Abstract
The HIV envelope glycoprotein (Env) trimer undergoes receptor-induced conformational changes that drive fusion of the viral and cellular membranes. Env conformational changes have been observed using low-resolution electron microscopy, but only large-scale rearrangements have been visible. Here, we use Hydrogen/Deuterium-exchange and oxidative labeling to gain a more precise understanding of the unliganded and CD4-bound forms of soluble Env trimers (SOSIP.664), including their glycan composition. CD4 activation induces reorganization of bridging sheet elements, V1/V2 and V3, much of the gp120 inner domain, and the gp41 fusion subunit. Two CD4 binding site-targeted inhibitors have substantially different effects: NBD-556 partially mimics CD4-induced destabilization of the V1/V2 and V3 crown, while BMS-806 only affects regions around the gp120/gp41 interface. The structural information presented here increases our knowledge of CD4- and small molecule-induced conformational changes in Env and the allosteric pathways that lead to membrane fusion.
Keywords: Env trimers, glycoprotein, glycoform, SOSIP, gp140, HD exchange, deuterium exchange, oxidative labeling, CD4, BMS-806, NBD-556
INTRODUCTION
The trimeric envelope glycoprotein (Env) complex on the surface of virions mediates HIV-1 entry and is the sole target for neutralizing antibodies (NAbs) that are induced during natural infection. A detailed understanding of Env structure, its conformational rearrangements, and how it presents NAb epitopes, is critical for guiding the design of both protein-based vaccines and Env-targeting entry inhibitors (Jardine et al., 2013; Walker and Burton, 2010). The 4.7 Å crystal and 5.8 Å cryo-EM structures of a soluble, cleaved form of the Env trimer (BG505 SOSIP.664) have unveiled novel aspects of its antigenicity and architecture (Julien et al., 2013a; Lyumkis et al., 2013). However, the complete organization of gp41 and the mechanism underlying CD4-induced structural reorganizations within the trimer remain unresolved. It also is necessary to understand how flexible elements, including gp120 glycans and surface loops, and the “breathing” of the overall Env complex modulate its recognition by antibodies and host factors (Davenport et al., 2013; Kwong et al., 2002; Myszka et al., 2000; Scanlan et al., 2007).
Characterizing conformational changes in multi-subunit glycoprotein complexes via classical structural methods can be very challenging. Here, we probe the structural dynamics of the Env trimer by using Hydrogen/Deuterium exchange (HDX) to measure the rates of deuterium incorporation into backbone amides under solution conditions. Unstructured or flexible regions undergo deuterium exchange rapidly, in marked contrast to ones involved in stable hydrogen bonding networks as part of the protein secondary structure, or those that are highly occluded from solvent. Coupled with mass spectrometry (MS), HDX information can be used to measure conformation dynamics with sequence-specific detail, localize protein-protein interactions, and detect ligand-induced conformational changes (Marcsisin and Engen, 2010). As a complementary approach, we performed oxidative labeling using short exposures to synchrotron radiation to generate hydroxyl radicals that modify protein side chains. As solvent-occluded side chains react less readily than exposed ones, this method allows solvent accessibility to be estimated, in some cases with single amino-acid resolution (Tong et al., 2008; Wang and Chance, 2011).
We have used the above techniques to study soluble, cleaved SOSIP.664 trimers based on the subtype A sequences, KNH1144 and BG505. The sequence modifications used to stabilize these trimers are described elsewhere and summarized in Figure 1A (Binley et al., 2000; Binley et al., 2002; Sanders et al., 2013; Sanders et al., 2002b). The resulting homogeneous trimers closely resemble native Env on virions, both antigenically and structurally (Harris et al., 2011; Khayat et al., 2013; Sanders et al., 2013). Here we report a structural analysis of the BG505 SOSIP.664 trimer in its unliganded pre-fusion and CD4-bound states. The HDX results reveal the nature of rearrangements in the gp120 receptor binding subunit and the allosteric changes in the gp41 fusion subunit. The new sequence-specific details complement electron microscopy structures of pre-fusion and CD4-bound trimers (Harris et al., 2011; Liu et al., 2008; Tran et al., 2012).
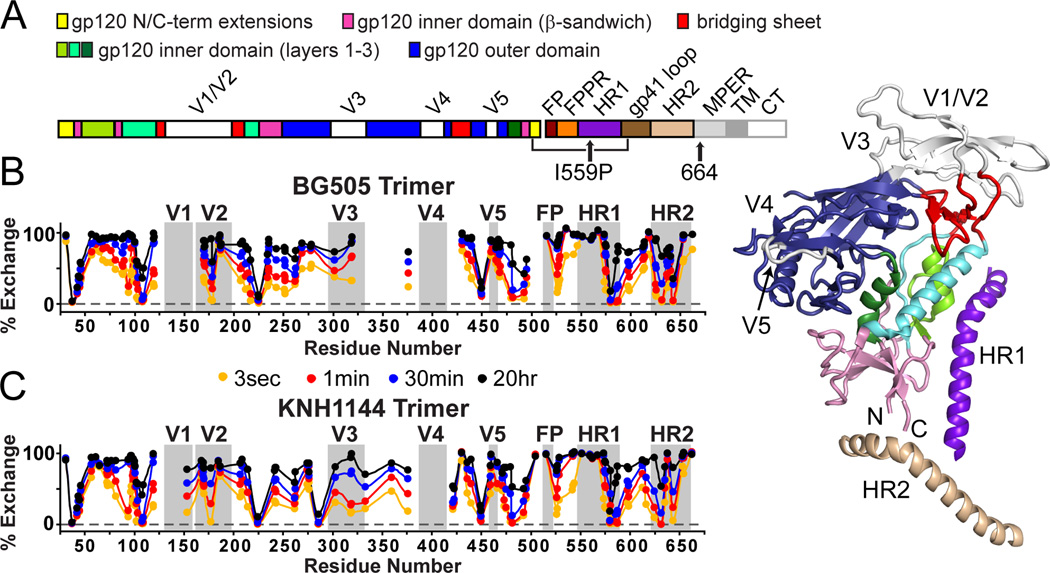
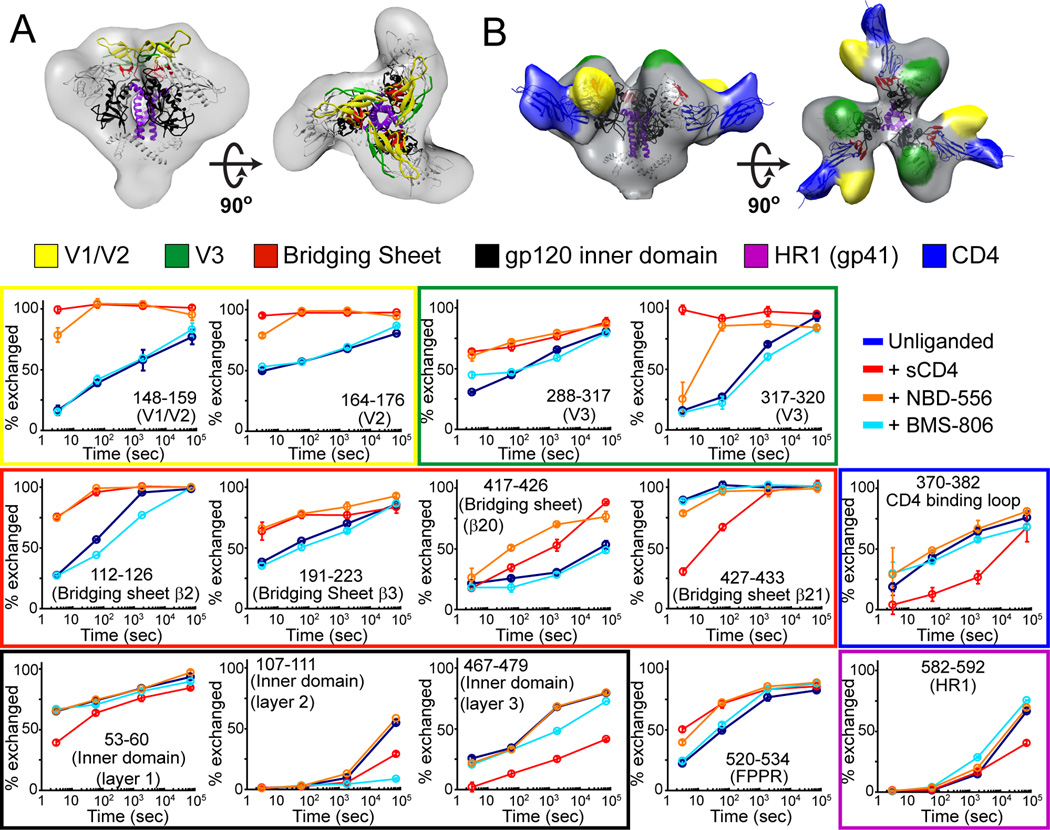
Figure 1. HDX profile of SOSIP.664 trimers.
(A) The sequence of mature, full-length Env is shown with the SOSIP modifications indicated. To the right, structural elements including variable loops 1–5, N/C termini, and heptad repeats of gp41 (HR1 and HR2) are mapped onto the ribbon diagram for one protomer from the BG505 SOSIP.664 trimer crystal structure (PDB 4NCO) (Julien et al., 2013a). (B,C) The H/D-exchange profiles are shown for unliganded BG505 and KNH1144 SOSIP.664 trimers. Percent exchange is shown after 3 s, 1 min, 30 min and 20 h for all observable peptic fragments at the midpoint of their primary sequence. For example, the exchange profile of fragment 105–111 is plotted at position 108. Individual exchange plots with errors are shown in Figures S2 and S3.
