Abstract
It is now clear that there is a diversity of circular RNAs in biological systems. Circular RNAs can be produced by the direct ligation of 5′ and 3′ ends of linear RNAs, as intermediates in RNA processing reactions, or by “backsplicing,” wherein a downstream 5′ splice site (splice donor) is joined to an upstream 3′ splice site (splice acceptor). Circular RNAs have unique properties including the potential for rolling circle amplification of RNA, the ability to rearrange the order of genomic information, protection from exonucleases, and constraints on RNA folding. Circular RNAs can function as templates for viroid and viral replication, as intermediates in RNA processing reactions, as regulators of transcription in cis, as snoRNAs, and as miRNA sponges. Herein, we review the breadth of circular RNAs, their biogenesis and metabolism, and their known and anticipated functions.
Keywords: circRNA, circular RNA, backsplicing, splicing, intron, intermediates, nonlinear
INTRODUCTION
In addition to the classic tRNA, mRNA, and rRNAs, cells contain a striking diversity of additional RNA types including miRNAs, lncRNAs, piRNAs, siRNAs, tmRNAs, sRNAs, tiRNAs, eRNAs, snoRNAs, snRNAs, and other noncoding RNAs. A growing component of this family of diverse RNA molecules is the increasing numbers and types of circular RNAs.
Five general types of RNA circles have been identified including viroids, intermediates in RNA processing reactions, and products of spliceosomal backsplicing of normal nuclear pre-mRNAs in eukaryotes (see Table 1, discussed below in detail). RNAs are circularized through 5′–3′ linkages as well as through 2′–5′ linkages formed by branchpoint nucleophilic attack during splicing.
TABLE 1.
Types and characteristics of RNA circles
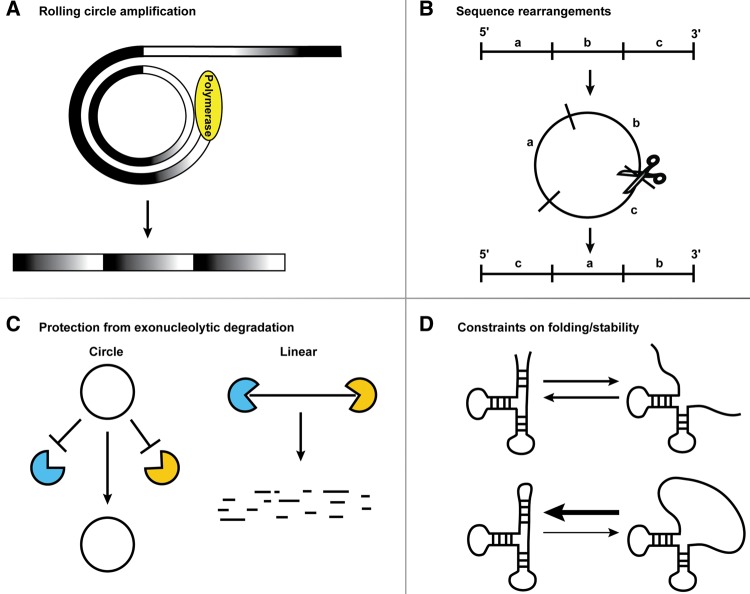
RNA circles may be prevalent in biology because they have distinct properties that can be advantageous (Fig. 1). For example, RNA circles can serve as templates for rolling circle replication, which is a highly efficient manner of generating many copies of a specific RNA. Circular RNAs also have the ability to rearrange the order of genetic information present in the DNA genome. Additionally, RNA circles can be highly stable due to their inaccessibility to exonucleases. Finally, circular RNAs can create a constraint on RNA folding, which may be beneficial in certain contexts.
FIGURE 1.
Properties of circles. Circular RNAs can serve as a template for rolling circle amplification (A), provide a means to rearrange sequences (B), evade exonucleolytic degradation (C), and constrain the RNA folding and structural stability (D).
In this review, we discuss the diversity of circular RNAs, their modes of biogenesis and how their circularity may affect their function.
DIFFERENT TYPES OF RNA CIRCLES
Circular RNA genomes—viroid and hepatitis delta virus circles
One class of circular single-stranded RNAs (ssRNAs) are viroids and the hepatitis delta virus (HDV). Circular RNAs are functionally important for viroids and HDV because circularity allows for rolling circle RNA replication wherein multiple genomic copies are produced from a single initiation event.
Viroids are small (∼250–400 bases) nonencapsulated circular ssRNAs that do not encode any proteins, and infect and replicate in plants as free RNA molecules (Ding 2010; Flores et al. 2011; Navarro et al. 2012). The hepatitis delta virus (HDV) also has a circular ssRNA genome. It encodes a single protein, and is a subviral satellite of hepatitis B virus (HBV) that gets encapsulated within HBV virions and thereby coinfects with HBV (Flores et al. 2011; Abbas and Afzal 2013; Alves et al. 2013).
Although there are some differences between how different viroids and HDV replicate, the common theme is that the viroid or HDV RNA recruits a host DNA-dependent RNA polymerase to initiate rolling circle replication of the RNA genome (Fig. 1). Following the production of linear concatemers, these RNAs are separated into monomer lengths by either ribozyme or enzymatic cleavage, leaving 5′-hydroxyl and 2′,3′-cyclic phosphate termini (Flores et al. 1999). Finally, a key step is ligation of the 5′ and 3′ ends of these linear monomeric molecules to produce negative stranded circular RNAs. The negative strand circles then undergo rolling circle amplification to produce a large number of positive strand circular genomes, which then propagate the infection. The exact mechanism of ligation is unclear; it could be a reversal of the ribozyme cleavage reaction in some cases, or it could be due to the action of a host cellular ligase that joins the 3′ and 5′ ends of the RNA.
Circular RNA from introns
Sequences excised from precursor RNA in the cases of group I introns, group II introns, spliceosomal splicing, and tRNA splicing can form circular molecules. Some of these excised introns may be functional RNAs.
The first discovery of a circular excised intron was the rRNA intervening sequence of Tetrahymena (Grabowski et al. 1981; Kruger et al. 1982; Zaug et al. 1983). These, and other group I introns, can form circular RNAs through autocatalytic ribozyme action requiring a guanosine cofactor (Nielsen et al. 2003). The circles can comprise the full-length intron when formed by hydrolysis at the 3′ splice site in conjunction with transesterification at the 5′ splice site, but more often the circles are truncated at the 5′ end of the intron when the excised linear intron undergoes a further internal transesterification reaction. More recently, circular products have been found that contain an additional residue, originating from the cofactor, at the site of circularization (Vicens and Cech 2009). Although circular forms of group I introns can be detected in vivo (e.g., Dalgaard and Garrett 1992; Vader et al. 1999, 2002), whether the circular form of the intron has any functional significance remains to be established.
Group II introns are self-splicing ribozymes found in bacteria and in some eukaryotic organelle genes. Transesterification reactions result in intron lariat products that are circularized via 2′–5′ linkages. These lariat RNAs can undergo reverse splicing, inserting into both DNA and RNA, which may contribute to their genetic mobility (Mörl and Schmelzer 1990; Lehmann and Schmidt 2003). Group II introns can also generate full-length circles in vivo in what might be an alternate splicing pathway initiated by a trans reaction involving two introns to first release the 3′ splice site for subsequent transesterification with the 5′ splice site (e.g., Murray et al. 2001; Li-Pook-Than and Bonen 2006; Molina-Sánchez et al. 2006).
