Abstract
Abscisic acid (ABA) is involved in a number of critical processes in normal growth and development as well as in adaptive responses to environmental stresses. For correct and accurate actions, a physiologically active ABA level is controlled through fine-tuning of de novo biosynthesis and catabolism. The hydroxylation at the 8′-position of ABA is known as the key step of ABA catabolism, and this reaction is catalyzed by ABA 8′-hydroxylase, a cytochrome P450. Here, we demonstrate CYP707As as the P450 responsible for the 8′-hydroxylation of (+)-ABA. First, all four CYP707A cDNAs were cloned from Arabidopsis and used for the production of the recombinant proteins in insect cells using a baculovirus system. The insect cells expressing CYP707A3 efficiently metabolized (+)-ABA to yield phaseic acid, the isomerized form of 8′-hydroxy-ABA. The microsomes from the insect cells exhibited very strong activity of 8′-hydroxylation of (+)-ABA (Km = 1.3 μm and kcat = 15 min−1). The solubilized CYP707A3 protein bound (+)-ABA with the binding constant Ks = 3.5 μm, but did not bind (−)-ABA. Detailed analyses of the reaction products confirmed that CYP707A3 does not have the isomerization activity of 8′-hydroxy-ABA to phaseic acid. Further experiments revealed that Arabidopsis CYP707A1 and CYP707A4 also encode ABA 8′-hydroxylase. The transcripts of the CYP707A genes increased in response to salt, osmotic, and dehydration stresses as well as ABA. These results establish that the CYP707A family plays a key role in regulating the ABA level through the 8′-hydroxylation of (+)-ABA.
The plant hormone abscisic acid (ABA) regulates many important physiological and developmental processes in plants as well as adaptive responses to environmental stresses (Zeevaart and Creelman, 1988). During seed development and dormancy, ABA content increases as a potential signal to organize expression of many embryo-specific genes. The ABA level is also elevated in response to various environmental stresses such as drought, high salinity, and low temperature conditions. These physiological processes controlled by ABA are primarily regulated by bioactive ABA pool size, which is thought to be maintained through fine-tuning the rates of de novo biosynthesis and catabolism. Thus, to understand the molecular mechanism that controls the ABA contents in plant tissues, the genes and enzymes in biosynthesis and catabolism of ABA must be studied in detail.
ABA is a sesquiterpene, and the first committed step in ABA biosynthesis is the oxidative cleavage of a 9-cis-epoxycarotenoid (C40) to form a xanthoxal (C15). The xanthoxal is oxidized to form abscisic aldehyde, and then further oxidized to ABA. To date, most of the genes in the ABA biosynthetic pathway have been isolated through identification and characterization of ABA deficient mutants from several plant species (Schwartz et al., 2003), and molecular studies on ABA biosynthesis are now possible to address the complex regulatory mechanisms of ABA level in detail (Seo and Koshiba, 2002; Xiong and Zhu, 2003).
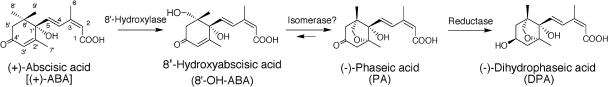
In contrast to ABA biosynthesis, the genes and enzymes in ABA catabolism as well as their regulation remain largely unknown (Cutler and Krochko, 1999; Zeevaart, 1999). In most plant tissues, inactivation of ABA occurs via the oxidative pathway as shown in Figure 1. The key step of ABA inactivation is the hydroxylation of the 8′-methyl group of ABA, which is catalyzed by ABA 8′-hydroxylase to yield 8′-hydroxy-ABA (8′-OH-ABA). 8′-OH-ABA is unstable and spontaneously (and/or enzymatically) isomerizes to form phaseic acid (PA). Bioactivity of PA is weak compared with that of ABA. The key enzyme, ABA 8′-hydroxylase, has been shown to be a cytochrome P450 (P450) by using the microsomal fraction of maize (Zea mays) suspension cultured cells (Krochko et al., 1998). Hydroxylation activity was induced by the substrate ABA treatment in maize suspension cells (Cutler et al., 1997) and also increased in response to a water deficit in maize kernels (Wang et al, 2002). The metabolite PA is further reduced at the 4′-position to form the biologically inactive dihydrophaseic acid (DPA). In some cases, ABA is metabolized to form its glucosyl ester (Zeevaart and Creelman, 1988), and ABA glucosyltransferase from adzuki bean (Vigna angularis) has been cloned recently (Xu et al., 2002). Several reports have described that the predominant pathway of ABA catabolism is via 8′-hydroxylase; however, the gene encoding ABA 8′-hydroxylase has not been identified yet. Because ABA 8′-hydroxylase catalyzes the first step in the oxidative inactivation of ABA and is thought to be the pivotal enzyme controlling the rate of ABA catabolism, the identification of the P450 gene encoding ABA 8′-hydroxylase will give us new insight into ABA homeostasis and the manipulation of ABA and/or metabolite levels.
Figure 1.
The oxidative pathway of ABA catabolism in higher plants.
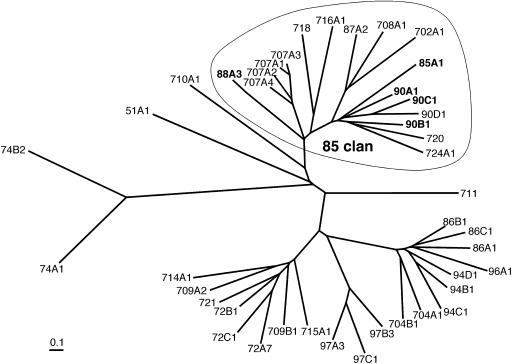
Complete sequencing of Arabidopsis genome has revealed 246 full-length P450 genes distributed into 45 families (http://drnelson.utmem.edu/Arablinks.html). Arabidopsis P450s are divided into two main classes of A-type and non-A-type genes (Durst and Nielsen, 1995; Schuler and Werck-Reichhart, 2003). About 60% of Arabidopsis P450s are grouped as the A-type and are primarily involved in unique plant biosynthetic pathways of various secondary metabolites such as phenylpropanoids, terpenoids, glucosinolates, and phytoalexins. On the other hand, the non-A-type P450s (Fig. 2) show similarities to P450s from other kingdoms (mammal, yeast, fungi, etc.) and often represent more fundamental enzymes involved in biosynthesis of sterols, oxygenated fatty acid, and phytohormones.
Figure 2.
Phylogenetic tree of the non-A-type P450s from Arabidopsis. The tree was built using a single gene of each subfamily, except for all four CYP707As. The 85-clan is circled, and the P450 families involved in brassinosteroid and gibberellin biosynthesis are shown in bold.
Aiming at elucidating P450s involved in essential reactions for plant growth, we have been focusing on biochemical characterization of P450 genes in the 85-clan within the non-A type P450s. The 85-clan (Fig. 2, circled), one of the clades in the non-A-type P450s, contains several P450s (CYP85, 88, and 90s) involved in brassinosteroid (BR) and gibberellin (GA) biosynthesis (Szekeres et al., 1996; Choe et al., 1998; Helliwell et al., 2001; Shimada et al., 2001). Recently, rice (Oryza sativa) genome has been sequenced completely, and classification of the P450 gene family in rice genome has also been completed (http://drnelson.utmem.edu/ricefams.html). In the 85-clan of rice P450s, there are no P450 genes belonging to the CYP702, 708, 716, 718, and 720 families found in Arabidopsis. In other words, it is possible that these P450s may function in metabolic pathways unique to Arabidopsis. Among the 85-clan P450s overlapping between Arabidopsis and rice, we selected CYP707A as the first target for our biochemical approach to identify the P450 function, because the CYP707A family is most closely related to CYP88As encoding ent-kaurenoic acid hydroxylase in the early steps of GA biosynthesis. The CYP707A family in Arabidopsis consists of four genes, implicating the possibility that the physiological function of this family may not be characterized by standard mutant screening due to their functional redundancy. Thus, biochemical characterization of the CYP707A family is crucial for identifying their enzymatic and physiological functions.
In this study, we isolated the cDNAs of four CYP707A genes in Arabidopsis and characterized the biochemical properties of the recombinant CYP707A proteins expressed in the baculovirus system as well as the expression patterns of the CYP707A genes in vivo. The results have demonstrated that Arabidopsis CYP707As encode (+)-ABA 8′-hydroxylase.
