Abstract
Plasma membrane (PM)-bound GTPase Rap1 recruits the Rap1-interacting-adaptor-molecule (RIAM), which in turn recruits talin to bind and activate integrins. However, it is unclear how RIAM recruits talin and why its close homolog lamellipodin does not. Here we report that, although RIAM possesses two talin-binding sites (TBS1 and TBS2), only TBS1 is capable of recruiting cytoplasmic talin to the PM, and the R8 domain is the strongest binding site in talin. Crystal structure of an R7R8:TBS1 complex reveals an unexpected kink in the TBS1 helix that is not shared in the homologous region of lamellipodin. This kinked helix conformation is required for the co-localization of RIAM and talin at the PM and proper activation of integrin. Our findings provide the structural and mechanistic insight into talin recruitment by RIAM that underlies integrin activation and explain the differential functions of the otherwise highly homologous RIAM and lamellipodin in integrin signaling.
Keywords: RIAM, lamellipodin, talin, integrin, inside-out signaling, crystal structure
INTRODUCTION
Integrin signaling promotes cell proliferation, migration and adhesion, and regulates cell survival via crosstalk with receptor tyrosine kinases (Clemmons and Maile, 2005; Hood et al., 2003; Menter and Dubois, 2012; Parsons, 2003; Ratnikov et al., 2005). The ectodomains of activated integrin bind the extracellular matrix (ECM) and trigger the outside-in signaling pathway that activates the FAK-Src signaling cascade (Arias-Salgado et al., 2003; Huang et al., 1993). Integrins are activated primarily by the cytoskeletal protein talin (Tadokoro et al., 2003; Wegener et al., 2007). Talin also plays an important role in promoting cancer progression and metastasis (Desiniotis and Kyprianou, 2011). In particular, the expression of talin is up-regulated in many primary tumors, and its overexpression correlates with metastases of oral squamous cell carcinomas and prostate cancers (Lai et al., 2011; Sakamoto et al., 2010). Therefore, talin may serve as a prognostic marker and/or drug target for cancer therapeutics.
Vertebrates possess two isoforms of talin, talin1 and talin2. Talin1 is ubiquitously expressed in mammalian cells, whereas talin2 is primarily expressed in the heart (Monkley et al., 2001). Talin1 is also more important in activating integrin in platelets (Nieswandt et al., 2007; Petrich et al., 2007). Activation of integrins by talin is initiated by a Rap1-induced signaling pathway known as the inside-out pathway (Banno and Ginsberg, 2008; Bivona et al., 2004; Bos et al., 2003; Lee et al., 2009; Wynne et al., 2012). Activated Rap1 recruits an effector protein, Rap1-interacting adaptor molecule (RIAM), which in turn recruits talin to the plasma membrane (PM) (Lafuente et al., 2004; Lee et al., 2009; Wynne et al., 2012). Thus, the specific binding to both Rap1 and talin allows RIAM to play a key role in this pathway by linking cytoplasmic talin to the PM-anchored Rap1 (Lee et al., 2009; Wynne et al., 2012; Zhang et al., 2014). Talin is composed of a 4-domain “head” region and a “rod” region consisting of 13 helical bundle domains (R1-R13) (Fig. 1A) (Goult et al., 2013b; Moser et al., 2009; Ye et al., 2014). Following its recruitment to the PM by RIAM, talin binds to the intracellular tail of the integrin β subunit via the F3 domain from the head region and, in doing so, switches integrin from a low- to a high-affinity state (Wegener et al., 2007). This activity is suppressed by a competitive intramolecular autoinhibitory interaction between the F3 domain in the head region and the R9 domain in the rod region. Talin also recruits other cytoskeletal proteins, such as actin and vinculin, to stabilize the integrin-mediated focal adhesions (Fig. 1A) (Humphries et al., 2007).
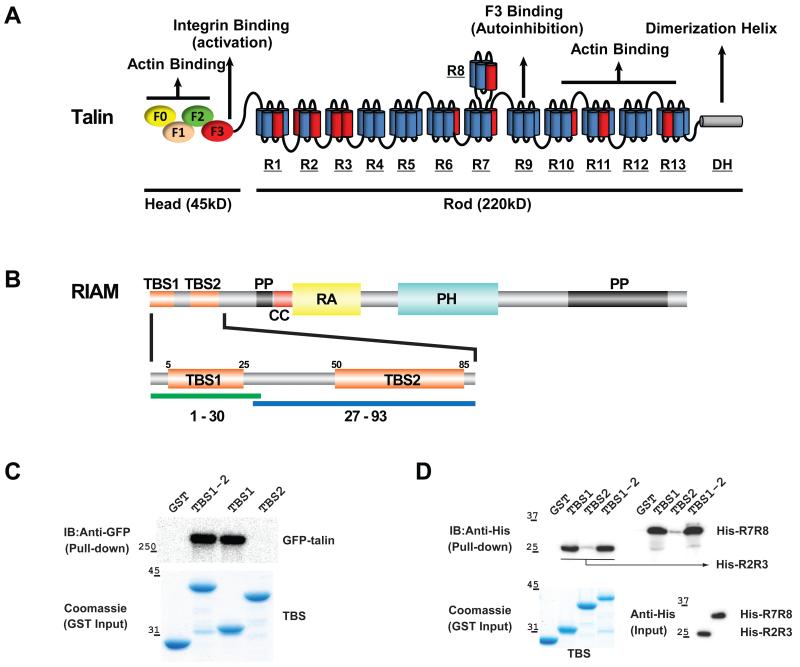
Figure 1.
(A) Schematic diagrams of talin. The head region possesses four N-terminal FERM domains (ovals), and the rod region contains 13 helical bundle domains (R1-R13, each cylinder represents one helix) and a dimerization helix (DH). Vinculin binding helices are colored in red. (B) Schematic diagrams of RIAM. Talin-binding sites (TBS1 and TBS2) are colored in orange; coiled-coil (CC) is in red; poly-proline (PP) is in black; the Ras-association domain (RA) is in yellow; and Pleckstrin-homology domain (PH) is in cyan. Fragments containing residues 1-30 and 27-93 were used in the biochemical characterization and functional assays to represent the TBS1 and TBS2 regions, respectively. (C) GST-tagged TBS1, TBS2, and TBS1-2 (residues 1-93) were used as bait to pull down GFP-talin expressed in HEK293 cells. Bound GFP-talin was analyzed by Western blotting. Input GST-tagged RIAM protein levels were shown by Coomassie staining. (D) GST-tagged TBS1, TBS2, and TBS1-2 were used as a bait to pull down purified recombinant His6-tagged R2R3 and R7R8 proteins. Bound His-R2R3 and His-R7R8 proteins were analyzed by Western blotting (upper). Input GST-tagged RIAM protein levels were shown by Coomassie staining (lower).
RIAM recruits talin to the PM via RIAM’s N-terminal region (Goult et al., 2013b; Lee et al., 2009). RIAM translocates to the PM via interactions of its Ras-associating domain (RA) with Rap1 GTPase and its pleckstrin-homology domain (PH) with the membrane lipid PI(4,5)P2 (Fig. 1B) (Wynne et al., 2012). Multiple interacting sites have been identified in RIAM and in the talin rod domains (Goult et al., 2013b; Lee et al., 2009). The first identified talin-binding site in RIAM, known as TBS1 (residues 1-30), may interact with several talin rod domains (Goult et al., 2013b). Interestingly, TBS1 also interacts with the vinculin domain 1 (Vd1), and TBS1 and Vd1 bind to talin in a mutually exclusive manner due to steric conflict (Goult et al., 2013b). A recent study revealed that a second talin-binding site known as TBS2 (residues 50-85) in RIAM could also interact with the R2 and R3 domains of talin (Goult et al., 2013b). However, it is unclear how TBS1 and TBS2 both function in the recruitment of talin to the PM. Furthermore, the TBS1 sequence is highly conserved in lamellipodin (Lpd), a paralogue of RIAM capable of binding to both talin and Rap1 (Chang et al., 2013; Krause et al., 2004; Lee et al., 2009). Unlike RIAM, Lpd exhibits little or even opposing effects on inside-out integrin signaling (Colo et al., 2012; Lafuente et al., 2004; Watanabe et al., 2008). Thus, the structural and functional characterization of the binding specificity of the RIAM:talin association is required to elucidate the molecular mechanisms by which Rap1-induced integrin activation is regulated by RIAM.
