Background: Identifying host factors used by influenza can aid in the defense against pandemics that threaten public health.
Results: Phospholipase D (PLD) contributes to viral infection and innate immune evasion strategies.
Conclusion: Inhibition of PLD activity reduces influenza reproduction.
Significance: PLD inhibition presents a novel approach to restrict influenza infection and viral escape.
Keywords: Antiviral Agent, Cell Signaling, Enzyme Inhibitor, Influenza Virus, Phospholipase D, Phospholipid, Inhibitors
Abstract
Lipid metabolism plays a fundamental role during influenza virus replication, although key regulators of lipid-dependent trafficking and virus production remain inadequately defined. This report demonstrates that infection by influenza virus stimulates phospholipase D (PLD) activity and that PLD co-localizes with influenza during infection. Both chemical inhibition and RNA interference of PLD delayed viral entry and reduced viral titers in vitro. Although there may be contributions by both major isoenzymes, the effects on viral infectivity appear to be more dependent on the PLD2 isoenzyme. In vivo, PLD2 inhibition reduced virus titer and correlated with significant increases in transcription of innate antiviral effectors. The reduction in viral titer downstream of PLD2 inhibition was dependent on Rig-I (retinoic acid-inducible gene-1), IRF3, and MxA (myxovirus resistance gene A) but not IRF7. Inhibition of PLD2 accelerated the accumulation of MxA in foci as early as 30 min postinfection. Together these data suggest that PLD facilitates the rapid endocytosis of influenza virus, permitting viral escape from innate immune detection and effectors that are capable of limiting lethal infection.
Introduction
Lipid species play integral roles in cellular signaling and intracellular trafficking. Influenza viruses exploit these fundamental processes within the host cell to facilitate entry and subsequently drive virus replication. The cycle begins with the viral surface protein hemagglutinin (HA) binding to an epithelial cell surface sialic acid moiety. Typically, this triggers endocytosis, allowing the virus entry inside the host cell where it incorporates itself within the host membrane. As the endosome matures, acidification initiates a conformational change in the HA, allowing it to fuse with the host endosomal membrane and release the ribonucleoprotein complexes, which are trafficked to the nucleus where RNA-dependent polymerases replicate the viral genome and make the mRNA for viral proteins. After proteins are synthesized, they accumulate at the cell membrane, complex with viral genomes, and bud off the cell surface, forming the viral envelope from the host cell membrane. Lipids are fundamentally involved in both the entry and egress of the virus (1).
In addition to playing structural roles in viral entry and budding, lipid species also serve as intra- and extracellular signals as part of the innate immune response. 25-Hydroxycholesterol is secreted by macrophages in response to the Stat1/IFN pathway and can promote resistance to multiple viruses (2). Protectin D1 was identified as promoting an anti-inflammatory state that alleviated severe disease in an H5N1 infection (3). The 12/15-lipoxygenase pathway that generates protectin D1 was identified in another screen as being associated with mild infection in both mice and humans (4). PI3K has been demonstrated to have a role in influenza infection and host defense (5). The lipid-binding protein Epsin-1 is a regulator of clathrin-mediated endocytosis of influenza virus (6). Lipid rafts also are involved in influenza virus (7) and HIV replication (8). Clearly, lipid species are influential during viral pathogenesis and present a ripe opportunity for further research.
Despite recent findings on the participation of sterols, arachidonic acid-derived species, and lipid structures (such as rafts) in the regulation of influenza infection, the roles of glycerophospholipids have been ill defined. PLD3 enzymes play critical roles in generating lipid signaling molecules and contributing to membrane structure. The primary lipid product of PLD, phosphatidic acid (PA), is an important bioactive lipid that is involved in membrane biogenesis and curvature in addition to being a second messenger in signaling pathways (9, 10). Specifically, PA can be converted into diacylglycerol by phosphatidic acid phosphohydrolase, which subsequently activates protein kinase C (PKC) and other signaling proteins. Prior studies demonstrated that inhibiting PKC activity during influenza infection interferes with intracellular trafficking of the virus and protects cells from infection (11). Thus, PLD appears to be a potential targetable candidate as a host restriction factor of influenza virus replication.
In mammals, two predominant isoenzymes, PLD1 and PLD2, are well characterized. Recently, PLD isoform-preferring inhibitors have been designed, synthesized, and characterized (12, 13). The development of these PLD inhibitors has facilitated the interrogation of the specific role of isoenzymes in various cellular processes (14). Utilizing these novel synthetic compounds and other biochemical approaches, the up-regulation of PLD activity during influenza infection was demonstrated, and evidence that PLD inhibition delays virus entry, allowing for a more robust innate antiviral response to be mounted in the infected host cell and leading to a significant reduction in viral titer, was obtained. Consequently, PLD is a targetable host restriction factor for influenza viruses that facilitates rapid endosomal trafficking and escape from innate immune detection in the host cell.
EXPERIMENTAL PROCEDURES
Chemical Synthesis and Purification
PLD isoenzyme-preferring inhibitors were synthesized and characterized as described in detail previously (12, 13).
Mass Spectrometry-based Lipid Analysis
For analysis of cellular lipids, A549 cells were serum-starved for 1 h, infected at 1 m.o.i. with influenza A/California/04/2009 or other indicated strains, and then maintained in 6-well plates for the indicated treatment times. Mock-infected and influenza A-infected cells were harvested at the indicated times after infection and administration of PLD inhibitors. To extract phospholipids, ∼1 × 106 cells were washed with ice-cold PBS and scraped into 1 ml of ice-cold PBS, and aliquots were taken for protein analysis. After centrifugation and PBS aspiration, the pellet was extracted by a modified Bligh and Dyer (15) method using 800 μl of ice-cold 0.1 n HCl, CH3OH (1:1) and 400 μl of ice-cold CHCl3. Following vortexing for 1 min at 4 °C, phases were separated by centrifugation (18,000 × g for 5 min at 4 °C). The lower organic layer was isolated, synthetic odd-carbon phospholipids (four per phospholipid class) were added as standards, and the solvent was evaporated. The resulting lipid film was dissolved in 100 μl of 58:40:2 isopropanol, hexane, 100 mm NH4COOH (aqueous) (mobile phase A) (16). Samples containing n-butyl alcohol were extracted similarly, and 5 μl of 10 μg/ml 32:0 phosphatidylmethanol was added as an internal standard. Mass spectrometric analysis and quantitation were performed essentially as previously described (17–19). An MDS SCIEX 4000QTRAP hybrid triple quadrupole/linear ion trap mass spectrometer (Applied Biosystems, Foster City, CA) coupled to a Shimadzu HPLC system (Shimadzu Scientific Instruments, Inc., Columbia, MD) consisting of an SCL 10 APV controller, two LC 10 ADVP pumps, and a CTC HTC PAL autosampler (Leap Technologies, Carrboro, NC) was utilized for the analyses. Phospholipids were separated on a Phenomenex Luna Silica column (Phenomenex, Torrance, CA) (2 × 250 mm, 5-μm particle size) using a 20-μl sample injection. A binary gradient consisting of 58:40:2 isopropanol, hexane, 100 mm NH4COOH (aqueous) (mobile phase A) and 50:40:10 isopropanol, hexane, 100 mm NH4COOH (aqueous) (mobile phase B) was used for the separation. Instrumentation parameters and solvent gradient were described previously (18). Time courses were performed in two independent experiments, each in triplicate. Statistical analysis was performed by two-way ANOVA with Bonferroni's post-test.