A similar class of circular introns are those derived from eukaryotic spliceosomal splicing. Circular intronic RNAs (ciRNAs) are intron lariats that have been circularized at the branchpoint 2′–5′ linkage, degraded from the 3′ end up to the branchpoint, but somehow escaped debranching and degradation, and are thus stabilized (Fig. 2; Zhang et al. 2013). Such stable lariat introns were first described on a gene by gene basis in the early 1990s and include the nuclear accumulation of introns from the Delta transcript in Drosophila (Kopczynski and Muskavitch 1992) and the T cell receptor-β mRNA in T cells (Qian et al. 1992). More recently, deep sequencing has revealed that a subset of introns is strikingly over-represented and presumably has features blocking the normal degradation pathway of debranching and exonuclease digestion. For example, analysis of mammalian ciRNAs indicates that debranching can be inhibited by specific sequences near the 5′ splice site and branchpoint, which possibly form a structure that limits access of the debranching enzyme to the branchpoint (Zhang et al. 2013).
FIGURE 2.
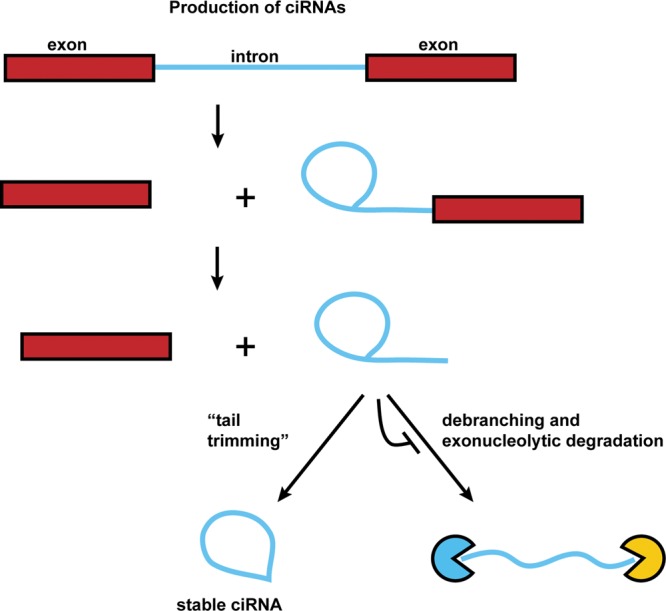
Production of ciRNAs. Circular intronic RNAs are produced by eukaryotic spliceosome-mediated splicing. The lariat intron generated from the splicing reaction evades normal debranching and degradation, and instead the 3′ “tail” downstream from the branchpoint is trimmed resulting in a stable ciRNA.
The functions of ciRNAs remain to be fully understood. However, one recent study argues that some ciRNAs enhance the transcription of the genes from which they are produced. The key observations are that antisense oligos against a ciRNA intron reduce expression of the resident gene, while oligos against other introns, or the region between the branchpoint and the 3′ splice site of the ciRNA intron, which would be lacking in the stable ciRNA intron, do not have an effect (Zhang et al. 2013). Moreover, ciRNAs can be shown to associate with RNA polymerase II (Zhang et al. 2013). Taken together, these observations suggest that ciRNAs might modulate RNA polymerase II in cis and thereby alter the expression of their gene. Consistent with this idea, ciRNAs accumulate in the nucleus and, where examined, can be localized to their sites of transcription (Kopczynski and Muskavitch 1992; Qian et al. 1992; Zhang et al. 2013).
Circular RNA intermediates in RNA processing reactions
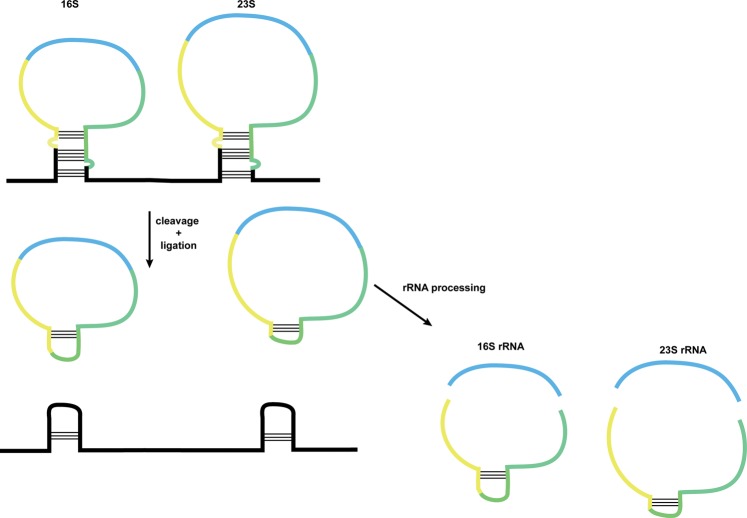
Circular RNAs can be produced as intermediates in RNA processing reactions. For example, in some archaea the 16S and 23S rRNAs are excised from a precursor as linear molecules that are then circularized by a ligase, much like the tRNA introns, followed by further RNA processing reactions to produce the mature rRNAs (Fig. 3; Tang et al. 2002; Danan et al. 2012). More strikingly, in some algae and archaea circular RNAs are critical intermediates in the biogenesis of permuted tRNA genes where the order of the “5′” and “3′” regions of the gene are reversed in the genome (Fig. 1B; Soma et al. 2007; Soma 2014). In order to correctly produce mature tRNA, the tRNA precursor is first circularized by a cellular ligase, followed by cleavage between the mature 5′ and 3′ ends.
FIGURE 3.
In some archaea, circular RNA is formed as an intermediate in ribosomal RNA processing. Ribosomal RNA precursors containing the bulge–helix–bulge motif are cleaved and ligated, forming circular intermediates which are further processed to release the 16S and 23S rRNA. rRNA regions are indicated by light blue; upstream and downstream regions contained within the circular intermediate are yellow and green, respectively.
The benefit of circularization in these types of reactions is that it allows elasticity in RNA sequence order. The arrangement of elements can be altered through circularization of a linear RNA and subsequent relinearization at a different position (Fig. 1B). The existence of RNA processing reactions that ligate 5′ and 3′ ends followed by internal cleavage of such an RNA circle raises the intriguing possibility that there are additional yet-to-be-described RNA processing pathways that produce RNAs with reordered segments as compared with the genomic template.
Circular, noncoding RNAs in archaea with snRNP functions
In some archaeal species, circular RNAs have been described for RNase P RNA and for some snoRNAs, which can be produced from excised circularized tRNA introns (Lykke-Andersen et al. 1997; Salgia et al. 2003; Starostina et al. 2004; Danan et al. 2012). Circularization of functional noncoding RNAs may be beneficial in that it protects them from exonucleases and could also increase proper folding by imposing structural constraints on the RNA (Fig. 1C,D). Such circularization of functional RNAs might be particularly important in thermophiles where circularization could limit thermal denaturation of a terminal helix between the 5′ and 3′ ends of an RNA.