RESULTS
Isolation of CYP707A1 and CYP707A3 cDNAs
Several P450 genes in the 85-clan of the non-A-type P450s have already been shown to function in phytohormone biosynthesis, and therefore we expected that other 85-clan members might be involved in metabolic pathways related to plant bioactive compounds. During the course of our systematic biochemical characterization of the 85-clan P450s, we investigated Arabidopsis CYP707A family members consisting of four genes (CYP707A1, A2, A3, and A4). First, we isolated the full-length cDNAs for CYP707A1 and CYP707A3 from a cDNA library of Arabidopsis seedlings. The CYP707A1 protein sequence is 87% identical to CYP707A3, while it is only about 60% identical to CYP707A2 and CYP707A4. A BLAST search with CYP707A1 and CYP707A3 revealed that these P450s show the highest identities (35%) to Arabidopsis CYP88A3 and CYP88A4 of ent-kaurenoic acid hydroxylase in the GA biosynthetic pathway, and 30% to 35% identities to CYP85A1, CYP90A1, CYP90B1, and CYP90C1, which are involved in BR biosynthesis (Fig. 2). In rice genome, two CYP707A genes (CYP707A5 and CYP707A6) have been identified (http://drnelson.utmem.edu/biblioD.html). The scanning of the CYP707A ESTs available in the TIGR database (http://www.tigr.org/tdb/tgi/plant.shtml) also indicated that the CYP707A-like genes occur in a variety of monocotyledonous and dicotyledonous plants. These results suggested that the CYP707A family functions in a fundamental metabolism common among higher plants.
CYP707As also share about 30% identities to CYP725A1, A2, and A3 from Taxus cuspidate, which are also in the 85-clan and involved in the biosynthesis of a diterpenoid, taxol (Jennewein et al., 2001, 2003; Schoendorf et al., 2001). Interestingly, CYP707As exhibit significant sequence identities to CYP120 of unknown function from Synechocystis (30%), and also to mammalian CYP26 (29%) involved in retinoic acid catabolism (Fujii et al., 1997; White et al., 1997). These results suggested the possibility that the CYP707A genes encode a P450 acting on terpenoids in plants.
Gene Expression Profiles in Response to ABA, GA, and BR
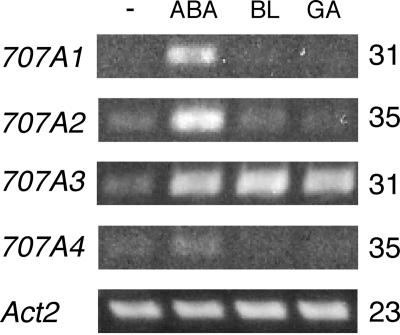
Plant hormones such as GA and BR negatively regulate their own biosynthetic genes and positively regulate their own catabolic genes in feed-back and feed-forward mechanisms, respectively. It should be noted that ABA is inactivated through the hydroxylation of the 8′-methyl group by a P450 (Krochko et al., 1998) and that ABA enhanced the enzyme activity of ABA 8′-hydroxylation in cultured maize cells (Cutler et al., 1997) as well as in suspension-cultured cells of Arabidopsis (Windsor and Zeevaart, 1997). The gene expression of the Arabidopsis CYP707A family in response to ABA, GA, and BR was analyzed by reverse transcription (RT)-PCR (Fig. 3). The transcript accumulation of all CYP707A genes greatly increased by 1 μm (±)-ABA. The CYP707A3 transcript level was also elevated by GA and BR, whereas virtually no significant transcript accumulation of the other CYP707A members was observed under the same experimental conditions.
Figure 3.
Expression patterns of CYP707As in response to ABA, BR, and GA. Arabidopsis plants (2 weeks old) cultured in GM liquid medium were treated with 1 μm of (±)-ABA (ABA), brassinolide (BL), and gibberellin (GA3), and incubated for 6 h. Total RNA isolated from the plants was subjected to RT-PCR using gene specific primers. The Actin2 (Act2) RT-PCR was included as a constitutive control. −, control; ABA, (±)-ABA; BL, brassinolide; GA, GA3. The number indicated on the right is the number of PCR cycles required to amplify the DNA fragment of each CYP707A transcript.
Heterologous Expression in Insect Cells
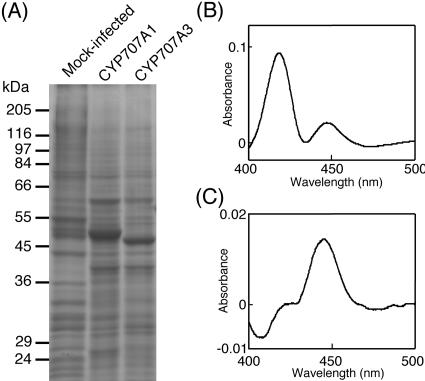
In order to characterize the enzymatic properties of the CYP707A family, we produced the recombinant CYP707A1 and CYP707A3 proteins in insect cells using a baculovirus system. SDS-PAGE analysis showed that the new intense bands of 53 and 52 kD appeared in the microsomal fractions of the insect cells upon infection with the recombinant viruses of CYP707A1 and CYP707A3, respectively (Fig. 4A). The microsomes were solubilized with 1% (w/v) Emulgen 913, and the resultant solubilized fractions were analyzed for P450 contents. The reduced-CO difference spectrum of the solubilized CYP707A1 fractions showed two Soret absorption peaks at 420 and 450 nm (Fig. 4B), indicating that the recombinant CYP707A1 would be easily inactivated. On the other hand, the solubilized CYP707A3 fractions showed a clear absorption peak at 450 nm (Fig. 4C). The solubilized fractions from the mock-infected cells did not give the 450-nm peak (data not shown). Thus, a part of the recombinant CYP707A1 and most of the recombinant CYP707A3 were expressed in insect cells as active forms.
Figure 4.
Heterologous expression of CYP707A1 and CYP707A3 by using the baculovirus-insect cell system. A, SDS-PAGE of the microsomes prepared from the cells without baculovirus infection (mock-infected) or the cells expressing CYP707A1 and CYP707A3. Fifteen micrograms of proteins were loaded per lane. Reduced CO-difference spectra of the recombinant CYP707A1 (B), CYP707A3 (C). The spectra were recorded using the solubilized fractions of the CYP707A1 and CYP707A3 microsomes.
ABA 8′-Hydroxylase Activity
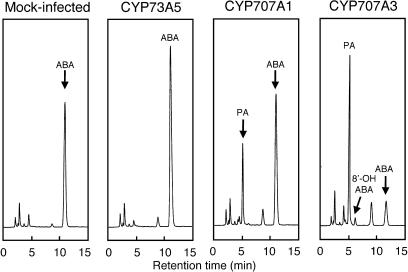
We studied ABA 8′-hydroxylase activity using the insect cell culture by adding (+)-ABA directly into the culture medium. The insect cells expressing either CYP707A1 or CYP707A3 were incubated in the presence of 200 μm (+)-ABA for 24 h at 27°C. HPLC analyses revealed that the ABA concentration (the peak at 12.1 min) was greatly decreased in the culture medium from the cells expressing CYP707A3 (Fig. 5). On the other hand, two new compounds, 1 and 2, were detected in the same analysis at 5.8 and 7.1 min, respectively, with a ratio of 98:2. These new peaks were identical in the retention times to authentic standards of PA (5.8 min) and 8′-OH-ABA (7.1 min), respectively. The assay for CYP707A1 also gave the similar results (Fig. 5). Compounds 1 and 2 were also detected inside the cells, but the amounts were much lower than those from the medium (data not shown). Compounds 1 and 2 were not in the assays of either the mock-infected Sf9 cells (Fig. 5) or cells expressing the Arabidopsis CYP73A5 cDNA encoding a trans-cinnamic acid 4-hydroxylase (Mizutani et al., 1997; Fig. 5).
Figure 5.
HPLC analysis of reaction products of ABA metabolized by insect cells expressing either the CYP707A1 or CYP707A3 proteins. The suspension cultures of Sf9 insect cells were infected with the recombinant baculovirus containing each P450 cDNA on a rotary shaker (150 rpm) at 27°C for 72 h. After 72-h culture, 200 μm (+)-ABA was added to the culture medium, and the cells were further incubated at 27°C for 24 h. The culture medium was collected by centrifugation and extracted four times with an equal volume of ethyl acetate. After evaporation, the ethyl acetate extracts were resuspended in 1.2 mL of methanol, and 1 μL of the sample was analyzed by HPLC. Mock-infected, the cells without baculovirus infection; CYP73A5, Arabidopsis cinnamate 4-hydroxylase.
To identify compounds 1 and 2, we carried out a large-scale expression of the recombinant CYP707A3 protein. The suspension-cultured Sf9 cells (60 mL) expressing the CYP707A3 protein were incubated with 200 μm (+)-ABA for 24 h, and compounds 1 and 2 were collected by preparative HPLC of the extract from the culture medium. Compound 1 was identified as (−)-PA by comparison of its spectral data with those of an authentic sample (data are presented in “Materials and Methods”). In this assay, 36% of the applied ABA (3.17 mg) was converted to PA (1.15 mg). Most of the formed PA was secreted into the medium, and only 4% (48 μg) of the formed PA was found within the cells. The collected material corresponding to compound 2 was a mixture of compounds 1 and 2 with a ratio 40:60 on HPLC analysis, showing that compound 2 partially isomerized to PA during isolation. The 1H NMR data of the mixture indicated the signals assignable to 8′-OH-ABA in addition to PA, and the mass spectrum of methyl ester of the mixture coincided with that of the methyl ester of PA. These data identified compound 2 as 8′-OH-ABA. PA and 8′-OH-ABA are in equilibrium with a final ratio 98:2 at 25°C (Todoroki et al., 2000), strongly suggesting that PA in the extracts from the reaction mixtures was formed from 8′-OH-ABA by spontaneous isomerization during extraction and preservation. This meant that CYP707A3 possesses at least the activity of (+)-ABA 8′-hydroxylase, although it was unclear whether CYP707A3 possesses the activity of the 8′-OH-ABA isomerization as well.
