Abstract
Transcription-coupled DNA supercoiling has been shown to be an important regulator of transcription that is broadly present in the cell. Here we review experimental work which shows that RNA polymerase is a powerful torsional motor that can alter DNA topology and structure, and DNA supercoiling in turn directly affects transcription elongation.
Keywords: E. coli RNA polymerase, transcription, DNA supercoiling, torque, single molecule, angular optical trap, torsional motor, twin supercoiled domain model
During transcription, RNA polymerase (RNAP) tracks the DNA helical groove, which requires RNAP to rotate relative to DNA. The rotation of RNAP may be hindered due to a large viscous drag from a long RNA transcript and its associated factors such as ribosomes in prokaryotes or spliceosomes in eukaryotes, as well as by its possible tethering to cell membranes and other cellular structures.1,2 Therefore DNA frequently needs to rotate as well. Because DNA is also constrained by viscous drag and cellular constraints, transcription induces (+) (over-wound) DNA supercoiling in front of, and (−) (under-wound) DNA supercoiling behind, RNAP, as described in the twin supercoiled domain model.1
Early in vitro and in vivo biochemical studies demonstrated that, in a torsionally-constrained and topoisomerase-deficient system, transcription can accumulate significant torsional stress in DNA and greatly alter its topology.3-6 Subsequently, Kouzine et al.7,8 showed that even in a linear DNA system with topoisomerases present, transcription-induced DNA supercoiling and dynamic torsional stress can still exist and play an essential role in gene regulation. Matsumoto et al.9 used fluorescence to visualize transcription-coupled (−) DNA supercoiling at approximately 150 loci on polytene chromosomes of D. melanogaster, demonstrating that transcription-coupled (−) DNA supercoiling exists widely within a cell, even in the presence of active topoisomerases. More recently, the dynamic (−) supercoiling generated by transcription in human Burkitt’s lymphoma cells was found to be present genome-wide and, on average, spread over ~1.5 kbp upstream of a start site of an active gene.10 Nahghton et al.11 also mapped out both the (+) and (−) supercoiling domains (median size of ~100 kbp, flanked by GC-AT boundaries) in human chromosome 11 and showed that these domains are formed and remodeled by transcription and topoisomerases activities. This cohort of experimental results suggests that transcription-coupled DNA supercoiling occurs more broadly than previously thought.
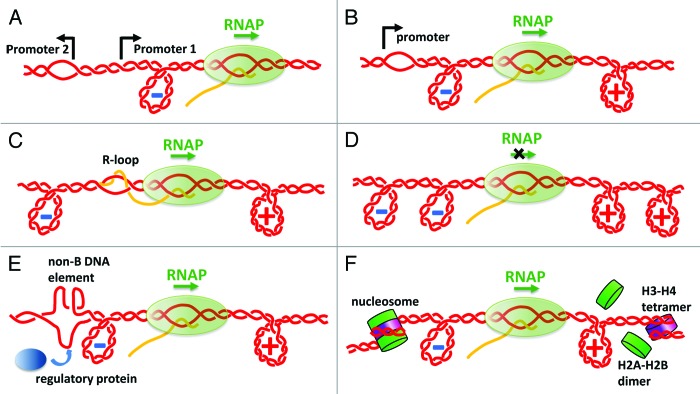
In addition, the role of DNA supercoiling in gene regulation has gradually come to light. It has been shown that transcription activities from various promoters are sensitive to the degree of DNA supercoiling.12,13 Experimental evidence13,14 has revealed that (−) supercoiling can facilitate transcription initiation, either by helping RNAP to form an open complex (for most genes in prokaryotes) or by helping to recruit transcription factors such as TATA binding protein (TBP) (for most eukaryotic genes). On the other hand, at certain promoters, (+) supercoiling can prevent transcription initiation15 and greatly diminish mRNA synthesis.16 It was also found that different promoters respond to DNA supercoiling in different ways.13,14 Thus, DNA supercoiling could serve as a flexible, global control for gene regulation. The supercoiling generated by the transcription at one promoter can ultimately affect the transcription at a distant promoter (Fig. 1A), and this is termed “topological promoter coupling”17,18. Transcription generated (−) supercoiling potentially could also encourage subsequent transcription from the same promoter (Fig. 1B). A recent theoretical work has suggested that bursts of transcription in bacteria could stem from dynamic supercoiling buildup inhibiting transcription initiation.19 These results thus support that transcription generated supercoiling can be used as a remote control and allow long range communication in gene regulation. Transcription coupled (−) supercoiling can also encourage non-B DNA formation, which can be recognized by DNA structure-sensitive regulatory proteins,7,8 and, sometimes, can also lead to R-Loop formation (Fig. 1C), which could hinder the further translocation of an elongating RNAP.20
Figure 1. Examples of regulatory roles of transcription-coupled DNA supercoiling. (A) (−) supercoiling facilitates promoter coupling and divergent transcription. (B) (−) supercoiling contributes to the melting of the same promoter and encourages subsequent transcription. (C) (−) supercoiling promotes the formation of an R-Loop (extensive RNA-DNA hybrid). (D) Accumulated (−) supercoiling upstream and (+) supercoiling downstream can cause RNAP to stall. (E) (−) supercoiling upstream facilitates the formation of non-B DNA structures which can attract regulatory proteins. (F) (+) supercoiling downstream can destabilize nucleosome structures and (−) supercoiling upstream can facilitate the re-assembly of nucleosome.