We also use HDX to compare the actions of two entry inhibitors that target the CD4 binding site: BMS-806 and NBD-556. The binding of NBD-556 to a monomeric gp120 core fragment has been studied via thermodynamic measurements and x-ray crystallography (Kwon et al., 2012; Schon et al., 2006; Zhao et al., 2005). NBD-556 binding has a major effect at the apex of the SOSIP.664 trimer, which is substantially different than what was seen using monomeric gp120. In contrast, BMS-806 does not effect of the apex of the trimer, consistent with it inhibiting CD4-induced conformational changes (Guo et al., 2003; Lin et al., 2003; Madani et al., 2004). Overall, our studies of how the HIV-1 Env trimer interacts with CD4, NBD-556 and BMS-806 provide insights into the allosteric networks that are involved in the receptor-mediated activation of this type-1 fusion protein.
RESULTS
HDX-MS profile of SOSIP.664 trimers
The KNH1144 and BG505 SOSIP.664 trimers were analyzed by HDX-MS. After optimizing pepsin digestion conditions, sequence coverage of 92% (89% of gp120 and 100% of gp41) from 103 unique KNH1144 peptides and 82% (76% of gp120 and 100% of gp41) from 120 unique BG505 peptides was achieved by MS (coverage maps are shown in Figure S2, S3). Expression of both trimers in N-acetylglucosaminyl transferase I deficient (GnTI−/−) 293S aided detection of glycosylated peptides during MS analysis, as the presence of only high-mannose type glycoforms reduced trimer heterogeneity (Table 1). Peptic fragments from V4 or the N-terminal half of V1 could not be monitored by HDX-MS. Their absence was presumably due to dense glycosylation, as V1 and V4 fragments were observable after the peptic fragments were deglycosylated with EndoH or PNGaseF.
Table 1.
Glycoforms observed in SOSIP.664 trimers.
| Glycosylation sites* | KNH1144 | BG505 (293S) | BG505 (293F) |
|---|---|---|---|
| N88 | Man5 | Man5 | Man5 & Complex |
| N156 | Man8,9 | - | - |
| N188/190 & N190c | Man5a | Man5a,b | Complex |
| N197 | Man5–9 | Man5–9 | Man5–9 |
| N234 | Man6–9a | Man8,9 | Man8,9 |
| N241 | Man6–9a | - | - |
| N262 | Man8,9 | Man8,9 | Man8,9 |
| N276 | Man7,8 | Man5–8 | Man5–8 |
| N295 | - | Man8b | Man8a |
| N301 | Man7–8 | Man8b | Man8a |
| N332 & N339 | Man8,9a | - | - |
| N356 & N363 | Man7a | - | - |
| N448 | Man8,9 | Man8,9 | Man8,9 |
| N463/462 | Man5 | Man5 | Complex |
| N611 & N616/618 | Man5a,b | Man5b | Complex |
| N625 | Man5b | Man5b | Man5b |
| N637 | Man5,6 | Man5–8 | Man5–8 |
N-linked glycosylation site(s) where some peptides contained more than one glycan (denoted by +); KNH1144/BG505 position
Data were obtained from peptides bearing two glycans; therefore, only the average for the total combined glycosylation is reported.
Glycan occupancy was observed at less than 90% as assessed by the relative abundance of non-glycosylated and deglycosylated signal after PNGaseF digestion (see Tables S1 and S2).
The exchange profiles for the KNH1144 and BG505 SOSIP.664 trimers are shown in Figure 1; the exchange data for each peptide are plotted along the primary protein sequence (all individual exchange plots are shown in figures S2 and S3). The patterns of protection were very similar for both trimers. Regions of high protection were seen throughout the gp120 subunits, as was in previous studies of gp120 monomers (Davenport et al., 2013; Guttman et al., 2012) and gp140 monomers (Guttman and Lee, 2013). However, we now also observed extensive protection within the variable loops of the gp120 subunits in the trimers. For example, a highly protected region spanned the N-terminal half of V2 (residues 165–181). Although the region was not covered in the BG505 dataset, there was strong protection between V1 and V2 up to residue 159 in the KNH1144 dataset. Hence the region around N160, a central feature of the bNAb PG9/16 epitope (McLellan et al., 2011; Walker et al., 2009), has a stable secondary structure in the unliganded trimer. In contrast, the rest of V2 (residues 184–190) undergoes rapid exchange, implying that this relatively short segment may be the only true flexible “loop” portion of the V2 region. The peptides spanning V3 were also significantly protected, indicating that in the unliganded trimer the V3 loop either has a constrained secondary structure or is solvent inaccessible.
A complex profile of protection throughout the gp41 subunit provides new information about its organization (Figure 1, S2, S3). The second half of HR1 (residues 569–592) is substantially protected, particularly so for residues 579–592. In contrast, the N-terminal half of HR1 (residues 546–568), which contains the I559P substitution integral to SOSIP.664 trimers, was not protected, indicating that this region is not involved in a stable secondary structure. A moderate level of exchange protection was seen within the fusion peptide proximal region (FPPR, residues 520–537), whereas the fusion peptide itself (FP, as probed via residues 512–519) was fully exchanged within seconds. The sequence following HR1 (residues 593–604), including the disulfide loop and the immunodominant epitope cluster I, was also substantially protected. Immediately C-terminal to the disulfide loop, there was only moderate protection for residues 603–622 spanning the N611 and N616/618 (KNH1144/BG505) glycosylation sites. The rapid exchange observed in a smaller, overlapping BG505 peptide indicates that residues 615–622 do not have a stable secondary structure. In HR2, segments 623–634 and 641–646 are very strongly protected in the pre-fusion form of the trimer, while the intervening segment 635–640 (including glycosylation site N637) is much less well protected. The second half of HR2 (residues 648–664) undergoes rapid exchange and appears to be highly dynamic in solution. Almost all of the SOSIP.664 peptides exhibited unimodal exchange profiles signifying conformational homogeneity within the samples, without any evidence of slow, large-scale, correlated protein motions (EX1 exchange kinetics). The only exception was fragment 538–547, which exhibited bimodal behavior at early time points in both the KNH1144 and BG505 datasets (see Figure S4).
Effect of glycosylation on underlying protein structural dynamics
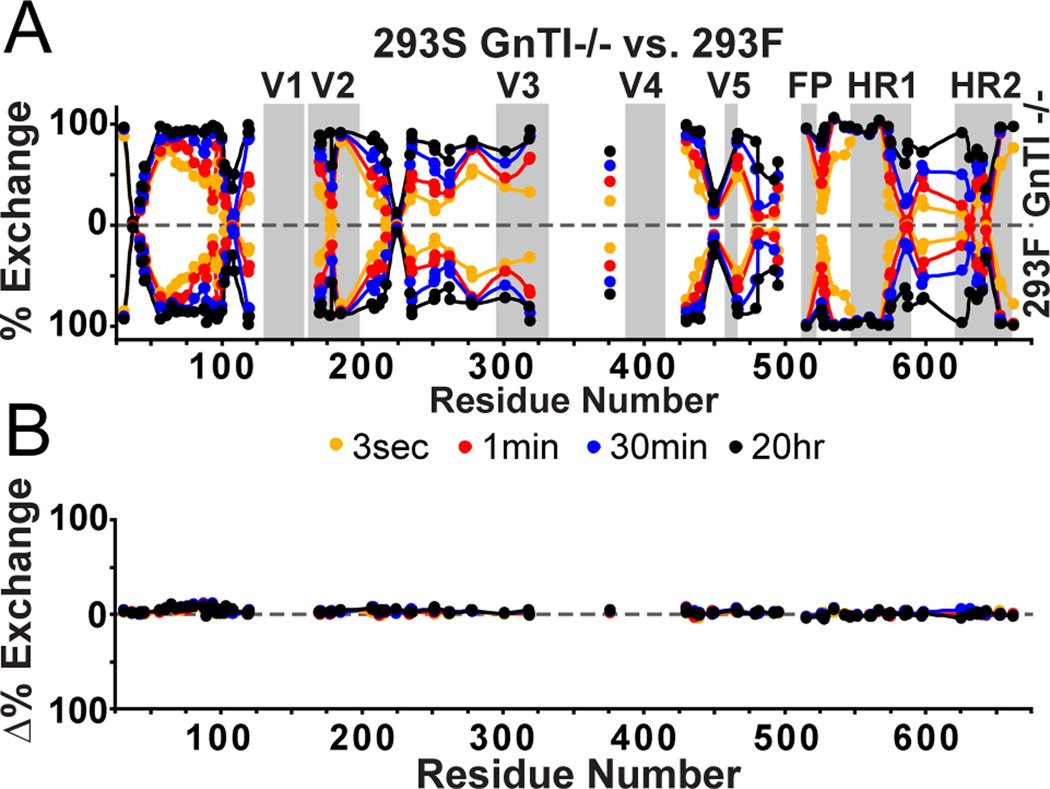
We addressed whether differences in trimer glycosylation affect the behavior of the underlying protein structure. To do so, we compared HDX profiles for BG505 SOSIP.664 trimers expressed in 293F cells (capable of making complex glycans) and 293S GnTI−/− cells (only high-mannose glycans present). The differences in glycosylation between expression systems were apparent by SDS and BN-PAGE (Figure S1). The peptic digest showed that the majority of observable glycopeptides from 293F–trimers were still in the high-mannose form, and only glycans at sites N88 and N462 had been processed to complex type (Table 1). Since complex glycoforms are generally harder to detect due to their lower ionization efficiency and microheterogeneity (Stavenhagen et al., 2013), we also examined the relative intensities of matched glycopeptides derived under identical conditions from BG505 SOSIP.664 trimers that had been produced in the two different cell types for a more direct assessment of glycosylation differences (Figure S5). The signals for the high-mannose glycopeptides at N276, N197, N262, N234, N448, N625 and N637 were relatively consistent between 293F and 293S data sets, suggesting that high mannose glycans are indeed present at all these sites irrespective of the cell type used to make the trimers. In contrast, the signal loss for the high mannose glycoforms at N88, N190, N462, and N611/N618 in the 293F dataset indicates that the glycans at these sites had probably been processed to complex types. Despite the glycosylation differences, the HDX profiles for the 293F and 293S material were very similar (Figure 2), suggesting that the conformational dynamics of the underlying protein structure is largely insensitive to any differences in glycan processing.