To examine whether the other two members of the Arabidopsis CYP707A family also have the same catalytic activity, the cDNAs for CYP707A2 and CYP707A4 were isolated by RT-PCR, and the recombinant protein of either CYP707A2 or CYP707A4 was expressed in insect cells. The cells expressing CYP707A4 were able to convert (+)-ABA to PA, while the cells expressing CYP707A2 did not metabolize (+)-ABA to PA (data not shown). This was probably ascribed to the instability of the recombinant CYP707A2 protein, which was found in spectral analysis only as the inactive form with the 420-nm Soret absorption peak (data not shown).
Biochemical Characterization of CYP707A Microsomes
Enzymatic properties were studied using the microsomal fractions from the insect cells expressing CYP707A3. The CYP707A3 microsomes were incubated with 100 μm (+)-ABA and 100 μm NADPH at 30°C for 10 min, and the reaction products were analyzed by HPLC (Table I). The CYP707A3 microsomes produced PA, indicating that endogenous NADPH-P450 reductase in the insect cells was able to support the electron transfer to the CYP707A3 protein to some extent. When the recombinant Arabidopsis NADPH-P450 reductase (AR1; Mizutani and Ohta, 1998) was added to the reaction mixture, the activity for PA production was increased by about 1.7 times compared to that observed without the AR1 addition. The reaction was completely dependent on NADPH, and NADH did not substitute for NADPH at all. The activity was completely inhibited by the addition of 100 μm cytochrome c, which interrupts the electron transfer to P450 protein by directly interacting with NADPH-P450 reductase. These results indicate that the electron transfer from NADPH-P450 reductase is essential for the ABA 8′-hydroxylase activity by CYP707A3. The microsomes from the cells expressing CYP707A1 also gave similar properties to the CYP707A3 microsomes (data not shown). On the other hand, no oxidative products of ABA were detected with the microsomes of the mock-infected Sf9 insect cells and the cells expressing CYP73A5 (data not shown).
TABLE I.
ABA 8′-hydroxylase acitivity of the recombinant CYP707A3 microsomes
| Assay conditions | ABA 8′-Hydroxylase Activity PA + 8′-OH-ABAb | Relative Activity |
|---|---|---|
| ng | % | |
| Completea | 197 ± 15 | 100 |
| - NADPH | n.d.c | - |
| - NADPH + 100 μM NADH | n.d. | - |
| + AR1 (NADPH-P450 reductase) | 338 ± 6 | 172 |
| +100 μM cytochrome c | n.d. | - |
| - (+)-ABA + 100 μM (-)-ABA | n.d. | - |
The complete reaction mixture contained 50 mm potassium phosphate (pH 7.25), 50 μg/mL recombinant CYP707A3 microsomes, 100 μm NADPH, and 100 μm (+)-ABA.
Reactions were initiated by addition of NADPH and carried out in the reaction mixture at 30°C for 10 min. The amounts of the reaction products (PA and 8′-OH-ABA) were estimated as the relative response to standard PA. Values are mean ± sd of three separate determinations.
Not detected. The detection limit for the total amounts of PA and 8′-OH-ABA in the reaction mixtures was 50 ng.
Substrate Binding
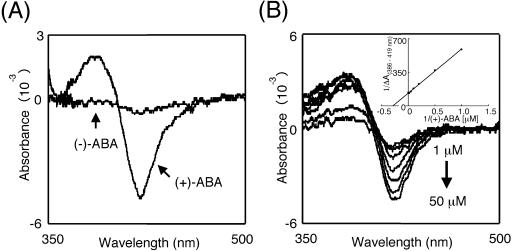
Most P450 enzymes are present in a low-spin state in the absence of substrate. Upon substrate binding, the heme electronic state is modified, and the heme iron spin state shifts from low-spin state to high-spin state. The recombinant CYP707A3 was solubilized with 1% (w/v) sodium cholate and used for substrate binding experiments with (+)-ABA and (−)-ABA at a final concentration of 100 μm. (+)-ABA produced a Type I difference spectrum with a maximum at 386 nm and a minimum at 419 nm (Fig. 6A). The amplitude of this difference was proportional to increasing amounts of (+)-ABA, and the binding constant was determined to be the Ks value of 3.5 ± 0.1 μm (Fig. 6B). This value is closely similar to that of the mung bean cinnamate 4-hydroxylase with trans-cinnamic acid (Ks = 2.8 μm; Mizutani et al., 1993). On the other hand, (−)-ABA did not induce significant spectral shift, indicating that (−)-ABA did not bind to CYP707A3.
Figure 6.
Substrate binding spectra of the CYP707A3 protein. The recombinant CYP707A3 microsomes were solubilized with 1.0% (w/v) sodium cholate, and the solubilized CYP707A3 was obtained by centrifugation at 100,000g. A, Type I difference spectra of CYP707A3. Either (+)-ABA or (−)-ABA was added to the solubilized CYP707A3 at a final concentration of 100 μm. B, Spectrophotometric titration of CYP707A3 with (+)-ABA. (+)-ABA was added to the solubilized CYP707A3 at concentrations ranging from 1 to 50 μm, and difference spectra were recorded between 500 and 350 nm. The spectral dissociation constant (Ks) was calculated from a double reciprocal plot of absorbance difference, ΔA(386–419 nm) versus the substrate concentration (inset).
Characteristics of the Reactions
Kinetic parameters of the CYP707A3 reaction were determined using the CYP707A3 microsomes. The Km value for (+)-ABA was estimated to be 1.3 ± 0.3 μm, indicating a high affinity of CYP707A3 for (+)-ABA. This value was comparable to the binding constant Ks determined above. Cutler et al. (2000) previously reported that (+)-ABA 8′-hydroxylase in the microsomes from suspension-cultured maize cells showed the Km value as 13.8 ± 1.1 μm. The substrate affinity of Arabidopsis CYP707A3 was 10-fold higher than that of the maize enzyme. The turnover rate kcat was also calculated to be 15 nmol min−1 nmol−1 P450. Thus, the CYP707A3 protein very actively catalyzed the 8′-hydroxylation of (+)-ABA.
Although (+)-7′-hydroxy-ABA has been shown to be produced as a minor metabolite in some plant species (Lehmann and Schwenen, 1988; Loveys and Milborrow, 1991; Hampson et al., 1992), we did not detect (+)-7′-hydroxy-ABA in our assay conditions. It has also been reported that (−)-ABA, an enantiomer at C-1′ of natural (+)-ABA, was metabolized by suspension-cultured maize cells to unnatural phaseic acid, (+)-PA, and (−)-7′-hydroxy-ABA (Balsevich et al., 1994). The CYP707A3 microsomes did not metabolize (−)-ABA at all, and this was consistent with the finding that (−)-ABA did not bind to the solubilized CYP707A3 protein.
CYP707A3 Does Not Isomerize 8′-OH-ABA
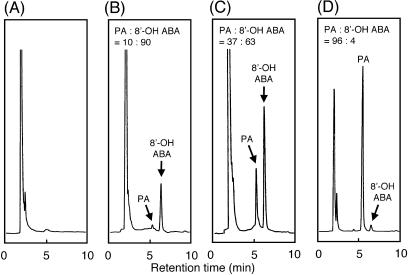
In the cell assays of the CYP707A enzymes, overnight culture with ABA resulted in the detection of PA as a major product (98%) and only a very small amount of 8′-OH-ABA (2%; Fig. 5). It was not clear whether the detected PA was formed by either spontaneous isomerization of 8′-OH-ABA or CYP707A enzyme-dependent isomerization. At the final equilibrium at 25°C, the PA to 8′-OH-ABA ratio should be 98:2 (Todoroki et al., 2000). In our cell assays, the products were present in the insect medium (pH 6.8) at 27°C for 24 h, and the ratio between 8′-OH-ABA and PA should be close to the final equilibrium by spontaneous isomerization. In order to prevent possible spontaneous isomerization during assay and extraction, the reaction mixture with the CYP707A3 microsomes was immediately acidified to pH 2 and directly subjected to HPLC analysis (Fig. 7). After a 10-min reaction, 8′-OH-ABA was detected as a major product, and the ratio of PA to 8′-OH-ABA was 10:90. After a 60-min reaction, the ratio decreased to 37:63, indicating the spontaneous cyclization of 8′-OH-ABA to PA. When the authentic PA was incubated under identical conditions, the ratio was 4:96, which is very close to the final equilibrium ratio. In these experiments, we were able to distinguish the reactions of the 8′-hydroxylation of ABA from the isomerization to PA. These results clearly indicate that CYP707A3 catalyzes the 8′-hydroxylation of ABA but does not catalyze the isomerization of 8′-OH-ABA to PA.
