Plk3 phosphorylates CtIP in G1 in a damage-inducible manner and is required with CtIP for the repair of complex double-strand breaks and regulation of resection-mediated end-joining pathways.
Abstract
DNA double-strand breaks (DSBs) are repaired by nonhomologous end joining (NHEJ) or homologous recombination (HR). The C terminal binding protein–interacting protein (CtIP) is phosphorylated in G2 by cyclin-dependent kinases to initiate resection and promote HR. CtIP also exerts functions during NHEJ, although the mechanism phosphorylating CtIP in G1 is unknown. In this paper, we identify Plk3 (Polo-like kinase 3) as a novel DSB response factor that phosphorylates CtIP in G1 in a damage-inducible manner and impacts on various cellular processes in G1. First, Plk3 and CtIP enhance the formation of ionizing radiation-induced translocations; second, they promote large-scale genomic deletions from restriction enzyme-induced DSBs; third, they are required for resection and repair of complex DSBs; and finally, they regulate alternative NHEJ processes in Ku−/− mutants. We show that mutating CtIP at S327 or T847 to nonphosphorylatable alanine phenocopies Plk3 or CtIP loss. Plk3 binds to CtIP phosphorylated at S327 via its Polo box domains, which is necessary for robust damage-induced CtIP phosphorylation at S327 and subsequent CtIP phosphorylation at T847.
Introduction
DNA double-strand breaks (DSBs) represent biologically important lesions because incorrectly repaired DSBs can lead to translocations and other genomic rearrangements, driving forces during carcinogenesis (van Gent et al., 2001; Jackson and Bartek, 2009; Bunting and Nussenzweig, 2013; Panier and Durocher, 2013). Two major DSB repair pathways exist, canonical nonhomologous end-joining (NHEJ; c-NHEJ) and homologous recombination (HR; Lukas et al., 2011b; Polo and Jackson, 2011; Chapman et al., 2012; Davis and Chen, 2013). NHEJ repairs the majority of ionizing radiation (IR)–induced DSBs and functions throughout the cell cycle (Rothkamm et al., 2003; van Gent and van der Burg, 2007). In contrast to NHEJ, HR is restricted to the S and G2 phases of the cell cycle, in which homologous sequences on the sister chromatid serve as a template for repair (Moynahan and Jasin, 2010). HR is initiated by C terminal binding protein–interacting protein (CtIP)–dependent resection to create 3′ overhangs at the DSB ends (Sartori et al., 2007). DSB repair can also occur by an alternative NHEJ mechanism, termed alt-NHEJ (Wang et al., 2005, 2006; Nussenzweig and Nussenzweig, 2007).
In addition to its role in promoting resection during HR, increasing evidence suggests that CtIP can also function during NHEJ. CtIP-dependent microhomology-mediated NHEJ occurs in wild-type (wt) chicken cells (Yun and Hiom, 2009), and short single-stranded DNA regions exposed by CtIP-dependent resection facilitate rejoining during class switch recombination in mammalian cells (Lee-Theilen et al., 2011). Moreover, a CtIP-dependent process exposes microhomologies and causes translocations from restriction enzyme-induced DSBs (Zhang and Jasin, 2011). CtIP also has end-processing functions in G1, which are important to remove topoisomerase II from the DSB site before NHEJ can ensue (Nakamura et al., 2010; Quennet et al., 2011). Finally, CtIP can promote hairpin opening and resection during variable (diversity) joining recombination in G1-phase lymphocytes devoid of H2AX (Helmink et al., 2011).
CtIP is regulated during the cell cycle by Cdks and is a substrate of DNA damage-induced phosphorylation by ATM and ATR (Yu and Chen, 2004; Huertas et al., 2008; Peterson et al., 2013). Two Cdk sites, S327 and T847, regulate resection in S and G2 (Yu et al., 2006; Huertas and Jackson, 2009; Yun and Hiom, 2009), although the role of the S327 modification during HR has been questioned (Nakamura et al., 2010; Reczek et al., 2013). Five additional Cdk sites in the central domain of CtIP have been identified and found to interact with NBS1, promoting ATM-dependent CtIP phosphorylation to activate resection and HR (Wang et al., 2013). However, in contrast to CtIP’s role in HR in G2, the mechanism regulating CtIP in G1 is unknown.
Polo-like kinases (Plks) are serine/threonine kinases. Similar to Cdks, they phosphorylate substrates containing an [S/T]-P motif (Elia et al., 2003), whereas ATM and ATR phosphorylate [S/T]-Q sites (Kim et al., 1999). Plks contain a highly conserved N-terminal kinase domain and a C-terminal substrate-binding domain, termed the Polo box domain (PBD). Of the five Plks identified in mammalian cells, Plk1 is the best studied and regulates mitosis and cytokinesis (Golsteyn et al., 1995; de Cárcer et al., 2011). Plk3 is required for S-phase entry, and protein as well as mRNA levels are highest in G1 (Anger et al., 2003; Zimmerman and Erikson, 2007). Plk3 has been previously implicated in various stress responses (Bahassi et al., 2002), but a role in DSB repair was hitherto unknown.
Here, we show that Plk3 phosphorylates CtIP in G1-phase cells at T847 and S327 in a damage-inducible manner. We show that Plk3 and CtIP significantly contribute to the formation of translocations and other genomic rearrangements. Although Plk3 and CtIP are not essential for DSB repair in G1 wt cells, they are required for alt-NHEJ processes that operate in G1 Ku−/− mutants. We show that complex DSBs undergo resection and repair in G1 via a process requiring Plk3 and its target sites on CtIP. Thus, we identify Plk3 as a novel regulator of an error-prone end-joining process in G1.
Results
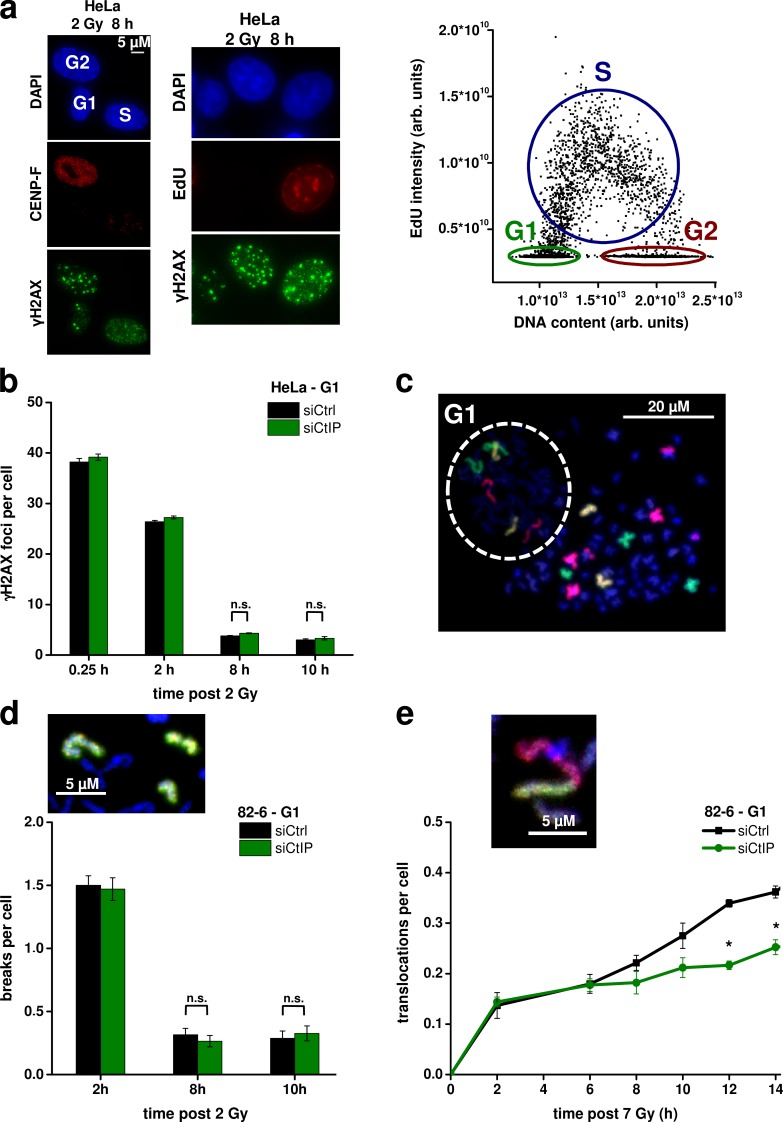
CtIP enhances translocation formation in G1
Repair of IR-induced DSBs in G2 involves CtIP-dependent resection and HR, but it is unclear whether and how CtIP impacts on repair in G1. In one approach, we analyzed DSB repair kinetics in G1 cells by measuring γ-H2AX foci after IR using a semiautomated microscopic approach. We incubated the cells with nocodazole (a spindle inhibitor) and 5-ethynyl-2′-deoxyuridine (EdU; a thymidine analogue) 30 min before IR and during the entire repair period and scanned them, after immunofluorescence labeling, under the microscope. The EdU signal was plotted against the DAPI signal, and G1 cells were discriminated from G2 cells based on their DNA content and from S-phase cells by the absence of EdU (Fig. 1 a, right). Control experiments confirmed that all cells identified as G1 were negative for the S/G2 marker CENP-F (Fig. 1 a, left). Cells identified as G1 cells were marked in the histogram and automatically relocated for manual foci enumeration (Fig. 1 a). The addition of nocodazole prevented G2-irradiated cells from progressing into G1 during repair incubation. Therefore, the identified G1 cells were irradiated in G1 and maintained in G1 during the entire repair period. Thus, this EdU-based approach allows us to measure cell cycle–specific DSB repair kinetics and avoids the usage of aphidicolin, which we used in previous studies to exclude S-phase cells from the analysis (Löbrich et al., 2010). To study the impact of CtIP on DSB repair in G1, we depleted CtIP by siRNA, which reduces Rad51 foci formation and phosphorylation of the RPA2 (replication protein A subunit 2) after IR in G2, demonstrating the efficiency of the knockdown approach (Fig. S1 a). At all time points analyzed, we observed the same level of γ-H2AX foci in CtIP-depleted, G1-phase HeLa cells as in control cells, suggesting that CtIP is not required for DSB repair in G1 (Fig. 1 b). The same result was obtained using G1-phase 82-6 fibroblasts depleted for CtIP (Fig. S1 b).
Figure 1.
CtIP enhances translocation formation in G1. (a) Identification of cell cycle phases (see first paragraph of the Results for description). arb. units, arbitrary units. (b) γ-H2AX foci in G1-phase HeLa cells after CtIP siRNA. (c) PCC spread from G1-phase 82-6 fibroblasts fused with mitotic HeLa cells. FISH staining was performed with chromosome probes 1, 2, and 4. The one-chromatid G1 PCC spread is encircled with a dashed line; the two-chromatid mitotic HeLa cell is outside this area. (d) Chromosome breaks in G1-phase 82-6 fibroblasts after CtIP siRNA. The image shows a chromosome break indicated by the presence of an additional fragment of the stained chromosome. (e) Translocations in G1-phase 82-6 fibroblasts after CtIP siRNA. The image shows a translocation indicated by the color junction (±SEM from at least three experiments). siCtrl, control siRNA; siCtIP, CtIP siRNA. *, P < 0.05.
We consolidated this finding by investigating chromosome breaks in G1-phase cells by using premature chromosome condensation (PCC), which involves fusing irradiated fibroblasts with mitotic HeLa cells. Condensed fibroblast chromosomes in G1 could be distinguished from condensed fibroblast chromosomes in G2 and from mitotic HeLa chromosomes by their one-chromatid morphology (Gotoh and Durante, 2006). FISH was used to visualize chromosomes 1, 2, and 4 in the condensed G1-phase fibroblast chromosomes (Fig. 1 c). Cell fusion was performed at varying times after IR to monitor the kinetics of repair. At all time points analyzed, CtIP-depleted cells showed similar levels of chromosome breaks as wt cells (Fig. 1 d).
Because CtIP depletion has been reported to reduce translocation and excision frequencies in reporter systems (Rass et al., 2009; Zhang and Jasin, 2011), we speculated that it might also reduce translocation levels after IR and investigated the formation of chromosome translocations in G1 cells by PCC/FISH analysis as described in the previous paragraph. To enhance sensitivity, we used a higher radiation dose (7 Gy); at this dose, break repair kinetics were still unaffected by CtIP depletion (Fig. S1 c). Some translocations in wt cells formed with fast kinetics being detectable within 2 h after IR (the earliest time point that could be investigated as a result of the time needed for the PCC protocol). Between 2 and 6 h after IR, only a few additional translocations arose before a second component of translocation formation occurred at times >6 h after IR (Fig. 1 e). Of note, CtIP depletion strongly diminished translocation levels at later (>6 h) but not at earlier times (Fig. 1 e). We analyzed G2-irradiated cells with a two-chromatid morphology as a control and observed translocation levels substantially lower than in G1-irradiated cells. Moreover, CtIP depletion did not affect translocation formation in G2-irradiated cells (Fig. S1 d). We conclude that CtIP functions in G1 and enhances translocation levels after IR, although the absence of CtIP does not alter DSB and chromosome break repair kinetics.
