Abstract
Salmonella enterica subspecies 1 serovar Typhimurium is a common cause of gastrointestinal infections. The host's innate immune system and a complex set of Salmonella virulence factors are thought to contribute to enteric disease. The serovar Typhimurium virulence factors have been studied extensively by using tissue culture assays, and bovine infection models have been used to verify the role of these factors in enterocolitis. Streptomycin-pretreated mice provide an alternative animal model to study enteric salmonellosis. In this model, the Salmonella pathogenicity island 1 type III secretion system has a key virulence function. Nothing is known about the role of other virulence factors. We investigated the role of flagella in murine serovar Typhimurium colitis. A nonflagellated serovar Typhimurium mutant (fliGHI) efficiently colonized the intestine but caused little colitis during the early phase of infection (10 and 24 h postinfection). In competition assays with differentially labeled strains, the fliGHI mutant had a reduced capacity to get near the intestinal epithelium, as determined by fluorescence microscopy. A flagellated but nonchemotactic cheY mutant had the same virulence defects as the fliGHI mutant for causing colitis. In competitive infections, both mutants colonized the intestine of streptomycin-pretreated mice by day 1 postinfection but were outcompeted by the wild-type strain by day 3 postinfection. Together, these data demonstrate that flagella are required for efficient colonization and induction of colitis in streptomycin-pretreated mice. This effect is mostly attributable to chemotaxis. Recognition of flagellar subunits (i.e., flagellin) by innate immune receptors (i.e., Toll-like receptor 5) may be less important.
Salmonella enterica serovar Typhimurium is a common cause of bacterial gastroenteritis in both developing and industrialized countries. This organism possesses a number of mechanistically different virulence factors. Two main type III secretion systems (TTSS) encoded in Salmonella pathogenicity island 1 (SPI-1) and SPI-2 play a fundamental role during infection of the animal host. The SPI-1 TTSS mediates invasion of epithelial cells and is involved in evoking enterocolitis (3, 52). The SPI-2 TTSS is required for intracellular survival and replication of Salmonella during systemic stages of infection (20, 51). Besides the TTSS, serovar Typhimurium harbors a multitude of virulence factors (for a review see reference 17), including fimbriae (48), lipopolysaccharide (LPS) (46), superoxide dismutases (14, 42, 47), the large virulence plasmid (Spv) (38), and flagella (40, 53). A variety of in vitro and in vivo models have been developed to better characterize the molecular mechanisms of the infection process (23). In humans and cattle, serovar Typhimurium causes gastrointestinal disease (49, 55). However, in mice serovar Typhimurium infection leads to a typhoid-like disease, and a specific type of intestinal pathology with pronounced monocyte infiltrates develops only during the final stages of the lethal infection (31).
Recently, we found that streptomycin-pretreated mice provide an animal model that can be used to study serovar Typhimurium colitis (3). The intestinal pathology characterized by epithelial damage, polymorphonuclear granulocyte (PMN) infiltration, and edema is strongly dependent on SPI-1-mediated protein translocation (3, 18). These results are consistent with previous findings obtained with the bovine enterocolitis model. However, it remains to be seen whether other serovar Typhimurium virulence factors also contribute to the pathology in this model.
Flagella are known to play a role in virulence in many bacterial pathogens (27). For Salmonella spp. the role of motility and chemotaxis in virulence has been extensively studied in vitro. Four different types of flagellum mutants with mutations that affect either the flagellar apparatus (app), the flagellum (fla), motility (mot), or chemotaxis (che) have been distinguished. Chemotaxis mutants (app+ fla+ mot+ che) assemble functional motile flagella but cannot move in response to chemotactic stimuli; there are “tumbly” and “smooth-swimming” chemotaxis mutants (26). Motility mutants (app+ fla+ mot che) assemble flagella but are deficient in motility. Flagellin mutants (app+ fla mot che) possess a functional flagella secretion apparatus but cannot assemble flagellar filaments. And flagellar apparatus mutants (app fla mot che) have mutations in genes encoding subunits of the flagellar apparatus or regulators of the flagellar apparatus assembly process.
The assembly of Salmonella serovar Typhimurium flagella is a tightly regulated process. Genes coding for different components of the flagellar apparatus (the flagellum regulon) are clustered in several operons on the Salmonella chromosome. Expression of these genes is organized in a hierarchical manner (class I, II, and III genes) (1, 35). Class I genes encode the master regulator proteins FlhDC, which are responsible for expression of the entire regulon. The class II gene products include structural components of the hook basal body, the TTSS, and regulatory proteins required for class III gene expression. Disruption of class II genes (i.e., fliGHI) prohibits the expression of class III genes, which encode the flagellar filament (i.e., FliC), the motor force generators, and the chemosensory machinery.
Flagella and chemotaxis allow Salmonella to respond to attractant and repellent gradients. It has been found that flagella play a role in attachment to and invasion of various cultured cells (10, 12, 26, 34, 40). The SPI-1-mediated induction of membrane ruffling in epithelial cells has also been reported to be reduced upon infection with a flagellum mutant compared to the induction observed with a wild-type strain (33). Recently, it was found that flagellin itself can play a proinflammatory role via Toll signaling. Salmonella flagellin interacts with Toll-like receptor 5 (TLR5) on human model epithelia, and this leads to activation of NF-κB and interleukin-8 secretion (16, 41, 54). Due to pleiotropic defects (movement to the cell and interaction with it), it has not always been easy to decipher the role of flagella in additional processes, such as attachment to cells, host cell invasion, induction of inflammation, and signaling via the TLRs. There have been only a few studies of the in vivo function of motility during enterocolitis. In bovine ileal loops a Salmonella strain (flhD) lacking the entire flagellar apparatus was significantly attenuated in terms of causing fluid secretion and PMN influx. An fliC fljB flagellin double mutant was slightly attenuated in terms of causing inflammation, although the results were not statistically significant (40).
Using the streptomycin-pretreated mouse model, we found in this study that motility and chemotaxis are important for the induction of colits in mice. We obtained evidence that the virulence defect of nonmotile and nonchemotactic Salmonella mutants can be attributed mainly to a less efficient interaction with the cecal epithelium.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Serovar Typhimurium strains were grown for 12 h at 37°C in Luria-Bertani broth containing 0.3 M NaCl, diluted 1:20 in fresh medium, and subcultured for 4 h with mild aeration. For infection experiments, bacteria were washed twice with ice-cold phosphate-buffered saline (PBS) and resuspended in cold PBS (5 × 107 CFU/50 μl). For competitive infection experiments, identical volumes of subcultures were mixed prior to washing with PBS.
The serovar Typhimurium SL1344 mutants used in this study were constructed by phage transduction. To construct M913, the fliGHI::Tn10 allele was transduced from a lysate of SB245 (ΔsipA ΔiagP ΔsicP ΔsptP::aphT fliGHI::Tn10; K. Kaniga and J. E. Galan, unpublished data), and transductants were selected for growth on tetracycline. The fliGHI::Tn10 allele originated from X3420 (34). The chemotaxis mutant M935 was constructed by P22-mediated transduction of the cheY::Tn10 allele of SJW589 (45), which was kindly provided by Shigeru Yamaguchi.
SPI-1 mutant strains SB225 (sipA::aphT) (30), SB220 (sipC::aphT), SB169 (sipB::aphT) (30), and SB302 (invJ::aphT) (7) were used as control strains for analysis of supernatant proteins.
All strains were tested by stab inoculation of semisolid agar plates (10% tryptone, 5% NaCl, 0.3% agar) and incubation at 37°C for 5 to 8 h until bacterial halos that had radii of 1 to 2 cm were visible with wild-type Salmonella serovar Typhimurium strain SL1344. In addition, the motility of all strains was analyzed microscopically.
Construction of fluorescent protein expression plasmids.
Cloning of DNA fragments was performed by using standard protocols (39).
Plasmid pM979 (a pBR322 derivative) for expression of green fluorescent protein (GFP) under control of the constitutive rpsM promoter was constructed as follows. The coding sequence of GFPmut3b was amplified from pJBA27 (a kind gift from Christoph Jacobi) by PCR performed with primers 5′ GCA GAA TTC AGG AAA CAG TAT TCA TGC GTA AAG GAG AAG AAC TTT TC 3′ and 5′ CTG GAA TTC TTA TTT GTA TAG TTC ATC CAT GCC 3′ and was cloned into pBluescript (Stratagene) by using EcoRI to obtain pM900. pM935 was created by cloning GFPmut3b and its Shine-Dalgarno sequence by using XbaI and HindIII from pM900 into pWKS30 (50). The lac promoter was subsequently removed from pM935 by partially digesting it with BglII; this was followed by Acc651 digestion, filling in with the Klenow fragment, and religation to obtain pM946. pM958 was constructed by XbaI/BamHI digestion of pM946 and introduction of a fragment containing 536 bp upstream of rpsM (designated prpsM) amplified by PCR from SL1344 genomic DNA with primers 5′ CAT TCT AGA CGA TAA AGT AAT GAC CCG CCT 3′ and 5′ CAT GGA TCC GGG CCA CTA TGC ACT CCT ACT 3′. GFPmut2 was then PCR amplified from pKEN2 (9) with primers 5′ CAT GAA TTC AGG AGG TAG TAT TGA TGA GTA AAG GAG AAG AAC TTT 3′ and 5′ CAT GAA TTC GAA GAC TTA TTT GTA TAG TTC ATC CAT GCC 3′ and introduced into EcoRI-digested pM958, and the product was designated pM965. pM966 was created by subcloning GFPmut2 from pM965 into pBluescript by using EcoRI. GFPmut3b was cloned from pJBA27 into pM136 by using XbaI and HindIII (44) to obtain pM183. pM963 was constructed by removing sopE-GFP-M45 from pM183 by Eco47III/HindIII digestion and replacing it with prpsM-GFPmut3b (NotI/EcoRV) from pM958. prpsM-GFPmut3b was replaced in pM963 by GFPmut2 from pM966 by NotI/EcoRV digestion, and the resulting plasmid was designated pM968 (Table 1). The rpsM-promoter was recovered from pM963 by XbaI/BamHI digestion and introduced into pM968 to obtain pM979 (Table 1).
