Abstract
Craniofrontonasal syndrome (CFNS) is an X-linked developmental disorder that shows paradoxically greater severity in heterozygous females than in hemizygous males. Females have frontonasal dysplasia and coronal craniosynostosis (fusion of the coronal sutures); in males, hypertelorism is the only typical manifestation. Here, we show that the classical female CFNS phenotype is caused by heterozygous loss-of-function mutations in EFNB1, which encodes a member of the ephrin family of transmembrane ligands for Eph receptor tyrosine kinases. In mice, the orthologous Efnb1 gene is expressed in the frontonasal neural crest and demarcates the position of the future coronal suture. Although EFNB1 is X-inactivated, we did not observe markedly skewed X-inactivation in either blood or cranial periosteum from females with CFNS, indicating that lack of ephrin-B1 does not compromise cell viability in these tissues. We propose that in heterozygous females, patchwork loss of ephrin-B1 disturbs tissue boundary formation at the developing coronal suture, whereas in males deficient in ephrin-B1, an alternative mechanism maintains the normal boundary. This is the only known mutation in the ephrin/Eph receptor signaling system in humans and provides clues to the biogenesis of craniosynostosis.
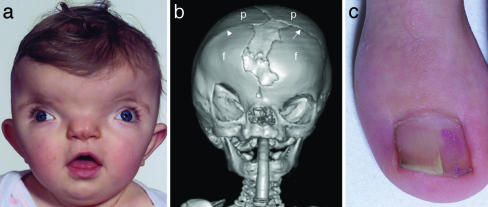
Craniofrontonasal syndrome (CFNS) is usually easy to diagnose in females, in whom typical manifestations are severe hypertelorism with a central nasal groove (Fig. 1a) associated with coronal craniosynostosis present either unilaterally (Fig. 1b) or bilaterally. Common extracranial features are sloping shoulders with dysplastic clavicles, mild cutaneous syndactyly, and characteristic longitudinal splitting of the nails (Fig. 1c). More occasionally, cleft lip and palate, duplication of the first digit, diaphragmatic hernia, and agenesis of the corpus callosum occur (1–10). The pattern of inheritance has been controversial because of the lack of a distinctive phenotype in males. Several three-generation pedigrees have shown transmission from a typically affected grandmother, through a mildly affected son, to all of his daughters but none of his sons, strongly suggesting X-linked inheritance (2, 3, 5, 11, 12). However, these obligate transmitting males manifest hypertelorism and occasional cleft lip only, indicating a paradoxical reversal in phenotypic severity between the sexes. Proposed explanations have included disruptive interaction between wild-type and mutant alleles (“metabolic interference”), aberrant functional disomy of the X chromosome, and compensation by a Y-linked homologue (11, 13). By contrast, the description of affected females having recurrent early miscarriages raises the possibility of male lethality in some families (1, 4, 7, 8).
Fig. 1.
Clinical features of CFNS. (a) Facial view showing marked hypertelorism, divergent squint, and central nasal groove (subject ID, 1593; age, 1 year). (b) Three-dimensional computed tomographic skull reconstruction (subject ID, 3167; age, 8 months) showing right unicoronal synostosis, lateral displacement of orbits, and central defect between frontal bones. Note bony ridge at site of obliterated right coronal suture (arrowhead); the left coronal suture is patent (arrow). f, frontal bone; p, parietal bone. (c) Longitudinal splitting of the nails is frequent.
CFNS was initially mapped to a 13-centimorgan region of Xp22, based on analysis of 12 unrelated families, with a maximum multipoint logarithm of odds (lod) score of 5.08 (13). We failed to find mutations in 28 different genes in this region (R.K., S.R.F.T., and A.O.M.W., unpublished data). Recently, Xp22 was excluded in a single family segregating CFNS, with linkage (maximum two-point lod = 1.66) reported instead to the pericentromeric region of the X chromosome (12). This report led us to broaden our search of candidate genes outside Xp22.
The gene EFNB1, encoding ephrin-B1, maps at Xq13.1 within the new linkage interval. Vertebrate ephrins, membrane-anchored ligands for Eph receptor tyrosine kinases, are divided into two families: A and B. The B family, which is characterized by an intracellular region containing multiple tyrosine residues and a PDZ domain, has three members (B1, B2, and B3) in many vertebrate species (reviewed in ref. 14). Efnb1, the murine ortholog of EFNB1, is expressed in frontonasal neural crest (15), and mice engineered with a loss-of-function mutation of Efnb1 have cleft palate, shortening of the skull, sternal abnormalities, and omphalocele (16). Significantly, heterozygous female mice manifested additional phenotypes (preaxial polydactyly and syndactyly) and had lower viability (1–2%) compared with hemizygous males (15%), recalling the inheritance pattern of CFNS (16).
