Abstract
BACKGROUND
In previous clinical trials involving children with X-linked severe combined immunodeficiency (SCID-X1), a Moloney murine leukemia virus–based γ-retrovirus vector expressing interleukin-2 receptor γ-chain (γc) complementary DNA successfully restored immunity in most patients but resulted in vector-induced leukemia through enhancer-mediated mutagenesis in 25% of patients. We assessed the efficacy and safety of a self-inactivating retrovirus for the treatment of SCID-X1.
METHODS
We enrolled nine boys with SCID-X1 in parallel trials in Europe and the United States to evaluate treatment with a self-inactivating (SIN) γ-retrovirus vector containing deletions in viral enhancer sequences expressing γc (SIN-γc).
RESULTS
All patients received bone marrow–derived CD34+ cells transduced with the SIN-γc vector, without preparative conditioning. After 12.1 to 38.7 months of follow-up, eight of the nine children were still alive. One patient died from an overwhelming adenoviral infection before reconstitution with genetically modified T cells. Of the remaining eight patients, seven had recovery of peripheral-blood T cells that were functional and led to resolution of infections. The patients remained healthy thereafter. The kinetics of CD3+ T-cell recovery was not significantly different from that observed in previous trials. Assessment of insertion sites in peripheral blood from patients in the current trial as compared with those in previous trials revealed significantly less clustering of insertion sites within LMO2 , MECOM, and other lymphoid proto-oncogenes in our patients.
CONCLUSIONS
This modified γ-retrovirus vector was found to retain efficacy in the treatment of SCID-X1. The long-term effect of this therapy on leukemogenesis remains unknown. (Funded by the National Institutes of Health and others; ClinicalTrials.gov numbers, NCT01410019, NCT01175239, and NCT01129544.)
X-linked severe combined immunodeficiency (SCID-X1) is caused by mutations in the gene encoding the interleukin-2 receptor γ chain (IL2RG) that result in a lack of response to common γ-chain (γc)–dependent cytokines, an absence of T-cell and natural killer (NK)–cell development, and impairment of B-cell function.1,2 Death from community-acquired or opportunistic infection usually occurs before 1 year of age unless allogeneic hematopoietic stem-cell transplantation (HSCT) is performed. The immunologic defect in children with SCID-X1 obviates the requirement for a preparative regimen before transplantation.3–6 Standard allogeneic HSCT with matched-sibling donors is associated with an 85 to 90% overall survival rate.7–11 However, transplantation with the use of mismatched related, matched unrelated, or umbilical-cord-blood donors or transplantation in patients with ongoing infection is associated with lower survival rates and higher rates of complications, including graft-versus-host disease.8–10 For instance, mortality among patients with SCID who have severe protracted infection at the time of HSCT is approximately 50%.8,9
In previous gene-therapy trials, 20 infants with SCID-X1 who did not have matched family donors were treated with an infusion of autologous CD34+ bone marrow cells transduced with a first-generation Moloney murine leukemia virus vector expressing the γc complementary DNA (MFG-γc) and containing duplicated viral enhancer sequences within the long terminal repeats (LTRs). The therapy evaluated in these trials resulted in reconstitution of T cells and in disease-free survival similar to that associated with matched-sibling-donor allogeneic HSCT.12–16 However, insertional mutagenesis leading to T-cell acute lymphoblastic leukemia occurred in 5 of the 20 patients, with transactivation of the LMO2 or CCND2 pro-to-oncogenes in the five leukemias.17–19
To improve safety while maintaining immunologic efficacy, we developed a self-inactivating (SIN) γ-retroviral vector (pSRS11.EFS.IL2RG. pre*, abbreviated SIN-γc) in which the Moloney murine leukemia virus LTR U3 enhancer was deleted. The modified vector expressed the IL2RG complementary DNA from the eukaryotic human elongation factor 1α short promoter, which directs ubiquitous expression in mammalian cells, and has previously been shown to be less mutagenic in vitro.20,21 Here we present interim results from a study of patients with SCID-X1 treated with this enhancer-deleted SIN-γc.
METHODS
VECTOR PRODUCTION
A genetic map of the vector is shown in Figure S1 in the Supplementary Appendix, available with the full text of this article at NEJM.org. The design and in vitro testing of the expression and immunologic efficacy of the SIN-γc vector and the production of the vector have been described elsewhere.20–22
PATIENTS AND CLINICAL PROTOCOL
We enrolled nine boys with confirmed IL2RG mutations who had immunologic profiles characteristic of SCID-X1 in parallel phase 1/2 trials conducted in Paris (ClinicalTrials.gov number, NCT01410019; five patients), London (NCT01175239, no patients), and the United States (NCT01129544; two patients in Boston, one patient in Cincinnati, and one patient in Los Angeles). At all sites, enrollment was offered for infants and children who either lacked a HLA-identical related or unrelated donor or had an active, therapy-resistant infection. In Paris, because of the different requirements of the French regulatory agencies, the presence of a therapy-resistant infection was a requirement for study entry. Written informed consent was obtained from the guardians or parents of all patients in accordance with local institutional review board–approved protocols.
CD34+ cells were purified from bone marrow with the use of the CliniMACS system. The cells were then subjected to transduction with clinical-grade SIN-γc supernatant,22 with the use of published regimens13,14 that were nearly identical to those used in the previous SCID-X1 gene-therapy trials. Common standard operating procedures were used at all sites. Supportive clinical care was delivered in accordance with local institutional norms.
INSERTION-SITE AND CLONALITY ANALYSIS
For the analysis of integration sites, DNA was purified from samples of blood cells and analyzed with the use of methods described elsewhere23–26 (see the Supplementary Appendix). A total of 2.9×104 unique integration sites representing 2.8×105 sequence reads were obtained from patients in the current trial and were compared with 1.3×10 4 unique integration sites representing 2.7×105 sequence reads collected from patients in the two previous trials.15,19,27–29 The abundance of cell clones was assessed by counting the number of different random break points that gave rise to the capture of each integration site and was analyzed statistically by means of the sonic Length method.24 Analysis of integration-site genomic intervals, termed “integration clumps,” was performed with the use of scan statistics,25 which allows analysis without assumptions about the genomic length of clumps or the number of integration sites involved. All integration-site data sets have been deposited in the National Center for Biotechnology Information Sequence Read Archive under the accession number SRP046756.
STATISTICAL ANALYSIS
The coprimary end points of the trial were the reconstitution of CD3+ cell number and function and the occurrence of severe adverse events related to gene therapy. To compare the repeated measures of CD3+ cell recovery in this trial with the combined data from the two previous trials, a generalized-estimating-equation approach (with an exchangeable correlation structure) was applied,30 with the use of the quasi-likelihood under the independence-model-criterion statistic31 to identify the preferred model. A log10 transformation was applied to exponentially distributed CD3+ cell counts (with a continuity correction of 0.01 added if CD3+ cell counts equaled zero) to meet normality assumptions. At 3 and 6 months, a two-sided Wilcoxon rank-sum test was used for the comparison. Analyses were performed according to the intention-to-treat principle. P values were two-sided, and values of less than 0.05 were considered to indicate statistical significance.