TABLE 1.
Serovar Typhimurium strains and plasmids used in this study
| Strain or plasmid | Relevant genotype | Motility agar | Swimming (microscopy) | Reference |
|---|---|---|---|---|
| Serovar Typhimurium strains | ||||
| SL1344 | Wild type | + | + | 22 |
| SB161 | ΔinvG | + | + | 28 |
| M913 | fliGHI::Tn10 | − | − | This study |
| M935 | cheY::Tn10 | − | + | This study |
| SB225 | sipA::aphT | NDa | ND | 29 |
| SB220 | sipC::aphT | ND | ND | 30 |
| SB169 | sipB::aphT | ND | ND | 30 |
| SB302 | invJ::aphT | ND | ND | 8 |
| Plasmids | ||||
| pM979 | Ampr; oripBR322; carrying gfpmut2 coding region and rpsM promoter region | This study | ||
| pM968 | Ampr; oripBR322; carrying gfpmut2 coding region without promoter | This study | ||
| pDsRed | Kanr Cmr; derivative of pACYC184; carrying dsRedexpress coding region and lac promoter region | C. Jacobi, unpublished data |
ND, not determined.
pDsRed, a pACYC184-derived plasmid, which expresses DsRed-Express (Clontech) under the control of the lac promoter, was a kind gift from Christoph Jacobi.
Animal experiments.
Animal experiments were performed at the BZL (Universität Zürich) as described previously (3) by using specific-pathogen-free female C57BL/6 mice that were 6 to 9 weeks old and were obtained from Harlan (Horst, The Netherlands). Water and food were withdrawn 4 h prior to per os (p.o.) treatment with 20 mg of streptomycin. After this, water and food were provided ad libitum. Twenty hours after the streptomycin treatment, water and food were withdrawn again for 4 h before the mice were infected with 5 × 107 CFU of serovar Typhimurium (50 μl of a suspension in PBS p.o.). At the times postinfection (p.i.) indicated below, the mice were sacrificed by cervical dislocation, and tissue samples were removed from the intestinal tract, spleen, and liver for analysis. Animal experiments were approved by the Swiss authorities and were performed according to the legal requirements.
Analysis of serovar Typhimurium loads in the intestine, mLN, spleen, and liver.
To analyze colonization, the spleen, liver, and mesenteric lymph nodes (mLN) were removed aseptically and homogenized in PBS (containing 0.5% Tergitol and 0.5% bovine serum albumin) at 4°C as described previously (3). The bacterial loads were determined by plating samples on MacConkey agar plates containing 50 μg of streptomycin per ml. The minimal detectable level was 10 CFU/organ for the mLN, 20 CFU/organ for the spleen, and 100 CFU/organ for the liver.
Cecal contents were collected at different times p.i., and the bacterial loads were determined by plating. The minimum detectable level was 10 CFU per 25- to 150-mg sample of intestinal contents.
For coinfection experiments, the levels of colonization of mutant bacteria (carrying an appropriate antibiotic marker) and wild-type bacteria were determined by plating samples on MacConkey agar plates containing streptomycin or streptomycin and tetracycline. The values were confirmed by replica plating.
Histological procedures.
Tissue samples were embedded in O.C.T. (Sakura, Torrance, Calif.), snap frozen in liquid nitrogen, and stored at −80°C. Cryosections (5 μm) were mounted on glass slides, air dried for 2 h at room temperature, and stained with hematoxylin and eosin (HE).
Cecum pathology was independently evaluated by two pathologists in a blinded manner by using 5-μm-thick HE-stained sections and a histopathological scoring scheme described previously (3), as follows.
(i) Submucosal edema.
Submucosal edema (SE) was deduced from the extension of the submucosa, and values (expressed as percentages) were determined by morphometric analysis by using the following formula: SE = (b − a)/c, where a is the area enclosed by the mucosa (mucosa and intestinal lumen), b is the area enclosed by the borderline between the submucosa and tunica muscularis (submucosa, mucosa, and intestinal lumen), and c is the area enclosed by the outer edge of the tunica muscularis (tunica muscularis, submucosa, mucosa, and lumen; area of the whole cecal cross section). Submucosal edema was scored as follows: 0, no pathological changes; 1, detectable edema (submucosal edema, <10%); 2, moderate edema (submucosal edema, 10 to 40%); 3, profound edema (submucosal edema, ≥40%).
(ii) PMN infiltration into the lamina propria.
PMN in the lamina propria were enumerated by examining 10 high-power fields (magnification, ×400; field diameter, 420 μm), and the average number of PMN per high-power field was calculated. The results were scored as follows: 0, less than 5 PMN per high-power field; 1, 5 to 20 PMN per high-power field; 2, 21 to 60 PMN per high-power field; 3, 61 to 100 PMN per high-power field; 4, more than 100 PMN per high-power field.
(iii) Goblet cells.
The average number of goblet cells per high-power field (magnification, ×400) was calculated by examining 10 different regions of the cecal epithelium. A score of 0 indicated that there were more than 28 goblet cells per high-power field. In the cecum of the normal specific-pathogen-free mice we observed an average of 6.4 crypts per high-power field, and the average crypt consisted of 35 to 42 epithelial cells, 25 to 35% of which were differentiated into goblet cells. A score of 1 indicated that there were 11 to 28 goblet cells per high-power field; a score of 2 indicated that there were 1 to 10 goblet cells per high-power field; and a score of 3 indicated that there was less than 1 goblet cell per high-power field.
(iv) Epithelial integrity.
Epithelial integrity was scored as follows: 0, no pathological changes detectable in 10 high-power fields (magnification, ×400); 1, epithelial desquamation; 2, erosion of the epithelial surface (gaps of 1 to 10 epithelial cells per lesion); 3, epithelial ulceration (gaps of >10 epithelial cells per lesion; at this stage there was generally granulation tissue below the epithelium).
The combined pathological score for each tissue sample was determined by adding the averaged scores. The combined scores indicated the following conditions: 0, intestine intact without any signs of inflammation; 1 to 2, minimal signs of inflammation (frequently found in the ceca of specific-pathogen-free mice); 3 to 4, slight inflammation; 5 to 8, moderate inflammation; 9 to 13, profound inflammation.
Immunofluorescence experiments.
Streptomycin-pretreated animals were infected with 1:1 mixtures of wild-type and mutant bacteria carrying GFP or DsRed expression plasmids (pM979 and pDsred). Mice were killed on day 1 p.i., and cecal tissues were recovered and treated as described recently (4). Briefly, the tissues were fixed in 4% paraformaldehyde in PBS (pH 7.4) overnight at 4°C. The fixed tissue samples were washed with PBS and equilibrated in PBS containing 20% sucrose and 0.1% NaN3 overnight at 4°C. The tissues were then embedded in O.C.T. (Sakura), snap frozen in liquid nitrogen, and stored at −80°C. Cryosections (7 μm) were mounted on glass slides and air dried for 2 h at room temperature prior to immunostaining. Sections were fixed in 4% paraformaldehyde for 5 min, washed, and blocked with 10% (wt/vol) normal goat serum in PBS for 1 h. The sections were stained for 1 h with polyclonal rabbit anti-Salmonella O-antigen group B serum (factors 1, 4, 5, and 12; 1:500 in PBS containing 10% [wt/vol] goat serum; Difco). Rhodamine-conjugated goat anti-rabbit serum (Dianova) diluted 1:300 in PBS containing 10% (wt/vol) goat serum was used as the secondary antibody. DNA was stained with DAPI (4′ 6′-diamidino-2-phenylindole) (0.5 μg/ml; Sigma). F-Actin was visualized by staining with Alexa-647-conjugated phalloidin (Molecular Probes). Sections were mounted with 50% glycerol in PBS, and each cover glass was sealed with nail polish.
The relative localization of GFP-positive and GFP-negative bacteria was determined as follows. Optical fields showing bacteria and epithelium were chosen for analysis. Images were recorded at a magnification of ×630 by using a Perkin-Elmer Ultraview confocal imaging system and a Zeiss Axiovert 200 microscope. Red, green, and cyan fluorescence was recorded confocally, while DAPI fluorescence was recorded by epifluorescence microscopy. Images were combined by using Adobe Photoshop, version 7.0.1. For quantification of GFP-positive bacteria close to the epithelium, all anti-LPS-stained (red) bacteria within 20 μm of the epithelium were counted. All GFP-positive bacteria that were LPS positive (green and red) were counted. The level of GFP-positive bacteria was expressed as a percentage of the total LPS-positive Salmonella population and was calculated as follows: number of GFP-positive red cells/number of red cells × 100.
