Abstract
Active DNA demethylation in mammals involves TET-mediated iterative oxidation of 5-methylcytosine (5mC)/5-hydroxymethylcytosine (5hmC) and subsequent excision repair of highly oxidized cytosine bases 5-formylcytosine (5fC)/5-carboxylcytosine (5caC) by Thymine DNA glycosylase (TDG). However, quantitative and high-resolution analysis of active DNA demethylation activity remains challenging. Here we describe M.SssI methylase-assisted bisulfite sequencing (MAB-seq), a method that directly maps 5fC/5caC at single-base resolution. Genome-wide MAB-seq allows systematic identification of 5fC/5caC in Tdg-depleted embryonic stem cells, thereby generating a base-resolution map of active DNA demethylome. A comparison of 5fC/5caC and 5hmC distribution maps indicates that catalytic processivity of TET enzymes correlates with local chromatin accessibility. MAB-seq also reveals strong strand asymmetry of active demethylation within palindromic CpGs. Integrating MAB-seq with other base-resolution mapping methods enables quantitative measurement of cytosine modification states at key transitioning steps of active demethylation pathway, and reveals a regulatory role of 5fC/5caC excision repair in active DNA demethylation cascade.
DNA methyltransferases (DNMTs) chemically modify the genome by adding a methyl group to the 5-position of cytosines 1, generating an epigenetic mark (5mC) that has a profound impact on genome stability, transcription and development 2, 3. Dysregulation of 5mC patterns is frequently associated with human cancers 4. Compelling evidence now indicates that reversal of DNA methylation plays an important role in mammalian development and cell-type specific gene expression. In mammals, DNA demethylation (conversion of 5mC to unmodified C) can be achieved either passively through successive rounds of DNA replication in the absence of functional DNA methylation maintenance machinery or actively by Ten-eleven translocation (TET) family of 5mC-modifying enzymes 5, 6. TET proteins are Fe2+ and 2-oxoglutarate-dependent dioxygenases capable of successively oxidizing 5mC to 5hmC, 5fC and 5caC (Fig. 1a) 7–10. Oxidative modification of 5mC by TET enzymes promotes DNA demethylation by either replication-dependent dilution of oxidized cytosines 11 or TDG-mediated 5fC/5caC excision followed by base-excision repair (BER) 10, 12, 13. The active demethylation pathway involving generation and excision repair of 5fC/5caC is of particular interest as it may take place in a wide range of somatic cell-types including post-mitotic cells. Interestingly, only TDG, but not other members of the uracil DNA glycosylase (UDG) superfamily, possesses robust 5fC/5caC excision activity and is indispensable for embryonic development 14, 15, implicating the TET/TDG-dependent DNA demethylation pathway in regulating tissue-specific gene expression and development.
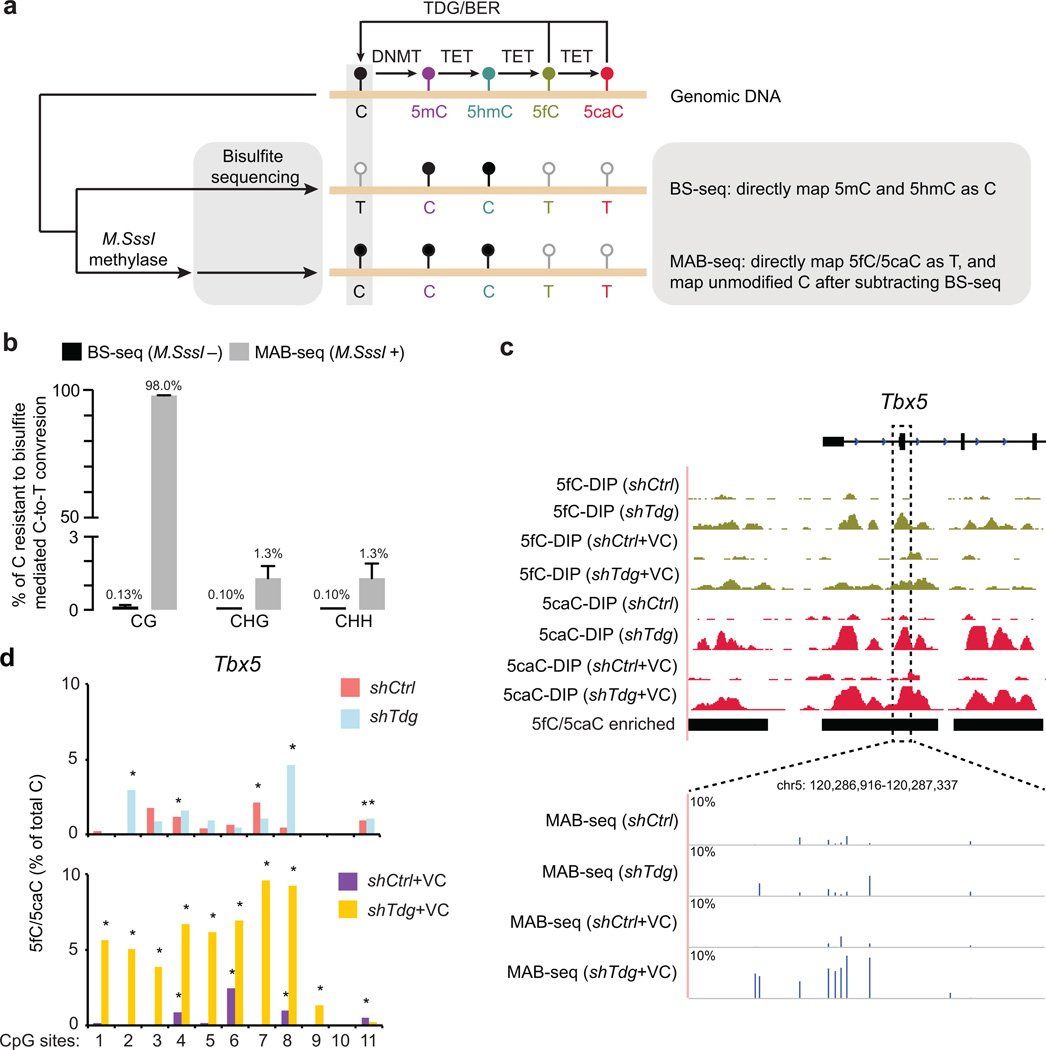
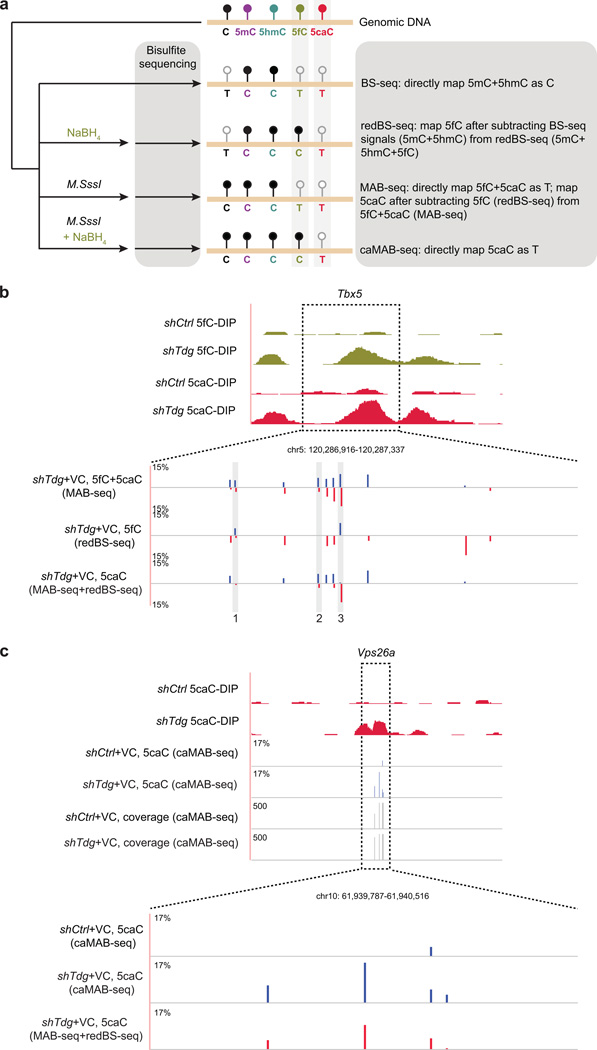
Figure 1. MAB-seq strategy and quantitative mapping of active DNA demethylation.
(a) Schematic diagram of MAB-seq. DNMT methylates unmodified C to generate 5mC, which can be successively oxidized by TET to generate 5hmC/5fC/5caC. Highly oxidized cytosine derivatives, 5fC and 5caC, are repaired by TDG/BER to regenerate unmodified C.
(b) M.SssI exhibits robust methylase activity towards unmodified cytosines within CpGs, but has significantly lower activity for cytosines in non-CpG contexts (CHG or CHH, H=A, T, C). M.SssI methylase activity was measured by MAB-seq analysis (Illumina deep sequencing) of unmethylated lambda DNA. Standard bisulfite sequencing (BS-seq) confirmed nearly complete conversion of unmethylated C to T at CpG and non-CpG sites.
(c) Locus-specific MAB-seq analysis of 5fC/5caC at Tbx5 by Illumina sequencing in control (shCtrl) and Tdg knockdown (shTdg) mouse ESCs. For comparison, also shown are 5fC/5caC antibody DNA immunoprecipitation (DIP) based maps of 5fC and 5caC in control and Tdg-depleted mouse ESCs. DIP-seq tracks are represented in normalized read density (reads per 10 million reads, rp10m) and the vertical axis range of all DIP-seq tracks is from 1 to 25. Black horizontal bars denote 5fC/5caC-enriched regions identified by 5fC/5caC DIP-seq. The level of 5fC/5caC (only Watson strand shown) is displayed as the percentage of total C modified as 5fC/5caC, and background signals detected in Tet1/2−/− mouse ESCs was subtracted.
(d) Statistical calling of 5fC/5caC-modified CpGs in locus-specific MAB-seq analysis. Shown are 5fC/5caC levels (background corrected using raw MAB-seq signals in Tet1/2−/−) of 11 CpG sites from the Tbx5 locus in shCtrl, shCtrl+VC, shTdg and shTdg+VC mouse ESCs. shTdg was compared with shCtrl while shTdg+VC was compared with shCtrl+VC. An asterisk indicates that a CpG is statistically enriched for 5fC/5caC (multiple comparison corrected P<0.05, Fisher’s exact test).
The observation that 5hmC can accumulate to a relatively high level in diverse cell-types, particularly in adult neurons 16, 17, raises the possibility that TET proteins tend to stall at 5hmC and that further oxidation of 5hmC to 5fC/5caC is a rate-limiting step. Therefore, identifying cytosines that are committed to active DNA demethylation requires methods that permit quantitative measurement of TDG-mediated excision of 5fC/5caC at high resolution. In addition, because both 5fC and 5caC are selectively recognized and excised by TDG, determining the strand-specific preference of active DNA demethylation activity requires the ability to simultaneously map both 5fC and 5caC. Recent studies using either 5fC/5caC-specific antibodies or chemical tagging of 5fC have shown that genomic regions containing cytosines undergoing TET/TDG-dependent active demethylation can be identified by analyzing ectopic 5fC/5caC accumulation in Tdg-depleted cells 18, 19. However, 5fC and 5caC maps generated by affinity-enrichment methods are of limited resolution (several hundred base-pairs), only represent relative enrichment, and lack strand distribution information. To address these limitations, we have developed a method named MAB-seq, which allows simultaneous and quantitative mapping of both 5fC and 5caC at base-resolution. Furthermore, our genome-wide MAB-seq analysis of mouse ESCs has provided new insights into catalytic processivity, strand asymmetry and feedback regulation of the TET/TDG-mediated active DNA demethylation pathway.