In this study, we investigate RIAM-mediated inside-out integrin signaling using talin1 and a platelet integrin αIIbβ3. We demonstrate that the TBS1 region, but not the TBS2 region, interacts with full-length talin in the cytoplasm, and that the R8 domain of talin is the strongest binding site for the TBS1 region. The TBS2 region only interacts with isolated talin rod domains weakly, and its role in talin recruitment and integrin activation is insignificant. The crystal structure of a TBS1 peptide in complex with the R7R8 double domains of talin was determined to 1.5-Å resolution. The structure reveals that TBS1 binds to the R8 domain via both hydrophobic and electrostatic interactions. Unexpectedly, this association also requires a unique, kinked helical conformation of the bound TBS1 peptide. Mutations of TBS1 that disrupt the kinked conformation or the side-chain interactions significantly diminished its association with talin, membrane co-clustering of RIAM with talin, and integrin activation. We also identified a highly conserved TBS1-binding surface in the talin R3 domain via a structural alignment with the R8 domain and demonstrated that disrupting binding sites in both the R3 and the R8 domains abolished the RIAM:talin interaction. Moreover, our analyses suggest that the talin-binding site in Lpd corresponding to the TBS1 region lacks the helical kink, leading to a low talin-binding affinity. These results suggest that only the TBS1 region, but not the TBS2 region, functions in promoting the talin:integrin interaction, and elucidate a structural basis for the specific recruitment of talin by RIAM.
RESULTS
Comparison of RIAM TBS1 and TBS2 regions in binding to cytoplasmic talin
The N-terminal flexible region of RIAM preceding the RA domain (residues 1-178) contains at least three predicted helical sequences (Fig. 1B: TBS1, TBS2, and CC) (Wynne et al., 2012; Zhang et al., 2014). Two of these sequences exhibit binding affinities to various talin domains and are named TBS1 and TBS2 (Goult et al., 2013b; Lee et al., 2009). Prior to its translocation to the PM, the homodimeric talin protein adopts a highly compacted autoinhibitory conformation (Goksoy et al., 2008; Goult et al., 2013a; Song et al., 2012). This conformation may partially mask the RIAM-binding domains. To assess the interaction of TBS1 and TBS2 with full-length cytoplasmic talin, we performed a pull-down assay using GST-tagged RIAM fragments (TBS1: residues 1-30; TBS2: residues 27-93; and TBS1-2: residues 1-93) with full-length talin expressed in mammalian cells. Both TBS1 and TBS1-2 exhibit a strong association with full-length talin at similar levels. However, the association of TBS2 with talin is undetectable in this binding assay (Fig. 1C).
Characterization of the primary RIAM binding sites in talin
Several talin rod domains including the R3 and R8 domains have been shown to interact with RIAM TBS1 (Goult et al., 2013b). Because it is suggested that the four helical-bundle R3 domain is intrinsically unstable and undergoes unfolding in response to force exerted on talin (Goult et al., 2013b), we assessed the binding affinities of RIAM TBS1, TBS2, and TBS1-2 fragments with talin R2R3 and R7R8 fragments. Recombinant R2R3 and R7R8 proteins were examined by CD spectra to confirm proper folding (Fig. S1A). The TBS1 and TBS1-2 fragments bind to both R2R3 and R7R8 in the in vitro binding assays, whereas the interactions of the TBS2 fragment with R2R3 and R7R8 are much weaker (Fig. 1D). We then assessed the binding affinities of TBS1 or TBS1-2 with R2R3 or R7R8 using quantitative pull-down assays. While the TBS1-binding affinities of R2R3 and R7R8 are both in the low micromolar range, TBS1 binds to R7R8 more strongly than R2R3, and the binding affinities of TBS1-2 with R7R8 or with R2R3 are similar to those of TBS1 (Fig. S1B-E). These results confirm that both R3 and R8 domains directly bind the TBS1 fragment and suggest that the R8 domain (in the form of R7R8) is a stronger TBS1-binding site.
Structure of the RIAM TBS1 in complex with the talin R7R8
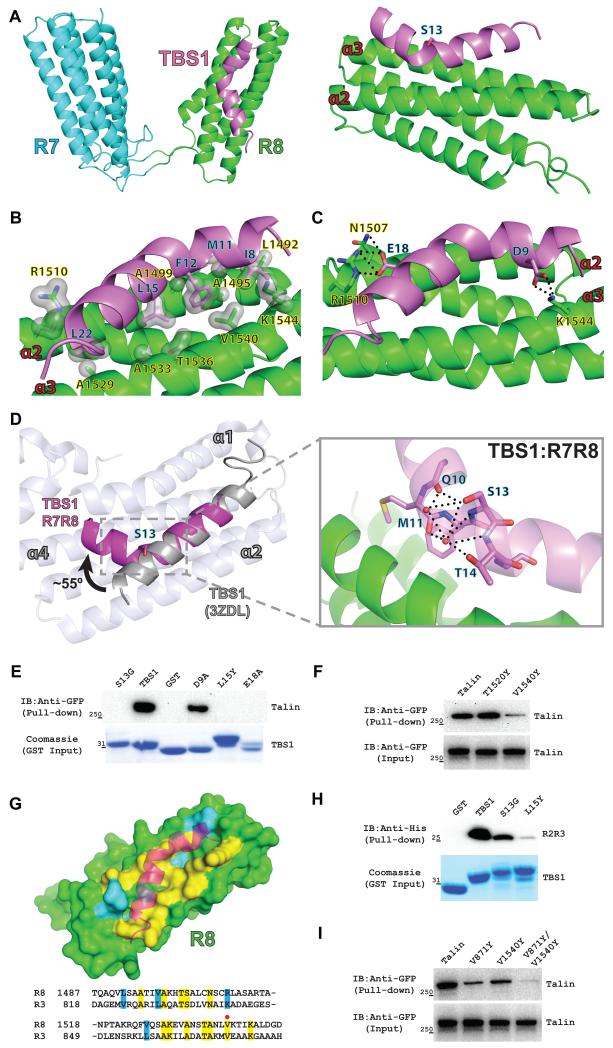
To better understand the structural basis for the interaction between RIAM and talin, we determined the crystal structure of a RIAM TBS1 peptide (residues 5-25) in complex with the talin R7R8 domains (residues 1357-1657) at 1.5 Å resolution (Table 1). The asymmetric unit possesses one R7R8 molecule with a well-defined TBS1 fragment (Fig. S2A). The TBS1 peptide interacts with the talin R8 domain, but not with the R7 domain (Fig. 2A). Although TBS1 also forms hydrogen bonds with the symmetrically related R7 domain (Fig. S2B), removal of the R7 domain did not affect the association of TBS1 with the R8 domain, suggesting that these hydrogen bonds are likely the result of crystal packing. The TBS1 peptide binds to the R8 domain at the α2 and α3 helices via both hydrophobic and electrostatic interactions. The association is mediated primarily through a large hydrophobic contact interface formed by multiple side chains (Ile8, Met11, Phe12, Leu15, and Leu22 in RIAM TBS1 and Leu1492, Ala1495, Ala1499, Ala1529, Ala1533, Thr1536, Val1540, Cα of Arg1510 and Lys1544 in the R8 domain) (Fig. 2B), and is further fortified by several electrostatic interactions (Asp9RIAM-Lys1544talin, Glu18RIAM-Arg1510talin, and Glu18RIAM-Asn1507talin) (Fig. 2C).
Table 1.