RPA loading and phosphorylation occurs in G1 at complex DSBs
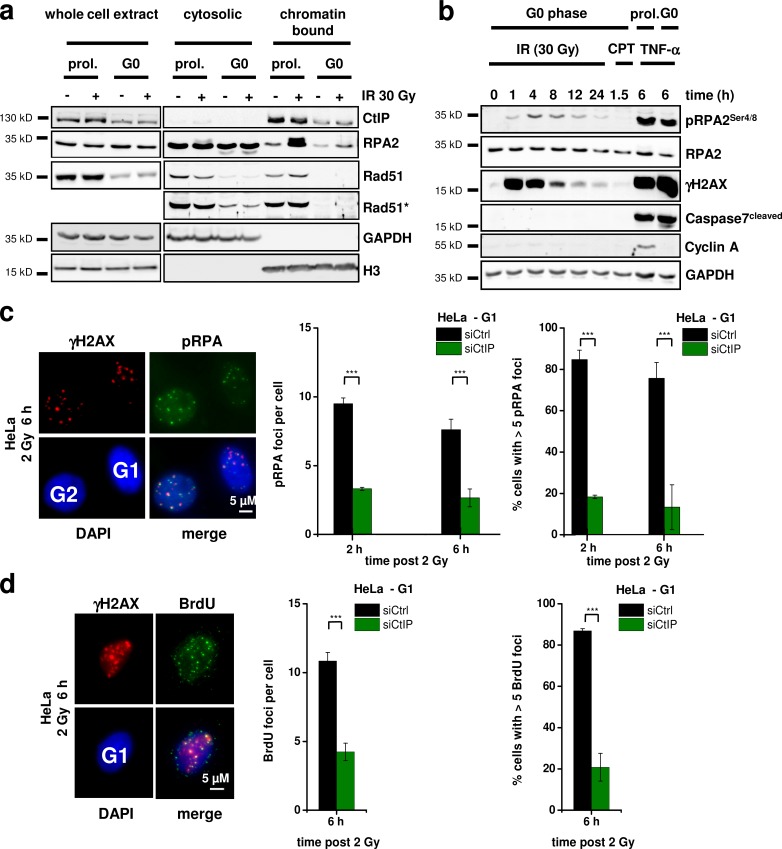
After having shown that CtIP impacts on translocation formation in G1, we investigated whether the function of CtIP in G1 involves resection. For this, we first examined RPA2 binding in 82-6 wt fibroblasts by Western blotting. To exclude any contribution from S or G2 cells, we used cell populations that were maintained in confluency for >4 wk, which resulted in >99.9% G0 cells (assessed by BrdU incorporation using FACS and immunofluorescence). We analyzed cytosolic and chromatin-bound protein levels and used proliferating 82-6 cell cultures as a control (Fig. 2 a). CtIP was present in the chromatin-bound fraction of confluent G0 cells, although at lower levels compared with proliferating cells (Fig. 2 a). The cytosolic Rad51 was strong in proliferating cells but weak in confluent G0 cells, consistent with a previous study (Chen et al., 1997). Chromatin-bound Rad51 was detectable in proliferating cells and increased after IR as described previously (Mladenov et al., 2006), but it was undetectable in confluent G0 cells. In contrast, cytosolic RPA2 was similarly present in proliferating and confluent cells. After IR, RPA2 levels strongly increased in the chromatin-bound fraction of proliferating cells. Significantly, RPA2 was also detectable and increased after IR in the chromatin-bound fraction of confluent cells (Fig. 2 a). The absence of chromatin-bound Rad51 in confluent cells confirmed the absence of contaminating S/G2-phase cells (Fig. 2 a). However, the RPA2 signal was much weaker in confluent compared with proliferating cells.
Figure 2.
RPA loading and phosphorylation occurs in G1 at complex DSBs. (a) Detection of cytosolic and chromatin-bound CtIP, RPA2, and Rad51 in proliferating (prol.) and confluency-arrested (G0) 82-6 fibroblasts 4 h after IR. The Rad51 signals are shown in two intensity settings (Rad51*, stronger intensity setting). GAPDH and H3 were used as markers for the cytosolic and chromatin-bound fraction, respectively. (b) Time course for the formation and disappearance of pRPA2 in whole cell extracts from confluency-arrested 82-6 fibroblasts. Caspase7, which provides a pronounced signal in cells treated with the apoptosis induction factor TNF and cycloheximide, was used to control for apoptosis. 1 µM camptothecin (CPT) treatment (for 1 h) was used to control for the presence of S-phase cells. (c and d) Detection of pRPA (c) and BrdU (d) foci in G1-phase HeLa cells treated with CtIP siRNA and irradiated with 2 Gy α particles. The mean number of foci per G1 cell (middle) and the fraction of G1 cells with more than five foci (right) are shown (±SEM from at least three experiments). siCtrl, control siRNA; siCtIP, CtIP siRNA. ***, P < 0.001.
We next wished to investigate RPA phosphorylation in G0, which is known to occur in G2 at chromatin-bound RPA2 after damage induction (Anantha et al., 2007; Stephan et al., 2009) in a manner dependent on CtIP (Fig. S1 a). We observed a weak but significant IR-induced pRPA2 signal in whole cell extracts from confluent 82-6 wt fibroblasts (Fig. 2 b). Treatment with the S-phase damaging agent camptothecin (CPT) produced no detectable signal, confirming that the IR-induced pRPA2 signal is specific for G0 cells and cannot be attributed to contaminating S/G2-phase cells. Moreover, the confluent cell cultures were devoid of any CyclinA signal, a marker for S/G2 phase (Fig. 2 b). We also investigated the kinetics for the formation and disappearance of the pRPA2 signal. pRPA2 increased in the first hours after IR, consistent with the time needed for resection (Fig. 2 b). At later times, pRPA2 decreased in parallel to the γ-H2AX signal, demonstrating that it represents repair occurring with slow kinetics instead of cell degeneration arising as a result of IR (Fig. 2 b). The latter notion was further supported by the lack of any apoptotic signal, consistent with a previous study showing that human fibroblasts remain metabolically active even if irradiated with a dose of 80 Gy (Rief and Löbrich, 2002). Of note, this biochemical analysis provides evidence for DSB resection in G0, although robust detection of RPA2 binding and phosphorylation requires a high radiation dose of 30 Gy. At doses of 15 Gy and below, only very faint pRPA2 signals could be detected (Fig. S1 e). We also wished to assess pRPA2 levels in G1 instead of G0 cells and, therefore, trypsinized and reseeded G0 cells before IR. Previous experiments had shown that G0 cells enter G1 and then progress into S phase at ∼12–16 h after trypsinization (Deckbar et al., 2010). At 4 h after trypsinization, cells were still in G1 as demonstrated by the lack of any pRPA2 signal after treatment with CPT. Such cells showed significantly higher pRPA2 levels than G0 cells but still lower levels compared with proliferating cells (Fig. S1, f and g). We conclude from this biochemical analysis that resection does occur in G0/1 but is much more limited than in G2.
We next established detection of RPA phosphorylation at DSBs in G1 cells by immunofluorescence analysis because this allowed us to use siRNA and complementation approaches, which were difficult to perform with confluent cell cultures. First, we used x-irradiation and failed to detect robust pRPA foci formation in G1 cells, consistent with literature data. We then applied α-particle irradiation, which induces DSBs that are more complex than x ray–induced DSBs and have a higher propensity to undergo resection in G2 (Shibata et al., 2011). Distinct pRPA foci, which colocalize with γ-H2AX foci, could readily be detected in G1 and G2 cells, although the signal intensity was significantly lower in G1 (Fig. 2 c, left). pRPA foci formation was maximal between 2 and 6 h after IR and then decreased as a result of slow DSB repair (Fig. S1 h). Of note, robust foci formation was observed in nearly all G1 cells identified by the approach described in Fig. 1 a (Fig. 2 c). Most importantly, pRPA foci levels were strongly reduced after CtIP siRNA, confirming that they represent DSBs that were resected in a manner dependent on CtIP (Fig. 2 c). We also directly analyzed the presence of single-stranded DNA by the formation of BrdU foci (Beucher et al., 2009) and observed BrdU foci after α-particle irradiation in a manner dependent on CtIP (Fig. 2 d). We conclude that CtIP impacts on DSB resection in G1, although robust evidence for resection is only observed after high radiation doses or the induction of complex DSBs, both of which enhances the level of resection that otherwise might escape detection by pRPA analysis.
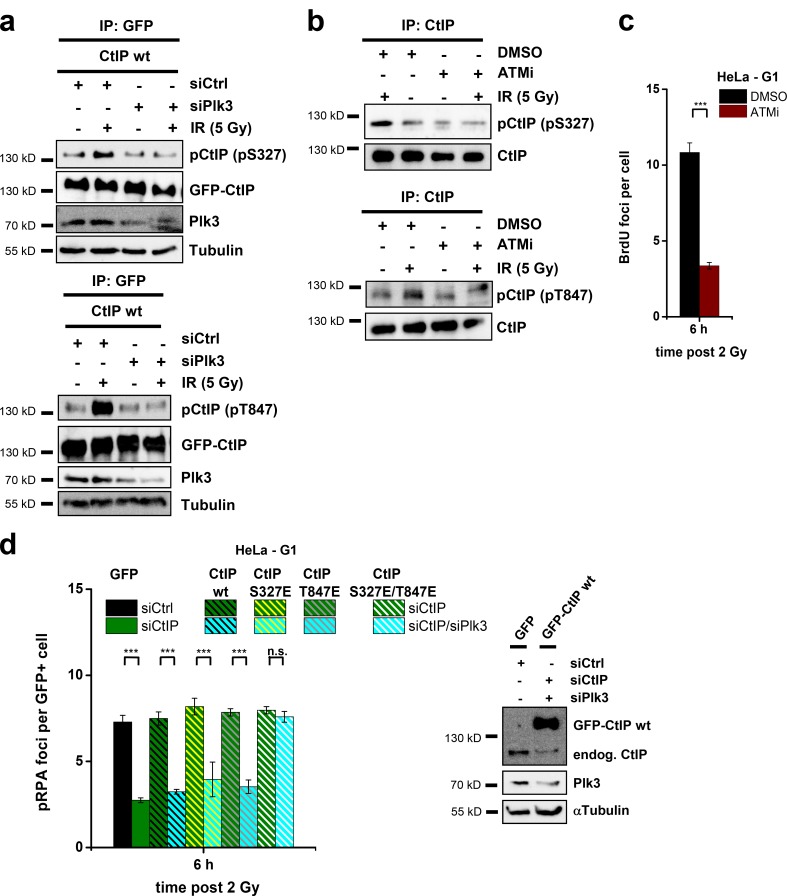
CtIP function in G1 requires T847 and S327 phosphorylation
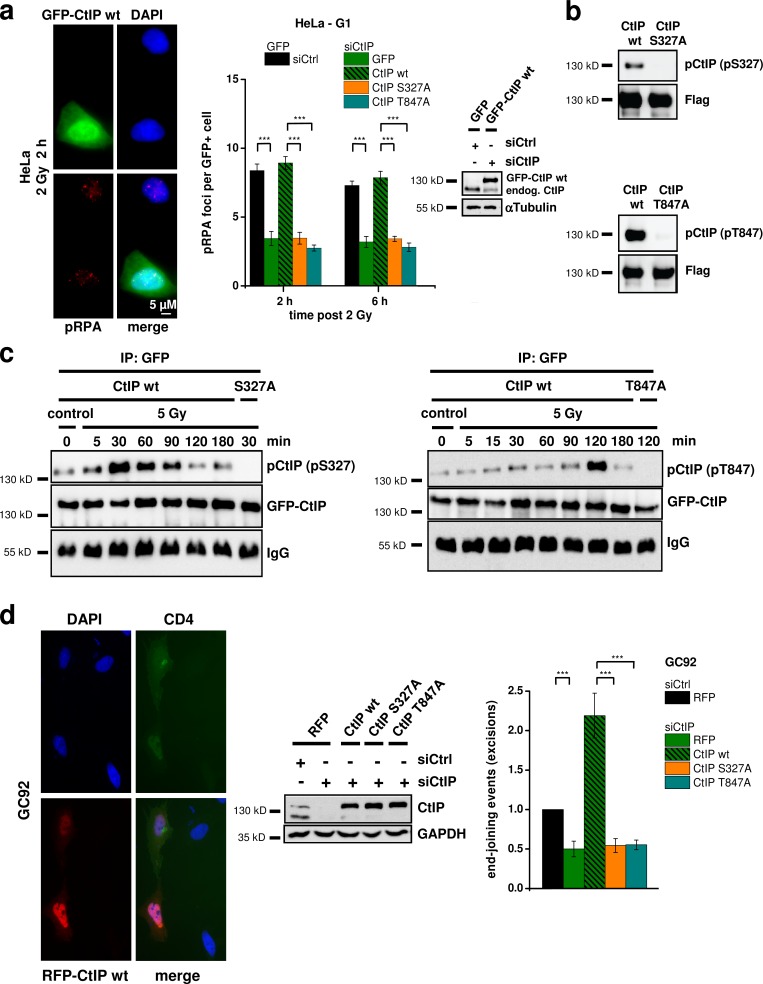
CtIP’s function during HR in G2 requires phosphorylation at the Cdk sites T847 and S327 (Yu et al., 2006; Huertas and Jackson, 2009; Yun and Hiom, 2009), although the role of the S327 modification during HR has been questioned (Nakamura et al., 2010; Reczek et al., 2013). To investigate whether these sites are required for CtIP’s resection function in G1, we transfected HeLa cells with wt or mutated GFP-CtIP plasmids (Fig. 3 a). For this and all other transfection experiments, we used siRNA-resistant plasmids and depleted the endogenous protein by siRNA. We investigated pRPA foci induced by α particles and observed robust foci levels in cells transfected with CtIP-wt. In contrast, cells transfected with CtIP-S327A or CtIP-T847A showed strongly diminished foci levels (Figs. 3 a and S2 a). These findings demonstrate that phosphorylations at the two Cdk sites T847 and S327 are necessary for CtIP’s resection function in human G1 cells.
Figure 3.