RESULTS
CHARACTERISTICS OF THE PATIENTS
All nine boys enrolled had a profound deficiency of autologous T cells. Patients 3, 5, 7, and 9 had maternal T-cell engraftment; in Patient 3, maternal engraftment was at a high level (i.e., >95% of T cells were of maternal origin). Eight of the nine patients had no expression of γc on the cell surface of peripheral-blood mononuclear cells. Eight patients had SCID-related infections, including three with disseminated bacille Calmette–Guérin infection, one with Epstein–Barr virus–driven lymphoproliferation, and one with severe systemic adenoviral disease (Table 1).
Table 1.
Characteristics of the Patients.*
| Patient No. |
Age at Gene Therapy |
Mutation and Predicted Effect |
γc | Maternal T-Cell Engraftment |
CD3+ T-Cell Count |
Infection History | CD34+ T-Cell Dose |
VCN of Graft |
Follow- up |
Status |
|---|---|---|---|---|---|---|---|---|---|---|
| mo |
cells/ mm3 |
cells/kg body weight |
copies/ cell |
mo | ||||||
| 1 | 8.3 | c.236G→C p.Trp74Cys |
No | No | 0 | Disseminated BCG,† pneumonitis |
7.6×106 | 1.9 | 38.7 | Alive, infections resolved |
| 2 | 5.8 | c.266A→G p.Tyr89Cys |
No | No | 1 | Oral ulcers during continuous acyclovir treatment† |
7.3×106 | 2.82 | 38.7 | Alive, infections resolved |
| 3 | 5.5 | c.676C→T p.Arg226Cys |
No | Yes | 8051‡ | RSV,† disseminated BCG,† CMV,† EBV LPD† |
10.0×106 | 1.2 | 35.9 | Alive, infections resolved |
| 4 | 6.8 | c.854G→A p.Arg285Gln |
No | No | 0 | Disseminated BCG,† Pneumocystis jiroveci pneumonia |
7.8×106 | 1.1 | 29.1 | Alive, infections resolved |
| 5 | 9.0 | c.674G→T p.Ser225Ile |
No | Yes | 44 | Systemic severe adenovirus with hepatitis† |
10.0×106 | 0.3 | Died | Died from infection |
| 6 | 10.5 | c.185G→C p.Cys62Ser |
No | No | 11 | Disseminated BCG† | 11.7×106 | 2.92 | 21.6 | Alive, receiving BCG suppressive medication |
| 7 | 3.9 | c.396T→G p.Leu132Arg |
No | Yes | 60§ | None | 9.1×106 | 0.35 | Off trial at 6 mo |
Alive after cord-blood transplantation |
| 8 | 8.2 | c.961_962insC p.Leu321fsX327 |
Yes | No | 0 | P. jiroveci pneumonia | 3.7×106¶ | 0.53 | 20 | Alive, infections resolved |
| 9 | 8.0 | c.202G→A p.Glu68Lys |
No | Yes | 42 |
P. jiroveci pneumonia, rotavirus intestinal infection† |
5.3×106 7.3×106║ |
0.25, 0.74 | 12.1 | Alive, infections resolved |
BCG denotes bacille Calmette–Guérin, CMV cytomegalovirus, EBV Epstein–Barr virus, γc γ-chain expression, LPD lymphoproliferative disease, and VCN vector copy number.
Infection was ongoing at the time of gene therapy.
The cell count was measured after treatment with fludarabine.
The cell count was measured after treatment with antithymocyte globulin.
Patient 8 received a second infusion 17.5 months after the first infusion; the CD34+ T-cell dose of the second infusion was 20×106 cells per kilogram, and the VCN was 0.51 copies per cell. Data for this patient beyond the time of the second infusion were not included in the analysis.
The interval between the first and second infusions was 1.2 months.
TRANSDUCTION AND PRODUCT CHARACTERISTICS
The median age at infusion of autologous bone marrow–derived CD34+ cells transduced with the SIN-γc vector was 8.0 months. Two patients received treatment for maternal engraftment that was high level, symptomatic, or both. Patient 2 received two doses of fludarabine (total, 80 mg per square meter of body-surface area), on days −3 and −2 before receiving transduced cells, which transiently reduced the massive number of maternally engrafted T cells. Patient 7 received three doses of rabbit antithymocyte globulin (total, 13 mg per kilogram of body weight), on days −23, −13, and −11, for treatment of symptomatic maternal graft-versus-host disease, which was manifested as diffuse erythroderma, without hepatic or gastrointestinal involvement. No myelosuppressive chemotherapy was given to any of the patients. The median dose of CD34+ cells infused was 7.8×106 per kilogram (range, 3.7×106 t o 11.7×106) with vector copy numbers ranging from 0.25 to 2.92 copies per cell (Table 1). Patient 9 received a second infusion of transduced cells 1 month after the first infusion, because of the low vector copy number of the initial product. Patient 8, who received cells with a low vector copy number, did not reach the primary end point of the trial and received a second infusion of transduced cells 17.5 months after the initial infusion, after full review by the data and safety monitoring board of the U.S. sites, which recommended proceeding to additional therapy. The family of Patient 8 gave consent for treatment in accordance with an amended protocol approved by the institutional review board of that site and reviewed by the Food and Drug Administration, allowing for a second infusion of transduced cells. Data on this patient are included in the insertion-site analysis and the analysis of adverse events up to the time of the second infusion of transduced cells but not afterward, as is appropriate for an intention-to-treat analysis of the first infusion.