RESULTS
Experimental strategy and validation of MAB-seq
Recognizing the limitation of affinity-enrichment methods and several recently developed chemical modification-assisted BS-seq methods that require subtraction of BS-seq signals to indirectly map 5fC or 5caC 19–21 (Supplementary Fig. 1a), we have explored MAB-seq, an approach that aims to achieve direct and simultaneous measurement of 5fC and 5caC at single-base resolution (Fig. 1a). In standard BS-seq, C/5fC/5caC react with sodium bisulfite and are efficiently deaminated to uracil (C/5fC) or 5caU (5caC), both of which are sequenced as thymine (T), whereas 5mC and 5hmC are resistant to this chemical conversion and sequenced as C (Fig. 1a). In MAB-seq, genomic DNA is first treated with the bacterial DNA CpG methyltransferase M.SssI, an enzyme that is originally isolated from Spiroplasma sp. strain MQ1 and is known to efficiently methylate cytosines within CpG dinucleotides 22. Bisulfite conversion of M. SssI-treated DNA may therefore only deaminate 5fC and 5caC; originally unmodified C within CpGs is protected as 5mC. Subsequent sequencing would reveal 5fC and 5caC as T, whereas C/5mC/5hmC as C (Supplementary Fig. 2a). Notably, MAB-seq is unable to distinguish 5fC/5caC from unmodified C within non-CpG context due to the poor activity of M. SssI towards C within non-CpG context. This limitation does not affect the application of this technique for two reasons: first, 5hmC is almost exclusively found in the CpG context (>99% in CpGs), even in mouse ESCs and neurons where non-CpG methylation is prevalent 17, 23; second, recent structural and biochemical analyses indicate that TET proteins have a strong preference for oxidizing 5mC in CpG sites than in non-CpG context 24, 25. Thus, oxidative modification of 5mC by TET proteins occurs predominantly in the CpG context, and MAB-seq may provide a quantitative measurement of the abundance of 5fC/5caC within CpG dyads.
Successful detection of 5fC/5caC using MAB-seq requires complete conversion of C to 5mC by M. SssI as well as efficient bisulfite conversion of 5fC/5caC. We first optimized the reaction conditions and achieved nearly complete (99.2%) conversion of unmodified CpGs to 5mCpGs by M. SssI methylase measured by Sanger sequencing (Supplementary Fig. 1b). Next, we performed high-throughput BS-seq analysis of a synthetic double-stranded DNA (dsDNA) containing CpGs with specific cytosine modifications (5hmC/5fC/5caC). Consistent with previous reports 21, our analysis showed that 5fC (84.7%) and 5caC (99.5%), but not 5hmC (3.3%), are efficiently deaminated by bisulfite treatment and read as T (Supplementary Fig. 2b). In addition to unmodified CpGs (C:C), asymmetrically-modified CpGs (5mC/5hmC/5fC/5caC:C) may be present at low levels in the genome 23. We thus tested MAB-seq in analyzing asymmetrically modified dsDNA (5hmC/5fC/5caC:C). This analysis demonstrated that M. SssI methylase is capable of efficiently methylating unmodified C in hemi-modified CpG dyads (Supplementary Fig. 2c), validating the capability of MAB-seq in mapping asymmetrically modified 5fC/5caC in a strand-specific manner.
We next performed BS-seq and MAB-seq analysis of unmethylated lambda phage genome (6,224 CpGs within 48,502 bp) using Illumina high-throughput sequencing and sequenced to an average depth of 239x and 305x per cytosine, respectively. In BS-seq, a nearly complete C-to-T conversion within CpG sites was observed (99.9 +/− 0.06%, n=3 experiments) contrasted to a low conversion rate in MAB-seq (2.04 +/− 0.14%, n=9) (Fig. 1b and Supplementary Fig. 3a). To test whether unprotected CpGs in MAB-seq experiments exhibit random distribution, we analyzed the sequences immediately flanking 67 CpGs (mean methylation: 94.1%) that are not efficiently methylated by M. SssI (Supplementary Fig. 3b–c). We found that these 67 CpGs are not associated with any specific sequences (Supplementary Fig. 3d), suggesting that M. SssI has minimal sequence preference for catalyzing CpG methylation reactions. Consistent with previous findings 22, we found that M. SssI only methylates a small fraction of cytosines (1.3%) within non-CpG context (Fig. 1b and Supplementary Fig. 3a).
To test MAB-seq in analyzing mammalian genomic DNA, we applied this method to examine four 5fC/5caC-enriched loci (Tbx5, Vps26a, Ace, and Slc2a12) that were previously identified in Tdg-depleted mouse ESCs by affinity-enrichment methods 18, 19 (Fig. 1c, Supplementary Fig. 4 and 5a). Using mouse ESCs deficient for both Tet1 and Tet2 (largely absent of 5fC/5caC) as a negative control to correct background signals, locus-specific MAB-seq analysis not only located individual CpGs associated with significant levels of 5fC/5caC (Fig. 1d and Supplementary Fig. 6; P<0.05, Fisher’s exact test), but also revealed the absolute level of 5fC/5caC at each CpG (Fig. 1c and Supplementary Fig. 4). In contrast, active promoter of Rest, where 5fC/5caC is undetectable by affinity methods, is largely absent of MAB-seq signals (left panel in Supplementary Fig. 5a). This analysis also confirmed the positive effect of vitamin C (VC) on the catalytic activity of TET enzymes 26, 27, as VC-treated Tdg-depleted mouse ESCs (shTdg+VC) display even higher level of 5fC/5caC compared to shTdg cells (Fig. 1c and Supplementary Fig. 4, 6). Notably, a small number of 5fC/5caC-modified CpGs were also identified in the shCtrl and shCtrl+VC samples (Fig. 1d and Supplementary Fig. 6). Although the levels of 5fC/5caC in shCtrl are much lower than those in shTdg, the results suggest that MAB-seq is highly sensitive and may identify rare 5fC/5caC modifications in wild-type cells. Similar 5fC/5caC distribution patterns were detected for biologically independent replicates, demonstrating the reproducibility and robustness of MAB-seq (Supplementary Fig. 5b).
Genome-wide MAB-seq analysis of mouse ESCs
Having validated MAB-seq using locus-specific analysis, we next applied MAB-seq to identify cytosines undergoing active DNA demethylation at a genome-scale. Affinity-enrichment-based studies of 5fC/5caC distributions in mouse ESCs have shown that a large cohort of 5fC and 5caC peaks overlap with genomic regions enriched for H3K4me1 18, 19, a histone mark broadly associated with active/poised enhancers, flanking regions of active/poised gene promoters, and intragenic regions of actively transcribed genes 28. Because H3K4me1-marked regions enrich for 5fC/5caC signals, we focused our initial genome-scale MAB-seq analysis on H3K4me1-enriched genomic regions captured through chromatin immunoprecipitation (termed H3K4me1-MAB-seq) (Supplementary Fig. 7a). To establish the baseline of false positive 5fC/5caC signals, we performed H3K4me1-MAB-seq analysis of mouse ESCs deficient for all three Dnmt enzymes (Dnmt1/3a/3b−/−) or Tet1/2 proteins (Tet1/2−/−). We found that median false positive signals in these mutant ESCs, in which both 5fC and 5caC are virtually absent, are very close to the error rate of M. SssI observed in methylating lambda DNA (dashed line in Fig. 2b). In comparison to control knockdown (shCtrl), Dnmt1/3a/3b−/−, Tet1/2−/− ESCs, 5fC/5caC is present at significantly higher levels in Tdg knockdown (shTdg) mouse ESCs (P<2.2×10−16, Wilcoxon rank sum test), and is further increased in VC-treated cells (shTdg+VC in Fig. 2b).
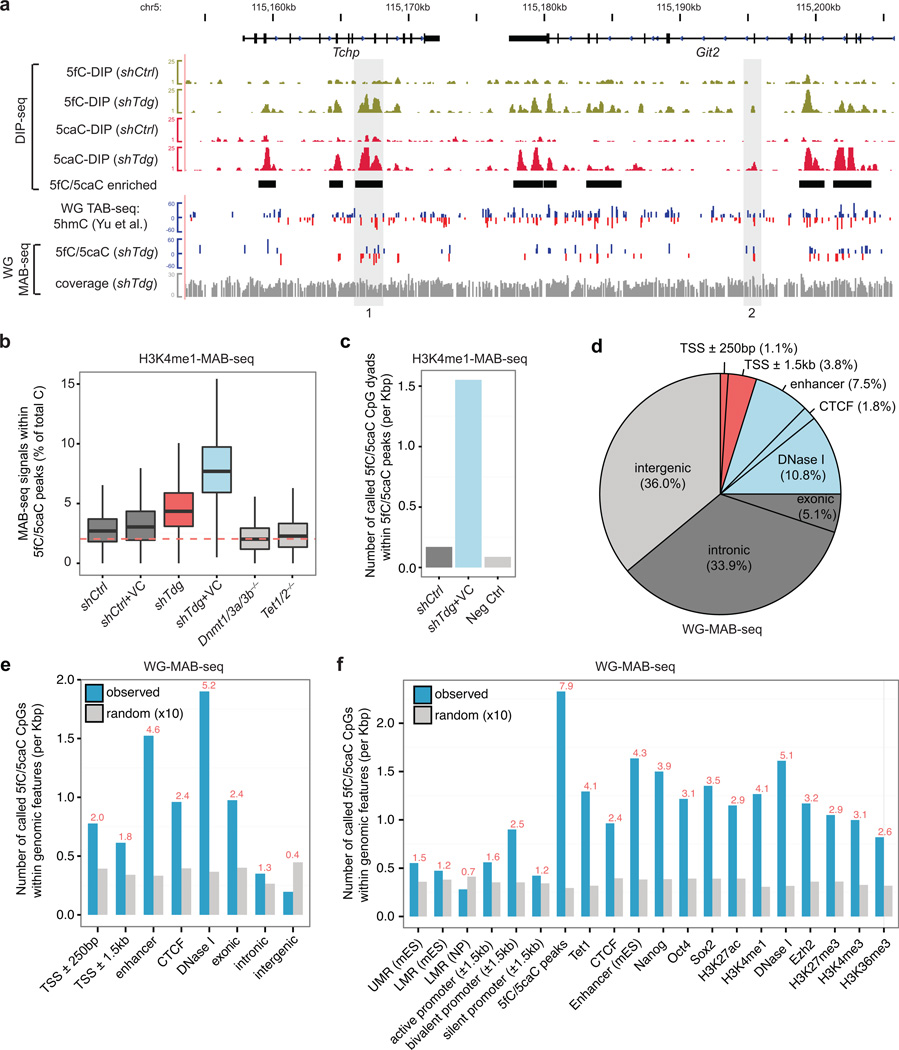
Figure 2. Genome-scale MAB-seq analysis of the mouse genome.