Figure 7.
HPLC analysis of reaction products from the recombinant CYP707A3 microsomes. The recombinant CYP707A3 microsomes were incubated without (+)-ABA (A), with 200 μm (+)-ABA for 10 min (B), or for 60 min (C), and with 200 μm PA (D). Reactions were carried out at 30°C, and stopped by acidification to pH 2 with 1 n HCl. A portion of reaction mixture (10 μL) was directly injected into the YMC AQ-311 column (6 × 100 mm, ODS). The reaction mixture contained 50 mm potassium phosphate (pH 7.25), 2 mg/mL recombinant CYP707A3 microsomes, and 200 μm NADPH.
Expression Patterns of CYP707A Family Genes in Arabidopsis
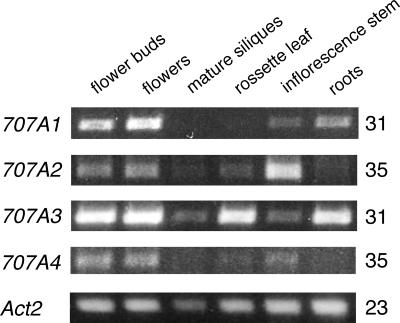
Firstly, we determined the expression profiles of the CYP707A genes in various tissues of Arabidopsis by RT-PCR (Fig. 8). The expression of the CYP707A genes was ubiquitous in various organs with different transcript accumulation levels. The PCR amplification with 31 cycles gave clear amplified bands for the CYP707A1 and CYP707A3 transcripts, while the number of PCR cycles had to be increased to 35 to obtain the signals for CYP707A2 and CYP707A4 gene expression, indicating that overall expression levels of CYP707A1 and CYP707A3 are much higher than those of CYP707A2 and CYP707A4. Flower buds and flowers were relatively abundant in the transcripts of all CYP707As compared with the other tissues, while mature siliques showed lower expression levels of CYP707As. In rosette leaves, the CYP707A3 mRNA was the major transcript, while CYP707A2 was abundantly expressed in inflorescence stems. In roots, CYP707A1 and CYP707A3 were moderately expressed, whereas weak expression of CYP707A2 and no expression of CYP707A4 were observed. These results indicated that the expression of the CYP707A genes is differently regulated in each organ.
Figure 8.
Tissue specific expression of CYP707As. Total RNA was extracted from various tissues in Arabidopsis plants (4 weeks old) except for root (2 weeks old). RT-PCR was performed using gene specific primers. The Act2 RT-PCR was included as a constitutive control. The number indicated on the right is the number of PCR cycles required to amplify the DNA fragment of each CYP707A cDNA.
It is known that environmental stresses such as high salinity and drought stresses induce the accumulation of ABA in various plant species. We studied how the expression of the CYP707A genes might respond to salt, osmotic, and drought stresses by semiquantitative RT-PCR (Fig. 9). When liquid-cultured Arabidopsis seedlings were stressed by adding 250 mm NaCl, strong induction of CYP707A1 and CYP707A4 expression and moderate increase of the CYP707A2 and CYP707A3 transcript levels were observed (Fig. 9A). The osmotic stress by 400 mm mannitol also had similar effects on the expression of the CYP707A genes (Fig. 9A). The expression patterns of the CYP707A genes in response to these stresses were quite similar to those in response to ABA. On the other hand, the expression patterns of the CYP707A genes under the drought stress were relatively complicated (Fig. 9B). CYP707A3 was rapidly induced within 30 min of the drought stress and the induced expression level was retained thereafter. The expression of CYP707A1 also increased in 30 min after the stress but reduced to a basal level at 1 h. The CYP707A1 transcript level increased again 3 h after treatment. CYP707A4 gene expression was slowly induced 3 h after treatment, while CYP707A2 expression was reduced at the early stages (30 min and 1 h) of the stress and returned to a basal level 3 h after treatment.
Figure 9.
Expression patterns of CYP707As in response to stresses. A, Expression patterns of CYP707As in response to ABA and stresses of high salinity and mannitol. Arabidopsis plants (2 weeks old) grown on GM plates were cultured in GM liquid medium for 3 d and treated with 100 μm ABA, 250 mm NaCl, and 400 mm mannitol for 6 h. B, Expression patterns of CYP707As in response to drought stress. Arabidopsis plants (2 weeks old) grown on GM plates were transferred onto a dried filter paper in a sealed petri dish and harvested after 0, 0.5, 1, 3, and 6 h. The Act2 RT-PCR was included as a constitutive control. The number indicated on the right is the number of PCR cycles required to amplify the DNA fragment of each CYP707A cDNA.
DISCUSSION
CYP707A Family Encodes ABA 8′-Hydroxylase
In this study, we have cloned the cDNAs of all four genes in the Arabidopsis CYP707A family, and CYP707A1, A3, and A4 have been functionally expressed in the baculovirus system. The enzymatic characterization of these recombinant P450s provides convincing evidence that the CYP707A family encodes (+)-ABA 8′-hydroxylase. First, CYP707A3 catalyzed hydroxylation at C-8′ methyl group of (+)-ABA but not at C-7′ position. Second, CYP707A3 bound (+)-ABA with a high affinity (Ks = 3.5 μm) but did not bind and hydroxylate (−)-ABA. Also, we demonstrated that CYP707A3 catalyzed the 8′-hydroxylation of (+)-ABA with a high affinity and efficiency (Km = 1.3 μm and kcat = 15 min−1). These enzymatic properties of CYP707A3 are quite consistent with those of ABA 8′-hydroxylase activity found in the microsomes from suspension-cultured maize cells (Krochko et al., 1998; Cutler et al., 2000). Although in vivo feeding experiments (Lehmann and Schwenen, 1988; Loveys and Milborrow, 1991; Hampson et al., 1992; Balsevich et al., 1994) indicated the presence of 7′-hydroxylation activity for both (+)-ABA and (−)-ABA in various plant species, these activities were probably due to the catalysis of enzymes other than (+)-ABA 8′-hydroxylase. Cutler et al. (2000) demonstrated that a suicide substrate for ABA 8′-hydroxylase inhibited 8′-hydroxylation of (+)-ABA but not 7′-hydroxylation of (−)-ABA, indicating that different enzymes were involved in the 7′- and 8′-hydroxylations.
We have also investigated whether PA was produced by either spontaneous isomerization of 8′-OH-ABA or CYP707A enzyme-dependent isomerization (Fig. 7). The isomerization of 8′-OH-ABA to PA is a cyclization reaction triggered by the intramolecular conjugated nucleophilic addition of 8′-hydroxy group oxygen into the electron-deficient enone at C-2′ position. This reaction is likely promoted by deprotonation of the 8′-hydroxy group, and the reaction rate can be affected by the pH of the medium. At 25°C, the PA to 8′-OH-ABA ratio was 98:2 at the final equilibrium, and the half-life of 8′-OH-ABA was determined to be 30 h at pH 3, 4 h at pH 7, and shorter than 1 min at pH 10 (Todoroki et al., 2000). When we minimized the effect of the spontaneous isomerization by acidification of the reaction mixtures and shortened the sample preparation time for the HPLC analysis, 8′-OH-ABA was detected as a major product after the 10-min reaction. This result clearly indicates that CYP707A3 catalyzes 8′-hydroxylation of (+)-ABA but does not catalyze isomerization of 8′-OH-ABA to PA. If no enzyme is involved in this isomerization step, the 8′-OH-ABA formed by CYP707As catalysis will be present for several hours under physiological conditions (pH 5–7 at 25°C). It has been reported that 3-hydroxy-3-methylglutaryl conjugate of 8′-OH-ABA accumulated in immature seeds of Robinia pseudo-acacia (Hirai et al., 1978), providing evidence for the transient presence of 8′-OH-ABA as an intermediate in the seeds. Although it is not clear whether 8′-OH-ABA spontaneously or enzymatically isomerizes PA in vivo (Milborrow et al., 1988), current results have established that 8′-hydroxylation by CYP707As is clearly the key step in the conversion of (+)-ABA to PA in the oxidative catabolic pathway.
CYP707A Genes in Vivo
Several T-DNA-inserted knockout lines for CYP707A1, A2, and A3 genes are found at the Arabidopsis Biological Resource Center (ABRC; Alonso et al., 2003). Under normal growth conditions, however, no obvious phenotypes were observed even in the homozygous lines (M. Mizutani, unpublished data). This was probably due to compensation by the functional redundancy of the CYP707A genes. Since the CYP707A genes are differently regulated during development and under environmental stresses, phenotypic alterations of each CYP707A mutant may be observed in association with specific physiological events such as seed germination, seed maturation, and development of stress tolerance.