Figure 2. HDX-MS comparisons of 293F vs. 293S GnTI−/− expression system.
A) The butterfly plot shows the exchange profiles of BG505 SOSIP.664 trimers from 293S GnTI−/− cells (all high mannose glycoforms, top) vs. 293F cells (contains complex glycosylation, bottom). B) The differences at each time point are plotted in the difference plot underneath, revealing no major changes attributed to differences in glycosylation types.
CD4-induced changes within the trimer
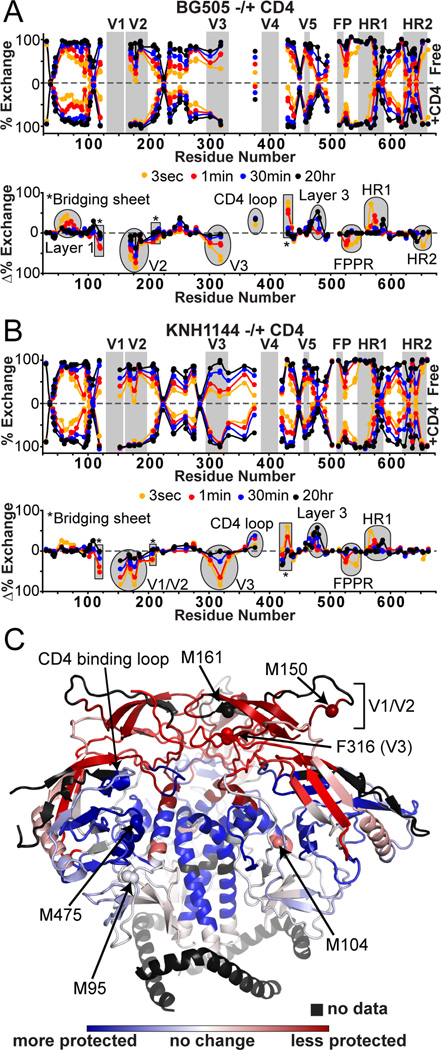
The binding of soluble, two-domain CD4 leads to dramatic conformational changes that are virtually identical for BG505 and KNH1144 SOSIP.664 trimers, as monitored by HDX-MS (Figure 3). The most striking changes occurred within V1/V2 and V3, which lost all protection upon CD4 binding. As expected, the CD4 binding region itself was more protected in the bound state. Major changes also occurred within regions that comprise the bridging sheet; strand β21 became more protected, while the other 3 strands (β2, β3 and β20) and the segment directly C-terminal to β21 was less protected. There was also an increase in protection within all three layers of the gp120 inner domain. Within gp41, the FPPR became less protected while the converse was seen for the C-terminal half of HR1. The slight protection of the second half of HR2 in the unliganded BG505 trimer was lost upon CD4 binding. A minor fraction the trimer-CD4 complex showed dimerization when analyzed on BN-PAGE gels for the BG505, but not the KNH1144 construct (Figure S1). However, the HDX data yielded no indications of the presence of multiple Env species, as assessed by the lack of bimodal distributions in the mass envelopes (Guttman et al., 2013; Weis et al., 2006). Thus, if some degree of oligomerization did occur in solution, it did not affect the HDX profile of the BG505 trimer-CD4 complex.
Figure 3. Changes upon CD4 binding by HDX.
Butterfly plots comparing the exchange profiles for SOSIP.664 trimers unliganded (top) and in complex with sCD4 (bottom) for KNH1144 (A) and BG505 (B). The corresponding difference plots below highlight the regions that gain and lose protection upon sCD4 binding, plotted above and below the axis, respectively. (C) Regions that are more protected (blue) or less protected (red) upon CD4 binding are mapped on the BG505 SOSIP.664 trimer crystal structure (PDB 4NCO) (Julien et al., 2013a). KNH1144 sequence data were used for these heat maps due to the greater coverage, although BG505 shows nearly identical trends for the regions covered. Individual exchange plots with errors are shown in Figures S2 and S3. Labeled spheres highlight the specific residues monitored by oxidative labeling.
Oxidative labeling with mass spectrometry was used as a complementary approach to track changes in the side-chain solvent accessibility of BG505 SOSIP.664 trimers upon CD4 binding. Although oxidative labeling was highly reproducible (Figure 4A), the sample-to-sample variability in digestion efficiency significantly affected the signal intensities of individual peptides, such that only residues highly susceptible to oxidation could be monitored reliably (see Methods). We specifically examined residues M95, M104, M150, M161, F316, M475 and M626, as well as leucine residues L660, L661 and L663 near the C-terminus of gp41 in this construct (Figure 3C). While M104, M95 and the triad of gp41 leucines exhibited little response to CD4 binding, significant changes were observed for M150 and M161 in V1/V2 and F316 in V3. These residues were all oxidized much more rapidly after CD4 bound, indicative of a marked increase in their solvent accessibility (Figure 4B). There was also a slight increase in the oxidation rate for M626 in gp41 HR2. In contrast, the oxidation rate at M475 in layer 3 of the gp120 inner domain was decreased, suggesting that this residue becomes more buried following CD4 binding.
Figure 4. Solvent accessibility changes upon CD4 binding monitored by oxidative labeling.
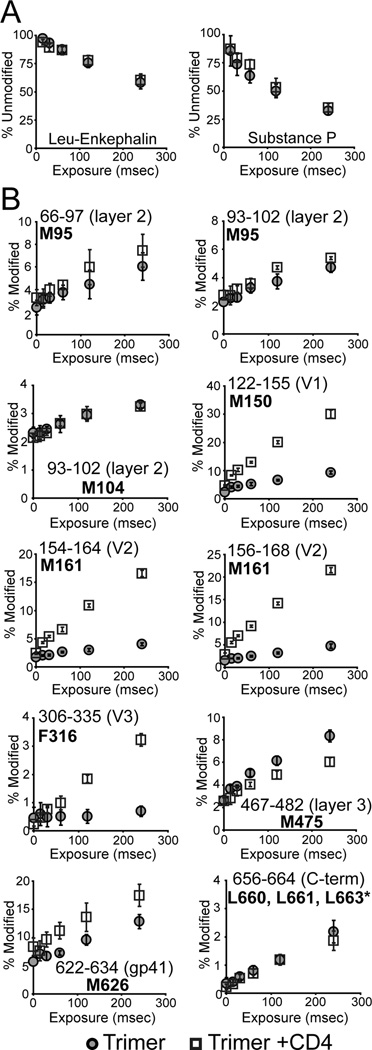
(A) Modification rates of peptides used as “dosimeters” for ensuring that both the unliganded and CD4-bound data sets were exposed to similar amounts of oxidative labeling. The signal for the unmodified form (normalized to a non-irradiated sample) of Leucine Enkephalin (left) and Substance P (right) are shown as a function of exposure to synchrotron radiation. (B) The degree of oxidation is shown as a function of radiation exposure for various regions of SOSIP.664 (BG505) in the unliganded (gray circles) or CD4-bound (squares) state. Percent modified refers to the signal intensity of the oxidized from (+16 Da) relative to the sum of signals for the modified and unmodified peptide, with the sites of oxidation shown in bold. Error bars represent the standard deviation between duplicate measurements. Examples of mass spectra revealing changes in oxidation rates are shown in Figure S6. The oxidation sites are illustrated on the trimer in Figure 3C.
*Data for peptide 656–664 encompass oxidation at residues L660, L661, and L663.
