799:a@massive.ucsd.edu.
Supplementary Material
Acknowledgments
We thank L. Ma and T. Shibue for critical review of the manuscript, R. Bronson for evaluating the histopathology, D.R. Mani and P. Thiru for statistical analyses and C. Baty for assistance in imaging. We also thank A. Oberle, J. Karlsson, E. Procopio for technical assistance.
This research was supported by the MIT Ludwig Center for Molecular Oncology and by grants from the Breast Cancer Research Foundation and National Institute of Health (NIH) (R01-CA078461, P01-CA080111 and U54-CA163109) to R.A.W. Additional support comes in part from the Department of Defense (BC032981 and BC044784), NIH (P30CA047904), the Hillman Foundation and the Glimmer of Hope Foundation to V.S.D. This work was also supported in part by the Broad Institute of MIT and Harvard, and by grants from NIH (U24CA160034) to S.A.C. R.A.W. is an American Cancer Society and Daniel K. Ludwig Foundation Cancer Research Professor. Postdoctoral fellowship supports for H.L. were from Cancer Research Institute (New York, NY) and the MIT Ludwig Center for Molecular Oncology.
References
- 1.Huber MA, Kraut N, Beug H. Molecular requirements for epithelial-mesenchymal transition during tumor progression. Curr Opin Cell Biol. 2005;17:548–558. doi: 10.1016/j.ceb.2005.08.001. [DOI] [PubMed] [Google Scholar]
- 2.Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139:871–890. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 3.Guo W, et al. Slug and Sox9 cooperatively determine the mammary stem cell state. Cell. 2012;148:1015–1028. doi: 10.1016/j.cell.2012.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mani SA, et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133:704–715. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Morel AP, et al. Generation of breast cancer stem cells through epithelial-mesenchymal transition. PLoS One. 2008;3:e2888. doi: 10.1371/journal.pone.0002888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buck E, et al. Loss of homotypic cell adhesion by epithelial-mesenchymal transition or mutation limits sensitivity to epidermal growth factor receptor inhibition. Mol Cancer Ther. 2007;6:532–541. doi: 10.1158/1535-7163.MCT-06-0462. [DOI] [PubMed] [Google Scholar]
- 7.Creighton CJ, et al. Residual breast cancers after conventional therapy display mesenchymal as well as tumor-initiating features. Proc Natl Acad Sci U S A. 2009;106:13820–13825. doi: 10.1073/pnas.0905718106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gupta PB, et al. Identification of selective inhibitors of cancer stem cells by high-throughput screening. Cell. 2009;138:645–659. doi: 10.1016/j.cell.2009.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li HJ, Reinhardt F, Herschman HR, Weinberg RA. Cancer-stimulated mesenchymal stem cells create a carcinoma stem cell niche via prostaglandin E2 signaling. Cancer Discov. 2012;2:840–855. doi: 10.1158/2159-8290.CD-12-0101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Malanchi I, et al. Interactions between cancer stem cells and their niche govern metastatic colonization. Nature. 2011;481:85–89. doi: 10.1038/nature10694. [DOI] [PubMed] [Google Scholar]
- 11.Merlos-Suarez A, et al. The intestinal stem cell signature identifies colorectal cancer stem cells and predicts disease relapse. Cell Stem Cell. 2011;8:511–524. doi: 10.1016/j.stem.2011.02.020. [DOI] [PubMed] [Google Scholar]
- 12.Wynn TA, Chawla A, Pollard JW. Macrophage biology in development, homeostasis and disease. Nature. 2013;496:445–455. doi: 10.1038/nature12034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Coussens LM, Zitvogel L, Palucka AK. Neutralizing tumor-promoting chronic inflammation: a magic bullet? Science. 2013;339:286–291. doi: 10.1126/science.1232227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gyorki DE, Asselin-Labat ML, van Rooijen N, Lindeman GJ, Visvader JE. Resident macrophages influence stem cell activity in the mammary gland. Breast Cancer Res. 2009;11:R62. doi: 10.1186/bcr2353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Elenbaas B, et al. Human breast cancer cells generated by oncogenic transformation of primary mammary epithelial cells. Genes Dev. 2001;15:50–65. doi: 10.1101/gad.828901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ong SE, et al. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol Cell Proteomics. 2002;1:376–386. doi: 10.1074/mcp.m200025-mcp200. [DOI] [PubMed] [Google Scholar]