It has been reported that ABA negatively regulates ABA accumulation by enhancing the ABA 8′-hydroxylase level (Cutler and Krochko, 1999), and it is well known that ABA biosynthetic genes are up-regulated by environmental stresses as well as ABA (Seo and Koshiba, 2002; Xiong and Zhu, 2003). In this study, we demonstrated that the transcript levels of all four CYP707A genes increased in response to ABA (Figs. 3 and 9A) and under various stress conditions (Fig. 9). In other words, ABA activates its own inactivation to PA via transcriptional activation of CYP707A gene expression. The enhanced expression of the CYP707A genes explains the previous finding that PA as well as ABA accumulated in response to dehydration stress (Qin and Zeevaart, 2002; Wang et al., 2002). Thus, bioactive ABA concentrations are maintained through a fine balance between biosynthesis and catabolism.
This raises the question regarding what signals activate the CYP707 genes in response to environmental stresses such as high salinity, osmotic, and dehydration stresses. These stresses may be direct signals for CYP707A induction. It is also possible that the ABA level elevated by the stresses may indirectly induce CYP707A gene expression. To address the question, it should be investigated how CYP707A gene expression could be induced in ABA deficient and insensitive mutants. In the ABA insensitive mutant abi1-1 seedlings, induction of CYP707A1 and CYP707A3 as well as many ABA responsive genes in response to ABA treatment was abolished (Hoth et al., 2002). It is therefore possible that protein phosphorylation/dephosphorylation events mediated by ABI1 protein phosphatase may be involved in CYP707A expression in response to ABA.
ABA Catabolism
The conversion of ABA to DPA is the primary inactivation pathway of ABA. 8′-OH-ABA still has biological activity similar to that of ABA in stimulating biosynthesis of a very long chain of fatty acids in Brassica napus embryos (Zou et al., 1995). It has also been reported that a stable analog of 8′-OH-ABA, (+)-3′-fluoro-8′-hydroxy-ABA, had significant biological activity (Todoroki et al., 1995; Arai et al., 1999). These reports suggest that 8′-OH-ABA is biologically active and keeps its biological activity until isomerization is completed. On the other hand, PA shows about 10% of the biological activity of ABA in several bioassays (Walton, 1983), and DPA is almost inactive in all of the bioassays (Walton, 1983). Thus, it is likely that ABA is inactivated in a stepwise manner to DPA and that PA 4′-reductase as well as ABA 8′-hydroxylase is a potentially important enzyme for ABA catabolism. Our finding, together with the identification of PA 4′-reductase, will allow us to describe the whole picture of the ABA catabolic pathways in plants, and this sheds light on the concerted mechanism that integrates and organizes various physiological phenomena through fine regulation of biosynthesis and catabolism of ABA. (+)-ABA 8′-hydroxylase will be a good target to manipulate ABA content and enhance stress tolerance by blocking its activity with either chemical inhibitors or transgenic plants.
MATERIALS AND METHODS
Instruments
Nuclear magnetic resonance (NMR) spectra were recorded with a Bruker (Billerica, MA) ARX500 instrument (500 MHz for 1H) at 300 K, using tetramethylsilane as the internal standard. Electron impact ionization mass spectrometry (EIMS) measurements were carried out with a JEOL JMS-600H mass spectrometer set at an electron potential of 70 eV, the temperature of the direct probe being increased from 30°C to 450°C at a rate of 128°C/min. UV spectra and optical rotation were measured with a UV 2200AI instrument (Shimadzu, Kyoto) and a DIP-1000 polarimeter (JASCO, Tokyo), respectively. The other all spectrophotometric determinations were carried out at room temperature with a UV-3101 spectrophotometer (Shimadzu).
Chemicals
(±)-ABA and (+)-ABA were purchased from Wako Pure Chemical Industries (Osaka) and BAL Planning Co. (Ichinomiya, Aichi, Japan), respectively. (−)-ABA was prepared by optical resolution of (±)-ABA (10.5 mg) by HPLC: column, Daicel Chiralcel OD (Osaka; 250-mm length × 4.6 mm i.d.); solvent, i-propanol-n-hexane (20:80) containing 0.1% (v/v) acetic acid; flow rate, 1.0 mL/min; detection at 254 nm. (+)-ABA and (−)-ABA were eluted at a retention time of 7.5 and 8.6 min, respectively, and the latter was collected to give 4.5 mg of colorless solids. For preparation of PA, β-hydroxy-β-methylglutaryl ester of 8′-OH-ABA was extracted and purified from immature seeds of Robinia pseudo-acacia as described previously (Hirai et al., 1978). The ester (43 mg) was dissolved in a mixture of 1 mL of methanol and 2 mL of 2 n NaOH aqueous solution, and then left at room temperature for 5 h. The solution was diluted with 20 mL of water, and partitioned with 10 mL of ethyl acetate four times at pH 2. The organic layers were combined, washed with water, dried over Na2SO4, and filtered. The filtrate was concentrated and subjected to silica gel (13 g) chromatography, using mixtures of toluene and ethyl acetate as the eluent. The materials eluted with 50% (v/v) and 60% (v/v) ethyl acetate were combined and concentrated to give PA (31 mg).
Plant Materials
Arabidopsis ecotype Columbia (Col) was grown under continuous light conditions at 22°C. Various organs such as buds, flowers, mature siliques, rosette leaves and inflorescence stems were obtained from nonsterilized plants grown in soil for 4 weeks. To isolate total RNA from roots, Arabidopsis seedlings were grown for 2 weeks under a sterile condition on 0.8% (w/v) agar plates containing GM medium (Valvekens et al., 1988) supplemented with 1 × Murashige and Skoog salt, 1% (w/v) Suc, 0.5 g/L MES (pH 5.7), 100 mg/L myoinositol, and GM vitamins (1 mg/L thiamin hydrochloride, 0.5 mg/L pyridoxin hydrochloride, and 0.5 mg/L nicotinic acid). For phytohormone treatment, the 1-week-old seedlings grown under a sterile condition were transferred to GM liquid medium and grown in a rotary shaker at 110 rpm for 1 week. The liquid cultures were then treated for 6 h with 1 μm of (±)-ABA, GA3, and brassinolide. For gene expression studies, the 2-week-old seedlings grown under a sterile condition were transferred to GM liquid medium for 3 d, and the seedlings were treated with either 100 μm (±)-ABA, 250 mm NaCl, or 400 mm mannitol in a rotary shaker at 110 rpm for 6 h. For drought stress, the 2-week-old seedlings grown under a sterile condition were transferred onto dried filter paper in a sealed petri dish, and harvested after 0, 0.5, 1, 3, and 6 h.
Cloning of CYP707A1 and 707A3 cDNAs
A cDNA library of Arabidopsis was prepared using total RNA from 7-d-old seedlings treated with 1 μm uniconazole for 3 d, using Superscript Lambda system (Invitrogen, Carlsbad, CA). A DNA fragment of the CYP707A1 gene was amplified from genomic DNA of Arabidopsis by PCR with a set of primers; 707A1N: 5′-ATGGATATCTCCGCCTTGTTTCTC-3′, and 707A1C: 5′-TTCTGTCATTCTACACTTCGATCT-3′. The PCR fragments were gel-purified, labeled with an AlkPhos direct labeling system (Amersham Biosciences, Piscataway, NJ), and used for the probe to isolate the CYP707A cDNAs. The cDNA library (200,000 plaques) was screened, and nine positive clones were isolated. The pZL1 plasmids containing the cDNA inserts were excised by in vivo excision according to the manufacturer's instruction, and the partial cDNA sequences were determined. Two of nine positive clones were found to be full-length CYP707A1 cDNAs, and the other seven clones were full-length CYP707A3 cDNAs. The CYP707A1 and CYP707A3 cDNAs were completely sequenced (DDBJ accession numbers, AB122149 and AB122150, respectively).
Cloning of CYP707A2 and CYP707A4 cDNAs
The cDNA containing an entire open-reading frame of either CYP707A2 or CYP707A4 was amplified by RT-PCR. Nucleotide sequences of gene-specific primers were as follows; CYP707A2: 5′-GGATCCATGCAAATCTCATCTTCATCGTCTTCAAATTTC-3′ (BamHI site in italic and start codon is underlined) and 5′-GTCGACTTAGGCTTAAATCGGGGTTACTCTTATTGG-3′ (SalI site in italic and stop codon is underlined); CYP707A4: 5′-GGATCCATGGCTGAAATTTGGTTCTTGGTTGTACCA-3′ (BamHI site in italic and start codon is underlined) and 5′-CTCGAGCTAAAGAGAATGTCGACGAAATGTAGCGGG-3′ (XhoI site in italic and stop codon is underlined). First-strand cDNA was synthesized with an oligo(dT) primer and a ReverTra Ace reverse transcriptase (Toyobo, Osaka) in a 10-μL reaction mixture containing 0.6 μg of total RNA from (±)-ABA treated Arabidopsis seedlings, and the RT reactions were carried out at 42°C for 30 min, 99°C for 5 min, and chilled to 5°C for 5 min. One μL of the reaction products was used as a template for the PCR reaction in a 10-μL reaction mixture containing 2.5 mm MgCl2, 10 mm Tris-HCl, 50 mm KCl, 0.25 unit of KOD-plus Taq DNA polymerase (Toyobo), 2 mm of dNTP, and 0.5 μm of gene-specific primers described above. Addition of 3′-A-overhangs was done by incubation of the PCR reaction with 1 unit of Taq DNA polymerase at 72°C for 10 min. The PCR product was gel-purified and cloned into the TA cloning vector pCR2.1 using a TOPO TA cloning kit (Invitrogen). The cloned inserts were sequenced to confirm no PCR errors in the inserts.