X-ray data collection and refinement statistics
| R7R8-TBS1 | |
|---|---|
| Data collection | |
| Space group | P21212 |
| Cell Dimensions | |
| a, b, c (Å) | 59.7, 105.9, 49.0 |
| α, β, γ (°) | 90.0, 90.0, 90.0 |
| Resolution (Å) | 50.0-1.50 (1.53-1.50) |
| Completeness (%) | 98.6 (96.9) |
| Rsym (%) | 5.1 (55.8) |
| I / σ(I) | 47.3 (2.9) |
| Unique reflections | 49933 (2414) |
| Redundancy | 5.7 (4.5) |
| Refinement | |
| Resolution (Å) | 50.0-1.50 |
| Rwork(%) | 22.3 (28.0) |
| Rfree (%) | 24.2 (30.3) |
| RMSD bonds (Å) | 0.005 |
| RMSD angle (°) | 0.934 |
| Protein atoms | 2244 |
| Peptide atoms | 162 |
| Solvent atoms | 305 |
| Total residues | 322 |
| Average B-factors (Å2) | |
| Protein | |
| Main chain atoms | 20.6 |
| Side chain atoms | 22.9 |
| Peptide | |
| Main chain atoms | 18.4 |
| Side chain atoms | 19.0 |
| Solvent | 36.5 |
| Ramachandran | |
| Favored regions (%) | 99.7 |
| Allowed regions (%) | 100.0 |
Rsym Σ|Iobs − Iavg|/ΣIavg; Rwork = Σ||Fobs − Fcalc||/ΣFobs; Rfree was calculated using 5% of the data and the same sums. Values in parentheses are for highest-resolution shell.
Figure 2.
(A) Ribbon diagram representation of the complex structure of the talin R7R8 domains and the RIAM TBS1 peptide. R7 domain is colored in cyan; R8 domain is in green; and the TBS1 peptide is in purple. Left: Top view of the R7R8-TBS1 complex. Right: Only the R8 domain and the TBS1 peptide are shown in the side view. The Ser13 residue located at the kink is shown as a stick representation. The two TBS1-interacting helices in R8 are labeled as α2 and α3. (B) Hydrophobic interactions are represented by a light gray surface. Residues of TBS1 are labeled in black and blue, and residues of the R8 domain are labeled in black and yellow. (C) Hydrogen bonds are denoted by a dotted line. (D) Superposition of the TBS1 peptide (residues 14-25) from the TBS1:R7R8 structure (purple) and the structure of TBS1 in complex with the vinculin Vd1 domain (PDB ID 3ZDL) shows a ~55° kink at Ser13. Vd1 helices (ivory) that interact with TBS1 are labeled (α1, α2, and α4). The hydrogen bond network mediated by the side chain of Ser13 is illustrated in the close-in view (E) GST-tagged TBS1 mutations (D9A, S13G, L15Y and E18A) were used as bait to pull down GFP-talin. Bound GFP-talin was analyzed by Western blotting. Input GST-TBS1 (wild type and mutants) levels were shown by Coomassie staining. (F) Pull-down assay using full-length talin (WT, T1520Y and V1540Y) and the RIAM TBS1 fragment. Input GFP-talin (wild type and mutants) levels were shown in the lower blot. (G) Surface representation of the R8 domain and cartoon diagram of the bound TBS1 peptide. TBS1 interacting residues that are identical in the R3 domain are highlighted in yellow, and similar residues are highlighted in blue. Conserved residues are highlighted in yellow in the structure-based sequence alignment of the R8 domain and the R3 domain. V1540 in the R8 domain and V871 in the R3 domain are indicated by a red dot. (H) Pull-down of His-tagged talin R2R3 domains by GST-TBS1 and its mutants (S13G and L15Y). Input GST-TBS1 (wild type and mutants) levels were shown by Coomassie staining. (I). Pull-down of full-length talin and its mutants (V871Y, V1540 and V871Y/V1540Y) with GST-TBS1. Input GFP-talin (wild type and mutants) levels were shown by Western blotting.
Strikingly, in the structure of TBS1:R7R8 complex, the TBS1 peptide adopts an unexpected helical conformation with an approximately 55° kink at the backbone of residue Ser13 (Fig. 2D). An earlier study reported a structure of the RIAM TBS1 in complex with the vinculin Vd1 domain, in which a conventional α-helical TBS1 interacted with the α1, α2 and α4 helices of the Vd1 domain (Fig. 2D) (Goult et al., 2013b). The side-chain hydroxyl group of Ser13 in the R7R8-bound TBS1 forms a hydrogen bond with the backbone of Gln10 and a water-mediated hydrogen bond network with the backbones of Met11 and Thr14 (Fig. 2D). These interactions stabilize the helical kink at Ser13 in the TBS1:R7R8 complex. The kinked TBS1 helix allows additional hydrophobic side chains proximal to the N-terminus, including Ile8, Met11, and Phe12, to participate in the hydrophobic interactions with the R8 domain. This conformation also positions the Asp9 residue towards the α3 helix of the R8 domain to form the Asp9RIAM-Lys1544talin salt bridge. As a result, the average B-factor for the R7R8-bound TBS1 is 18.4 Å2 for main chain atoms (18.7 Å2 for all atoms), representing a much more stable conformation than that of the Vd1-bound TBS1 (average B-factor is 56.1 Å2 for main chain atoms and 57.1 Å2 for all atoms) (Fig. S2C). The shape complementary of the TBS1:R7R8 interaction (0.773) is also higher than that of the TBS1:Vd1 interaction (0.714) (Lawrence and Colman, 1993). The refined model was analyzed by MolProbity (Chen et al., 2010). 99.7% of all residues are in the favored regions, and 100% of all residues are in the allowed regions of the Ramachandran plot. The Clashscore is 1.24 (99th percentile) and the overall MolProbity score is 0.84 (100th percentile). Thus, the crystal structure of the R7R8:TBS1 complex suggests that binding to R7R8 induces a more stable kinked helical conformation of the TBS1 fragment, thus enhancing the TBS1:talin interaction.
Identification of the residues critical for TBS1:talin binding
To validate the side chain interactions observed in the crystal structure, we constructed several single point mutations in the RIAM TBS1 region (D9A, L15Y, and E18A) and a surface mutation V1540Y in the R8 domain, and evaluated their effect on the association of the RIAM TBS1 region with full-length talin. Additionally, to assess the contribution of the helical kink to the interaction, we also mutated Ser13 to a glycine to destabilize the kink in the RIAM TBS1 region. The D9A mutation in the RIAM TBS1 fragment and the V1540Y mutation in the R8 domain diminished the TBS1:talin association, and the other TBS1 mutations (L15Y, E18A, and S13G) completely abolished the interaction with full-length talin in the pull-down assays (Fig. 2E). In contrast, another talin surface mutation at a non-interacting residue (T1520Y) in the same helix as V1540Y exhibited no reduction in RIAM TBS1 binding (Fig. 2F). These data demonstrate that not only the hydrophobic and electrostatic interactions, but also the helical kink, are required for the high affinity TBS1:talin association.
The R3 domain may bind to RIAM TBS1 via a highly conserved interface
Although both the talin R3 and R8 domains contain four helical bundles, their primary sequences are significantly different with a sequence identity of less than 20%. Surprisingly, structure-based sequence alignment of the two domains reveals that the residues that interact with RIAM TBS1 in the R8 domain are completely conserved (75% identical and 25% similar) in the R3 domain (Fig. 2G). To examine whether the R3 domain also interacts with TBS1 via this conserved surface, we assessed the effects of the corresponding mutations on the association of TBS1 with the R2R3 domains and with full-length talin. Both single mutations, S13G and L15Y of TBS1, significantly diminished the TBS1:R2R3 interaction (Fig. 2H). Furthermore, the V871Y mutation in the R3 domain, equivalent to the V1540Y mutation in the R8 domain, also diminished the interaction of RIAM TBS1 with the full-length talin. Strikingly, the V871Y/1540Y mutant abolished the interaction (Fig. 2I), suggesting that the interactions of TBS1 with other talin rod domains are insignificant. These results suggest that RIAM TBS1 interacts with the talin R3 domain in a mode highly similar to the TBS1:R8 interaction via a conserved interface.