Spatial Infection Model
A spatial infection model for testing compound efficacy and influenza replication was adapted from Lam et al. (20). Monolayers of A549 cells were grown on chamber slides. A semisolid overlay made of agar and growth medium was allowed to cure on top of the monolayer. A Pasteur pipette was used to create a consistent reservoir in the overlay, and influenza virus was introduced through this reservoir. Cultures were then incubated at 37 °C for the duration of the infection.
RNA Interference
A549 cells were transfected with 100 nm siRNA (Ambion) specific to PLD isoforms using NeoFX (Ambion) and were subsequently infected with 1 m.o.i. influenza A/Brisbane/59/2007 (H1N1) for 24 h. Innate immune proteins were knocked down using 100 nm siRNA and the Neon Transfection System (Invitrogen). Knockdown was confirmed with gene-specific TaqMan assays and the 2ΔΔCt method using GAPDH for normalization on Western blots to confirm loss of protein.
Immunofluorescence and Live Cell Imaging
Samples were fixed in 4% formaldehyde, permeabilized with 0.3% Triton X-100, and then exposed to antisera targeting proteins of interest and corresponding fluorescent secondary antibodies alongside DAPI to visualize nuclei. Spatial infections were imaged and processed using Nikon C1Si and NIS Elements software. Confocal images were captured with a Zeiss LSM 510 NLO Meta and analyzed with Zeiss Zen 2011 and NIH ImageJ software. To determine colocalization, the PSC (Pearson-Spearman Correlation) colocalization plug-in was used to calculate Pearson's and Spearman's coefficients. Briefly, nucleoprotein (NP) and PLD2 signal channels were merged. Each Z-slice was projected to maximum intensity. NP-positive cells were masked, and the mask was used to determine whether positive signal for each channel colocalized. Each mask was in excess of 20,000 pixels to ensure a robust data set. Live cell imaging was performed using spinning disc laser-scanning confocal microscopy carried out on a Marianas spinning disk confocal imaging system (Intelligent Imaging Innovations/3i, Denver, CO) comprising a CSU22 confocal scan head (Yokogowa Electric Corp.) and solid state lasers with wavelengths of 488 and 658 nm configured on a motorized Axio Observer Z1 inverted microscope (Carl Zeiss MicroImaging) equipped with a Definite Focus system (Carl Zeiss) and spherical aberration correction optics. Time-lapsed three-dimensional imaging was performed at 37 °C in 5% (v/v) CO2 in a humidified atmosphere using an environmental control chamber (Intelligent Imaging Innovations/3i), and images were acquired using a Plan-Apochromat 63× 1.4 numerical aperture oil objective on an Evolve 512 EMCCD camera (Photometrics, Tucson, AZ) using Slidebook 5.5 software (Intelligent Imaging Innovations/3i).
Animal Infection
All animal studies were approved by the St. Jude Children's Research Hospital Animal Care and Use Committee (Protocol number 098) following the guidelines established by the Institute of Laboratory Animal Resources approved by the Governing Board of the United States National Research Council. Female C57BL/6 mice (8–10 weeks old) were anesthetized and infected with the indicated doses and strains of influenza A virus. Mice were weighed and monitored daily; tissues were collected at the specified times and kept at −80 °C until analysis. For drug treatment, mice were administered 13 mg/kg VU0364739 or vehicle (10% DMSO, 90% PEG) every 8 h from day −1 to day 3 after infection or every 12 h during the H7N9 study. All procedures were performed according to an institutionally approved Institutional Animal Care and Use Committee protocol, which includes a requirement for daily observation and euthanasia upon detection of severe moribundity.
Titering
Infected animal lungs were titered using a standard plaque assay. Supernatant from infected cultures was titered using a traditional TCID50 assay, and immunofluorescence was used to enumerate the number of infected cells in cultured samples.
Host Gene Expression
RNA was isolated from lungs and used in reverse transcription-PCR. The cDNA was then used in gene-specific TaqMan assays to determine host gene expression, and the differences in expression were quantified using the 2ΔΔCt method. The same amount of RNA was used in each reaction, and samples were run in triplicate.
Statistical Analysis
Quantitative data are presented as mean ± S.E. of at least three independent experiments.
RESULTS
Influenza Infection Potentiates PLD Catalytic Activity, and That Activity Is Attenuated by VU0364739 Treatment
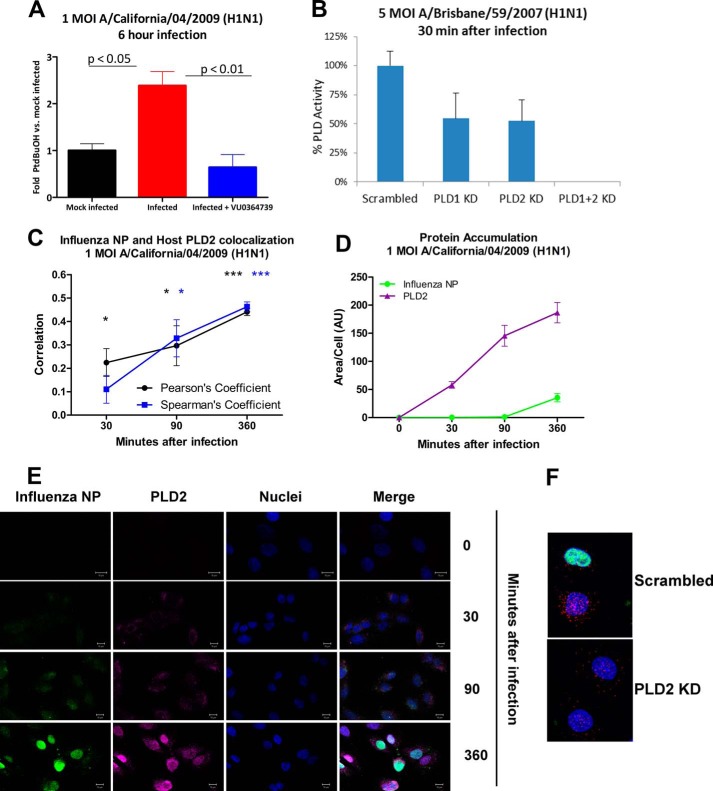
To determine the effect of influenza infection on PLD activity, human adenocarcinomic alveolar basal epithelial cells (A549) were infected with 1 m.o.i. human influenza strain A/California/04/2009 (H1N1) for 6 h in the presence of 0.6% n-butyl alcohol. Instead of the typical biological nucleophile water, in the presence of a primary alcohol, PLD will utilize the alcohol to produce a metabolically stable transphosphatidylation product, phosphatidylalcohol, as an alternative to hydrolysis producing PA. Fig. 1A illustrates that influenza infection markedly stimulated PLD activity as measured by the increase in phosphatidylbutanol production compared with cells that were not exposed to influenza virus (mock-infected). Treatment with a PLD2-preferring inhibitor, VU0364739, was sufficient to block the influenza-mediated PLD activity increase. As shown in Fig. 1B, both PLD1 and PLD2 were active during the viral entry phase, and complete ablation of catalytic activity required knockdown of both PLD1 and -2. This result suggests independent roles for both isoenzymes in entry.
FIGURE 1.