- 17.Udeshi ND, et al. Methods for quantification of in vivo changes in protein ubiquitination following proteasome and deubiquitinase inhibition. Mol Cell Proteomics. 2012;11:148–159. doi: 10.1074/mcp.M111.016857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barker TH, Hagood JS. Getting a grip on Thy-1 signaling. Biochim Biophys Acta. 2009;1793:921–923. doi: 10.1016/j.bbamcr.2008.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wetzel A, et al. Human Thy-1 (CD90) on activated endothelial cells is a counterreceptor for the leukocyte integrin Mac-1 (CD11b/CD18) J Immunol. 2004;172:3850–3859. doi: 10.4049/jimmunol.172.6.3850. [DOI] [PubMed] [Google Scholar]
- 20.Lim E, et al. Transcriptome analyses of mouse and human mammary cell subpopulations reveal multiple conserved genes and pathways. Breast Cancer Res. 2010;12:R21. doi: 10.1186/bcr2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shipitsin M, et al. Molecular definition of breast tumor heterogeneity. Cancer Cell. 2007;11:259–273. doi: 10.1016/j.ccr.2007.01.013. [DOI] [PubMed] [Google Scholar]
- 22.Dontu G, et al. In vitro propagation and transcriptional profiling of human mammary stem/progenitor cells. Genes Dev. 2003;17:1253–1270. doi: 10.1101/gad.1061803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tsuchiya S, et al. Establishment and characterization of a human acute monocytic leukemia cell line (THP-1) Int J Cancer. 1980;26:171–176. doi: 10.1002/ijc.2910260208. [DOI] [PubMed] [Google Scholar]
- 24.Donnenberg VS, et al. Localization of CD44 and CD90 positive cells to the invasive front of breast tumors. Cytometry B Clin Cytom. 2010;78:287–301. doi: 10.1002/cyto.b.20530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Donnenberg AD, Zimmerlin L, Landreneau RJ, Luketich JD, Donnenberg VS. KIT (CD117) expression in a subset of non-small cell lung carcinoma (NSCLC) patients. PLoS One. 2012;7:1–11. doi: 10.1371/journal.pone.0052885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Medrek C, Ponten F, Jirstrom K, Leandersson K. The presence of tumor associated macrophages in tumor stroma as a prognostic marker for breast cancer patients. BMC Cancer. 2012;12:306. doi: 10.1186/1471-2407-12-306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tsutsui S, et al. Macrophage infiltration and its prognostic implications in breast cancer: the relationship with VEGF expression and microvessel density. Oncol Rep. 2005;14:425–431. [PubMed] [Google Scholar]
- 28.Ch’ng ES, Tuan Sharif SE, Jaafar H. In human invasive breast ductal carcinoma, tumor stromal macrophages and tumor nest macrophages have distinct relationships with clinicopathological parameters and tumor angiogenesis. Virchows Archiv: an international journal of pathology. 2013;462:257–267. doi: 10.1007/s00428-012-1362-4. [DOI] [PubMed] [Google Scholar]
- 29.Qian BZ, Pollard JW. Macrophage diversity enhances tumor progression and metastasis. Cell. 2010;141:39–51. doi: 10.1016/j.cell.2010.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Murray PJ, Wynn TA. Protective and pathogenic functions of macrophage subsets. Nat Rev Immunol. 2011;11:723–737. doi: 10.1038/nri3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lin EY, Nguyen AV, Russell RG, Pollard JW. Colony-stimulating factor 1 promotes progression of mammary tumors to malignancy. J Exp Med. 2001;193:727–740. doi: 10.1084/jem.193.6.727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ohri CM, Shikotra A, Green RH, Waller DA, Bradding P. The tissue microlocalisation and cellular expression of CD163, VEGF, HLA-DR, iNOS, and MRP 8/14 is correlated to clinical outcome in NSCLC. PLoS One. 2011;6:e21874. doi: 10.1371/journal.pone.0021874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gabrilovich DI, Ostrand-Rosenberg S, Bronte V. Coordinated regulation of myeloid cells by tumours. Nat Rev Immunol. 