We report here that heterozygous EFNB1 mutations, predicted to cause loss of ephrin-B1 function, are present in all 20 unrelated females with CFNS in our study group. We show that the expression pattern of murine Efnb1 is consistent with an important role for this gene in defining the position of the coronal suture. Finally, we demonstrate that, despite causing loss of ephrin-B1 function, X-inactivation in CFNS females is not highly skewed in blood or cranial periosteum. We discuss the implications of these findings for the development of the coronal suture and the unusual inheritance pattern of CFNS.
Materials and Methods
Mutation Screening of EFNB1 and X-Inactivation Analysis. The study was approved by the Oxfordshire Clinical Research Ethics Committee, and informed consent was obtained before performing sampling procedures. DNA was obtained from whole blood samples, cultured fibroblasts, lymphoblastoid cell lines, or cranial periosteum by phenol-chloroform extraction. The following GenBank accession nos. were used for EFNB1 primer design: cDNA, NM_004429; genomic, AL136092. Standard PCRs (30 μl) contained 15 mM Tris·HCl (pH 8.0), 50 mM KCl, 2.5 mM MgCl2, 100 μM dNTP (each), 0.5 μM primers, 0.75 units of Amplitaq Gold polymerase (Applied Biosystems), and 0.15 units of Pwo polymerase (Roche). Amplicons were analyzed by denaturing HPLC (dHPLC) on a Wave 3500HT (Transgenomic, Omaha, NE). Primer sequences, PCR amplification, and dHPLC analysis conditions are provided in Table 2, which is published as supporting information on the PNAS web site. The coding regions of all five exons of EFNB1 were analyzed by dHPLC in every affected subject. Amplicons exhibiting aberrant migration patterns were sequenced by using BigDye (version 3) on a 3100 DNA sequencer (Applied Biosystems). Mutations were confirmed either by restriction enzyme digestion or by allele-specific oligonucleotide hybridization (see Table 3, which is published as supporting information on the PNAS web site). Correct biological relationships between children and their parents were inferred from the concordant segregation of at least eight microsatellites of 78% average heterozygosity, each located on different chromosomes. The minimum probability of correct paternity in these trios, based on the conservative assumption that the father's alleles corresponded to those of highest frequency in a search of The Genome Database (April 8, 2004; http://gdb.wehi.edu.au/gdb), was 99.1%.
In addition to the analysis of 20 CFNS families reported here, 155 further unrelated patients with craniosynostosis of unknown cause (mutations in FGFR1, FGFR2, FGFR3, TWIST, MSX2, and ALX4 having been excluded) were screened for mutations in EFNB1. Samples from males were doped with an approximately equal amount of PCR product from a control male before heteroduplexes were made. A C→T variant (dbSNP: rs2230423) was encountered in 16 of 230 chromosomes, but no pathogenic changes were found.
X-inactivation analysis of the androgen receptor (AR) locus was undertaken, as described (17, 18). The positive control sample was from a female with Melnick–Needles syndrome (18). Measurements were made in triplicate for samples from blood and in duplicate for samples from cranial periosteum.
RNA in Situ Hybridization. Whole-embryo in situ hybridization was performed as described (19). The Efnb1 probe (15) was characterized by sequence analysis. Expression was visualized by means of alkaline phosphatase-conjugated antidigoxygenin antibody. Embryos were subsequently embedded in Cryo-M-Bed compound (Bright Instruments, Huntingdon, England) and cut on a cryostat at 15 μm. Specimens were viewed and photographed by using a MP3 dissecting microscope (Wild, Heerbrugg, Switzerland) and a DMRBE light microscope with Photoautomat (Leica, Deerfield, IL).