Heterologous Expression Using Baculovirus-Insect Cell System
The full-length cDNAs of CYP707A1 and CYP707A3 in pZL1 plasmid vector were excised with the restriction enzymes, SalI and NotI, and were purified by 1% (w/v) agarose gel electrophoresis. The cDNAs of CYP707A2 and CYP707A4 in pCR2.1 were digested with restriction enzymes, BamHI and SalI for CYP707A2, and BamHI and XhoI for CYP707A4, and agarose gel-purified. These cDNAs were then ligated into pFastBac1 vector (Invitrogen) digested with the same sets of the restriction enzymes. The pFastBac1-CYP707A constructs were used for the preparation of recombinant Bacmid DNA by transformation of Escherichia coli strain DH10Bac (Invitrogen). Spodoptera furugiperda 9 (Sf9) cells were maintained at 27°C as a monolayer culture in Grace's insect cell medium (Invitrogen) supplemented with 10% (v/v) fetal bovine serum. For large-scale expression, Sf9 cells were propagated as suspension cultures in Grace's insect medium containing 0.1% (w/v) Pluronic F-68 (Invitrogen), and incubated in a rotary shaker at 27°C and 150 rpm. For expression of recombinant CYP707A proteins, Sf9 cells were cultured in the above Grace's insect medium supplemented with 100 μm 5-aminolevulinic acid and 100 μm ferrous citrate to compensate for the low heme synthetic capacity of the insect cells.
Cell Assay
The suspension cultures of Sf9 insect cells were infected with the recombinant baculovirus each containing CYP707A cDNA on a rotary shaker (150 rpm) at 27°C for 72 h. After 72-h culture, 200 μm (+)-ABA was added to the culture medium, and the cells were further incubated at 27°C for 24 h. The cells and the culture medium were separately collected by centrifugation at 1,000g for 5 min and were extracted four times with an equal volume of ethyl acetate. After evaporation, the ethyl acetate extracts were resuspended in 1.2 mL of methanol, and 1 μL of the sample was subjected to HPLC: column, YMC AQ-311 (Kyoto; ODS, 100-mm length × 6 mm i.d.); solvent, 45% (v/v) methanol in water containing 0.1% (v/v) acetic acid; flow rate, 1.0 mL/min; detection at 254 nm. Retention times of authentic samples were 5.8 min for PA, 9.6 min for 2E-ABA, and 12.1 min for ABA. The detection limit for these three compounds was 5 ng.
Microsomal Assay
For preparation of microsomes, the cells (250 mL of suspension-cultured cells) expressing either CYP707A1 or CYP707A3 were washed with PBS and suspended in buffer A consisting of 20 mm potassium phosphate (pH 7.25), 20% (w/v) glycerol, 1 mm EDTA, and 1 mm dithiothreitol. The cells were sonicated, and cell debris was removed by centrifugation at 10,000g for 15 min. The supernatant was further centrifuged at 100,000g for 1 h, and the pellet was homogenized with buffer A to provide microsomal fractions. The complete reaction mixture (2 mL) contained 50 mm potassium phosphate (pH 7.25), 50 μg/mL recombinant CYP707A3 microsomes, 100 μm NADPH, and 100 μm (+)-ABA. Reactions were initiated by the addition of NADPH and carried out at 30°C for 10 min. After stopping the reactions by adding 200 μL of 1 n HCl, the reaction products were extracted four times with an equal volume of ethyl acetate. The organic layers were combined, washed with a small amount of water twice, dried over Na2SO4, filtered, and concentrated to give ethyl acetate-soluble materials. The materials were dissolved in 50 μL of methanol, and 5 μL of the sample was subjected to HPLC in the same conditions as those described in “Cell Assay.” For kinetic analysis, CYP707A3 was assayed using the microsomes (25 μg/mL) and (+)-ABA at concentrations ranging from 1 to 32 μm. Reactions were initiated by the addition of NADPH and carried out at 30°C for 10 min. The kinetic constants were calculated from triplicated data sets. Km and Vmax were calculated from a double reciprocal plot of the initial velocity (v0) versus substrate concentration.
Assay of 8′-Hydroxylase Activity
The reaction mixture (100 μL) contained 50 mm potassium phosphate (pH 7.25), 2 mg/mL recombinant CYP707A3 microsomes, 200 μm NADPH, and 200 μm (+)-ABA. Reactions were carried out at 30°C and stopped by acidification to pH 2 with 5 μL of 1 n HCl. The reaction mixtures were briefly centrifugated at 3,000g, and a portion of each reaction mixture (10 μL) was directly subjected to HPLC in the same conditions as those described in “Cell Assay.”
Identification of PA and 8′-OH-ABA
Sf9 cells expressing CYP707A3 were cultured in 60 mL of a medium containing (+)-ABA (3.17 mg) at 27°C for 24 h in the darkness. The cell suspension was centrifuged at 1,000g for 10 min. The supernatant was acidified to pH 2 with 1 n HCl and partitioned with 30 mL of ethyl acetate four times. The organic layers were combined, washed with a small amount of water, dried over Na2SO4, filtered, and concentrated to give solids (7.9 mg). The solids dissolved in 1.2 mL of methanol were subjected to preparative HPLC in the same conditions as those described in “Cell Assay.” The materials eluted at the retention times 5.8 min and 7.1 min were collected separately and concentrated to give compounds 1 (1.15 mg) and 2 (48 μg), respectively. Compound 1 (PA): 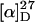 −30° (c 0.056, methanol); UV λmax nm (methanol): 256 (ɛ 18,400); 1H-NMR (500 MHz, CD3OD): δ 1.01 (3H, s, H-9′), 1.21 (3H, s, H-7′), 2.04 (3H, d, J = 1.2 Hz, H-6), 2.38 (1H, dd, J = 18.1 and 2.6 Hz, H-5′proR), 2.45 (1H, dd, J = 18.0 and 2.6 Hz, H-3′proS), 2.70 (1H, dd, J = 18.1 and 2.6 Hz, H-5′proS), 2.80 (1H, d, J = 18.0 Hz, H-3′proR), 3.66 (1H, d, J = 7.6 Hz, H-8′proS), 3.94 (1H, dd, J = 7.6 and 2.9 Hz, H-8′proR), 5.80 (1H, brs, H-2), 6.40 (1H, d, J = 15.9 Hz, H-5), 8.06 (1H, d, J = 15.9 Hz, H-4). Compound 1 (0.89 mg) was methylated with ethereal diazomethane to give its methyl ester. EIMS (probe) m/z (relative intensity): 294 [M]+ (24), 276 [M-H2O]+ (18), 263 (12), 244 (13), 233 (9), 217 (11), 177 (22), 167 (30), 163 (31), 154 (31), 139 (37), 135 (39), 125 (100), 122 (68). Compound 2 (a mixture of PA and 8′-OH-ABA): 1H-NMR (500 MHz, CD3OD): signals corresponding to 8′-OH-ABA;
−30° (c 0.056, methanol); UV λmax nm (methanol): 256 (ɛ 18,400); 1H-NMR (500 MHz, CD3OD): δ 1.01 (3H, s, H-9′), 1.21 (3H, s, H-7′), 2.04 (3H, d, J = 1.2 Hz, H-6), 2.38 (1H, dd, J = 18.1 and 2.6 Hz, H-5′proR), 2.45 (1H, dd, J = 18.0 and 2.6 Hz, H-3′proS), 2.70 (1H, dd, J = 18.1 and 2.6 Hz, H-5′proS), 2.80 (1H, d, J = 18.0 Hz, H-3′proR), 3.66 (1H, d, J = 7.6 Hz, H-8′proS), 3.94 (1H, dd, J = 7.6 and 2.9 Hz, H-8′proR), 5.80 (1H, brs, H-2), 6.40 (1H, d, J = 15.9 Hz, H-5), 8.06 (1H, d, J = 15.9 Hz, H-4). Compound 1 (0.89 mg) was methylated with ethereal diazomethane to give its methyl ester. EIMS (probe) m/z (relative intensity): 294 [M]+ (24), 276 [M-H2O]+ (18), 263 (12), 244 (13), 233 (9), 217 (11), 177 (22), 167 (30), 163 (31), 154 (31), 139 (37), 135 (39), 125 (100), 122 (68). Compound 2 (a mixture of PA and 8′-OH-ABA): 1H-NMR (500 MHz, CD3OD): signals corresponding to 8′-OH-ABA; 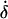 1.08 (3H, s, H-9′), 1.92 (3H, s, H-7′), 1.93 (3H, s, 6-H), 2.37 (1H, d, J = 17.1 Hz, H-5′proR), 2.48 (1H, d, J = 17.1 Hz, H-5′proS), 3.70 (1H, d, J = 11.0 Hz, H-8′proS), 5.83 (1H, brs, H-2), 5.90 (1H, s, H-3′), 5.93 (1H, d, J = 16.1 Hz, H-5), 7.58 (1H, d, J = 16.1 Hz, H-4), and the signal of H-8′proR overlapped with solvent signals; the other signals coincided with those of compound 1. Compound 2 (28 μg) was methylated with ethereal diazomethane to give its methyl ester. EIMS (probe) m/z (relative intensity): 294 [M]+ (20), 276 [M-H2O]+ (22), 244 (12), 233 (11), 217 (13), 177 (25), 167 (36), 163 (33), 154 (31), 139 (39), 135 (41), 125 (100), 122 (76).