Influenza infection stimulates PLD activity. A, A549 cells treated for 1 h with or without 10 μm PLD inhibitor VU0364739 were then infected for 1 h with 1 m.o.i. influenza A/California/04/2009 (or mock-infected) after which the inoculum was removed. The cells were cultured for 6 h in the presence of 0.6% n-butyl alcohol. PLD activity as a measure of phosphatidylbutanol (PtdBuOH) was determined in cell lysates by mass spectrometric analysis. B, RNAi of PLD1, PLD2, or PLD1 and -2. A549 cells were transfected with isoform-specific siRNA 1 day before a 5 m.o.i. infection of influenza A/Brisbane/59/2007 (H1N1). 30 min after infection, PLD catalytic activity was measured in the same manner as in A. C, D, and E, A549 cells were infected with 1 m.o.i. influenza A/California/04/2009 (H1N1), and samples were fixed and probed for influenza NP and PLD2 at 0, 30, 90, and 360 min after infection. Z-stacks of the infected cultures were collected using confocal microscopy, and ImageJ was used to determine PLD and influenza NP colocalization (C) during infection as well as protein accumulation (D). Quantification of the degree of correlation between NP and PLD shows that they significantly colocalize as early as 30 min postinfection, and the colocalization has the highest correlation coefficient 360 min postinfection. PLD2 signal begins to intensify 30 min after infection and continues to increase in both size and intensity throughout the duration of the infection. NP signal lags behind that of PLD2 but begins to intensify 90 and 360 min postinfection. E, representative images from a 1 m.o.i. influenza A/California/04/2009 (H1N1) infection taken 0, 30, 90, and 360 min postinfection. The green signal is influenza (NP), the magenta signal is PLD2, and the blue signal is DAPI staining. Scale bars, 10 μm. As the infection progresses, PLD2 staining begins to accumulate at the cell periphery, then intensifies, and finally moves to a perinuclear region. NP staining first appears at the cell periphery and then intensifies in the nucleus. F, representative images from PLD2 monoclonal antibody validation. A549 cells were electroporated with 100 nm scrambled control or PLD2 siRNA and left to rest for 24 h. Cells were fixed 8 h after a 1 m.o.i. A/Brisbane/59/2007 (H1N1) infection. Green signal is influenza NP, red signal is PLD2, and blue signal is nuclei. All data are mean ± S.E. (error bars). *, p < 0.05; ***, p < 0.001. KD, knockdown.
Confocal microscopy-based experiments were conducted to assess changes in the accumulation and localization of PLD during an influenza infection. A549 cells were grown on chamber slides and infected with 1 m.o.i. influenza A/California/04/2009 (H1N1). Samples were fixed at 0, 30, 90, and 360 min postinfection and probed for influenza NP and PLD2. PLD2 began to accumulate at the periphery of the cell as early as 30 min postinfection (Fig. 1, C, D, and E). At the same time point, very low levels of NP, also occurring at the outer reaches of the cell (Fig. 1E), were detected. PLD2 staining continued to intensify 90 and 360 min postinfection (Fig. 1D), and the observed PLD2 signal was increasing as well as moving from the cytoplasm to a perinuclear region (Fig. 1E). Influenza NP signal also intensified 360 min postinfection, and NP was moving from the cytoplasm (30 and 90 min postinfection) toward the nucleus (Fig. 1, D and E). During the infection, PLD2 and NP were trafficking to similar subcellular locations. The extent of this colocalization was quantified. PLD2 and NP were increasingly colocalized at 30, 90, and 360 min postinfection (Fig. 1C) as measured by both Pearson's coefficient and rank correlation (Spearman). To further confirm that siRNA treatment was knocking down levels of PLD2 and that the PLD2 antibody was specific, we probed for PLD2 expression after infection of cells electroporated with control or PLD2 siRNA. We imaged more than 75 cells for each condition and found a consistent loss of 50% or more of stainable PLD2 in the siRNA-treated cells (1F). Together these data suggest that PLD activity is stimulated by influenza infection, endogenous PLD is redistributed during the infection, and the accumulation of PLD is occurring in the same subcellular location as influenza NP.
PLD Is a Targetable Host Factor That Facilitates Efficient Influenza Infection
To determine the role of PLD-mediated PA production in influenza infection, we used a spatial infection model in the presence of 0.6% n-butyl alcohol with tert-butyl alcohol used as a negative control. This assay utilizes the use of primary alcohols as preferred nucleophiles in a PLD transphosphatidylation reaction as an assessment of exclusively PLD-produced PA.
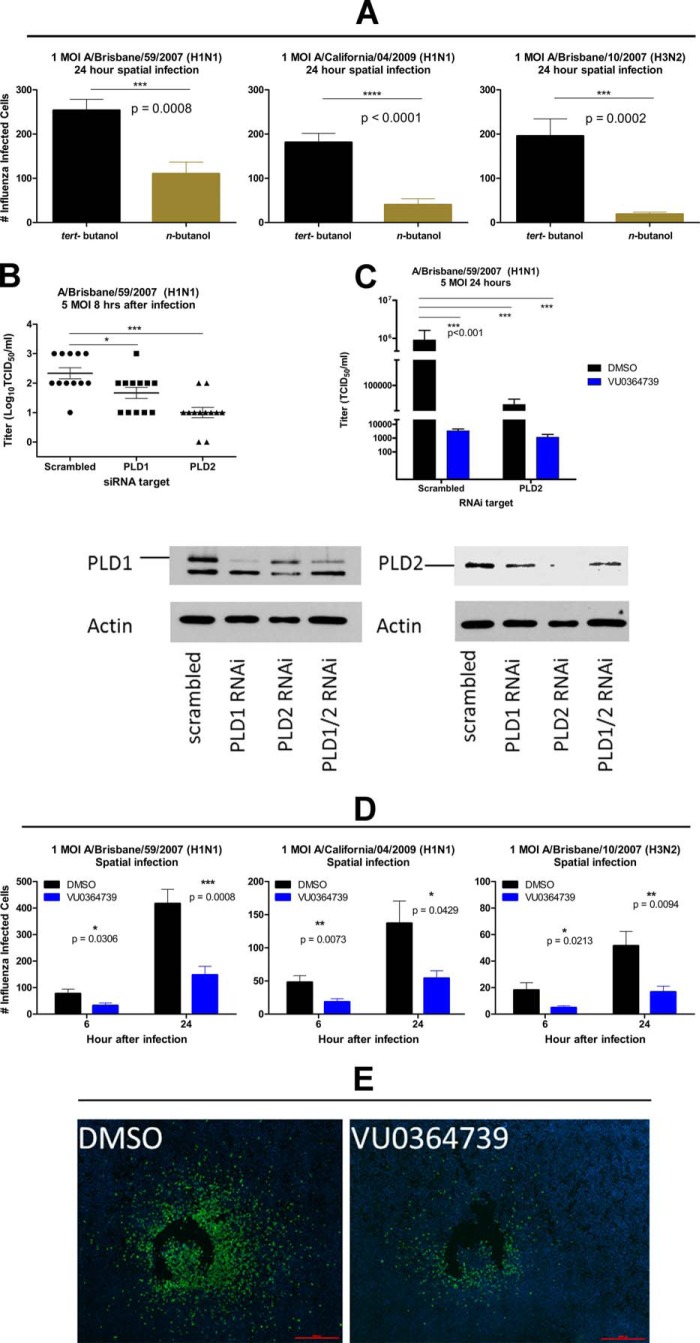
Twenty-four hours postinfection, infected cells were counted by anti-influenza NP staining (Fig. 2A). Infection with influenza strain A/Brisbane/59/2007 (H1N1), A/California/04/2009 (H1N1), or A/Brisbane/10/2007 (H3N2) resulted in fewer infected cells in the presence of n-butyl alcohol, indicating that blocking PLD production of PA substantially reduces the rate of cell to cell transmission of infection. Notably, averting the production of PLD-generated PA to PLD-generated phosphatidylbutanol by use of primary alcohol did not entirely prevent influenza infection but did significantly decrease the rate of infectious spread within the cultures.
FIGURE 2.