CtIP function in G1 requires S327 and T847 phosphorylation. (a) pRPA foci in GFP-positive (GFP+), G1-phase HeLa cells irradiated with 2 Gy α particles. Endogenous CtIP was depleted by siRNA, and cells were transfected with various GFP-CtIP plasmids. Knockdown efficiency for CtIP and transfection efficiencies for GFP-CtIP were confirmed by Western blotting. endog., endogenous. (b) Insect cell–purified CtIP-wt, CtIP-S327A, and CtIP-T847A proteins were incubated with recombinant Cdk2/CyclinA and analyzed with phosphospecific antibodies for CtIP-pS327 or CtIP-pT847. (c) Phosphorylation of CtIP in vivo. HEK293T cells were treated with CtIP siRNA, transfected with GFP-CtIP-wt, GFP-CtIP-S327A, or GFP-CtIP-T847A plasmids, enriched in G1 (≥90%), and irradiated with 5 Gy. GFP-CtIP was obtained by IP and analyzed for pS327 or pT847 by immunoblotting. (d) Analysis of excision events in GC92 cells containing an NHEJ reporter substrate. Excision formation from the repair of two I-SceI–induced DSBs results in a CD4-positive signal. Cells were treated with CtIP siRNA and transfected with various RFP-CtIP plasmids. Excisions were measured by the fraction of RFP-positive cells that exhibited a CD4-positive signal relative to all RFP-positive cells. Results were normalized to control siRNA-treated cells that were transfected with an RFP empty vector. Knockdown efficiencies for CtIP and transfection efficiencies for RFP-CtIP plasmids were confirmed by Western blotting (±SEM from at least three experiments). siCtrl, control siRNA; siCtIP, CtIP siRNA. ***, P < 0.001.
We then generated phosphospecific antibodies for detecting CtIP-S327 and CtIP-T847 phosphorylations (CtIP-pS327 and CtIP-pT847). CtIP proteins were expressed in insect cells, purified, and phosphorylated using Cdk2/CyclinA. CtIP-wt but not CtIP-S327A proteins showed a strong signal with the pS327 antibody, and CtIP-wt but not CtIP-T847A proteins showed a strong signal with the pT847 antibody (Fig. 3 b). Using these antibodies, we investigated the in vivo phosphorylation of CtIP in G1-enriched HEK293T cell populations. For this, we transfected the cells with GFP-CtIP using magnet-assisted transfection (MATra), which provided in this and all subsequent experiments ≥90% transfected G1 cells (Fig. S2 b). We immunoprecipitated GFP-CtIP and observed for both antibodies weak CtIP phosphorylation signals, which strongly increased after irradiation. Of note, S327 phosphorylation increased strongly at 30 min after IR and then decreased to background values at 2 h after irradiation. T847 phosphorylation, in contrast, was very weak at earlier times and sharply peaked at 2 h after IR (Fig. 3 c). To exclude the possibilities that the observed phosphorylation pattern is a peculiarity of transformed cells and that S or G2 cells contribute to the signal, we investigated nontransformed confluent 82-6 fibroblasts. We observed IR-induced CtIP phosphorylation of S327 at 30 min and of T847 at 2 h after IR (Fig. S2 c). Thus, CtIP phosphorylation occurs at both sites in vivo with a defined time course upon damage induction.
Finally, we wished to examine whether phosphorylation of the two phosphorylation sites T847 and S327 is important for the generation of genomic rearrangements. However, because of the limited transfection efficiency of CtIP plasmids, it was difficult to measure chromosomal translocations in CtIP complemented cells by the PCC/FISH approach described in Fig. 1 e. We, therefore, used a reporter system, which detects the formation of excisions (deletions) from the misrejoining of two I-SceI–induced DSBs (Fig. S2 d; Rass et al., 2009). Using this approach, we were able to measure excisions in cells treated with CtIP siRNA and efficiently complemented with various RFP-CtIP plasmids (Fig. 3 d). Of note, CtIP depletion lead to a reduction in excisions by ∼50%, demonstrating that a significant proportion of such erroneous end-joining events arise from CtIP function. Cells transfected with CtIP-wt showed highly elevated excision levels, likely caused by CtIP overexpression (Fig. 3 d). But most importantly, cells expressing CtIP-S327A or CtIP-T847A showed the same low level of excisions as CtIP-depleted cells without complementation (Fig. 3 d). The limitation of this and other reporter systems is that cells cannot be maintained with sufficient accuracy in G1 for the entire time period necessary for I-SceI expression and excision formation (72 h). To investigate in which cell cycle phase such excisions are formed, we treated the cells with roscovitine, an inhibitor of Cdk1/2 that is required for CtIP-dependent resection in S/G2. Of note, roscovitine did not affect the excision levels but did diminish gene conversion frequencies in a classical HR reporter assay (Fig. S2 e). The same result was obtained after depletion of Cdk2 (Fig. S2 e). Therefore, the observed events likely reflect CtIP function during NHEJ in G1. In summary, the CtIP sites S327 and T847 are required for resection in G1, are phosphorylated in G1 in a damage-inducible manner, and mediate the formation of genomic rearrangements.
Plk3 is required for resection in G1 and phosphorylates CtIP in vitro
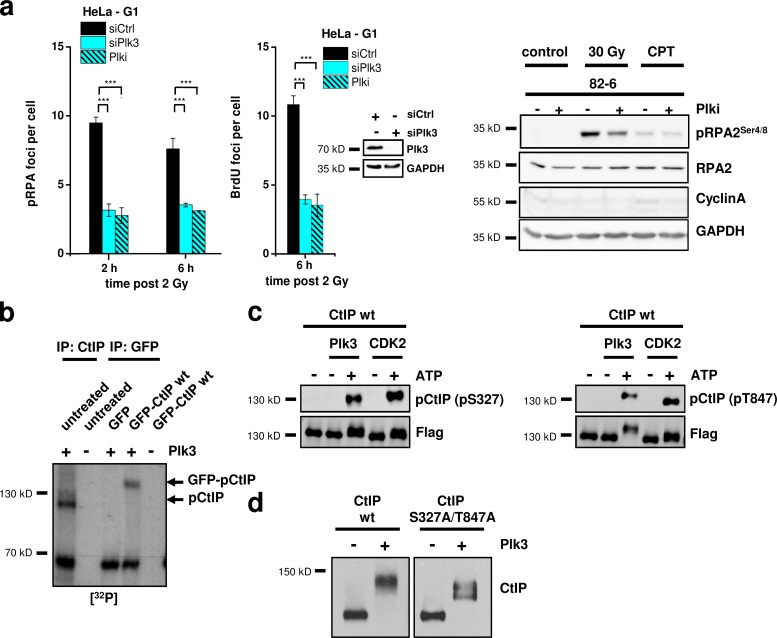
The Cdk1/2 inhibitor, roscovitine, diminished pRPA foci formation in G2 cells but did not affect pRPA foci levels in G1 cells (Fig. S3 a), suggesting that T847 and S327 phosphorylation in G1 is not mediated by Cdk1/2. Overexpression of the budding yeast homologue of the Plk family, Cdc5, leads to hyperphosphorylation of Sae2 (Donnianni et al., 2010), which is the homologue of CtIP (Sartori et al., 2007). In human cells, several Plk family members exist, of which Plk3 is required for S-phase entry (Anger et al., 2003; Zimmerman and Erikson, 2007). Of note, Plk3 is phosphorylated and activated after IR in a manner dependent on ATM (Bahassi et al., 2002). We, therefore, investigated whether Plk3 is involved in CtIP phosphorylation in G1. We observed that pRPA and BrdU foci after α-particle irradiation and the x ray–induced pRPA2 signal on a Western blot were reduced after treatment with Plk3 siRNA or a Plk1/3 small molecule inhibitor (Plk inhibitor [Plki]; Figs. 4 a and S3, a and b; Lansing et al., 2007), demonstrating that Plk activity is required for efficient resection in G1. As a control, we measured pRPA foci formation in G2 cells and observed the same foci level with and without Plk3 siRNA (Fig. S3 a). Additionally, Plk1 siRNA did not affect pRPA foci formation in G1 or G2 (Fig. S3 a). This suggests that the kinase requirement for resection is distinct in G1 (Plk3 dependent) versus G2 (Cdk1/2 dependent).
Figure 4.
Plk3 is required for resection in G1 and phosphorylates CtIP in vitro. (a) pRPA (left) and BrdU (middle) foci in G1-phase HeLa cells treated with Plk3 siRNA or Plki and irradiated with 2 Gy α particles. Knockdown efficiency for Plk3 was confirmed by Western blotting. Detection of pRPA2 at 4 h after IR or CPT (1 µM for 1 h) in whole cell extracts from confluency-arrested 82-6 fibroblasts with and without Plki (right). GAPDH was used as a loading control, and CyclinA was used as a marker for S/G2 cells. (b) Endogenous CtIP obtained by IP of CtIP from untransfected HeLa cells or GFP-CtIP obtained by IP of GFP from transfected HeLa cells was incubated in vitro with recombinant, constitutively active Plk3. Phosphorylation was measured by 32P incorporation. The bands representing pCtIP or GFP-pCtIP are indicated. (c) Insect cell purified CtIP-wt protein was incubated in vitro with Plk3 or Cdk2/CyclinA and analyzed with phosphospecific antibodies for CtIP-pT847 or CtIP-pS327. (d) Insect cell purified CtIP-wt and CtIP-S327A/T847A proteins were incubated in vitro with Plk3 and analyzed with an antibody against CtIP (±SEM from at least three experiments). siCtrl, control siRNA; siPlk3, Plk3 siRNA. ***, P < 0.001.
To provide evidence that Plk3 phosphorylates CtIP, we first performed in vitro kinase assays. We used HeLa cells either with or without GFP-CtIP expression, immunoprecipitated either endogenous CtIP or GFP-CtIP, and added recombinant, constitutively active Plk3 protein. Phosphorylation was measured by 32P incorporation, and phosphorylation signals were obtained at molecular weights corresponding to those of CtIP or GFP-CtIP, demonstrating phosphorylation of CtIP and not of a CtIP-binding partner (Fig. 4 b). Moreover, the CtIP phosphorylation signal was diminished for CtIP mutated at the Cdk sites S327 and T847 but not for CtIP mutated at the ATM-dependent phosphorylation sites S664 and S745 (Li et al., 2000), consistent with the notion that Plk3 phosphorylates [S/T]-P but not [S/T]-Q sites (Fig. S3 c). We also used the phosphospecific antibodies to investigate CtIP phosphorylation by Plk3 in vitro. For this, we used insect cell purified CtIP, incubated it with recombinant Plk3 protein, and obtained robust phosphorylation signals at CtIP-S327 and CtIP-T847 (Fig. 4 c). Next, we studied CtIP phosphorylation in a gel shift experiment. Insect cell purified CtIP-wt protein incubated with recombinant Plk3 showed a substantial level of phosphorylation, which was diminished but not absent in the CtIP protein mutated at S327 and T847 to unphosphorylatable alanine (Fig. 4 d). This shows that Plk3 phosphorylates CtIP in vitro at S327, T847, and likely also at other sites. Finally, we immunoprecipitated GFP-CtIP or S protein–CtIP from HEK293T cells and incubated it in vitro with Plk3. We observed CtIP-S327 and CtIP-T847 phosphorylation in the absence of Plk3, which increased after Plk3 incubation (Fig. S3 d). Analysis of these samples by mass spectrometry confirmed the presence of multiple phosphorylations in CtIP from undamaged cells and revealed additional Plk3 phosphorylation sites (Table 1).
Table 1.
Mass spectrometry reveals CtIP phosphorylation at S327 and T847 after incubation with Plk3
| Site | CtIP | CtIP + Plk3 |
| S163 | P | P |
| S171 | P | P |
| T190 | N | U |
| S197 | N | P |
| S231 | P | P |
| S233 | P | P |
| T245 | P | N |
| S298 | N | U |
| T299 | N | U |
| T302 | N | P |
| S305 | P | P |
| S309 | U | U |
| S311 | P | P |
| T312 | U | U |
| S313 | U | N |
| T315 | P | P |
| T323 | P | P |
| S326 | U | P |
| S327 | U | P |
| T333 | N | P |
| T344 | P | P |
| S345 | P | P |
| S347 | P | P |
| S349 | N | P |
| T361 | N | P |
| T367 | N | U |
| S377 | P | U |
| S379 | P | U |
| S382 | U | P |
| T386 | U | U |
| S389 | N | P |
| S413 | N | P |
| S415 | N | P |
| S439 | P | P |
| T450 | P | N |
| S454 | N | U |
| T520 | N | P |
| T527 | U | U |
| S539 | P | P |
| T544 | U | P |
| S549 | P | U |
| S555 | P | P |
| S568 | P | P |
| S593 | P | P |
| T596 | U | P |
| S627 | N | P |
| S649 | P | U |
| T671 | N | P |
| S679 | U | N |
| T687 | N | P |
| T693 | N | P |
| S713 | N | P |
| S714 | N | P |
| S723 | P | P |
| T731 | U | N |
| S743 | P | N |
| S745 | P | N |
| Y780 | N | P |
| T788 | N | U |
| S789 | N | U |
| T847 | N | P |
| T859 | N | U |
| Y867 | N | P |
| S889 | P | P |
We analyzed CtIP immunoprecipitated and purified from stably transfected HEK293T cells with or without in vitro incubation with recombinant Plk3. All observed phosphorylation sites are shown. Statistical analysis (Ascore calculation) was performed at the Taplin Mass Spectrometry Facility (Harvard Medical School; Beausoleil et al., 2006). The sites were classified as P = phosphorylation (Ascore ≥ 19), U = unlikely phosphorylation (Ascore 0–18), and N = no phosphorylation. CtIP phosphorylation (Ascore ≥ 19) at S327 and T847 was detected with, but not without, Plk3 incubation. 17 additional CtIP sites were phosphorylated with Plk3 but not phosphorylated without Plk3 incubation. Note the high basal CtIP phosphorylation level without Plk3 incubation.