Statistical analysis.
Statistical analyses of the individual pathological scores for submucosal edema, PMN infiltration, loss of goblet cells, and epithelial integrity and of the combined pathological score were performed by using the exact Mann-Whitney U test and the SPSS software, version 11.0, as described previously (3). P values of <0.05 were considered statistically significant. Bacterial colonization was analyzed in a similar manner. To allow statistical analysis of the bacterial loads, the values used for animals that yielded no CFU were the minimal detectable levels (mLN, 10 CFU; spleen, 20 CFU; liver, 100 CFU; intestinal contents, between 67 and 400 CFU [see above]). After this, the median values were calculated by using Microsoft Excel XP, and a statistical analysis was performed by using the exact Mann-Whitney U test and the SPSS software, version 11.0. P values of <0.05 were considered statistically significant.
Analysis of proteins from serovar Typhimurium culture supernatants.
Overnight cultures of serovar Typhimurium strains were grown in Luria-Bertani medium without antibiotics. Bacteria were pelleted by centrifugation at 8,000 × g for 20 min. Then 1.6 ml of each supernatant was filtered through 0.22-μm-pore-size low-protein-binding filters (Millex GV; Millipore). Proteins were precipitated for 2 h on ice by addition of 375 μl of trichloroacetic acid, followed by centrifugation at 14,000 × g for 30 min. Each precipitate was washed in ice-cold acetone and centrifuged. The resulting pellet was resuspended in sample buffer (50 mM Tris-Cl [pH 6.8], 100 mM dithiothreitol, 2% sodium dodecyl sulfate, 0.1% bromphenol blue, 10% glycerol). The proteins were boiled for 5 min, separated on sodium dodecyl sulfate-10% polyacrylamide gels, and visualized with Coomassie brilliant blue R250 stain.
Preparation of chromosomal DNA and Southern blotting.
For Southern blot analysis chromosomal DNA of serovar Typhimurium strains were prepared by using standard methods (39). DNA was digested with EcoRV and HindIII, electrophoresed on a 1% agarose gel, and transferred to a ZETA-Probe BT blotting membrane with a vacuum blotter (Bio-Rad Laboratories). Southern hybridization was performed overnight at 58°C. Synthesis of fluorescein-labeled probes and detection were performed by using the protocols of the manufacturer (random prime labeling system, version II; Amersham, Little Chalfont, England). Probes used for detection of insertions in Salmonella genes fliGHI and cheY were amplified by PCR performed with primers fliGHI-fwd (5′GCC AGT GGA TGA GTA ACG AT 3′), fliGHI-rev (5′ CGT CGC CCT CCT GCT TTA T 3′), cheY-fwd (5′ TGC GCG TTA GTA AAG CTG GTA 3′), and cheY-rev (5′CCA GTC CGG CAG TGA TTA TTA 3′).
RESULTS
Construction and verification of flagellum mutants.
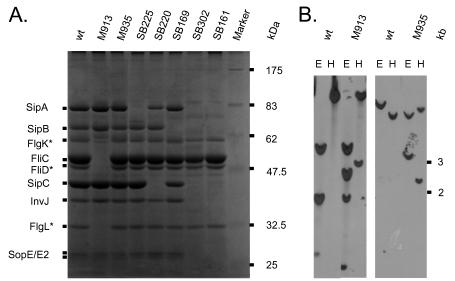
To construct an isogenic serovar Typhimurium mutant that lacked flagella, we transduced the fliGHI::Tn10 allele of SB245 into wild-type serovar Typhimurium strain SL1344. The resulting strain was designated M913. fliGHI code for class II genes of the flagellum regulon, the rotor/switch protein (fliG), the ATPase that drives flagellar export (fliI), and the negative regulator of fliI (fliH). Deletion of fliG is known to lead to a nonflagellated phenotype (24). Thus, M913 was nonmotile on motility agar (Table 1). Furthermore, M935, a mutant defective in chemotaxis, was created by P22 transduction of the cheY::Tn10 allele of SJW589 into SL1344. Correct insertion of the Tn10 insertion into the two mutants was verified by Southern blot hybridization by using probes specific for fliGHI and cheY, respectively (Fig. 1B). It is well established that flagellated serovar Typhimurium sheds a significant proportion of its flagellar subunits into the culture supernatant when it is grown in vitro. FliC (H1 flagellin; 52 kDa) is the most abundant secreted protein under these conditions (32). Therefore, we analyzed the secreted proteins of M913 and M935. To facilitate identification of flagellar subunits by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, several mutants with deletions in the SPI-1 TTSS apparatus (ΔinvG, invJ::aphT) and effector protein genes sipA, sipB and sipC were also included. As expected, M913 did export SPI-1 effector proteins but no flagellar subunits into the culture supernatant (Fig. 1A). Although an isogenic cheY::Tn10 mutant (M935) was unable to move in a chemotactic manner, it was able to assemble functional flagella (Table 1). These data confirmed that M913 and M935 could be used to assess the global importance of flagella and the specific role of chemotaxis in serovar Typhimurium colitis.
FIG. 1.
Characterization of flagellum mutants. (A) Supernatant proteins from overnight Salmonella cultures were prepared as described in Material and Methods. They were separated by 10% polyacrylamide gel electrophoresis and stained with Coomassie brilliant blue. Major bands resulting from secreted proteins via the flagellum apparatus or the SPI-1 TTSS are indicated. M913, fliGHI::Tn10; M935, cheY::Tn10; SB225, sipA::aphT; SB220, sipC::aphT; SB169, sipB::aphT; SB302, invJ::aphT; SB161, ΔinvG. (B) Southern blot analysis of wild-type Salmonella, M913, and M935. Chromosomal DNA was digested with EcoRV (lanes E) or HindIII (lanes H), and the resulting fragments were separated on a 1% agarose gel prior to Southern blotting. Hybridization was performed with fluorescein-labeled probes corresponding to the fliGHI (right panel) and cheY (left panel) regions (see Materials and Methods). Asterisks indicate identities tentatively assigned based on the findings of Komoriya et al. (32). wt, wild type.
Mouse virulence of a nonflagellated mutant 2 days p.i.
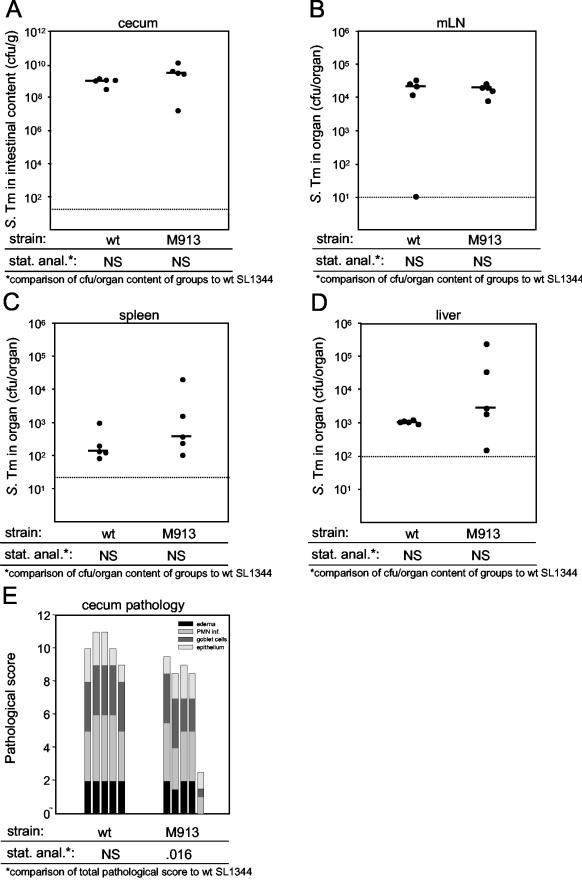
First we investigated the role of flagella in murine serovar Typhimurium colitis. Two groups of five streptomycin-pretreated C57BL/6 mice were infected intragastrically with 5 × 107 CFU of mutant M913 lacking flagella or wild-type strain SL1344. At day 2 p.i., the animals were killed, and we analyzed colonization of the cecum, spleen, liver, and mLN, as well as pathological changes in the cecal tissue. We found that the levels of colonization of the cecum and mLN by the two strains were not significantly different (Fig. 2A and B). The levels of mutant M913 were slightly but not significantly higher in the liver and spleen (P ≥ 0.05) (Fig. 2C and D). This finding is consistent with the findings of previous studies of the relevance of flagella for systemic infection in the murine typhoid model (40). Histopathological analysis revealed that there was a small but significant difference in the total pathological scores between M913 and the isogenic wild-type strain (Fig. 2E) (P = 0.016, as determined by the Mann-Whitney U test), but the differences between the scores for the individual parameters were not significant. The inflammation in mice infected with the nonflagellated strain appeared to be slightly weaker than the inflammation in mice infected with the wild-type strain 2 days p.i. even though the two groups of mice had equal numbers of bacteria in their cecal contents and mLN. This indicates that flagella may play a role in the initiation of colitis in streptomycin-pretreated mice.
FIG. 2.