PLD2 is required for efficient influenza infection, and inhibition of PLD-generated PA by primary alcohol, RNAi, or small molecule VU0364739 dramatically hinders cell to cell spread of influenza. A, number of influenza-infected cells in cultures of A549 cells treated with 0.6% tert-butyl alcohol or n-butyl alcohol for 1 h prior to spatial infection with the indicated influenza virus strain at 1 m.o.i. for 24 h. Differences were assessed by t test. B, upper panel, RNAi of PLD isoforms. A549 cells were transfected with siRNA targeting PLD1 or PLD2 24 h prior to infection with 5 m.o.i. influenza A/Brisbane/59/2007 (H1N1); 8 h postinfection, influenza replication was measured by TCID50 assay. Lower panel, immunoblot of human PLD1 and PLD2 from A549 cells following RNAi treatment. Lysates were separated on a 10% polyacrylamide gel, transferred to nitrocellulose overnight, and incubated with primary antibodies for phosphatidylcholine-PLD1 (Santa Cruz Biotechnology sc-25512) or PLD2 (Abgent AT3337a). Immunoblots were developed with ECL Western blotting substrate (Pierce catalog number 32106). C, A549 cells were transiently transfected with PLD2-specific siRNA or scrambled control siRNA 24 h before infection. An hour before infection, the cells were treated with DMSO or VU0364739 (10 μm). The cells were infected with 5 m.o.i. A/Brisbane/59/2007 (H1N1), and influenza replication was measured by TCID50 assay 24 h after infection. Differences between amounts of infected cells as well as influenza replication were compared using a one-way ANOVA and Dunnett's post-test. D, influenza spatial infection (following 1-h 10 μm PLD2-preferring inhibitor VU0364739 pretreatment) with clinically relevant strains of influenza at 1 m.o.i. At both 6 and 24 h postinfection, significantly fewer numbers of influenza-infected cells were counted in VU0364739-pretreated samples. Data were analyzed by t test. E, representative fluorescent photomicrograph mosaics following a 24-h spatial infection of A549 cells with influenza A/Brisbane/59/2007 (H1N1). Green signal is influenza-infected cells, and blue signal is DAPI staining. Scale bars, 10 μm. All data are mean ± S.E. (error bars). *, p < 0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001.
To determine whether PLD1 or PLD2 was preferentially required for efficient influenza infection, RNAi was used to selectively knock down individual PLD isoforms. A549 cells were transiently transfected with siRNA that targeted PLD1 or PLD2 24 h prior to a 5 m.o.i. influenza infection of A/Brisbane/59/2007 (H1N1). Twenty-four hours postinfection, a TCID50 assay was used to measure influenza virus titers in culture supernatants (Fig. 2B). RNAi of either PLD1 or PLD2 inhibited influenza replication. However, the magnitude of the effect was greater with PLD2 knockdown. Although each isoform appears to contribute to efficient influenza replication, the stronger inhibitory effects seen after PLD2 knockdown led us to focus on PLD2 inhibition and its effect on the host and viral replication for subsequent studies.
The development and optimization of the PLD inhibitors have been described (12) with the PLD2-preferring inhibitor VU0364739 having a 75-fold selectivity for PLD2 over PLD1 (13). Based on previous studies, VU0364739 was used at 10 μm in all cell-based assays unless otherwise indicated (21). To demonstrate the importance of PLD2 during influenza replication, another RNAi experiment was conducted while treating the PLD2-deficient cells with PLD2 inhibitor VU0364739. Viral titer was measured by a TCID50 assay after a 24-h, 5 m.o.i. influenza A/Brisbane/59/2007 (H1N1) infection. Pretreatment with VU0364739 for 1 h caused a dramatic decrease in viral replication (Fig. 2C). Similar reductions in titer were also observed when PLD2 was knocked down by RNAi in vehicle-treated samples. Finally, the specificity of this compound was confirmed by combining the apparent marginal, non-statistically significant additive nature of RNAi with VU0364739 treatment.
These observations were extended to establish the importance of PLD2 as a host factor required for infection by other clinically relevant strains of influenza. Pretreating cells for 1 h with VU0364739 resulted in a significant decrease in the number of infected cells measured in the spatial infection assay with multiple viral strains at both 6 and 24 h postinfection (Fig. 2D). Representative images of the spatial infection model assay (Fig. 2E) illustrate the reduction in influenza spread observed after VU0364739 treatment. PLD2 inhibition by VU0364739 effectively protected A549 cells from cell to cell spread of influenza, further demonstrating that PLD2 is required for efficient influenza infection and spread. A time-of-addition study was conducted where inhibitor was added over a time course spanning 2 h before infection to 4 h postinfection. Protection from infection was observed at all times of PLD inhibitor addition (data not shown).
Inhibition of PLD Activity Dramatically Reduces Influenza Replication
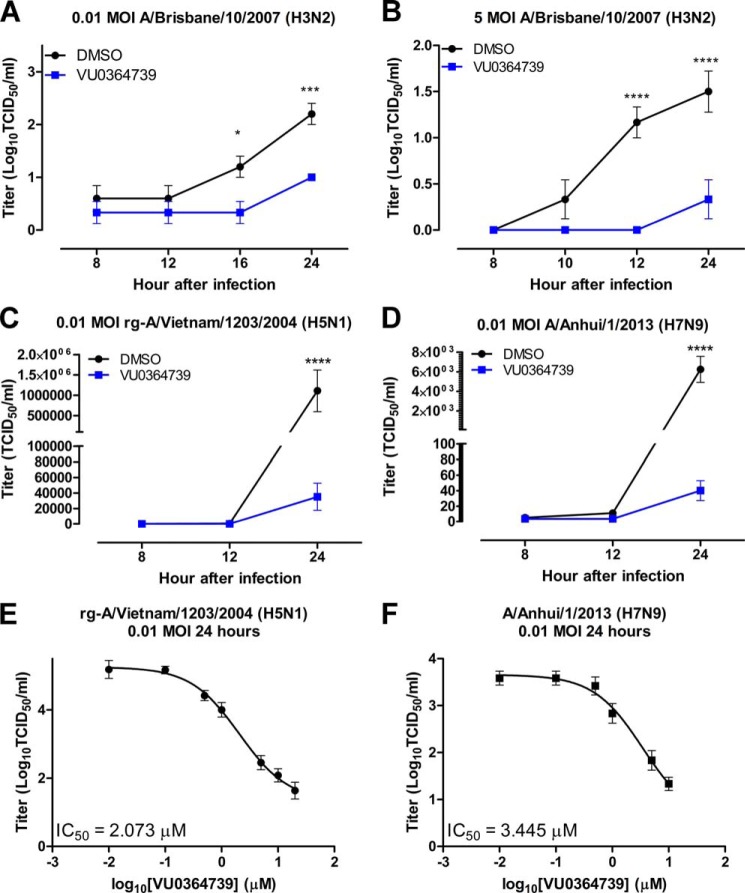
The spatial infection model assay data were validated using the more traditional TCID50 assay to assess viral reproduction in vitro. A549 cells were treated with 10 μm VU0364739 for 1 h before infection, and the cells were then infected with the indicated doses and strains of influenza. At the indicated time points, infectious supernatant was removed from the A549 cells and titrated on Madin-Darby canine kidney cells to measure viral reproduction. Using either a low m.o.i. (Fig. 3A) or a high m.o.i. (Fig. 3B) of influenza A/Brisbane/10/2007 (H3N2), a lower viral reproduction was observed when PLD2 was inhibited. In the case of the low m.o.i. infection, viral titers were first noted to be lower at 16 h postinfection, and the effect was sustained 24 h postinfection. Using the high m.o.i. model, when A549 cells were treated with VU0364739, a significant reduction in viral reproduction was noted 12 h postinfection and lasted through at least 24 h.
FIGURE 3.
