Abstract
Flaviviruses, the human pathogens responsible for dengue fever, West Nile fever, tick-borne encephalitis and yellow fever, are endemic in tropical and temperate parts of the world. The flavivirus non-structural protein 1 (NS1) functions in genome replication as an intracellular dimer and in immune system evasion as a secreted hexamer. We report crystal structures for full-length, glycosylated NS1 from West Nile and dengue viruses. The NS1 hexamer in crystal structures is similar to a solution hexamer visualized by single-particle electron microscopy. Recombinant NS1 binds to lipid bilayers and remodels large liposomes into lipoprotein nanoparticles. The NS1 structures reveal distinct domains for membrane association of the dimer and interactions with the immune system, and are a basis for elucidating the molecular mechanism of NS1 function.
Flaviviruses have a positive-sense RNA genome that encodes a single viral polyprotein. The polyprotein is inserted into the ER membrane through several signal sequences and processed by viral and host proteases into three structural and seven non-structural proteins (NS1, NS2A, NS2B, NS3, NS4A, NS4B, NS5) (1). Six of the non-structural proteins (NS2A – NS5) form a replication complex on the cytoplasmic side of the ER membrane where the NS3 and NS5 enzymes function at a scaffold created by the other four trans-membrane proteins. The remaining protein, the conserved glycosylated NS1, is associated with lipids, both early in infection, where intracellular dimeric NS1 localizes on the ER membrane at the site of viral RNA replication, and late in infection, where secreted hexameric NS1 lipo-protein particles interact with components of the complement-mediated immune system (2, 3). NS1 is essential for replication of the flavivirus genome (4–8), possibly through interactions with transmembrane proteins NS4A and NS4B (5, 9). Electron microscopy (EM) studies of secreted NS1 (sNS1) identified a symmetric barrel-shaped hexamer that carries a cargo of ~70 lipid molecules (10, 11). NS1 interacts with multiple components of both the innate and adaptive immune systems, (12–14) is involved in immune system evasion and pathogenesis, (12–15) and is the major antigenic marker of viral infection (16). While the role of NS1 in multiple stages of the virus life cycle is well established, little is known of the molecular mechanisms of its various functions. The lack of sequence identity to any protein of known structure and the difficulty of producing pure, stable protein have hindered progress in understanding the roles and mechanisms of NS1. An understanding of NS1 structure will help to sort out these contradictory results and will facilitate more efficient vaccine development.
We produced recombinant, full-length, glycosylated West Nile virus (WNV) and dengue virus type 2 (DEN2) NS1 in insect cells using a baculovirus expression system. Despite the presence of a secretion signal in the expression construct, nearly all NS1 was retained by cells and partitioned with the membrane fraction. Soluble NS1 was released by mild detergent treatment of the membrane fraction and appeared as a dimer by gel filtration chromatography, in agreement with direct visualization of particles by negative-stain electron microscopy (EM) (Fig. S1A, B). Multiple chromatography steps without detergent shifted the oligomeric state to a hexamer, presumably due to removal of bound detergent (Fig. S1C, D). WNV NS1 crystallized in two forms and DEN2 NS1 in one form. The WNV NS1 dimer structure was solved from the anomalous scattering of the native sulfur atoms (12 Cys and 5 Met per subunit) using high-multiplicity (~100 fold) data acquired from 18 crystals of form 1 (Table S1, Fig. S2A, B). The twelve cysteines form six disulfide bonds within the NS1 monomer. Three asparagines are glycosylated (Asn130, Asn175 and Asn207), each with clear electron density for one to five sugar residues (Fig S2C). The structure is complete for all amino acids with the exception of one internal loop (amino acids 108 – 128). Identical dimer structures occur in WNV and DEN2 NS1 (0.58 Å root-mean-square deviation of 576 Cα atoms).