(a) Snapshot of base-resolution 5fC/5caC maps (whole-genome [WG] in shTdg+VC) and affinity-based 5fC/5caC maps at the Tchp-Git2 locus in wild-type or Tdg-depleted mouse ESCs compared to the base-resolution 5hmC map (TAB-seq)23 in wild-type ESCs. For comparison, 5fC/5caC-enriched regions (shTdg-specific) identified by DIP-seq methods are highlighted by black horizontal bars. For base-resolution maps, positive values (blue) indicate cytosines on the Watson strand, whereas negative values (red) indicate cytosines on the Crick strand. For base-resolution maps of 5hmC and 5fC/5caC, the vertical axis limits are −60% to +60%. Only cytosines sequenced to depth ≥5 are shown. Cytosines associated with statistically significant level of 5hmC (FDR=5%) and 5fC/5caC (FDR=5%) are shown in separate tracks. Sequencing coverage for WG-MAB-seq experiments is shown in gray.
(b) Genome-scale H3K4me1-MAB-seq analysis confirms that TDG inactivation and/or VC treatment resulted in higher 5fC/5caC levels. Shown are boxplots of raw MAB-seq signals [percentage of T/(C+T)] mapped to 5fC/5caC-enriched regions (n= 51,235; covered cytosines≥20; identified by 5fC/5caC antibody DIP-seq). Dnmt1/3a/3b−/− and Tet1/2−/− mouse ESCs serve as negative controls. The dash line denotes the error rate for M.SssI methylase (2.04%). shCtrl, control knockdown mouse ESCs; shCtrl+VC, control knockdown and VC treated mouse ESCs; shTdg, TDG knockdown mouse ESCs; shTdg+VC, TDG knockdown and VC treated mouse ESCs.
(c) Statistical analysis of H3K4me1-MAB-seq datasets (depth ≥5, FDR<10%) identifies specific CpG dyads enriched for 5fC/5caC. Shown are the number of called cytosines within 5fC/5caC-enriched regions for control, shTdg+VC, and Neg Ctrl. shCtrl, denotes merged MAB-seq datasets from shCtrl and shCtrl+VC samples. Neg Ctrl denotes merged MAB-seq datasets from Dnmt1/3a/3b−/− and Tet1/2−/− samples.
(d) Pie chart showing overlap of 5fC/5caC-modified CpGs (FDR=5% in WG-MAB-seq analysis of shTdg+VC sample) with genomic elements. Genic features were extracted from the UCSC the RefGene database (mm9). Promoter-distal regulatory regions (e.g. enhancers, CTCF binding sites, and DNase I hypersensitive regions) were experimentally mapped by ChIP-seq and DNase-seq experiments of the ENCODE project30. Each 5fC/5caC is counted only once: the overlap of a genomic region excludes all previously overlapped cytosines clockwise from proximal promoters (TSS±250 bp). Red, promoter-proximal regions; blue, promoter-distal regulatory elements; dark gray, genic regions; light gray, intergenic regions.
(e) The relative enrichment of 5fC/5caC-modified CpGs at genomic elements (blue) and corresponding randomly shuffled control regions (gray), normalized to the total size of the element type (per kilo base-pairs [Kbp]). Random consists of 10 random sampling of specific genomic elements in the mouse genome. The ratio between observed and random for each genomic element is shown on the top (red).
(f) The relative enrichment of 5fC/5caC-modified CpGs at specific gene regulatory regions (blue) and corresponding randomly shuffled control regions (gray). 5fC/5caC peaks, 5fC/5caC enriched regions identified by DIP-seq. UMR, unmethylated region, typically marking transcriptionally active CpG-rich promoters; LMR, low methylation region. mES, mouse ESCs; NP, neural progenitors.
Given that 5fC/5caC is absent in Dnmt/Tet mutant (denoted as ‘Neg Ctrl’ thereafter) mouse ESCs, MAB-seq signals in these mutant cells may provide an empirical estimate of false discovery rate (FDR). Because the probability that a CpG can be confidently identified as 5fC/5caC-modified is governed by the sequencing depth and abundance of the modification at the cytosine, we modeled the largely stochastic event of M.SssI failure in CpG methylation with a binomial distribution [N as the depth of sequencing at the cytosine and p (2.04%) as the error rate of M. SssI]. Application of this statistical strategy to H3K4me1-MAB-seq datasets identified a total of 127,576 (7.6% out of 1,670,036 CpG dyads with N≥10) 5fC/5caC-modified CpG dyads in VC-treated, Tdg-depleted mouse ESCs (shTdg+VC) with an empirical FDR of 5%. Using a less stringent cutoff (N≥5, FDR<10%), we have identified 267,325 (8.0% out of 3,326,034 CpG dyads) 5fC/5caC-modified CpGs. There are 17.6 times as many 5fC/5caC-modified CpGs overlapping with affinity-identified regions as expected by chance (Fig. 2c), demonstrating the effectiveness of the empirical FDR-based statistical filter.
We next analyzed genomic DNA from mouse ESCs by whole genome (WG)-MAB-seq. Since VC is present at a relatively high level during early development in vivo29, base-resolution 5fC/5caC map in VC-treated cells may represent a more physiologically relevant profile of TET/TDG-dependent active demethylation activity. We therefore focused our WG-MAB-seq analysis on shTdg+VC mouse ESCs and sequenced the sample to an average depth of 28.4x per CpG dyad (covering 95.2% of all CpG dyads). Locus-specific analysis of non-enriched and H3K4me1-enriched DNA suggests that 5fC/5caC levels at specific CpGs within selected loci are largely comparable (Supplementary Fig. 7b, c). Further comparative analysis of both H3K4me1-MAB-seq and WG-MAB-seq experiments indicates that MAB-seq signals (T/C+T) are highly similar in both experiments at a wide range of promoter-proximal and distal genomic elements (Supplementary Fig. 8a), supporting the validity of using the statistical filter established for H3K4me1-MAB-seq to analyze WG-MAB-seq dataset. Using the empirical P value cutoff established for H3K4me1-MAB-seq datasets with comparable sequencing depth, we identified a total of 675,325 5fC/5caC-modified CpGs (out of 24,872,637 CpGs [N≥10]) in shTdg+VC mouse ESCs through whole genome MAB-seq analysis (with an empirical FDR of 5%). Identified 5fC/5caC-modified CpGs in Tdg-depleted cells correlate well with peaks of 5fC and 5caC enrichment identified by the antibody-based DNA immunoprecipitation (DIP) approach (region 1 in Fig. 2a). Compared to random controls, 5fC/5caC-CpGs significantly overlap with 5fC/5caC-enriched peaks (7.9 times as many as expected by chance). Furthermore, 83.6% of affinity-enrichment identified regions (n=50,923 out of 60,912 covered by WG-MAB-seq) overlap with at least one 5fC/5caC-modified CpGs. In contrast, 80.7% of 5fC/5caC-modified CpGs are not recovered by 5fC/5caC DIP-seq (region 2 in Fig. 2a), suggesting that MAB-seq has a markedly increased sensitivity.
Genomic distribution of active DNA demethylation activity
Previous studies using affinity-enrichment based methods have shown that 5fC/5caC are enriched at poised/active enhancers, Polycomb group protein (PcG) repressed promoters, and gene bodies 18, 19. However, the true abundance of 5fC/5caC cannot be determined from affinity-based approaches, thus precluding quantitative analysis of 5fC/5caC-excision-dependent DNA demethylation events at these gene regulatory elements. In shTdg+VC mouse ESCs, we found that a significant fraction of 5fC/5caC (20.1%) reside in distal regulatory regions (Fig. 2d). Analysis of both raw MAB-seq signals and relative enrichment of called 5fC/5caC at each class of genomic features indicates that 5fC/5caC are more enriched at DNase I hypersensitive sites (observed/random [obs/rand]=5.2), predicted enhancers (obs/rand=4.6), and CTCF-biding sites (obs/rand=2.4) relative to promoter-proximal regions and other genic regions (Fig. 2e and Supplementary Fig. 8c–d). In support of this observation, regions enriched for pluripotency-related transcription factors (Oct4, Nanog, and Sox2) as well as active enhancers (marked by H3K27ac and/or H3K4me1) are also more enriched with 5fC/5caC than with other genomic elements (Fig. 2f and Supplementary Fig. 8b). Consistent with previous reports 18, 19, we found that 5fC/5caC is more enriched at mouse ESC-specific distal enhancers than at other tissue-specific enhancers (Supplementary Fig. 8e–f), supporting the notion that CpGs within or surrounding active cell-type-specific enhancers tend to undergo 5fC/5caC-excision-dependent active DNA demethylation.
Distinct processivity of TET enzymes at different cytosines
The ability of identifying individual CpGs undergoing 5fC/5caC-excision dependent DNA demethylation offers an opportunity for investigating the processivity of TET-mediated 5mC oxidation. We thus focused on CpGs sufficiently covered by both the whole-genome 5fC/5caC map (in shTdg+VC) and 5hmC map generated by TAB-seq 23 (Fig. 2a). Comparative analysis of TAB-seq and WG-MAB-seq signals (depth≥10) allows us to estimate the number of CpGs associated with 5fC/5caC-alone (5hmC−, 5fC/5caC+; n=508,261), 5hmC-alone (5hmC+, 5fC/5caC−; n=1,454,388), and both (5hmC+, 5fC/5caC+; n=117,327) (Fig. 3a). The median raw MAB-seq signal at 5fC/5caC-alone CpGs is 23.1%, significantly higher than 5.6% detected for 5hmC-alone CpGs (Supplementary Fig. 9a). Notably, 5hmC and 5fC/5caC largely exist at different cytosines (Fig. 3a). Only 7.5% of 5hmC-modified CpGs (depth≥10) also have significant level of 5fC/5caC (corresponding to 18.8% of all 5fC/5caC-modified CpGs, FDR=5%), whereas the majority of 5hmC-modified CpGs appear to represent 5hmC stably accumulated in wild-type cells without being efficiently further oxidized by TET proteins to 5fC/5caC (pair #1 in Supplementary Fig. 9b). Additional analysis using CpGs with higher sequencing depth (depth≥20) reached similar conclusion (pair #2 in Supplementary Fig. 9b). In contrast, affinity-based 5hmC/5fC/5caC mapping methods suggest a much higher overlapping percentage (from 41.3% to 77.7%) between 5hmC-enriched regions and 5fC/5caC peaks (pair #3–5 in Supplementary Fig. 9b), probably due to their lower resolution.