Effect of small compound compounds BMS-806 and NBD-556
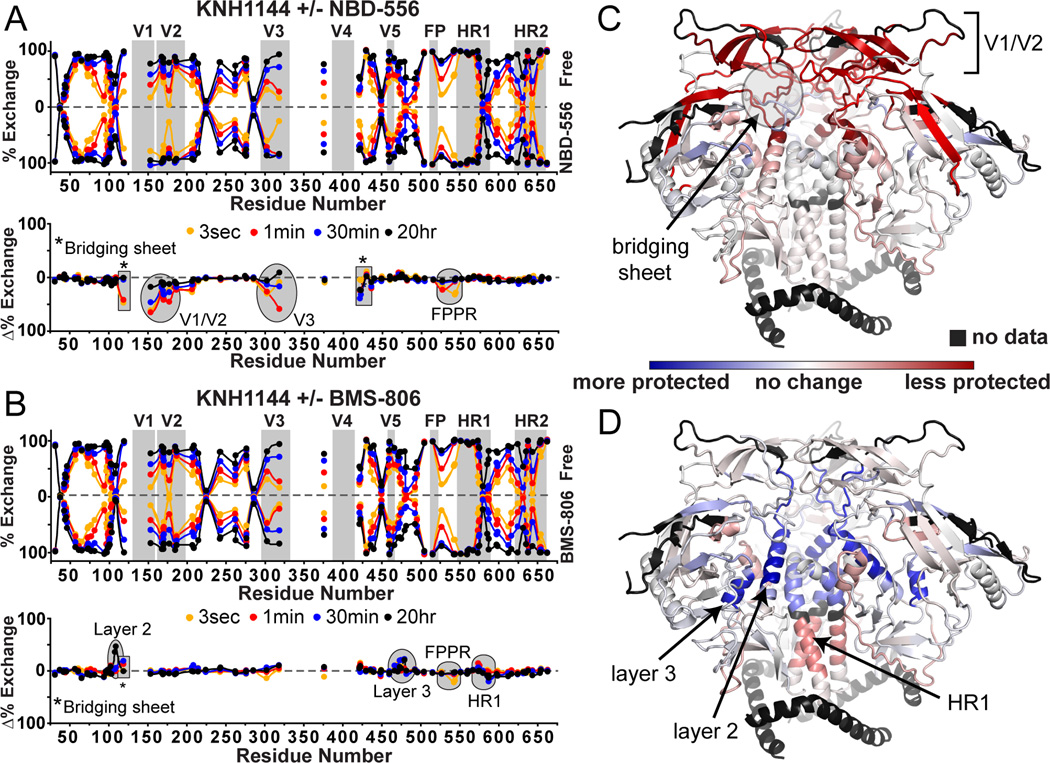
To further investigate ligand-induced allosteric changes in the structure of SOSIP.664 trimers, we used HDX to examine the effects of two small molecule compounds that target the CD4 binding site and inhibit a subset of HIV-1 isolates (Zhao et al., 2005). NBD-556 binds to the Phe43 cavity in gp120 (Kwon et al., 2012) and induces conformational changes similar to CD4, at least in the context of monomeric gp120 (Schon et al., 2006). BMS-806 is also thought to bind at the CD4 binding site, but has been shown to block these same conformational changes (Guo et al., 2003; Madani et al., 2004). Reported IC50 values for NBD-556 are generally in the µM range (Schon et al., 2006; Zhao et al., 2005), while for BMS-806 are in the nM range (Madani et al., 2004; Si et al., 2004). Thus, HDX experiments with the KNH1144 SOSIP.664 trimers were performed in the presence of 100 µM of each compound to ensure saturating occupancy, and all buffers contained 2% DMSO to maintain the compounds in solution. Under these conditions, the binding of antibody 17b, which recognizes a CD4-induced epitope overlapping the co-receptor binding site (Thali et al., 1993), to the trimers was enhanced by NBD-556 but hindered by BMS-806 (Figure S7A). An additional unliganded HDX dataset derived using the same buffer showed that the inclusion of 2% DMSO had minimal effects (Figure S7B).
The conformational changes induced in the KNH1144 SOSIP.664 trimer by NBD-556 binding were in part similar to those seen upon CD4 binding (Figure 5A, cf. Figure 3A). For example, destabilization occurred within V1/V2 and V3, throughout the bridging sheet, and even at the FPPR within gp41. However, in contrast to CD4, NBD-556 binding did not lead to observable changes in the gp120 inner domain or in HR1 of gp41 (Figure 5B). In contrast, the binding of BMS-806 altered the trimer in a way that is substantially different to the effect of CD4, or of NBD-556 (Figure 5C, D). When BMS-806 bound, layers 2 and 3 of the gp120 inner domain became more protected with only modest effects on gp41 FPPR and HR1 and, notably, no major changes in V1/V2 or V3.
Figure 5. Changes upon NBD-556 and BMS-806 binding by HDX.
Butterfly and difference plots showing the effects of NBD-556 (A), and BMS-806 (B) on KNH1144 SOSIP.664 trimers. Differences upon binding NBD-556 (C) and BMS-806 (D) are plotted on the trimer crystal structure (PDB 4NCO), revealing regions that are more protected (blue) or less protected (red) upon CD4 binding. All of the major changes in the highlighted regions are significant as assessed by the experimental error (individual plots shown in Figure 6).
DISCUSSION
Structural organization of Env trimers in solution
Cleaved, stabilized SOSIP gp140s are presently the only form of recombinant soluble trimers with a native-like structure, as exemplified by the KNH1144 (Bartesaghi et al., 2013; Harris et al., 2011) and BG505 SOSIP trimers (Julien et al., 2013a; Julien et al., 2013b; Khayat et al., 2013; Lyumkis et al., 2013; Sanders et al., 2013). The data presented here provide additional evidence that these SOSIP trimers have conformations consistent with the functional, pre-fusion Env structure. In contrast, the alternative way to make soluble gp140s, by knocking-out the inter-subunit cleavage site, yields non-native proteins with heterogeneous, splayed-out configurations of the gp120 heads held together by the gp41 stem, which has probably adopted a post-fusion (i.e., 6-helix bundle) state (Guttman and Lee, 2013; Ringe et al., 2013).
The present analysis gives insight into the underlying structural dynamics in the unliganded trimer while also providing sequence-specific information about elements that could not be resolved in the recent ~5 Å structures. Both the cryo-EM trimer structure in complex with antibody PGV04 and the crystal structure in complex with antibody PGT122 were obtained using the same BG505 SOSIP.664 construct studied here (Julien et al., 2013a; Lyumkis et al., 2013). These structures revealed how the V1/V2 and V3 variable regions form intimate interactions at the trimer crown. We find that several regions of V1/V2 do indeed have a high degree of HDX protection, which is consistent with the extensive β-sheet secondary structure present in the Greek key motif seen in the PG9-bound scaffolded V1/V2 structure (McLellan et al., 2011) and in the SOSIP.664 trimer structures (Julien et al., 2013a; Lyumkis et al., 2013). Similarly, the V3 region of the trimer is extensively protected before, but not after, CD4 binding (Figure 3A, B). No such protection of the V1/V2 or V3 regions was seen in our previous studies of monomeric gp120 (Davenport et al., 2013; Guttman et al., 2012) or monomeric, uncleaved gp140 (Guttman and Lee, 2013) implying that these regions of Env are only well-ordered in pre-fusion trimers. The slow oxidative labeling kinetics seen for residues M161 and F316 reinforces the assessment that V1/V2 and V3 segments are largely inaccessible to the solvent in the pre-fusion form of the BG505 SOSIP.664 trimer (Figure 4B). Thus, while V3 is well exposed on gp120 monomers, our results reinforce its sequestration within the trimer structure (Julien et al., 2013a; Lyumkis et al., 2013).
While the available structures reveal the overall organization of the pre-fusion form of gp41, for example highlighting a central helical bundle, much of this subunit has not yet been interpreted in atomic detail (Julien et al., 2013a; Lyumkis et al., 2013). The new HDX-MS data provide additional sequence-specific information for gp41 structural order. The N-terminal half of HR1 (residues 550–568), which contains the I559P gp41-stabilizing modification, is in rapid exchange and, hence, is not involved in any stable secondary structure. In contrast, the rest of HR1 (TVWGIKQLQARVLAVERYLRDQQL; residues 569–592) is extensively protected (Figure 1B, C). We infer that the long central helices observed in the trimer structures (Bartesaghi et al., 2013; Julien et al., 2013a; Lyumkis et al., 2013) correspond to residues 569–592, rather than the entirety of HR1 (546–586) that is observed as a helix in the post-fusion gp41 structure (Chan et al., 1997).
In both the crystal and EM structures of the BG505 SOSIP.664 trimer, HR2 is proposed to form a long helix. We now find, via HDX, that the C-terminal helical portion of HR2 (residues 649–664) is only modestly protected. This region thus appears to be more dynamic in solution than is implied by the static structures in the crystal and in vitreous ice. In contrast, significant segments in the N-terminal portion of HR2 and the preceding gp41 loop region are highly protected and, hence, likely to be engaged in a stable secondary structure. These elements of the trimer are situated close to the gp120/gp41 interface as they are linked, via the engineered SOS disulfide bond (C501-C605), to the C-terminal region of gp120 (Binley et al., 2000). Of note is that the N- and C-terminal extensions of gp120 are also highly protected in the trimer but not in gp120 (Davenport et al., 2013; Guttman et al., 2012), again consistent with their involvement in gp41 association (Helseth et al., 1991).
The N-terminal portion of the FP was protected negligibly, which is surprising since this hydrophobic region might be anticipated to be solvent-occluded in the pre-fusion state. In a similar study with influenza hemagglutinin, also a type I fusion protein, the FP was indeed moderately protected in a way attributable to either solvent occlusion or hydrogen bonding with residues lining the FP pocket (Garcia et al., unpublished data; Wilson et al., 1981). In Env trimers, the FP may be only loosely buried, without any participation of its backbone in hydrogen bonds. It is also possible that the SOSIP modifications partially disrupt the packing of the FP. Alternatively, the MPER, which is absent from the SOSIP.664 construct, may play a role in occluding the FP in full-length, functional trimers.