Binding determinants in the TBS1:R7R8 complex and the helical kink in TBS1 are required for RIAM:talin co-localization at the PM and for integrin activation
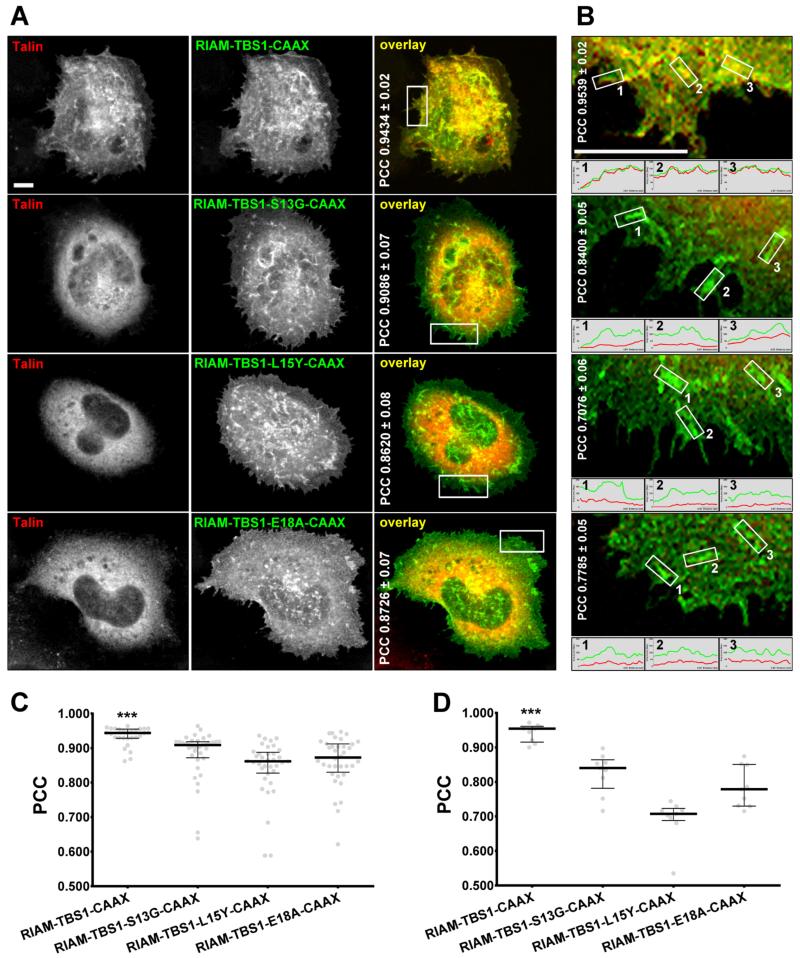
It has been shown that co-transfection of talin and RIAM TBS1, containing a C-terminal CAAX sequence directing it to the PM, in stably expressing αIIbβ3-integrin A5 cells, bears the capacity to both activate integrins and facilitate the cell surface co-clustering of these overexpressed proteins (Lee et al., 2009). To assess the role of TBS1 single mutants in regulating talin recruitment in integrin signaling, we compared the TBS1 single mutants S13G, L15Y, and E18A to GFP-RIAM-TBS1-CAAX control for their ability to co-cluster with mCherry-talin. As expected, wild type TBS1 containing CAAX and talin co-localize and form co-clusters at the lamellae, whereas RIAM TBS1 single mutants S13G, L15Y, and E18A failed to recruit talin into clusters (Fig. 3A,B). General Pearson’s correlation coefficient (PCC) showed modest, but significant higher correlations for wild type: RIAM-TBS1-CAAX (0.9434 ± 0.02) compared to S13G (0.9086 ± 0.07), L15Y (0.8620 ± 0.08) and E18A (0.8726 ± 0.07) (Fig. 3C). Nonetheless, when calculated solely at the cell’s lamellae (see Materials and Methods for details), differences in observed PCC values were more substantial: RIAM-TBS1-CAAX (0.9539 ± 0.02) compared to S13G (0.8400 ± 0.05), L15Y (0.7076 ± 0.06) and E18A (0.7785 ± 0.05) (Fig. 3D). These results, in full concordance with our crystal structure and with the in vitro binding studies, suggest that binding determinants and the helical kink in TBS1 are required for TBS1:talin co-clustering.
Figure 3. Binding determinants and the helical kink are required for the co-clustering of RIAM and talin at the PM.
A5 cells were transiently co-transfected with mCherry-talin and GFP-RIAM-TBS1-CAAX constructs as indicated. 24 hrs post-transfection, the cells were plated on fibrinogen-coated coverslips and allowed to adhere and spread for 1 hr. (A) Fixed cells were imaged using a spinning-disk confocal microscope and representative monochromatic and color-merged images are shown. Note that yellow colors in merged images represent areas of common red and green localizations. Numbers in white correspond to unrefined median PCCs ± SD. (B) Enlarged lamellae sections correspond to the white rectangles marked in A. Median PCC values are shown. Note that small histogram graphs in B are representative of intensity levels of the assorted overexpressed proteins. Bar scales in A and B represent 5 μm. Graphs in C and D respectively correspond to A and B PCC medians (± interquartile range) values. (C) Data calculated from three experimental repetitions counting with 10 images per condition. (D) Data included 9 lamella regions selected from 3 representative cell bodies per condition. *** P < 0.001.
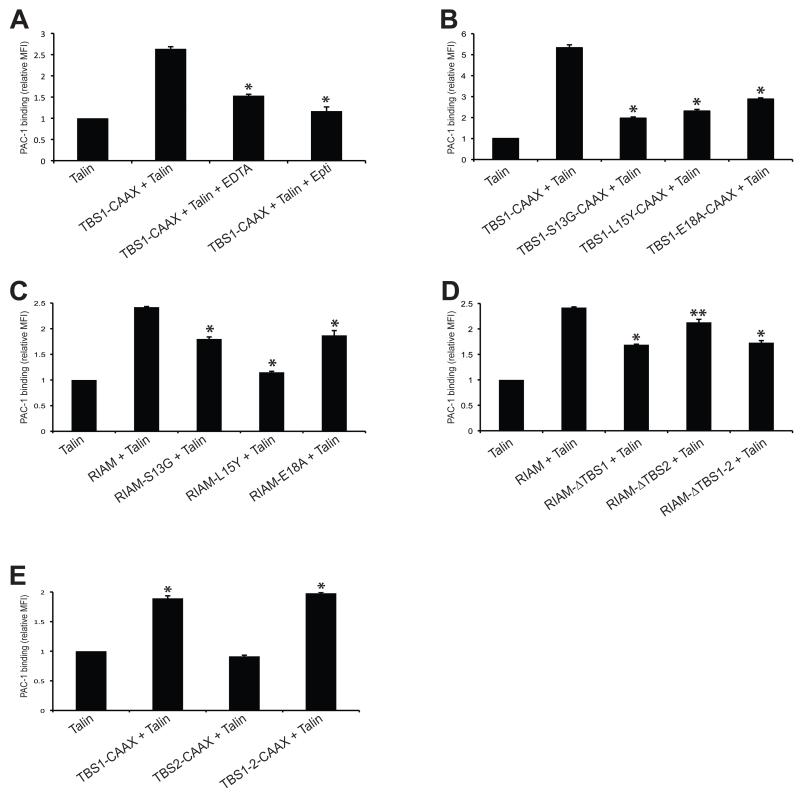
We then examined the effect of the TBS1 mutations on integrin activation in a well-accepted fluorescence-activated cell sorting (FACS) assay. Co-transfection of RIAM TBS1-CAAX and talin in A5 cells promotes activation of αIIbβ3 integrins, and this effect can be inhibited by EDTA and an αIIbβ3 integrin-specific inhibitor, Eptifibatide (Fig. 4A). The TBS1 mutants, including S13G, L15Y, and E18A, significantly diminish integrin activation (Fig. 4B). Full-length RIAM bearing these mutations also exhibit impaired function in promoting integrin activation when co-expressed with talin (Fig. 4C). To compare the effect of TBS1, TBS2, and TBS1-2 on mediating integrin activation, we deleted TBS1, TBS2, or both (ΔTBS1, ΔTBS2, and ΔTBS1-2) in RIAM and assessed their effects on integrin activity when co-expressed with full-length talin. Deletion of TBS1 and TBS1-2 leads to significant loss of integrin activity, whereas the effect of ΔTBS2 is much weaker on altering integrin activity (Fig. 4D). Furthermore, TBS1-CAAX and TBS1-2-CAAX, but not TBS2-CAAX, are capable of promoting the inside-out integrin activation (Fig. 4E). Together, our results suggest that binding determinants in the TBS1:R7R8 complex and the helical kink in the RIAM TBS1 are required for integrin activation, and TBS1, but not TBS2, is essential for talin recruitment in inside-out integrin signaling.
Figure 4. Integrin activity analyses for TBS1 and TBS2.