Analysis of Salmonella wild-type and fliGHI mutant at 48 h p.i. Two groups of five streptomycin-pretreated mice were infected for 2 days with 5 × 107 CFU of serovar Typhimurium strain SL1344 (wild type) or M913 (fliGHI). (A to D) Bacterial loads in the cecal contents (A), the mLN (B), the spleen (C), and the liver (D). The dotted lines indicate the limits of detection, and the solid horizontal lines indicate the medians. (E) Histopathological analyses. HE-stained sections of cecal tissue were scored for edema in the submucosa (black bars), PMN infiltration (inf.) (medium gray bars), reductions in the numbers of goblet cells (dark grey bars), and desquamation, erosion, and ulceration of the epithelial layer (light gray bars) (see Materials and Methods). The scores are expressed as stacked vertical bars. The total pathological score (sum of the separate scores) was statistically analyzed by using the exact Mann-Whitney U test (in comparison to SL1344). S. Tm, Salmonella serovar Typhimurium; wt, wild type; stat. anal., statistical analysis; NS, not statistically significant (P ≥ 0.05).
Time course of cecum pathology.
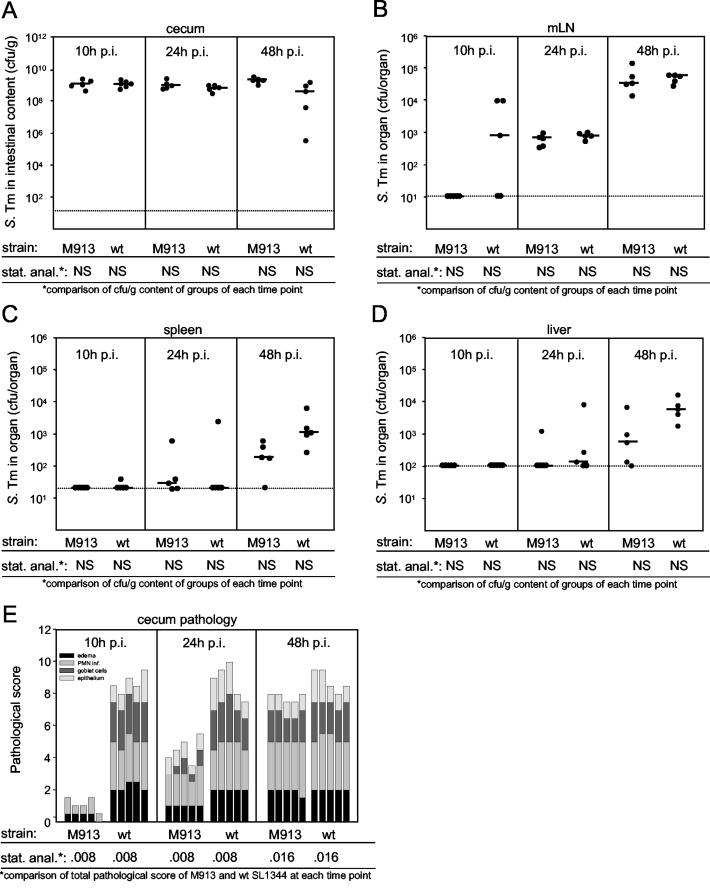
At 2 days p.i., the colitis caused by a nonflagellated serovar Typhimurium strain was slightly attenuated. It has been shown that streptomycin-pretreated mice develop colitis as early as 8 h p.i. (3). This raised the possibility that flagella might play a more pronounced role in the early phase of infection. To test this hypothesis, we infected 15 streptomycin-pretreated mice with wild-type serovar Typhimurium and 15 streptomycin-pretreated mice with nonflagellated strain M913. At 10, 24, and 48 h p.i. five mice from each group were killed and analyzed as described above.
Both strains efficiently colonized the cecum (108 to 109 CFU/g) within 10 h p.i., and the level of colonization remained constant throughout the rest of the experiment (Fig. 3A). The level of colonization of the mLN of three of five animals infected with the wild-type serovar Typhimurium strain was low, while no bacteria were detected in the mLN of animals infected with nonflagellated mutant M913. However, the difference was not statistically significant (Fig. 3B). Moreover, no differences between colonization of the mLN by M913 and colonization of the mLN by wild-type serovar Typhimurium were observed at later times. In accordance with previous data (3), colonization of the spleen and liver was apparent between 24 and 48 h p.i. While we recovered slightly higher numbers of wild-type bacteria than of the nonmotile mutant, the differences were not statistically significant (Fig. 3C and D). In spite of the similar colonization levels, M913 caused significantly reduced levels of inflammation (Fig. 3E and Table 2). In keeping with our first observation, the difference was small but significant at 48 h p.i. but much more pronounced at earlier times (Fig. 3E). At 10 h p.i., the ceca of M913-infected mice showed almost no signs of inflammation, such as PMN influx, edema, or epithelial damage. In contrast, mice infected with wild-type serovar Typhimurium had already developed severe colitis at this time. At 24 h p.i., mice infected with the fliGHI mutant showed the first symptoms of inflammation, and at day 2 p.i., there was only a small difference between the pathological scores for the two groups. This implies that the onset of inflammation is delayed upon infection with a nonmotile serovar Typhimurium mutant.
FIG. 3.
Time course of Salmonella wild-type and fliGHI mutant infection. Two groups of 15 streptomycin-pretreated mice were treated with 10% bicarbonate p.o. prior to infection with 5 × 107 CFU of serovar Typhimurium strains SL1344 (wild type) and M913 (fliGHI). After 10, 24, and 48 h five mice from each group were sacrificed. (A to D) Bacterial loads in the cecal contents (A), the mLN, (B), the spleen, (C), and the liver (D). The dotted lines indicate the limits of detection, and the solid horizontal lines indicate the medians. (E) Histopathological analysis. HE-stained sections of cecal tissue were scored for edema in the submucosa (black bars), PMN infiltration (medium gray bars), reductions in the numbers of goblet cells (dark grey bars), and desquamation, erosion, and ulceration of the epithelial layer (light gray bars) (see Materials and Methods). The scores are expressed as stacked vertical bars. The total pathological score (sum of the separate scores) and the bacterial load were statistically analyzed by using the exact Mann-Whitney U test (in comparison to wild-type strain SL1344). S. Tm, Salmonella serovar Typhimurium; wt, wild type; stat. anal., statistical analysis; NS, not statistically significant (P ≥ 0.05).
TABLE 2.
| Comparison |
P for comparison with wild-type strain SL1344
|
||
|---|---|---|---|
| M913 (10 h) | M913 (24 h) | M913 (48 h) | |
| Combined score | 0.008 | 0.008 | 0.016 |
| Edema | 0.008 | 0.016 | NS |
| PMN infiltration | 0.008 | 0.008 | NS |
| Goblet cells | 0.008 | 0.095 | NS |
| Epithelium | 0.008 | 0.008 | NS |
The data in Fig. 3E were analyzed by using the Mann-Whitney U test (see Materials and Methods). NS, not significant (P ≥ 0.05).
Localization of the fliGHI mutant.
The experiments described above demonstrated that flagella are required for efficient induction of serovar Typhimurium colitis in streptomycin-pretreated mice. However, the bacterial loads in the intestinal lumen do not account for the difference observed. Therefore, we reasoned that less efficient penetration of the mucus layer and reduced access to the intestinal epithelium might provide a plausible explanation. To test this hypothesis, we performed competition experiments. In this type of experiment, streptomycin-pretreated animals are infected with a mixture of wild-type and mutant bacteria. If each strain is labeled with a different fluorescent marker (GFP or DsRed), fluorescence microscopy can be used to compare the relative capacities of the two strains to interact with the intestinal wall.
M913 and the isogenic wild-type strain were transformed with GFP (pM979) and DsRed (pDsRed) expression plasmids. Preliminary infection experiments confirmed that the plasmids were stably propagated by both strains in the murine intestine for at least 2 days. Less than 5% of all serovar Typhimurium cells recovered from the cecum contents had lost the plasmids (data not shown).
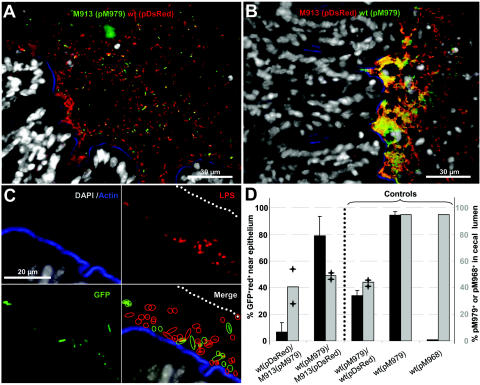
We then performed competitive infection experiments (Fig. 4). Three streptomycin-pretreated mice were infected with a 1:1 mixture of wild-type strain SL1344(pDsRed) and strain M913(pM979) (Fig. 4A). Three other mice were infected with a 1:1 mixture of wild-type strain SL1344(pM979) and strain M913(pDsRed) (Fig. 4B). At 24 h p.i., the animals were sacrificed, and cecum tissue samples were processed and stained for immunofluorescence microscopy (see Materials and Methods); DNA was stained with DAPI and was grey, and f-actin was stained with Alexa-647-phalloidin and was blue. As DsRed fluorescence was too weak for reliable detection, we stained all serovar Typhimurium cells in the cecum sections with a rabbit anti-LPS antibody and an anti-rabbit serum-rhodamine conjugate (which was red) (see Materials and Methods). In the first group of mice, red bacteria (wild type) were more abundant than yellow-green bacteria [M913(pM979)] at the epithelial surface (Fig. 4A). In contrast, roughly equal numbers of the competing strains were present in the cecal lumen (Fig. 4D). Very similar observations were made with the second group of mice; yellow-green bacteria (wild type) were more abundant than red bacteria [M913(pDsred)] at the epithelial surface (Fig. 4B), but equal numbers of the two strains were present in the cecal lumen (Fig. 4D). These findings indicate that flagella are required by serovar Typhimurium to efficiently contact the intestinal epithelium.