Influenza replication is severely reduced when PLD2 is inhibited by VU034739. A549 cells were pretreated with 10 μm VU0364739 or DMSO for 1 h and then infected with either low 0.01 m.o.i. H3N2 influenza (A), high 5.0 m.o.i. H3N2 influenza (B), 0.01 m.o.i. H5N1 influenza (C), or 0.01 m.o.i. H7N9 influenza (D) for an hour at 4 °C, and then the infectious supernatant containing the virus was removed and titrated on Madin-Darby canine kidney cells to assess viral production at the indicated times postinfection. Under PLD2 inhibitor treatment, poor viral replication in all influenza strains tested by 24 h postinfection was observed, and the viral output defect was noted as early as 12 h postinfection in the case of H3N2 and H7N9. Differences were assessed using a two-way ANOVA and Bonferroni's post-test where * represents p < 0.05, *** represents p < 0.001, and **** represents p < 0.0001. E and F, dose-response curves to determine the IC50 of VU0364739 on influenza titer after a 24-h infection with rg-A/Vietnam/1204/2004 (H5N1) (E) or A/Anhui/1/2013 (H7N9) (F). A549 cells were treated with the indicated concentration of VU0364739 for 1 h before infection, and a TCID50 assay was used to measure viral load. All data are mean ± S.E. (error bars).
The H3N2 strain used in our previous experiments is considered a low pathogenicity strain of influenza. To determine whether host PLD is required for high pathogenicity and quickly replicating strains of influenza, VU0364739-treated A549 cells were infected with 0.01 m.o.i. influenza rg-A/Vietnam/1203/2004 (H5N1), and viral reproduction was assessed during a more severe infection. Twenty-four hours postinfection, a massive decrease in viral titer was observed when PLD2 was inhibited during H5N1 infection (Fig. 3C). Subsequent investigation to determine whether host PLD2 activity is required for infection was conducted using a recently emergent virus with pandemic potential, influenza strain A/Anhui/01/2013 (H7N9). After 1 h of 10 μm VU0364739 pretreatment, viral reproduction was effectively blocked 24 h postinfection (Fig. 3D). Viral titer was near the limit of detection when PLD catalytic activity was inhibited, consistent with PLD2 being a host factor required for low and high pathogenicity influenza infections.
Using the same model, in vitro dose-response experiments were performed using VU0364739 to determine the efficacy of the PLD2 inhibitor during H5N1 or H7N9 infection. A549 cells were treated for 1 h with varying concentrations of VU0364739 before a 0.01 m.o.i. infection of either influenza rg-A/Vietnam/1204/2004 or A/Anhui/1/2013. Supernatant was then used to measure viral reproduction in a TCID50 assay 24 h postinfection. In the case of the H5N1 infection, the IC50 of VU0364739 was calculated by non-linear regression analysis to be 2.1 μm (Fig. 3E). Similarly, the IC50 of VU0364739 was found to be 3.4 μm when cells were infected with an H7N9 influenza strain (Fig. 3F). These IC50 values are consistent with in vitro dose-response experiments using influenza A/Brisbane/59/2007 (H1N1), A/Brisbane/10/2007 (H3N2), and A/California/04/2009 (H1N1) as well (data not shown). Based on these results, it was determined that inhibition of PLD2 can significantly lower influenza reproduction in vitro and that the decrease in viral titer occurs in a dose-dependent fashion.
PLD2-preferring Inhibitor VU0364739 Reduced Viral Titer in Mouse Lungs and Delayed Mortality during Lethal H7N9 Influenza Infection
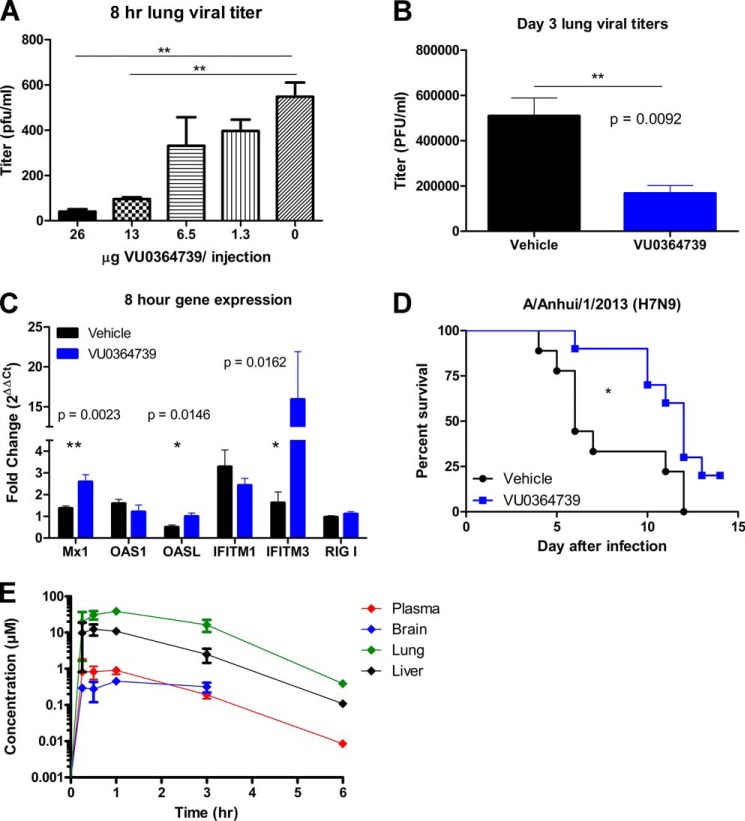
Having shown that abrogation of PLD2 activity leads to a decrease in viral spread and reproduction in vitro, we wanted to determine whether the loss of PLD2 could reduce viral titer in vivo. Female C57BL/6 (B6) mice were treated intraperitoneally with dilutions of PLD2-preferring inhibitor VU0364739 every 8 h from day −1 to 8 h after infection with 1 LD50 (4000 EID50) influenza A/Puerto Rico/8/1934 (H1N1) (PR8) administered intranasally on day 0. Because of solubility issues and observed acute vehicle toxicity, a long term survival study with optimal dosing to achieve the therapeutic in vivo concentration continuously was not feasible at this time, and viral titer was used as a readout to determine the role of PLD2 in a mouse model of influenza infection. Viral titer in lungs decreased significantly with PLD2 inhibitor VU0364739 treatment (Fig. 4A), and these protective effects were dose-dependent. Additionally, to determine whether PLD2 inhibition could lead to lower viral titers in a more chronic situation, mice were given 13 mg/kg VU0364739 every 8 h intraperitoneally from day −1 to day 3 after infection. Animals treated with VU0364739 had significantly less viral replication in their lungs (Fig. 4B) on day 3. Concomitant with the decrease in viral titers, 8 h after PR8 influenza infection, significant up-regulation of the innate immune proteins Mx1, OASL (2′-5′-oligoadenylate synthetase-like protein), and IFITM3 was observed when PLD2 was inhibited (Fig. 4C), indicating that the early immune response may be an important part of the mechanism of protection when mice are treated with VU0364739. IFITM3 has recently been described as a human restriction factor for influenza infection (22).
FIGURE 4.