(A) A5 cells were co-transfected with HA-talin and GFP-tagged RIAM-TBS1-CAAX. Non-specific inhibitor of integrin (EDTA) and specific inhibitor of integrin αIIbβ3 (Eptifibatide) were added into the transfected cells before adding PAC-1 antibody. Integrin activation was detected by PAC-1 binding using FACS. The MFI of cells co-transfected with GFP and HA-Talin was defined as 1. *P < 0.001 compared with RIAM-TBS1-CAAX. (B) A5 cells were co-transfected with HA-talin and indicated mutants of GFP-tagged RIAM-TBS1-CAAX. PAC-1 binding was detected as described in A. *P < 0.001 compared with wild type RIAM-TBS1-CAAX. (C) A5 cells were co-transfected with HA-talin and indicated mutants of GFP-tagged RIAM. *P < 0.001 compared with wild type RIAM. (D) PAC-1 binding was detected using GFP-RIAM full length or RIAM with TBS regions deleted constructs. *P < 0.001, ** P < 0.01 compared with full length RIAM. (E) PAC-1 binding was detected in cells expressing TBS1, TBS2 and TBS1-2 constructs fused with a C-terminal CAAX tail. *P < 0.001 compared with co-transfection of GFP and talin. Data shown in A-E are means ± SD, n = 3.
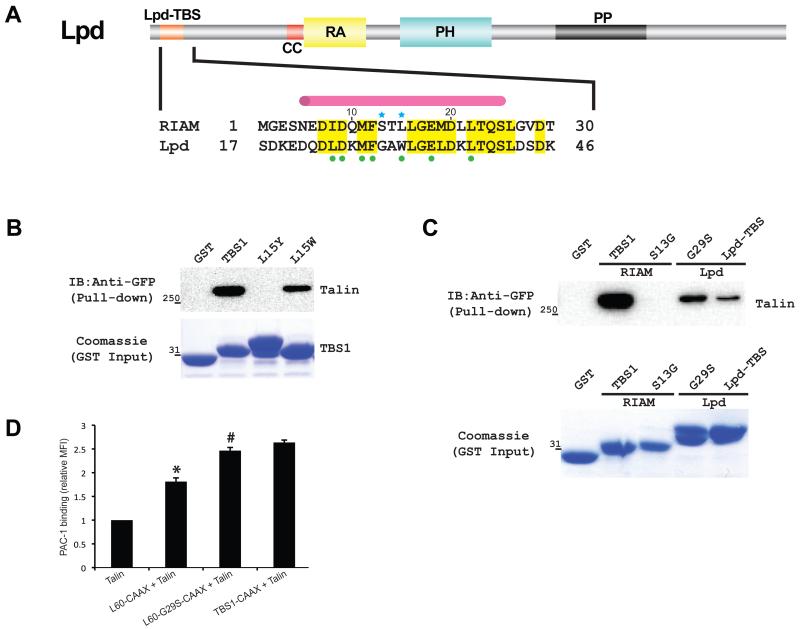
Substitution of RIAM TBS1 with corresponding residues in Lpd reduces talin binding and impairs integrin activation
RIAM and Lpd affect cell adhesion differently despite their similar structural architecture with 59% sequence identity in the TBS1-2 and the RA-PH regions (Krause et al., 2004; Lafuente et al., 2004). Lpd has been identified as an M-Ras effector protein but retains a moderate Rap1-binding affinity owing to an RA-PH functional module highly similar to that of RIAM (Lafuente et al., 2004; Tasaka et al., 2012; Yoshikawa et al., 2007; Zhang et al., 2014). Lpd also possesses all R8-interacting residues in RIAM TBS1 in its N-terminal talin-binding site (Lpd-TBS) except a single substitution of Leu15RIAM with Trp31Lpd (Fig. 5A). Interestingly, the Lpd-like L15W mutant retains substantial binding capacity to talin compared with the L15Y mutant (Fig. 5B). However, the association of RIAM TBS1 with cytoplasmic talin is much stronger than that of Lpd-TBS and talin (Fig. 5C). This result is also confirmed by an in vitro pull-down using a purified talin protein containing R7, R8, and R9 domains (R7R8R9) (Fig. S3A). Furthermore, the effect of Lpd-TBS-CAAX on promoting integrin activity is also much weaker than that of RIAM TBS1-CAAX (Fig. 5D). These results suggest that Lpd-TBS lacks other crucial factors for a tight talin association. We have shown that the Ser13 residue in RIAM stabilizes the helical kink in the R8-bound TBS1. This residue is substituted by a glycine residue in Lpd. The TBS1 fragment (residues 5-25) alone does not possess a stable secondary structure, and the S13G mutation does not alter the conformation of the isolated TBS1 fragment (Fig. S3B). However, the glycine residue is unable to form the hydrogen bond network that stabilizes the helical kink in the R8-bound TBS1. To test whether the lack of a stable helical kink leads to the weak association of talin with Lpd-TBS, we produced a gain-of-function mutation by mutating the corresponding Gly29 residue to serine in the Lpd-TBS fragment. Indeed, Lpd-TBS-G29S enhances the interaction of Lpd-TBS and talin (Fig. 5C). This mutation also improves integrin activation mediated by Lpd-TBS (Fig. 5D). Together, these results suggest that talin preferentially binds to RIAM over Lpd via a kinked helical configuration and favorable side chains of the TBS1 site.
Figure 5.
(A) Schematic representation of Lpd domain organization (Lpd-TBS: talin-binding region, orange; CC: coiled-coil region, red; PP: poly-proline region, black; RA: Ras-association domain, yellow; and PH: Pleckstrin homology domain, cyan). Lpd-TBS shares conserved talin-binding residues (highlighted in yellow) with RIAM TBS1. Talin interacting residues are depicted as green dots. A pink cylinder indicates the helical region in TBS1. Blue stars indicate residues that define the binding specificity in RIAM. (B) The L15Y mutant that disrupts TBS1-talin binding and the L15W mutant that mimics Lpd were assessed for talin-binding by pull-down. Bound GFP-talin was analyzed by Western blot. Input GST-TBS1 (wild type and mutants) levels were shown by Coomassie staining. (C) TBS1-S13G of RIAM and Lpd-TBS-G29S of Lpd were assessed for talin-binding by pull-down. Bound GFP-talin was analyzed by Western blot. Input GST-tagged protein levels were shown by Coomassie staining. (D) PAC-1 binding was detected in cells expressing TBS1-CAAX and Lpd-TBS (L60)-CAAX or G29S mutant. *P < 0.01 compared with co-transfection of GFP and talin. #P < 0.01 compared with L60-CAAX.
DISCUSSION
Talin recruitment to the PM by RIAM is a key step in inside-out integrin signaling that promotes the direct interaction between talin and the C-terminal tail of the integrin β chain (Lee et al., 2009; Wegener et al., 2007). However, how each of the interacting sites in RIAM and talin function together to recruit and activate talin still remains unclear. We demonstrate that cytoplasmic talin only interacts with TBS1, whereas TBS2 does not interacts with talin nor does it facilitate the TBS1:talin interaction. Although weak binding was observed between TBS2 with isolated R2R3 or R7R8 domains of talin, our functional studies confirm that TBS1, but not TBS2, plays an essential role in recruiting talin to the PM that activates integrins (Goult et al., 2013b). Furthermore, TBS1 adopts two distinct helical conformations when bound to talin or vinculin. It has been proposed that the TBS1:vinculin interaction may play an inhibitory role for integrin activation (Goult et al., 2013b). Nevertheless, the B-factors for the R7R8-bound TBS1 and Vd1-bound TBS1 are strikingly different, suggesting that the TBS1 fragment prefers the more stable kinked helical conformation. Talin also possesses 11 vinculin binding sites (VBS) that bind to the Vd1 domain with much higher affinity than that of the Vd1 and TBS1 (Gingras et al., 2005; Goult et al., 2013b; Rees et al., 1990; Yogesha et al., 2011). Thus, it appears that the interaction of RIAM and vinculin is rather transient and the main function of RIAM TBS1 is to recruit cytoplasmic talin to the PM.