Results
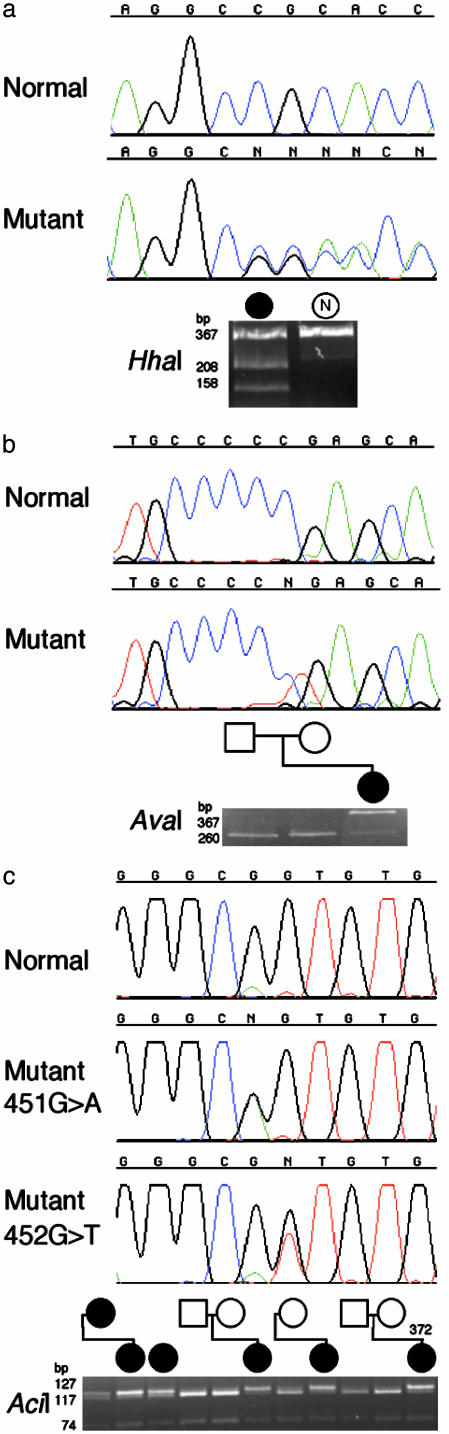
Heterozygous Mutations of EFNB1 in CFNS. We PCR amplified EFNB1, which comprises five exons and encodes a protein of 346 aa, and analyzed the gene in 24 affected females from 20 unrelated families. We identified pathogenic mutations in every case (Table 1). There were 17 distinct mutations, one of which was a frameshifting single-nucleotide deletion (Fig. 2a), the remainder being single-nucleotide substitutions that either alter a gt/ag splice site (three mutations), predict nonsense codons (two mutations) (Fig. 2b), or encode missense changes in the amino acid sequence (11 mutations). One mutation, 451G→A (encoding Gly151Ser) was recurrent (four families); this transition arises in a CpG doublet (Fig. 2c). Nine of these different mutations were shown to have arisen de novo from the unaffected biological parents (Table 1), demonstrating their pathogenic nature conclusively. The remaining mutations, for which both parents were not available, either grossly disrupt the EFNB1 sequence or, in the case of missense changes, occur at highly conserved positions (see below) and were not observed in 180 normal control chromosomes from the same population. Transmission of the mutation was observed in the four affected mother–daughter pairs available for analysis. We conclude that EFNB1 mutations cause CFNS, and we find no evidence of genetic heterogeneity in affected females.
Table 1. Clinical features of CFNS and mutations of EFNB1.
| Additional clinical features
|
Mutation
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Proband | Coronal craniosynostosis | Cleft lip and/or palate | Duplex thumb hallux | Agenesis of corpus callosum | Other | DNA | Exon (intron) | Protein | Familial or de novo | Confirmation | Ref.* |
| 1219 | r, l | — | — | — | — | 1A→G | 1 | M1V | d | BslI (+) | — |
| 353 | r | — | — | — | — | 57G→A | 1 | W19X | u | AfeI (+) | 10 [6] |
| 656 | — | P | — | — | Sprengel shoulders | 185T→C | 2 | I62T | d | ASO | — |
| 344 | l | — | — | — | — | 196C→T | 2 | R66X | d | AvaI (-) | 10 [5] |
| 369 | r, l | L, P | — | ± | — | 246delG | 2 | P83fsX75 | f | HhaI (+) | 10 [4] |
| 2613 | r, l | — | — | + | Lower-limb asymmetry | 293T→C | 2 | L98S | d | BstXI (-) | — |
| 723 | — | — | rH | + | — | 344A→C | 2 | Q115P | d | MspI (+) | — |
| 3167 | r | — | — | — | — | 355C→A | 2 | P119T | f | ASO | 10 [10] |
| 387† | r | — | — | + | — | 356C→A | 2 | P119H | u | BstXI (-) | 10 [9] |
| 2301 | — | — | — | + | — | 406+1G→A | (2) | sp | u | ASO | — |
| 347 | r, l | L, P | — | + | — | 407-1G→A | (2) | sp | d | PstI (-) | 10 [7] |
| 350 | r | — | — | — | — | 451G→A | 3 | G151S | d | AciI (-) | 10 [3] |
| 1257 | r, l | L, P | — | — | — | 451G→A | 3 | G151S | f | AciI (-) | 6 |
| 738 | r, l | — | — | — | — | 451G→A | 3 | G151S | u | AciI (-) | — |
| 373 | r, l | — | — | — | — | 451G→A | 3 | G151S | u | AciI (-) | 10 [2] |
| 372 | r | P | — | — | — | 452G→T | 3 | G151V | d | AciI (-) | 10 [1] |
| 1593 | r, l | — | rT | ± | I diaphragmatic hernia | 463A→C | 3 | T155P | d | ASO | — |