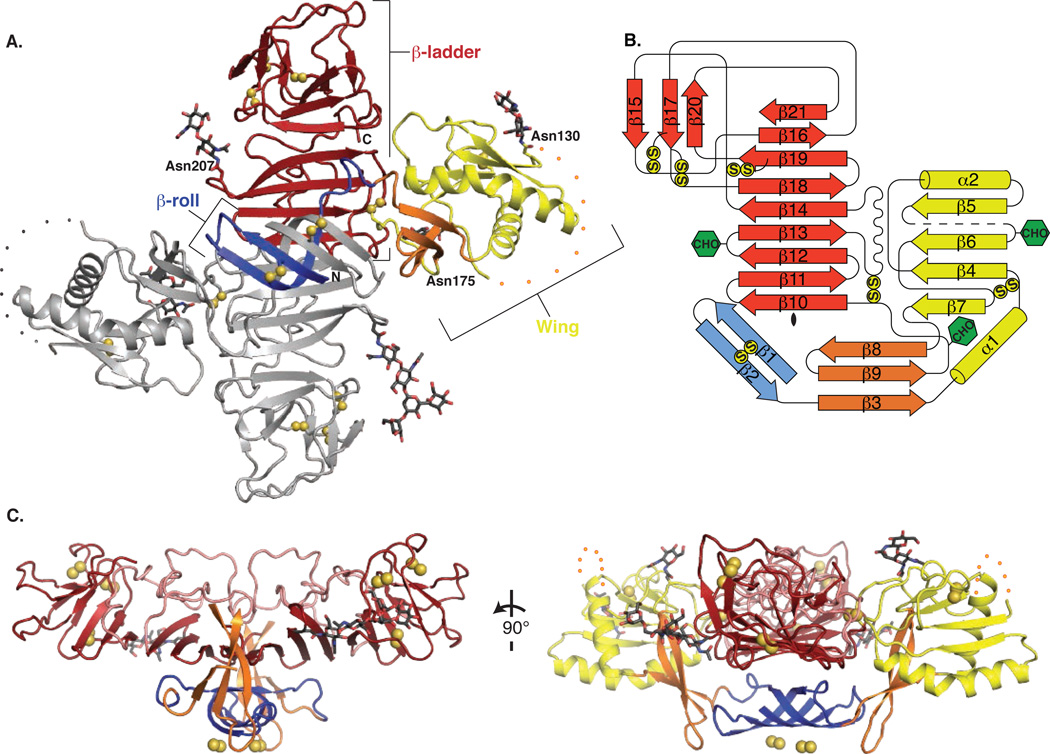
The NS1 dimer is constructed around an extended central β-sheet domain (Fig. 1A, B, Fig S3). Each monomer has three domains. A small “β-roll” dimerization domain (amino acids 1-29) is a mini domain-swap structure of two β-hairpins, each stabilized by a disulfide (Cys4-Cys15). The β-hairpins extend across the dimer axis and intertwine to form a four-stranded β-sheet that curves into a roll-like structure (Fig. 1A, blue; Fig. S2D, E). The second domain (amino acids 30-180) of each monomer protrudes from the central β-domain like a wing (Fig. 1A, yellow). Each “wing” domain contains two glycosylation sites (Asn130, Asn175), an internal disulfide (Cys55-Cys143), and two discreet subdomains. An α/β subdomain (amino acids 38 – 151) comprises a four-stranded β-sheet, two α-helices and a disordered distal tip (amino acids 108 – 128; Fig. 1A, dotted line). A discontinuous connector subdomain,(amino acids 30 – 37 and 152 –180) packs against the β-roll (Fig. 1A, orange) and also links the wing to the central β-sheet domain through a disulfide (Cys179-Cys223). The predominant structural feature of NS1 is the third domain, a continuous β-sheet that extends along the length of the dimer with its 18 β-strands arranged like the rungs of a ladder (Fig. 1A, red). This core “β-ladder” domain is formed by the C-terminal half of NS1 (amino acids 181 – 352), in an arrangement where each monomer contributes nine rungs to the anti-parallel β-ladder. In a simple (+1) topology, the first five β-strand rungs of each monomer begin at the dimer interface and proceed sequentially towards the end of the ladder (Fig. 1B). Most of the inter-strand loops are short with the notable exception of a long “spaghetti loop” between β13 and β14, (amino acids 219 – 272) that lacks secondary structure but is ordered by 57 hydrogen bonds (Fig. 1C, pink). A conserved tip region (Fig. S3) at each end of the β-ladder domain (amino acids 278 – 352) contains four strands of the central β-ladder, a small three-stranded β-sheet and three disulfides.