IMMUNOLOGIC AND CLINICAL FOLLOW-UP
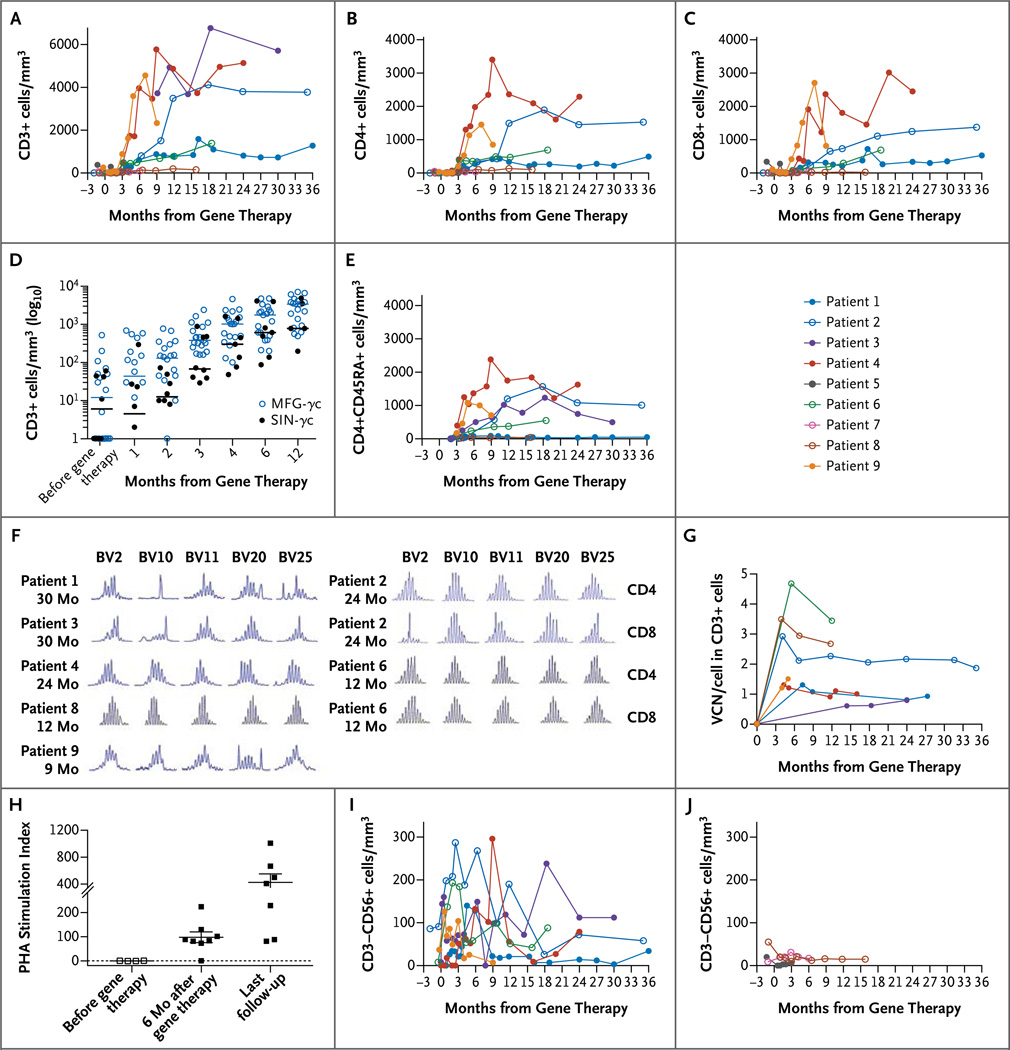
Eight of the nine children have survived, with a median follow-up of 29.1 months (range, 12.1 to 38.7). Preexisting infections were resolved in all patients except Patient 5, who died from a preexisting disseminated adenovirus infection 4 months after gene therapy and before full reconstitution of the T-cell compartment with gene-modified cells (Table S1 in the Supplementary Appendix). Patient 7, who received a graft with a low vector copy number (Table 1), had no evidence of gene marking (i.e., detection of vector genomic DNA by means of quantitative polymerase chain reaction) in T cells; therefore, he received a mismatched umbilical-cord-blood transplant 8 months after gene therapy. At the most recent follow-up assessment, 15 months after transplantation, he was doing well. CD3+, CD4+, and CD8+ T-cell recovery was achieved and sustained in six of the seven remaining patients (Fig. 1A, 1B, and 1C and Table 2). There was no significant difference in the kinetics of T-cell reconstitution between the patients in the current trial and those in the previous trials, either at 6 months or during the first year (P = 0.39 and P = 0.28, respectively) (Fig. 1D).
Figure 1. Immune Reconstitution and Gene Marking after Gene Therapy.
Panels A, B, and C show changes over time in the numbers of CD3+, CD4+, and CD8+ lymphocytes, respectively. Panel D shows CD3+ cell counts at the indicated times after gene therapy, compared between the 20 patients enrolled in previous trials using the MFG-γc vector (open blue circles) and the 8 patients enrolled in the current trials using self-inactivating γ-retrovirus (SIN-γc) (solid black circles) (P = 0.28). Patient 3 was excluded because of high-level maternal engraftment. Panel E shows changes over time in naive CD4+CD45RA+ lymphocyte numbers. Panel F shows T-cell receptor diversity in T cells of 7 patients after gene therapy. The length of complementarity-determining region 3 (CDR3) in each indicated T-cell receptor beta chain variable (TCRBV) gene family was measured after amplification with family-specific primers. The horizontal axis represents CDR3 length, and the vertical axis represents the frequency of sequences with a given CDR3 length; a Gaussian distribution of CDR3 lengths is indicative of normal diversity. Panel G shows that vector copy number (VCN) in peripheral-blood CD3+ lymphocytes was detectable in 7 patients and was sustained over time. Panel H shows T-cell proliferation in response to phytohemagglutinin (PHA), measured 6 months after gene therapy and at the last follow-up. Panels I and J show increases in CD3–CD56+ (natural killer) lymphocyte numbers in patients with CD34+ cells infused with a VCN of at least 0.7 copies per cell (Panel I) but not in those infused with a VCN of less than 0.7 copies per cell (Panel J).
Table 2.
Cellular Immunologic Outcome after Gene Therapy.*
| Patient No. |
Time of Assessment |
CD3+ | CD4+ | CD8+ | CD4+ CD45RA+ |
CD4+ CD45RA+ CD31+ |
Phytohemagglutinin | TREC | Proliferation, Antigen- Specific |
|
|---|---|---|---|---|---|---|---|---|---|---|
|
mo after gene therapy |
cells/mm3 | counts/min | SI |
no./105 PBMCs |
counts/min | |||||
| 1 | 36 | 1275 | 493 | 527 | 49 | 19 | 16,500† | 10 | 16 | 2,600,†‡ 10,500§ |
| 2 | 35 | 3774 | 1527 | 1374 | 1009 | 765 | 219,836 | 1007 | 997 | 7,552†‡ |
| 3 | 30 | 6141 | 1246 | 3916 | 498 | 361 | 81,500 | 81 | 204 | 62,000¶ |
| 4 | 24 | 5135 | 2291 | 2449 | 1627 | 893 | 75,000║ | 499 | 663 | 20,500,** 19,000† |
| 6 | 18 | 1389 | 689 | 689 | 252 | 237 | 118,586†† | 408†† | 1374 | ND |
| 8 | 15 | 154 | 102 | 25 | 41 | 58 | 104,122 | 666 | 191 | 2,237†‡ |
| 9 | 9 | 2331 | 851 | 814 | 706 | 485 | 88,000‡‡ | 88 | 716 | 2,200†‡§§ |
Patient 5 died, and Patient 7 did not have reconstitution with gene-marked cells and was not included in the trial after 6 months. ND denotes not done, PBMCs peripheral-blood mononuclear cells, SI stimulation index, and TREC T-cell receptor excision circle.