Although initial experiments have successfully linked DNA supercoiling to gene regulation, most focused on the effect of (−) DNA supercoiling on transcription initiation. The influence of both (−) and (+) DNA supercoiling on transcription elongation as well as the torque RNAP can generate remained elusive until a recent study that quantitatively examined transcription under DNA supercoiling. Ma et al. developed a novel single molecule assay to monitor transcription-generated DNA supercoiling and torque buildup in real-time using an angular optical trap (AOT).21 They directly followed movement by E. coli RNAP under a defined torque, and determined the torque required to stall an elongating RNAP, i.e., “stall torque.” The stall torque characterizes how powerful RNAP is in terms of torque generation.
These experiments showed that as transcription proceeds, RNAP may be stalled by torsional stress accumulated in (+) supercoiled downstream DNA or in (−) supercoiled upstream DNA (Fig. 1D). The measured “stall torques” (i.e., the torques that RNAP can generate) had an average of ~11 pN⋅nm, for both cases. This torque value is twice the lower bound of the maximum torque (~5 pN⋅nm) estimated from a previous single-molecule study.22 This amount of torque in the (−) supercoiled upstream DNA is capable of melting DNA, of arbitrary sequence,23 not just AT-rich sequences which are prone to melting24 or GC repeats which are prone to form right-handed Z DNA.24 These non-B DNA structures can be further used as a distinct target to attract regulatory proteins in gene regulation (Fig. 1E).7,8 The torque RNAP generated in (+) supercoiled downstream DNA and/or (−) supercoiled upstream DNA could also be utilized to affect the stability of bound proteins. Recently, in a single molecule study also using an AOT, Sheinin et al.25 have shown that a torque from (+) DNA supercoiling comparable to the measured RNAP stall torque can destabilize nucleosome structures and cause a preferential loss of H2A/H2B dimers in a nucleosome (Fig. 1F). It is also possible that the transcription-generated (−) DNA supercoiling could facilitate nucleosome re-assembly in the upstream DNA (Fig. 1F).26 Finally, the measured stall torques of both (−) and (+) supercoiling are sufficient to convert DNA into the plectonemic state and to substantially modify chromatin topology.27 All these results suggest that RNAP is a power torsional motor and the torque‐generating capacity of RNAP may have been tuned to important transitions in DNA or chromatin structures.
Ma et al.21 also found that, upon torque release, ~50% of stalled RNAPs were able to resume transcription within ~90 s. This suggests that, in vivo, stalled RNAPs can be rescued if torsional stress is released in a timely fashion, either by topoisomerases, or via DNA and/or RNAP rotation. This may be beneficial to the cell ‘economy’ by enabling the cell to make good use of unfinished transcripts. Otherwise, the stalled RNAPs could become roadblocks that would hinder other vital cellular processes. Dutta et al.28 have shown that collision between a replication fork and a stalled RNAP could cause DNA breakage and induce genome instability. Stalled RNAPs thus could be lethal to the cell.
Ma et al.21 further found that an elongating RNAP is resilient to short (< 0.5 s) torque pulses, but not to long ones (> 5 s). Therefore, transcription is minimally perturbed by transient torque fluctuations. In the cell, the torsional stress on DNA changes dynamically due to various processes such as a spontaneous loss of torsional constraints in DNA, the action of topoisomerases or other motors, or sudden environmental condition changes. The resilience to short torsional fluctuations ensures that transcription elongation processes can be immune to the “high-frequency” noise in the complex environment of cell.
Finally, Ma et al.21 showed that torque can directly regulate transcription speed. A resisting torque slows down transcription, while an assisting torque does the opposite. Moreover, a resisting torque will increase the chance for an elongating RNAP to enter into and dwell in a paused state.