2012;12:253–268. doi: 10.1038/nri3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hartman ZC, et al. Growth of Triple-Negative Breast Cancer Cells Relies upon Coordinate Autocrine Expression of the Proinflammatory Cytokines IL-6 and IL-8. Cancer Res. 2013 doi: 10.1158/0008-5472.CAN-12-4524-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Korkaya H, Liu S, Wicha MS. Regulation of cancer stem cells by cytokine networks: attacking cancer’s inflammatory roots. Clin Cancer Res. 2011;17:6125–6129. doi: 10.1158/1078-0432.CCR-10-2743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wyckoff J, et al. A paracrine loop between tumor cells and macrophages is required for tumor cell migration in mammary tumors. Cancer Res. 2004;64:7022–7029. doi: 10.1158/0008-5472.CAN-04-1449. [DOI] [PubMed] [Google Scholar]
- 37.Tam WL, et al. Protein Kinase C alpha Is a Central Signaling Node and Therapeutic Target for Breast Cancer Stem Cells. Cancer Cell. 2013;24:347–364. doi: 10.1016/j.ccr.2013.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thomson S, Petti F, Sujka-Kwok I, Epstein D, Haley JD. Kinase switching in mesenchymal-like non-small cell lung cancer lines contributes to EGFR inhibitor resistance through pathway redundancy. Clinical & experimental metastasis. 2008;25:843–854. doi: 10.1007/s10585-008-9200-4. [DOI] [PubMed] [Google Scholar]
- 39.Gentile A, Lazzari L, Benvenuti S, Trusolino L, Comoglio PM. Ror1 is a pseudokinase that is crucial for Met-driven tumorigenesis. Cancer Res. 2011;71:3132–3141. doi: 10.1158/0008-5472.CAN-10-2662. [DOI] [PubMed] [Google Scholar]
- 40.Pasquale EB. Eph receptors and ephrins in cancer: bidirectional signalling and beyond. Nat Rev Cancer. 2010;10:165–180. doi: 10.1038/nrc2806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lamberto I, et al. Distinctive binding of three antagonistic peptides to the ephrin-binding pocket of the EphA4 receptor. Biochem J. 2012;445:47–56. doi: 10.1042/BJ20120408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhou L, et al. EphA4 signaling regulates phospholipase Cgamma1 activation, cofilin membrane association, and dendritic spine morphology. J Neurosci. 2007;27:5127–5138. doi: 10.1523/JNEUROSCI.1170-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Griner EM, Kazanietz MG. Protein kinase C and other diacylglycerol effectors in cancer. Nat Rev Cancer. 2007;7:281–294. doi: 10.1038/nrc2110. [DOI] [PubMed] [Google Scholar]
- 44.Kalo MS, Pasquale EB. Signal transfer by eph receptors. Cell Tissue Res. 1999;298:1–9. [PubMed] [Google Scholar]
- 45.Lu ZG, Liu H, Yamaguchi T, Miki Y, Yoshida K. Protein kinase Cdelta activates RelA/p65 and nuclear factor-kappaB signaling in response to tumor necrosis factor-alpha. Cancer Res. 2009;69:5927–5935. doi: 10.1158/0008-5472.CAN-08-4786. [DOI] [PubMed] [Google Scholar]
- 46.Hoffmann E, Dittrich-Breiholz O, Holtmann H, Kracht M. Multiple control of interleukin-8 gene expression. J Leukoc Biol. 2002;72:847–855. [PubMed] [Google Scholar]
- 47.Sasaki CY, Ghosh P, Longo DL. Recruitment of RelB to the Csf2 promoter enhances RelA-mediated transcription of granulocyte-macrophage colony-stimulating factor. J Biol Chem. 2011;286:1093–1102. doi: 10.1074/jbc.M110.119438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yoon S, et al. NF-kappaB and STAT3 cooperatively induce IL6 in starved cancer cells. Oncogene. 2012;31:3467–3481. doi: 10.1038/onc.2011.517. [DOI] [PubMed] [Google Scholar]
- 49.Ryan KM, Ernst MK, Rice NR, Vousden KH. Role of NF-kappaB in p53-mediated programmed cell death. Nature. 2000;404:892–897. doi: 10.1038/35009130. [DOI] [PubMed] [Google Scholar]