The R8 domain that binds to RIAM TBS1 is adjacent to the autoinhibitory R9 domain. This raises the question of whether the PM recruitment of talin via RIAM TBS1 may also contribute to the dissociation of the R9:F3 interdomain complex via steric conflict. To test this, we generated two talin constructs F0R8 (residues 1-1653) and F0R9 (residues 1-1848) that represent the active form and the auto-inhibited form of talin. No significant difference is observed between the two forms in binding to TBS1 (Fig. S4A). We then titrated purified R7R8 domains or R9 domain of talin into the F0R8 or F0R9-expressing cell lysate (Fig. S4B, C). As expected, the R7R8 domains compete with F0R8 and F0R9 for TBS1 binding. However, the R9 domain has no effect on the binding of TBS1 with F0R8 or F0R9. Furthermore, although the E1770A mutation in the R9 domain disrupts the autoinhibitory configuration of talin (Goult et al., 2009; Song et al., 2012), this mutant retains similar binding capacity to TBS1 compared to the wild type talin (Fig. S4D). Together, these data suggest that the autoinhibitory configuration of talin via the F3:R9 interaction does not conflict with the binding to RIAM TBS1. Nevertheless, a recent study of talin revealed that the R1-R2-R3 region also interacts with the F2F3 region (Goult et al., 2013a). Whether this interaction also contributes to talin autoinhibition and how it may be affected by TBS1 binding require further investigation.
As the closest paralogue of RIAM, Lpd is unable to promote integrin-mediated cell adhesion (Lafuente et al., 2004). Our structural and functional studies of the RIAM:talin association provided more insight into the unique role of RIAM in mediating inside-out signaling. We have previously revealed the structural basis of the specific binding of RIAM and Rap1 and demonstrated that Lpd also interacts with Rap1 but with a much lower affinity due to unfavorable side-chain interactions (Zhang et al., 2014). Here we showed that talin binds to RIAM much more strongly than to Lpd. Despite its low affinity to talin, Lpd overexpression may still induce integrin activation in cells overwhelmed by exogenous talin and integrin, suggesting that the low affinity of Lpd to its signaling partners may be compensated by a high concentration of these components (Lee et al., 2009). Nevertheless, the binding specificity required for the Rap1 association and talin recruitment allows only RIAM, but not Lpd, to properly mediate Rap1-induced integrin activation under physiological conditions. Furthermore, Lpd has also been shown to negatively affect inside-out integrin signaling (Lafuente et al., 2004). What is the possible molecular mechanism for this inhibitory effect? We have previously shown that the PM translocation of RIAM may be autoinhibited by its N-terminal region that overlaps with the TBS1-2 region (Wynne et al., 2012). Interestingly, in addition to the Lpd-TBS region that corresponds to the RIAM TBS1, Lpd also possesses another helical region (residues 72-107) similar to the RIAM TBS2 region (Fig. S3C). It is possible that this TBS2-like region, together with the Lpd-TBS region, also inhibits PM translocation of RIAM by mimicking the RIAM TBS1-2 region. Regardless, future efforts to elucidate the molecular basis of RIAM autoinhibition are required to assess this mechanism.
MATERIALS AND METHODS
Plasmid construction and protein purification
The TALIN rod domains were cloned into the modified pET28a expression vector with a His6-tag and a TEV protease cleavage site. The RIAM talin-binding sites were cloned into the pGEX-5X-1 expression vector with a GST-tag. RIAM talin-binding site 1 (TBS1) consists of residues 1 to 30 and talin-binding site 2 (TBS2) consists of residues 27 to 93. Talin and RIAM point mutations were constructed using a site-directed mutagenesis method. Plasmids were transformed into Escherichia coli BL21(DE3) for protein expression. Cells were grown in LB medium containing kanamycin or ampicillin at 37°C until the A600 reached 0.6-0.7, and induced by adding 0.4 mM IPTG with incubation continued overnight at 20°C for talin and for 3 hrs at 37°C for RIAM. Protein purification was carried out at 4°C. The cell pellets were resuspended in 20 mM Tris pH 7.5 with 500 mM NaCl for His-tagged proteins and 50 mM Tris pH 7.5, 150 mM NaCl, and 5 mM DTT for GST-tagged proteins. Cells were then lysed with an EmulsiFlex-C3 homogenizer (AVESTIN, Inc.). Protein samples were extracted from the supernatants using either a HisTrap FF or GSTrap FF column (GE Healthcare). For crystallization, the His-tag was removed via incubation with TEV protease and then further purified using a HiTrap Desalting column and a HisTrap FF column (GE Healthcare).
GST pull-down and Western blotting
For in vitro pull-down assays, purified GST-RIAM proteins were immobilized on glutathione agarose beads and then incubated with purified His-tagged talin proteins in reaction buffer (20 mM Tris-HCl pH 7.5, 100 mM NaCl, and 2 mM DTT) to a total volume of 250 μl on a rotator for 1 hr at 4°C. Binding curves were fit to a single-site (saturating) binding model using SigmaPlot (Systat Software). For cell lysate pull-down assays, HEK 293T cells were transfected with various GFP-talin constructs using Lipofectamine 2000 reagent (Invitrogen) and lysed 48 hrs post-transfection with lysis buffer (20 mM Tris-HCl pH 7.5, 150 mM NaCl, 5 mM MgCl2, 1% (v/v) Triton X-100, 1 mM DTT, and complete protease inhibitor cocktail (Roche)). The cell lysates (400 μg per reaction) were clarified by centrifugation, and supernatants were mixed with the purified GST-RIAM proteins immobilized on 50 μl of 50% slurry of glutathione agarose beads and incubated on a rotator for 1 hr at 4°C. The bound proteins were washed three times in 500 μl of the lysis buffer and were eluted using an elution buffer (20 mM Tris-HCl pH 7.5, 100 mM NaCl, 2 mM DTT, and 10 mM reduced glutathione) at 4°C. The proteins were resolved by SDS-PAGE and detected by Coomassie staining or Western blotting. The Immobilon-P transfer membranes (EMD Millipore) were blocked with TBST buffer (20 mM Tris-HCl pH 7.5, 150 mM NaCl, 0.1% (v/v) Tween 20) containing 5% (w/v) BSA for 1 hour and then incubated with anti-His (Sigma) or anti-GFP antibody (Clontech) for 1 hour at room temperature followed by a second incubation with HRP-conjugated secondary antibody (Santa Cruz Biotechnology). The blots were visualized with the SuperSignal West Pico Chemiluminescent Substrate (Thermo Scientific) and detected using the FluorChem E imager (ProteinSimple).
X-ray crystallography
The RIAM peptide consisting of residues 5 to 25 (Genemed Synthesis, Inc.) was dissolved in 100 mM Tris pH 8.0 with 100 mM NaCl and the final pH was adjusted to pH 7-8. Talin R7R8 (11 mg/mL) was incubated with the RIAM peptide on ice at a 1:3 molar ratio prior to the crystallization setup. The talin R7R8 domains-RIAM peptide complex was crystallized using the hanging-drop vapor diffusion method at room temperature, with the droplets containing equal volumes of protein and reservoir solution. The crystals were grown in 100 mM NaCl, 20% (w/v) polyethylene glycol 3350 and 20% (v/v) ethylene glycol from microseeding. The crystals were grown for 2-3 days and then flash-frozen in liquid nitrogen prior to the diffraction experiments.
Final X-ray diffraction data were collected using beamline X29 of the National Synchrotron Light Source at the Brookhaven National Laboratory (Upton, NY). Data were processed using the HKL-2000 package (Otwinowski and Minor, 1997). The complex structure of the talin R7R8 domains and the RIAM TBS1 was determined by molecular replacement using talin rod residues 1359-1659 (PDB ID 2X0C) as the initial search model (Gingras et al., 2010). Structural refinement was performed using REFMAC (Murshudov et al., 1997) and Coot (Emsley and Cowtan, 2004). The models were validated with SFCHECK (Vaguine et al., 1999) and MolProbity (http://molprobity.biochem.duke.edu/). Data collection and refinement statistics are listed in Table 1. Structural figures were generated using the PyMOL program suite (http://pymol.sourceforge.net). The final atomic model contains residues 1357-1651 of chain A (talin) and residues 5-25 of chain B (RIAM). The atomic coordinates and structure factor have been deposited in the Protein Data Bank under the accession number 4W8P.