The interplay between DNA supercoiling and transcription is clearly important in gene regulation. Recent experiments provide a quantitative framework for understanding this relationship, and also establish E. coli RNAP as a powerful torsional motor with the ability to greatly alter DNA topology. As DNA topology also plays important roles in other vital cellular processes,2,29 such as replication and DNA recombination, it would be interesting to see how transcription generated DNA supercoiling could affect all these processes. Additionally, although the stall torque for E. coli RNAP has been measured, the torque that an eukaryotic RNA polymerase such as Pol II can generate still remains unknown. It has long been hypothesized that the torque generated by Pol II during (+) supercoiling could be utilized to destabilize nucleosome structures. Indeed recent experiments have shown that torque from (+) supercoiling can destabilize a nucleosome.25 It would be intriguing to see what torque Pol II is capable of generating, and to what degree it can alter chromatin topology. Interestingly, Bancaud et al.27 have shown that, compared with naked DNA, a nucleosome array is highly resilient and is able to accommodate a much larger amount of supercoiling. Chromatin may thus serve as a topological buffer, allowing RNAP to transcribe a longer gene under a torsionally-constrained condition without help from topoisomerases. Finally, in vivo, there are a large number of transcription factors, some of which can help RNAP translocate and generate larger force.30,31 It would be interesting to examine if these factors could also help RNAP generate larger torque. We believe that with the advent of new techniques such as the AOT, we will gain a greater understanding of the role that DNA supercoiling plays in transcription and other vital cellular processes.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank members of the Wang lab for critical reading of the manuscript. We wish to acknowledge support to MDW. from the National Science Foundation grant (MCB-0820293).
References
- 1.Liu LF, Wang JC. Supercoiling of the DNA template during transcription. Proc Natl Acad Sci U S A. 1987;84:7024–7. doi: 10.1073/pnas.84.20.7024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Koster DA, Crut A, Shuman S, Bjornsti M-A, Dekker NH. Cellular strategies for regulating DNA supercoiling: a single-molecule perspective. Cell. 2010;142:519–30. doi: 10.1016/j.cell.2010.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wu H-Y, Shyy SH, Wang JC, Liu LF. Transcription generates positively and negatively supercoiled domains in the template. Cell. 1988;53:433–40. doi: 10.1016/0092-8674(88)90163-8. [DOI] [PubMed] [Google Scholar]
- 4.Tsao Y-P, Wu H-Y, Liu LF. Transcription-driven supercoiling of DNA: direct biochemical evidence from in vitro studies. Cell. 1989;56:111–8. doi: 10.1016/0092-8674(89)90989-6. [DOI] [PubMed] [Google Scholar]
- 5.Krasilnikov AS, Podtelezhnikov A, Vologodskii A, Mirkin SM. Large-scale effects of transcriptional DNA supercoiling in vivo. J Mol Biol. 1999;292:1149–60. doi: 10.1006/jmbi.1999.3117. [DOI] [PubMed] [Google Scholar]
- 6.Samul R, Leng F. Transcription-coupled hypernegative supercoiling of plasmid DNA by T7 RNA polymerase in Escherichia coli topoisomerase I-deficient strains. J Mol Biol. 2007;374:925–35. doi: 10.1016/j.jmb.2007.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kouzine F, Liu J, Sanford S, Chung H-J, Levens D. The dynamic response of upstream DNA to transcription-generated torsional stress. Nat Struct Mol Biol. 2004;11:1092–100. doi: 10.1038/nsmb848. [DOI] [PubMed] [Google Scholar]
- 8.Kouzine F, Sanford S, Elisha-Feil Z, Levens D. The functional response of upstream DNA to dynamic supercoiling in vivo. Nat Struct Mol Biol. 2008;15:146–54. doi: 10.1038/nsmb.1372. [DOI] [PubMed] [Google Scholar]
- 9.Matsumoto K, Hirose S. Visualization of unconstrained negative supercoils of DNA on polytene chromosomes of Drosophila. J Cell Sci. 2004;117:3797–805. doi: 10.1242/jcs.01225. [DOI] [PubMed] [Google Scholar]
- 10.Kouzine F, Gupta A, Baranello L, Wojtowicz D, Ben-Aissa K, Liu J, Przytycka TM, Levens D. Transcription-dependent dynamic supercoiling is a short-range genomic force. Nat Struct Mol Biol. 2013;20:396–403. doi: 10.1038/nsmb.2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Naughton C, Avlonitis N, Corless S, Prendergast JG, Mati IK, Eijk PP, Cockroft SL, Bradley M, Ylstra B, Gilbert N. Transcription forms and remodels supercoiling domains unfolding large-scale chromatin structures. Nat Struct Mol Biol. 2013;20:387–95. doi: 10.1038/nsmb.2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hirose S, Matsumoto K. Possible Roles of DNA Supercoiling in Transcription. In: Ohyama T, ed. DNA conformation and transcription: Springer, 2005:138-43. [Google Scholar]