The value was subnormal for proliferation in response to PHA or to antigens as measured at each institution.
The test was for tetanus toxoid antigen.
The test was for varicella–zoster virus antigen.
The test was for tuberculin antigen.
The assay was performed at 20 months.
The test was for Candida albicans antigen.
The assay was performed at 12 months.
The assay was performed at 7 months.
The assay was performed at 4 months.
At the most recent assessment of T cells (at a median of 24 months), these seven patients had evidence of naive CD4+CD45RA+ T-cell generation, recent thymic emigrants (CD4+CD45RA+CD31+), and T-cell–receptor excision circles (Fig. 1E and Table 2). T-cell diversity was polyclonal in most patients, with some showing skewed distribution in certain T-cell–receptor beta variable (TCRBV) gene families, as was previously observed in patients who had disseminated bacille Calmette– Guérin infection (Fig. 1F). All seven remaining patients had gene marking in T cells and a return of phytohemagglutinin-induced T-cell proliferation to the normal range, as well as antigen-induced T-cell proliferation after infection or immunization (Fig. 1G and 1H and Table 2).
Higher vector copy numbers were correlated with successful T-cell engraftment. The six patients with a vector copy number in the CD34+ graft of at least 0.7 copies per cell had T-cell reconstitution, which was also heralded by an early rise in CD56+ NK cells (Fig. 1I). Patient 8, with a vector copy number of 0.53 copies per cell in the infused cells, had responsiveness to phytohemagglutinin after receiving gene-modified cells, but his T-cell numbers never exceeded 300 per cubic millimeter. He received a second infusion of gene-modified cells at 17.5 months after the first infusion and has remained clinically well.
Gene marking in CD3+ T cells (Fig. 1G) was sustained over time. Patients who had recovery of NK cell numbers also had gene marking in CD56+ NK cells (Fig. 1I, and Fig. S2 in the Supplementary Appendix). In contrast, as expected in the absence of conditioning, minimal and variable levels of gene marking were detected in the B-cell or granulocyte lineages (0 to 0.17 and 0 to 0.11 copies per cell, respectively), and a very low percentage of CD27+ memory B cells was detected at the most recent follow-up. The majority of circulating B cells had an immature naive IgD+CD27− phenotype (Table 3). Among the seven patients, three had normal serum IgA levels for their age, and four had normal serum IgM levels for their age (Table 3). All patients currently continue to be treated with intravenous immune globulin. On the basis of historical data obtained in studies of other series of patients treated with gene therapy, we conclude that the SIN-γc vector and the MFG-γc vector have similar efficacy in the reconstitution of T cells, T-cell proliferative responses, and immunity.
Table 3.
Humoral Immunologic Outcome after Gene Therapy.*
| Patient No. |
Time of Assessment |
CD19+ | CD19+ CD27+ |
CD27+ | IgA | IgM |
|---|---|---|---|---|---|---|
|
mo after gene therapy |
cells/mm3 | % | mg/dl | |||
| 1 | 19 | 391† | 22 | 2.0 | 206 | 109 |
| 2‡ | 35 | 2118 | 155 | 7.3 | 139 | 180 |
| 3 | 30 | 2670 | 53 | 2.0 | <5 | 91 |
| 4 | 24 | 2686 | — | — | 15 | 24 |
| 6 | 18 | 891 | 127 | 14.3 | <7§ | 27§ |
| 8 | 12 | 381¶ | 64 | 8.8 | 10 | 85 |
| 9 | 9 | 1332 | — | — | <5 | 100 |
All patients received immune globulin replacement therapy. Patient 5 died, and Patient 7 did not have reconstitution with gene-marked cells and was not included in the trial after 6 months.
The cell count was measured at 36 months.
The patient was tested for tetanus antibody response after suspension of immune globulin replacement for 3 months and three rounds of vaccination; the results were negative.
The assay was performed at 12 months.
The cell count was measured at 15 months.
SAFETY ANALYSIS
No severe adverse events related to the gene-transfer vector or cell manipulations have been reported to date in any of the children. The results of tests for the presence of replication-competent retrovirus have been uniformly negative. In the previous two trials, in which the MFG-γc vector was used, leukemia had a latency period of 2.0 to 5.5 years.17–19 Our patients have been followed for 12.1 to 38.7 months, with a median of 29.1 months of follow-up, and no leukemia has occurred to date. As a surrogate for leukemia at this earlier time point, we analyzed the genome for vector insertions affecting enhanced reconstitution, termed “clonal skewing.”32 We analyzed integration-site distributions from a total of 125 serial sorted peripheral-blood samples from eight patients enrolled in the current trial (Patient 5 was excluded) and compared the results with published data from patients in the first trials (pooled samples collected from 2 to more than 100 months after treatment). Data from the patients in the two previous trials showed no major differences and were therefore pooled.
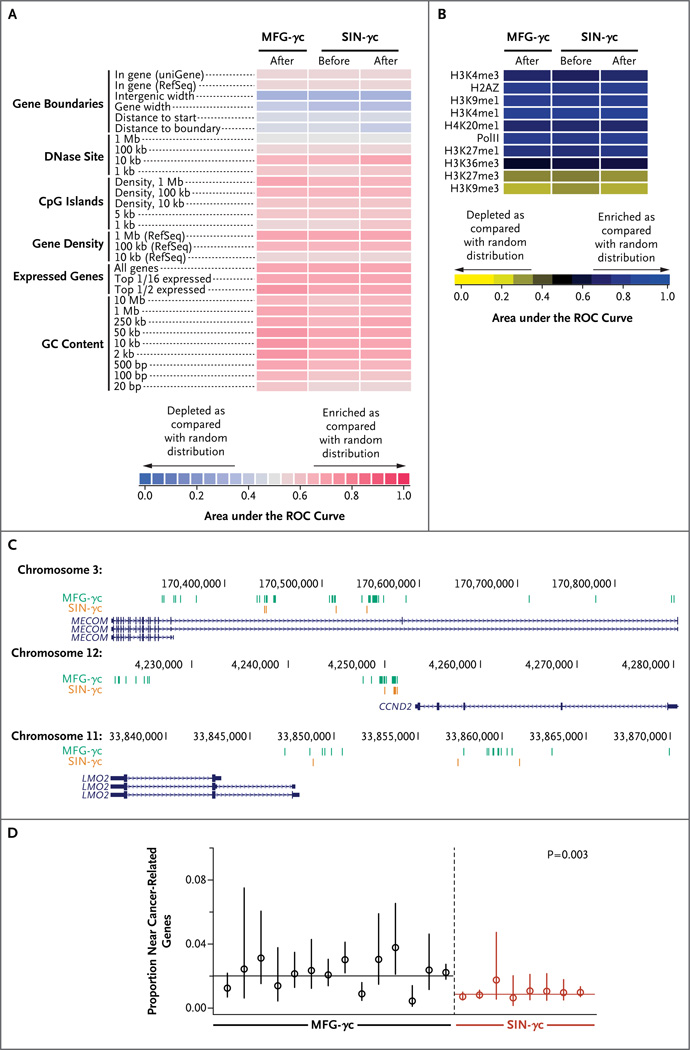
The global distributions of integration sites were similar between the MFG-γc trials and the current trial, showing elevated frequency near transcription start sites, gene-dense regions, and epigenetic marks associated with active transcription units (e.g., H3K4me1 and H3K4me3, H3K27me1, and RNA polymerase II) (Fig. 2A and 2B), and integration-site diversity was in the same range for all three trials (Fig. S3 in the Supplementary Appendix). Integration sites near gene-sparse regions and heterochromatic marks (H3K27me3 and H3K9me3) were recovered less frequently. Thus, the alterations to the SIN-γc vector did not have major effects on global associations of integration sites with these genomic features.