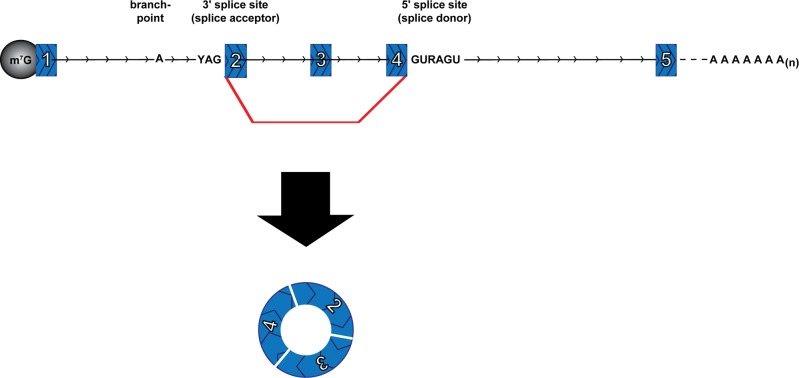
Circular RNA from eukaryotic ‘backsplicing’
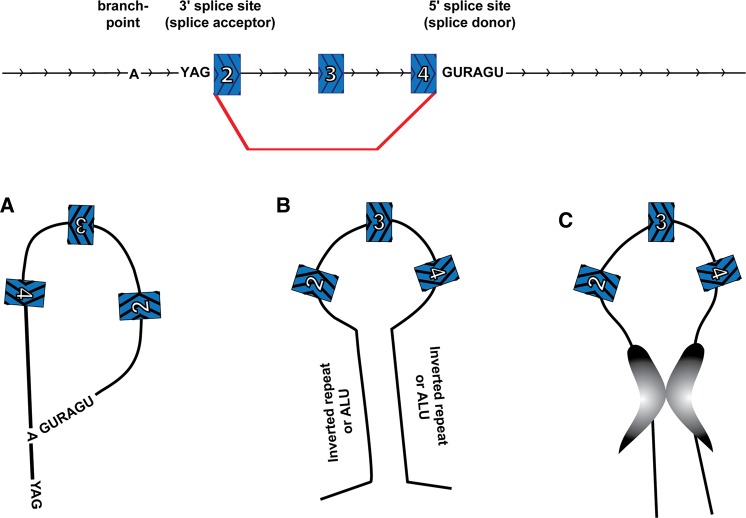
Another type of RNA circles in eukaryotes comes from spliceosome-mediated pre-mRNA splicing wherein a downstream 5′ splice site (splice donor) is joined to an upstream 3′ splice site (splice acceptor), thereby generating a circular product (circRNA) (Fig. 4). This process is referred to as backsplicing. Backspliced RNAs were first detected on specific genes as cases wherein the exons were joined together in something other than an expected linear order, so-called “scrambled exons” or “exon shuffling” (Nigro et al. 1991; Cocquerelle et al. 1992, 1993; Capel et al. 1993; Zaphiropoulos 1996, 1997). Genome-wide RNA-sequencing analysis has now suggested there are a staggering number of circles produced by backsplicing (Table 2; Salzman et al. 2012, 2013; Jeck et al. 2013; Memczak et al. 2013; Ashwal-Fluss et al. 2014; Guo et al. 2014; Zhang et al. 2014). Recent studies have estimated that there are upward of 25,000 different circRNAs in human cells, derived from >14% of transcribed genes in fibroblasts (Jeck et al. 2013).
FIGURE 4.
A circRNA is formed by a backsplice event. Spliceosome-mediated splicing joins a 5′ splice site (splice donor) of a downstream exon with a 3′ splice site (splice acceptor) of an upstream exon to yield a circular RNA with a “scrambled exon” junction between exon 4 and exon 2. (Red line) Backsplice and (arrowheads) 5′–3′ direction.
TABLE 2.
Evidence for human circRNAs (transcriptome-wide)
IDENTIFICATION AND VALIDATION OF BACKSPLICED CIRCLES
Since most circles arising from backsplicing are identified based on the interpretation of RNA-seq reads, an important issue is how many of these “circles” really are circular RNAs. As detailed in Tables 2 and 3, a variety of different assays are used to identify and validate RNA circles.
TABLE 3.
Evidence for eukaryotic circRNAs (gene-specific)
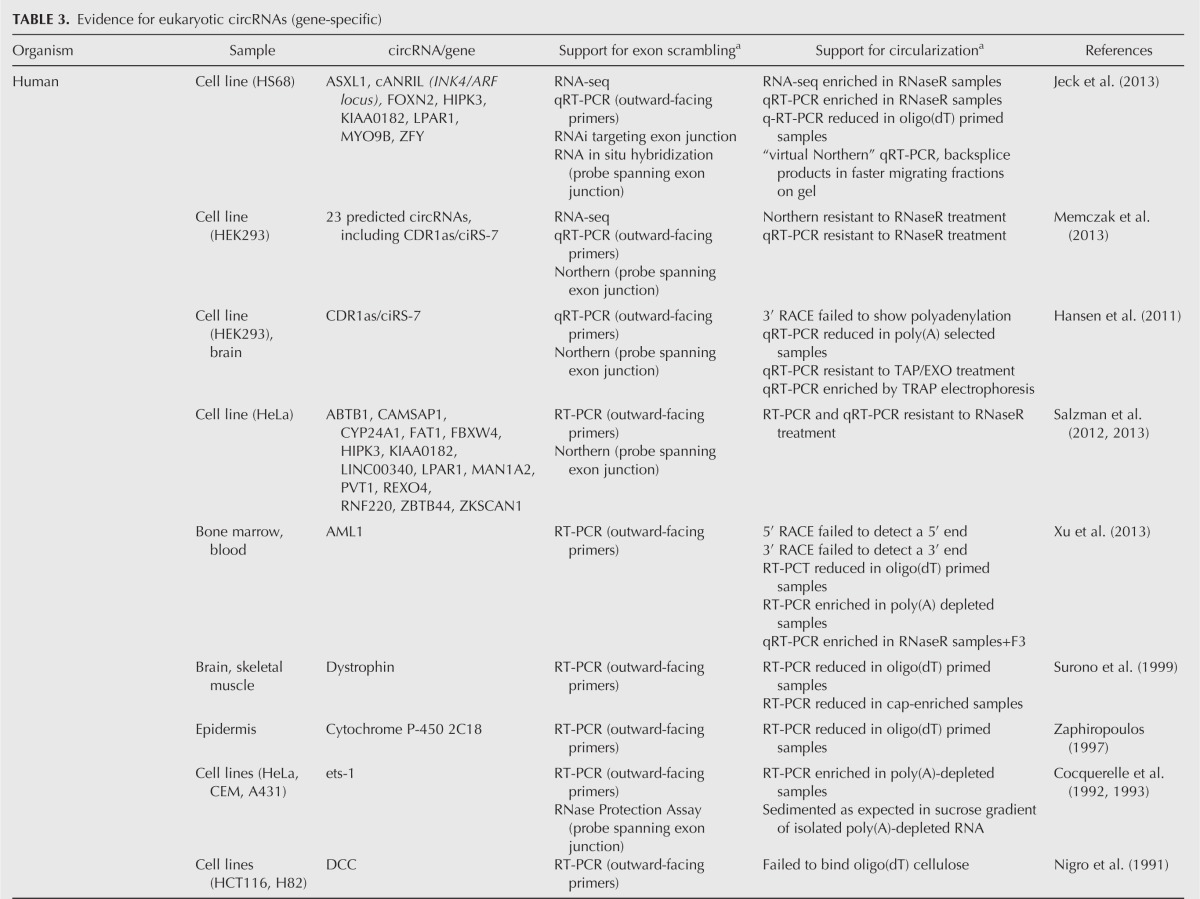
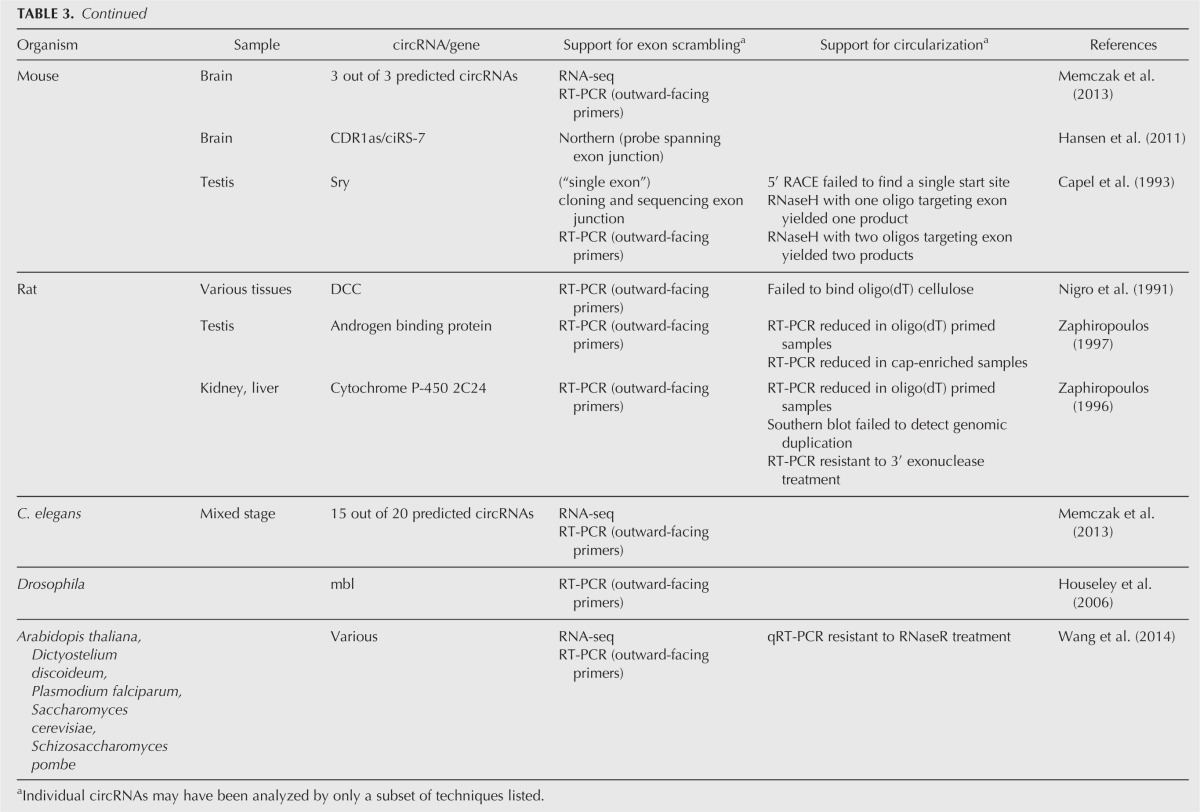
Scrambled exons
One criterion for a circRNA is the presence of scrambled exons, where RNA sequences are out of order relative to the corresponding genome due to the joining of downstream and upstream splice sites. On individual genes, scrambled exons are detected by RT-PCR using outward-facing primers for a specific gene (Fig. 5) or by Northern blot using a probe spanning the exon junction. A variety of computational approaches have now been used to identify vast numbers of scrambled sequences in genome-wide RNA-seq data sets. For example, Salzman et al. (2012) initially detected thousands of potential circRNAs by using paired-end RNA-seq reads that could not be aligned to a canonical transcriptome database but could align to a custom database of all possible exon–exon pair junctions. Memczak et al. (2013) designed a computational pipeline that mapped read ends from a genomic anchor position to a “breakpoint” flanked by GU/AG sequence, indicating exon junctions. Guo et al. (2014) similarly mapped reads with a “dual alignment” and GU/AG junctions. Jeck et al. (2013) used MapSplice (Wang et al. 2010), an algorithm that segments reads allowing detection of noncanonical junctions and fusions, and identified the “backsplice” events where spliced junctions occurred in a nonlinear order. These methods allowed identification of candidate circRNAs by detection of scrambled exons in RNA-seq data as well as calculations of relative abundance compared with corresponding nonscrambled transcripts.