FIG. 4.
fliGHI mutant cannot efficiently reach the cecal epithelium. Three mice were inoculated with 10% bicarbonate prior to infection for 24 h with 5 × 107 CFU of M913(pM979) plus 5 × 107 CFU of SL1344(pDsRed), 5 × 107 CFU of SL1344(pM979) plus 5 × 107 CFU of M913(pDsRed), 5 × 107 CFU of SL1344(pM979) plus 5 × 107 CFU of SL1344(pDsRed), 5 × 107 CFU of SL1344(pM979), or 5 × 107 CFU of SL1344(pM968). Cecal tissue sections were stained with DAPI (grey), Alexa-647-phalloidin (blue), a rabbit anti-LPS antibody, and a goat anti-rabbit serum-rhodamine conjugate (red) (see Materials and Methods). Samples of cecal contents from two mice per group were plated to determine the fraction of pM979 or pM968 carrying bacteria (see Materials and Methods) (panel D, grey columns). (A) Cecum section from a mouse infected with M913(pM979) plus SL1344(pDsRed). (B) Cecum section from a mouse infected with SL1344(pM979) plus M913(pDsRed). (C) Strategy for quantification of GFP-positive red Salmonella cells (green circles) and red Salmonella cells (red circles) within 20 μm (dotted line) of the cecal epithelium (blue). (D) Presence of GFP-positive red Salmonella cells within 20 μm of the cecal epithelium (dotted line) as determined by fluorescence microscopy (black bars; average) (scale on the left side). The fraction of serovar Typhiumurium cells carrying pM979 or pM968 in the cecal contents was determined by replica plating (grey bars; average) (scale on the right side). The error bars indicate standard deviations. The plus signs indicate results for the two individual measurements. wt, wild type.
In order to quantify the defect in M913, we determined the numbers of wild-type and mutant bacteria present within 20 μm of the epithelial surface (Fig. 4C) (see Materials and Methods). The data were obtained by scoring at least 400 bacteria from different optical fields of cecum sections from all three mice from each group. The results were expressed as percentages of the GFP-positive red bacteria (number of GFP-positive red bacteria/number of red bacteria × 100). In this way we verified that the fliGHI mutant M913 was deficient in terms of approaching the cecal epithelium (Fig. 4D). The results of all of the control experiments performed were consistent with this conclusion (Fig. 4D). In mice infected with a 1:1 mixture of wild-type strain SL1344(pM979) and wild-type strain SL1344(pDsRed), roughly equal numbers of the two strains were present in the cecal lumen and close to the epithelial surface (Fig. 4D). Infections with wild-type SL1344(pM979) alone and infections with a strain carrying a control vector (pM968; no GFP expression) yielded >95% and <5% GFP-positive red bacteria, respectively, at the epithelium (Fig. 4D); >95% of both strains recovered from the intestinal lumen harbored the appropriate plasmid (Fig. 4D). These observations demonstrated that the GFP vector allowed reliable detection and discrimination between wild-type and mutant strains. Altogether, these data provide strong evidence that serovar Typhimurium requires flagella to approach the intestinal epithelium efficiently.
A chemotaxis mutant is attenuated for causing enterocolitis.
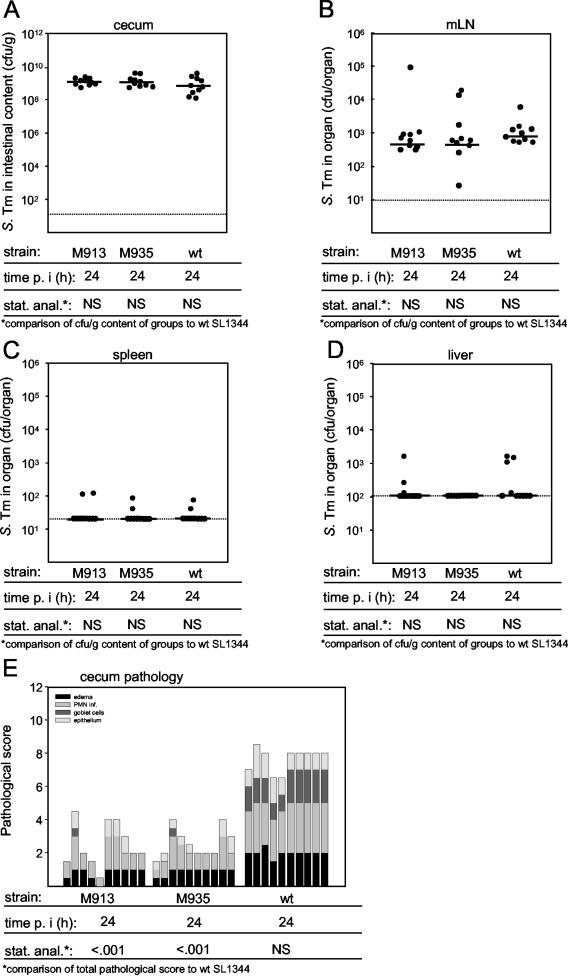
The data presented above demonstrated that the Salmonella mutant M913 lacking the entire flagellum secretion apparatus is impaired in terms of causing colitis and that this virulence defect may be linked to a reduced ability to approach the epithelial surface. In order to examine whether the virulence defect might be attributed to a chemotaxis defect, we constructed M935, a cheY transposon insertion mutant of serovar Typhimurium, by phage transduction (see Materials and Methods). This mutant was able to assemble functional flagella and secrete flagellin (FliC) (Fig. 1A), but it was not able to swim in a directed manner (45). On motility agar M935 formed no halo (Table 1). We performed two independent experiments with five streptomycin-pretreated mice per group. The animals were infected with 5 × 107 CFU of serovar Typhimurium M913, M935, or wild-type strain SL1344, and we analyzed colonization and cecal inflammation 24 h p.i. The results were identical in both experiments and are summarized in Fig. 5. fliGHI mutant M913, cheY mutant M935, and the isogenic wild-type strain had the same capacity to colonize the cecal lumen and the mLN (Fig. 5A and B). No significant colonization of the liver and spleen was detected in any of the experimental groups. cheY mutant M935 and the nonflagellated mutant M913 induced significantly less inflammation than the wild-type strain induced. There was no significant difference in pathology between M913 and M935 (P ≥ 0.05) (Fig. 5E and Table 3). These data were consistent with the results described above and suggested that chemotactic movement through the mucus layer is required for efficient induction of colitis.
FIG. 5.
Colitis caused by the Salmonella wild-type strain, an fliGHI mutant, and a cheY mutant. Three groups of 10 streptomycin-pretreated mice were treated with 10% bicarbonate p.o. prior to infection for 24 h with 5 × 107 CFU of serovar Typhimurium strains SL1344 (wild type), M913 (fliGHI), and M935 (cheY). The data were obtained from two independent experiments performed with groups of five mice. (A to D) Bacterial loads in the cecal contents (A), the mLN, (B), the spleen, (C), and the liver (D). The dotted lines indicate the limits of detection, and the solid horizontal lines indicate the medians. (E) Histopathological analysis. HE-stained sections of cecal tissue were scored for edema in the submucosa (black bars), PMN infiltration (PMN inf.; medium gray bars), reductions in the numbers of goblet cells (dark grey bars), and desquamation, erosion, and ulceration of the epithelial layer (light gray bars) (see Materials and Methods). The scores are expressed as stacked vertical bars. The total pathological score (sum of the separate scores) and the bacterial load were statistically analyzed by using the exact Mann-Whitney U test (in comparison to wild-type strain SL1344). S. Tm, Salmonella serovar Typhimurium; wt, wild type; stat. anal., statistical analysis; NS, not statistically significant (P ≥ 0.05).
TABLE 3.
| Comparison |
P for comparison with wild-type strain SL1344
|
P for comparison with M913
|
||
|---|---|---|---|---|
| M913 | M935 | M935 | SL1344 | |
| Combined score | <0.001 | <0.001 | NS | <0.001 |
| Edema | <0.001 | <0.001 | NS | <0.001 |
| PMN infiltration | <0.001 | <0.001 | NS | <0.001 |
| Goblet cells | <0.001 | <0.001 | NS | <0.001 |
| Epithelium | 0.005 | <0.001 | NS | 0.005 |
The data in Fig. 5E were analyzed by using the Mann-Whitney U test (see Materials and Methods). NS, not significant (P ≥ 0.05).
Competitive infection experiments to assess colonization defects of cheY and fliGHI mutants.