Figure 1.
NS1 dimer structure. (A) NS1 dimer with one subunit in gray and the other colored by domain (blue β-roll, yellow wing with orange connector sub-domain, red central β-ladder). Disulfides are shown as yellow spheres and N-linked glycosylation sites as sticks with black C. A 20-residue disordered region is indicated with dotted lines. (B) Topology diagram for NS1 monomer, colored in blue, yellow, orange and red as in A. Glycosylation sites are indicated with green hexagons and disulfides with yellow circles. (C) Perpendicular views of NS1 from the edge (left) and the end (right) of the β-ladder. The β-roll (blue) and β-connector subdomain (orange) of the wing form a protrusion on one face of the β-ladder with the spaghetti loop (pink) and glycosylation sites on the other face. The wing domain is omitted from the left image for clarity.
The overall dimensions of the NS1 dimer are 90 Å along the length of the β-ladder, 90 Å in width from wingtip to wingtip and 40 Å in the third dimension (Fig. 1A). The β-ladder defines a plane through the NS1 dimer with the β-roll domain on one side (Fig. 1C). On the other side of the plane are the spaghetti loop, the glycosylation sites, the wing domain disordered loop, and the C-terminus, which, prior to proteolytic cleavage, is fused to the >20-residue lumen-side N-terminus of viral protein NS2A.
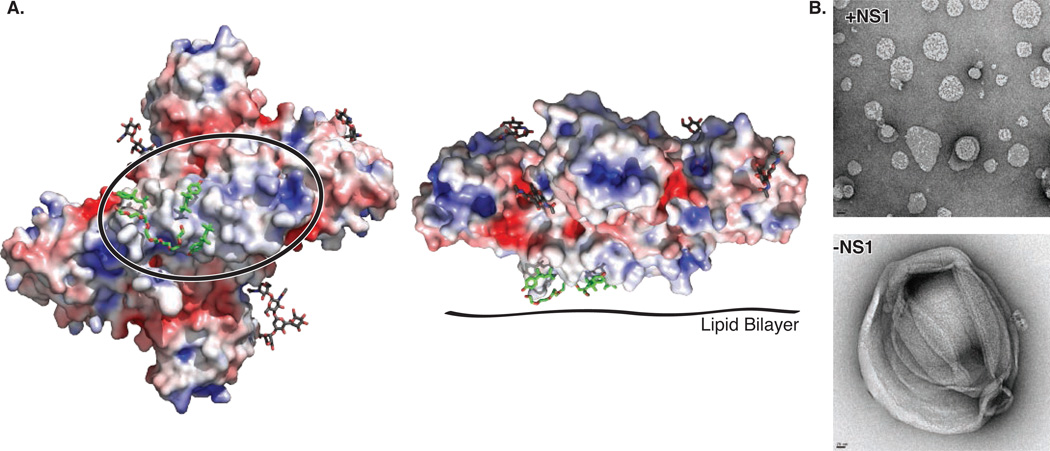
The β-roll and connector subdomain of the wing create a protrusion with a strikingly hydrophobic surface on one face of the dimer (Figs 1C, 2A). Extra electron densities at this surface were evident in the earliest maps, and were fit as parts of three detergent molecules after all regions of the polypeptide had been assigned (Fig. S2D, E). The hydrophobic character of the β-roll/connector protrusion is conserved (Fig. S3, S4). Furthermore, a dipeptide (Arg10-Gln11) implicated in interaction with the transmembrane protein NS4B (9) is located at the periphery of the hydrophobic surface in a loop of the β-roll (Fig. S2E). Thus the hydrophobic protrusion is a strong candidate for dimeric NS1 interaction with the ER membrane and with the replication complex through transmembrane proteins NS4A/B, where NS1 plays a poorly defined but essential role in viral replication.