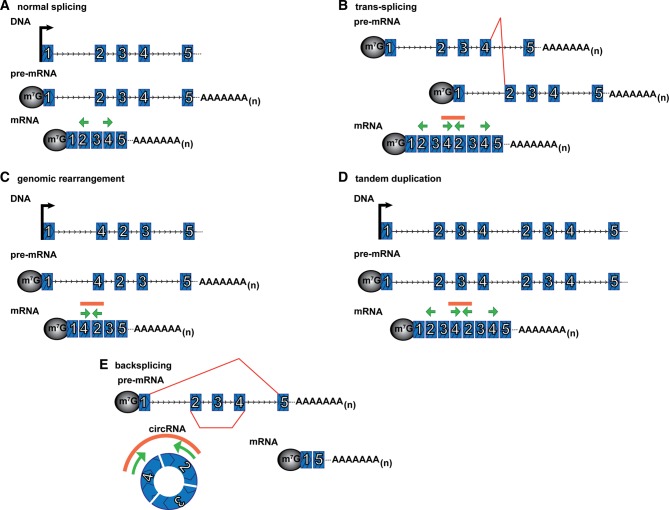
FIGURE 5.
Scrambled exon junctions can be formed from different mechanisms. Outward-facing primers in exons 2 and 4 (green arrows) will not yield an RT-PCR product if the exons are ordered linearly (A), but will yield a product, indicated by orange line above primers, if the exons are scrambled in the mRNA. Scrambled exons could occur through trans-splicing (B), genomic rearrangements (C), including tandem duplication within the DNA (D), or backsplicing (E). (Red lines) Unusual (nonsequential) splicing, (arrowheads) 5′–3′ direction, (green arrows) outward-facing PCR primers, and (orange line) RT-PCR product.
It is notable that many of the circRNAs annotated by each individual group are not detected by the other groups (Guo et al. 2014). This may be because each method uses a different set of thresholds and criteria to define a circRNA, and therefore may result in a different list of circRNAs. For example, whereas Memczak et al. (2013) required at minimum two independent junction-spanning reads, Guo et al. (2014) required at least 10% abundance of the circular fraction. Jeck et al. (2013) required enrichment in the RNase R sample, and read fusions within 2 Mb, but did not require splice sites.
Scrambled exon–exon junctions can also originate by alternative mechanisms including DNA rearrangements, tandem duplications in the DNA, and trans-splicing (Fig. 5). Additionally, an apparently scrambled sequence could be generated by artifacts of RT-PCR or RNA-seq. In principle, scrambled exons arising from trans-splicing or genomic rearrangements can at least sometimes be identified computationally if the mate-pair of a read spanning a scrambled exon–exon junction maps outside the region that would be included within the circle of those exons (Salzman et al. 2012; Guo et al. 2014). Therefore scrambled exons are consistent with a circRNA but not sufficient to prove circularity.
Additional criteria for circularity
Due to the absence of 5′ and 3′ ends, exonuclease resistance has emerged as a common method for enriching for circRNAs. For example, resistance to RNase R, which degrades most linear RNAs without destroying circular molecules, is used as a criterion for circularity (Suzuki et al. 2006; Vincent and Deutscher 2006; Suzuki and Tsukahara 2014). Some groups used RNase R enrichment prior to RNA-seq as a criteria for identifying global circRNAs (Jeck et al. 2013; Ashwal-Fluss et al. 2014; Zhang et al. 2014). Alternatively, treatment with TAP (tobacco acid pyrophosphatase), which removes the 5′ cap, and a 5′–3′ exonuclease also depletes samples of linear mRNAs.
Additional support for the circularity of scrambled exon transcripts can include the absence of linear mRNA characteristics. Linear mRNA, but not circRNA, transcripts are typically capped and polyadenylated. Therefore RT-PCR reactions or RNA-seq of cap-enriched, poly(A) selected, or oligo(dT) reverse-transcription-primed samples preferentially detect linear (including those from trans-splicing), but not circular, scrambled exon RNAs (Fig. 5B). Methods which ligate adaptor molecules onto RNA ends also favor linear transcript detection. Additionally, scrambled exons resulting from DNA rearrangements or tandem duplications would not only produce linear RNAs, but would be detectable in genomic DNA PCR reactions (Fig. 5C,D). Together, scrambled exon enrichment in an RNase R treated sample and depletion in a linear-selected (i.e., poly(A) or cap selected) sample provides reasonable confidence of a circRNA.