SOSIP.664 trimers have a much higher content of high mannose glycans than was found in a previous study of monomeric gp120 (Leonard et al., 1990). This finding is consistent with previous reports that quaternary interactions within Env trimers occlude several glycan chains from glycosidases and therefore maintain their high mannose form (Doores et al., 2010). Positions N276, N197 and N301, which bear complex glycans in the context of gp120 monomers and uncleaved gp140 constructs (Go et al., 2008), show little mannose trimming and are present as predominantly Man9 and Man8 glycoforms in SOSIP.664 trimers, even from 293F cells. As none of these glycans are part of the 2G12 epitope, the use of a 2G12 affinity column to purify the trimers is unlikely to skew the observed glycosylation profiles (Sanders et al., 2002a). Collectively, these observations may be relevant to immunogen design as several of these glycans are involved in bNAb epitopes (Jardine et al., 2013; Pancera et al., 2013; Pejchal et al., 2011; Walker et al., 2011).
CD4-induced transitions in Env trimers
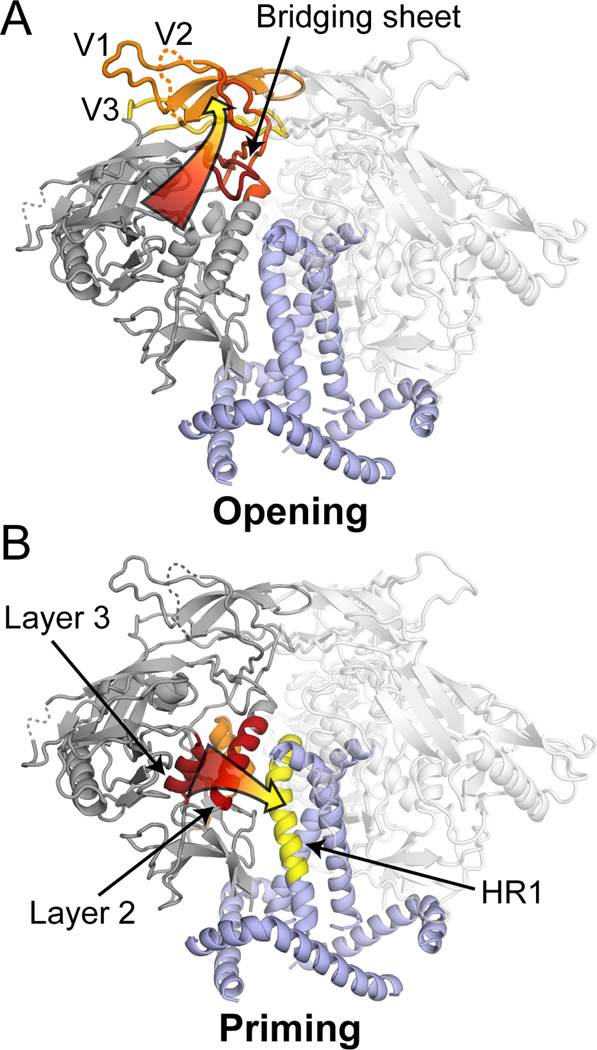
These new data provide a detailed glimpse into the CD4-induced reorganization of the trimer from the closed to the open form, a process that has been imaged at low-resolution by cryo-EM (Harris et al., 2011; Liu et al., 2008; Tran et al., 2012). In the closed, pre-fusion form, the V1/V2 region is involved in inter- and intra-protomer interactions that form a cap at the trimer apex (Figure 6A). Likewise, the V3 and the bridging sheet segments, which form the co-receptor binding site, are also constrained by their involvement in the quaternary structure within the pre-fusion trimer. Upon CD4 binding, the structure at the crown is disrupted and the V1/V2 and V3 regions become disordered (Figure 3C). The oxidation rates for V2 residues M150, M161 and V3 residue F316 are also drastically increased when CD4 binds (Figure 4B). This increase in the local solvent accessibility is again consistent with an unraveling of the V1/V2 and V3 interactions at the trimer apex as the entire subdomain structure opens up. Overall, our data illustrate how CD4 binding acts to disrupt key quaternary interactions and thereby expose the elements necessary for co-receptor binding and the subsequent conformational changes that drive fusion. Moreover, these same conformational changes uncover various NAb epitopes that are efficiently shielded on the pre-fusion form of the trimer, such as those associated with the co-receptor binding site, including V3 elements.
Figure 6. Key CD4-induced changes in trimeric SOSIP.664 monitored by HDX-MS.
(A) The SOSIP.664 trimer crystal structure (PDB 4NCO) (Julien et al., 2013a) was modeled into the EM density of unliganded Env on virions (EMD 5019; (Liu et al., 2008): V1/V2 (yellow) and V3 (green), the bridging sheet (red), HR1 (purple) and gp120 inner domain (black). (B) The sCD4-bound gp120 core structure (PDB 3JWD; (Pancera et al., 2010) was modeled into the sCD4-bound electron density map (EMD 5455) (Tran et al., 2012) with CD4 shown in blue. In the CD4-bound state, the bridging sheet is reorganized, positioning V1/V2 away from the trimeric interface and exposing the V3 loop. Individual exchange profiles of the key regions of interest are shown for unliganded trimer (blue), sCD4 bound (red), NBD-556 bound (orange) and BMS-806 bound (cyan). Error bars represent standard deviations from duplicate measurements. EM renderings were made with Chimera (Pettersen et al., 2004).
The effects of CD4 binding extend beyond the immediate contact zone to the gp120 inner domain as well as to previously unresolved elements in gp41. The increased protection of the inner domain, along with a decrease in solvent accessibility (as probed by the oxidation rate of M475), is not surprising as these regions reside at the interface with CD4. More unexpected was the large allosteric effect observed in gp41 FPPR and HR1. Upon CD4 binding, the FPPR becomes less protected, alleviating constraints around the FP while the center of HR1 becomes more protected, possibly a reflection of enhanced packing of the central helical core. For the BG505 trimers, a slight decrease in protection at the C-terminal half of HR2 was also evident (Figure 3B). These alterations to gp41 might serve as an initial priming event prior to the full activation of Env by co-receptor binding.
CD4 binding site targeted ligands reveal distinct allosteric networks in Env
A comparison of the effects imparted on Env trimers by the ligands examined here suggests that two allosteric networks are engaged when CD4 activates the trimer: one involves “opening” of the Env crown, the other leads to “priming” of gp41. The network attributable to “opening” involves rearrangements of the bridging sheet elements and the disruption of inter-protomer interactions mediated by V1/V2 and V3, and culminates in the formation/exposure of the co-receptor binding site (Figure 7A). The “priming” network links the CD4 binding site to HR1 through layers 1–3 of the gp120 inner domain (Figure 7B).
Figure 7. Allosteric networks in Env trimers.
CD4 binding leads to two allosteric effects within Env trimers (red to yellow pathways). Pathways are highlighted on a single protomer of the trimer with neighboring protomers rendered transparent. (A) Repositioning of the elements of the bridging sheet leads to the disruption of the trimeric interactions within V1/V2 and V3, while somehow also affecting the FPPR, whose location and relation to the crown region have yet to be interpreted in high resolution structures. (B) The conformational changes induced within three layers of the inner domain also influence HR1 within gp41.
While CD4 binding triggers both networks, NBD-556 leads only to the opening of Env and does not appear to prime gp41 (Figure 5, Figure 6). This mode of action was not evident from earlier structural studies that used a truncated gp120 core, which adopts a receptor-bound conformation with an ordered inner domain that is nearly identical in structure to the CD4-bound core (Kwon et al., 2012). Conversely, BMS-806 appears to influence the priming network, without affecting the opening network. Based on our interpretation that residues 569–592 correspond to the central gp41 helix, the BMS-806 induced changes in HR1 appear to be driven through the α1 helix in gp120 layer 2, which like layer 3 also becomes substantially more ordered (Figures 5D, 7B).
The gp120 inner domain changes that we implicate as part of the priming pathway are consistent with mutagenesis studies that probe the transduction of CD4-induced conformational changes through the inner domain to gp41 (Desormeaux et al., 2013; Finzi et al., 2010). It is surprising perhaps that the FPPR in gp41 responds to both CD4 and NBD-556 whereas the rest of gp41 appears unchanged by NBD-556 binding. The position of the FPPR was not interpreted in the existing SOSIP.664 trimer structures, so its linkage to the opening network is not yet clear.
Overall, our present HDX-MS analysis reveals how CD4 and CD4 mimetics induce profound conformational changes in the trimer by activating distinct networks that open the trimer crown, unmask key elements of the co-receptor binding site, and prime the fusogenic properties of the gp41 subunit. Further studies will be required to address how the remaining unmapped elements of the trimer, and the co-receptor binding event, are orchestrated to drive the membrane fusion stages of virus entry.