1.08 (3H, s, H-9′), 1.92 (3H, s, H-7′), 1.93 (3H, s, 6-H), 2.37 (1H, d, J = 17.1 Hz, H-5′proR), 2.48 (1H, d, J = 17.1 Hz, H-5′proS), 3.70 (1H, d, J = 11.0 Hz, H-8′proS), 5.83 (1H, brs, H-2), 5.90 (1H, s, H-3′), 5.93 (1H, d, J = 16.1 Hz, H-5), 7.58 (1H, d, J = 16.1 Hz, H-4), and the signal of H-8′proR overlapped with solvent signals; the other signals coincided with those of compound 1. Compound 2 (28 μg) was methylated with ethereal diazomethane to give its methyl ester. EIMS (probe) m/z (relative intensity): 294 [M]+ (20), 276 [M-H2O]+ (22), 244 (12), 233 (11), 217 (13), 177 (25), 167 (36), 163 (33), 154 (31), 139 (39), 135 (41), 125 (100), 122 (76).
Spectrophotometric Analysis
P450 was estimated from the CO-difference spectrum using an extinction coefficient (Σ = 91 mm−1 cm−1; Omura and Sato, 1964). NADPH-P450 reductase was assayed by measuring its NADPH-cytochrome c reductase activity, and the rate of cytochrome c reduction was calculated from the A550 change using an extinction coefficient (Σ = 21 mm−1 cm−1; Imai, 1976). The protein content was measured by the Bradford method using a Coomassie protein assay reagent (Pierce, Rockford, IL). For the analysis of the CO-difference spectra, the recombinant CYP707A proteins were solubilized from the microsomes with buffer A supplemented with 1% (w/v) Emulgen 913 (Kao Chemicals, Tokyo; buffer B) for 1 h. After centrifugation at 10,000g for 10 min, the supernatant was used for further P450 assays.
Spectral Analysis of Substrate Binding
The microsomes from the cells expressing the recombinant CYP707A3 were solubilized with 1% (w/v) sodium cholate for 1 h, and centrifugated at 100,000g for 1 h. The supernatant was used as the solubilized recombinant CYP707A3. Either (+)-ABA or (−)-ABA was added to the solubilized CYP707A3 at a final concentration of 100 μm, and type I difference spectra were recorded between 500 and 350 nm. For the reference, dimethylsulfoxide, which is the solvent for 20 mm ABA stock solution, was added at a final concentration of 1% (v/v). For spectrophotometric titration, (+)-ABA was added to the solubilized CYP707A3 at concentrations ranging from 1 to 50 μm, and difference spectra were recorded between 500 and 350 nm. The spectral dissociation constant (Ks) was calculated from a double reciprocal plot of absorbance difference ΔA(386–419 nm) versus substrate concentration.
RT-PCR Analysis
Total RNA from Arabidopsis plants was isolated with a MagExtractor-RNA (Toyobo). RT-PCR was performed using a ReverTra Dash RT-PCR kit (Toyobo) according to the manufacturer's instruction. First-strand cDNA was synthesized in a 10-μL reaction mixture containing 0.6 μg of total RNA with an oligo(dT) primer and a ReverTra Ace reverse transcriptase (Toyobo), and the RT reactions were carried out at 42°C for 30 min, 99°C for 5 min, and chilling to 5°C for 5 min. The reaction mixture was diluted 2 times, and 0.5 μL of aliquot was used as a template for each of the PCR amplifications. The PCR reaction was carried out in a 10-μL reaction mixture containing 2.5 mm MgCl2, 10 mm Tris-HCl, 50 mm KCl, 0.25 unit of KOD dash Taq DNA polymelase (Toyobo), 2 mm of dNTP, and 0.5 μm of each gene-specific primer as shown below. Nucleotide sequences of gene-specific primers were as follows; CYP707A1, 707A1-F: 5′-CTTTTTCCAATCAAAACAGAAAAGGTATGG-3′ and 707A1-R: 5′-TCGCTCCAACAATTGACCAACTGTACTTGG-3′; CYP707A2, 707A2-F: 5′-CACTTTTTCATAAGTCCATGAAGGCAAGAA-3′ and 707A2-R: 5′-TACGTGTAAGGTTTTGGTGCCACCTCGAAT-3′; CYP707A3, 707A3-F: 5′-TGTTCTTTGCAGCAAAACAGAGAAGATACG-3′ and 707A3-R: 5′-TAGGCCCTACGATTGACCATCTGTACTTAG-3′; CYP707A4, 707A4-F: 5′-CCTACCAGGAGATGAAGAAGTTCGCCTTTG-3′ and 707A4-R: 5′-CTCCTCCCTTCACTTCCCATCGGAAATTGG-3′; Actin2, act2-F: 5′-GTGAAGGCTGGATTTGCAGGA-3′ and act2-R: 5′-AACCTCCGATCCAGACACTGT-3′. The RT sample was first denatured at 94°C for 2 min, and the PCR amplification was performed in following conditions; CYP707A1 and CYP707A3: 29, 31, 33, and 35 cycles of 94°C for 30 s, 54°C for 2 s, and 74°C for 45 s; CYP707A2: 31, 33, 35, and 37 cycles of 94°C for 30 s, 60°C for 2 s, and 74°C for 45 s; CYP707A4: 31, 33, 35, and 37 cycles of 94°C for 30 s, 54°C for 2 s, and 74°C for 45 s; and Actin2: 19, 21, 23, and 25 cycles of 94°C for 30 s, 54°C for 2 s, and 74°C for 45 s by using GeneAmp PCR system 9700 (Applied Biosystems, Norwalk, CT). The PCR products were analyzed with a 1% (w/v) agarose gel containing 20 ng/mL of ethydium bromide.
Sequence data from this article have been deposited with the EMBL/GenBank data libraries under accession numbers AB122149 and AB122150.
This work was supported by a part of the R&D Project of Industrial Science and Technology Frontier Program supported by NEDO (New Energy and Industrial Technology Development Organization), Japan (to D.O.).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.103.037614.