Plk3 phosphorylates CtIP in vivo
We then investigated CtIP phosphorylation in vivo using the phosphospecific antibodies and G1-enriched HEK293T populations. For both antibodies, we observed a strong IR-induced increase in the pCtIP level, which was abolished in cells treated with Plk3 siRNA (Figs. 5 a and S4 a). These results were confirmed with confluent 82-6 fibroblasts (Fig. S4 b). Because Plk3 is phosphorylated and activated after IR in a manner dependent on ATM (Fig. S4 c; Bahassi et al., 2002), we investigated whether CtIP phosphorylation by Plk3 depends on ATM activity. Of note, confluent 82-6 fibroblasts treated with an ATM inhibitor (ATMi) did not show the IR-induced increase in CtIP phosphorylation (Fig. 5 b). The same was observed with G1-enriched HEK293T cells transfected with GFP-CtIP and treated with ATM siRNA (Fig. S4 d). Based on this observation, we predicted that ATM is needed for efficient resection and investigated BrdU foci formation after α-particle irradiation (because ATM is involved in RPA phosphorylation, the pRPA foci assay could not be applied to investigate the requirement of ATM for resection). Notably, cells treated with ATMi showed strongly diminished levels of BrdU foci compared with control cells (Fig. 5 c). Collectively, these data establish that CtIP phosphorylation at S327 and T847 represents a damage-inducible process dependent on Plk3.
Figure 5.
Plk3 phosphorylates CtIP in vivo. (a) HEK293T cells were treated with Plk3 and CtIP siRNAs, transfected with GFP-CtIP plasmids, enriched in G1 (≥90%), and irradiated with 5 Gy. GFP-CtIP was obtained by IP and analyzed for pS327 at 30 min after IR or pT847 at 2 h after IR by immunoblotting. (b) Confluent 82-6 fibroblasts were treated with ATMi and irradiated with 5 Gy. Endogenous CtIP was obtained by IP and analyzed for pS327 at 30 min after IR or pT847 at 2 h after IR by immunoblotting. (c) BrdU foci in ATMi-treated, G1-phase HeLa cells irradiated with 2 Gy α particles. (d) pRPA foci in GFP-positive (GFP+), G1-phase HeLa cells after 2 Gy α-particle irradiation. Cells were treated with CtIP and Plk3 siRNAs and transfected with GFP-CtIP plasmids. Knockdown efficiencies for CtIP and Plk3 as well as transfection efficiencies for GFP-CtIP were confirmed by Western blotting (±SEM from three experiments). endog., endogenous; siCtrl, control siRNA; siCtIP, CtIP siRNA; siPlk3, Plk3 siRNA. ***, P < 0.001.
Finally, we asked whether CtIP with phosphomimic substitutions at T847 and S327 can overcome the requirement for Plk3. We enumerated pRPA foci in G1-phase HeLa cells that were codepleted for Plk3 and CtIP and transfected with CtIP-wt or CtIP with phosphomimic substitutions at S327 and/or T847 (S327E, T847E, or S327E/T847E). Cells transfected with CtIP-wt, with CtIP-S327E, or with CtIP-T847E but not cells transfected with CtIP-S327E/T847E showed diminished pRPA foci formation (Figs. 5 d and S4 e), demonstrating that only cells with phosphomimic substitutions at both CtIP sites can overcome the requirement for Plk3. Thus, this result provides strong evidence that Plk3 functions during CtIP-dependent resection in G1 by phosphorylating CtIP at S327 and T847.
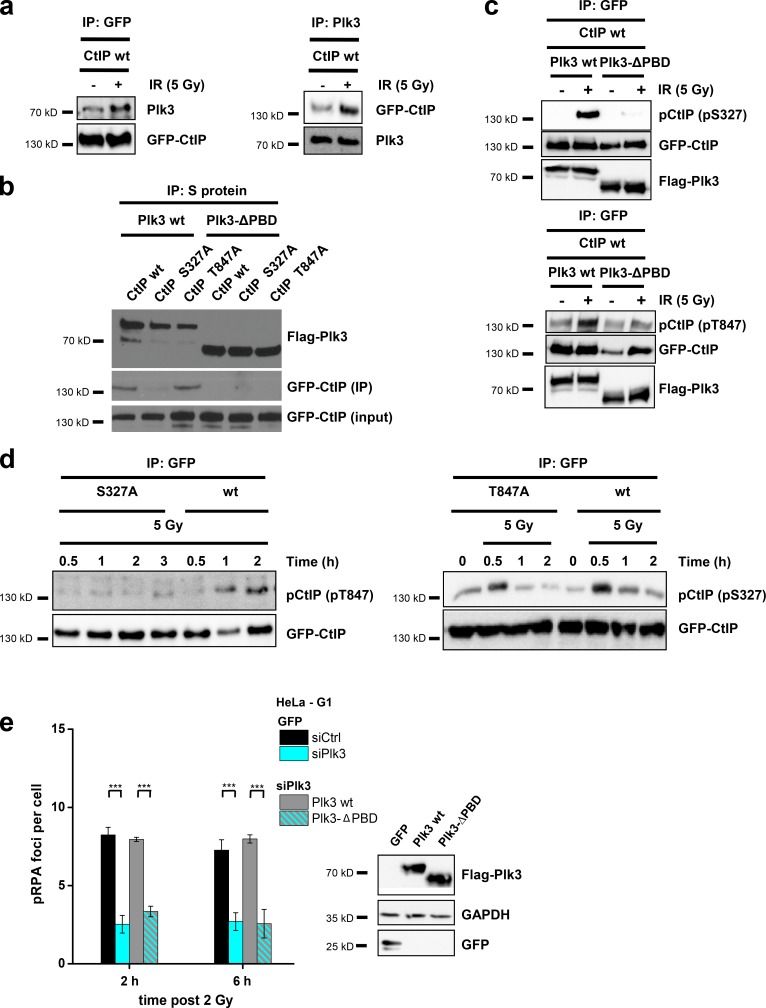
Plk3 interacts with CtIP to promote resection in G1
Plks have been reported to bind to their targets in a phosphorylation-dependent manner using a C-terminal PBD. This priming event activates Plks, facilitating the phosphorylation of Plk targets at neighboring sites (Elia et al., 2003). We performed coimmunoprecipitation (IP; co-IP) experiments and observed interaction of endogenous Plk3 with GFP-CtIP in transfected HEK293T cells, which was increased after irradiation (Fig. 6 a). We then transfected HEK293T cells with Flag-tagged Plk3 and GFP-tagged CtIP plasmids and performed co-IP experiments 30 min after IR. We detected robust interaction between Plk3 and CtIP-wt and between Plk3 and CtIP-T847A mutants but not between Plk3 and CtIP-S327A mutants (Fig. 6 b). Moreover, Plk3 with deleted PBDs (Plk3-ΔPBD) was unable to interact with any CtIP proteins (Fig. 6 b). The same result is obtained with Plk3 and CtIP precipitated from unirradiated cells (Fig. S5 a). These data demonstrate that Plk3 interacts via its PBDs with CtIP in a manner that is dependent on CtIP phosphorylation at S327. We then examined whether Plk3 binding to CtIP is required for CtIP phosphorylation at T847. First, we observed that cells with Plk3-ΔPBD do not show the IR-induced increase in CtIP-S327 and -T847 phosphorylation (Fig. 6 c). Second, cells transfected with CtIP-T847A show phosphorylation at S327, but CtIP-S327A mutants fail to show phosphorylation at T847 (Fig. 6 d). Finally, we investigated whether Plk3-ΔPBD can efficiently promote resection and observed that Plk3-wt but not Plk3-ΔPBD is able to restore pRPA foci levels in Plk3 siRNA–treated cells (Fig. 6 e). Collectively, this shows that Plk3 binds via its PBDs to CtIP phosphorylated at S327, which primes further CtIP phosphorylation at S327 and is necessary for subsequent CtIP phosphorylation at T847 and efficient resection.
Figure 6.
Plk3 interacts with CtIP to promote resection in G1. (a) Interaction of GFP-CtIP and Plk3 after 5 Gy IR. HEK293T cells were treated with CtIP siRNA and transfected with a GFP-CtIP plasmid. GFP-CtIP or Plk3 were obtained by IP, and the precipitates were analyzed by immunoblotting. (b) Interaction of Plk3-ΔPBD with CtIP-S327A and CtIP-T847A at 30 min after 5 Gy. HEK293T cells were treated with CtIP and Plk3 siRNAs and transfected with GFP-CtIP and SFB-Plk3 plasmids. Plk3 was obtained by IP, and the precipitates were analyzed by immunoblotting. (c) CtIP phosphorylation in a Plk3-ΔPBD mutant in vivo. HEK293T cells were treated with CtIP and Plk3 siRNAs, transfected with GFP-CtIP and SFB-Plk3 plasmids, and irradiated with 5 Gy. GFP-CtIP was obtained by IP and analyzed for pS327 at 30 min after IR or pT847 at 2 h after IR by immunoblotting. (d) HEK293T cells were treated with CtIP siRNA, transfected with GFP-CtIP plasmids, and irradiated with 5 Gy. GFP-CtIP was obtained by IP and analyzed for pS327 or pT847 by immunoblotting. (e) pRPA foci in Flag-positive (Flag+), G1-phase HeLa cells after 2 Gy α-particle irradiation. Cells were treated with Plk3 siRNA and transfected with SFB-Plk3 plasmids. Transfection efficiencies were confirmed by Western blotting (±SEM from three experiments). siCtrl, control siRNA; siPlk3, Plk3 siRNA. ***, P < 0.001.
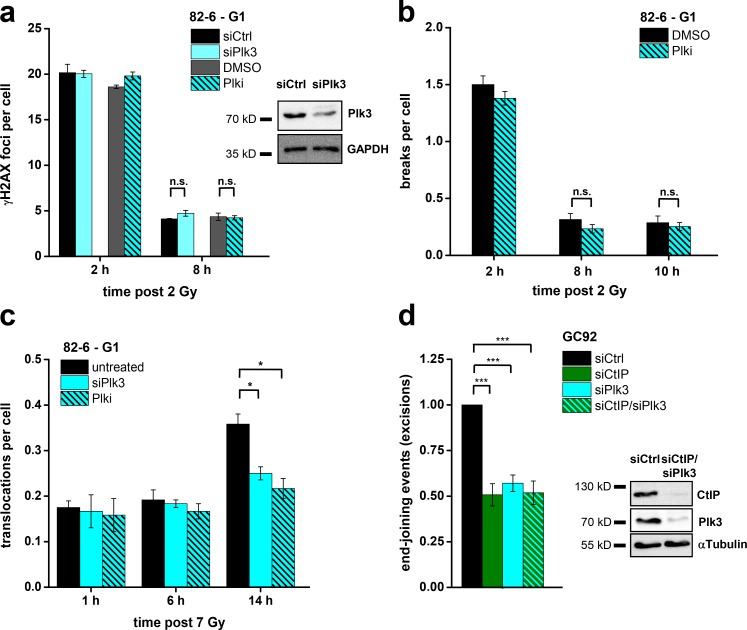
Plk3 enhances the formation of translocations and genomic rearrangements in G1
Having established that CtIP’s function in G1 is regulated by Plk3, we sought to investigate the impact of Plk3 depletion on the formation of chromosome translocations. Addition of Plki as well as Plk3 siRNA did not affect the level of unrepaired DSBs or chromosome breaks (Fig. 7, a and b), consistent with the notion that CtIP is not required for break repair in G1. However, when we analyzed the formation of chromosome translocations after Plki and Plk3 siRNA by PCC/FISH analysis as in Fig. 1 d, we observed diminished translocation levels at late but not at early times (Fig. 7 c). As a control, we analyzed G2-irradiated cells and did not observe an impact of Plki or Plk3 siRNA on translocation formation (Fig. S5 b). This result is again in full agreement with the impact of CtIP siRNA on translocation formation. Finally, we analyzed the formation of excisions in Plk3-depleted cells using the reporter system described in Fig. 3 d. We observed a reduction in excision events after Plk3 siRNA or Plki by ∼50% (Figs. 7 d and S5 c), similar to the reduction after CtIP siRNA. Dual depletion of Plk3 and CtIP had no further impact, confirming that both factors operate in the same error-prone repair pathway (Fig. 7 d). In contrast, Plki did not impact on gene conversion frequencies measured with an HR reporter assay, supporting the notion that Plk3 regulation of CtIP is specific for G1 (Fig. S5 c).
Figure 7.
Plk3 enhances the formation of translocations and genomic rearrangements in G1. (a) γ-H2AX foci in G1-phase 82-6 fibroblasts treated with Plk3 siRNA or Plki. Plk3 knockdown efficiency was confirmed by Western blotting. (b) Chromosome breaks in G1-phase 82-6 fibroblasts treated with Plki. (c) Translocations in G1-phase 82-6 fibroblasts treated with Plk3 siRNA or Plki. (d) Analysis of excision events in GC92 cells. Cells were treated with CtIP and/or Plk3 siRNA, and excisions were measured by the fraction of cells that exhibited a CD4-positive signal relative to all cells. Results were normalized to control siRNA-treated cells. The knockdown efficiencies for CtIP and Plk3 were confirmed by Western blotting (±SEM from at least three experiments). siCtrl, control siRNA; siCtIP, CtIP siRNA; siPlk3, Plk3 siRNA. *, P < 0.05; ***, P < 0.001.