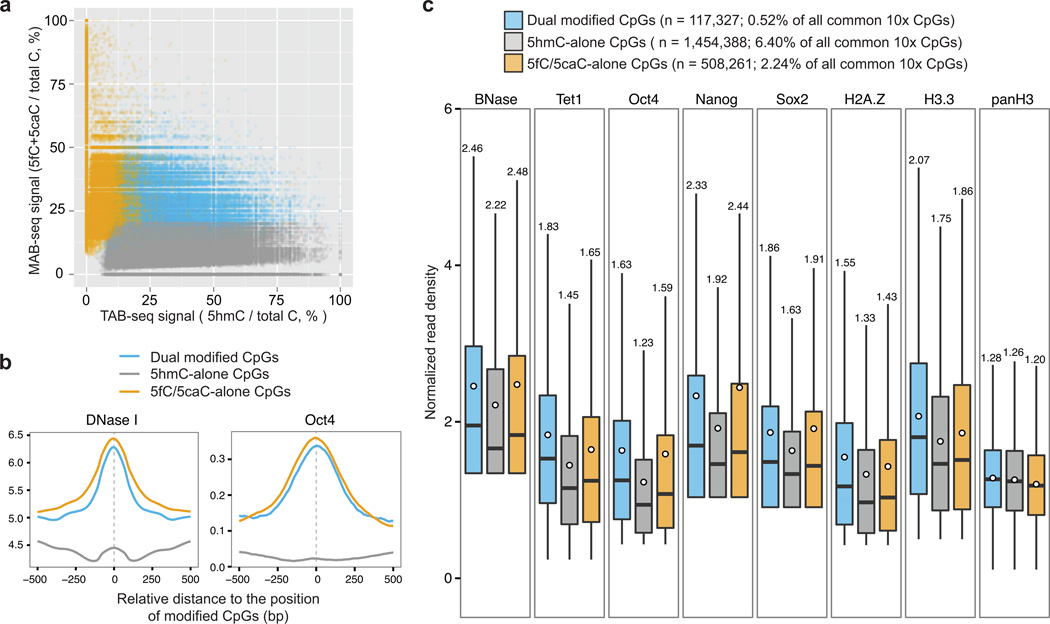
Figure 3. Identification of CpGs associated with distinct processivity of TET-mediated oxidation.
(a) Comparative analysis of base-resolution 5fC/5caC (WG-MAB-seq in shTdg+VC) and 5hmC maps (TAB-seq in wild-type)23 identifies CpGs associated only with 5fC/5caC (5fC/5caC-alone; n=508261, FDR=5%), 5hmC (5hmC-alone; n=1454388, FDR=5%), and both (dual modified with 5hmC and 5fC/5caC; n=117327, FDR=5%). 5fC/5caC-alone CpGs (yellow), which are only associated with generation and excision repair of 5fC/5caC, represent cytosines undergoing active DNA demethylation; 5hmC-alone CpGs (gray), which are only associated with relatively stable 5hmC in wild-type cells, represent cytosines where TET proteins tend to stall at 5hmC; dual modified CpGs (blue), which are associated with active demethylation but 5hmC also accumulates to detectable level in wild-type cells. The levels of MAB-seq and TAB-seq signals are depicted in the scatter plot for all CpGs (n= 22,730,906) that are covered by both MAB-seq (depth≥10) and TAB-seq (depth≥10).
(b) Averaged read density of DNase I hypersensitivity signals (left panel; averaged from two DNase-seq replicates) and Oct4 ChIP-seq (right panel; whole cell extract [WCE] control ChIP-seq signal subtracted) around 5hmC-alone CpGs, 5fC/5caC-alone and dual-modified CpGs (±500 bp).
(c) The levels of chromatin accessibility signals (BNase), TET1 occupancy, pluripotency transcriptional factors (Nanog, Oct4 and Sox2), and histone variants (H2A.Z and H3.3) around the genomic position (±50 bp) of indicated groups of modified CpGs. The black bars and white circles in boxplots denote median and mean of normalized read density (reads per 10 million reads [rp10m]). The mean of each boxplot is shown on the top.
Given that 5fC/5caC are preferentially enriched at active/poised gene regulatory regions where chromatin is generally more accessible, we reasoned that local chromatin structure may influence the occupancy level and/or processivity (the ability to further oxidize 5hmC to 5fC/5caC) of TET enzymes. We first analyzed DNase I hypersensitivity (measured by DNase-seq 30) at genomic regions (±50 bp) immediately flanking 5fC/5caC-alone, 5hmC-alone and dual modified CpGs. This analysis reveals that 5fC/5caC-alone and dual modified CpGs are associated with significantly higher level of DNase I hypersensitivity signals than 5hmC-alone CpGs (Fig. 3b and Supplementary Fig. 9c–d). Further analysis indicates that as compared to 5hmC-alone CpGs, 5fC/5caC-alone and dual modified CpGs are associated with higher levels of Benzonase (BNase) sensitivity signals 31, histone variants (H2A.Z and H3.3) known to destabilize nucleosome structure 31, 32, TET1 occupancy 33 and pluripotency-related transcription factors (TFs; Oct4, Nanog, and Sox2) 34 (Fig. 3c and Supplementary Fig. 9d). In contrast, comparable levels of general histone H3 were detected at distinct groups of CpGs (Fig. 3c). These results suggest that TET proteins by default tend to stall at the 5hmC step at most CpGs, but exhibit higher processivity at CpGs associated with a more accessible chromatin state.
Strand asymmetry of 5fC/5caC in palindromic CpGs
While cytosine methylation in palindromic CpG dyads is generally symmetric and exhibits very high heritability upon DNA replication 2, previous studies suggest that >80% of steady-state 5hmC in human and mouse ESCs are asymmetrically modified 23. This prompted us to examine whether 5fC/5caC-modified CpGs also show strand biases (Fig. 4a). Focusing on CpG dyads with both strands sufficiently covered by WG-MAB-seq (n=9,261,306 CpG dyads and depth≥10 on both Watson and Crick strands), we found that only 4.97% of called CpG dyads (22,590 out of 454,400 called CpG dyads, FDR=5%) are symmetrically modified with 5fC/5caC. Further analysis focusing on CpG dyads with both strands covered by higher sequencing depth (≥20) reached a similar conclusion (7.92% symmetrically modified with 5fC/5caC). However, because the abundance of 5fC/5caC is low at any given CpG dyad, it is still possible that sequencing depth in WG-MAB-seq might not be sufficient to identify all 5fC/5caC, leading to an underestimation of symmetrically-modified CpGs. To address this issue, we pooled MAB-seq signals of all called 5fC/5caC-modified CpGs and compared the called strand to the opposite strand. The average abundance of 5fC/5caC at the called CpGs is 26.9%, while that of the opposite CpGs is only 8.5% (Fig. 4a). To further confirm the observed strand asymmetry of 5fC/5caC-modified CpG dyads, we analyzed the difference in 5fC/5caC levels of called and opposite strands for each called CpG dyads (FDR=5%, depth≥10). For asymmetrically-modified CpGs, the mean difference in 5fC/5caC levels between called and opposite cytosines is on average 18.6% at each CpG dyad (or 14.4% for CpG dyads with depth≥20) (Fig. 4b). Visualizing the absolute levels of 5fC/5caC on both called and opposite strands in a two-dimensional histogram plot showed a clear shift toward the called cytosines (Fig. 4c). Notably, similar analysis of H3K4me1-MAB-seq datasets also supports the observed strand asymmetry of 5fC/5caC-modified CpG dyads (Supplementary Fig. 6d–f). Further analysis showed no strand bias for M.SssI in methylating lambda phage genome (97.97% for Watson strand versus 97.90% for Crick strand) or genomic DNA of Dnmt1/3a/3b−/− cells (97.82% for Watson strand and 97.80% for Crick strand). Consistent with genome-scale MAB-seq analysis, strand-specific analysis of Tbx5 and Vps26a loci also revealed that the majority of 5fC/5caC-modified CpG dyads exhibit strand asymmetry [asymmetric (blue)+partially asymmetric (yellow): 86.7% in shTdg and 60% in shTdg+VC] (Fig. 4d–e). Locus-specific MAB-seq analysis of biological replicates suggests that the strand-specific distribution of 5fC/5caC detected at each CpG sites is not entirely stochastic (Supplementary Fig. 5b), suggesting that strand-specific 5fC/5caC generation and excision at individual CpGs is a regulated process.
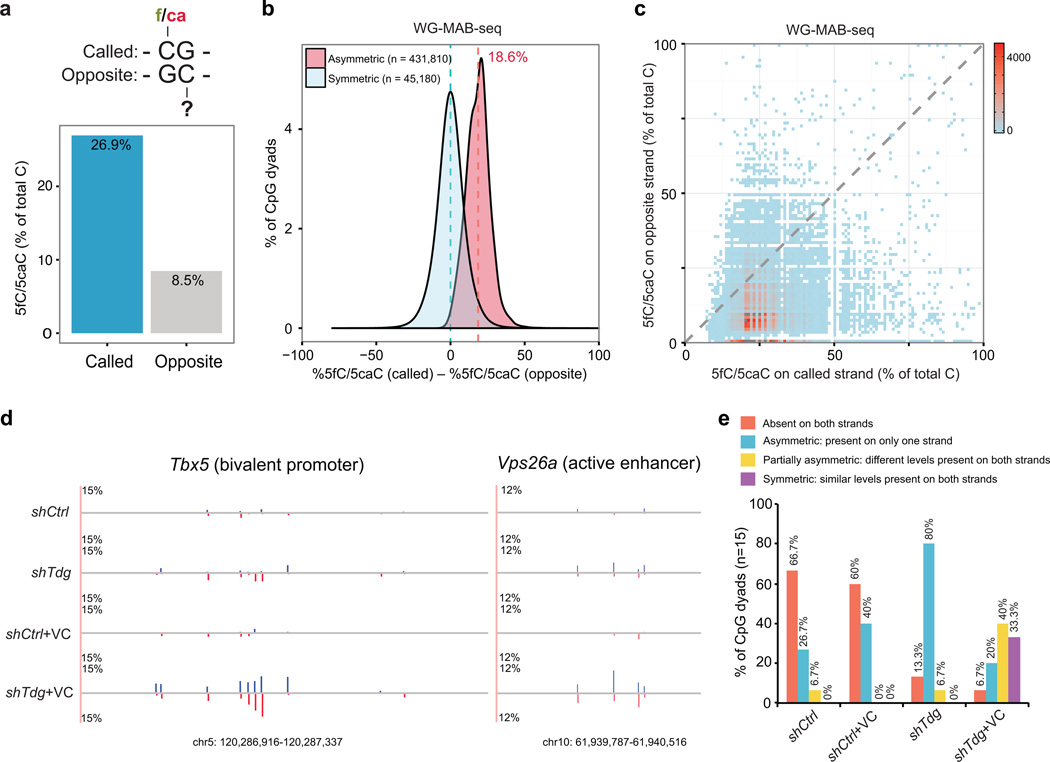
Figure 4. Strand asymmetry of TET/TDG-dependent active DNA demethylation.
(a) The average 5fC/5caC level on the called cytosines (n= 454,400, identified in WG-MAB-seq; FDR=5%; sequencing depth on both strands≥10) is significantly higher than that of cytosines on the opposite strand. Called, cytosines enriched for 5fC/5caC in shTdg+VC; opposite, cytosines on the opposite strand.
(b) Distribution of differences of WG-MAB-seq signals (in shTdg+Vc) between called and opposite cytosines. Symmetric, both strands are called for enriching 5fC/5caC in shTdg+VC; Asymmetric, only one strand is called.
(c) Two-dimensional density-plot of WG-MAB-seq signals at each called/opposite CpG dyad reveals the strand asymmetry of TET/TDG-dependent active DNA demethylation.
(d) Locus-specific MAB-seq analysis of two representative loci illustrating the asymmetric distribution of 5fC/5caC. 5fC/5caC levels for the Watson strand are in blue, whereas those for the Crick strand are in red.