References
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P,, Cheuk R, et al (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Arai S, Todoroki Y, Ibaraki S, Naoe Y, Hirai N, Ohigashi H (1999) Synthesis and biological activity of 3′-chloro, -bromo, and -iodoabscisic acids, and biological activity of 3′-fluoro-8′-hydroxyabscisic acid. Phytochemistry 52: 1185–1193 [Google Scholar]
- Balsevich JJ, Cutler AJ, Lamb N, Friesen LJ, Kurz EU, Perras MR, Abrams SR (1994) Response of cultured maize cells to (+)-abscisic acid, (−)-abscisic acid, and their metabolites. Plant Physiol 106: 135–142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe SW, Dilkes BP, Fujioka S, Takatsuto S, Sakurai A, Feldmann KA (1998) The DWF4 gene of Arabidopsis encodes a cytochrome P450 that mediates multiple 22 α-hydroxylation steps in brassinosteroid biosynthesis. Plant Cell 10: 231–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutler AJ, Krochko JE (1999) Formation and breakdown of ABA. Trends Plant Sci 4: 472–478 [DOI] [PubMed] [Google Scholar]
- Cutler AJ, Rose PA, Squires TM, Loewen MK, Shaw AC, Quail JW, Krochko JE, Abrams SR (2000) Inhibitors of abscisic acid 8′-hydroxylase. Biochemistry 39: 13614–13624 [DOI] [PubMed] [Google Scholar]
- Cutler AJ, Squires TM, Loewen MK, Balsevich JJ (1997) Induction of (+)-abscisic acid 8′-hydroxylase by (+)-abscisic acid in cultured maize cells. J Exp Bot 48: 1787–1795 [Google Scholar]
- Durst F, Nielsen DR (1995) Diversity and evolution of plant P450 and P450-reductases. Drug Metab Drug Interact 12: 189–206 [DOI] [PubMed] [Google Scholar]
- Fujii H, Sato T, Kaneko S, Gotoh O, Fujii-Kuriyama Y, Osawa K, Kato S, Hamada H (1997) Metabolic inactivation of retinoic acid by a novel P450 differentially expressed in developing mouse embryos. EMBO J 16: 4163–4173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampson CR, Reaney MJT, Abrams GD, Abrams SR, Gusta LV (1992) Metabolism of (+)-abscisic acid to (+)-7′-hydroxyabscisic acid by bromegrass cell cultures. Phytochemistry 31: 2645–2648 [Google Scholar]
- Helliwell CA, Chandler PM, Poole A, Dennis ES, Peacock WJ (2001) The CYP88A cytochrome P450, ent-kaurenoic acid oxidase, catalyzes three steps of the gibberellin biosynthesis pathway. Proc Natl Acad Sci USA 98: 2065–2070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirai N, Fukui H, Koshimizu K (1978) A novel ABA metabolite from seeds of Robinia pseudacacia. Phytochemistry 17: 1625–1627 [Google Scholar]
- Hoth S, Morgant M, Sanchez JP, Hanafey MK, Tingey SV, Chua NH (2002) Genome-wide gene expression profiling in Arabidopsis thaliana reveals new targets of abscisic acid and largely impaired gene regulation in the abi1-1 mutant. J Cell Sci 115: 4891–4900 [DOI] [PubMed] [Google Scholar]
- Imai Y (1976) The use of 8-aminooctyl Sepharose for separation of some components of the hepatic microsomal electron transfer system. J Biochem (Tokyo) 80: 267–276 [DOI] [PubMed] [Google Scholar]
- Jennewein S, Rithner CD, Williams RM, Croteau RB (2001) Taxol biosynthesis: taxane 13 α-hydroxylase is a cytochrome P450-dependent monooxygenase. Proc Natl Acad Sci USA 98: 13595–13600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jennewein S, Rithner CD, Williams RM, Croteau RB (2003) Taxoid metabolism: Taxoid 14β-hydroxylase is a cytochrome P450-dependent monooxygenase. Arch Biochem Biophys 413: 262–270 [DOI] [PubMed] [Google Scholar]
- Krochko JE, Abrams GD, Loewen MK, Abrams SR, Cutler AJ (1998) (+)-Abscisic acid 8′-hydroxylase is a cytochrome P450 monooxygenase. Plant Physiol 118: 849–860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann H, Schwenen L (1988) Nigellic acid—an endogenous abscisic acid metabolite from Vicia faba leaves. Phytochemistry 27: 677–678 [Google Scholar]
- Loveys B, Milborrow VB (1991) Hydroxylation of methyl abscisate and the formation of three small β-glucosides. Phytochemistry 31: 67–72 [Google Scholar]
- Milborrow BV, Carrington NJ, Vaughan GT (1988) The cyclization of 8′-hydroxy abscisic acid to phaseic acid in vivo. Phytochemistry 27: 757–759 [Google Scholar]
- Mizutani M, Ohta D, Sato R (1993) Purification and characterization of a cytochrome P450 (trans-cinnamic acid 4-hydroxylase) from etiolated mung bean seedlings. Plant Cell Physiol 34: 481–488 [Google Scholar]
- Mizutani M, Ohta D, Sato R (1997) Isolation of a cDNA and a genomic clone encoding cinnamate 4-hydroxylase from Arabidopsis and its expression manner in planta. Plant Physiol 113: 755–763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizutani M, Ohta D (1998) Two isoforms of NADPH: cytochrome P450 reductase in Arabidopsis thaliana. Gene structure, heterologous expression in insect cells, and differential regulation. Plant Physiol 116: 357–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omura T, Sato R (1964) The carbon monoxide binding pigment of liver microsomes. I. Evidence for its hemoprotein nature. J Biol Chem 239: 2370–2378 [PubMed] [Google Scholar]
- Qin X, Zeevaart JAD (2002) Overexpression of a 9-cis-epoxycarotenoid dioxygenase gene in Nicotiana plumbaginifolia increases abscisic acid and phaseic acid levels and enhances drought tolerance. Plant Physiol 128: 544–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoendorf A, Rithner CD, Williams RM, Croteau RB (2001) Molecular cloning of a cytochrome P450 taxane 10 beta-hydroxylase cDNA from Taxus and functional expression in yeast. Proc Natl Acad Sci USA 98: 1501–1506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuler MA, Werck-Reichhart D (2003) Functional genomics of P450s. Annu Rev Plant Biol 54: 629–667 [DOI] [PubMed] [Google Scholar]
- Schwartz SH, Qin X, Zeevaart JA (2003) Elucidation of the indirect pathway of abscisic acid biosynthesis by mutants, genes, and enzymes. Plant Physiol 131: 1591–1601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo M, Koshiba T (2002) Complex regulation of ABA biosynthesis in plants. Trends Plant Sci 7: 41–48 [DOI] [PubMed] [Google Scholar]
- Shimada Y, Fujioka S, Miyauchi N, Kushiro M, Takatsuto S, Nomura T, Yokota T, Kamiya Y, Bishop GJ, Yoshida S (2001) Brassinosteroid-6-oxidases from Arabidopsis and tomato catalyze multiple C-6 oxidations in brassinosteroid biosynthesis. Plant Physiol 26: 770–779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szekeres M, Nemeth K, Koncz-Kalman Z, Mathur J, Kauschmann A, Altmann T, Redei GP, Nagy F, Schell J, Koncz C (1996) Brassinosteroids rescue the deficiency of CYP90, a cytochrome P450, controlling cell elongation and de-etiolation in Arabidopsis. Cell 85: 171–182 [DOI] [PubMed] [Google Scholar]
- Todoroki Y, Hirai N, Ohigashi H (1995) Synthesis, biological activity and metabolism of (S)-(+)-3′-fluoroabscisic acid. Tetrahedron 51: 6911–6926 [Google Scholar]
- Todoroki Y, Hirai H, Ohigashi H (2000) Analysis of isomerization process of 8′-hydroxyabscisic acid and its 3′-fluorinated analog in aqueous solutions. Tetrahedron 56: 1649–1653 [Google Scholar]
- Valvekens D, Montagu MV, Lijsebettens MV (1988) Agrobacterium tumefaciens-mediated transformation of Arabidopsis thaliana root explants by using kanamycin selection. Proc Natl Acad Sci USA 85: 5536–5540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walton DC (1983) Structure-activity relationships of abscisic acid analogs and metabolites. In FT Addicott, ed, Abscisic Acid. Praeger, New York pp 113–146
- Wang Z, Mambelli S, Setter TL (2002) Abscisic acid catabolism in maize kernels in response to water deficit at early endosperm development. Ann Bot (Lond) 90: 623–630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White JA, Beckett-Jones B, Guo YD, Dilworth FJ, Bonasoro J, Jones G, Petkovich M (1997) cDNA cloning of human retinoic acid-metabolizing enzyme (hP450RAI) identifies a novel family of cytochromes P450. J Biol Chem 272: 18538–18541 [DOI] [PubMed] [Google Scholar]
- Windsor ML, Zeevaart JA (1997) Induction of ABA 8′-hydroxylase by (+)-S-, (−)-R- and 8′-8′-8′-trifluoro-S-abscisic acid in suspension cultures of potato and Arabidopsis. Phytochemistry 45: 931–934 [DOI] [PubMed] [Google Scholar]
- Xiong L, Zhu JK (2003) Regulation of abscisic acid biosynthesis. Plant Physiol 133: 29–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu ZJ, Nakajima M, Suzuki Y, Yamaguchi I (2002) Cloning and characterization of the abscisic acid-specific glucosyltransferase gene from adzuki bean seedlings. Plant Physiol 129: 1285–1295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeevaart JAD (1999) Abscisic acid metabolism and its regulation. In MA Hooykaas, MA Hall, KR Libbenga, eds, Biochemistry and Molecular Biology of Plant Hormones. Elsevier Science Publishing, New York, pp 189–207
- Zeevaart JAD, Creelman RA (1988) Metabolism and physiology of abscisic acid. Annu Rev Plant Physiol Plant Mol Biol 39: 439–473 [Google Scholar]
- Zou J, Abrams GD, Barton DL, Taylor DC, Pomeroy MK, Abrams SR (1995) Induction of Lipid and Oleosin Biosynthesis by (+)-abscisic acid and its metabolites in microspore-derived embryos of Brassica napus L.cv Reston (biological responses in the presence of 8′-hydroxyabscisic acid). Plant Physiol 108: 563–571 [DOI] [PMC free article] [PubMed] [Google Scholar]