Figure 2. Analysis of Integration-Site Distributions in Patients Treated with the SIN Vector.
Panel A shows a heat map summarizing the placement of integration sites (data sets in columns) relative to mapped genomic features (in rows). Each column summarizes results for a pool of unique integration sites from all patients in each trial or the pretransplantation pools from patients in the current trial. Each row indicates a form of genomic annotation; relevant databases are given in parentheses. Some associations were measured with the use of a sliding window of defined length; because the most meaningful length for comparison was often not known in advance, comparisons over multiple different lengths are shown (indicated by numbers on the left). Colors indicate departures from random distributions; darker shades of red indicate more strongly positive associations, and darker shades of blue indicate more strongly negative associations, as compared with the random distribution. Associations were summarized with the use of the receiver-operating-characteristic (ROC) method. The integration frequency relative to gene activity (“Expressed genes”) was quantified as for gene density, but only genes in the 1/16 highest expression category or top half expression category in 1-Mb intervals were counted. Panel B shows the distribution of integration sites relative to published mapped sites of epigenetic modification in hematopoietic progenitor cells (CD34+CD133+ cells).33 Darker shades of blue indicate more strongly positive associations, and darker shades of yellow indicate more strongly negative associations, as compared with the random distribution. Panel C shows the three top-scoring clumps of integration sites, all of which were enriched in the trials with the MFG-γc vector relative to the current trial (a detailed description is provided in the report in the Supplementary Appendix). The numbers denote coordinates on the indicated chromosome; blue lines denote mapped transcripts, and vertical lines indicate the positions of integration sites from each trial (the MFG-γc trial in green and the SIN-γc trial in orange). Clumps were found at MECOM (top), CCND2 (middle), and LMO2 (bottom). Panel D shows the comparison of the frequencies of integration sites near the lymphoid proto-oncogenes. Each point represents an individual patient who has undergone gene therapy, and the P value of 0.003 is for the comparison between trials on a per-patient basis (i.e., each patient was analyzed as a single data point). The integration-site data sets are catalogued in the Methods section of the Supplementary Appendix.
To investigate the potential influence of integration-associated enhanced proliferation in vivo, we used scan statistics25 to assess the possible enrichment of integration clumps near genes such as LMO2 and CCND2 (a detailed report is provided in the Supplementary Appendix). In data from the first SCID-X1 trials with LTR-intact vectors, clusters of integration sites were found near LMO2 and CCND2 , which suggested that integration of the vector near these genes promoted clonal expansion.17,19,27–29 Twenty-one clumps were found that differed between the MFG-γc trials and the current trial (false discovery rate, 0.041). The three highest-scoring clumps contained more integration sites in samples from the MFG-γc trials. These highest-scoring clumps were located near the 5′ ends of MECOM (EVI1 and MDS1 complex), CCND2 , and LMO2 (Fig. 2C), all of which have been involved in adverse events in human gene therapy. Several additional clumps near cancer-associated genes, including the HOXB gene cluster, JARID2 , HMGA2, and MCL1, were also found to be enriched in samples from the MFG-γc trials. Therefore, clumps enriched in integration sites upstream of cancer-associated genes were significantly more frequent in samples from the MFG-γc trials than in samples from the current trial (P<0.001), which is consistent with the idea that integration near these genes promoted clonal expansion or persistence.
The proportions of integration sites near other cancer-associated genes were also compared for each patient, and the mean proportions were compared between trials. A highly significant difference was seen for comparisons of the proportions of integration sites near human lymphoid cancer genes (Fig. 2D; the full list is provided in the Supplementary Appendix), with enrichment in the first SCID-X1 trials using MFG-γc both within genes and near transcription start sites (P≤0.006 for both comparisons). Comparison with a second, broader list of cancer genes (not focused on lymphoid genes) also showed significant enrichment in samples from the first trial (P = 0.01 for enrichment within transcription units; the difference was not significant for enrichment near transcription start sites). We conclude that integration-site distributions were globally similar between the MFG-γc trials and the current trial, but peripheral-blood cell clones with vectors integrated near relevant cancer-associated genes were significantly more common in the first trial.
DISCUSSION
In this trial, in which an SIN γ-retrovirus vector with deletion of strong viral enhancers was used to treat SCID-X1, eight of nine patients survived, and seven patients had clearance of infection, recovery of gene-marked T cells, evidence of naive T-cell generation, and a diverse T-cell receptor repertoire. The kinetics of the recovery of T-cell numbers was similar to that in previous trials in which LTR-based vectors were used. Future studies could compare T-cell recovery in the full cohort with that among recipients of alternative types of allogeneic transplants, to determine whether either approach leads to more rapid recovery of immunity.
Insertion analysis showed a polyclonal integration profile with reduced numbers of clones near known lymphoid proto-oncogenes and genes implicated in serious adverse events in previous genetherapy trials.13–16 However, since leukemia was seen at a median of 33 months after gene therapy in the initial trial, the absence of leukemia must be considered a preliminary finding, because the majority of patients included in the analysis to date have been followed for less than 33 months. The distribution of insertion sites in circulating T cells reflects the composite of initial insertion sites in transduced CD34+ cells and in vivo selection. However, because the use of the SIN-γc vector did not have major effects on global associations of integration sites, the comparative results for integration-site profiles reported here suggest that the viral enhancer contained in the MFG-γc vector influenced either engraftment of T-cell progenitors or in vivo selection of T-cell clones containing insertions near proto-oncogenes.