(e) Bar-graph of relative percentage of CpG dyads (n=15, analyzed by locus-specific MAB-seq) that are associated with different degrees of strand asymmetry of active DNA demethylation activity in shCtrl, shCtrl+VC, shTdg, shTdg+VC mouse ESCs.
Base-resolution mapping of 5fC and 5caC separately
Base-resolution mapping of 5fC has recently been demonstrated using subtraction-based, chemical modification-assisted BS-seq methods such as fCAB-seq 19 and redBS-seq 21 (Supplementary Fig. 1a). However, relatively low protection rate of 5caC deamination (50–60%) reported in a similar subtraction-based 5caC mapping approach (caCAB-seq) suggests that further optimization is required 20. To achieve 5caC mapping at base-resolution, we combined MAB-seq with the 5fC mapping method, redBS-seq 21. After validating redBS-seq using synthetic oligonucleotides (Supplementary Fig. 10a), we performed locus-specific redBS-seq and BS-seq to quantify 5fC at 73 CpG sites from four loci enriched for 5fC and/or 5caC (Tbx5, Vps26a, Slc2a12 and Ace). The abundance and position of 5caC at these sites can then be determined by subtracting 5fC signals (derived from the difference between redBS-seq and BS-seq) from the levels of 5fC+5caC measured by MAB-seq (Fig. 5b and Supplementary Fig. 10c–d). Using this integrative approach to map 5fC and 5caC separately, we observed that 5fC and 5caC display largely distinct distribution patterns at individual CpGs within these loci. For instance, within exon2 of Tbx5, a region enriched for both 5fC and 5caC, some CpG sites are only modified with 5fC (CpG #1 at Tbx5 in Fig. 5b), while some others are 5caC-only (CpG #2 at Tbx5 in Fig. 5b). More strikingly, as exemplified by CpG #3 at Tbx5 in Fig. 5b, CpG sites associated with both 5fC and 5caC may exhibit non-overlapping, strand-specific 5fC/5caC distribution. This conclusion is further supported by analysis of an active enhancer within the Vps26a gene (Supplementary Fig. 10d). Such diverse distribution patterns of 5fC and 5caC indicate distinct processivity of TETs and/or substrate preference of TDG (5fC versus 5caC) at individual CpGs. Notably, most 5fC-modified CpGs (20 out of 24) identified by redBS-seq are also detected by MAB-seq as 5fC/5caC-modified (Supplementary Fig. 10c). The few inconsistent sites between MAB-seq and redBS-seq probably arise from the different principles that the two methods are based upon.
Figure 5. Base-resolution mapping analysis reveals distinct distribution patterns of 5fC and 5caC.
(a) Schematic diagram of combining BS-seq, redBS-seq, MAB-seq and caMAB-seq to map 5fC and 5caC individually at single-base resolution.
(b) Locus-specific analysis of 5fC and 5caC at the Tbx5 locus indicates that 5fC and 5caC are largely non-overlapping and both modifications exhibit strong strand asymmetry. For comparison, affinity-based 5fC and 5caC maps are shown on the top. In enlarged views, base-resolution maps of 5fC+5caC (measured by MAB-seq), 5fC (measured by redBS-seq) and 5caC (subtraction between MAB-seq and redBS-seq) are shown. Signals for the Watson strand are in blue, whereas those for the Crick strand are in red.
(c) Comparative locus-specific analysis of 5caC at the Vps26a locus by two base-resolution methods: indirect mapping through subtraction between MAB-seq and redBS-seq (red), direct mapping by caMAB-seq (blue). For comparison, also shown are DIP-seq based maps of 5caC in control and Tdg-depleted mouse ESCs. The level of 5caC (only Watson strand shown) is displayed as the percentage of total C modified as 5caC. Corresponding caMAB-seq sequencing depth (vertical axis limits are 0 to 500) at each CpG was also shown.
Base-resolution mapping 5caC through integrating MAB-seq and redBS-seq requires two rounds of subtractions, representing a technical challenge for genome-scale analysis. Thus, we explored a subtraction-independent 5caC mapping strategy by taking advantage of the fact that 5fC can be selectively reduced by sodium borohydride (NaBH4) to 5hmC 35. In this modified version of MAB-seq (termed caMAB-seq), M.SssI-treated DNA was further incubated with NaBH4 so that only 5caC is read as T in bisulfite sequencing (Fig. 5a). Analyses of defined sequences (lambda DNA or synthetic oligonucleotides) or 5caC-enriched genomic loci (e.g. Vps26a) demonstrated the validity of caMAB-seq in direct base-resolution mapping of 5caC (Fig. 5c and Supplementary Fig. 10e).
Integrative analysis of unmodified C, 5mC/5hmC and 5fC/5caC
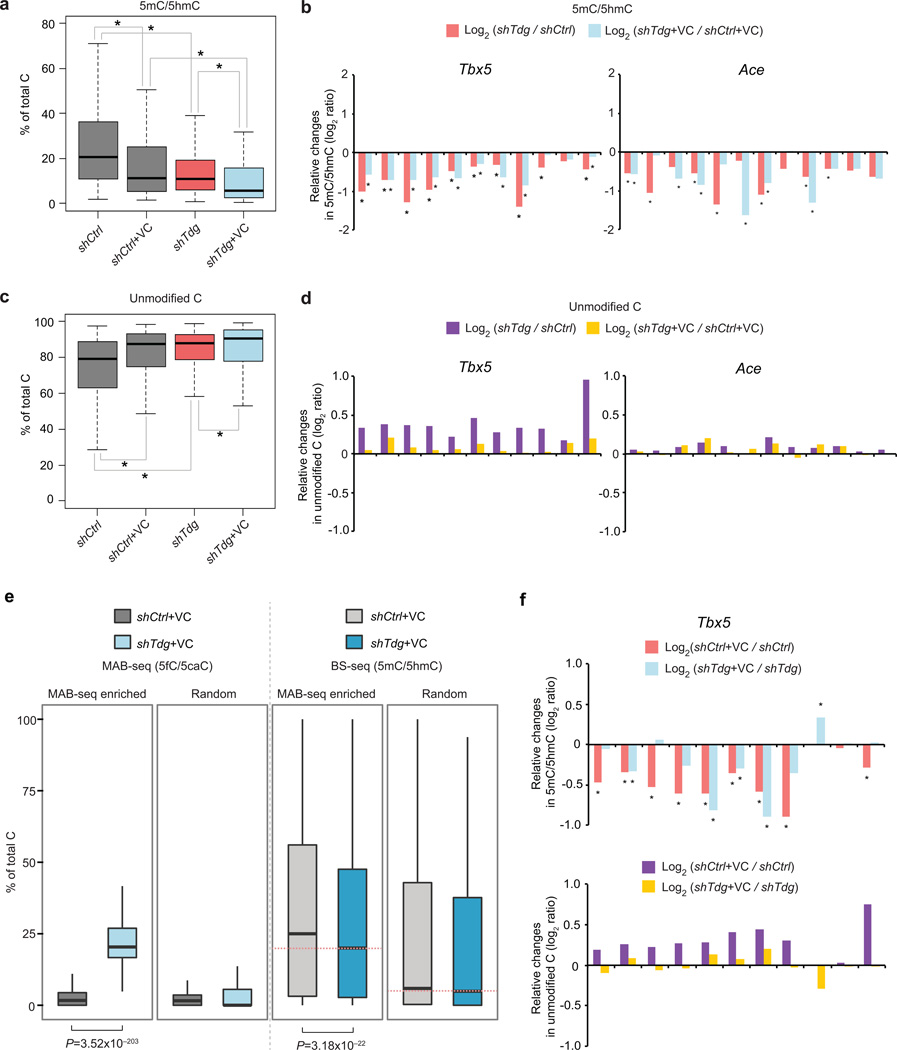
Standard BS-seq cannot distinguish unmodified C from 5fC/5caC. When combined with BS-seq, MAB-seq is able to quantify the true abundance of unmodified CpG at base-resolution (Fig. 1a), providing a unique opportunity for exploring the potential role of 5fC/5caC excision in regulating upstream (5mC+5hmC) and downstream (unmodified C) steps in the TET/TDG-dependent active DNA demethylation pathway. By performing locus-specific BS-seq and MAB-seq through Illumina deep sequencing, we quantified the levels of unmodified C, 5mC/5hmC and 5fC/5caC at 73 CpGs within four 5fC/5caC-enriched regions. BS-seq analysis showed that Tdg-depletion induces significant decrease in 5mC/5hmC within these loci (shTdg versus shCtrl or shTdg+VC versus shCtrl+VC in Fig. 6a). Further analysis shows that significant decrease in 5mC/5hmC was detected at multiple CpGs within each locus (Fig. 6a–b and Supplementary Fig. 11a). In contrast to 5mC/5hmC, integrative analysis of BS-seq and MAB-seq results revealed that unmodified C is significantly increased in response to TDG depletion (Fig. 6c–d and Supplementary Fig. 11b).
Figure 6. Base-resolution mapping of unmodified C and 5mC/5hmC reveals that TDG-mediated 5fC/5caC excision affects active demethylation dynamics.
(a) Tdg knockdown and/or VC treatment results in a significant decrease in BS-seq signals (5mC+5hmC). Shown are boxplots summarizing the levels of 5hmC+5mC at 73 CpG sites from Ace, Slc2a12, Tbx5 and Vps26a loci. An asterisk indicates a statistically significant difference between the two indicated groups, as determined by Wilcoxon signed-rank test for matched pairs (multiple comparison corrected P-value<0.05).
(b) Shown are relative changes in 5mC+5hmC levels (log2 ratio) in response to Tdg knockdown at individual CpG sites from Ace and Tbx5 loci. shTdg was compared with shCtrl while shTdg+VC was compared with shCtrl+VC. An asterisk indicates a statistically significant change determined by Fisher’s exact test (multiple comparison corrected P-value<0.05).
(c) Tdg knockdown or VC treatment leads to a significant increase in the level of unmodified C (measured by subtracting C signals in BS-seq from those of MAB-seq). Shown are boxplots summarizing the levels of unmodified C at 73 CpG sites from Ace, Slc2a12, Tbx5 and Vps26a loci.
(d) Shown are relative changes in unmodified C levels (log2 ratio) in response to Tdg knockdown at individual CpG sites from Ace and Tbx5 loci. shTdg was compared with shCtrl while shTdg+VC was compared with shCtrl+VC.
(e) Genome-scale H3K27me3-MAB-seq and H3K27me3-BS-seq analyses in control or Tdg-depleted mouse ESCs show that 5m/5hmC levels are significantly decreased within genomic regions enriched for 5fC/5caC. Boxplots depicting MAB-seq and BS-seq signals (binned to 500bp intervals) are shown for 5fC/5caC-enriched regions. Equal number of randomly sampled regions that are covered by both BS-seq and MAB-seq are analyzed.
(f) Relative changes (log2 ratio) in 5mC+5hmC levels (top panels) and unmodified C levels (bottom panels) in response to VC treatment. shCtrl+VC was compared with shCtrl while shTdg+VC was compared with shTdg. For relative changes in 5mC+5hmC levels, an asterisk indicates a statistically significant change determined by Fisher’s exact test (multiple comparison corrected P-value<0.05).