Another means of verifying the circularity of nucleic acid molecules is “TRAP electrophoresis.” Loading samples in melted agarose into the well of a preformed gel can selectively trap circular molecules by electrophoresis, allowing linear ones to migrate into the gel matrix. Following “TRAP electrophoresis” RNA can be recovered by phenol–chloroform extraction and ethanol precipitation (Schindler et al. 1982; Akerman and Cole 2002; Hansen et al. 2011).
Circles can also be distinguished by characteristic differences in mobility on a gel compared with their corresponding linear molecules (Jeck et al. 2013; Memczak et al. 2013) (although this can be due to differences in number of included nucleotides). Indeed, group I intron circles from Tetrahymena rRNA were initially detected as a distinct band migrating aberrantly on nondenaturing and denaturing electrophoresis gels (Grabowski et al. 1981). One- and 2D polyacrylamide gel electrophoresis using different percentage gels can also reveal characteristic migration patterns of circular RNAs (Ruskin et al. 1984; Tabak et al. 1988). Single-hit nicking or targeted RNase H cleavage should convert the circle band to a single linear species with expected electrophoretic mobility. Conversely, cleavage of a linear RNA would result in two products on a gel. Finally, electron microscopy may be used to visualize RNA circles under irreversibly denaturing conditions to remove secondary structure (Sanger et al. 1976; Arnberg et al. 1980; Halbreich et al. 1980; Grabowski et al. 1981).
PROPERTIES OF EUKARYOTIC circRNAS
There are several noteworthy properties of circRNAs produced by backsplicing. First, there is an incredible diversity of such circRNAs. They are encoded by a variety of different genes, of different sizes and expression levels, with different sized introns flanking the backsplice sites. They can comprise a single exon or multiple exons. The size of the spliced circle molecule can range from under 100 nt to over 4 kb. A gene that gives rise to a circRNA could encode a single isoform, or many (Salzman et al. 2013; Zhang et al. 2014). Some, but not all, are associated with exon skipping alternative splicing of the corresponding linear transcript (Zaphiropoulos 1996). Some circRNAs contain some of the same exons as annotated mRNAs, and some are from noncoding RNAs. Some are antisense to annotated genes.
Second, the existence of circRNAs is a widespread phenomenon and may be a ubiquitous feature of pre-mRNA splicing (Jeck et al. 2013; Memczak et al. 2013; Salzman et al. 2013; Guo et al. 2014). Publicly available data sets from a wide variety of cell types and organisms can be searched on the circBase website (circbase.org) developed by the Rajewsky laboratory (Glažar et al. 2014). Nearly 2000 circRNAs have been predicted in mouse sequences, and over 700 in C. elegans (Memczak et al. 2013). Drosophila has RNA sequencing evidence for over 800 scrambled exon spliced junctions (Salzman et al. 2013). Additionally, circRNAs have been recently reported in plants (Arabidopsis thaliana), yeast (Schizosaccharomyces pombe and Saccharomyces cerevisiae), and protists (Plasmodium falciparum and Dictyostelium discoideum) (Wang et al. 2014).
Some circRNAs are conserved between different species. For example, genes encoding circles in one species are more likely to also encode circles in the other (Salzman et al. 2013; Jeck et al. 2013; Memczak et al. 2013). Intriguingly, Jeck et al. (2013) identified 69 circRNAs that contained precisely conserved backsplice junctions between mouse and human. Whether conservation of specific circRNAs reveals functional roles, or conserved features of pre-mRNAs that also promote circRNA formation remains to be determined.
Fourth, circRNAs are expressed at a variety of different levels, and at altered levels in different cell types. Attempts to calculate this have indicated that the majority of circRNAs are present in quite low abundance (Jeck et al. 2013; Salzman et al. 2013; Guo et al. 2014). For most genes with both linear and circular RNA species, the amount of circRNAs is between 0.1% and 10% of the linear amounts, with most <1%. However, for at least 50 genes the circular form is thought to be more abundant that the linear RNAs (Jeck et al. 2013). Additionally, there are genes with scrambled exons/predicted circles with no corresponding forward splicing at the backsplice sites. Given the wide variation in circRNA levels, one anticipates that low abundance circRNAs may primarily arise due to errors in pre-mRNA splicing, while cells have also taken advantage of backsplicing both to regulate alternative splicing and to produce functional circRNAs.
Biogenesis, transport, and degradation of circRNAs
circRNA biogenesis
The features of circRNA biogenesis, subcellular localization, and degradation are just beginning to emerge. Two features of pre-mRNAs have been suggested to enhance the formation of circRNAs by spliceosomal backsplicing. First, an intron lariat generated by skipping alternative exons may also bring nonlinear splice sites together (shown in Fig. 6A). Indeed, there is some correlation between exons detected in circularized transcripts and those skipped in mature mRNAs (Fig. 5E; Zaphiropoulos 1996, 1997; Surono et al. 1999). In fact Jeck et al. (2013) identified splicing sequencing reads corresponding to the exon skipping events of 45% of the circRNAs. This implies that a single pre-mRNA transcript can yield both a linear spliced mRNA and a circRNA of the skipped exons, and that either splicing event may influence the other. This illustrates that backsplicing may itself be modulated by the efficiency of any given linear splicing event and alternative splicing choices. As such, backsplicing will be subject to all of the same regulatory inputs as alternative splicing, which may provide an explanation for the cell-type expression of different circRNAs (Salzman et al. 2013).
FIGURE 6.
Possible mechanisms for promoting circularization by bringing backsplice sites into close proximity. (A) An intron lariat resulting from exon skipping. (B) Flanking inverted repeats or ALU elements forms an extended base-paired structure. (C) Interactions between RNA-binding proteins form a bridge between the flanking introns. (Red line) backsplice and (arrowheads) 5′–3′ direction.
A second feature that can promote backsplicing is structure of the pre-mRNA that brings the two splice sites into close proximity. Bioinformatic analysis demonstrated that circularized exons are more likely to contain inverted ALU repeat elements in the flanking introns than noncircularized exons (Jeck et al. 2013). These ALU repeats likely base-pair in the RNA transcript, and bring the backsplice splice sites into proximity with one another, facilitating circularization (Fig. 6B). Inverted repeats can allow spliced circularization in cell culture (Dubin et al. 1995; Hansen et al. 2013a; Zhang et al. 2014) and cell lysates (Pasman et al. 1996). One anticipates that there will be additional mechanisms to facilitate nonlinear splice site pairings, and these may include other cis-acting RNA sequences or trans-acting factors (Fig. 6C). Recently, it was demonstrated that MBL can enable backsplicing of an exon within its own pre-mRNA gene (Ashwal-Fluss et al. 2014).
Cellular localization of circRNAs
Following biogenesis, circRNAs appear to be efficiently transported to the cytosol since the endogenous circRNAs that have been analyzed are cytoplasmic (Nigro et al. 1991; Capel et al. 1993; Cocquerelle et al. 1993; Salzman et al. 2012; Hansen et al. 2013a; Jeck et al. 2013; Memczak et al. 2013). The manner by which circRNAs are tagged for export is not yet clear but may involve the deposition of an exon-junction complex during splicing, which could then recruit mRNA export factors (Le Hir et al. 2001).