Our observations are consistent with a model in which the integration of the enhancer-containing MFG-γc vector in the earlier trials increased expression at specific loci, resulting in outgrowth or persistence of specific cell clones. Subsequent additional secondary genetic events may lead to leukemogenesis, as noted in the previous SCID-X1 trials.17–19,34,35 Alternative explanations are possible. Initial integration targeting could differ in the long-term repopulating cells; this cannot be assessed directly, because these cells are a minority component of the initially transduced cell product. Although clumping was mostly in place in the patients in the first trials by 18 to 30 months after transplantation,29 clustering of integration sites in the patients in the current trial might not be evident in the time frame of the current analysis, even at more than 3 years since the infusion of cells. Long-term follow-up will clarify whether the integration-site profiles reported here are indeed correlated with improved safety for patients.13−16
In conclusion, the SIN γ-retrovirus vector was compatible with high-titer vector production in a clinical setting, with good transduction efficiencies overall, leading to transgene expression that restored immunity in the majority of patients treated in this trial. Specifically, we found that a modified γ-retrovirus vector retained efficacy in the treatment of SCID-X1 through the generation of a functional polyclonal T-cell repertoire. If long-term safety is confirmed, consideration might be given to the inclusion of low-dose conditioning, to enhance the likelihood of sustained B-cell and NK-cell reconstitution.36,37 Our data also provide a comparison of integration sites of γ-retrovirus vectors with and without the U3 enhancer in otherwise identical human trials and will be informative with respect to integration data from trials in which lentivirus vectors are used.
Supplementary Material
Acknowledgments
Supported by grants from the National Institutes of Health (U01 AI087628-05, to Drs. Williams, Pai, Notarangelo, Filipovich, Bushman, and Kohn; HL 073104, to Dr. Kohn; and AI 082020, to Dr. Bushman) and the Production Assistance for Cellular Therapy (PACT) program, National Heart, Lung, and Blood Institute (HHSN268201000009C), a Translational Investigator Service Award (to Dr. Pai), and grants from the Wellcome Trust (to Dr. Thrasher), the Great Ormond Street Hospital Biomedical Research Centre and the Great Ormond Street Hospital Children’s Charity (both to Drs. Thrasher and Gaspar), the European Union Seventh Framework Program for Research (CELL-PID 261387, to Drs. Thrasher, Gaspar, Baum, Cavazzana, Hacein-Bey-Abina, and Fischer), the German Research Foundation for the Cluster of Excellence REBIRTH (EXC 62/1, to Drs. Baum and Schambach), the National Institute of Health Research (to Dr. Thrasher), Programme Hospitalier de Recherche Clinique of the Health Ministry, Assistance Publique–Hôpitaux de Paris (PHRC national 2008 00-64), and the European Research Council (ERC PIDIMMUN 249816, to Dr. Fischer) and ERC Regenerative Therapy (269037, to Dr. Cavazzana).
We thank the medical monitor, Linda M. Griffith (Division of Allergy, Immunology, and Transplantation, National Institute of Allergy and Infectious Diseases, National Institutes of Health); the members of the Transatlantic Gene Therapy Consortium; other members of the steering committee, as well as members of the data and safety monitoring board in the United States (Malcolm Brenner [cochair], Adil Shamoo, Lynette Westfall, Kenneth Weinberg [chair], and Donn Young) and the data and safety monitoring board in France (Michel Broyer [chair], Sabine Sarnacki, and Yves Bertrand); Colleen Dansereau and Lucinda Williams for project management in the United States; the staff of the Vector Production Facility at Cincinnati Children’s Hospital Medical Center for production, holding, and distribution of the clinical vector; Ken Cornetta and the National Gene Vector Laboratory for performing replication-competent retrovirus testing; Sabine Charrier from Genethon for quantitative polymerase-chain-reaction development; Manfred Schmidt and Christoph von Kalle for integration-site data from the previous London SCID-X1 gene therapy trial; the many clinical staff members, research nurses, clinical research assistants, and data managers involved from the Harvard Catalyst Clinical and Translational Research Center, the Clinical and Translational Investigation Program and Connell O’Reilly Cell Manipulation Core Facility of Dana–Farber Cancer Institute, the Cellular Processing and Manipulation Laboratory of Cincinnati Children’s Hospital, and the UCLA Human Gene Medicine Program and Broad Stem Cell Research Center; Maria Suarez and Natasha Rossi for administrative assistance; the patients’ families for their continuous support of the study; and the medical and nursing staff of the Immunology and Pediatric Hematology Department, Hôpital Necker, Paris, Dana–Farber/Children’s Hospital Center for Cancer and Blood Disorders, Boston, and the Cancer Blood and Disease Institute, Cincinnati Children’s Hospital, Cincinnati, and UCLA Mattel Children’s Hospital, Los Angeles, for patient care.
APPENDIX
The authors’ full names and academic degrees are as follows: Salima Hacein-Bey-Abina, Pharm.D., Ph.D., Sung-Yun Pai, M.D., H. Bobby Gaspar, M.R.C.P., Ph.D., Myriam Armant, Ph.D., Charles C. Berry, Ph.D., Stephane Blanche, M.D., Jack Bleesing, M.D., Ph.D., Johanna Blondeau, M.S., Helen de Boer, B.S., Karen F. Buckland, Ph.D., Laure Caccavelli, Ph.D., Guilhem Cros, M.D., Satiro De Oliveira, M.D., Karen S. Fernández, M.D., Dongjing Guo, M.P.H., Chad E. Harris, M.S., Gregory Hopkins, B.S., Leslie E. Lehmann, M.D., Annick Lim, M.S., Wendy B. London, Ph.D., Johannes C.M. van der Loo, Ph.D., Nirav Malani, M.S., Frances Male, B.A., Punam Malik, M.D., M. Angélica Marinovic, M.D., Anne-Marie McNicol, Ph.D., Despina Moshous, M.D., Ph.D., Benedicte Neven, M.D., Ph.D., Matías Oleastro, M.D., Capucine Picard, M.D., Ph.D., Jerome Ritz, M.D., Christine Rivat, Ph.D., Axel Schambach, M.D., Ph.D., Kit L. Shaw, Ph.D., Eric A. Sherman, B.A., Leslie E. Silberstein, M.D., Emmanuelle Six, Ph.D., Fabien Touzot, M.D., Ph.D., Alla Tsytsykova, Ph.D., Jinhua Xu-Bayford, Dip.H.E., Christopher Baum, M.D., Frederic D. Bushman, Ph.D., Alain Fischer, M.D., Ph.D., Donald B. Kohn, M.D., Alexandra H. Filipovich, M.D., Luigi D. Notarangelo, M.D., Marina Cavazzana, M.D., Ph.D., David A. Williams, M.D., and Adrian J. Thrasher, M.B., B.S., Ph.D.