EXPERIMENTAL PROCEDURES
Sample preparation
KNH1144 and BG505 SOSIP.664 were expressed and purified by 2G12 affinity chromatography as described previously (Iyer et al., 2007). A final round of size exclusion chromatography over a Superdex 200 column (GE Healthcare) in PBS (20mM Sodium Phosphate pH 7.4, 150mM NaCl, 1mM EDTA, 0.02% sodium azide) was performed prior to HDX to remove Env aggregates, dimers and monomers. Proteins were concentrated with VivaSpin spin filters (GE Healthcare). Sample purity was assessed with reducing and non-reducing SDS-PAGE and BN-PAGE (Figure S1). The complex with sCD4 was formed by overnight incubation with a threefold molar excess of two-domain human sCD4 at 4°C. sCD4 was obtained from the NIH AIDS reagents program (Garlick et al., 1990). BMS-806 was purchased from SelleckChem. NBD-556 was synthesized and purified as described previously (Schon et al., 2006).
Hydrogen/Deuterium-Exchange Mass Spectrometry
A 10 µg (80 pmol) aliquot of SOSIP.664 (per time point) was incubated in deuterated buffer (85% D2O) at 22°C for 3 s, 1 min, 30 min, and 20 h. Samples were added to an equal volume (100uL) of ice-cold quench solution (200mM TCEP, 0.02% formic acid) with 10 µL of a 3 mg/mL pepsin solution in 100 mM Na3PO4 pH 4.0, to a final pH of 2.5. After 5 minutes of pepsin digestion on ice, samples were flash frozen in liquid nitrogen and stored at −80°C until analysis. Deglycosylated peptic digests were prepared by treatment with either: 0.5 mU of N-glycanase (Prozyme) (pH 7.0) or 0.1 mU EndoH (Prozyme) (pH 5.5) for 2 h at 37°C. Compounds NBD-556 and BMS-806 were resuspended in DMSO (>99.9% Sigma-Aldrich) and pre-incubated with SOSIP.664 at a final concentration of 100 µM in a buffer containing 2% DMSO. Identical compound and DMSO concentrations were included in the deuterium incubations.
Deuterated samples were analyzed by LC-MS as previously described (Guttman and Lee, 2013). Peptides were identified by exact mass and MS/MS spectra with the aid of protein prospector (Baker et al., 2011). Identification of glycopeptides was aided by MS/MS spectra of the enzymatically deglycosylated pepsin digests. Deuterium shifts were calculated with using HX-Express v2 (Guttman et al., 2013; Weis et al., 2006) to monitor the mass envelope widths for possible bimodal behavior. The percentage exchange for each fragment was calculated relative to zero and fully deuterated standards, as described previously (Guttman et al., 2012). Heat maps were made using PyMOL (DeLano, 2002).
Since additional GlcNAc groups in complex type glycan chains can contribute to the observed deuterium uptake kinetics, comparisons of the exchange kinetics of glycopeptides with different glycan compositions may be misleading (Guttman et al., 2011). For this reason, the HDX comparisons between 293S GnTI−/−. and 293F expressed material were made only with data from the high mannose glycoforms within the 293F dataset, which were a minor species for sites showing complex type glycosylation. While this partially limits the scope of the comparison, if altered glycosylation does have structural or dynamic effects beyond the glycosylation site, it is likely that differences in the HDX profiles of peripheral regions will be seen.
Oxidative labeling
X-ray irradiation experiments were performed at Beamline 4-2 of the Stanford Synchrotron Radiation Lightsource. The X-ray beam (11 keV with a ring current of 495–500 mA) was attenuated to give dose responses in the appropriate range for oxidative labeling as assessed on-site using samples containing 80 µM Alexafluor-488 dye in an identical phosphate buffer and Ribonuclease A at 1 mg/mL, to mimic sample conditions (Xu and Chance, 2007). Samples of unliganded and CD4-bound trimer (with a threefold molar excess of sCD4 relative to each SOSIP.664 protomer) in PBS were all prepared at a total protein concentration of 1 mg/mL to avoid differences in scavenging due to protein concentration effects (Tong et al., 2008). 330 ng of Leucine Enkephalin and 580 ng of Substance P (Sigma Aldrich) were included in all samples as internal peptide dosimeters to ensure consistency of labeling between sample sets (Tong et al., 2008). Samples (20 µL) were loaded with an autosampler using a Microlab 560 syringe pump (Hamilton) and irradiated as they passed through a quartz capillary. The flow rates during irradiation were adjusted to achieve exposure times ranging from 15 to 240 msec. Immediately after exposure the samples were dispensed and mixed into a tube containing 2.5 µL of 200 mM methionine to prevent secondary oxidation (Xu et al., 2005), and stored at −20°C. Samples with no irradiation were collected under identical conditions to serve as the non-irradiated standards. All experiments were performed in duplicate.
Irradiated samples were mixed 1:1 with a solution of 6 M Guanidine HCl, 20 mM DTT, 50mM Tris pH 8.0 and denatured at 85°C for 30 min. Cysteines were then alkylated by incubation in the dark for 1 h with 20 mM iodoacetamide, followed by an addition of 10 mM DTT to quench the reaction. After dilution with 20 mM Tris pH 8.0 to achieve a final Gnd-HCl concentration of 0.5 M, samples were treated with 0.5 mU of N-glycanase (Prozyme), at 37°C for 90 min to achieve full enzymatic deglycosylation. LysC and GluC (Promega) were then added at ratios of 6:1 and 3:1 (substrate:enzyme) and left at 37°C for 18 h. Samples were analyzed by LC-MS on a Synapt Q-TOF mass spectrometer coupled to a Aquity UPLC system (Waters). Peptides were loaded onto a Hypersil 1×50mm 2.1 µm C18 column (Thermo Scientific) and resolved with a gradient of 0 to 32% B over 15 min (A: 3% Acetonitrile, 0.1% formic acid; B: 100% acetonitrile, 0.1% formic acid). The GluC and LysC digests were run in alternating order to minimize any effects of sample carryover. Chromatographic peaks for the most abundant and non-overlapped isotopic peaks were integrated with MassLynx (Waters).
Oxidation rates were initially calculated by monitoring the net signal intensity of the unmodified peptide as a function of irradiation dose. Although this type of analysis was useful for monitoring the oxidation rates of the internal dosimeter peptides, it was not suitable for the analysis of the Env derived proteolytic fragments. The high variability in digestion efficiency during sample processing led to significant variability in the resulting peptide signal intensities. Instead, the intensity of each modified form of a peptide (+16 Da) was measured relative to the sum of the modified and unmodified signal intensities (Chen et al., 2012), a metric that should be independent of digestion efficiency. This approach provided precise, residue-specific information, but limited the scope of the analysis since only peptides with a sufficient degree of modification could be analyzed. Peptides and oxidation sites were identified by manual inspection of MS/MS spectra aided by Protein Prospector (Baker et al., 2011).
Supplementary Material
Highlights.
-
-
CD4 binding disrupts quaternary interactions at the Env trimer apex
-
-
CD4 binding induces dynamic changes within portions of gp41
-
-
NBD-556 and BMS-806 have drastically different effects on Env
-
-
Different glycosylation profiles have little effect on Env’s conformational dynamics
ACKNOWLEDGMENTS
We thank Matthew McDonald for assistance with the synthesis of NBD-556 and Asim Debnath, Per Johan Klasse, Max Crispin, and Andrew B. Ward for valuable discussions. Two-domain sCD4-183 (Pharmacia, Inc.) was obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH. This work was supported by NIH grant F32-GM097805 (M.G.), T32-GM007750 (N.K.G.), R00-GM080352 (K.K.L.), R01-GM099989 (K.K.L.), P01 AI82362 (J.P.M., I.A.W.), R01 AI084817 (I.A.W.), NIH R37 AI36082 (J.P.M.), NIH R01 AI41420 (J.P.M.), the Bill and Melinda Gates Foundation Collaboration for AIDS Vaccine Discovery (CAVD) grant OPP1033102 (M.G., K.K.L.), UAB CFAR grant (P30-AI027767) through the CNIHR program for new HIV investigators (K.K.L.) as well as the International AIDS Vaccine Initiative Neutralizing Antibody Consortium and Center (I.A.W., J.P.M), CHAVI-ID UM1 AI100663 (I.A.W.), the Hope Barns Fellowship (N.K.G.), a Vidi grant from the Netherlands Organization for Scientific Research (NWO) (R.W.S.), a Starting Investigator Grant from the European Research Council ERC-StG-2011-280829-SHEV (R.W.S.), and a Canadian Institutes of Health Research fellowship (J.P.J.). Portions of this research were carried out at the Stanford Synchrotron Radiation Lightsource. The SSRL Structural Molecular Biology Program is supported by the Department of Energy’s Office of Biological and Environmental Research and by the National Institutes of Health’s National Institute of General Medical Sciences (Grant P41GM103393) and National Center for Research Resources (Grant P41RR001209).
Abbreviations
- HDX-MS
hydrogen deuterium exchange coupled with mass spectrometry
- DMSO
dimethyl sulfoxide
- Env
HIV envelope glycoprotein
- FP
Fusion peptide
- FPPR
fusion peptide proximal region
- HR
heptad repeat domain
- MPER
membrane proximal external region
- TM
transmembrane domain
- CT
cytoplasmic tail
- BN-PAGE
Blue-native polyacrylamide gel electrophoresis
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author contributions:
MG, NKG, TM conducted the experiments. AC, JPJ and RWS provided essential reagents. MG and KKL analyzed the data and made the figures. MG, NKG, JPJ, JPM, RWS, IAW and KKL wrote the manuscript.