Once in the cytosol, the circRNAs analyzed are not associated with ribosomes (Capel et al. 1993; Jeck et al. 2013; Guo et al. 2014) suggesting that circRNAs are not generally translated. Two experiments using RNA fluorescence in situ hybridization argue that circRNAs can accumulate in RNP granules such as P-bodies (Hansen et al. 2013a; Jeck et al. 2013). For example, the ciRS-7 circRNA, which appears to modulate miRNA function (see below), was seen in cytoplasmic foci that colocalize with the P-body marker Dcp1a in a manner dependent on miR-7. Given that Argonaut proteins (components of the RISC complex) can localize to P-bodies (Liu et al. 2005), it makes sense that circRNAs bound to miRNAs would be also found in P-bodies. Whether additional circRNAs are targeted to specific locations in the cytosol remains to be determined.
circRNA degradation
circRNAs would be predicted to escape the predominant mechanisms of mRNA degradation, which are based on exonucleolytic degradation from either the 3′ or 5′ end. In addition, quality control systems that trigger endonuclease cleavage on mRNAs with premature nonsense codons, or strong translation stalls, would not be expected to work on circRNAs since they are not translated. Consistent with these generalities, specific endogenous circRNAs that have been analyzed are quite stable (Cocquerelle et al. 1993; Jeck et al. 2013). This high stability of circRNAs suggests that circRNAs may be actually produced at very low rates but then accumulate to detectable levels due to their high stability. One anticipates that there will be degradation pathways for at least some circRNAs, presumably involving endonuclease cleavage. Indeed, the miRNA sponge ciRS-7/CDR1as is subject to the slicer activity of Ago2 in a miR-671-dependent manner (Hansen et al. 2011).
circRNA function
circRNAs can function to modulate miRNA activity
The diversity, abundance, and conservation of circRNAs suggest that at least some circRNAs will have biological functions. To date, this has only been clearly demonstrated for circRNA ciRS-7/CDR1as (for circular RNA sponge for miR-7 or CDR1 antisense) and Sry, which function as miRNA sponges (Hansen et al. 2013a; Memczak et al. 2013). The circRNA formed from the transcript antisense to the CDR1 gene contains over 70 conserved seed matches for the miRNA miR-7. Although this circRNA is densely Ago-protein-bound, the seed matches are limited in complementarity, thereby preventing bound miR-7 from causing slicing of the circRNA. This circRNA has a single target site match for a different miRNA, with extended and near-perfect complementarity. This miRNA, miR-671, can direct miRNA-mediated cleavage of ciRS-7/CDR1as (Hansen et al. 2011). Two groups demonstrated that circRNA ciRS-7/CDR1as was conserved, stable, and modulated the activity of the associated miR-7 (Hansen et al. 2013a; Memczak et al. 2013). This represents a newly discovered way to regulate miRNA activity, through sequestering specific miRNA complexes and potentially by releasing them again when the circRNA is cleaved (Hansen et al. 2013b). Although the idea of competing endogenous RNA as a regulator is not novel (Ebert and Sharp 2010; Bak and Mikkelsen 2014), albeit controversial (Denzler et al. 2014), the stability and other properties of circular RNAs may afford these sponges additional advantages.
Murine Sry (Sex-determining region Y), the gene responsible for mammalian sex determination, can encode a testis-specific circular transcript (Capel et al. 1993). This 1.2-kb single-exon circRNA contains 16 miR-138 target sites, and likely also functions as a micro RNA sponge (Hansen et al. 2013a). Although both the circRNAs ciRS-7/CDR1as and Sry function as competing endogenous RNAs (sponges), the role is based on specific RNA sequences and repeats. Therefore, it remains to be determined whether or not this is a general principle of the thousands of circRNAs or a gene-specific function.
Production of circRNAs could function to regulate alternative splicing
The production of circRNAs is almost certain to also play a role in the regulation of alternative splicing. Such a role is simply due to the fact that the splicing pattern of any pre-mRNA is dictated by competitions between alternative pairing of 5′ and 3′ splice sites (Ashwal-Fluss et al. 2014; Zhang et al. 2014). In this case once a backsplice occurs, it removes the internal exons and dictates that the remaining pre-mRNA splice in an alternative manner, or potentially be degraded (see Fig. 5E). In this manner, the production of circRNAs plays a role in the regulation of alternative splicing as a process, although the resulting circRNAs may not be functional.
Other possible functions of circRNAs
A variety of other functions have been proposed for circRNAs (Hentze and Preiss 2013), including “sponging” other factors, such as RNA-binding proteins or RNPs (ribonucleoprotein complexes). CircRNAs could bind and sequester such factors, preventing them from acting, or perhaps provide a protective reservoir of these molecules. This could be especially potent if the binding sites on the circRNA are repeated as is the case for miR-7 on ciRS-7/CDR1as. However, given the low abundance of most circRNAs, this may not substantially affect target molecule levels (Denzler et al. 2014).
CircRNAs could be used to bind and store components, to sort and deliver factors to particular subcellular locations, or as scaffolds for the assembly of other complexes or reactions. Although some circRNAs have start codons and/or reasonable open reading frames, and translation can occur on engineered circular RNAs containing IRESes in vitro (Chen and Sarnow 1995), none of the endogenous circRNAs analyzed are associated with ribosomes and therefore evidence for translation of circRNA is, at this time, still absent (Capel et al. 1993; Jeck et al. 2013; Guo et al. 2014).
circRNAs in disease
It is possible that circRNAs will affect human disease. For example, changes in the level of ciRS-7/CDR1as (the miR-7 circRNA sponge) alters the levels of miR-7 target genes (both endogenous and reporter) (Hansen et al. 2013a; Memczak et al. 2013). This is relevant since miR-7 has been characterized as having both oncogenic and tumor suppressive properties, and its function has been implicated in Parkinson disease, stress handling, brain development, and cellular proliferation (Hansen et al. 2013b; Memczak et al. 2013). Additionally, this circRNA is less abundant in sporadic Alzheimer's disease CA1 hippocampal regions relative to age-matched controls (Lukiw 2013). Finally, expression of ciRS-7/CDR1as is induced by stable overexpression of the PrPC prion protein, leading to speculation regarding its potential involvement in prion disease (Hansen et al. 2013b).
At least one other (non-miRNA-sponge) circRNA is known to correlate with a disease phenotype. ANRIL is a long noncoding RNA near the INK4/ARF locus that encodes multiple RNA isoforms, including circular variants. Intriguingly, expression of circular ANRIL (cANRIL) correlates with atherosclerotic vascular disease risk (Burd et al. 2010).
Unanswered questions
The field of circular RNAs has a wealth of unanswered questions. While it is clear that natural circRNAs are an abundant, diverse, and conserved class of RNA molecules, it is not yet apparent what, if any, biological role the majority of circRNAs play. Very little is known about the control of the backsplicing events that generate circRNAs, and there is limited information about the localization and degradation of most circRNAs.
Given the paucity of information on circRNA function, it is possible that circRNAs, sponges, and nonsponges, will emerge with roles in both disease phenotypes and disease therapeutics. Considering the stability and cytoplasmic localization of circRNAs, engineered circRNAs could be exploited for a range of molecular tools or therapies. Circular RNA constructs have been engineered both in vitro and in vivo using group I permuted intron–exon (PIE) sequences (Puttaraju and Been 1992; Ford and Ares 1994; Perriman and Ares 1998), by including complementary sequences in the sequences flanking spliceosomal backsplice sites (Pasman et al. 1996; Hansen et al. 2013a; Ashwal-Fluss et al. 2014; Zhang et al. 2014), as well as through splinted RNA ligation (Chen and Sarnow 1995). Conceivably, this technology could be applied to effectively sequester not only microRNAs or other RNAs of choice, but any RNA-binding protein with known sequence or structure specificity. Perhaps one could also control the release of these stored molecules through directed cleavage of the circRNA. Controlled inhibition and subsequent derepression of any cellular process could be an incredibly valuable tool for research in diverse areas.
ACKNOWLEDGMENTS
We thank Anne Webb for help in figure preparation and Carolyn Decker for critical reading of the manuscript. This work was supported by NIH grant GM045443 and funds from the Howard Hughes Medical Institute.
Footnotes
Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.047126.114.