Figure 2.
Hydrophobic protrusion for membrane interaction. (A) NS1 electrostatic surface potential at pH 6.5 colored from electropositive in blue (+5 kT) to electronegative in red (-5 kT) with bound detergent (sticks with green C) and glycosylation sites (sticks with black C), viewed on the left as in Fig. 1A with the β-roll circled and facing the reader, and on the right as in Fig. 1C (right panel). (B) Effect of WNV NS1 on liposome structure. The negative-stain EM images show how NS1 treatment remodels liposomes (10% cholesterol, 90% phosphatidyl choline) into nano-particles, leaving almost no free NS1 when mixed in a 585:1 ratio of lipid:NS1 hexamer. (Control images in Fig. S5).
We investigated the ability of recombinant WNV NS1 to interact with membranes by incubating purified protein with liposomes and imaging the mixture by negative-stain EM. Upon exposure to NS1, the large heterogeneous liposomes were not only coated with NS1, presumably in its dimeric form, but also were converted into much smaller lipid-protein nano-particles (Fig. 2B, Fig. S5). This unexpected ability to interact with and re-model membranes may have implications for the role of NS1 in organizing replication complexes or in conversion from a membrane-associated dimer in the ER lumen to a secreted proteolipid hexamer.
A “greasy finger” loop on the connector subdomain forms a prominent part of the hydrophobic protrusion and is mobile, as it is disordered in the DEN2 NS1 structure. Mutations to this conserved region (Gly159-Phe-Gly-Val162) were deleterious to virus replication as measured by plaque assay (Table S2). Substitution of charged amino acids in DEN2 NS1 (F160D and V162D) or the double substitution G159A/F160A resulted in no detectable plaques. A single substitution (F160A) impaired virus viability and RNA synthesis (Fig. S6). Purified NS1 variants with these substitutions re-modeled liposomes similarly to the wild type (Fig. S6D), suggesting that the observed phenotype is due to loss of effective interactions with transmembrane proteins of the replication complex.
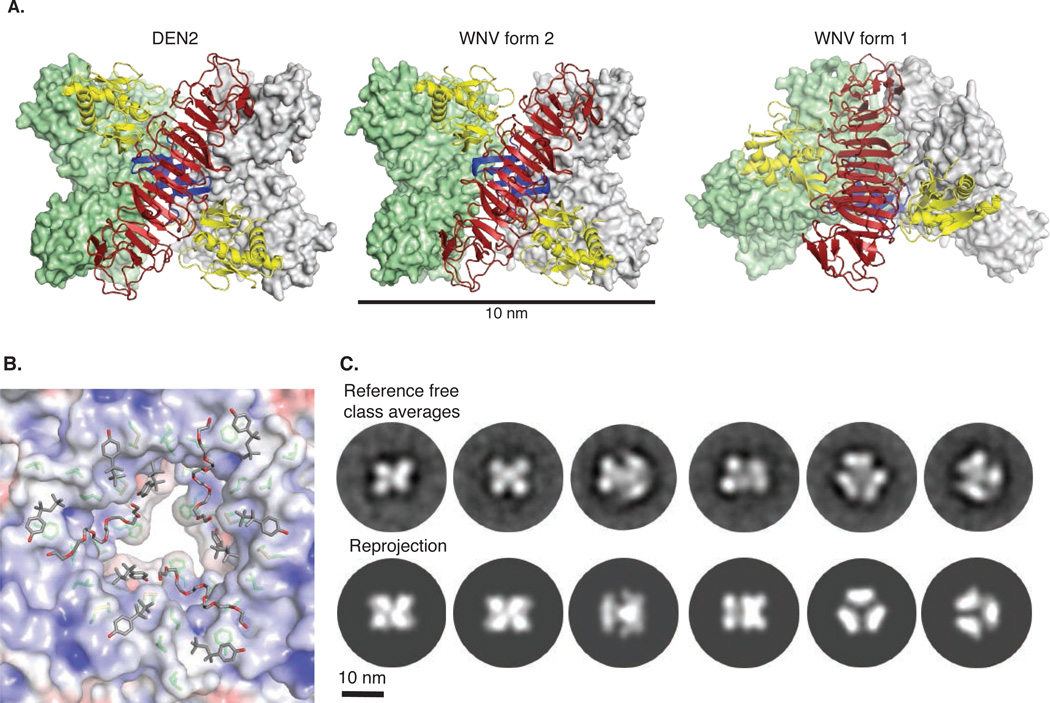