REFERENCES
- Baker PR, Trinidad JC, Chalkley RJ. Modification site localization scoring integrated into a search engine. Mol Cell Proteomics. 2011;10:M111. doi: 10.1074/mcp.M111.008078. 008078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartesaghi A, Merk A, Borgnia MJ, Milne JL, Subramaniam S. Prefusion structure of trimeric HIV-1 envelope glycoprotein determined by cryo-electron microscopy. Nat Struct Mol Biol. 2013;20:1352–1357. doi: 10.1038/nsmb.2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binley JM, Sanders RW, Clas B, Schuelke N, Master A, Guo Y, Kajumo F, Anselma DJ, Maddon PJ, Olson WC, Moore JP. A recombinant human immunodeficiency virus type 1 envelope glycoprotein complex stabilized by an intermolecular disulfide bond between the gp120 and gp41 subunits is an antigenic mimic of the trimeric virion-associated structure. J Virol. 2000;74:627–643. doi: 10.1128/jvi.74.2.627-643.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binley JM, Sanders RW, Master A, Cayanan CS, Wiley CL, Schiffner L, Travis B, Kuhmann S, Burton DR, Hu SL, et al. Enhancing the proteolytic maturation of human immunodeficiency virus type 1 envelope glycoproteins. J Virol. 2002;76:2606–2616. doi: 10.1128/JVI.76.6.2606-2616.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan DC, Fass D, Berger JM, Kim PS. Core structure of gp41 from the HIV envelope glycoprotein. Cell. 1997;89:263–273. doi: 10.1016/s0092-8674(00)80205-6. [DOI] [PubMed] [Google Scholar]
- Chen J, Rempel DL, Gau BC, Gross ML. Fast photochemical oxidation of proteins and mass spectrometry follow submillisecond protein folding at the amino-acid level. J Am Chem Soc. 2012;134:18724–18731. doi: 10.1021/ja307606f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davenport TM, Guttman M, Guo W, Cleveland B, Kahn M, Hu SL, Lee KK. Isolate-specific differences in the conformational dynamics and antigenicity of HIV-1 gp120. J Virol. 2013;87:10855–10873. doi: 10.1128/JVI.01535-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLano WL. The PyMOL molecular graphics system. San Carlos, CA, USA: DeLano Scientific; 2002. [Google Scholar]
- Desormeaux A, Coutu M, Medjahed H, Pacheco B, Herschhorn A, Gu C, Xiang SH, Mao Y, Sodroski J, Finzi A. The highly-conserved layer 3 component of the HIV-1 gp120 inner domain is critical for CD4-required conformational transitions. J Virol. 2013;87:2549–2562. doi: 10.1128/JVI.03104-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doores KJ, Bonomelli C, Harvey DJ, Vasiljevic S, Dwek RA, Burton DR, Crispin M, Scanlan CN. Envelope glycans of immunodeficiency virions are almost entirely oligomannose antigens. Proc Natl Acad Sci U S A. 2010;107:13800–13805. doi: 10.1073/pnas.1006498107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finzi A, Xiang SH, Pacheco B, Wang L, Haight J, Kassa A, Danek B, Pancera M, Kwong PD, Sodroski J. Topological layers in the HIV-1 gp120 inner domain regulate gp41 interaction and CD4-triggered conformational transitions. Mol Cell. 2010;37:656–667. doi: 10.1016/j.molcel.2010.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garlick RL, Kirschner RJ, Eckenrode FM, Tarpley WG, Tomich CS. Escherichia coli expression, purification, and biological activity of a truncated soluble CD4. AIDS Res Hum Retroviruses. 1990;6:465–479. doi: 10.1089/aid.1990.6.465. [DOI] [PubMed] [Google Scholar]
- Go EP, Irungu J, Zhang Y, Dalpathado DS, Liao HX, Sutherland LL, Alam SM, Haynes BF, Desaire H. Glycosylation site-specific analysis of HIV envelope proteins (JR-FL and CON-S) reveals major differences in glycosylation site occupancy, glycoform profiles, and antigenic epitopes' accessibility. J Proteome Res. 2008;7:1660–1674. doi: 10.1021/pr7006957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Q, Ho HT, Dicker I, Fan L, Zhou N, Friborg J, Wang T, McAuliffe BV, Wang HG, Rose RE, et al. Biochemical and genetic characterizations of a novel human immunodeficiency virus type 1 inhibitor that blocks gp120-CD4 interactions. J Virol. 2003;77:10528–10536. doi: 10.1128/JVI.77.19.10528-10536.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Kahn M, Garcia NK, Hu SL, Lee KK. Solution structure, conformational dynamics, and CD4-induced activation in full-length, glycosylated, monomeric HIV gp120. J Virol. 2012;86:8750–8764. doi: 10.1128/JVI.07224-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Lee KK. A functional interaction between gp41 and gp120 is observed for monomeric but not oligomeric, uncleaved HIV-1 Env gp140. J Virol. 2013;87:11462–11475. doi: 10.1128/JVI.01681-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Scian M, Lee KK. Tracking hydrogen/deuterium exchange at glycan sites in glycoproteins by mass spectrometry. Anal Chem. 2011;83:7492–7499. doi: 10.1021/ac201729v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Weis DD, Engen JR, Lee KK. Analysis of overlapped and noisy hydrogen/deuterium exchange mass spectra. J Am Soc Mass Spectrom. 2013;24:1906–1912. doi: 10.1007/s13361-013-0727-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris A, Borgnia MJ, Shi D, Bartesaghi A, He H, Pejchal R, Kang YK, Depetris R, Marozsan AJ, Sanders RW, et al. Trimeric HIV-1 glycoprotein gp140 immunogens and native HIV-1 envelope glycoproteins display the same closed and open quaternary molecular architectures. Proc Natl Acad Sci U S A. 2011;108:11440–11445. doi: 10.1073/pnas.1101414108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helseth E, Olshevsky U, Furman C, Sodroski J. Human immunodeficiency virus type 1 gp120 envelope glycoprotein regions important for association with the gp41 transmembrane glycoprotein. J Virol. 1991;65:2119–2123. doi: 10.1128/jvi.65.4.2119-2123.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer SP, Franti M, Krauchuk AA, Fisch DN, Ouattara AA, Roux KH, Krawiec L, Dey AK, Beddows S, Maddon PJ, et al. Purified, proteolytically mature HIV type 1 SOSIP gp140 envelope trimers. AIDS Res Hum Retroviruses. 2007;23:817–828. doi: 10.1089/aid.2006.0261. [DOI] [PubMed] [Google Scholar]
- Jardine J, Julien JP, Menis S, Ota T, Kalyuzhniy O, McGuire A, Sok D, Huang PS, MacPherson S, Jones M, et al. Rational HIV immunogen design to target specific germline B cell receptors. Science. 2013;340:711–716. doi: 10.1126/science.1234150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julien JP, Cupo A, Sok D, Stanfield RL, Lyumkis D, Deller MC, Klasse PJ, Burton DR, Sanders RW, Moore JP, et al. Crystal structure of a soluble cleaved HIV-1 envelope trimer. Science. 2013a;342:1477–1483. doi: 10.1126/science.1245625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julien JP, Lee JH, Cupo A, Murin CD, Derking R, Hoffenberg S, Caulfield MJ, King CR, Marozsan AJ, Klasse PJ, et al. Asymmetric recognition of the HIV-1 trimer by broadly neutralizing antibody PG9. Proc Natl Acad Sci U S A. 2013b;110:4351–4356. doi: 10.1073/pnas.1217537110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khayat R, Lee JH, Julien JP, Cupo A, Klasse PJ, Sanders RW, Moore JP, Wilson IA, Ward AB. Structural characterization of cleaved, soluble HIV-1 envelope glycoprotein trimers. J Virol. 2013;87:9865–9872. doi: 10.1128/JVI.01222-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon YD, Finzi A, Wu X, Dogo-Isonagie C, Lee LK, Moore LR, Schmidt SD, Stuckey J, Yang Y, Zhou T, et al. Unliganded HIV-1 gp120 core structures assume the CD4-bound conformation with regulation by quaternary interactions and variable loops. Proc Natl Acad Sci U S A. 2012;109:5663–5668. doi: 10.1073/pnas.1112391109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwong PD, Doyle ML, Casper DJ, Cicala C, Leavitt SA, Majeed S, Steenbeke TD, Venturi M, Chaiken I, Fung M, et al. HIV-1 evades antibody-mediated neutralization through conformational masking of receptor-binding sites. Nature. 2002;420:678–682. doi: 10.1038/nature01188. [DOI] [PubMed] [Google Scholar]
- Leonard CK, Spellman MW, Riddle L, Harris RJ, Thomas JN, Gregory TJ. Assignment of intrachain disulfide bonds and characterization of potential glycosylation sites of the type 1 recombinant human immunodeficiency virus envelope glycoprotein (gp120) expressed in Chinese hamster ovary cells. J Biol Chem. 1990;265:10373–10382. [PubMed] [Google Scholar]