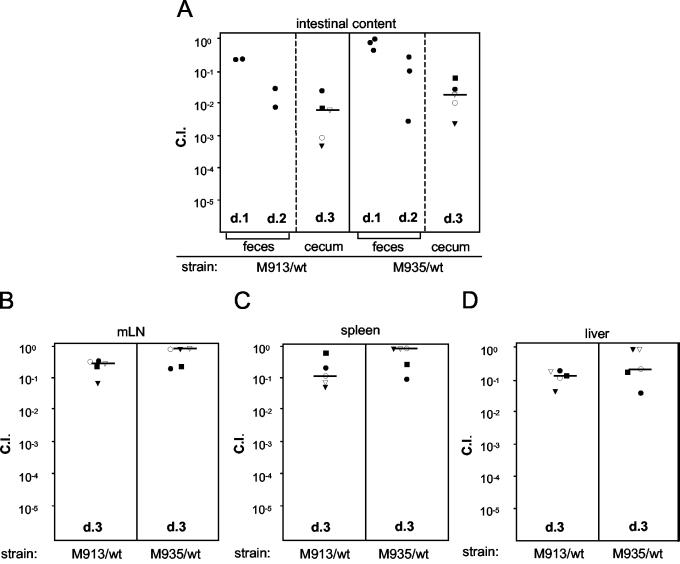
A second type of competition experiment was performed to examine whether motility and chemotaxis contribute to intestinal colonization in the streptomycin-pretreated mouse model. In the previous experiments we analyzed just one strain per mouse (Fig. 2, 3, and 5). This did not reveal any major colonization defect of nonmotile or nonchemotactic strains in the cecal lumen. However, a competition experiment could allow detection of even subtle defects. In this experiment, two groups of five streptomycin-pretreated mice were infected with a 1:1 mixture (total amount, 5 × 107 CFU) of wild-type strain SL1344 and M913 or wild-type strain SL1344 and M935. The ratio of the two strains in the feces was analyzed at days 1 and 2 p.i. The mice were sacrificed at day 3 p.i., and we analyzed the bacterial loads and the ratios of the two strains in the cecal contents, mLN, spleen, and liver.
In the feces from each of the groups, equal numbers of the two strains were found at day 1 p.i. (competitive indices [CI], ∼0.3 to 1.2). The CI values for the nonmotile and nonchemotactic strains decreased at day 2 p.i., and the CI for M913 and M935 in the cecal contents at day 3 p.i. was significantly lower (median, 10−2) (Fig. 6A).
FIG. 6.
CI for intestinal colonization of fliGHI and cheY mutants. Two groups of five streptomycin-pretreated mice were infected for 3 days with 5 × 107 CFU of a 1:1 mixture of serovar Typhimurium strains SL1344 (wild type) and M913 (fliGHI) or strains SL1344 and M935 (cheY). The exact ratio of the wild-type strain to the mutant strain in each inoculum was determined by replica plating (ratioinput). The ratios of the wild-type strain to the mutant strain in fecal pellets on days 1 and 2 p.i. (two or three pellets) and in cecal contents on day 3 p.i. (A), in mLN (B), in the spleen (C), and in the liver (D) were determined by replica plating (ratiooutput). CI were calculated with the formula CI = ratiooutput/ratioinput. The solid squares, open circles, solid circles, open triangles, and solid triangles indicate the individual mice in each group. The CI for fecal pellets on days 1 and 2 p.i. are indicated by solid circles because data could not be assigned to individual mice. d, day; wt, wild type.
The median CI for M913 and M935 in the mLN, spleen, and liver ranged from 1 to 0.1 (Fig. 6B, C, and D). These data suggest that flagella and, more specifically, chemotaxis contribute to serovar Typhimurium colonization of the murine intestine but not to the spread to systemic sites.
DISCUSSION
The role of flagella in Salmonella serovar Typhimurium virulence has been extensively studied in vitro. The in vivo function of flagella is less well understood, and it was unclear whether the flagella play a role in murine serovar Typhimurium colitis. We found that flagella and, more specifically, chemotaxis are required for efficient induction of murine Salmonella-induced colitis. In contrast, the systemic infection which occurred parallel to the enterocolitis did not depend significantly on flagellum function.
The role of motility during systemic stages of serovar Typhimurium infection in typhoid mouse models has been somewhat controversial. Carsiotis et al. found that a nonflagellated strain was attenuated compared to a wild-type strain in oral, intraperitoneal, and intravenous infections (5). However, nonmotile and nonchemotactic mutants that both possessed intact flagella were not attenuated. Similarly, Weinstein et al. showed that a Salmonella wild-type strain survived longer than a nonflagellated mutant survived in macrophage infection assays and exhibited faster net growth in spleens of infected mice (53). During systemic infection Salmonella cells are thought to reside and replicate mainly inside phagocytic cells (21, 25). Interestingly, expression analyses have demonstrated that flagellar genes are downregulated inside J774-A.1 macrophages, which may indicate that flagella are not required during the intracellular stages of the Salmonella life cycle (13). It has been reported by other workers that fla+ mot mutants, in contrast to fla mot mutants, are slightly attenuated in mice upon oral infection but not when the mice are infected by the intraperitoneal route (34). In another study an flhD mutant, but not an fliC fljB flagellum mutant, was described as having a significantly lower 50% lethal dose in the mouse typhoid model for Salmonella serovar Typhimurium and exhibited faster net growth in mouse macrophages. This effect was attributed to uncharacterized regulatory functions of flhD (40).
The initiation of a systemic infection (mLN, liver, spleen) was not significantly attenuated in fliGHI and cheY mutants. This contrasts with the significant role of flagella in the early phase of murine colitis, but it is consistent with previous studies performed with the murine typhoid model (40). This obvious discrepancy might be explained by peculiar features of the Peyer's patches of the distal ileum, which are thought to function as the port of entry to systemic sites (6). The structure of the follicle-associated epithelium, which contains specialized M cells, is different in many ways from the structure of the surrounding villous epithelium. For example, it lacks the overlying mucus layer (36). Hence, flagella might not be required for the interaction with M cells, and this could explain the similar organ counts for flagellum mutants and wild-type Salmonella serovar Typhimurium in our experiments.
Little is known about the role of flagella in intestinal salmonellosis. Robertson et al. investigated S. enterica serovar Enteritidis-induced salmonellosis in the rat. This serovar is monophasic, and deletion of the only flagellar subunit gene (fliC) attenuated intestinal inflammation. The levels of several inflammatory parameters measured in the small intestine contents of infected rats were reduced in the early stages of infection (1 and 2 days p.i.) with an fliC flagellum mutant compared to the levels in rats infected with the wild type (37). In bovine ligated ileal loops the enteropathogenic response to Salmonella serovar Typhimurium flhD or fliC fljB mutant strains was decreased (40). Here, the flhD mutant provoked a statistically reduced secretory response and PMN influx. These inflammation parameters were also attenuated upon infection with the fliC fljB double mutant, although the differences were not statistically significant. In line with these observations in the bovine model, we found that an fliGHI mutant is impaired in terms of eliciting colitis in streptomycin-pretreated mice. This effect is very pronounced at earlier time points (10 or 24 h p.i.), while the mutant catches up later, causing severe colitis to almost the same level as wild-type serovar Typhimurium at 48 h p.i. This suggests that flagella are required for establishing an infection in streptomycin-pretreated mice.
In spite of many similarities to the bovine model, there are several aspects which need to be kept in mind when data from the murine colitis model are compared with data obtained with bovine ligated ileal loops. Both models suffer from the absence of or a severe reduction in the level of a complete adult-type commensal flora. Both models are characterized by mucosal edema, PMN influx, destruction of the epithelial cell layer, and fast tissue regeneration. However, in the bovine system there is massive luminal fluid secretion which is not observed in the murine model. The reason for this is currently unknown.
In several studies the workers have addressed the role of flagella in invasion of cultured epithelial cells. Various mutants that lacked flagella or had impaired motility or chemotaxis were less invasive or had a defect in attachment (10, 26, 33, 34, 40). Jones et al. found that an fla mutant and a smooth-swimming chemotaxis mutant were not attenuated for invasion of murine ligated ileal loops, while a mot mutant and a tumbly che mutant were less invasive than the wild-type strain (26). Altogether, these results demonstrate that functional flagella are necessary to efficiently invade epithelial cells, a process which also requires the SPI-1 TTSS. However, it is still a matter of dispute whether flagella merely allow the bacteria to approach the host cell in order to position the SPI-1 TTSS optimally for injection or whether flagella might have additional functions. The similar levels of attenuation of fliGHI (no flagella) and cheY (flagellated but non chemotactic) mutants observed in our experiments suggest that chemotactic movement is the major virulence function of flagella in the streptomycin-pretreated mouse model.
Could flagella have additional functions in this model? Flagellin has been shown to elicit innate immune responses. This phenomenon has been studied extensively in tissue culture. Flagellin (FliC) binds TLR5, activates NF-κB, and induces proinflammatory responses (11, 41, 54). Similar observations have been made in vivo. After intraperitoneal injection into mice, Salmonella flagellin leads to systemic release of interleukin-6, a key mediator of inflammation (19). This indicates that the manner in which murine tissues respond to flagellin is similar to the manner in which human cells respond to flagellin. So far, no TLR5 knockout mice have been described. However, knockout mice lacking a central element of TLR signaling cascades (MyD88) do not respond to flagellin (19). These observations suggest that flagellin could contribute to the inflammatory response in the streptomycin-pretreated mouse model.