| 1041 | l | — | — | ± | — | 472A→G | 3 | M158V | f | NlaIII (-) | — |
| 1818 | r, l | — | — | — | — | 474G→T | 3 | M158I | f | MseI (+) | — |
| 355 | — | — | — | — | Sacrococcygeal teratoma | 629-2A→G | (4) | sp | u | BslI (+) | 10 [8] |
r, Right side affected; l, left side affected; L, cleft lip; P, cleft palate; T, duplex thumb; H, duplex hallux; ±, partial agenesis of corpus callosum; sp, splicing mutation; d, proven de novo mutation; f, familial mutation; u, de novo mutation on clinical history but not molecularly proven; ASO, allele-specific oligonucleotide hydridization.
Case numbers for patients reported in ref. 10 are given in brackets.
An additional rare variant, 461G→A (R154H), present on the opposite allele, was inherited from the unaffected mother.
Fig. 2.
Selected heterozygous mutations of EFNB1 in females with CFNS. Each figure part shows, from top to bottom, a normal DNA sequence chromatogram, mutant DNA sequence chromatogram(s), and corresponding restriction digest confirmation. Pedigree symbols (shown in black for affected individuals) are aligned vertically with corresponding lanes of the gel. N, normal control. (a) Frame-shifting deletion 246delG in subject 369 (sequenced with reverse primer) and confirmation by HhaI digest, showing creation of a new restriction site. (b) De novo nonsense mutation 196C→ T in subject 344, which abolishes an AvaI site. (c) The recurrent mutation 451G→ A, present in four unrelated families, is transmitted by an affected mother to her daughter and has arisen de novo in one parent–child trio. Also shown is the adjacent mutation 452G→ T in subject 372 that abolishes the same AciI site.
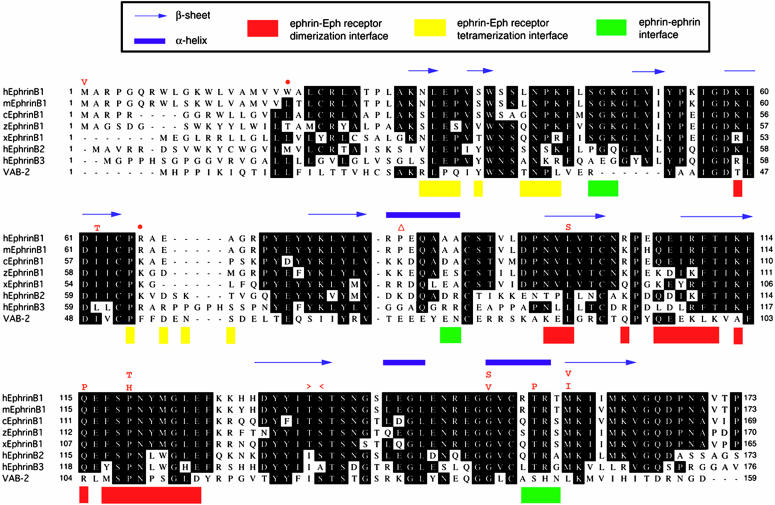
Mutations Predict Complete or Partial Loss of Ephrin-B1 Function. The location of the mutations in ephrin-B1 in relation to known structural motifs and selected homologous sequences is shown in Fig. 3. The extracellular portion of murine ephrin-B2 consists of an eight-stranded β-barrel crystal structure with Greek key folding topology (20). The frameshift, splicing, and nonsense mutations truncate the ephrin-B1 protein in the extracellular region, suggesting complete loss of function, and the 1A→G (Met1Val) mutation should prevent normal translation initiation. The remaining missense mutations occur (with the exception of Ile62Thr, which arose de novo) at positions that are identical in the three human B-type ephrins, and they all are located in structurally important regions (see Fig. 3 legend) (21). Additional evidence that these are loss-of-function mutations is provided by genetic and biochemical data from the Caenorhabditis elegans ephrin-A homologue VAB-2. The VAB-2 mutations Met1Leu and Pro108Leu, which occur at positions equivalent to Met-1 and Pro-119 in human ephrin-B1 (Fig. 3), were described, respectively, as showing weak and intermediate defects in epithelial and neuronal morphogenesis (22). Loss of binding of VAB-2 to its Eph receptor VAB-1 was observed for the Pro108Leu substitution (22).