Tdg-depletion induced changes in 5mC/5hmC and unmodified C appear to be more pronounced at Tbx5, Ace and Slc2a12 loci when compared to Vps26a locus (an active enhancer), suggesting that active DNA demethylation dynamics at these transcriptionally repressed/poised regions (previously determined by ChIP-seq) is modulated by the generation and/or excision of 5fC/5caC. To test this possibility at a genome-scale, we captured genomic regions marked by H3K27me3, a repressive histone modification associated with transcriptionally poised gene promoters of lineage-specific TFs (e.g. Tbx5), and subjected H3K27me3-enriched DNA to genome-scale MAB-seq and BS-seq analyses. H3K27me3-MAB-seq identified 1,231 genomic intervals enriched for 5fC/5caC signals in shTdg+VC cells, and H3K27me3-BS-seq showed that there was a significant decrease in 5mC/5hmC levels within these 5fC/5caC-enriched regions (P= 3.18×10−22, Wilcoxon) (“MAB-seq enriched” in Fig. 6e). For 1,231 randomly sampled regions where 5fC/5caC signals are largely absent, the change in 5mC/5hmC is much less pronounced (“random” in Fig. 6e). Consistent with results of locus-specific analysis, genome-scale analysis supports the notion that inhibition of TDG-mediated 5fC/5caC or 5fC/5caC accumulation itself leads to dysregulation of upstream steps of the cytosine-modifying cascade.
Notably, VC treatment alone can substantially reduce the level of 5mC/5hmC (shCtrl+VC versus shCtrl or shTdg+VC versus shTdg in Fig. 6a and upper panels in Fig. 6f), possibly due to its positive effect on stimulating TET catalytic activity (oxidizing 5mC/5hmC). In addition, VC treatment can induce a significant increase in the level of unmodified C (shCtrl+VC versus shCtrl or shTdg+VC versus shTdg in Fig. 6c). However, VC-induced increase in unmodified C is more pronounced in shCtrl (purple in lower panel of Fig. 6f) than in shTdg (yellow in lower panel of Fig. 6f), indicating that accumulation of unmodified C by VC treatment requires TDG/BER-mediated 5fC/5caC excision step. Together, these results suggest that interfering 5fC/5caC excision step by depleting TDG proteins may result in dysregulation of both upstream (5mC/5hmC generation or oxidization) and downstream (generation and/or methylation of unmodified C) steps of the DNMT/TET/TDG-dependent cytosine-modifying cascade.
DISCUSSION
In this study, we have established an approach, MAB-seq, for quantitative measurement of 5fC/5caC excision repair-dependent active DNA demethylation activity at base-resolution, providing a tool that has broad application in the study of active DNA demethylation. A major advantage of MAB-seq over other subtraction-based 5fC or 5caC mapping methods is the ability to directly determine the location and abundance of 5fC and 5caC in a single experiment. MAB-seq can therefore achieve similar sensitivity for detecting 5fC/5caC with lower sequencing depth and simplifies the computational procedure to confidently identify CpGs marked 5fC/5caC. When combined with Tdg depletion, simultaneous mapping 5fC and 5caC enables quantitative measurement of the generation and excision of 5fC/5caC, providing a direct readout of the TET/TDG-mediated active DNA demethylation activity.
Application of whole genome MAB-seq analysis to mouse ESCs allowed us to investigate the genomic architecture and dynamics of active DNA demethylation activity at single-base resolution across the genome of this stem cell population. While affinity-based 5fC and 5caC mapping methods suggest that 5hmC and 5fC/5caC enriched regions are largely overlapped 18, 19, comparative analysis of base-resolution 5fC/5caC map and 5hmC map reveals that a large fraction of 5fC/5caC and 5hmC are associated with distinct CpGs within 5hmC/5fC/5caC-enriched regions. These results support a model in which TET proteins tend to stall at 5hmC step at most CpGs, but exhibit higher processivity to further oxidize 5hmC to 5fC/5caC at CpGs with higher chromatin accessibility. Moreover, integrating MAB-seq with 5fC-mapping (e.g. redBS-seq) or 5caC-mapping (e.g. caMAB-seq) method not only provides a base-resolution approach to map 5fC and 5caC separately, but also reveals that 5fC and 5caC are frequently non-overlapped at individual CpGs. These findings suggest the possibility that TET exhibits distinct processivity depending on local chromatin accessibility or other yet to be identified regulatory processes. It is also possible that TDG may exhibit distinct activity or substrate preference at different CpGs. In addition, simultaneous mapping of 5fC/5caC by MAB-seq reveals that TET/TDG-dependent active DNA demethylation activity preferentially targets palindromic CpG dyads asymmetrically, supporting the recently proposed asymmetric base-flipping model 24, 36. Lastly, we demonstrate that integrative analysis of MAB-seq and BS-seq datasets not only allows quantitative measurement of cytosine derivatives at all major steps of TET/TDG-mediated active demethylation pathway [iterative oxidation (5mC/5hmC), excision repair (5fC/5caC) and regeneration (unmodified C)], but also reveals that 5fC/5caC excision by TDG may act as a regulatory checkpoint of the active DNA demethylation cascade.
In addition to a robust base-resolution mapping method for 5fC/5caC, we provide here a whole-genome base-resolution map of TET/TDG-dependent active DNA demethylation activity in the mammalian genome. Given the simplicity and cost-effectiveness of MAB-seq, we anticipate that genome-scale MAB-seq analysis can be applied to analyze diverse cell-types in future studies to provide new insights into the mechanism and function of the DNMT-TET-TDG/BER cytosine-modifying cascade. Thus, the 5fC/5caC mapping technology described in this study and other base-resolution mapping methods set the stage for systematic investigation of the functional significance of active DNA demethylation in mammalian development and human diseases.
Methods
Mouse ESC cultures, lentiviral knockdown of TDG and vitamin C treatment
V6.5 (control and Tdg knockdown), E14Tg2A (control, Tdg knockdown and Tet1/2−/−), and J1 (Dnmt1/3a/3b−/−) mouse ESC lines were cultured in feeder-free gelatin-coated plates in Dulbecco’s Modified Eagle Medium (DMEM) (GIBCO, 11995) supplemented with 15% FBS (GIBCO), 2 mM L-glutamine (GIBCO), 0.1 mM 2-mercaptoethanol (Sigma), nonessential amino acids (GIBCO), 1,000 units/ml LIF (Millipore, ESG1107). The culture was passaged every 2–3 days using 0.05% Trypsin (GIBCO). Lentivirus-mediated Tdg knockdown in mouse ESCs were performed as previously described 18. For vitamin C (Sigma, A8960) treatment, control and Tdg knockdown mouse ESCs were treated with 100 µg/ml of vitamin C for 60 hrs.
Chromatin immunoprecipitation for genome-scale MAB-seq/BS-seq
To capture H3K4me1- or H3K27me3-enriched chromatin for genome-scale MAB-seq analysis, 1–2×107 cells were cross-linked with 1% formaldehyde for 10 minutes at room temperature followed by exposure to 0.125 M glycine. After two washes with cold PBS, cells were collected and stored at −80 °C before use. Nuclei were extracted and lysed sequentially with lysis buffer 1 (LB1, 50 mM Hepe2-KOH, pH7.5, 140 mM NaCl, 1 mM EDTA, 10% glycerol, 0.5% NP40, 0.25% Triton X-100), lysis buffer 2 (LB2, 10 mM Tris-HCl, pH8.0, 200 mM NaCl, 1 mM EDTA, 0.5 mM EGTA), and lysis buffer 3 (LB3, 10 mM Tris-HCl, pH8.0, 100 mM NaCl, 1 mM EDTA, 0.5 mM EGTA, 0.1% Na-Deoxycholate, 0.5% N-lauroylsarcosine). Chromatin was sonicated using a microtip (Branson sonifier 450) until the DNA fragments were reduced to 200–1000 bp in length. 10 µg antibodies were immobilized with 100 µl Dynal protein-G beads (Invitrogen) for at least 6 hours. Immunoprecipitation was performed overnight at 4 °C with antibody-conjugated protein-G beads. DNA/protein complexes were washed with RIPA buffer (50 mM Hepes-KOH, pH7.6, 500 mM LiCl, 1 mM EDTA, 1% NP-40, 0.7% Na-Deoxycholate) for five times and reverse cross-linked at 65 °C overnight. The DNA was treated sequentially with RNase A and proteinase K, and purified by phenol/chloroform extraction and ethanol precipitation. Following antibodies were used in ChIP assays: anti-H3K4me1 (ab8895, Abcam) and anti-H3K27me3 (07–449, Millipore).
Library generation for genome-scale MAB-seq/BS-seq
1 ug unenriched genomic DNA (whole-genome) or 200–500 ng of ChIP DNA (H3K4me1- or H3K27me3-enriched) was first spiked-in with unmethylated lambda DNA (1:400), which was then repaired and ligated to methylated (5mC) custom adapters (forward 5’-ACACTCTTTCCCTACACGACGCTCTTCC GATC*T-3’; reverse 5’-/5Phos/GATCGGAAGAGCACACGTCTGAACTCCAGTC-3’; the asterisk denotes phosphorothioate bond) with the NEBNext Ultra DNA Library Prep kit from Illumina (NEB). Adaptor-ligated DNA was then purified with 1.2x AMPure XP beads (Beckman Coulter). For BS-seq, methylated-adpator-ligated DNA was directly subjected to bisulfite conversion using Qiagen EpiTect DNA Bisulfite Kit (Qiagen, 59104) per manufacturer’s instructions, except that the thermal cycle was repeated twice. For MAB-seq, methylated adpator-ligated DNA was treated by M.SssI (New England Biolabs, M0226M) in a 50-µL reaction for two rounds. In each round of treatment, DNA was first incubated with 1.0 unit/µL M.SssI methylase (New England Biolabs, M0226M) for 4 hours in 25-µL reaction [1.25 µL of 20 unit/µL M.SssI and 0.5 µL of 32 mM SAM (final concentration: 640 µM)], and additional 25-µL containing same concentration of M.SssI (1.0 unit/µL) and SAM (640 µM) was supplemented to treat DNA for another 8 hours (in 50-µL). Of note, the first round of M.SssI treatment was performed in Mg++-free reaction buffer [10 mM Tris-HCl (pH 8.0), 50 mM NaCl, 10 mM EDTA], while the second round was carried out with NEB buffer #2 [10 mM Tris-HCl (pH 7.9), 50 mM NaCl, 10 mM MgCl2, 1 mM DTT]. DNA was purified by sequential Phenol/Chloroform/Isoamyl Alcohol (PCI, 25:24:1) extraction and ethanol precipitation after each round of M.SssI treatment. M.SssI-treated, methylated adaptor-ligated DNA was then subjected to bisulfite conversion using the EpiTect DNA Bisulfite Kit (Qiagen) as described above. Bisulfite-treated DNA was pre-amplified for 5 cycles using KAPA HiFi Uracil+ HotStart ReadyMix (KAPA) with indexed and universal primers from NEBNext Multiplex Oligos for Illumina (Index Primers Set 1). The optimal PCR cycle numbers required to generate the final libraries were then determined by quantitative PCR. Final libraries were generated by scaled-up PCR reactions using the cycles determined above, and purified with 1.2x AMPure XP beads. Libraries were sequenced on an Illumina Hiseq 2500 (single-end, 60 or 76 bp).