Intracellular NS1 is thought to be predominantly dimeric whereas secreted NS1 is a soluble, hexameric proteolipid particle (3, 10, 11). Lipid-free recombinant NS1 occurred in both dimeric and hexameric forms in solution (Fig. S1), but in all three crystal structures, NS1 exists in a hexameric arrangement of three dimers (Fig. 3A). The three β-rolls face the interior of the hexamer while the outer surface contains the spaghetti loops, glycosylation sites and disordered wing-domain loop. The DEN2 NS1 dimer and those in WNV NS1 crystal form 2 are arranged as a loose, open hexamer with full D3 symmetry and dimensions ~80 Å along the central three-fold axis and ~110 Å in diameter (Fig. 3A, left and center panels) whereas the WNV NS1 form 1 hexamer is splayed open at one end and lacks full hexameric symmetry (Fig. 3A, right panel). The inner-facing hydrophobic protrusions create a conserved hydrophobic interior hexamer surface of diameter ~20 Å (Fig. 3B, Fig. S3, S4). We compared the D3 hexamers in crystal structures with EM images of the NS1 hexamer in solution (Fig. 3C). Two-dimensional EM class averages of NS1 embedded in negative stain reveal particle projections that are remarkably similar to reprojections of the hexamer assemblies observed in crystal structures of DEN2 NS1 and in WNV NS1 form 2. The symmetric hexamers in our crystal structures and the EM images, although without lipid, have similar overall dimensions to those of lipid-bound sNS1 secreted from virus-infected cells (10, 11). Given the hydrophobic interior of the crystallized hexamer, we conclude that the secreted lipid-NS1 particle is organized similarly with β-rolls facing inward and the spaghetti loop, glycosylation sites and disordered loop facing outward.
Figure 3.
NS1 hexamer association. (A) NS1 hexamers: the symmetric hexamers in DEN2 NS1 (left) and WNV NS1 form 2 (center) and the splayed hexamer in WNV NS1 crystal form 1 (right). Molecular surfaces are green and white for two dimers. The third, front-most dimer has a red β-ladder, blue β-roll and yellow wings. The β-roll is entirely inside the hexamer. (B) Association of hydrophobic protrusions at the center of the NS1 hexamer: The electrostatic surface potential illustrates the hydrophobicity of the surfaces. Conserved side chains that form this surface are shown as sticks with green C. Fragments of the detergent molecules (Triton X-100) bound to the hydrophobic surface are shown as sticks with gray C. For clarity, the splayed WNV NS1 form 1 viewed from the open end. (C) Comparison of WNV NS1 hexamers in solution with the symmetric NS1 hexamer in crystals. Two-dimensional class averages (top row) are very similar to reprojections calculated from the WNV NS1 hexamer in crystal form 2 (bottom row).