Fig. 3.
Comparative amino acid sequence alignment of human (h), mouse (m), chick (c), zebrafish (z), and Xenopus (x) ephrin-B1; human ephrin-B2 and ephrin-B3; and C. elegans VAB-2. Above the human ephrin-B1 sequence, the identities of missense substitutions are shown in red single-letter codes, together with the positions of the single-nucleotide deletion (▵), nonsense (•), and donor and acceptor splice site mutations (> and <, respectively). Regions of secondary structure in murine ephrin-B2 are indicated at the top of each sequence block. Interacting regions in the complex with murine EphB2 (21) are color-coded at the bottom of each sequence block. Note that the targets of missense mutations at L98, Q115, and P119 are amino acids that are predicted to interact with the Eph receptor at the major (dimerization) interface: L98 and Q115 form hydrogen bonds while P119 inserts into a hydrophobic pocket, forming both van der Waals contacts and a hydrogen bond (21).
Although CFNS subjects differed in the extent of craniosynostosis and occurrence of additional clinical features, we did not observe any distinct genotype–phenotype correlation in the heterozygous females between truncating and missense mutations (Table 1), suggesting that all mutations cause a comparable disturbance of ephrin-B1 function. At this stage, we cannot exclude the possibility that a genotype–phenotype correlation exists for hemizygous males, in whom viability might depend on residual ephrin-B1 activity.
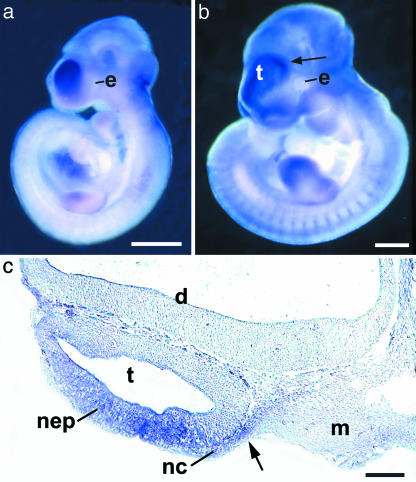
A Boundary of Efnb1 Expression Corresponds to the Position of the Future Coronal Suture. We examined the RNA expression of murine Efnb1 at embryonic day (E)9.5 and E10.5, the time at which the frontal-parietal boundary is established. Confirming and extending previous work (15), in situ hybridization shows that the most rostral Efnb1 expression domain corresponds specifically to the neural crest-derived, future frontal bone territory and underlying telencephalic vesicle neuroepithelium but is absent from the mesenchyme that is destined to form the parietal bone (Fig. 4).
Fig. 4.
RNA in situ hybridization of Efnb1 in mouse embryos at E9.5 (a) and E10.5 (b). Arrow in b marks the boundary between a high level of expression in the region of the telencephalon compared with the adjacent diencephalon, and it indicates the level of the section shown in c. e, eye. (c) Section of an E10.5 embryo showing high Efnb1 transcript levels in the neuroepithelium (nep) of the telencephalon and adjacent neural crest-derived mesenchyme (nc) but not in the diencephalon (d) or cranial mesoderm (m). e, eye. Arrow shows the boundary between neural crest-derived and mesoderm-derived cranial mesenchyme, which marks the position of the future coronal suture (23). Scale bars indicate 1 mm (a and b) and 200 μm(c).