Interestingly, bacteria of the normal gut flora are known to produce and release a wide variety of TLR ligands (pathogen-associated molecular patterns [PAMPs], including flagellin) that can induce innate immune responses (43). However, these ligands normally do not lead to intestinal inflammation. This apparent discrepancy has been resolved by evidence which suggests that TLRs are localized on the basolateral side of the intestinal epithelium (16). Hence, bacterial flagellin released in the intestinal lumen cannot readily come in contact with TLR5.
Do serovar Typhimurium flagellin-TLR5 interactions contribute to colitis in streptomycin-pretreated mice? This question is quite difficult to answer as flagella in this case would serve two different functions, innate immune signaling and chemotaxis. In vitro, purified Salmonella flagellin can induce inflammatory responses when it is added to tissue culture cells (11, 41, 54). Therefore, interactions between flagellin released from bacteria and TLR5 might be involved in murine serovar Typhimurium colitis. The SPI-1 TTSS is a key virulence factor in this model (3). Serovar Typhimurium mutants with a disrupted SPI-1 TTSS do not cause significant inflammation in streptomycin-pretreated mice even though they efficiently colonize the murine large intestine and have a fully functional flagellar system (3, 18). Thus, the mere presence of serovar Typhimurium flagella (or other PAMPs released by serovar Typhimurium) in the intestinal lumen is not enough to induce colitis. However, in the wild-type infection, SPI-1-induced processes (i.e., disruption of the epithelial barrier) might facilitate access of flagellin to TLR5 receptors. In this case, the flagellin-TLR5 interactions might contribute to the SPI-1-dependent inflammation observed with wild-type serovar Typhimurium. We did not observe a significant difference between the (delayed) inflammatory responses elicited by fliGHI (no flagellin secretion) and cheY (flagellated or secreted flagellin but nonchemotactic) mutants. Therefore, the flagellin-TLR5 interaction does not seem play a major role in murine serovar Typhimurium colitis. However, the effects caused by the flagellin-TLR5 interaction might simply be masked by other inflammatory stimuli attributable to other serovar Typhimurium virulence factors or PAMPs.
The competition experiments indicated that chemotaxis is required for efficient colonization of the murine intestine. This is in line with previous observations made with the chicken model (2). Overall, these observations suggest that Salmonella spp. actively move through the mucus layer in a chemotactic manner towards the epithelium in order to inject effector proteins via the SPI-1 TTSS. In experiments with Vibrio cholerae, Freter et al. demonstrated that motile bacteria penetrated the intestinal mucus layer in mice more efficiently than either nonmotile or nonchemotactic mutants penetrated the intestinal mucus layer (15). Interestingly, these authors found that nonchemotactic mutants of V. cholerae still invaded the mucus at a low rate similar to the rate of inert particles. A similar mechanism could explain the delayed onset of disease symptoms with the nonmotile mutant in our experiments. We suppose that fliGHI and cheY mutants might contact the epithelium less efficiently than the wild-type strain and therefore translocate SPI-1 effector proteins at a lower frequency. Additional work is required to address this possible correlation between motility and the efficiency of SPI-1 effector protein translocation.
In summary, the data presented here clearly show that motility and chemotaxis are required for the efficient induction of colitis in streptomycin-pretreated mice. Further studies are needed to identify the other Salmonella virulence factors involved and to elucidate the inflammatory cascades leading to colitis in streptomycin-treated mice.
Acknowledgments
We are grateful to Kristin Ehrbar for providing Southern hybridization probes and to Shin Numao, Cosima Pelludat, and Gerald Weidinger for discussions and critical reading of the manuscript.
This work was supported by grant 3100A0-100175/1 to W.-D.H. from the Swiss National Foundation.
Editor: J. B. Bliska
REFERENCES
- 1.Aldridge, P., and K. T. Hughes. 2002. Regulation of flagellar assembly. Curr. Opin. Microbiol. 5:160-165. [DOI] [PubMed] [Google Scholar]
- 2.Allen-Vercoe, E., and M. J. Woodward. 1999. Colonisation of the chicken caecum by afimbriate and aflagellate derivatives of Salmonella enterica serotype Enteritidis. Vet. Microbiol. 69:265-275. [DOI] [PubMed] [Google Scholar]
- 3.Barthel, M., S. Hapfelmeier, L. Quintanilla-Martinez, M. Kremer, M. Rohde, M. Hogardt, K. Pfeffer, H. Russmann, and W. D. Hardt. 2003. Pretreatment of mice with streptomycin provides a Salmonella enterica serovar Typhimurium colitis model that allows analysis of both pathogen and host. Infect. Immun. 71:2839-2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bongaerts, R. J., I. Hautefort, J. M. Sidebotham, and J. C. Hinton. 2002. Green fluorescent protein as a marker for conditional gene expression in bacterial cells. Methods Enzymol. 358:43-66. [DOI] [PubMed] [Google Scholar]
- 5.Carsiotis, M., D. L. Weinstein, H. Karch, I. A. Holder, and A. D. O'Brien. 1984. Flagella of Salmonella typhimurium are a virulence factor in infected C57BL/6J mice. Infect. Immun 46:814-818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carter, P. B., and F. M. Collins. 1974. The route of enteric infection in normal mice. J. Exp. Med. 139:1189-1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Collazo, C. M., and J. E. Galan. 1996. Requirement for exported proteins in secretion through the invasion-associated type III system of Salmonella typhimurium. Infect. Immun. 64:3524-3531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Collazo, C. M., M. K. Zierler, and J. E. Galan. 1995. Functional analysis of the Salmonella typhimurium invasion genes invl and invJ and identification of a target of the protein secretion apparatus encoded in the inv locus. Mol. Microbiol. 15:25-38. [DOI] [PubMed] [Google Scholar]
- 9.Cormack, B. P., R. H. Valdivia, and S. Falkow. 1996. FACS-optimized mutants of the green fluorescent protein (GFP). Gene 173:33-38. [DOI] [PubMed] [Google Scholar]
- 10.Dibb-Fuller, M. P., E. Allen-Vercoe, C. J. Thorns, and M. J. Woodward. 1999. Fimbriae- and flagella-mediated association with and invasion of cultured epithelial cells by Salmonella enteritidis. Microbiology 145:1023-1031. [DOI] [PubMed] [Google Scholar]
- 11.Eaves-Pyles, T., K. Murthy, L. Liaudet, L. Virag, G. Ross, F. G. Soriano, C. Szabo, and A. L. Salzman. 2001. Flagellin, a novel mediator of salmonella-induced epithelial activation and systemic inflammation: I kappa B alpha degradation, induction of nitric oxide synthase, induction of proinflammatory mediators, and cardiovascular dysfunction. J. Immunol. 166:1248-1260. [DOI] [PubMed] [Google Scholar]
- 12.Eichelberg, K., and J. E. Galan. 2000. The flagellar sigma factor FliA (σ28) regulates the expression of Salmonella genes associated with the centisome 63 type III secretion system. Infect. Immun. 68:2735-2743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eriksson, S., S. Lucchini, A. Thompson, M. Rhen, and J. C. Hinton. 2003. Unravelling the biology of macrophage infection by gene expression profiling of intracellular Salmonella enterica. Mol. Microbiol. 47:103-118. [DOI] [PubMed] [Google Scholar]
- 14.Fang, F. C., M. A. DeGroote, J. W. Foster, A. J. Baumler, U. Ochsner, T. Testerman, S. Bearson, J. C. Giard, Y. Xu, G. Campbell, and T. Laessig. 1999. Virulent Salmonella typhimurium has two periplasmic Cu,Zn-superoxide dismutases. Proc. Natl. Acad. Sci. USA 96:7502-7507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Freter, R., P. C. O'Brien, and M. S. Macsai. 1981. Role of chemotaxis in the association of motile bacteria with intestinal mucosa: in vivo studies. Infect. Immun. 34:234-240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gewirtz, A. T., T. A. Navas, S. Lyons, P. J. Godowski, and J. L. Madara. 2001. Cutting edge: bacterial flagellin activates basolaterally expressed TLR5 to induce epithelial proinflammatory gene expression. J. Immunol. 167:1882-1885. [DOI] [PubMed] [Google Scholar]
- 17.Groisman, E. A., and H. Ochman. 1997. How salmonella became a pathogen. Trends Microbiol. 5:343-349. [DOI] [PubMed] [Google Scholar]
- 18.Hapfelmeier, S., K. Ehrbar, B. Stecher, M. Barthel, M. Kremer, and W. D. Hardt. 2004. Role of the Salmonella pathogenicity island 1 effector proteins SipA, SopB, SopE, and SopE2 in Salmonella enterica subspecies 1 serovar Typhimurium colitis in streptomycin-pretreated mice. Infect. Immun. 72:795-809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hayashi, F., K. D. Smith, A. Ozinsky, T. R. Hawn, E. C. Yi, D. R. Goodlett, J. K. Eng, S. Akira, D. M. Underhill, and A. Aderem. 2001. The innate immune response to bacterial flagellin is mediated by Toll-like receptor 5. Nature 410:1099-1103. [DOI] [PubMed] [Google Scholar]