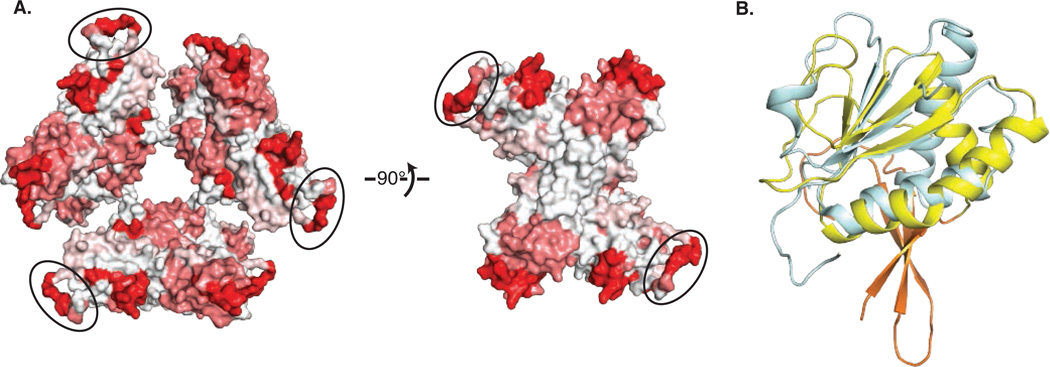
Secreted NS1 is a diagnostic marker for flavivirus infection in serum (17) where immune system proteins encounter the sNS1 hexamer as a proteolipid particle. We identified 108 NS1 linear epitopes elicited in response to immune stimulation by virus or full-length NS1 (18), and mapped these onto the hexamer structure (Fig. 4A). The epitopes localize to a few hot spots, including the wing domain, the C-terminal tip of the β-ladder and the β-roll. The most frequently identified epitopes are the most accessible parts of the NS1 hexamer: the wing domain disordered loop and the C-terminal tip of the β-ladder. The wing domain epitopes are concentrated at a highly conserved Gly-Trp-Lys-Ala-Trp-Gly peptide (amino acids 114-119) (Fig. S3), with invariant tryptophans and conserved lysine. The hydrophobic protrusion is another frequently identified epitope, implying that the inside of the hexameric proteolipid is accessible to the immune recognition system at some point, either before hexamer formation or by hexamer dissociation. Anti-NS1 antibodies have been implicated in immune pathogenesis in dengue virus (19–23), and antibodies with cross-reactivity to human proteins have been mapped to the conserved wing peptide (23) and to the conserved tip of the β-ladder (19).
Figure 4.
NS1 and the immune system (A) Linear epitopes to NS1 mapped on the structure. The molecular surface of hexameric NS1 is colored from white to red based on the frequency of 108 NS1-mapped epitopes (http://www.iedb.org). The disordered loop at the periphery of the wing domain, modeled as poly-alanine and circled, is a frequent epitope. (B) Similarity of the NS1 wing α/β subdomain to the RIG-I family of innate immune proteins. An SF2 helicase domain of RIG-I (blue, 3TBK(25)) is superimposed on the WNV NS1 wing domain (yellow).
Production of interferons and cytokines by the innate immune system is an important host defense against flaviviruses, particularly through the pattern recognition receptors RIG-I, MDA5 and TLR3 (2, 24). Intriguingly, the α/β subdomain of the NS1 wing resembles a helicase domain of RIG-I (25) and MDA5 (26) (Fig. 4B). These cytoplasmic helicase domains recognize pathogenic RNAs and trigger the antiviral response. As viral mimicry of host proteins is a well established method to thwart the immune system, it is tempting to propose such a role for the NS1 wing domain. However, the NS1 wing-domain surface analogous to the RIG-I and MDA5 RNA-binding surface is negatively charged and in contact with the central β-ladder domain. Moreover, NS1 is not known to localize to the cytoplasm where RIG-I or MDA5 detect RNA. Thus this intriguing similarity to RIG-I/MDA5 adds yet another enigma to the NS1 story.