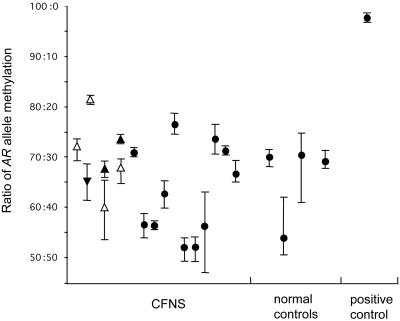
X-Inactivation in CFNS Females. Expression data from somatic cell hybrids indicate that EFNB1 is normally subject to X-inactivation (24). We investigated X-inactivation in DNA extracted from whole blood in 18 affected females informative for the AR microsatellite assay, a widely used method for identifying skewed X-inactivation in X-linked disorders (17, 18, 25). We did not observe extreme skewing (>90:10) in any individual, and all but one sample showed skewing within the normal range (50:50 to <80:20) (Fig. 5). In four mother–daughter pairs, we could deduce (directly in the daughters and by virtue of close genetic linkage between AR and EFNB1, which are separated by only ≈1.3 Mb, in three of the mothers) whether the bias of X-inactivation was toward or against the mutant allele; in six of seven cases, there was a weak bias toward preferential inactivation of the mutant allele (Fig. 5). Because the X-inactivation status in blood is likely to differ from that in the coronal suture, we obtained samples of cranial periosteum overlying the entire length of both coronal sutures from a familial case (subject ID 3167) at the time of reconstructive surgery. The level of inactivation of the mutant X chromosome in eight evenly spaced specimens varied from 48.6% to 84.3% (data not shown). We conclude that normal ephrin-B1 function is not required for cell survival either in blood or in cranial periosteum.
Fig. 5.
X-inactivation patterns in the AR gene of CFNS subjects. Mean and range of values obtained from three independent PCRs is shown. Pairs of affected mothers and daughters are aligned at the same position on the horizontal axis and indicated by ▴ and ▵ for mothers and daughters, respectively. Triangles point up or down, respectively, according to whether the mutant or the wild-type allele is preferentially inactivated. •, Phase-unknown cases.
Discussion
The identification of significant mutations in all 20 unrelated CFNS females studied, including nine different de novo mutations, provides convincing evidence that EFNB1 is the major CFNS locus. The diversity of the mutations, the disruptive nature of several of them, the location of missense mutations in regions of secondary structure or receptor binding, and the lack of genotype–phenotype correlation indicate reduction or loss of ephrin-B1 function. Our findings are consistent with recent linkage data for one family (12) but are difficult to reconcile with an earlier report that mapped CFNS to Xp22 (13). None of the patients that we investigated were included in either of these studies, so mutation analysis of EFNB1 in patients from the linkage panels should establish whether the previous findings are explained by genetic heterogeneity, coincidence, or other factors. Our findings are of particular interest in relation both to the pathogenesis of the component malformations of CFNS, especially craniosynostosis, and because of the apparent reversal in phenotypic severity between the sexes for an X-linked disorder.
Craniosynostosis affects ≈1 in 2,500 births and previously identified single-gene mutations in the FGFR1, FGFR2, FGFR3, TWIST, and MSX2 genes (26) account for ≈25% of cases overall (A.O.M.W. and S.A.W., unpublished data). Efforts to elucidate the pathogenesis of craniosynostosis in mice have focused pre-dominantly on the E14–E16 period, when the orthologous genes are expressed in the maturing cranial sutures (27, 28). CFNS characteristically affects the coronal suture, which separates the frontal and parietal bones (Fig. 1b); recent evidence from mice indicates that these bones have distinct embryological origins (frontal bone from neural crest and parietal bone from mesoderm) (23). This finding raises the question of what molecular signals mark the separate identities of these tissues at the time that the position of the future coronal suture is established. Ephrins are good candidates for such signals because the spatial distribution of their biological activity (within the A and B classes) is often complementary to their cognate Eph receptors (29), and this interaction appears to mediate repulsive signaling between distinct cell types (reviewed in refs. 30 and 31). The pattern of Efnb1 expression is exactly as expected for a molecule required for boundary formation at the coronal suture (compare Fig. 4c with Fig. 3D in ref. 23), and the demonstration of EFNB1 mutations in CFNS provides genetic confirmation of the importance of ephrin-B1 in this process. An important focus for future work may be to define further the molecular processes that lead to neural crest/mesoderm boundary formation and, hence, coronal suture positioning. Interestingly, there is evidence for direct interactions between the ephrin-B1 and fibroblast growth factor signaling pathways (32, 33), suggesting a possible mechanism by which boundary formation might lead to the onset of growth-related signaling in the coronal suture.