- 50.Li S, et al. TWIST1 associates with NF-kappaB subunit RELA via carboxyl-terminal WR domain to promote cell autonomous invasion through IL8 production. BMC Biol. 2012;10:73. doi: 10.1186/1741-7007-10-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gisina AM, et al. Detection of minor subpopulations of colorectal adenocarcinoma cells expressing cancer stem cell markers. Bull Exp Biol Med. 2011;151:234–238. doi: 10.1007/s10517-011-1297-9. [DOI] [PubMed] [Google Scholar]
- 52.He J, et al. CD90 is identified as a candidate marker for cancer stem cells in primary high-grade gliomas using tissue microarrays. Mol Cell Proteomics. 2012;11 doi: 10.1074/mcp.M111.010744. M111 010744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jiang J, et al. Trastuzumab (herceptin) targets gastric cancer stem cells characterized by CD90 phenotype. Oncogene. 2012;31:671–682. doi: 10.1038/onc.2011.282. [DOI] [PubMed] [Google Scholar]
- 54.Yang ZF, et al. Significance of CD90+ cancer stem cells in human liver cancer. Cancer Cell. 2008;13:153–166. doi: 10.1016/j.ccr.2008.01.013. [DOI] [PubMed] [Google Scholar]
- 55.Oki M, et al. Overexpression of the receptor tyrosine kinase EphA4 in human gastric cancers. World J Gastroenterol. 2008;14:5650–5656. doi: 10.3748/wjg.14.5650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Oshima T, et al. Overexpression of EphA4 gene and reduced expression of EphB2 gene correlates with liver metastasis in colorectal cancer. Int J Oncol. 2008;33:573–577. [PubMed] [Google Scholar]
- 57.Binda E, et al. The EphA2 receptor drives self-renewal and tumorigenicity in stem-like tumor-propagating cells from human glioblastomas. Cancer Cell. 2012;22:765–780. doi: 10.1016/j.ccr.2012.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gyorffy B, et al. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Res Treat. 2010;123:725–731. doi: 10.1007/s10549-009-0674-9. [DOI] [PubMed] [Google Scholar]
- 59.Orimo A, et al. Stromal fibroblasts present in invasive human breast carcinomas promote tumor growth and angiogenesis through elevated SDF-1/CXCL12 secretion. Cell. 2005;121:335–348. doi: 10.1016/j.cell.2005.02.034. [DOI] [PubMed] [Google Scholar]
- 60.Yu M, et al. A developmentally regulated inducer of EMT, LBX1, contributes to breast cancer progression. Genes Dev. 2009;23:1737–1742. doi: 10.1101/gad.1809309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shibue T, Weinberg RA. Integrin beta1-focal adhesion kinase signaling directs the proliferation of metastatic cancer cells disseminated in the lungs. Proc Natl Acad Sci U S A. 2009;106:10290–10295. doi: 10.1073/pnas.0904227106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tergaonkar V, Pando M, Vafa O, Wahl G, Verma I. p53 stabilization is decreased upon NFkappaB activation: a role for NFkappaB in acquisition of resistance to chemotherapy. Cancer Cell. 2002;1:493–503. doi: 10.1016/s1535-6108(02)00068-5. [DOI] [PubMed] [Google Scholar]
- 63.Stewart SA, et al. Lentivirus-delivered stable gene silencing by RNAi in primary cells. RNA. 2003;9:493–501. doi: 10.1261/rna.2192803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Han J, et al. Tbx3 improves the germ-line competency of induced pluripotent stem cells. Nature. 2010;463:1096–1100. doi: 10.1038/nature08735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hu Y, Smyth GK. ELDA: extreme limiting dilution analysis for comparing depleted and enriched populations in stem cell and other assays. J Immunol Methods. 2009;347:70–78. doi: 10.1016/j.jim.2009.06.008. [DOI] [PubMed] [Google Scholar]
- 66.Hammond SL, Ham RG, Stampfer MR. Serum-free growth of human mammary epithelial cells: rapid clonal growth in defined medium and extended serial passage with pituitary extract. Proc Natl Acad Sci U S A. 1984;81:5435–5439. doi: 10.1073/pnas.81.17.5435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Naba A, et al. The matrisome: in silico definition and in vivo characterization by proteomics of normal and tumor extracellular matrices. Mol Cell Proteomics. 2012;11 doi: 10.1074/mcp.M111.014647. M111 014647. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.