Supplementary Material
Acknowledgments
We thank Donald Raymond for characterization of initial crystals, Gregory Dodge for assistance with protein purification and crystallization, and Annie Dosey for assistance with electron microscopy. Supported by a grant from the National Institutes of Health (P01AI055672) to RJK and JLS, by the Martha L. Ludwig Professorship of Protein Structure and Function to JLS, by the Pew Scholar Program in Biomedical Sciences to GS, and by a Perrigo Undergraduate Summer Fellowship to TJJ. Beamlines of GM/CA@APS were supported by the National Institute of General Medical Sciences (“GM”, Y1-GM-1104) and the National Cancer Institute (“CA”, Y1-CO-1020). Atomic coordinates and structure factor files have been deposited in the RCSB Protein Data Bank (PDB) database under the accession codes 4O6B for DEN2 NS1, 4O6C for WNV NS1 crystal form 2 and 4O6D for WNV NS1 crystal form 1.
References and Notes
- 1.Lindenbach BD, Rice CM. Molecular biology of flaviviruses. Adv Virus Res. 2003;59:23–61. doi: 10.1016/s0065-3527(03)59002-9. [DOI] [PubMed] [Google Scholar]
- 2.Suthar MS, Diamond MS, Gale M., Jr West Nile virus infection and immunity. Nat Rev Microbiol. 2013;11:115–128. doi: 10.1038/nrmicro2950. [DOI] [PubMed] [Google Scholar]
- 3.Muller DA, Young PR. The flavivirus NS1 protein: molecular and structural biology, immunology, role in pathogenesis and application as a diagnostic biomarker. Antiviral Res. 2013;98:192–208. doi: 10.1016/j.antiviral.2013.03.008. [DOI] [PubMed] [Google Scholar]
- 4.Khromykh AA, Sedlak PL, Westaway EG. cis- and trans-acting elements in flavivirus RNA replication. J Virol. 2000;74:3253–3263. doi: 10.1128/jvi.74.7.3253-3263.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lindenbach BD, Rice CM. Genetic interaction of flavivirus nonstructural proteins NS1 and NS4A as a determinant of replicase function. J Virol. 1999;73:4611–4621. doi: 10.1128/jvi.73.6.4611-4621.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Westaway EG, Mackenzie JM, Kenney MT, Jones MK, Khromykh AA. Ultrastructure of Kunjin virus-infected cells: colocalization of NS1 and NS3 with double-stranded RNA, and of NS2B with NS3, in virus-induced membrane structures. J Virol. 1997;71:6650–6661. doi: 10.1128/jvi.71.9.6650-6661.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lindenbach BD, Rice CM. trans-Complementation of yellow fever virus NS1 reveals a role in early RNA replication. J Virol. 1997;71:9608–9617. doi: 10.1128/jvi.71.12.9608-9617.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mackenzie JM, Jones MK, Young PR. Immunolocalization of the dengue virus nonstructural glycoprotein NS1 suggests a role in viral RNA replication. Virology. 1996;220:232–240. doi: 10.1006/viro.1996.0307. [DOI] [PubMed] [Google Scholar]
- 9.Youn S, et al. Evidence for a genetic and physical interaction between nonstructural proteins NS1 and NS4B that modulates replication of West Nile virus. J Virol. 2012;86:7360–7371. doi: 10.1128/JVI.00157-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gutsche I, et al. Secreted dengue virus nonstructural protein NS1 is an atypical barrelshaped high-density lipoprotein. Proc Natl Acad Sci U S A. 2011;108:8003–8008. doi: 10.1073/pnas.1017338108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Muller DA, et al. Structure of the dengue virus glycoprotein non-structural protein 1 by electron microscopy and single-particle analysis. J Gen Virol. 2012;93:771–779. doi: 10.1099/vir.0.039321-0. [DOI] [PubMed] [Google Scholar]