Confocal imaging & Pearson’s correlation coefficient analyses
Co-clustering of RIAM and talin was performed as previously described (Zhang et al., 2014). Briefly, CHO cells stably expressing αIIβ3 integrins (A5 cells) were co-transfected with mCherry-talin and either GFP-RIAM-TBS1-CAAX or the assorted mutants. 24 hrs post co-transfection, the cells were allowed to adhere and spread on glass cover slips coated with 10 μg/mL fibrinogen for 1 hr at 37°C and then fixed with 4% (w/v) paraformaldehyde for 30 mins at room temperature. The fixed cells were washed with PBS buffer and mounted with ProLong Gold antifade reagent (Molecular Probes). Images were acquired using a 60X 1.45 Pan APO TIRF oil immersion objective using a Nikon TE-2000U microscope (Optical Apparatus Co., Ardmore, PA) equipped with the Ultraview spinning-disc confocal (Perkin-Elmer Life Sciences, Boston, MA). Using Velocity 6.3, the software was instructed to acquire double channel simultaneous sequential images at 488nm (GFP) and 568nm (mCherry) wavelengths. Image files were captured as 0.1 μm-thick Z-slices and processed to obtain maximum projections. Ten cells per condition were analyzed corresponding to three independent experimental repetitions. Pearson’s correlation coefficients (PCCs) were obtained from whole cells as well as lamellar locations using MetaMorph offline 7.8 software (Molecular Devices, Downingtown, PA). Cellular lamellae areas were selected using RIAM’s GFP intensities to detect the cell edge, while cellular centroids were identified using the same MetaMorph software. The outermost 5 microns measured from the cell’s centroid constituted lamella positive areas.
Integrin activation assay
A5 cells were co-transfected with HA-talin and GFP-tagged RIAM or Lpd vectors. 24 hrs after transfection, the cells were resuspended by trypsinization and washed with Tyrode’s buffer (136.9 mM NaCl, 10 mM HEPES, 5.5 mM glucose, 11.9 nM NaHCO3, 2.7 mM KCl, 0.5 mM CaCl2, 1.5 mM MgCl2, and 0.4 mM NaH2PO4, pH 7.4). PAC-1 mAb (Becton Dickinson Immunocytometry System), which specifically recognizes activated integrin, was then added to the cell suspension and incubated for 1 hr at 4°C. The cells were washed by Tyrode’s buffer and incubated with Alexa Fluor 647 conjugated goat anti-mouse IgM antibody for 1 hr on ice and resuspended in cold PBS. To specifically inhibit integrin activation, 10 mM EDTA or 10 μM Eptifibatide was incubated with cell suspension for 20 mins before adding PAC1 antibody. Stained cells were analyzed using the LSRII FACS instrument (BD Scientific). The collected data were processed using the FlowJo software package and expression levels of talin were examined by Western blot (Fig. S5). Only GFP positive cells were gated on to analyze PAC-1 binding. The mean fluorescence intensity (MFI) of PAC-1 channel of each transfectant was normalized to that of the GFP empty vector and HA-talin co-transfection to obtain the relative MFI. The data were represented by the means ± SD from at least three experiments. The equal expression of talin under each condition was verified by Western blot using anti-HA antibody (Fig. S5). The unpaired t test was performed to calculate the P value using GraphPad software.
CD spectroscopy
CD spectra were recorded in a 0.1-cm pathlength quartz cell on a CD spectrometer (model 62A DS; Aviv Associates, Lakewood, NJ). Proteins (R2R3 and R7R8) or peptides (TBS1 and TBS1-S13G) were measured at 0.5 mg/mL concentration. Spectra were recorded at 25°C over a wavelength of 200–300 nm.
Supplementary Material
ACKNOWLEDGEMENTS
We thank M. Xu and H. Roder for collecting the CD spectra and discussion, J.R. Peterson for manuscript comments, and the beamline staff of X29 at the National Synchrotron Light Source, Brookhaven National Laboratory for technical support. This work was supported by NIH core grant CA006927 (To Fox Chase Cancer Center). Y.C.C. performed the crystallographic studies and a portion of the biochemistry experiments. H.Z. performed most of the biochemistry and in-cell experiments. M.L.B. and T.P. performed protein production and contributed to the crystallographic studies. J.F.B. and E.C. performed the fluorescent imaging analysis. Y.C.C, H.Z., M.L.B., and E.C. contributed to manuscript preparation. J.W. supervised the project and was the principal manuscript author.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Arias-Salgado EG, Lizano S, Sarkar S, Brugge JS, Ginsberg MH, Shattil SJ. Src kinase activation by direct interaction with the integrin beta cytoplasmic domain. Proc Natl Acad Sci U S A. 2003;100:13298–13302. doi: 10.1073/pnas.2336149100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banno A, Ginsberg MH. Integrin activation. Biochem Soc Trans. 2008;36:229–234. doi: 10.1042/BST0360229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bivona TG, Wiener HH, Ahearn IM, Silletti J, Chiu VK, Philips MR. Rap1 up-regulation and activation on plasma membrane regulates T cell adhesion. J Cell Biol. 2004;164:461–470. doi: 10.1083/jcb.200311093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bos JL, de Bruyn K, Enserink J, Kuiperij B, Rangarajan S, Rehmann H, Riedl J, de Rooij J, van Mansfeld F, Zwartkruis F. The role of Rap1 in integrin-mediated cell adhesion. Biochem Soc Trans. 2003;31:83–86. doi: 10.1042/bst0310083. [DOI] [PubMed] [Google Scholar]
- Chang YC, Zhang H, Brennan ML, Wu J. Crystal structure of Lamellipodin implicates diverse functions in actin polymerization and Ras signaling. Protein Cell. 2013;4:211–219. doi: 10.1007/s13238-013-2082-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen VB, Arendall WB, 3rd, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemmons DR, Maile LA. Interaction between insulin-like growth factor-I receptor and alphaVbeta3 integrin linked signaling pathways: cellular responses to changes in multiple signaling inputs. Mol Endocrinol. 2005;19:1–11. doi: 10.1210/me.2004-0376. [DOI] [PubMed] [Google Scholar]
- Colo GP, Lafuente EM, Teixido J. The MRL proteins: Adapting cell adhesion, migration and growth. Eur J Cell Biol. 2012 doi: 10.1016/j.ejcb.2012.03.001. [DOI] [PubMed] [Google Scholar]
- Desiniotis A, Kyprianou N. Significance of talin in cancer progression and metastasis. Int Rev Cell Mol Biol. 2011;289:117–147. doi: 10.1016/B978-0-12-386039-2.00004-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Gingras AR, Bate N, Goult BT, Patel B, Kopp PM, Emsley J, Barsukov IL, Roberts GC, Critchley DR. Central region of talin has a unique fold that binds vinculin and actin. J Biol Chem. 2010;285:29577–29587. doi: 10.1074/jbc.M109.095455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gingras AR, Ziegler WH, Frank R, Barsukov IL, Roberts GC, Critchley DR, Emsley J. Mapping and consensus sequence identification for multiple vinculin binding sites within the talin rod. J Biol Chem. 2005;280:37217–37224. doi: 10.1074/jbc.M508060200. [DOI] [PubMed] [Google Scholar]