PLD2 inhibitor decreased early viral titer in a dose-dependent manner and delayed mortality in a lethal H7N9 model. A, mice treated with VU0364739 displayed lung viral titers that were drastically reduced 8 h after 4000 EID50 PR8 influenza infection, and the reduction occurred in a dose-dependent manner. Data were compared by ANOVA and Bonferroni's post-test with n ≥ 5 mice used for each dose. **, p < 0.01. B, viral replication was similarly inhibited 3 days after 4000 EID50 PR8 influenza infection in mice treated with 13 mg/kg VU0364739 three times a day from day −1 to day 3 postinfluenza infection. Data were analyzed by t test. **, p < 0.01. C, RNA from PR8 influenza-infected lungs was isolated, and gene expression was measured using TaqMan-based quantitative PCR 8 h after infection. Innate immune proteins Mx1, OASL (2′-5′-oligoadenylate synthetase-like protein), and IFITM3 were significantly up-regulated as early as 8 h after infection as measured by t test. *, p < 0.05. D, mice receiving 13 mg/kg VU0364739 twice a day from day −1 to day 5 postinfection were inoculated with 103.5 TCID50 (1 LD50) influenza A/Anhui/1/2013 (H7N9). The PLD2 inhibitor conferred a substantial delay in mortality and a 20% survival advantage, a significant benefit as measured by either log rank test (p = 0.0194) or Gehan-Breslow-Wilcoxon test (p = 0.0165). *, p < 0.05. E, plasma, brain, lung, and liver exposures following a single 10 mg/kg intraperitoneal dose of the PLD inhibitor VU0364739 in mouse. Samples were stored at −80 °C until extraction and LC-MS/MS analysis (n = 2 samples per time point). Data are means ± S.E. (error bars).
To identify survival benefits conferred by the observed reduction in viral titers, a study was conducted dosing mice with 13 mg/kg VU0364739 every 12 h from day −1 to day 3 of a lethal influenza A/Anhui/1/2013 (H7N9) infection (Fig. 4D). Relative concentrations of the PLD inhibitor in various tissues of interest over time are presented in Fig. 4E. Administration of the PLD2-preferring inhibitor resulted in a modest yet significant increase in survival and in addition delayed mortality (Fig. 4D). Although 80% of the mice administered VU0364739 eventually succumbed, death occurred considerably later after infection (compared with influenza-infected mice administered vehicle), clearly providing an extended window for further supportive therapy in a clinical setting. This demonstrates that by inhibiting PLD activity in the mouse viral replication is decreased and that antiviral responses are up-regulated, leading to some protective benefits during a lethal influenza infection. No significant toxicity or neurological impairment was noted in rats receiving identical treatment (Tables 1 and 2). This inhibitor is a preclinical compound that has not been optimized for pharmacokinetic or pharmacodynamic properties, and yet it demonstrates robust effects on viral spread and reproduction.
TABLE 1.
Rat liver toxicity panel shows little effect of PLD inhibitor VU0364739
Only globulin and total protein showed higher levels with inhibitor treatment (n = 5–6 rats per treatment; p < 0.05 by Mann-Whitney test), and both were within normal clinical ranges for both the vehicle control and PLD inhibitor treatment groups. Principal component analysis of the z-score standardized values across the screening panel displayed no clustering of the multivariate results by treatment, and none of the principal components differed significantly (p > 0.05 by t test).
| Means ± S.E. |
Units | p value | ||
|---|---|---|---|---|
| Vehicle | VU0364739 | |||
| Albumin | 3.6 ± 0.1 | 3.6 ± 0.0 | g/dl | 0.5368 |
| Alkaline phosphatase | 212 ± 11 | 207 ± 13 | units/liter | 0.7922 |
| Alanine aminotransferase | 102 ± 8 | 113 ± 5 | units/liter | 0.5368 |
| Amylase | 1118 ± 59 | 1142 ± 41 | units/liter | 0.6623 |
| Blood urea nitrogen | 12.0 ± 2.0 | 15.6 ± 1.5 | mg/dl | 0.0823 |
| Calcium | 10.8 ± 0.2 | 11.0 ± 0.2 | mg/dl | 0.3290 |
| Creatinine | 0.33 ± 0.01 | 0.42 ± 0.04 | mg/dl | 0.1255 |
| Globulin | 2.3 ± 0.1 | 2.6 ± 0.1 | g/dl | 0.0303 |
| Glucose | 143 ± 7 | 141 ± 5 | mg/dl | 0.7922 |
| Potassium | 5.72 ± 0.42 | 5.72 ± 0.15 | mmol/liter | 0.6623 |
| Sodium | 139 ± 2 | 142 ± 1 | mmol/liter | 0.3290 |
| Phosphorus | 8.8 ± 0.2 | 9.0 ± 0.2 | mg/dl | 0.5368 |
| Total bilirubin | 0.22 ± 0.07 | 0.16 ± 0.01 | mg/dl | 0.7922 |
| Total protein | 5.9 ± 0.1 | 6.3 ± 0.1 | g/dl | 0.0303 |
TABLE 2.
Modified Irwin Neurological Battery with VU0364739
PLD inhibitor VU0364739 is without effect in a Modified Irwin Neurological Battery in rats. Changes in the Modified Irwin Neurological Battery were evaluated using a rating scale from 0 to 2: 0, no effect; 1, modest effects; 2, robust effect. Male Harlan Sprague-Dawley rats (n = 6; approximately 250 g) were pretreated with vehicle alone or a 13 mg/kg intraperitoneal dose of VU0364739 and then tested in the Irwin battery at 30 min after treatment and subsequently monitored for 8 h.
| Time |
|||||
|---|---|---|---|---|---|
| 30 min | 1 h | 2 h | 4 h | 8 h | |
| Autonomic nervous system | |||||
| Ptosis | 0 | 0 | 0 | 0 | 0 |
| Exophthalmus | 0 | 0 | 0 | 0 | 0 |
| Miosis | 0 | 0 | 0 | 0 | 0 |
| Mydriasis | 0 | 0 | 0 | 0 | 0 |
| Corneal reflex loss | 0 | 0 | 0 | 0 | 0 |
| Pinna reflex loss | 0 | 0 | 0 | 0 | 0 |
| Piloerection | 0 | 0 | 0 | 0 | 0 |
| Respiratory rate | 0 | 0 | 0 | 0 | 0 |
| Writhing | 0 | 0 | 0 | 0 | 0 |
| Tail erection | 0 | 0 | 0 | 0 | 0 |
| Lacrimation | 0 | 0 | 0 | 0 | 0 |
| Salivation | 0 | 0 | 0 | 0 | 0 |
| Vasodilatation | 0 | 0 | 0 | 0 | 0 |
| Skin color | 0 | 0 | 0 | 0 | 0 |
| Irritability | 0 | 0 | 0 | 0 | 0 |
| Loose stool | 0 | 0.25 | 0.5 | 0.4 | 0 |
| Rectal temperature (°C) | 37 | 37.3 | 37.4 | 37.2 | 37 |
| Somatomotor systems | |||||
| Motor activity | 0 | 0 | 0 | 0 | 0 |
| Convulsions | 0 | 0 | 0 | 0 | 0 |
| Arch/roll | 0 | 0 | 0 | 0 | 0 |
| Tremors | 0 | 0 | 0 | 0 | 0 |
| Leg weakness | 0 | 0 | 0 | 0 | 0 |
| Rigid stance | 0 | 0 | 0 | 0 | 0 |
| Spraddle | 0 | 0 | 0 | 0 | 0 |
| Placing loss | 0 | 0 | 0 | 0 | 0 |
| Grasping loss | 0 | 0 | 0 | 0 | 0 |
| Righting loss | 0 | 0 | 0 | 0 | 0 |
| Catalepsy | 0 | 0 | 0 | 0 | 0 |
| Tail pinch | 0 | 0 | 0 | 0 | 0 |
| Escape loss | 0 | 0 | 0 | 0 | 0 |
PLD2 Inhibition Alters Endocytosis Kinetics and Aggregation of Endocytic Proteins
Inhibition of PLD function has been shown to decrease uptake of ligands in various systems (9, 23, 24). Given that perturbation of influenza virus trafficking during the early infection process can lead to degradation of the entering virus (11), it was hypothesized that PLD inhibition during influenza infection led to alterations of entry events that occur in the first minutes of infection. To support this hypothesis, the effect of PLD2 inhibition on normal endocytosis rates was assessed using the established transferrin uptake model, a classic demonstration of a clathrin-dependent trafficking process.