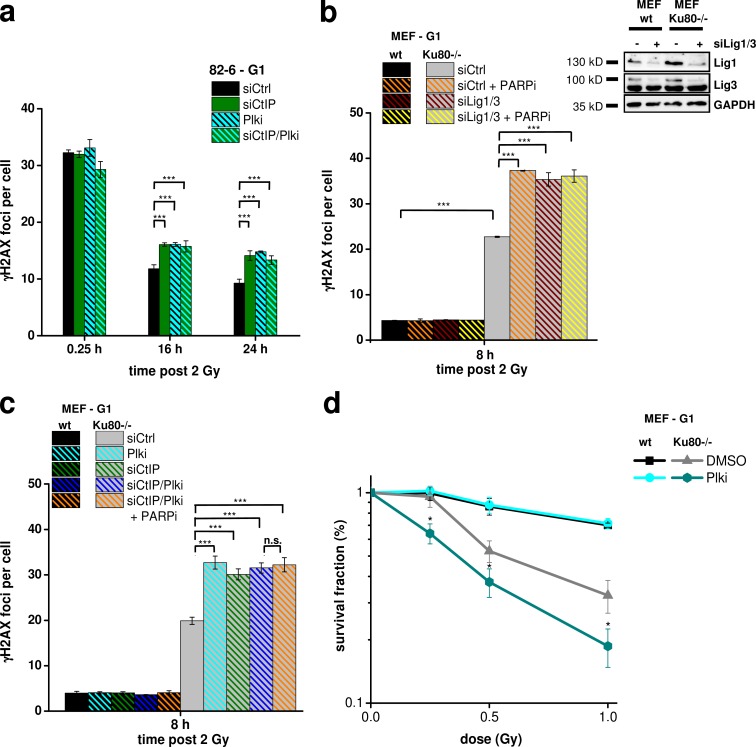
Plk3 is required for the repair of complex DSBs and DSBs in G1-phase Ku−/− MEFs
Although Plk3 and CtIP were not essential for the repair of x ray–induced DSBs in wt cells, we reasoned that they might be required under special conditions. Because resection in G1 could be readily detected at α particle–induced DSBs, we speculated that the repair of such complex breaks might require Plk3 and CtIP and enumerated γ-H2AX foci in G1 cells at various times after IR as described in Fig. 1 a. Of note, we observed similar initial foci levels but a substantial and epistatic repair defect after Plk3 and CtIP siRNA (Fig. 8 a). Thus, repair of complex DSBs with multiple lesions in close proximity requires Plk3/CtIP.
Figure 8.
Plk3 is required for the repair of complex DSBs and DSBs in G1-phase Ku−/− MEFs. (a) γ-H2AX foci in G1-phase 82-6 fibroblasts treated with CtIP siRNA and/or Plki and irradiated with 2 Gy α particles. (b) γ-H2AX foci in G1-phase wt and Ku80−/− MEFs treated with Lig1/3 siRNA and/or PARP inhibition (PARPi). Knockdown efficiencies were confirmed by Western blotting. (c) γ-H2AX foci in G1-phase wt and Ku80−/− MEFs treated with CtIP siRNA and/or Plki and PARP inhibition. (d) Survival of G1-phase wt and Ku80−/− MEFs after Plki. G1 cells were obtained by mitotic shake-off followed by 2-h incubation before IR (±SEM from at least three experiments; Jackman and O’Connor, 2001). siCtrl, control siRNA; siCtIP, CtIP siRNA; siLig1/3, Lig1/3 siRNA. *, P < 0.05; ***, P < 0.001.
Another condition when DSBs cannot easily be repaired without resection occurs in c-NHEJ mutants, which repair DSBs by alt-NHEJ. To investigate whether this process requires Plk3/CtIP, we used Ku−/− mouse embryonic fibroblasts (MEFs) and analyzed γ-H2AX foci in G1 cells that were identified as in Fig. 1 a. Consistent with the described role of poly (ADP-ribose) polymerase (PARP) in alt-NHEJ (Wang et al., 2006), chemical inhibition of PARP increased the residual level of DSBs in Ku80−/− MEFs but did not affect the repair capacity of wt MEFs (Fig. 8 b). To verify that PARP operates in alt-NHEJ, we codepleted Lig1 and 3, which are also implicated in alt-NHEJ (Wang et al., 2005). Dual depletion of Lig1/3 increased residual DSB levels in Ku80−/− MEFs similar to PARP inhibition but had no effect on wt cells (Fig. 8 b). PARP inhibition in Lig1/3-depleted cells conferred no greater impact, suggesting that PARP inhibition and Lig1/3 depletion impede the same alt-NHEJ pathway (Fig. 8 b). We then inhibited Plk3 and CtIP in Ku80−/− MEFs and observed elevated unrepaired DSB levels similar to PARP inhibition. Inhibition of Plk3, CtIP, and PARP together did not further increase the residual DSB level, suggesting that Plk3 and CtIP function during PARP-dependent alt-NHEJ process (Fig. 8 c). As a control, we analyzed γ-H2AX foci in G2-irradiated Ku80−/− MEFs and observed that CtIP siRNA but not Plki increased residual foci levels (Fig. S5 d). Finally, we investigated clonogenic cell survival in G1-irradiated wt and Ku80−/− MEFs. wt MEFs were not affected by Plki, but Ku mutants, which were already quite radiosensitive without Plki, were even more radiosensitive after Plki (Fig. 8 d). Thus, these cell survival data fully recapitulate the DSB repair data in G1. We conclude that Plk3 and CtIP represent essential factors for the process of alt-NHEJ in G1.
Discussion
Mounting evidence suggests that CtIP is active in G1 and supports various important cellular processes. However, how CtIP is regulated during G1 phase is completely unknown. Here, we show that Plk3 phosphorylates CtIP in G1 in a damage-inducible manner to promote error-prone DSB repair. Hence, Plk3 represents a novel DSB response factor. We show that Plk3 impacts on various cellular processes. First, we show that Plk3 enhances the formation of IR-induced translocations in G1-phase wt cells similar to CtIP; second, it promotes error-prone excision events from two restriction enzyme-induced DSBs in wt cells in a manner epistatic with CtIP; third, it is required for activating CtIP to support resection and repair of complex DSBs in G1; and finally, it is required for rejoining IR-induced DSBs in the absence of Ku by an alt-NHEJ process. In the following paragraphs, we discuss the significance of the observation that Plk3 mediates the activation of CtIP during these processes in a damage-induced manner.
Long-standing questions in cancer research concern the mechanisms underlying chromosome translocation formation (Nussenzweig and Nussenzweig, 2010). Translocations have been proposed to arise via NHEJ (Bunting and Nussenzweig, 2013). However, c-NHEJ mutants show enhanced instead of reduced chromosomal translocations, suggesting that, in the absence of c-NHEJ factors, translocations are formed by alt-NHEJ processes (Weinstock et al., 2007; Boboila et al., 2010). A recent study has provided evidence that a CtIP-dependent process exposes microhomologies and causes translocations from restriction enzyme-induced DSBs (Zhang and Jasin, 2011). Here, we have focused our analysis on G1-phase cells because most cells in a human body are in G0/G1 phase, and we have used x rays to induce DSBs because radiation exposure, even at low doses (e.g., after computer tomography scans), can cause cancer (Pearce et al., 2012). We show that a substantial fraction of IR-induced translocations in G1 arise from a Plk3- and CtIP-dependent process. Moreover, also, genomic rearrangements arising from the misrejoining of two restriction enzyme–induced DSBs are promoted by Plk3 and CtIP. Thus, Plk3 represents a novel DSB response factor, which is involved in error-prone DSB repair processes underlying the formation of chromosome translocations and other genomic rearrangements.
Translocation formation ≤6 h after IR in G1 occurs in a manner independent of CtIP, with most translocations in this component arising during the first 2 h. This suggests that translocations can form very quickly after damage induction, an observation consistent with previous findings (Darroudi et al., 1998). However, we observed a second component of translocation formation at >6 h, which is dependent on Plk3 and CtIP. The occurrence of Plk3/CtIP-dependent translocations with slow kinetics is consistent with the delayed phosphorylation and activation of CtIP. Conceptually, this delay provides a sensible mechanism to initiate CtIP-dependent processing only if the cells fail to repair the DSBs by a CtIP-independent process. This is the case in Ku-deficient cells, in which repair ensues by alt-NHEJ, but also occurs in wt cells at complex DSBs. Thus, the model emerges that cells first try to repair DSBs without CtIP function and, only if this fails, activate CtIP by Plk3. We would like to point out in this context that we did not investigate the issue of pathway usage for DSBs that are resected in G1 wt cells by Plk3/CtIP. Although such breaks clearly undergo repair by alt-NHEJ in Ku-deficient cells, c-NHEJ as well as alt-NHEJ might be capable of repairing resected DSBs in wt cells. Because Plk3 is required for G1–S transition in unperturbed cells (Zimmerman and Erikson, 2007) and is itself activated by IR (Bahassi et al., 2002), this kinase seems to be perfectly suited to activate this process, which might be regarded as the last resort to complete DSB repair before S-phase entry commences. This last resort, however, comes along with an increased likelihood for generating translocations. Of note, conceptually similar last resort mechanisms have been proposed for cells progressing though mitosis, where resolvases are activated late to process joint DNA molecules in mitosis (Lukas et al., 2011a; Naim et al., 2013; Ying et al., 2013).
CtIP function during G1 requires CtIP phosphorylation on T847 and S327, similar to CtIP’s function during HR in G2. Our analysis of the time course of CtIP phosphorylation has revealed that S327 phosphorylation peaks sharply at 30 min and that T847 phosphorylation peaks sharply at 120 min after IR in G1. We demonstrate that both sites need to be phosphorylated for CtIP function in G1 and reveal the defined time course of events. In S/G2 phase, Cdks phosphorylate these sites, and it’s possible that the time course is different to that in G1 phase, which may reflect the distinct outcomes in terms of repair. In G2, HR requires avid resection; in G1, resection is more limited, and repair occurs by end-joining processes that require little resection (Chapman et al., 2013; Di Virgilio et al., 2013; Escribano-Díaz et al., 2013; Feng et al., 2013; Zimmermann et al., 2013). Consistent with the more limited resection in G1, we failed to detect pRPA foci after x-irradiation. However, resection in G1 can be monitored after higher x-ray dose or by inducing complex DSBs, which have a higher propensity to undergo resection than x ray–induced DSBs (Shibata et al., 2011).
We have studied the mechanism of CtIP phosphorylation by Plk3 and investigated the temporal interplay of CtIP phosphorylation at S327 and T847. Consistent with the literature (Bahassi et al., 2002), we observed that Plk3 is phosphorylated after IR in an ATM-dependent manner. Furthermore, Plk3 binds to CtIP phosphorylated at S327 via its PBD domains, which is necessary for robust CtIP phosphorylation at S327 and subsequent CtIP phosphorylation at T847. The initial priming event (phosphorylation) before Plk3 binds at CtIP-pS327 via its PBD domains is either performed by Plk3 itself (a self-priming mechanism) or by a hitherto unknown kinase (a non–self-priming mechanism); both modes of action have been described for Plk1 and other factors regulating the cell cycle and damage response (Reinhardt and Yaffe 2013; Lee et al., 2014). In the latter case, it is possible that Cdk4/6 mediates the initial CtIP phosphorylation in G1 to prime the Plk3 response. Although we favor the notion that the Plk3-mediated response is G1 specific, we cannot eliminate the possibility that it might be activated also in S/G2 and thus represents a more general stress-induced Plk3 response. Of note, the amplification process at CtIP-S327 after Plk3 binding to CtIP-pS327 might involve the transphosphorylation of neighboring CtIP molecules by Plk3. Such a mechanism would provide a critical mass means for amplification of CtIP phosphorylation at a DSB site (Fig. S5 e, model of CtIP phosphorylation by Plk3). Moreover, because a phosphomimic CtIP-S327E/T847E double mutant but not a CtIP-T847E single mutant is able to rescue the resection defect of Plk3-deficient cells, CtIP-S327 phosphorylation not only serves to promote T847 phosphorylation but likely has another role in activating CtIP.
Our finding that CtIP phosphorylation on S327 and T847 promotes an error-prone DSB repair process in G1 might explain the observation that mice with unphosphorylatable CtIP-S327A do not display elevated cancer rates (Reczek et al., 2013), although it was previously shown that CtIP phosphorylation at this site is required for efficient HR (Yu et al., 2006; Yun and Hiom, 2009). Our finding that the kinase Plk3 regulates CtIP during this error-prone end-joining process in G1 might be clinically important because some tumors exhibit increased Plk3 levels (Weichert et al., 2004, 2005). Thus, Plk3 inhibition might have a beneficial effect during tumor treatment by lowering the capacity of a tumor cell to develop genomic instability. Moreover, Plk3 inhibition can sensitize tumors that rely on CtIP-dependent DSB repair in G1, such as tumors with defects in c-NHEJ factors.
Materials and methods
Cell lines and cell culture
Immortalized and transformed cell lines were 82-6 hTert (wt), HeLa, HeLa pGC, GC92, HEK293T, and wt and Ku80−/− MEFs. HeLa, HeLa pGC, GC92, HEK293T, and MEFs were cultured in DMEM with 10% FCS and 1% NEAA (plus 0.3 µg/ml puromycin for HeLa pGC or 2 mM l-glutamine for HEK293T cells), and 82-6 cells were cultured in MEM with 20% FCS and 1% NEAA. All cells were maintained at 37°C in a 5% CO2 incubator. HeLa pGC cells contained an HR substrate carrying an inactive GFP gene with an I-SceI recognition site and a truncated GFP gene that was positioned in the same orientation on the other side of a puromycin selection cassette (Mansour et al., 2008). The GC92 cell line was derived from SV40-transformed GM639 human fibroblasts and contained the NHEJ substrate described in Fig. S2 d (Rass et al., 2009). Ku80−/− MEFs (provided by G. Li, Memorial Sloan Kettering Cancer Center, New York, NY) were obtained from Ku80−/− mice.