From the Departments of Biotherapy (S.H.-B.-A., J. Blondeau, L.C., F.T., M.C.) and Immunology and Pediatric Hematology (S.B., G.C., D.M., B.N., C.P., F.T., A.F.) and the Centre d’Étude des Déficits Immunitaires (C.P.), Hôpital Necker–Enfants Malades, Assistance Publique–Hôpitaux de Paris (AP-HP), the Biotherapy Clinical Investigation Center, Groupe Hospitalier Universitaire Ouest, AP-HP, IN-SERM (S.H.-B.-A., J. Blondeau, L.C., F.T., M.C.), Unité de Technologies Chimiques et Biologiques pour la Santé, Centre National de la Recherche Scientifique, 8258–INSERM Unité 1022, Faculté des Sciences Pharmaceutiques et Biologiques, Université Paris Descartes (S.H.-B.-A.), Immunology Laboratory, Groupe Hospitalier Universitaire Paris-Sud, AP-HP, Le Kremlin-Bicêtre (S.H.-B.-A.), Imagine Institute, Paris Descartes–Sorbonne Paris Cité University (S.B., J. Blondeau, L.C., D.M., B.N., C.P., E.S., A.F., M.C.), INSERM Unités Mixtes de Recherche 1163, Laboratory of Human Lymphohematopoiesis (J. Blondeau, L.C., E.S., F.T., A.F., M.C.), Groupe Immunoscope, Immunology Department, Institut Pasteur (A.L.), and Collège de France (A.F.) — all in Paris; Division of Hematology–Oncology (S.-Y.P., H.B., D.G., C.E.H., G.H., L.E.L., W.B.L., D.A.W.) and Division of Immunology (L.D.N.), Boston Children’s Hospital, Department of Pediatric Oncology, Dana–Farber Cancer Institute (S.-Y.P., D.G., L.E.L., W.B.L., D.A.W.), Harvard Medical School (S.-Y.P., M.A., L.E.L., W.B.L., J.R., L.E.S., A.T., L.D.N., D.A.W.), Center for Human Cell Therapy, Program in Cellular and Molecular Medicine, Boston Children’s Hospital (M.A., J.R., L.E.S., A.T.), Division of Hematologic Malignancies, Dana–Farber Cancer Institute (J.R.), and the Manton Center for Orphan Disease Research (L.D.N.) — all in Boston; Great Ormond Street Hospital for Children NHS Foundation Trust (H.B.G., J.X.-B., A.J.T.) and Section of Molecular and Cellular Immunology, University College London Institute of Child Health (H.B.G., K.F.B., A.-M.M., C.R., A.J.T.), London; Division of Biostatistics and BioInformatics, Department of Family and Preventive Medicine, University of California at San Diego, La Jolla (C.C.B.); Division of Bone Marrow Transplantation and Immune Deficiency (J. Bleesing, A.H.F.) and Division of Experimental Hematology and Cancer Biology (J.C.M.L., P.M.), Cincinnati Children’s Hospital Medical Center, Cincinnati; Department of Pediatrics, Children’s Discovery and Innovation Institute, UCLA Mattel Children’s Hospital (S.D.O., D.B.K.), and Department of Microbiology, Immunology, and Molecular Genetics, David Geffen School of Medicine (K.L.S., D.B.K.), Los Angeles; University of Illinois College of Medicine at Peoria, Peoria (K.S.F.); Department of Microbiology, University of Pennsylvania School of Medicine, Philadelphia (N.M., F.M., E.A.S., F.D.B.); Clínica Santa María, Unidad de Inmunología y Alergias, Santiago, Chile (M.A.M.); Hospital Nacional de Pediatría Garrahan, Servicio de Inmunología y Reumatología, Buenos Aires (M.O.); and Institute of Experimental Hematology, Hannover Medical School, Hannover, Germany (A.S., C.B.).
Footnotes
Note added in proof: While this article was in production, the patients had been followed for a median of 33 months (range, 16 to 43). No leukemia had occurred in any patient (see Fig. S4 in the Supplementary Appendix).
Disclosure forms provided by the authors are available with the full text of this article at NEJM.org.
REFERENCES
- 1.Noguchi M, Yi H, Rosenblatt HM, et al. Interleukin-2 receptor gamma chain mutation results in X-linked severe combined immunodeficiency in humans. Cell. 1993;73:147–157. doi: 10.1016/0092-8674(93)90167-o. [DOI] [PubMed] [Google Scholar]
- 2.Puck JM, Deschênes SM, Porter JC, et al. The interleukin-2 receptor gamma chain maps to Xq13.1 and is mutated in X-linked severe combined immunodeficiency, SCIDX1. Hum Mol Genet. 1993;2:1099–1104. doi: 10.1093/hmg/2.8.1099. [DOI] [PubMed] [Google Scholar]
- 3.Gatti RA, Meuwissen HJ, Allen HD, Hong R, Good RA. Immunological reconstitution of sex-linked lymphopenic immunological deficiency. Lancet. 1968;2:1366–1369. doi: 10.1016/s0140-6736(68)92673-1. [DOI] [PubMed] [Google Scholar]
- 4.O’Reilly RJ, Dupont B, Pahwa S, et al. Reconstitution in severe combined immunodeficiency by transplantation of marrow from an unrelated donor. N Engl J Med. 1977;297:1311–1318. doi: 10.1056/NEJM197712152972403. [DOI] [PubMed] [Google Scholar]
- 5.Reisner Y, Kapoor N, Kirkpatrick D, et al. Transplantation for severe combined immunodeficiency with HLA-A,B,D,DR incompatible parental marrow cells fractionated by soybean agglutinin and sheep red blood cells. Blood. 1983;61:341–348. [PubMed] [Google Scholar]
- 6.Buckley RH, Schiff SE, Schiff RI, et al. Hematopoietic stem-cell transplantation for the treatment of severe combined immunodeficiency. N Engl J Med. 1999;340:508–516. doi: 10.1056/NEJM199902183400703. [DOI] [PubMed] [Google Scholar]
- 7.Myers LA, Patel DD, Puck JM, Buckley RH. Hematopoietic stem cell transplantation for severe combined immunodeficiency in the neonatal period leads to superior thymic output and improved survival. Blood. 2002;99:872–878. doi: 10.1182/blood.v99.3.872. [DOI] [PubMed] [Google Scholar]
- 8.Antoine C, Müller S, Cant A, et al. Long-term survival and transplantation of haemopoietic stem cells for immunodeficiencies: report of the European experience 1968–99. Lancet. 2003;361:553–560. doi: 10.1016/s0140-6736(03)12513-5. [DOI] [PubMed] [Google Scholar]
- 9.Gennery AR, Slatter MA, Grandin L, et al. Transplantation of hematopoietic stem cells and long-term survival for primary immunodeficiencies in Europe: entering a new century, do we do better? J Allergy Clin Immunol. 2010;126:602–610. doi: 10.1016/j.jaci.2010.06.015. [DOI] [PubMed] [Google Scholar]
- 10.Buckley RH. Transplantation of hematopoietic stem cells in human severe combined immunodeficiency: longterm outcomes. Immunol Res. 2011;49:25–43. doi: 10.1007/s12026-010-8191-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brown L, Xu-Bayford J, Allwood Z, et al. Neonatal diagnosis of severe combined immunodeficiency leads to significantly improved survival outcome: the case for newborn screening. Blood. 2011;117:3243–3246. doi: 10.1182/blood-2010-08-300384. [DOI] [PubMed] [Google Scholar]