Why do mutations in EFNB1 apparently lead to craniosynostosis only in females? Although mice harboring the Efnb1 null mutation were reported to have shortened skulls (16), no information is available regarding the presence of craniosynostosis in these animals. More instructively, heterozygous female Efnb1+/– mice exhibited additional limb phenotypes (preaxial polydactyly and syndactyly) that are not present in either Efnb1– males or Efnb1–/– females (16, 34). This appearance was demonstrated to correlate with a patchy distribution of Efnb1 expression in the limbs of Efnb1+/– females in a pattern reciprocal to the receptors EphA4 (34), EphB2, and Ephb3 (16). Moreover, there was marked up-regulation of Efnb1 expression in regions where the X chromosome bearing the wild-type allele was active (16). Our finding that X-inactivation is not highly skewed in CFNS females, either in blood or cranial periosteum (Fig. 5), is consistent with observations in mice and leads us to propose that the craniosynostosis results from disturbance in the formation of the normally sharp neural crest/mesoderm tissue boundary at the future coronal suture, caused by patchy abnormalities of signaling arising from random X-inactivation and subsequent sorting of ephrin-B1 positive and negative cells (processes that would be specific to females). The observation that males inheriting the CFNS mutation do not have craniosynostosis (2, 3, 5, 11, 12) suggests functional redundancy in the mechanisms of cranial suture determination, perhaps involving other members of the ephrin/EphR signaling system (29).
Supplementary Material
Acknowledgments
This article is dedicated to the memory of Professor Robin Winter, a pioneer in the genetics of craniosynostosis. We thank all family members who participated in the study; M. Barrow, J. Byren, D. Furniss, S. Greenwood, J. Hurst, D. Johnson, S. Mann, S. Rannan-Eliya, S. Smithson, G. Vittu, and I. Young for help in obtaining samples; I. Antonopoulou, S. Butler, K. Clark, and M. Oldridge for technical assistance; D. Wilkinson for the Efnb1 probe; R. Maxson for discussions; and L. Wang and F. Jin for supporting R.K.'s doctoral studies. This work was supported by the China Scholarship Council (R.K.), Nuffield Medical Fellowship (S.P.R.), the Edward Penley Abraham Research Fund (C.B. and G.M.M.-K.), and the Wellcome Trust (A.O.M.W.).
Abbreviations: CFNS, craniofrontonasal syndrome; En, embryonic day n.
References
- 1.Cohen, M. M., Jr. (1979) Birth Defects Orig. Artic. Ser. 15, 85–89. [PubMed] [Google Scholar]
- 2.Slover, R. & Sujansky, E. (1979) Birth Defects Orig. Artic. Ser. 15, 75–83. [PubMed] [Google Scholar]
- 3.Reynolds, J. F., Haas, R. J., Edgerton, M. T. & Kelly, T. E. (1982) J. Craniofac. Genet. Dev. Biol. 2, 233–238. [PubMed] [Google Scholar]
- 4.Sax, C. M. & Flannery, D. B. (1986) Clin. Genet. 29, 508–515. [DOI] [PubMed] [Google Scholar]
- 5.Morris, C. A., Palumbos, J. C. & Carey, J. C. (1987) Am. J. Med. Genet. 27, 623–631. [DOI] [PubMed] [Google Scholar]
- 6.Young, I. D. (1987) J. Med. Genet. 24, 193–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grutzner, E. & Gorlin, R. J. (1988) Oral Surg. Oral Med. Oral Pathol. 65, 436–444. [DOI] [PubMed] [Google Scholar]
- 8.Devriendt, K., Van Mol, C. & Fryns, J. P. (1995) Genet. Couns. (Geneva, Switzerland) 6, 361–364. [PubMed] [Google Scholar]
- 9.Saavedra, D., Richieri-Costa, A., Guion-Almeida, M. L. & Cohen, M. M., Jr. (1996) Am. J. Med. Genet. 61, 147–151. [DOI] [PubMed] [Google Scholar]
- 10.Orr, D. J. A., Slaney, S., Ashworth, G. J. & Poole, M. D. (1997) Br. J. Plast. Surg. 50, 153–161. [DOI] [PubMed] [Google Scholar]
- 11.Rollnick, B., Day, D., Tissot, R. & Kaye, C. (1981) Am. J. Hum. Genet. 33, 823–826. [PMC free article] [PubMed] [Google Scholar]
- 12.Wieland, I., Jakubiczka, S., Muschke, P., Wolf, A., Gerlach, L., Krawczak, M. & Wieacker, P. (2002) Cytogenet. Genome Res. 99, 285–288. [DOI] [PubMed] [Google Scholar]
- 13.Feldman, G. J., Ward, D. E., Lajeunie-Renier, E., Saavedra, D., Robin, N. H., Proud, V., Robb, L. J., Der Kaloustian, V., Carey, J. C., Cohen, M. M., Jr., et al. (1997) Hum. Mol. Genet. 6, 1937–1941. [DOI] [PubMed] [Google Scholar]
- 14.Holder, N. & Klein, R. (1999) Development (Cambridge, U.K.) 126, 2033–2044. [DOI] [PubMed] [Google Scholar]
- 15.Flenniken, A. M., Gale, N. W., Yancopoulos, G. D. & Wilkinson, D. G. (1996) Dev. Biol. 179, 382–401. [DOI] [PubMed] [Google Scholar]
- 16.Compagni, A., Logan, M., Klein, R. & Adams, R. H. (2003) Dev. Cell 5, 217–230. [DOI] [PubMed] [Google Scholar]
- 17.Allen, R. C., Zoghbi, H. Y., Moseley, A. B., Rosenblatt, H. M. & Belmont, J. W. (1992) Am. J. Hum. Genet. 51, 1229–1239. [PMC free article] [PubMed] [Google Scholar]
- 18.Robertson, S. P., Twigg, S. R. F., Sutherland-Smith, A. J., Biancalana, V., Gorlin, R. J., Horn, D., Kenwrick, S. J., Kim, C. A., Morava, E., Newbury-Ecob, R., et al. (2003) Nat. Genet. 33, 487–491. [DOI] [PubMed] [Google Scholar]
- 19.Wilkinson, D. G. (1992) in In Situ Hybridization: A Practical Approach, ed. Wilkinson, D. G. (IRL, Oxford), pp. 75–83.