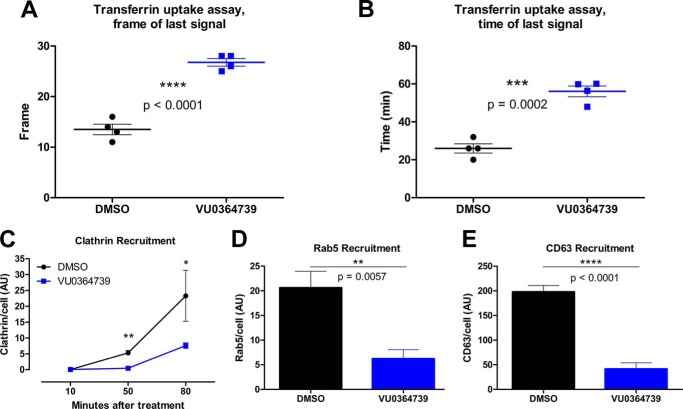
A549 cells pretreated with VU0364739 were labeled with fluorescent transferrin at 4 °C for 1 h and then placed into a heated microscope for live cell imaging. Images were recorded for 1 h, and the frame and time of the transferrin fluorescence disappearance was noted (Fig. 5, A and B). Cells treated with vehicle control were able to take up, traffic, and degrade transferrin by the 14th frame, corresponding to an average time of 26 min after warming. In contrast, cells treated with VU0364739 took ∼56 min to process the fluorescent ligand. These data indicate that inhibiting PLD2 activity with VU0364739 alters trafficking kinetics by extending the endocytosis process rather than acting as a strict blockade.
FIGURE 5.
PLD inhibition alters ligand trafficking kinetics and accumulation of endocytosis regulatory proteins. A and B, A549 cells were treated for 1 h with DMSO or VU0364739 (10 μm) prior to being labeled for 1 h at 4 °C with 100 nm Alexa Fluor 647-transferrin. Live cell imaging was used to assess the kinetics of transferrin uptake. To quantify this assay, both the frame (A) and the time (B) were used to determine when the fluorescent signal disappeared, signaling recycling of transferrin. Live cell imaging was performed using Slidebook imaging software, and frame time measurements were compared using t tests. Data are mean ± S.E. (error bars). C–E, A549 cells were treated for 1 h with DMSO or VU0364739 (10 μm) and then infected with 0.05 m.o.i. influenza A/California/04/2009 (H1N1). Cells were stained and examined by confocal microscopy for the accumulation of clathrin (C), Rab5 (early endosome) (D), and CD63 (late endosome) (E). Less clathrin is recruited at 50 and 80 min postinfection under inhibitor treatment, and less Rab5 (D) and CD63 (E) accumulate after 10 and 90 min postinfection. In these experiments, protein accumulation was defined by gating signal intensity as well as size such that higher y axis values represent bigger and brighter foci of signal. A two-way ANOVA with Bonferroni's post-test was used to examine data from C. t tests were used to analyze data from D and E with p values indicated. Data are mean ± S.E. (error bars). *, p < 0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001.
Influenza entry is not entirely dependent on clathrin-mediated endocytosis, but it is widely accepted that the majority of incoming viruses enter via clathrin-dependent events primarily by the de novo formation of clathrin-coated pits on the cell surface after exposure to influenza virus (25). After entry, influenza virus needs to be properly trafficked from the membrane to the nucleus. Along the way, the endosome is acidified (26), and the hemagglutinin protein undergoes a conformational change that creates a fusion pore between the viral and vesicle membranes through which viral ribonucleoproteins gain access to the cytoplasm, eventually entering the nucleus to initiate new virus production. The accumulation of trafficking-associated proteins was visualized by confocal microscopy in A549 cells during a 0.05 m.o.i. influenza A/California/04/2009 (H1N1) infection. Signal intensity and particle size were gated to determine protein accumulation. As proteins accumulate, the brightness and size of the fluorescence increases on a per cell basis. Clathrin recruitment was inhibited 50 min postinfection and remained significantly lower (up to 80 min postinfection) with VU0364739 administration (Fig. 5C). During the first phase of viral entry, Rab5 accumulated on the early endosomes; however, in the presence of VU0364739, much less Rab5 accumulated 10 min postinfection (Fig. 5D). Similarly, VU0364739 treatment led to a significant reduction in recruitment of the late endosome marker CD63 90 min postinfection (Fig. 5E). Vesicular trafficking is also important for key elements of the late stages of viral replication, and defects in Rab11 accumulation associated with viral protein trafficking to the membrane were consistently observed (data not shown). These data indicate that when PLD2 is inhibited during an infection the normal cascade of protein accumulation required for proper endosomal maturation is disrupted, leading to inefficient trafficking of incoming virus particles, indicating that PLD2 is a host factor required for the efficient trafficking of influenza virus once within the cell.
Protection by PLD2 Inhibition Requires Intact Innate Antiviral Signaling
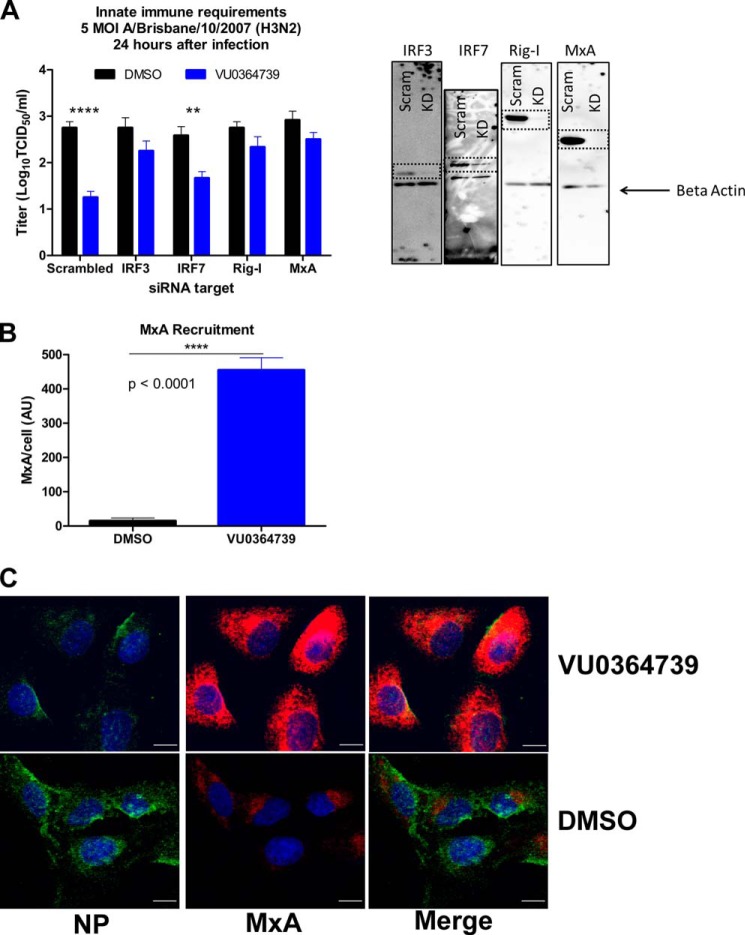
To assess whether the innate antiviral response was required for protection when cells were treated with the PLD2 inhibitor, an RNAi screen of essential innate immunity proteins in vitro was conducted. A549 cells were electroporated with 100 nm gene-specific siRNA and infected with 5 m.o.i. influenza A/Brisbane/10/2007 (H3N2) for 24 h; a decrease in protein expression was confirmed by Western blot analysis (data not shown). Viral titer was then measured using a traditional TCID50 assay. A549 cells treated with control siRNA were protected by VU0364739 administration (Fig. 6A), showing a decrease in titer of 1.5 log units. However, VU0364739 treatment was not sufficient to protect cells transfected with siRNA targeting IRF3, Rig-I, or MxA. These three proteins are known to be vital to the innate defense against influenza. Interestingly, cells transfected with siRNA targeting the transcription factor IRF7 were still protected by VU0364739 treatment but to a lower degree compared with cells transfected with control siRNA (0.9-log unit decrease in viral titer). These experiments demonstrate that altering trafficking kinetics with a PLD2 inhibitor alone was insufficient for complete protection from influenza infection. Cells need an intact antiviral response to attack and neutralize the invading virus, suggesting that PLD2 plays a critical role in the temporal-spatial regulation of endocytosis and innate antiviral sensing/defense during influenza infection. Disruption of PLD2 regulation of these events affords the host cell more time to detect the virus and mount an immune response.