- 12.Cavazzana-Calvo M, Hacein-Bey S, de Saint Basile G, et al. Gene therapy of human severe combined immunodeficiency (SCID)-X1 disease. Science. 2000;288:669–672. doi: 10.1126/science.288.5466.669. [DOI] [PubMed] [Google Scholar]
- 13.Hacein-Bey-Abina S, Le Deist F, Carlier F, et al. Sustained correction of X-linked severe combined immunodeficiency by ex vivo gene therapy. N Engl J Med. 2002;346:1185–1193. doi: 10.1056/NEJMoa012616. [DOI] [PubMed] [Google Scholar]
- 14.Gaspar HB, Parsley KL, Howe S, et al. Gene therapy of X-linked severe combined immunodeficiency by use of a pseudotyped gammaretroviral vector. Lancet. 2004;364:2181–2187. doi: 10.1016/S0140-6736(04)17590-9. [DOI] [PubMed] [Google Scholar]
- 15.Hacein-Bey-Abina S, Hauer J, Lim A, et al. Efficacy of gene therapy for X-linked severe combined immunodeficiency. N Engl J Med. 2010;363:355–364. doi: 10.1056/NEJMoa1000164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gaspar HB, Cooray S, Gilmour KC, et al. Long-term persistence of a polyclonal T cell repertoire after gene therapy for X-linked severe combined immunodeficiency. Sci Transl Med. 2011;3:97ra79. doi: 10.1126/scitranslmed.3002715. [DOI] [PubMed] [Google Scholar]
- 17.Hacein-Bey-Abina S, Von Kalle C, Schmidt M, et al. LMO2-associated clonal T cell proliferation in two patients after gene therapy for SCID-X1. Science. 2003;302:415–419. doi: 10.1126/science.1088547. [DOI] [PubMed] [Google Scholar]
- 18.Howe SJ, Mansour MR, Schwarzwaelder K, et al. Insertional mutagenesis combined with acquired somatic mutations causes leukemogenesis following gene therapy of SCID-X1 patients. J Clin Invest. 2008;118:3143–3150. doi: 10.1172/JCI35798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hacein-Bey-Abina S, Garrigue A, Wang GP, et al. Insertional oncogenesis in 4 patients after retrovirus-mediated gene therapy of SCID-X1. J Clin Invest. 2008;118:3132–3142. doi: 10.1172/JCI35700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thornhill SI, Schambach A, Howe SJ, et al. Self-inactivating gammaretroviral vectors for gene therapy of X-linked severe combined immunodeficiency. Mol Ther. 2008;16:590–598. doi: 10.1038/sj.mt.6300393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zychlinski D, Schambach A, Modlich U, et al. Physiological promoters reduce the genotoxic risk of integrating gene vectors. Mol Ther. 2008;16:718–725. doi: 10.1038/mt.2008.5. [DOI] [PubMed] [Google Scholar]
- 22.van der Loo JCM, Swaney WP, Grass-man E, et al. Critical variables affecting clinical-grade production of the self-inactivating gamma-retroviral vector for the treatment of X-linked severe combined immunodeficiency. Gene Ther. 2012;19:872–876. doi: 10.1038/gt.2012.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Berry C, Hannenhalli S, Leipzig J, Bushman FD. Selection of target sites for mobile DNA integration in the human genome. PLoS Comput Biol. 2006;2(11):e157. doi: 10.1371/journal.pcbi.0020157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Berry CC, Gillet NA, Melamed A, Gormley N, Bangham CR, Bushman FD. Estimating abundances of retroviral insertion sites from DNA fragment length data. Bioinformatics. 2012;28:755–762. doi: 10.1093/bioinformatics/bts004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Berry CC, Ocwieja KE, Malani N, Bushman FD. Comparing DNA integration site clusters with scan statistics. Bioinformatics. 2014;30:1493–1500. doi: 10.1093/bioinformatics/btu035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brady T, Roth SL, Malani N, et al. A method to sequence and quantify DNA integration for monitoring outcome in gene therapy. Nucleic Acids Res. 2011;39(11):e72. doi: 10.1093/nar/gkr140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Deichmann A, Hacein-Bey-Abina S, Schmidt M, et al. Vector integration is nonrandom and clustered and influences the fate of lymphopoiesis in SCID-X1 gene therapy. J Clin Invest. 2007;117:2225–2232. doi: 10.1172/JCI31659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schwarzwaelder K, Howe SJ, Schmidt M, et al. Gammaretrovirus-mediated correction of SCID-X1 is associated with skewed vector integration site distribution in vivo. J Clin Invest. 2007;117:2241–2249. doi: 10.1172/JCI31661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang GP, Berry CC, Malani N, et al. Dynamics of gene-modified progenitor cells analyzed by tracking retroviral integration sites in a human SCID-X1 gene therapy trial. Blood. 2010;115:4356–4366. doi: 10.1182/blood-2009-12-257352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hardin JW, Hilbe JM. Generalized estimating equations. Boca Raton, FL: Chapman & Hall/CRC Press; 2003. [Google Scholar]
- 31.Pan W. Akaike’s information criterion in generalized estimating equations. Biometrics. 2001;57:120–125. doi: 10.1111/j.0006-341x.2001.00120.x. [DOI] [PubMed] [Google Scholar]
- 32.Kustikova O, Fehse B, Modlich U, et al. Clonal dominance of hematopoietic stem cells triggered by retroviral gene marking. Science. 2005;308:1171–1174. doi: 10.1126/science.1105063. [DOI] [PubMed] [Google Scholar]
- 33.Cui K, Zang C, Roh TY, et al. Chromatin signatures in multipotent human hematopoietic stem cells indicate the fate of bivalent genes during differentiation. Cell Stem Cell. 2009;4:80–93. doi: 10.1016/j.stem.2008.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Coffin JM, Hughes SH, Varmus HE. Retroviruses. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1997. [PubMed] [Google Scholar]
- 35.Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA, Jr, Kinzler KW. Cancer genome landscapes. Science. 2013;339:1546–1558. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Aiuti A, Cattaneo F, Galimberti S, et al. Gene therapy for immunodeficiency due to adenosine deaminase deficiency. N Engl J Med. 2009;360:447–458. doi: 10.1056/NEJMoa0805817. [DOI] [PubMed] [Google Scholar]
- 37.Candotti F, Shaw KL, Muul L, et al. Gene therapy for adenosine deaminase-deficient severe combined immune deficiency: clinical comparison of retroviral vectors and treatment plans. Blood. 2012;120:3635–3646. doi: 10.1182/blood-2012-02-400937. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.