- 20.Toth, J., Cutforth, T., Gelinas, A. D., Bethoney, K. A., Bard, J. & Harrison, C. J. (2001) Dev. Cell 1, 83–92. [DOI] [PubMed] [Google Scholar]
- 21.Himanen, J.-P., Rajashankar, K. R., Lackmann, M., Cowan, C. A., Henkemeyer, M. & Nikolov, D. B. (2001) Nature 414, 933–938. [DOI] [PubMed] [Google Scholar]
- 22.Chin-Sang, I. D., George, S. E., Ding, M., Moseley, S. L., Lynch, A. S. & Chisholm, A. D. (1999) Cell 99, 781–790. [DOI] [PubMed] [Google Scholar]
- 23.Jiang, X., Iseki, S., Maxson, R. E., Sucov, H. M. & Morriss-Kay, G. M. (2002) Dev. Biol. 241, 106–116. [DOI] [PubMed] [Google Scholar]
- 24.Carrel, L., Cottle, A. A., Goglin, K. C. & Willard, H. F. (1999) Proc. Natl. Acad. Sci. USA 96, 14440–14444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Plenge, R. M., Stevenson, R. A., Lubs, H. A., Schwartz, C. E. & Willard, H. F. (2002) Am. J. Hum. Genet. 71, 168–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Muenke, M. & Wilkie, A. O. M. (2000) in The Metabolic and Molecular Bases of Inherited Disease, eds. Scriver, C. R., Beaudet, A. L., Sly, W. S. & Valle, D. (McGraw–Hill, New York), pp. 6117–6146.
- 27.Iseki, S., Wilkie, A. O. M. & Morriss-Kay, G. M. (1999) Development (Cambridge, U.K.) 126, 5611–5620. [DOI] [PubMed] [Google Scholar]
- 28.Rice, D. P. C., Åberg, T., Chan, Y., Tang, Z., Kettunen, P. J., Pakarinen, L., Maxson, R. E. Jr. & Thesleff, I. (2000) Development (Cambridge, U.K.) 127, 1845–1855. [DOI] [PubMed] [Google Scholar]
- 29.Gale, N. W., Holland, S. J., Valenzuela, D. M., Flenniken, A., Pan, L., Ryan, T. E., Henkemeyer, M., Strebhardt, K., Hirai, H., Wilkinson, D. G., et al. (1996) Neuron 17, 9–19. [DOI] [PubMed] [Google Scholar]
- 30.Wilkinson, D. G. (2001) Nat. Rev. Neurosci. 2, 155–164. [DOI] [PubMed] [Google Scholar]
- 31.Palmer, A. & Klein, R. (2003) Genes Dev. 17, 1429–1450. [DOI] [PubMed] [Google Scholar]
- 32.Chong, L. D., Park, E. K., Latimer, E., Friesel, R. & Daar, I. O. (2000) Mol. Cell. Biol. 20, 724–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moore, K. B., Mood, K., Daar, I. O. & Moody, S. A. (2004) Dev. Cell 6, 55–67. [DOI] [PubMed] [Google Scholar]
- 34.Davy, A., Aubin, J. & Soriano, P. (2004) Genes Dev. 18, 572–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.