RNA interference and plasmid transfection
In all transfection experiments, the endogenous protein was depleted by siRNA, and siRNA-resistant plasmids were used. siRNA treatment of HeLa, 82-6, and MEFs was performed using HiPerFect Transfection Reagent (QIAGEN) following the manufacturer’s instructions. Experiments were performed 48 h after siRNA treatment. 8 h after incubation with siRNA, HeLa cells were transfected with various GFP-tagged siRNA-resistant CtIP plasmids (Quennet et al., 2011). HeLa pGC and GC92 cells were treated with siRNA using HiPerFect Transfection Reagent and, 48 h later, transfected with the pBL464-pCBASce plasmid (I-SceI endonuclease) using jetPEI transfection reagent (Polyplus Transfection), and again treated with siRNA after another 6 h. GC92 cells were additionally transfected 24 h after the first siRNA treatment with various RFP-tagged siRNA-resistant CtIP plasmids using jetPEI transfection reagent. Transfection of HEK293T cells with various siRNA-resistant GFP-tagged CtIP plasmids and SFB-tagged Plk3 plasmids was performed using MATra (IBA) following the manufacturer’s instruction. This transfection protocol provided >90% G1 cells (Fig. S2 b). Human full-length Plk3 and a Plk3 deletion mutant (Plk3-ΔPBD; aa 1–480) were transferred into an N-terminal triple epitope tag destination vector (SFB [S protein tag, Flag epitope tag, and streptavidin-binding peptide tag]) using Gateway LR Clonase Enzyme mix kit (Life Technologies) following the manufacturer’s instruction. The siRNA-resistant CtIP plasmids were obtained by subcloning the human CtIP cDNA into a pEGFP-C1 or ptagRFP-C1 vector (Takara Bio Inc.) and changing three nucleotides in the CtIP_2 siRNA targeting region (C133T, A135G, and A138G). The plasmids expressing the phosphorylation mutants were generated by site-directed mutagenesis.
siRNA
siRNA sequences used were human ATM (5′-CACCGTTTTGTTAGTTTATTA-3′), human Cdk2 (SI02654631), human CtIP_1 (5′-TCCACAACATAATCCTAATAA-3′; used for chromosomal experiments), human CtIP_2 (5′-AAGCTAAAACAGGAACGAATC-3′; used for all other experiments), human Plk1_2 (SI00071624), human Plk1_6 (SI02223837), human Plk3 (5′-CTGCATCAAGCAGGTTCACTA-3′), human Plk3_1 (SI00059388), human Plk3_11 (SI05056450), control (5′-AATTCTCCGAACGTGTCACGT-3′), mouse CtIP (5′-AAACAGATACTTACAAATAAA-3′), mouse Lig1 (5′-CAGGAAAGTATCCTGACATTA-3′), and mouse Lig3 (5′-GCACAAAGATTGTCTGCTA-3′).
NHEJ and HR reporter assays
GC92 cells containing an NHEJ substrate (Rass et al., 2009) and HeLa pGC cells containing a HR substrate (Mansour et al., 2008) were fixed and stained 72 h after transfection with I-SceI. 15,000 cells per sample were analyzed with a microscope (Axiovert 200M; Carl Zeiss) and Metafer software (MetaSystems).
Irradiation and chemical treatment
If not mentioned otherwise, irradiation was performed with x rays at 90 kV and 19 mA. When stated, α-particle irradiation was performed with a 241Am source (Kühne et al., 2000). Chemical inhibitors were added 1 h before IR or 6 h after transfection with I-SceI and maintained during repair incubation. The ATMi Ku60019 (Tocris Bioscience), Cdk1/2 inhibitor roscovitine (Sigma-Aldrich), the Plki GW 843682X (half-maximal inhibitory concentration values of 2.2 and 9.1 nM for Plk1 and Plk3, respectively; Tocris Bioscience), and the PARP inhibitor PJ34 (EMD Millipore) were used at concentrations of 0.5, 25, 0.5, and 15 µM, respectively. Cells were treated with 1 µM CPT (Sigma-Aldrich) for 1 h.
Chromosomal analysis
For translocation and chromosome break analysis of G1 cells, exponentially growing 82-6 fibroblasts were irradiated and treated with 100 ng/ml nocodazole to prevent G2-irradiated cells from progressing into G1 during repair incubation. At the end of repair incubation, cells were mixed at a ratio of 1:1 with mitotic HeLa cells (enriched by treatment with colcemid for 20 h). After centrifugation, cell fusion was mediated by adding 0.25 ml polyethylene glycol 1500 (Roche) for 1 min to the cell pellet (Mosesso et al., 1999). For translocation measurements in G2-irradiated cells, exponentially growing 82-6 fibroblasts were irradiated, incubated in the presence of EdU for 14 h, and harvested by adding 50 ng/ml calyculin A for 30 min. Only EdU-negative G2 PCC spreads with two-chromatid morphology were evaluated. G1 and G2 PCC spreads were prepared by hypotonic treatment with 0.075 mM KCl and fixation using 3:1 methanol/acetic acid. FISH experiments using whole chromosome probes 1, 2, and 4 were performed following the manufacturer’s protocol (MetaSystems). Slides were processed using a microscope (Axioplan 2; Carl Zeiss), an EC Plan Neofluar (63×) with a numerical aperture of 1.25 (Carl Zeiss), and Metafer software. Translocations as well as chromosome breaks were scored in the stained chromosomes 1, 2, and 4. Color junctions between two stained chromosomes or between a stained and an unstained chromosome represent translocations. An additional fragment that is not connected to another stained or unstained chromosome is counted as a chromosome break. At least 200 PCC spreads were analyzed per data point.
Protein extracts, IP, and immunoblotting
For preparation of whole cell extracts, cells were resuspended in radioimmunoprecipitation assay buffer containing 50 mM Tris/HCl, pH 8, 150 mM NaCl, 0.5% natriumdesoxycholat, 1% Triton X-100, 0.1% SDS, freshly added protease inhibitor cocktail (1:25), and PhosSTOP (1:10) and sonicated three times for 1 min (Quennet et al., 2011). For chromatin fractionation, cells were resuspended in Chelsky buffer (10 mM Tris/HCl, pH 7.5, 10 mM NaCl, 3 mM MgCl2, and 30 mM sucrose) containing 0.5% NP-40 and centrifuged for 10 min at 400 g. The supernatant represented the cytosolic fraction. The pellet was washed once in Chelsky buffer containing 0.5% NP-40 and twice in Chelsky buffer containing 10 mM CaCl2 (10 min, 1500× g). Cell pellet was resuspended in 20 mM Tris/HCl, pH 7.9, 100 mM KCl, 0.2 mM EDTA, and 20% glycerol containing protease inhibitor and incubated 10 min on ice. After centrifugation (10 min at 1,500 g), chromatin fraction was lysed in radioimmunoprecipitation assay buffer. For IP, antibodies (2 µg) were linked to Dynabeads Protein G (Invitrogen), washed three times in 0.1% BSA/PBS, and then incubated with the cell extract at 4°C overnight. The membrane was blocked for 1 h in 5% low fat milk or 5% BSA in TBS/0.1% Tween 20. Immunoblotting was performed in TBS/0.1% Tween 20/1% low fat milk or 5% BSA overnight at 4°C or for 2 h at room temperature followed by HRP-conjugated secondary antibody incubation in PBS/0.1% Tween 20/1% low fat milk or 5% BSA for 1 h. Immunoblots were developed using ECL (Roche). Signal detection was performed with an image acquisition system (Chemi-Smart; Vilber Lourmat). For detection of pCtIP (T847 or S327), HEK293T cells transfected with GFP-CtIP plasmids were immunoprecipitated with mouse α-GFP (Santa Cruz Biotechnology, Inc.). Primary antibodies were rabbit α-ATM at 1:500 (Abcam), rabbit α–cleaved Caspase7 at 1:1,000 (Cell Signaling Technology), mouse α-Cdk2 at 1:1,000 (Santa Cruz Biotechnology, Inc.), mouse α-CtIP at 1:1,000 (Santa Cruz Biotechnology, Inc.), rabbit α-pCtIP (T847) at 1:200, rabbit α-pCtIP (S327) at 1:1,000, rabbit α-CyclinA at 1:1,000 (Santa Cruz Biotechnology, Inc.), mouse α-Flag (OctA) at 1:1,000 (Santa Cruz Biotechnology, Inc.), rabbit α-GAPDH at 1:1,000 (Santa Cruz Biotechnology, Inc.), rabbit α-GFP at 1:1,000 (Santa Cruz Biotechnology, Inc.), mouse α–γ-H2AX at 1:1,000 (EMD Millipore), mouse α-H3 at 1:2,000 (Abcam), rabbit α-Plk3 at 1:1,000 (Santa Cruz Biotechnology, Inc.), rabbit α-Rad51 at 1:2,000 (Abcam), mouse α-RPA2 at 1:1,000 (EMD Millipore), rabbit α-pRPA2 (S4/8) at 1:10,000 (Bethyl Laboratories, Inc.), and mouse α-tubulin at 1:2,000 (Santa Cruz Biotechnology, Inc.).
Immunofluorescence
Cells were grown on glass coverslips. 1 µM EdU was added before irradiation to label S-phase cells, and 100 ng/µl nocodazole was added immediately after IR to prevent G2-phase cells progressing into G1 during repair incubation (Löbrich et al., 2010). For α-particle irradiation, cells were grown on Mylar foil. Cells were fixed 15 min in PBS/2.5% formaldehyde, permeabilized 10 min in PBS/1% FCS/0.5% Triton X-100, and blocked 30 min in PBS/1% FCS/5% BSA. Samples were incubated with primary antibodies overnight at 4°C, washed three times in PBS/1% FCS, and incubated for 1 h at room temperature with Alexa Fluor 488– or Alexa Fluor 594–conjugated secondary antibodies (1:1,000; Invitrogen). After three washes in PBS, cells were DAPI (Sigma-Aldrich) stained and mounted using Vectashield mounting medium (Vector Laboratories; Quennet et al., 2011). For BrdU foci analysis, cells were preextracted 10 min with PBS/0.5% Triton X-100, fixed 20 min in PBS/2.5% formaldehyde, permeabilized 20 min in PBS/1% FCS/0.5% Triton X-100, and blocked 20 min in PBS/1% FCS/5% BSA. Antibodies were mouse α-BrdU at 1:200 (BD), mouse α–γ-H2AX at 1:2,000, rabbit α–γ-H2AX at 1:2,000 (Abcam), mouse α-GFP at 1:200 (Roche), rabbit α-tagRFP at 1:2,000 (Evrogen), rabbit α-pRPA (pT21) at 1:15,000 (Abcam), and rat α-mouse CD4-FITC at 1:100 (BioLegend). To stain EdU, the EdU staining kit (Invitrogen) was used following the manufacturer’s instructions. Foci counting was performed on a microscope (Axio Imager.M1; Carl Zeiss), and imaging was performed on an inverted microscope (Axiovert 200M; Carl Zeiss). Objectives used were an EC Plan Neofluar (100×) with a numerical aperture of 1.3 or a Plan Apochromat (20×) with a numerical aperture of 0.8 (Carl Zeiss). Imaging was performed at room temperature using a camera (AxioCam MRm; Carl Zeiss) and AxioVision acquisition software (Carl Zeiss). Images were processed using ImageJ (National Institutes of Health). Samples were evaluated blindly.
Human CtIP expression
An expression construct for CtIP-wt was generated by PCR from a pBSK-CtIP construct (gift from W.-H. Lee, University of California, Irvine School of Medicine, Irvine, CA) to generate an N-terminal, Flag-tagged wt allele in pFastBac1 (Invitrogen). S327A and T847A mutants were generated by mutagenesis (QuikChange; Agilent Technologies). Transfer vectors were converted into corresponding bacmids and were used to make the virus according to the manufacturer’s instructions for the baculovirus expression system (Bac-to-Bac; Invitrogen). The wt and mutant CtIP proteins were expressed in Sf21 insect cells.
CtIP protein purification
Cells were lysed in buffer A (20 mM Tris, pH 8.0, 100 mM NaCl, 10% glycerol, and 1 mM DTT) supplemented with 0.5% Triton X-100, 2.5 mM Na4P2O7, 1 mM glycerol-β-phosphate, and 5 mM PMSF. Lysate was sonicated on ice until homogenous, and insoluble material was removed by centrifugation (100,000 g) for 1 h at 4°C. Supernatant was bound to 2 ml anti-Flag M2 agarose resin slurry (Sigma-Aldrich) at 4°C for 1 h, and then, resin was washed with 20 ml buffer A followed by 10 ml of 0.5-M LiCl and 20 ml buffer A. The protein was eluted with 10 ml buffer A containing Flag peptide (Sigma-Aldrich) at a concentration of 100 µg/ml. Eluted protein was loaded onto a 1 ml HiTrap SP column (GE Healthcare), washed with 20 ml buffer A, and eluted with buffer A containing 0.6 M NaCl. The peak fractions were dialyzed twice against 300 ml of fresh buffer A for 1 h at 4°C and flash frozen in liquid nitrogen. Protein concentration was determined by Coomassie staining of the proteins separated by SDS-PAGE and compared with known standards.