REFERENCES
- Abbas Z, Afzal R 2013. Life cycle and pathogenesis of hepatitis D virus: a review. World J Hepatol 5: 666–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akerman B, Cole KD 2002. Electrophoretic capture of circular DNA in gels. Electrophoresis 23: 2549–2561. [DOI] [PubMed] [Google Scholar]
- Alves C, Branco C, Cunha C 2013. Hepatitis δ virus: a peculiar virus. Adv Virol 2013: 560105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnberg AC, Van Ommen GJ, Grivell LA, Van Bruggen EF, Borst P 1980. Some yeast mitochondrial RNAs are circular. Cell 19: 313–319. [DOI] [PubMed] [Google Scholar]
- Ashwal-Fluss R, Meyer M, Pamudurti NR, Ivanov A, Bartok O, Hanan M, Evantal N, Memczak S, Rajewsky N, Kadener S 2014. circRNA biogenesis competes with pre-mRNA splicing. Mol Cell 10.1016/j.molcel.2014.08.019. [DOI] [PubMed] [Google Scholar]
- Bak RO, Mikkelsen JG 2014. miRNA sponges: soaking up miRNAs for regulation of gene expression. Wiley Interdiscip Rev RNA 5: 317–333. [DOI] [PubMed] [Google Scholar]
- Burd CE, Jeck WR, Liu Y, Sanoff HK, Wang Z, Sharpless NE 2010. Expression of linear and novel circular forms of an INK4/ARF-associated non-coding RNA correlates with atherosclerosis risk. PLoS Genet 6: e1001233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capel B, Swain A, Nicolis S, Hacker A, Walter M, Koopman P, Goodfellow P, Lovell-Badge R 1993. Circular transcripts of the testis-determining gene Sry in adult mouse testis. Cell 73: 1019–1030. [DOI] [PubMed] [Google Scholar]
- Chen CY, Sarnow P 1995. Initiation of protein synthesis by the eukaryotic translational apparatus on circular RNAs. Science 268: 415–417. [DOI] [PubMed] [Google Scholar]
- Cocquerelle C, Daubersies P, Majérus MA, Kerckaert JP, Bailleul B 1992. Splicing with inverted order of exons occurs proximal to large introns. EMBO J 11: 1095–1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cocquerelle C, Mascrez B, Hétuin D, Bailleul B 1993. Mis-splicing yields circular RNA molecules. FASEB J 7: 155–160. [DOI] [PubMed] [Google Scholar]
- Dalgaard JZ, Garrett RA 1992. Protein-coding introns from the 23S rRNA-encoding gene form stable circles in the hyperthermophilic archaeon Pyrobaculum organotrophum. Gene 121: 103–110. [DOI] [PubMed] [Google Scholar]
- Danan M, Schwartz S, Edelheit S, Sorek R 2012. Transcriptome-wide discovery of circular RNAs in Archaea. Nucleic Acids Res 40: 3131–3142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denzler R, Agarwal V, Stefano J, Bartel DP, Stoffel M 2014. Assessing the ceRNA hypothesis with quantitative measurements of miRNA and target abundance. Mol Cell 54: 766–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding B 2010. Viroids: self-replicating, mobile, and fast-evolving noncoding regulatory RNAs. Wiley Interdiscip Rev RNA 1: 362–375. [DOI] [PubMed] [Google Scholar]
- Dubin RA, Kazmi MA, Ostrer H 1995. Inverted repeats are necessary for circularization of the mouse testis Sry transcript. Gene 167: 245–248. [DOI] [PubMed] [Google Scholar]
- Ebert MS, Sharp PA 2010. Emerging roles for natural microRNA sponges. Curr Biol 20: R858–R861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores R, Navarro JA, de la Peña M, Navarro B, Ambrós S, Vera A 1999. Viroids with hammerhead ribozymes: some unique structural and functional aspects with respect to other members of the group. Biol Chem 380: 849–854. [DOI] [PubMed] [Google Scholar]
- Flores R, Grubb D, Elleuch A, Nohales MÁ, Delgado S, Gago S 2011. Rolling-circle replication of viroids, viroid-like satellite RNAs and hepatitis δ virus: variations on a theme. RNA Biol 8: 200–206. [DOI] [PubMed] [Google Scholar]
- Ford E, Ares M Jr 1994. Synthesis of circular RNA in bacteria and yeast using RNA cyclase ribozymes derived from a group I intron of phage T4. Proc Natl Acad Sci 91: 3117–3121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glažar P, Papavasileiou P, Rajewsky N 2014. circBase: a database for circular RNAs. RNA 20: 1666–1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabowski PJ, Zaug AJ, Cech TR 1981. The intervening sequence of the ribosomal RNA precursor is converted to a circular RNA in isolated nuclei of Tetrahymena. Cell 23: 467–476. [DOI] [PubMed] [Google Scholar]
- Guo JU, Agarwal V, Guo H, Bartel DP 2014. Expanded identification and characterization of mammalian circular RNAs. Genome Biol 15: 409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halbreich A, Pajot P, Foucher M, Grandchamp C, Slonimski P 1980. A pathway of cytochrome b mRNA processing in yeast mitochondria: specific splicing steps and an intron-derived circular DNA. Cell 19: 321–329. [DOI] [PubMed] [Google Scholar]
- Hansen TB, Wiklund ED, Bramsen JB, Villadsen SB, Statham AL, Clark SJ, Kjems J 2011. miRNA-dependent gene silencing involving Ago2-mediated cleavage of a circular antisense RNA. EMBO J 30: 4414–4422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, Kjems J 2013a. Natural RNA circles function as efficient microRNA sponges. Nature 495: 384–388. [DOI] [PubMed] [Google Scholar]
- Hansen TB, Kjems J, Damgaard CK 2013b. Circular RNA and miR-7 in cancer. Cancer Res 73: 5609–5612. [DOI] [PubMed] [Google Scholar]
- Hentze MW, Preiss T 2013. Circular RNAs: splicing's enigma variations. EMBO J 32: 923–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houseley JM, Garcia-Casado Z, Pascual M, Paricio N, O'Dell KMC, Monckton DG, Artero RD 2006. Noncanonical RNAs from transcripts of the Drosophila muscleblind gene. J Hered 97: 253–260. [DOI] [PubMed] [Google Scholar]
- Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J, Marzluff WF, Sharpless NE 2013. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA 19: 141–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopczynski CC, Muskavitch MA 1992. Introns excised from the Δ primary transcript are localized near sites of Δ transcription. J Cell Biol 119: 503–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruger K, Grabowski PJ, Zaug AJ, Sands J, Gottschling DE, Cech TR 1982. Self-splicing RNA: autoexcision and autocyclization of the ribosomal RNA intervening sequence of Tetrahymena. Cell 31: 147–157. [DOI] [PubMed] [Google Scholar]
- Le Hir H, Gatfield D, Izaurralde E, Moore MJ 2001. The exon–exon junction complex provides a binding platform for factors involved in mRNA export and nonsense-mediated mRNA decay. EMBO J 20: 4987–4997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann K, Schmidt U 2003. Group II introns: structure and catalytic versatility of large natural ribozymes. Crit Rev Biochem Mol Biol 38: 249–303. [DOI] [PubMed] [Google Scholar]
- Li-Pook-Than J, Bonen L 2006. Multiple physical forms of excised group II intron RNAs in wheat mitochondria. Nucleic Acids Res 34: 2782–2790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Valencia-Sanchez MA, Hannon GJ, Parker R 2005. MicroRNA-dependent localization of targeted mRNAs to mammalian P-bodies. Nat Cell Biol 7: 719–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukiw WJ 2013. Circular RNA (circRNA) in Alzheimer's disease (AD). Front Genet 4: 307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lykke-Andersen J, Aagaard C, Semionenkov M, Garrett RA 1997. Archaeal introns: splicing, intercellular mobility and evolution. Trends Biochem Sci 22: 326–331. [DOI] [PubMed] [Google Scholar]
- Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer M, et al. 2013. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 495: 333–338. [DOI] [PubMed] [Google Scholar]