- Goksoy E, Ma YQ, Wang X, Kong X, Perera D, Plow EF, Qin J. Structural basis for the autoinhibition of talin in regulating integrin activation. Mol Cell. 2008;31:124–133. doi: 10.1016/j.molcel.2008.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goult BT, Bate N, Anthis NJ, Wegener KL, Gingras AR, Patel B, Barsukov IL, Campbell ID, Roberts GC, Critchley DR. The structure of an interdomain complex that regulates talin activity. J Biol Chem. 2009;284:15097–15106. doi: 10.1074/jbc.M900078200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goult BT, Xu XP, Gingras AR, Swift M, Patel B, Bate N, Kopp PM, Barsukov IL, Critchley DR, Volkmann N, et al. Structural studies on full-length talin1 reveal a compact auto-inhibited dimer: implications for talin activation. J Struct Biol. 2013a;184:21–32. doi: 10.1016/j.jsb.2013.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goult BT, Zacharchenko T, Bate N, Tsang R, Hey F, Gingras AR, Elliott PR, Roberts GC, Ballestrem C, Critchley DR, et al. RIAM and vinculin binding to talin are mutually exclusive and regulate adhesion assembly and turnover. J Biol Chem. 2013b;288:8238–8249. doi: 10.1074/jbc.M112.438119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hood JD, Frausto R, Kiosses WB, Schwartz MA, Cheresh DA. Differential alphav integrin-mediated Ras-ERK signaling during two pathways of angiogenesis. J Cell Biol. 2003;162:933–943. doi: 10.1083/jcb.200304105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang MM, Lipfert L, Cunningham M, Brugge JS, Ginsberg MH, Shattil SJ. Adhesive ligand binding to integrin alpha IIb beta 3 stimulates tyrosine phosphorylation of novel protein substrates before phosphorylation of pp125FAK. J Cell Biol. 1993;122:473–483. doi: 10.1083/jcb.122.2.473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphries JD, Wang P, Streuli C, Geiger B, Humphries MJ, Ballestrem C. Vinculin controls focal adhesion formation by direct interactions with talin and actin. J Cell Biol. 2007;179:1043–1057. doi: 10.1083/jcb.200703036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krause M, Leslie JD, Stewart M, Lafuente EM, Valderrama F, Jagannathan R, Strasser GA, Rubinson DA, Liu H, Way M, et al. Lamellipodin, an Ena/VASP ligand, is implicated in the regulation of lamellipodial dynamics. Dev Cell. 2004;7:571–583. doi: 10.1016/j.devcel.2004.07.024. [DOI] [PubMed] [Google Scholar]
- Lafuente EM, van Puijenbroek AA, Krause M, Carman CV, Freeman GJ, Berezovskaya A, Constantine E, Springer TA, Gertler FB, Boussiotis VA. RIAM, an Ena/VASP and Profilin ligand, interacts with Rap1-GTP and mediates Rap1-induced adhesion. Dev Cell. 2004;7:585–595. doi: 10.1016/j.devcel.2004.07.021. [DOI] [PubMed] [Google Scholar]
- Lai MT, Hua CH, Tsai MH, Wan L, Lin YJ, Chen CM, Chiu IW, Chan C, Tsai FJ, Jinn-Chyuan Sheu J. Talin-1 overexpression defines high risk for aggressive oral squamous cell carcinoma and promotes cancer metastasis. J Pathol. 2011;224:367–376. doi: 10.1002/path.2867. [DOI] [PubMed] [Google Scholar]
- Lawrence MC, Colman PM. Shape complementarity at protein/protein interfaces. J Mol Biol. 1993;234:946–950. doi: 10.1006/jmbi.1993.1648. [DOI] [PubMed] [Google Scholar]
- Lee HS, Lim CJ, Puzon-McLaughlin W, Shattil SJ, Ginsberg MH. RIAM activates integrins by linking talin to ras GTPase membrane-targeting sequences. J Biol Chem. 2009;284:5119–5127. doi: 10.1074/jbc.M807117200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menter DG, Dubois RN. Prostaglandins in cancer cell adhesion, migration, and invasion. Int J Cell Biol. 2012;2012:723419. doi: 10.1155/2012/723419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monkley SJ, Pritchard CA, Critchley DR. Analysis of the mammalian talin2 gene TLN2. Biochem Biophys Res Commun. 2001;286:880–885. doi: 10.1006/bbrc.2001.5497. [DOI] [PubMed] [Google Scholar]
- Moser M, Legate KR, Zent R, Fassler R. The tail of integrins, talin, and kindlins. Science. 2009;324:895–899. doi: 10.1126/science.1163865. [DOI] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- Nieswandt B, Moser M, Pleines I, Varga-Szabo D, Monkley S, Critchley D, Fassler R. Loss of talin1 in platelets abrogates integrin activation, platelet aggregation, and thrombus formation in vitro and in vivo. J Exp Med. 2007;204:3113–3118. doi: 10.1084/jem.20071827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Parsons JT. Focal adhesion kinase: the first ten years. J Cell Sci. 2003;116:1409–1416. doi: 10.1242/jcs.00373. [DOI] [PubMed] [Google Scholar]
- Petrich BG, Marchese P, Ruggeri ZM, Spiess S, Weichert RA, Ye F, Tiedt R, Skoda RC, Monkley SJ, Critchley DR, et al. Talin is required for integrin-mediated platelet function in hemostasis and thrombosis. J Exp Med. 2007;204:3103–3111. doi: 10.1084/jem.20071800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratnikov BI, Partridge AW, Ginsberg MH. Integrin activation by talin. J Thromb Haemost. 2005;3:1783–1790. doi: 10.1111/j.1538-7836.2005.01362.x. [DOI] [PubMed] [Google Scholar]
- Rees DJ, Ades SE, Singer SJ, Hynes RO. Sequence and domain structure of talin. Nature. 1990;347:685–689. doi: 10.1038/347685a0. [DOI] [PubMed] [Google Scholar]
- Sakamoto S, McCann RO, Dhir R, Kyprianou N. Talin1 promotes tumor invasion and metastasis via focal adhesion signaling and anoikis resistance. Cancer Res. 2010;70:1885–1895. doi: 10.1158/0008-5472.CAN-09-2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song X, Yang J, Hirbawi J, Ye S, Perera HD, Goksoy E, Dwivedi P, Plow EF, Zhang R, Qin J. A novel membrane-dependent on/off switch mechanism of talin FERM domain at sites of cell adhesion. Cell Res. 2012;22:1533–1545. doi: 10.1038/cr.2012.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tadokoro S, Shattil SJ, Eto K, Tai V, Liddington RC, de Pereda JM, Ginsberg MH, Calderwood DA. Talin binding to integrin beta tails: a final common step in integrin activation. Science. 2003;302:103–106. doi: 10.1126/science.1086652. [DOI] [PubMed] [Google Scholar]
- Tasaka G, Negishi M, Oinuma I. Semaphorin 4D/Plexin-B1-mediated M-Ras GAP activity regulates actin-based dendrite remodeling through Lamellipodin. J Neurosci. 2012;32:8293–8305. doi: 10.1523/JNEUROSCI.0799-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaguine AA, Richelle J, Wodak SJ. SFCHECK: a unified set of procedures for evaluating the quality of macromolecular structure-factor data and their agreement with the atomic model. Acta Crystallogr D Biol Crystallogr. 1999;55:191–205. doi: 10.1107/S0907444998006684. [DOI] [PubMed] [Google Scholar]
- Watanabe N, Bodin L, Pandey M, Krause M, Coughlin S, Boussiotis VA, Ginsberg MH, Shattil SJ. Mechanisms and consequences of agonist-induced talin recruitment to platelet integrin alphaIIbbeta3. J Cell Biol. 2008;181:1211–1222. doi: 10.1083/jcb.200803094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegener KL, Partridge AW, Han J, Pickford AR, Liddington RC, Ginsberg MH, Campbell ID. Structural basis of integrin activation by talin. Cell. 2007;128:171–182. doi: 10.1016/j.cell.2006.10.048. [DOI] [PubMed] [Google Scholar]
- Wynne JP, Wu J, Su W, Mor A, Patsoukis N, Boussiotis VA, Hubbard SR, Philips MR. Rap1-interacting adapter molecule (RIAM) associates with the plasma membrane via a proximity detector. J Cell Biol. 2012;199:317–330. doi: 10.1083/jcb.201201157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye F, Snider AK, Ginsberg MH. Talin and kindlin: the one-two punch in integrin activation. Front Med. 2014;8:6–16. doi: 10.1007/s11684-014-0317-3. [DOI] [PubMed] [Google Scholar]
- Yogesha SD, Sharff A, Bricogne G, Izard T. Intermolecular versus intramolecular interactions of the vinculin binding site 33 of talin. Protein Sci. 2011;20:1471–1476. doi: 10.1002/pro.671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshikawa Y, Satoh T, Tamura T, Wei P, Bilasy SE, Edamatsu H, Aiba A, Katagiri K, Kinashi T, Nakao K, et al. The M-Ras-RA-GEF-2-Rap1 pathway mediates tumor necrosis factor-alpha dependent regulation of integrin activation in splenocytes. Mol Biol Cell. 2007;18:2949–2959. doi: 10.1091/mbc.E07-03-0250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Chang YC, Brennan ML, Wu J. The structure of Rap1 in complex with RIAM reveals specificity determinants and recruitment mechanism. J Mol Cell Biol. 2014;6:128–139. doi: 10.1093/jmcb/mjt044. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.