Anti-pS327 and anti-pT847 antibody preparation
Phosphospecific antibodies were produced in rabbits against CtIP-pS327 and CtIP-pT847 (custom antibody service from PhosphoSolutions). The antigens were synthetic phosphopeptides corresponding to amino acids surrounding the phosphorylated S327 or T847 in the human CtIP sequence. To remove the cross-reacting antibody from the affinity-purified pT847 rabbit polyclonal antibody solution, the anti-pT847 antibody was incubated with equal amounts of Flag-tagged CtIP (T847A) protein overnight at 4°C in the presence of anti-Flag magnetic beads. Magnetic beads were separated from the supernatant, which was diluted to 10× the initial volume, and glycerol concentration was adjusted to 30%. The resulting solution of purified antibody was stored at −20°C.
Plk3 in vitro kinase assays
HeLa cells were transiently transfected with pEGFP-C1 and pEGFP-CtIP constructs, and proteins were obtained by IP against GFP (mouse α-GFP; Santa Cruz Biotechnology, Inc.). Endogenous CtIP was obtained by IP against CtIP (mouse α-CtIP). 0.32 µg Plk3 (Life Technologies) was diluted in 20 µl kinase buffer (25 mM Tris, pH 7.5, 10 mM MgCl2, 0.5 mM EGTA, 0.5 mM Na3VO4, 2.5 mM DTT, 0.01% Triton X-100, and 200 µM ATP) and incubated for 15 min at 30°C. The kinase buffer containing Plk3 was added to the immunoprecipitated CtIP, GFP, or GFP-CtIP. The 32P kinase assay was performed in the presence of 5 µCi [32P]ATP for 30 min at 30°C, and gels were analyzed by audiography after SDS-PAGE. The kinase assay using phosphospecific antibodies was performed in the presence of 400 µM ATP for 30 min at 30°C.
Cdk in vitro kinase assay
200 ng wt, S327A, or T847A recombinant CtIP protein was phosphorylated using Cdk2/CyclinA (P6025S; New England Biolabs, Inc.) in the presence of 1 mM ATP at 30°C for 1 h. Reactions were analyzed by SDS-PAGE and by immunoblotting using 1 µg/ml anti-pT847 antibody.
Mass spectrometry
HEK293T cells stably expressing SFB-tagged CtIP were selected by culturing in medium with 2 µg/ml puromycin. For affinity purification, HEK293T cells were lysed in NETN buffer (100 mM NaCl, 20 mM Tris-Cl, pH 8.0, 0.5 mM EDTA, and 0.5% [vol/vol] Nonidet P-40) with protease and phosphatase inhibitors. Lysates were incubated with S protein beads (EMD Millipore) for 2 h at 4°C. The beads were washed four times with NETN buffer, and the bound proteins were subjected to SDS-PAGE. Protein bands were excised and analyzed by mass spectrometry at the Taplin Mass Spectrometry Facility, Harvard Medical School (Beausoleil et al., 2006).
Colony formation assay
G1-phase MEFs were obtained by mitotic shake-off. For this, the cell culture flasks were softly knocked to collect mitotic cells, which were then reseeded in new dishes and irradiated after 2 h when they were in G1 (Jackman and O’Connor, 2001). 30 min before IR, cells were treated with Plki. 9 h after IR, the medium containing Plki was replaced by fresh medium without Plki. After 7 d, colonies were fixed and stained with 0.1% crystal violet in 25% ethanol. Colonies containing ≥30 cells were counted.
Statistical analysis
All data shown represent the mean value of at least three independent experiments. Background foci (<0.5 per cell), chromosome breaks (<0.1 per cell), and translocations (<0.01 per cell) were subtracted. The error bars show the SEM between the experiments. P-values were obtained by t test and represent a comparison of all cells analyzed in the indicated cell populations (for all foci and chromosomal experiments) or a comparison of the data mean (for data obtained with the NHEJ and HR reporter assay, the colony formation assay, and the 32P in vitro phosphorylation assay).
Online supplemental material
Fig. S1 (for Figs. 1 and 2) demonstrates the efficiency of the CtIP knockdown approach, shows that CtIP is not required for G1-phase DSB and chromosome break repair, and presents evidence for RPA phosphorylation in G0 and G1 cells using Western blotting and for pRPA foci formation in G1 cells using immunofluorescence analysis. Fig. S2 (for Fig. 3) shows that pRPA foci formation in G1 requires CtIP phosphorylation at S327 and T847, confirms that CtIP is phosphorylated in vivo at S327 and T847, and reveals that genomic rearrangements require CtIP but not Cdk2. Fig. S3 (for Fig. 4) shows that Plk3 is required for resection in G1 but not in G2, whereas roscovitine treatment affects resection in G2 but not in G1 and confirms that Plk3 phosphorylates CtIP in vitro at S327 and T847. Fig. S4 (for Fig. 5) shows that Plk3 phosphorylates CtIP at S327 and T847 in vivo, reveals that these phosphorylation events require ATM, and confirms that pRPA foci formation in G1 requires CtIP phosphorylation at S327 and T847 by Plk3. Fig. S5 (for Figs. 6, 7, and 8) confirms that Plk3 interacts with CtIP phosphorylated at S327 in a manner dependent on its PBD domains, demonstrates that genomic rearrangements but not gene conversion events require Plk3, and shows that Plk3 but not CtIP is dispensable for alt-NHEJ in G2-phase Ku80−/− MEFs. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201401146/DC1.
Supplementary Material
Acknowledgments
We thank Marco Durante for advice with the PCC approach.
Work in the M. Löbrich laboratory is supported by the Deutsche Forschungsgemeinschaft (Lo 677/4-3 and GRK1657), the Bundesministerium für Bildung und Forschung (02NUK001C), and the Bundesministerium für Umwelt, Energie und Reaktorsicherheit (3610S30015).
The authors declare no competing financial interests.
Footnotes
Abbreviations used in this paper:
- ATMi
- ATM inhibitor
- c-NHEJ
- canonical NHEJ
- CPT
- camptothecin
- CtIP
- C-terminal binding protein–interacting protein
- DSB
- double-strand break
- EdU
- 5-ethynyl-2′-deoxyuridine
- HR
- homologous recombination
- IP
- immunoprecipitation
- IR
- ionizing radiation
- MATra
- magnet-assisted transfection
- MEF
- mouse embryonic fibroblast
- NHEJ
- nonhomologous end joining
- PARP
- poly (ADP-ribose) polymerase
- PBD
- Polo box domain
- PCC
- premature chromosome condensation
- Plk
- Polo-like kinase
- Plki
- Plk inhibitor
- wt
- wild type
References
- Anantha, R.W., Vassin V.M., and Borowiec J.A.. 2007. Sequential and synergistic modification of human RPA stimulates chromosomal DNA repair. J. Biol. Chem. 282:35910–35923 10.1074/jbc.M704645200 [DOI] [PubMed] [Google Scholar]
- Anger, M., Kues W.A., Klima J., Mielenz M., Kubelka M., Motlik J., Esner M., Dvorak P., Carnwath J.W., and Niemann H.. 2003. Cell cycle dependent expression of Plk1 in synchronized porcine fetal fibroblasts. Mol. Reprod. Dev. 65:245–253 10.1002/mrd.10289 [DOI] [PubMed] [Google Scholar]
- Bahassi, M., Conn C.W., Myer D.L., Hennigan R.F., McGowan C.H., Sanchez Y., and Stambrook P.J.. 2002. Mammalian Polo-like kinase 3 (Plk3) is a multifunctional protein involved in stress response pathways. Oncogene. 21:6633–6640 10.1038/sj.onc.1205850 [DOI] [PubMed] [Google Scholar]
- Beausoleil, S.A., Villén J., Gerber S.A., Rush J., and Gygi S.P.. 2006. A probability-based approach for high-throughput protein phosphorylation analysis and site localization. Nat. Biotechnol. 24:1285–1292 10.1038/nbt1240 [DOI] [PubMed] [Google Scholar]
- Beucher, A., Birraux J., Tchouandong L., Barton O., Shibata A., Conrad S., Goodarzi A.A., Krempler A., Jeggo P.A., and Löbrich M.. 2009. ATM and Artemis promote homologous recombination of radiation-induced DNA double-strand breaks in G2. EMBO J. 28:3413–3427 10.1038/emboj.2009.276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boboila, C., Jankovic M., Yan C.T., Wang J.H., Wesemann D.R., Zhang T., Fazeli A., Feldman L., Nussenzweig A., Nussenzweig M., and Alt F.W.. 2010. Alternative end-joining catalyzes robust IgH locus deletions and translocations in the combined absence of ligase 4 and Ku70. Proc. Natl. Acad. Sci. USA. 107:3034–3039 10.1073/pnas.0915067107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunting, S.F., and Nussenzweig A.. 2013. End-joining, translocations and cancer. Nat. Rev. Cancer. 13:443–454 10.1038/nrc3537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman, J.R., Taylor M.R., and Boulton S.J.. 2012. Playing the end game: DNA double-strand break repair pathway choice. Mol. Cell. 47:497–510 10.1016/j.molcel.2012.07.029 [DOI] [PubMed] [Google Scholar]
- Chapman, J.R., Barral P., Vannier J.B., Borel V., Steger M., Tomas-Loba A., Sartori A.A., Adams I.R., Batista F.D., and Boulton S.J.. 2013. RIF1 is essential for 53BP1-dependent nonhomologous end joining and suppression of DNA double-strand break resection. Mol. Cell. 49:858–871 10.1016/j.molcel.2013.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, F., Nastasi A., Shen Z., Brenneman M., Crissman H., and Chen D.J.. 1997. Cell cycle-dependent protein expression of mammalian homologs of yeast DNA double-strand break repair genes Rad51 and Rad52. Mutat. Res. 384:205–211 10.1016/S0921-8777(97)00020-7 [DOI] [PubMed] [Google Scholar]
- Darroudi, F., Fomina J., Meijers M., and Natarajan A.T.. 1998. Kinetics of the formation of chromosome aberrations in X-irradiated human lymphocytes, using PCC and FISH. Mutat. Res. 404:55–65 10.1016/S0027-5107(98)00095-5 [DOI] [PubMed] [Google Scholar]
- Davis, A.J., and Chen D.J.. 2013. DNA double strand break repair via non-homologous end-joining. Transl. Cancer Res. 2:130–143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Cárcer, G., Manning G., and Malumbres M.. 2011. From Plk1 to Plk5: functional evolution of polo-like kinases. Cell Cycle. 10:2255–2262 10.4161/cc.10.14.16494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deckbar, D., Stiff T., Koch B., Reis C., Löbrich M., and Jeggo P.A.. 2010. The limitations of the G1-S checkpoint. Cancer Res. 70:4412–4421 10.1158/0008-5472.CAN-09-3198 [DOI] [PubMed] [Google Scholar]
- Di Virgilio, M., Callen E., Yamane A., Zhang W., Jankovic M., Gitlin A.D., Feldhahn N., Resch W., Oliveira T.Y., Chait B.T., et al. 2013. Rif1 prevents resection of DNA breaks and promotes immunoglobulin class switching. Science. 339:711–715 10.1126/science.1230624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnianni, R.A., Ferrari M., Lazzaro F., Clerici M., Tamilselvan Nachimuthu B., Plevani P., Muzi-Falconi M., and Pellicioli A.. 2010. Elevated levels of the polo kinase Cdc5 override the Mec1/ATR checkpoint in budding yeast by acting at different steps of the signaling pathway. PLoS Genet. 6:e1000763 10.1371/journal.pgen.1000763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elia, A.E.H., Rellos P., Haire L.F., Chao J.W., Ivins F.J., Hoepker K., Mohammad D., Cantley L.C., Smerdon S.J., and Yaffe M.B.. 2003. The molecular basis for phosphodependent substrate targeting and regulation of Plks by the Polo-box domain. Cell. 115:83–95 10.1016/S0092-8674(03)00725-6 [DOI] [PubMed] [Google Scholar]
- Escribano-Díaz, C., Orthwein A., Fradet-Turcotte A., Xing M., Young J.T., Tkáč J., Cook M.A., Rosebrock A.P., Munro M., Canny M.D., et al. 2013. A cell cycle-dependent regulatory circuit composed of 53BP1-RIF1 and BRCA1-CtIP controls DNA repair pathway choice. Mol. Cell. 49:872–883 10.1016/j.molcel.2013.01.001 [DOI] [PubMed] [Google Scholar]
- Feng, L., Fong K.W., Wang J., Wang W., and Chen J.. 2013. RIF1 counteracts BRCA1-mediated end resection during DNA repair. J. Biol. Chem. 288:11135–11143 10.1074/jbc.M113.457440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golsteyn, R.M., Mundt K.E., Fry A.M., and Nigg E.A.. 1995. Cell cycle regulation of the activity and subcellular localization of Plk1, a human protein kinase implicated in mitotic spindle function. J. Cell Biol. 129:1617–1628 10.1083/jcb.129.6.1617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotoh, E., and Durante M.. 2006. Chromosome condensation outside of mitosis: mechanisms and new tools. J. Cell. Physiol. 209:297–304 10.1002/jcp.20720 [DOI] [PubMed] [Google Scholar]