- Lin PF, Blair W, Wang T, Spicer T, Guo Q, Zhou N, Gong YF, Wang HG, Rose R, Yamanaka G, et al. A small molecule HIV-1 inhibitor that targets the HIV-1 envelope and inhibits CD4 receptor binding. Proc Natl Acad Sci U S A. 2003;100:11013–11018. doi: 10.1073/pnas.1832214100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Bartesaghi A, Borgnia MJ, Sapiro G, Subramaniam S. Molecular architecture of native HIV-1 gp120 trimers. Nature. 2008;455:109–113. doi: 10.1038/nature07159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyumkis D, Julien JP, de Val N, Cupo A, Potter CS, Klasse PJ, Burton DR, Sanders RW, Moore JP, Carragher B, et al. Cryo-EM structure of a fully glycosylated soluble cleaved HIV-1 envelope trimer. Science. 2013;342:1484–1490. doi: 10.1126/science.1245627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madani N, Perdigoto AL, Srinivasan K, Cox JM, Chruma JJ, LaLonde J, Head M, Smith AB, Sodroski JG., 3rd Localized changes in the gp120 envelope glycoprotein confer resistance to human immunodeficiency virus entry inhibitors BMS-806 and #155. J Virol. 2004;78:3742–3752. doi: 10.1128/JVI.78.7.3742-3752.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcsisin SR, Engen JR. Hydrogen exchange mass spectrometry: what is it and what can it tell us? Anal Bioanal Chem. 2010;397:967–972. doi: 10.1007/s00216-010-3556-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLellan JS, Pancera M, Carrico C, Gorman J, Julien JP, Khayat R, Louder R, Pejchal R, Sastry M, Dai K, et al. Structure of HIV-1 gp120 V1/V2 domain with broadly neutralizing antibody PG9. Nature. 2011;480:336–343. doi: 10.1038/nature10696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myszka DG, Sweet RW, Hensley P, Brigham-Burke M, Kwong PD, Hendrickson WA, Wyatt R, Sodroski J, Doyle ML. Energetics of the HIV gp120-CD4 binding reaction. Proc Natl Acad Sci U S A. 2000;97:9026–9031. doi: 10.1073/pnas.97.16.9026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pancera M, Majeed S, Ban YE, Chen L, Huang CC, Kong L, Kwon YD, Stuckey J, Zhou T, Robinson JE, et al. Structure of HIV-1 gp120 with gp41-interactive region reveals layered envelope architecture and basis of conformational mobility. Proc Natl Acad Sci U S A. 2010;107:1166–1171. doi: 10.1073/pnas.0911004107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pancera M, Shahzad-Ul-Hussan S, Doria-Rose NA, McLellan JS, Bailer RT, Dai K, Loesgen S, Louder MK, Staupe RP, Yang Y, et al. Structural basis for diverse N-glycan recognition by HIV-1-neutralizing V1-V2-directed antibody PG16. Nat Struct Mol Biol. 2013;20:804–813. doi: 10.1038/nsmb.2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pejchal R, Doores KJ, Walker LM, Khayat R, Huang PS, Wang SK, Stanfield RL, Julien JP, Ramos A, Crispin M, et al. A potent and broad neutralizing antibody recognizes and penetrates the HIV glycan shield. Science. 2011;334:1097–1103. doi: 10.1126/science.1213256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Ringe RP, Sanders RW, Yasmeen A, Kim HJ, Lee JH, Cupo A, Korzun J, Derking R, van Montfort T, Julien JP, et al. Cleavage strongly influences whether soluble HIV-1 envelope glycoprotein trimers adopt a native-like conformation. Proc Natl Acad Sci U S A. 2013;110:18256–18256. doi: 10.1073/pnas.1314351110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders RW, Derking R, Cupo A, Julien JP, Yasmeen A, de Val N, Kim HJ, Blattner C, de la Pena AT, Korzun J, et al. A next-generation cleaved, soluble HIV-1 Envtrimer, BG505 SOSIP.664 gp140, expresses multiple epitopes for broadly neutralizing but not non-neutralizing atibodies. PLoS Pathog. 2013;9:e1003618. doi: 10.1371/journal.ppat.1003618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders RW, Venturi M, Schiffner L, Kalyanaraman R, Katinger H, Lloyd KO, Kwong PD, Moore JP. The mannose-dependent epitope for neutralizing antibody 2G12 on human immunodeficiency virus type 1 glycoprotein gp120. J Virol. 2002a;76:7293–7305. doi: 10.1128/JVI.76.14.7293-7305.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders RW, Vesanen M, Schuelke N, Master A, Schiffner L, Kalyanaraman R, Paluch M, Berkhout B, Maddon PJ, Olson WC, et al. Stabilization of the soluble, cleaved, trimeric form of the envelope glycoprotein complex of human immunodeficiency virus type 1. J Virol. 2002b;76:8875–8889. doi: 10.1128/JVI.76.17.8875-8889.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scanlan CN, Offer J, Zitzmann N, Dwek RA. Exploiting the defensive sugars of HIV-1 for drug and vaccine design. Nature. 2007;446:1038–1045. doi: 10.1038/nature05818. [DOI] [PubMed] [Google Scholar]
- Schon A, Madani N, Klein JC, Hubicki A, Ng D, Yang X, Smith AB, Sodroski J, 3rd, Freire E. Thermodynamics of binding of a low-molecular-weight CD4 mimetic to HIV-1 gp120. Biochemistry. 2006;45:10973–10980. doi: 10.1021/bi061193r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Si Z, Madani N, Cox JM, Chruma JJ, Klein JC, Schon A, Phan N, Wang L, Biorn AC, Cocklin S, et al. Small-molecule inhibitors of HIV-1 entry block receptor-induced conformational changes in the viral envelope glycoproteins. Proc Natl Acad Sci U S A. 2004;101:5036–5041. doi: 10.1073/pnas.0307953101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stavenhagen K, Hinneburg H, Thaysen-Andersen M, Hartmann L, Varon Silva D, Fuchser J, Kaspar S, Rapp E, Seeberger PH, Kolarich D. Quantitative mapping of glycoprotein micro-heterogeneity and macro-heterogeneity: an evaluation of mass spectrometry signal strengths using synthetic peptides and glycopeptides. J Mass Spectrom. 2013;48:627–639. doi: 10.1002/jms.3189. [DOI] [PubMed] [Google Scholar]
- Thali M, Moore JP, Furman C, Charles M, Ho DD, Robinson J, Sodroski J. Characterization of conserved human immunodeficiency virus type 1 gp120 neutralization epitopes exposed upon gp120-CD4 binding. J Virol. 1993;67:3978–3988. doi: 10.1128/jvi.67.7.3978-3988.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong X, Wren JC, Konermann L. gamma-Ray-mediated oxidative labeling for detecting protein conformational changes by electrospray mass spectrometry. Anal Chem. 2008;80:2222–2231. doi: 10.1021/ac702321r. [DOI] [PubMed] [Google Scholar]
- Tran EE, Borgnia MJ, Kuybeda O, Schauder DM, Bartesaghi A, Frank GA, Sapiro G, Milne JL, Subramaniam S. Structural mechanism of trimeric HIV-1 envelope glycoprotein activation. PLoS Pathog. 2012;8:e1002797. doi: 10.1371/journal.ppat.1002797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker LM, Burton DR. Rational antibody-based HIV-1 vaccine design: current approaches and future directions. Curr Opin Immunol. 2010;22:358–366. doi: 10.1016/j.coi.2010.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker LM, Huber M, Doores KJ, Falkowska E, Pejchal R, Julien JP, Wang SK, Ramos A, Chan-Hui PY, Moyle M, et al. Broad neutralization coverage of HIV by multiple highly potent antibodies. Nature. 2011;477:466–470. doi: 10.1038/nature10373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker LM, Phogat SK, Chan-Hui PY, Wagner D, Phung P, Goss JL, Wrin T, Simek MD, Fling S, Mitcham JL, et al. Broad and potent neutralizing antibodies from an African donor reveal a new HIV-1 vaccine target. Science. 2009;326:285–289. doi: 10.1126/science.1178746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Chance MR. Structural mass spectrometry of proteins using hydroxyl radical based protein footprinting. Anal Chem. 2011;83:7234–7241. doi: 10.1021/ac200567u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weis DD, Engen JR, Kass IJ. Semi-automated data processing of hydrogen exchange mass spectra using HX-Express. J Am Soc Mass Spectrom. 2006;17:1700–1703. doi: 10.1016/j.jasms.2006.07.025. [DOI] [PubMed] [Google Scholar]
- Wilson IA, Skehel JJ, Wiley DC. Structure of the haemagglutinin membrane glycoprotein of influenza virus at 3 Å resolution. Nature. 1981;289:336–373. doi: 10.1038/289366a0. [DOI] [PubMed] [Google Scholar]
- Xu G, Chance MR. Hydroxyl radical-mediated modification of proteins as probes for structural proteomics. Chem Rev. 2007;107:3514–3543. doi: 10.1021/cr0682047. [DOI] [PubMed] [Google Scholar]
- Xu G, Kiselar J, He Q, Chance MR. Secondary reactions and strategies to improve quantitative protein footprinting. Anal Chem. 2005;77:3029–3037. doi: 10.1021/ac048282z. [DOI] [PubMed] [Google Scholar]
- Zhao Q, Ma L, Jiang S, Lu H, Liu S, He Y, Strick N, Neamati N, Debnath AK. Identification of N-phenyl-N’-(2,2,6,6-tetramethyl-piperidin-4-yl)-oxalamides as a new class of HIV-1 entry inhibitors that prevent gp120 binding to CD4. Virology. 2005;339:213–225. doi: 10.1016/j.virol.2005.06.008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.