- Molina-Sánchez MD, Martinez-Abarca F, Toro N 2006. Excision of the Sinorhizobium meliloti group II intron RmInt1 as circles in vivo. J Biol Chem 281: 28737–28744. [DOI] [PubMed] [Google Scholar]
- Mörl M, Schmelzer C 1990. Group II intron RNA-catalyzed recombination of RNA in vitro. Nucleic Acids Res 18: 6545–6551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray HL, Mikheeva S, Coljee VW, Turczyk BM, Donahue WF, Bar-Shalom A, Jarrell KA 2001. Excision of group II introns as circles. Mol Cell 8: 201–211. [DOI] [PubMed] [Google Scholar]
- Navarro B, Gisel A, Rodio M-E, Delgado S, Flores R, Di Serio F 2012. Viroids: how to infect a host and cause disease without encoding proteins. Biochimie 94: 1474–1480. [DOI] [PubMed] [Google Scholar]
- Nielsen H, Fiskaa T, Birgisdottir AB, Haugen P, Einvik C, Johansen S 2003. The ability to form full-length intron RNA circles is a general property of nuclear group I introns. RNA 9: 1464–1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nigro JM, Cho KR, Fearon ER, Kern SE, Ruppert JM, Oliner JD, Kinzler KW, Vogelstein B 1991. Scrambled exons. Cell 64: 607–613. [DOI] [PubMed] [Google Scholar]
- Pasman Z, Been MD, Garcia-Blanco MA 1996. Exon circularization in mammalian nuclear extracts. RNA 2: 603–610. [PMC free article] [PubMed] [Google Scholar]
- Perriman R, Ares M Jr 1998. Circular mRNA can direct translation of extremely long repeating-sequence proteins in vivo. RNA 4: 1047–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puttaraju M, Been MD 1992. Group I permuted intron–exon (PIE) sequences self-splice to produce circular exons. Nucleic Acids Res 20: 5357–5364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian L, Vu MN, Carter M, Wilkinson MF 1992. A spliced intron accumulates as a lariat in the nucleus of T cells. Nucleic Acids Res 20: 5345–5350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruskin B, Krainer AR, Maniatis T, Green MR 1984. Excision of an intact intron as a novel lariat structure during pre-mRNA splicing in vitro. Cell 38: 317–331. [DOI] [PubMed] [Google Scholar]
- Salgia SR, Singh SK, Gurha P, Gupta R 2003. Two reactions of Haloferax volcanii RNA splicing enzymes: joining of exons and circularization of introns. RNA 9: 319–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salzman J, Gawad C, Wang PL, Lacayo N, Brown PO 2012. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS One 7: e30733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salzman J, Chen RE, Olsen MN, Wang PL, Brown PO 2013. Cell-type specific features of circular RNA expression. PLoS Genet 9: e1003777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger HL, Klotz G, Riesner D, Gross HJ, Kleinschmidt AK 1976. Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc Natl Acad Sci 73: 3852–3856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindler CW, Krolewski JJ, Rush MG 1982. Selective trapping of circular double-stranded DNA molecules in solidifying agarose. Plasmid 7: 263–270. [DOI] [PubMed] [Google Scholar]
- Soma A 2014. Circularly permuted tRNA genes: their expression and implications for their physiological relevance and development. Front Genet 5: 63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soma A, Onodera A, Sugahara J, Kanai A, Yachie N, Tomita M, Kawamura F, Sekine Y 2007. Permuted tRNA genes expressed via a circular RNA intermediate in Cyanidioschyzon merolae. Science 318: 450–453. [DOI] [PubMed] [Google Scholar]
- Starostina NG, Marshburn S, Johnson LS, Eddy SR, Terns RM, Terns MP 2004. Circular box C/D RNAs in Pyrococcus furiosus. Proc Natl Acad Sci 101: 14097–14101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surono A, Takeshima Y, Wibawa T, Ikezawa M, Nonaka I, Matsuo M 1999. Circular dystrophin RNAs consisting of exons that were skipped by alternative splicing. Hum Mol Genet 8: 493–500. [DOI] [PubMed] [Google Scholar]
- Suzuki H, Tsukahara T 2014. A view of pre-mRNA splicing from RNase R resistant RNAs. Int J Mol Sci 15: 9331–9342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki H, Zuo Y, Wang J, Zhang MQ, Malhotra A, Mayeda A 2006. Characterization of RNase R-digested cellular RNA source that consists of lariat and circular RNAs from pre-mRNA splicing. Nucleic Acids Res 34: e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabak HF, Van der Horst G, Smit J, Winter AJ, Mul Y, Groot Koerkamp MJ 1988. Discrimination between RNA circles, interlocked RNA circles and lariats using two-dimensional polyacrylamide gel electrophoresis. Nucleic Acids Res 16: 6597–6605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang TH, Rozhdestvensky TS, d'Orval BC, Bortolin M-L, Huber H, Charpentier B, Branlant C, Bachellerie J-P, Brosius J, Hüttenhofer A 2002. RNomics in Archaea reveals a further link between splicing of archaeal introns and rRNA processing. Nucleic Acids Res 30: 921–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vader A, Nielsen H, Johansen S 1999. In vivo expression of the nucleolar group I intron-encoded I-dirI homing endonuclease involves the removal of a spliceosomal intron. EMBO J 18: 1003–1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vader A, Johansen S, Nielsen H 2002. The group I-like ribozyme DiGIR1 mediates alternative processing of pre-rRNA transcripts in Didymium iridis. Eur J Biochem 269: 5804–5812. [DOI] [PubMed] [Google Scholar]
- Vicens Q, Cech TR 2009. A natural ribozyme with 3′,5′ RNA ligase activity. Nat Chem Biol 5: 97–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent HA, Deutscher MP 2006. Substrate recognition and catalysis by the exoribonuclease RNase R. J Biol Chem 281: 29769–29775. [DOI] [PubMed] [Google Scholar]
- Wang K, Singh D, Zeng Z, Coleman SJ, Huang Y, Savich GL, He X, Mieczkowski P, Grimm SA, Perou CM, et al. 2010. MapSplice: accurate mapping of RNA-seq reads for splice junction discovery. Nucleic Acids Res 38: e178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang PL, Bao Y, Yee M-C, Barrett SP, Hogan GJ, Olsen MN, Dinneny JR, Brown PO, Salzman J 2014. Circular RNA is expressed across the eukaryotic tree of life. PLoS One 9: e90859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu A, Chen X, Tan Y, Qi X, Xu Z, Zhang L, Ren F, Bian S, Chen Y, Wang H 2013. Identification of a novel circularized transcript of the AML1 gene. BMB Rep 46: 163–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaphiropoulos PG 1996. Circular RNAs from transcripts of the rat cytochrome P450 2C24 gene: correlation with exon skipping. Proc Natl Acad Sci 93: 6536–6541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaphiropoulos PG 1997. Exon skipping and circular RNA formation in transcripts of the human cytochrome P-450 2C18 gene in epidermis and of the rat androgen binding protein gene in testis. Mol Cell Biol 17: 2985–2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaug AJ, Grabowski PJ, Cech TR 1983. Autocatalytic cyclization of an excised intervening sequence RNA is a cleavage-ligation reaction. Nature 301: 578–583. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Zhang X-O, Chen T, Xiang J-F, Yin Q-F, Xing Y-H, Zhu S, Yang L, Chen L-L 2013. Circular intronic long noncoding RNAs. Mol Cell 51: 792–806. [DOI] [PubMed] [Google Scholar]
- Zhang X-O, Wang H-B, Zhang Y, Lu X, Chen L-L, Yang L 2014. Complementary sequence-mediated exon circularization. Cell 99: 99–999. [DOI] [PubMed] [Google Scholar]