- Helmink, B.A., Tubbs A.T., Dorsett Y., Bednarski J.J., Walker L.M., Feng Z., Sharma G.G., McKinnon P.J., Zhang J., Bassing C.H., and Sleckman B.P.. 2011. H2AX prevents CtIP-mediated DNA end resection and aberrant repair in G1-phase lymphocytes. Nature. 469:245–249 10.1038/nature09585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huertas, P., and Jackson S.P.. 2009. Human CtIP mediates cell cycle control of DNA end resection and double strand break repair. J. Biol. Chem. 284:9558–9565 10.1074/jbc.M808906200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huertas, P., Cortés-Ledesma F., Sartori A.A., Aguilera A., and Jackson S.P.. 2008. CDK targets Sae2 to control DNA-end resection and homologous recombination. Nature. 455:689–692 10.1038/nature07215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackman, J., and O’Connor P.M.. 2001. Methods for synchronizing cells at specific stages of the cell cycle. Curr. Protoc. Cell Biol. Chapter 8:Unit 8.3. [DOI] [PubMed] [Google Scholar]
- Jackson, S.P., and Bartek J.. 2009. The DNA-damage response in human biology and disease. Nature. 461:1071–1078 10.1038/nature08467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, S.T., Lim D.S., Canman C.E., and Kastan M.B.. 1999. Substrate specificities and identification of putative substrates of ATM kinase family members. J. Biol. Chem. 274:37538–37543 10.1074/jbc.274.53.37538 [DOI] [PubMed] [Google Scholar]
- Kühne, M., Rothkamm K., and Löbrich M.. 2000. No dose-dependence of DNA double-strand break misrejoining following α-particle irradiation. Int. J. Radiat. Biol. 76:891–900 10.1080/09553000050050909 [DOI] [PubMed] [Google Scholar]
- Lansing, T.J., McConnell R.T., Duckett D.R., Spehar G.M., Knick V.B., Hassler D.F., Noro N., Furuta M., Emmitte K.A., Gilmer T.M., et al. 2007. In vitro biological activity of a novel small-molecule inhibitor of polo-like kinase 1. Mol. Cancer Ther. 6:450–459 10.1158/1535-7163.MCT-06-0543 [DOI] [PubMed] [Google Scholar]
- Lee, K.S., Park J.E., Kang Y.H., Kim T.S., and Bang J.K.. 2014. Mechanisms underlying Plk1 polo-box domain-mediated biological processes and their physiological significance. Mol. Cells. 37:286–294 10.14348/molcells.2014.0002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee-Theilen, M., Matthews A.J., Kelly D., Zheng S., and Chaudhuri J.. 2011. CtIP promotes microhomology-mediated alternative end joining during class-switch recombination. Nat. Struct. Mol. Biol. 18:75–79 10.1038/nsmb.1942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, S., Ting N.S., Zheng L., Chen P.L., Ziv Y., Shiloh Y., Lee E.Y., and Lee W.H.. 2000. Functional link of BRCA1 and ataxia telangiectasia gene product in DNA damage response. Nature. 406:210–215 10.1038/35018134 [DOI] [PubMed] [Google Scholar]
- Löbrich, M., Shibata A., Beucher A., Fisher A., Ensminger M., Goodarzi A.A., Barton O., and Jeggo P.A.. 2010. gammaH2AX foci analysis for monitoring DNA double-strand break repair: strengths, limitations and optimization. Cell Cycle. 9:662–669 10.4161/cc.9.4.10764 [DOI] [PubMed] [Google Scholar]
- Lukas, C., Savic V., Bekker-Jensen S., Doil C., Neumann B., Pedersen R.S., Grøfte M., Chan K.L., Hickson I.D., Bartek J., and Lukas J.. 2011a. 53BP1 nuclear bodies form around DNA lesions generated by mitotic transmission of chromosomes under replication stress. Nat. Cell Biol. 13:243–253 10.1038/ncb2201 [DOI] [PubMed] [Google Scholar]
- Lukas, J., Lukas C., and Bartek J.. 2011b. More than just a focus: The chromatin response to DNA damage and its role in genome integrity maintenance. Nat. Cell Biol. 13:1161–1169 10.1038/ncb2344 [DOI] [PubMed] [Google Scholar]
- Mansour, W.Y., Schumacher S., Rosskopf R., Rhein T., Schmidt-Petersen F., Gatzemeier F., Haag F., Borgmann K., Willers H., and Dahm-Daphi J.. 2008. Hierarchy of nonhomologous end-joining, single-strand annealing and gene conversion at site-directed DNA double-strand breaks. Nucleic Acids Res. 36:4088–4098 10.1093/nar/gkn347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mladenov, E., Anachkova B., and Tsaneva I.. 2006. Sub-nuclear localization of Rad51 in response to DNA damage. Genes Cells. 11:513–524 10.1111/j.1365-2443.2006.00958.x [DOI] [PubMed] [Google Scholar]
- Mosesso, P., Fonti E., Bassi L., Lorenti Garcia C., and Palitti F.. 1999. The involvement of chromatin condensation in camptothecin-induced chromosome breaks in G0 human lymphocytes. Mutagenesis. 14:103–105 10.1093/mutage/14.1.103 [DOI] [PubMed] [Google Scholar]
- Moynahan, M.E., and Jasin M.. 2010. Mitotic homologous recombination maintains genomic stability and suppresses tumorigenesis. Nat. Rev. Mol. Cell Biol. 11:196–207 10.1038/nrm2851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naim, V., Wilhelm T., Debatisse M., and Rosselli F.. 2013. ERCC1 and MUS81-EME1 promote sister chromatid separation by processing late replication intermediates at common fragile sites during mitosis. Nat. Cell Biol. 15:1008–1015 10.1038/ncb2793 [DOI] [PubMed] [Google Scholar]
- Nakamura, K., Kogame T., Oshiumi H., Shinohara A., Sumitomo Y., Agama K., Pommier Y., Tsutsui K.M., Tsutsui K., Hartsuiker E., et al. 2010. Collaborative action of Brca1 and CtIP in elimination of covalent modifications from double-strand breaks to facilitate subsequent break repair. PLoS Genet. 6:e1000828 10.1371/journal.pgen.1000828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nussenzweig, A., and Nussenzweig M.C.. 2007. A backup DNA repair pathway moves to the forefront. Cell. 131:223–225 10.1016/j.cell.2007.10.005 [DOI] [PubMed] [Google Scholar]
- Nussenzweig, A., and Nussenzweig M.C.. 2010. Origin of chromosomal translocations in lymphoid cancer. Cell. 141:27–38 10.1016/j.cell.2010.03.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panier, S., and Durocher D.. 2013. Push back to respond better: regulatory inhibition of the DNA double-strand break response. Nat. Rev. Mol. Cell Biol. 14:661–672 10.1038/nrm3659 [DOI] [PubMed] [Google Scholar]
- Pearce, M.S., Salotti J.A., Little M.P., McHugh K., Lee C., Kim K.P., Howe N.L., Ronckers C.M., Rajaraman P., Sir Craft A.W., et al. 2012. Radiation exposure from CT scans in childhood and subsequent risk of leukaemia and brain tumours: a retrospective cohort study. Lancet. 380:499–505 10.1016/S0140-6736(12)60815-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson, S.E., Li Y., Wu-Baer F., Chait B.T., Baer R., Yan H., Gottesman M.E., and Gautier J.. 2013. Activation of DSB processing requires phosphorylation of CtIP by ATR. Mol. Cell. 49:657–667 10.1016/j.molcel.2012.11.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polo, S.E., and Jackson S.P.. 2011. Dynamics of DNA damage response proteins at DNA breaks: a focus on protein modifications. Genes Dev. 25:409–433 10.1101/gad.2021311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quennet, V., Beucher A., Barton O., Takeda S., and Löbrich M.. 2011. CtIP and MRN promote non-homologous end-joining of etoposide-induced DNA double-strand breaks in G1. Nucleic Acids Res. 39:2144–2152 10.1093/nar/gkq1175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rass, E., Grabarz A., Plo I., Gautier J., Bertrand P., and Lopez B.S.. 2009. Role of Mre11 in chromosomal nonhomologous end joining in mammalian cells. Nat. Struct. Mol. Biol. 16:819–824 10.1038/nsmb.1641 [DOI] [PubMed] [Google Scholar]
- Reczek, C.R., Szabolcs M., Stark J.M., Ludwig T., and Baer R.. 2013. The interaction between CtIP and BRCA1 is not essential for resection-mediated DNA repair or tumor suppression. J. Cell Biol. 201:693–707 10.1083/jcb.201302145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhardt, H.C., and Yaffe M.B.. 2013. Phospho-Ser/Thr-binding domains: navigating the cell cycle and DNA damage response. Nat. Rev. Mol. Cell Biol. 14:563–580 10.1038/nrm3640 [DOI] [PubMed] [Google Scholar]
- Rief, N., and Löbrich M.. 2002. Efficient rejoining of radiation-induced DNA double-strand breaks in centromeric DNA of human cells. J. Biol. Chem. 277:20572–20582 10.1074/jbc.M200265200 [DOI] [PubMed] [Google Scholar]
- Rothkamm, K., Krüger I., Thompson L.H., and Löbrich M.. 2003. Pathways of DNA double-strand break repair during the mammalian cell cycle. Mol. Cell. Biol. 23:5706–5715 10.1128/MCB.23.16.5706-5715.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sartori, A.A., Lukas C., Coates J., Mistrik M., Fu S., Bartek J., Baer R., Lukas J., and Jackson S.P.. 2007. Human CtIP promotes DNA end resection. Nature. 450:509–514 10.1038/nature06337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata, A., Conrad S., Birraux J., Geuting V., Barton O., Ismail A., Kakarougkas A., Meek K., Taucher-Scholz G., Löbrich M., and Jeggo P.A.. 2011. Factors determining DNA double-strand break repair pathway choice in G2 phase. EMBO J. 30:1079–1092 10.1038/emboj.2011.27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephan, H., Concannon C., Kremmer E., Carty M.P., and Nasheuer H.P.. 2009. Ionizing radiation-dependent and independent phosphorylation of the 32-kDa subunit of replication protein A during mitosis. Nucleic Acids Res. 37:6028–6041 10.1093/nar/gkp605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Gent, D.C., and van der Burg M.. 2007. Non-homologous end-joining, a sticky affair. Oncogene. 26:7731–7740 10.1038/sj.onc.1210871 [DOI] [PubMed] [Google Scholar]
- van Gent, D.C., Hoeijmakers J.H., and Kanaar R.. 2001. Chromosomal stability and the DNA double-stranded break connection. Nat. Rev. Genet. 2:196–206 10.1038/35056049 [DOI] [PubMed] [Google Scholar]
- Wang, H., Rosidi B., Perrault R., Wang M., Zhang L., Windhofer F., and Iliakis G.. 2005. DNA ligase III as a candidate component of backup pathways of nonhomologous end joining. Cancer Res. 65:4020–4030 10.1158/0008-5472.CAN-04-3055 [DOI] [PubMed] [Google Scholar]
- Wang, H., Shi L.Z., Wong C.C., Han X., Hwang P.Y., Truong L.N., Zhu Q., Shao Z., Chen D.J., Berns M.W., et al. 2013. The interaction of CtIP and Nbs1 connects CDK and ATM to regulate HR-mediated double-strand break repair. PLoS Genet. 9:e1003277 10.1371/journal.pgen.1003277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, M., Wu W., Wu W., Rosidi B., Zhang L., Wang H., and Iliakis G.. 2006. PARP-1 and Ku compete for repair of DNA double strand breaks by distinct NHEJ pathways. Nucleic Acids Res. 34:6170–6182 10.1093/nar/gkl840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weichert, W., Denkert C., Schmidt M., Gekeler V., Wolf G., Köbel M., Dietel M., and Hauptmann S.. 2004. Polo-like kinase isoform expression is a prognostic factor in ovarian carcinoma. Br. J. Cancer. 90:815–821 10.1038/sj.bjc.6601610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weichert, W., Kristiansen G., Winzer K.J., Schmidt M., Gekeler V., Noske A., Müller B.M., Niesporek S., Dietel M., and Denkert C.. 2005. Polo-like kinase isoforms in breast cancer: expression patterns and prognostic implications. Virchows Arch. 446:442–450 10.1007/s00428-005-1212-8 [DOI] [PubMed] [Google Scholar]
- Weinstock, D.M., Brunet E., and Jasin M.. 2007. Formation of NHEJ-derived reciprocal chromosomal translocations does not require Ku70. Nat. Cell Biol. 9:978–981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying, S., Minocherhomji S., Chan K.L., Palmai-Pallag T., Chu W.K., Wass T., Mankouri H.W., Liu Y., and Hickson I.D.. 2013. MUS81 promotes common fragile site expression. Nat. Cell Biol. 15:1001–1007 10.1038/ncb2773 [DOI] [PubMed] [Google Scholar]
- Yu, X., and Chen J.. 2004. DNA damage-induced cell cycle checkpoint control requires CtIP, a phosphorylation-dependent binding partner of BRCA1 C-terminal domains. Mol. Cell. Biol. 24:9478–9486 10.1128/MCB.24.21.9478-9486.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, X., Fu S., Lai M., Baer R., and Chen J.. 2006. BRCA1 ubiquitinates its phosphorylation-dependent binding partner CtIP. Genes Dev. 20:1721–1726 10.1101/gad.1431006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun, M.H., and Hiom K.. 2009. CtIP-BRCA1 modulates the choice of DNA double-strand-break repair pathway throughout the cell cycle. Nature. 459:460–463 10.1038/nature07955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Y., and Jasin M.. 2011. An essential role for CtIP in chromosomal translocation formation through an alternative end-joining pathway. Nat. Struct. Mol. Biol. 18:80–84 10.1038/nsmb.1940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerman, W.C., and Erikson R.L.. 2007. Polo-like kinase 3 is required for entry into S phase. Proc. Natl. Acad. Sci. USA. 104:1847–1852 10.1073/pnas.0610856104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann, M., Lottersberger F., Buonomo S.B., Sfeir A., and de Lange T.. 2013. 53BP1 regulates DSB repair using Rif1 to control 5′ end resection. Science. 339:700–704 10.1126/science.1231573 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.