- 12.Avirutnan P, et al. Binding of flavivirus nonstructural protein NS1 to C4b binding protein modulates complement activation. J Immunol. 2011;187:424–433. doi: 10.4049/jimmunol.1100750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Avirutnan P, et al. Antagonism of the complement component C4 by flavivirus nonstructural protein NS1. J Exp Med. 2010;207:793–806. doi: 10.1084/jem.20092545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chung KM, et al. West Nile virus nonstructural protein NS1 inhibits complement activation by binding the regulatory protein factor H. Proc Natl Acad Sci U S A. 2006;103:19111–19116. doi: 10.1073/pnas.0605668103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Krishna VD, Rangappa M, Satchidanandam V. Virus-specific cytolytic antibodies to nonstructural protein 1 of Japanese encephalitis virus effect reduction of virus output from infected cells. J Virol. 2009;83:4766–4777. doi: 10.1128/JVI.01850-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Young PR, Hilditch PA, Bletchly C, Halloran W. An antigen capture enzyme-linked immunosorbent assay reveals high levels of the dengue virus protein NS1 in the sera of infected patients. J Clin Microbiol. 2000;38:1053–1057. doi: 10.1128/jcm.38.3.1053-1057.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Alcon-LePoder S, et al. Secretion of flaviviral non-structural protein NS1: from diagnosis to pathogenesis. Novartis Found Symp. 2006;277:233–247. doi: 10.1002/0470058005.ch17. discussion 247-253. [DOI] [PubMed] [Google Scholar]
- 18.Vita R, et al. The immune epitope database 2.0. Nucleic Acids Res. 2010;38:D854–D862. doi: 10.1093/nar/gkp1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cheng HJ, et al. Proteomic analysis of endothelial cell autoantigens recognized by antidengue virus nonstructural protein 1 antibodies. Exp Biol Med (Maywood) 2009;234:63–73. doi: 10.3181/0805-RM-147. [DOI] [PubMed] [Google Scholar]
- 20.Falconar AK. The dengue virus nonstructural-1 protein (NS1) generates antibodies to common epitopes on human blood clotting, integrin/adhesin proteins and binds to human endothelial cells: potential implications in haemorrhagic fever pathogenesis. Arch Virol. 1997;142:897–916. doi: 10.1007/s007050050127. [DOI] [PubMed] [Google Scholar]
- 21.Falconar AK. Monoclonal antibodies that bind to common epitopes on the dengue virus type 2 nonstructural-1 and envelope glycoproteins display weak neutralizing activity and differentiated responses to virulent strains: implications for pathogenesis and vaccines. Clin Vaccine Immunol. 2008;15:549–561. doi: 10.1128/CVI.00351-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Henchal EA, Henchal LS, Schlesinger JJ. Synergistic interactions of anti-NS1 monoclonal antibodies protect passively immunized mice from lethal challenge with dengue 2 virus. J Gen Virol. 1988;69(Pt 8):2101–2107. doi: 10.1099/0022-1317-69-8-2101. [DOI] [PubMed] [Google Scholar]
- 23.Liu IJ, Chiu CY, Chen YC, Wu HC. Molecular mimicry of human endothelial cell antigen by autoantibodies to nonstructural protein 1 of dengue virus. J Biol Chem. 2011;286:9726–9736. doi: 10.1074/jbc.M110.170993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wilson JR, de Sessions PF, Leon MA, Scholle F. West Nile virus nonstructural protein 1 inhibits TLR3 signal transduction. J Virol. 2008;82:8262–8271. doi: 10.1128/JVI.00226-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Civril F, et al. The RIG-I ATPase domain structure reveals insights into ATP-dependent antiviral signalling. EMBO Rep. 2011;12:1127–1134. doi: 10.1038/embor.2011.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Motz C, et al. Paramyxovirus V proteins disrupt the fold of the RNA sensor MDA5 to inhibit antiviral signaling. Science. 2013;339:690–693. doi: 10.1126/science.1230949. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.