Data processing of genome-scale MAB-seq/BS-seq
Raw sequencing reads were trimmed for low-quality bases and adaptor sequences using Trimmomatic 37, and the data quality was examined with FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/). The trimmed reads were mapped against the mouse genome (mm9 build) with Bismark 38. PCR duplicates were removed using the Picard program (http://picard.sourceforge.net/). Methylation call for each CpG was extracted using bismark methylation extractor script from Bismark. Cell line-specific SNPs overlapping with CpGs in the mouse genome (mm9) were filtered out with BisSNP 39. All programs were performed with default setting. For MAB-seq analysis, raw signals were calculated as % of T/(C+T) at each called CpG. For BS-seq analysis, raw signals were calculated as % of C/(C+T) at each called CpG. Statistics of all genome-scale sequencing libraries is summarized in Supplementary Table 1.
Statistical calling of 5fC/5caC and assessing false discovery rate (FDR) of MAB-seq
For each cytosine within CpG dinucleotides, we counted the number of “T” bases from MAB-seq reads as 5fC/5caC (denoted NT) and the number of “C” bases as other forms of cytosines (C/5mC/5hmC; denoted NC). Next, we used the binomial distribution (N as the sequencing coverage (NT + NC) and p as the error rate (2.04%) of M.SssI methylase) to assess the probability of observing NT or greater by chance. To estimate empirical FDR of calling 5fC/5caC-modified CpGs, we repeated the steps above on MAB-seq signals of merged negative control sample (Dnmt1/3a/3b−/− and Tet1/2−/−) in which 5fC/5caC is largely absent. The FDR for a given P-value cutoff of the binomial distribution is the number of 5fC/5caC-called CpGs divided by the number detected in the sample of Tdg-deficient VC-treated mouse ESCs. For calling 5fC/5caC-modified CpG dyads (Fig 2c), reads covering Watson strand and Crick strand of the same CpG dyads were combined. We restricted our analysis to CpG dyads covered by at least five reads. For Fig 3 and 4, strand-specific analysis was performed and we restricted our analysis to CpGs covered by at least ten reads. For Fig 3a, called 5fC/5caC-modified CpGs were visualized in Integrative Genomics Viewer (IGV).
Validation of MAB-seq by synthetic 38-bp oligonucleotides
To monitor the behavior of C and modified C in MAB-seq, 38-bp single-stranded DNA oligos were synthesized (forward strand: 5’-AGCCXGXGCXGXGCXGGTXGAGXGGCXGCTCCXGCAGC-3’, reverse (complementary) strand: 5’-GCTGXGGGAGXGGCXGCTXGACXGGXGXGGXGXGGGCT-3’, in which X is either unmodified C, 5hmC, 5fC or 5caC). To test whether M.sssI treatment alters the behavior of 5hmC, 5fC and 5caC during bisulfite sequencing, forward strands containing 5hmC, 5fC and 5caC were annealed to reverse strands containing the same modified Cs and ligated to methylated adaptors. The resulting oligos were treated by M.sssI using the same protocol used for locus-specific MAB-seq (described below), followed by bisulfite conversion and deep sequencing to determine their behavior. To test whether M.sssI functions at hemi-5hmC, 5fC and 5caC CpGs, top strands containing 5hmC, 5fC and 5caC were annealed to bottom strand containing unmodified C. The same experimental procedures were undertaken to assess the efficiency of M.sssI in these contexts.
Locus-specific MAB-seq of genomic DNA
1 µg genomic DNA was treated by M.sssI in a 50 µL reaction for four rounds. During each round of treatment, DNA was first treated by M.sssI for 2 hours (1.5 µL M.sssI and 1 µL SAM), and additional M.sssI and SAM were supplemented to treat DNA for another 4 hours (0.5 µL M.sssI, 1 µL SAM), increasing the total concentration of the enzyme to 0.8 unit/µL. DNA was purified by PCI after each round of treatment.
After M.sssI treatment, bisulfite conversion was performed as described above. Selected loci were amplified by PCR using KAPA HiFi Hotstart Uracil+ DNA polymerase (Kapa Biosystems, KK2801) followed by sonication by Bioruptor (Diagenode), library preparation using NEBNext DNA Library Prep Master Mix Set (New England Biolabs) and deep sequencing by illumina HiSeq 2500 sequencer. Alternatively, PCR amplified DNA was cloned into TOPO vectors (Zero Blunt TOPO Cloning Kit, Invitrogen) for standard Sanger sequencing. Primer sequences for all locus-specific MAB-seq experiments were summarized in Supplementary Table 2.
5fC/5caC calling for locus-specific MAB-seq
In a MAB-seq experiment, 5fC and 5caC are read as T while unmodified C, 5mC and 5hmC are read as C. For a CpG site, if we name the number of T reads as NT and the number of C reads as NC, 5fC+5caC level (MAB-seq raw signal before background subtraction) can be calculated as NT/(NT+NC). Because conversion of unmodified C to 5mC by M.sssI cannot reach 100%, some T reads may come from incomplete methylation rather than real 5fC/5caC, leading to an overestimation of 5fC/5caC level. To estimate the level of these background signals resulted from incomplete conversion, Tet1/2−/− mouse ESCs was examined by MAB-seq, and any 5fC/5caC signal detected in this negative control is treated as background signal (false positive).
To call 5fC/5caC-positive CpG sites in a locus-specific MAB-seq experiment, MAB-seq signals detected in shCtrl, shCtrl+VC, shTdg and shTdg+VC samples were compared to the background signals detected in Tet1/2−/− sample. For a CpG site in a tested sample, Fisher’s exact test was performed using NC and NT of the tested sample and Tet1/2−/− sample to determine whether MAB-seq signal at this CpG site is significantly different from the corresponding background signal. False discovery rate (FDR) control based on Benjamini–Hochberg procedure was then performed to correct the p values for multiple comparisons, and p<0.05 was used as a cut-off value to generate a list of CpG sites of which MAB-seq signals are significantly different from the background signals detected in Tet1/2−/− sample. After that, we further applied the numeric filter that real 5fC/5caC signals should be numerically higher than background signals, generating the list of CpG sites that are 5fC/5caC positive.
Calculating 5fC/5caC level in locus-specific MAB-seq
In Tet1/2−/− sample, certain background (false-positive) signals were indeed detected by MAB-seq. To counteract these noises introduced by the method, these background signals were subtracted from raw MAB-seq signals when calculating real 5fC/5caC levels. For example, if raw MAB-seq signal detected at a CpG site in our sample of interest is a% while the corresponding background signal detected in Tet1/2−/− sample is b%, and if a≥b, then (a–b)% will be the 5fC/5caC level. If a<b is observed (in some rare cases), then 5fC/5caC level at that CpG site in our sample of interest will be set as 0%. To determine whether the 5fC/5caC level at a group of CpG sites in one sample is different from that in another sample, Wilcoxon signed-rank test for matched pairs was performed.
Analysis of strand asymmetry of 5fC/5caC in locus-specific MAB-seq
For Fig 4d–e, 11 palindromic CpG dyads from Tbx5 locus and 4 palindromic CpG dyads from Vps26a locus were examined by locus-specific MAB-seq to determine whether strand asymmetry of 5fC/5caC exists. 5fC/5caC calling was achieved through performing Fisher’s exact test, and the 15 CpG dyads were first categorized into three groups: no significant 5fC/5caC signal on either the top and bottom strands; significant 5fC/5caC signal on one strand but not the other; significant 5fC/5caC signals on both strands. The existence of the second group is a direct support for strand asymmetry of 5fC/5caC. As for the third group of CpG dyads, Fisher’s exact test was performed to determine whether 5fC/5caC levels differ significantly between the top and bottom strands, and this group was further separated into two groups. For each CpG dyad, if 5fC/5caC levels differ significantly between strands, strand asymmetry of 5fC/5caC exists at this dyad. If not, other methods such as hairpin MAB-seq may help to conclusively determine the state of 5fC/5caC.
Data analysis of locus-specific BS-seq
In BS-seq experiment, unmodified C, 5fC and 5caC are read as T while 5mC and 5hmC are read as C. For a CpG site, if we name the number of T reads as NT and the number of C reads as NC, 5mC+5hmC level at this CpG site (before background subtraction) will be calculated as NT /(NT + NC). To determine whether the 5mC+5hmC level at a CpG site is altered by Tdg knockdown or vitamin C treatment, Fisher’s exact test was performed. To determine whether the 5mC+5hmC level at a group of CpG sites in one sample is different from that in another sample, Wilcoxon signed-rank test for matched pairs was performed.
Calculating the level of unmodified cytosines by combining locus-specific MAB-seq with BS-seq
For a CpG site in our sample of interest, the true level of 5fC+5caC was calculated by subtracting the background signal from the raw MAB-seq signal as described above. The level of 5hmC+5mC was measured by BS-seq. The level of unmodified C equals to 100% - (abundance of 5fC+5caC) - (abundance of 5hmC+5mC). To determine whether the unmodified C level at a group of CpG sites in one sample is different from that in another sample, Wilcoxon signed-rank test for matched pairs was performed.
Locus-specific redBS-seq of genomic DNA
redBS-seq was performed as previously described 21. In short, 5 µL freshly made sodium borohydride aquerous solution (1M) was added to 250 ng DNA diluted in 15 µL water. The reaction was placed in darkness for an hour, quenched by 10 µL sodium acetate (0.75M, pH=5) and purified by PCI extraction. Bisulfite conversion was then performed as described above, followed by PCR amplification of specific loci and deep sequencing analysis of PCR amplicons.
Analysis of locus-specific redBS-seq data
In a redBS-seq experiment, 5mC, 5hmC and 5fC are read as C while unmodified C and 5caC are read as T. For a CpG site, the difference between the levels of C signal measured by redBS-seq and BS-seq was calculated to represent the level of 5fC. In rare cases when negative values were obtained after this subtraction, 5fC levels were designated as 0%. To call CpG sites with significant levels of 5fC, Fisher’s exact test was performed to determine which CpG sites have significantly higher C signals in redBS-seq compared with BS-seq (FDR corrected p value less than 0.05). To calculate the level of 5caC, the level of 5fC measured by redBS-seq was subtracted from the level of 5fC+5caC measured by MAB-seq, and negative values resulted from the subtraction were designated as 0%. To reduce the noise resulted from the two subtractions (one for 5fC calculation and one for 5caC calculation), uncalled sites in redBS-seq or MAB-seq were regarded as having 0% 5fC or 5fC+5caC, respectively.
Locus-specific caMAB-seq and 5caC calling
To perform locus-specific caMAB-seq, genomic DNA was firstly treated by M.sssI as described above and purified through PCI extraction. M.sssI-treated DNA was then reduced by NaBH4 as described above in the redBS-seq protocol, followed by bisulfite conversion using Qiagen Epitect Bisulfite Kit. Selected loci were then amplified by PCR, and PCR amplicons were sequenced using Illumina deep sequencing. To examine whether unmodified C and 5fC are read as C during caMAB-seq, unmodified lambda DNA and synthesized 5fC oligo were also treated using the same protocol and analyzed by Illumina deep sequencing. In caMAB-seq, 5caC is read as T while C, 5mC, 5hmC and 5fC are read as C. To call 5caC-positive sites in a locus-specific caMAB-seq experiment, Fisher’s exact test was performed to determine whether a CpG site in a sample has significantly higher percentage of T compared with background T signals detected in Tet1/2−/− sample. To calculate the absolute level of 5caC in a locus-specific experiment, the background signal detected in Tet1/2−/− sample was subtracted from the raw signal detected in a tested sample.