- 20.Hensel, M. 2000. Salmonella pathogenicity island 2. Mol. Microbiol. 36:1015-1023. [DOI] [PubMed] [Google Scholar]
- 21.Hensel, M., J. E. Shea, C. Gleeson, M. D. Jones, E. Dalton, and D. W. Holden. 1995. Simultaneous identification of bacterial virulence genes by negative selection. Science 269:400-403. [DOI] [PubMed] [Google Scholar]
- 22.Hoiseth, S. K., and B. A. Stocker. 1981. Aromatic-dependent Salmonella typhimurium are non-virulent and effective as live vaccines. Nature 291:238-239. [DOI] [PubMed] [Google Scholar]
- 23.Hurley, B. P., and B. A. McCormick. 2003. Translating tissue culture results into animal models: the case of Salmonella typhimurium. Trends Microbiol. 11:562-569. [DOI] [PubMed] [Google Scholar]
- 24.Irikura, V. M., M. Kihara, S. Yamaguchi, H. Sockett, and R. M. Macnab. 1993. Salmonella typhimurium fliG and fliN mutations causing defects in assembly, rotation, and switching of the flagellar motor. J. Bacteriol. 175:802-810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jantsch, J., C. Cheminay, D. Chakravortty, T. Lindig, J. Hein, and M. Hensel. 2003. Intracellular activities of Salmonella enterica in murine dendritic cells. Cell. Microbiol. 5:933-945. [DOI] [PubMed] [Google Scholar]
- 26.Jones, B. D., C. A. Lee, and S. Falkow. 1992. Invasion by Salmonella typhimurium is affected by the direction of flagellar rotation. Infect. Immun. 60:2475-2480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Josenhans, C., and S. Suerbaum. 2002. The role of motility as a virulence factor in bacteria. Int. J. Med. Microbiol. 291:605-614. [DOI] [PubMed] [Google Scholar]
- 28.Kaniga, K., J. C. Bossio, and J. E. Galan. 1994. The Salmonella typhimurium invasion genes invF and invG encode homologues of the AraC and PulD family of proteins. Mol. Microbiol. 13:555-568. [DOI] [PubMed] [Google Scholar]
- 29.Kaniga, K., D. Trollinger, and J. E. Galan. 1995. Identification of two targets of the type III protein secretion system encoded by the inv and spa loci of Salmonella typhimurium that have homology to the Shigella IpaD and IpaA proteins. J. Bacteriol. 177:7078-7085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kaniga, K., S. Tucker, D. Trollinger, and J. E. Galan. 1995. Homologs of the Shigella IpaB and IpaC invasins are required for Salmonella typhimurium entry into cultured epithelial cells. J. Bacteriol. 177:3965-3971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kingsley, R. A., A. D. Humphries, E. H. Weening, M. R. De Zoete, S. Winter, A. Papaconstantinopoulou, G. Dougan, and A. J. Baumler. 2003. Molecular and phenotypic analysis of the CS54 island of Salmonella enterica serotype Typhimurium: identification of intestinal colonization and persistence determinants. Infect. Immun. 71:629-640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Komoriya, K., N. Shibano, T. Higano, N. Azuma, S. Yamaguchi, and S. I. Aizawa. 1999. Flagellar proteins and type III-exported virulence factors are the predominant proteins secreted into the culture media of Salmonella typhimurium. Mol. Microbiol. 34:767-779. [DOI] [PubMed] [Google Scholar]
- 33.La Ragione, R. M., W. A. Cooley, P. Velge, M. A. Jepson, and M. J. Woodward. 2003. Membrane ruffling and invasion of human and avian cell lines is reduced for aflagellate mutants of Salmonella enterica serotype Enteritidis. Int. J. Med. Microbiol. 293:261-272. [DOI] [PubMed] [Google Scholar]
- 34.Lockman, H. A., and R. Curtiss III. 1990. Salmonella typhimurium mutants lacking flagella or motility remain virulent in BALB/c mice. Infect. Immun. 58:137-143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Macnab, R. M. 2003. How bacteria assemble flagella. Annu. Rev. Microbiol. 57:77-100. [DOI] [PubMed] [Google Scholar]
- 36.Owen, R. L. 1999. Uptake and transport of intestinal macromolecules and microorganisms by M cells in Peyer's patches—a personal and historical perspective. Semin. Immunol. 11:157-163. [DOI] [PubMed] [Google Scholar]
- 37.Robertson, J. M., N. H. McKenzie, M. Duncan, E. Allen-Vercoe, M. J. Woodward, H. J. Flint, and G. Grant. 2003. Lack of flagella disadvantages Salmonella enterica serovar Enteritidis during the early stages of infection in the rat. J. Med. Microbiol. 52:91-99. [DOI] [PubMed] [Google Scholar]
- 38.Rotger, R., and J. Casadesus. 1999. The virulence plasmids of Salmonella. Int. Microbiol. 2:177-184. [PubMed] [Google Scholar]
- 39.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 40.Schmitt, C. K., J. S. Ikeda, S. C. Darnell, P. R. Watson, J. Bispham, T. S. Wallis, D. L. Weinstein, E. S. Metcalf, and A. D. O'Brien. 2001. Absence of all components of the flagellar export and synthesis machinery differentially alters virulence of Salmonella enterica serovar Typhimurium in models of typhoid fever, survival in macrophages, tissue culture invasiveness, and calf enterocolitis. Infect. Immun. 69:5619-5625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sierro, F., B. Dubois, A. Coste, D. Kaiserlian, J. P. Kraehenbuhl, and J. C. Sirard. 2001. Flagellin stimulation of intestinal epithelial cells triggers CCL20-mediated migration of dendritic cells. Proc. Natl. Acad. Sci. USA 98:13722-13727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sly, L. M., D. G. Guiney, and N. E. Reiner. 2002. Salmonella enterica serovar Typhimurium periplasmic superoxide dismutases SodCI and SodCII are required for protection against the phagocyte oxidative burst. Infect. Immun. 70:5312-5315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Smith, K. D., E. Andersen-Nissen, F. Hayashi, K. Strobe, M. A. Bergman, S. L. Barrett, B. T. Cookson, and A. Aderem. 2003. Toll-like receptor 5 recognizes a conserved site on flagellin required for protofilament formation and bacterial motility. Nat. Immunol. 4:1247-1253. [DOI] [PubMed] [Google Scholar]
- 44.Stender, S., A. Friebel, S. Linder, M. Rohde, S. Mirold, and W. D. Hardt. 2000. Identification of SopE2 from Salmonella typhimurium, a conserved guanine nucleotide exchange factor for Cdc42 of the host cell. Mol. Microbiol. 36:1206-1221. [DOI] [PubMed] [Google Scholar]
- 45.Togashi, F., S. Yamaguchi, M. Kihara, S. I. Aizawa, and R. M. Macnab. 1997. An extreme clockwise switch bias mutation in fliG of Salmonella typhimurium and its suppression by slow-motile mutations in motA and motB. J. Bacteriol. 179:2994-3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tsolis, R. M., L. G. Adams, T. A. Ficht, and A. J. Baumler. 1999. Contribution of Salmonella typhimurium virulence factors to diarrheal disease in calves. Infect. Immun. 67:4879-4885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Uzzau, S., L. Bossi, and N. Figueroa-Bossi. 2002. Differential accumulation of Salmonella [Cu, Zn] superoxide dismutases SodCI and SodCII in intracellular bacteria: correlation with their relative contribution to pathogenicity. Mol. Microbiol. 46:147-156. [DOI] [PubMed] [Google Scholar]
- 48.van der Velden, A. W., A. J. Baumler, R. M. Tsolis, and F. Heffron. 1998. Multiple fimbrial adhesins are required for full virulence of Salmonella typhimurium in mice. Infect. Immun. 66:2803-2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wallis, T. S., and E. E. Galyov. 2000. Molecular basis of Salmonella-induced enteritis. Mol. Microbiol. 36:997-1005. [DOI] [PubMed] [Google Scholar]
- 50.Wang, R. F., and S. R. Kushner. 1991. Construction of versatile low-copy-number vectors for cloning, sequencing and gene expression in Escherichia coli. Gene 100:195-199. [PubMed] [Google Scholar]
- 51.Waterman, S. R., and D. W. Holden. 2003. Functions and effectors of the salmonella pathogenicity island 2 type III secretion system. Cell. Microbiol. 5:501-511. [DOI] [PubMed] [Google Scholar]
- 52.Watson, P. R., E. E. Galyov, S. M. Paulin, P. W. Jones, and T. S. Wallis. 1998. Mutation of invH, but not stn, reduces Salmonella-induced enteritis in cattle. Infect. Immun. 66:1432-1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Weinstein, D. L., M. Carsiotis, C. R. Lissner, and A. D. O'Brien. 1984. Flagella help Salmonella typhimurium survive within murine macrophages. Infect. Immun. 46:819-825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zeng, H., A. Q. Carlson, Y. Guo, Y. Yu, L. S. Collier-Hyams, J. L. Madara, A. T. Gewirtz, and A. S. Neish. 2003. Flagellin is the major proinflammatory determinant of enteropathogenic salmonella. J. Immunol. 171:3668-3674. [DOI] [PubMed] [Google Scholar]
- 55.Zhang, S., L. G. Adams, J. Nunes, S. Khare, R. M. Tsolis, and A. J. Baumler. 2003. Secreted effector proteins of Salmonella enterica serotype Typhimurium elicit host-specific chemokine profiles in animal models of typhoid fever and enterocolitis. Infect. Immun. 71:4795-4803. [DOI] [PMC free article] [PubMed] [Google Scholar]