- 13.Lim HM, Lewis DEA, Lee HJ, Liu M, Adhya S. Effect of varying the supercoiling of DNA on transcription and its regulation. Biochemistry. 2003;42:10718–25. doi: 10.1021/bi030110t. [DOI] [PubMed] [Google Scholar]
- 14.Tabuchi H, Handa H, Hirose S. Underwinding of DNA on binding of yeast TFIID to the TATA element. Biochem Biophys Res Commun. 1993;192:1432–8. doi: 10.1006/bbrc.1993.1576. [DOI] [PubMed] [Google Scholar]
- 15.Revyakin A, Ebright RH, Strick TR. Promoter unwinding and promoter clearance by RNA polymerase: detection by single-molecule DNA nanomanipulation. Proc Natl Acad Sci U S A. 2004;101:4776–80. doi: 10.1073/pnas.0307241101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gartenberg MR, Wang JC. Positive supercoiling of DNA greatly diminishes mRNA synthesis in yeast. Proc Natl Acad Sci U S A. 1992;89:11461–5. doi: 10.1073/pnas.89.23.11461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lilley DMJ, Higgins CF. Local DNA topology and gene expression: the case of the leu-500 promoter. Mol Microbiol. 1991;5:779–83. doi: 10.1111/j.1365-2958.1991.tb00749.x. [DOI] [PubMed] [Google Scholar]
- 18.Rhee KY, Opel M, Ito E, Hung Sp, Arfin SM, Hatfield GW. Transcriptional coupling between the divergent promoters of a prototypic LysR-type regulatory system, the ilvYC operon of Escherichia coli. Proc Natl Acad Sci U S A. 1999;96:14294–9. doi: 10.1073/pnas.96.25.14294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mitarai N, Dodd IB, Crooks MT, Sneppen K. The generation of promoter-mediated transcriptional noise in bacteria. PLoS Comput Biol. 2008;4:e1000109. doi: 10.1371/journal.pcbi.1000109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Drolet M, Broccoli S, Rallu F, Hraiky C, Fortin C, Massé E, Baaklini I. The problem of hypernegative supercoiling and R-loop formation in transcription. Front Biosci. 2003;8:d210–21. doi: 10.2741/970. [DOI] [PubMed] [Google Scholar]
- 21.Ma J, Bai L, Wang MD. Transcription under torsion. Science. 2013;340:1580–3. doi: 10.1126/science.1235441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harada Y, Ohara O, Takatsuki A, Itoh H, Shimamoto N, Kinosita K., Jr. Direct observation of DNA rotation during transcription by Escherichia coli RNA polymerase. Nature. 2001;409:113–5. doi: 10.1038/35051126. [DOI] [PubMed] [Google Scholar]
- 23.Sheinin MY, Forth S, Marko JF, Wang MD. Underwound DNA under tension: structure, elasticity, and sequence-dependent behaviors. Phys Rev Lett. 2011;107:108102. doi: 10.1103/PhysRevLett.107.108102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Oberstrass FC, Fernandes LE, Lebel P, Bryant Z. Torque spectroscopy of DNA: base-pair stability, boundary effects, backbending, and breathing dynamics. Phys Rev Lett. 2013;110:178103. doi: 10.1103/PhysRevLett.110.178103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sheinin MY, Li M, Soltani M, Luger K, Wang MD. Torque modulates nucleosome stability and facilitates H2A/H2B dimer loss. Nat Commun. 2013;4:2579. doi: 10.1038/ncomms3579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Clark DJ, Felsenfeld G. Formation of nucleosomes on positively supercoiled DNA. EMBO J. 1991;10:387–95. doi: 10.1002/j.1460-2075.1991.tb07960.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bancaud A, Conde e Silva N, Barbi M, Wagner G, Allemand J-F, Mozziconacci J, Lavelle C, Croquette V, Victor JM, Prunell A, et al. Structural plasticity of single chromatin fibers revealed by torsional manipulation. Nat Struct Mol Biol. 2006;13:444–50. doi: 10.1038/nsmb1087. [DOI] [PubMed] [Google Scholar]
- 28.Dutta D, Shatalin K, Epshtein V, Gottesman ME, Nudler E. Linking RNA polymerase backtracking to genome instability in E. coli. Cell. 2011;146:533–43. doi: 10.1016/j.cell.2011.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Potaman VN, Sinden RR. DNA: Alternative conformations and biology. In: Ohyama T, ed. DNA conformation and transcription: Springer, 2005:3-17. [Google Scholar]
- 30.Herbert KM, Zhou J, Mooney RA, Porta AL, Landick R, Block SME. E. coli NusG inhibits backtracking and accelerates pause-free transcription by promoting forward translocation of RNA polymerase. J Mol Biol. 2010;399:17–30. doi: 10.1016/j.jmb.2010.03.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Galburt EA, Grill SW, Wiedmann A, Lubkowska L, Choy J, Nogales E, Kashlev M, Bustamante C. Backtracking determines the force sensitivity of RNAP II in a factor-dependent manner. Nature. 2007;446:820–3. doi: 10.1038/nature05701. [DOI] [PubMed] [Google Scholar]