Genome-wide 5mC/5hmC/5fC/5caC DIP-Seq
The antisera for 5fC and 5caC were previously described 40. For each DIP experiment, 10 µg of sonicated, adaptor ligated genomic DNA from control or Tdg knockdown mouse ESCs (V6.5) was used as input, and 5 µl of 5mC antibody (Eurogentec, BI-MECY-0500), 5 µL of 5hmC antibody (Active Motif, 39791), 1 µl of 5fC antiserum or 0.3 µl of 5caC antiserum was added to immunoprecipitate modified DNA. DNA and antibodies were incubated at 4 °C overnight in a final volume of 500 µL DIP buffer (10 mM sodium phosphate (pH 7.0), 140 mM NaCl, 0.05% Triton X-100) as previously described 41. After the DNA-antibody incubation, 30 µl of Protein G Dynabeads (Invitrogen) were added to the tube and incubated with the DNA-antibody mixture for 2 h at 4 °C. The beads were washed three times with 1 mL of DIP buffer, and then treated with proteinase K at 55 °C for 3 hours to elute the immunoprecipitated DNA, which was further amplified for high-throughput sequencing.
Published Data Sets
To calculate in Figure 2b and 3a, we used following published data sets: Tet1 (Wu et al. 2011) 33, 5hmC base-resolution map 23, H3K4me3, H3K36me3, and H3K27me3 28, H3K4me142, Ezh2 43, Oct4, Nanog, Sox2 34, bivalent/active/silent promoters 44, LMR and UMR 45, H3K27ac and p300 46, CTCF and tissue-specific enhancers 47, BNase, H2A.Z 31, H3.3, panH3 32, and DNase I hypersensitive sites (ENCODE project) 48.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Drs. Shinpei Yamaguchi and Falong Lu for generating and characterizing Tet mutant mouse ESCs. We also thank Drs. Luis M. Tuesta and Shinpei Yamaguchi for critical reading of the manuscript. This project is supported by NIH grant U01DK089565. H.W. was supported by a postdoctoral fellowship from the Jane Coffin Childs Memorial Fund for Medical Research and is currently supported by the National Human Genome Research Institute (K99HG007982). X.W. was supported by the Chinese Scholarship Council. Y.Z. is an investigator of the Howard Hughes Medical Institute.
Footnotes
Accession codes. GEO: GSE62631.
AUTHOR CONTRIBUTIONS
H.W. and Y.Z. conceived the project. H.W. and X.W. performed experiments and carried out data analysis. L.S. performed sequencing. H.W., X.W. and Y.Z. wrote the manuscript.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Bestor TH. The DNA methyltransferases of mammals. Hum Mol Genet. 2000;9:2395–2402. doi: 10.1093/hmg/9.16.2395. [DOI] [PubMed] [Google Scholar]
- 2.Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 3.Cedar H, Bergman Y. Programming of DNA methylation patterns. Annu Rev Biochem. 2012;81:97–117. doi: 10.1146/annurev-biochem-052610-091920. [DOI] [PubMed] [Google Scholar]
- 4.Baylin SB, Jones PA. A decade of exploring the cancer epigenome - biological and translational implications. Nat Rev Cancer. 2011;11:726–734. doi: 10.1038/nrc3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wu H, Zhang Y. Reversing DNA methylation: mechanisms, genomics, and biological functions. Cell. 2014;156:45–68. doi: 10.1016/j.cell.2013.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pastor WA, Aravind L, Rao A. TETonic shift: biological roles of TET proteins in DNA demethylation and transcription. Nature reviews. Molecular cell biology. 2013;14:341–356. doi: 10.1038/nrm3589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tahiliani M, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ito S, et al. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature. 2010;466:1129–1133. doi: 10.1038/nature09303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ito S, et al. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science. 2011;333:1300–1303. doi: 10.1126/science.1210597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.He YF, et al. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science. 2011;333:1303–1307. doi: 10.1126/science.1210944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Inoue A, Zhang Y. Replication-dependent loss of 5-hydroxymethylcytosine in mouse preimplantation embryos. Science. 2011;334:194. doi: 10.1126/science.1212483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maiti A, Drohat AC. Thymine DNA Glycosylase Can Rapidly Excise 5-Formylcytosine and 5-Carboxylcytosine: POTENTIAL IMPLICATIONS FOR ACTIVE DEMETHYLATION OF CpG SITES. The Journal of biological chemistry. 2011;286:35334–35338. doi: 10.1074/jbc.C111.284620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nabel CS, et al. AID/APOBEC deaminases disfavor modified cytosines implicated in DNA demethylation. Nature chemical biology. 2012;8:751–758. doi: 10.1038/nchembio.1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cortazar D, et al. Embryonic lethal phenotype reveals a function of TDG in maintaining epigenetic stability. Nature. 2011;470:419–423. doi: 10.1038/nature09672. [DOI] [PubMed] [Google Scholar]
- 15.Cortellino S, et al. Thymine DNA glycosylase is essential for active DNA demethylation by linked deamination-base excision repair. Cell. 2011;146:67–79. doi: 10.1016/j.cell.2011.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kriaucionis S, Heintz N. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science. 2009;324:929–930. doi: 10.1126/science.1169786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lister R, et al. Global epigenomic reconfiguration during Mammalian brain development. Science. 2013;341:1237905. doi: 10.1126/science.1237905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shen L, et al. Genome-wide analysis reveals TET- and TDG-dependent 5-methylcytosine oxidation dynamics. Cell. 2013;153:692–706. doi: 10.1016/j.cell.2013.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Song C-X, et al. Genome-wide Profiling of 5-Formylcytosine Reveals Its Roles in Epigenetic Priming. Cell. 2013;153:678–691. doi: 10.1016/j.cell.2013.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lu X, et al. Chemical Modification-Assisted Bisulfite Sequencing (CAB-Seq) for 5-Carboxylcytosine Detection in DNA. J Am Chem Soc. 2013;135:9315–9317. doi: 10.1021/ja4044856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Booth MJ, Marsico G, Bachman M, Beraldi D, Balasubramanian S. Quantitative sequencing of 5-formylcytosine in DNA at single-base resolution. Nature Chemistry. 2014 doi: 10.1038/nchem.1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Renbaum P, et al. Cloning, characterization, and expression in Escherichia coli of the gene coding for the CpG DNA methylase from Spiroplasma sp. strain MQ1(M.SssI) Nucleic Acids Res. 1990;18:1145–1152. doi: 10.1093/nar/18.5.1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yu M, et al. Base-resolution analysis of 5-hydroxymethylcytosine in the mammalian genome. Cell. 2012;149:1368–1380. doi: 10.1016/j.cell.2012.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hu L, et al. Crystal Structure of TET2-DNA Complex: Insight into TET-Mediated 5mC Oxidation. Cell. 2013;155:1545–1555. doi: 10.1016/j.cell.2013.11.020. [DOI] [PubMed] [Google Scholar]
- 25.Hashimoto H, et al. Structure of a Naegleria Tet-like dioxygenase in complex with 5-methylcytosine DNA. Nature. 2013 doi: 10.1038/nature12905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yin R, et al. Ascorbic Acid Enhances Tet-Mediated 5-Methylcytosine Oxidation and Promotes DNA Demethylation in Mammals. Journal of the American Chemical Society. 2013 doi: 10.1021/ja4028346. [DOI] [PubMed] [Google Scholar]
- 27.Blaschke K, et al. Vitamin C induces Tet-dependent DNA demethylation and a blastocyst-like state in ES cells. Nature. 2013:1–7. doi: 10.1038/nature12362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mikkelsen TS, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sotiriou S, et al. Ascorbic-acid transporter Slc23a1 is essential for vitamin C transport into the brain and for perinatal survival. Nature medicine. 2002;8:514–517. doi: 10.1038/0502-514. [DOI] [PubMed] [Google Scholar]
- 30.Encode Project Consortium. A user's guide to the encyclopedia of DNA elements (ENCODE) PLoS Biol. 2011;9:e1001046. doi: 10.1371/journal.pbio.1001046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hu G, et al. H2A.Z facilitates access of active and repressive complexes to chromatin in embryonic stem cell self-renewal and differentiation. Cell Stem Cell. 2013;12:180–192. doi: 10.1016/j.stem.2012.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Banaszynski LA, et al. Hira-dependent histone H3.3 deposition facilitates PRC2 recruitment at developmental loci in ES cells. Cell. 2013;155:107–120. doi: 10.1016/j.cell.2013.08.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu H, et al. Dual functions of Tet1 in transcriptional regulation in mouse embryonic stem cells. Nature. 2011;473:389–393. doi: 10.1038/nature09934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Whyte WA, et al. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell. 2013;153:307–319. doi: 10.1016/j.cell.2013.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Booth MJ, et al. Quantitative sequencing of 5-methylcytosine and 5-hydroxymethylcytosine at single-base resolution. Science. 2012;336:934–937. doi: 10.1126/science.1220671. [DOI] [PubMed] [Google Scholar]
- 36.Hashimoto H, et al. Structure of a Naegleria Tet-like dioxygenase in complex with 5-methylcytosine DNA. Nature. 2014;506:391–395. doi: 10.1038/nature12905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014 doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Krueger F, Andrews SR. Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics. 2011;27:1571–1572. doi: 10.1093/bioinformatics/btr167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu Y, Siegmund KD, Laird PW, Berman BP. Bis-SNP: Combined DNA methylation and SNP calling for Bisulfite-seq data. Genome Biol. 2012;13:R61. doi: 10.1186/gb-2012-13-7-r61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Inoue A, Shen L, Dai Q, He C, Zhang Y. Generation and replication-dependent dilution of 5fC and 5caC during mouse preimplantation development. Cell Res. 2011;21:1670–1676. doi: 10.1038/cr.2011.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wu H, et al. Genome-wide analysis of 5-hydroxymethylcytosine distribution reveals its dual function in transcriptional regulation in mouse embryonic stem cells. Genes Dev. 2011;25:679–684. doi: 10.1101/gad.2036011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Meissner A, et al. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature. 2008;454:766–770. doi: 10.1038/nature07107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ku M, et al. Genomewide analysis of PRC1 and PRC2 occupancy identifies two classes of bivalent domains. PLoS Genet. 2008;4:e1000242. doi: 10.1371/journal.pgen.1000242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Marson A, et al. Connecting microRNA genes to the core transcriptional regulatory circuitry of embryonic stem cells. Cell. 2008;134:521–533. doi: 10.1016/j.cell.2008.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stadler MB, et al. DNA-binding factors shape the mouse methylome at distal regulatory regions. Nature. 2011;480:490–495. doi: 10.1038/nature10716. [DOI] [PubMed] [Google Scholar]
- 46.Creyghton MP, et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci U S A. 2010;107:21931–21936. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shen Y, et al. A map of the cis-regulatory sequences in the mouse genome. Nature. 2012;488:116–120. doi: 10.1038/nature11243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Consortium EP. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.