FIGURE 6.
Innate immune factors are required for PLD2 inhibitor-mediated protection from influenza. A, A549 cells were transfected with siRNA targeting effectors or transcription factors involved in the type I IFN pathway and then subsequently infected with 5 m.o.i. influenza A/Brisbane/10/2007 (H3N2) for 24 h, and supernatant was titered on Madin-Darby canine kidney cells to measure viral reproduction. As a control, viral replication was significantly reduced in cells transfected with scrambled siRNA and treated with PLD2 inhibitor. When cells lack IRF3, Rig-I, or MxA, treatment with VU0364739 cannot protect against influenza infection. The innate requirements are restricted to IRF3-mediated signaling as protection was afforded to cells with IRF7 knocked down. Representative Western blots are presented to confirm loss of protein expression after RNAi. B, to demonstrate the robust innate antiviral response to influenza infection after PLD2 inhibition, A549 cells were infected with 1 m.o.i. influenza A/California/04/2009 (H1N1), and levels of MxA were assessed by confocal microscopy 30 min postinfection. C, MxA accumulates much more rapidly and to a much greater magnitude when PLD2 is inhibited. The dramatic MxA activity is visualized in representative images where green signal marks influenza NP, red signal is MxA, and blue signal is DAPI. Scale bars, 10 μm. A two-way ANOVA with Bonferroni's post-test was used to examine data from A. **, p < 0.01; ****, p < 0.0001. A t test was used to analyze data from B. ****, p < 0.0001. Data are mean ± S.E. (error bars).
One potential benefit of the delayed trafficking kinetics is the greater window allowed for innate immune interference during influenza infection. A key component of the innate response in the respiratory epithelium is MxA, a cytoplasmic dynamin-like GTPase, which is transcriptionally induced to high levels as part of the type I IFN response but is also found at basal levels in resting cells. MxA can interfere with early virus replication events by binding to viral NP (27). MxA recruitment was measured in A549 cells postinfection by the same manner used to measure the endosomal markers. When PLD2 was inhibited during infection, MxA was observed to accumulate in foci at significant levels as early as 30 min postinfection (Fig. 6, B and C). These data support a model whereby the slower rate of endocytosis following PLD2 inhibition allows cells more time to detect and respond to incoming influenza virus. Afforded this additional time, PLD inhibitor-treated host cells were able to mount an immune response of greater magnitude and efficacy to neutralize the incoming virus, thus promoting survival of the cell and protection from degradation of the invading virus.
DISCUSSION
PLD has been identified as a host restriction factor for influenza virus replication, and PLD catalytic activity is stimulated by influenza infection that colocalizes with NP-positive-staining structures over time. In addition, PLD2 functions to facilitate rapid endosomal trafficking, allowing the virus to escape an otherwise effective innate antiviral response. This response depends on Rig-I, IRF3, and the well known antiviral protein MxA but is largely independent of the antiviral targets of IRF7. Importantly, this work identified a host cell process exploited by the virus to maximize its reproductive capacity and accelerate life cycle kinetics, allowing the pathogen to escape innate immune detection and neutralization.
The roles of PLD in endosomal trafficking are well documented (9, 10). One advantage of targeting the PLD isoenzymes of host cells as an experimental antiviral strategy is that minimal evolution pressure would be exerted on the virus that would otherwise encourage mutation and variant viral escape. Importantly, these enzymes have been shown to be nonessential to animal survival when either the PLD1 or PLD2 genes are knocked out (28, 29). Although the amount of PA generated by PLD2 alone may not be a significant fraction of the total cellular PA pool, discrete processes are likely to be linked to individual sources of PA-PLD2. For example, Yang et al. (30) reported that PLD2 was involved in opioid receptor endocytosis via a PA-p38 MAPK pathway. This may explain the negligible levels of toxicity observed using VU0364739 in vivo.
PA serves a multitude of roles in membrane biogenesis and curvature as well as cellular signaling and has the potential to facilitate viral entry, replication, and egress. A detailed lipidomic analysis demonstrated that cytomegalovirus infection elicited a robust generation of PA in human fibroblasts (16). It is also interesting to note that PKC is a direct regulator of PLD activity, and the role of PKC in influenza virion transport (11) through the late endosome may include some as yet unelucidated role for PLD. A more recently described PLD isoform known as PLD3 or HuK4 is related by sequence homology to p37, an essential protein in vaccinia virus. Although this PLD isoenzyme is not as well characterized, genetic variants in PLD3 were recently found to be of predictive value for risk in Alzheimer disease (31). Overexpression of PLD3 leads to a significant decrease in intracellular amyloid-β precursor protein and extracellular Aβ42 and Aβ40. Although human PLD3 cannot complement a p37-deficient vaccinia virus (32), primary alcohols similarly inhibit its function and its localization in the endoplasmic reticulum, suggesting some conservation of function. These recent discoveries provide further evidence that lipid signaling is a central process in many disease states. The identification of a role for PLD in influenza entry and escape from innate immune detection expands our understanding that host cell signaling lipases have diverse and essential roles in viral infection and potentially offer a new series of therapeutic targets for pharmacological intervention.
Influenza virus and potentially other enveloped viruses hijack the endosomal compartments of infected host cells to hide from detection and to safely mature to a state that is favorable for efficient viral infection. Inhibition of PLD2 appears to alter the temporal spatial kinetics of this process. Despite the mode of entry, there is consensus that influenza virus must gain access to an endosome and be sorted to late endosomes with lower pH environments to undergo viral membrane-host membrane fusion. Our data suggest that when PLD2 is inhibited endosomal sorting occurs more slowly than normal. We chose to stain clathrin, Rab5, and CD63 to elucidate well known markers of intracellular trafficking. Taken together the results suggest that PLD2 inhibition perturbs multiple pathways of influenza entry. The findings suggest that these proteins accumulate more slowly when PLD2 is inhibited. Interestingly, PLD has been noted to be involved in various trafficking paradigms including clathrin, caveolin, and macropinocytosis (33–35). By shifting the kinetics of influenza trafficking, much less viral spread was observed using the spatial infection model method and reduced viral replication was observed using the traditional TCID50 and plaque assays. Concomitant with the shift in influenza trafficking, accumulation of antiviral sensors and effectors was observed. The protective mechanism was dependent on the innate immune system, indicating that the invading virus can be detected and the immune response mounted before the virus can begin an efficient infection. The inhibition of PLD effectively alters the host factor required by influenza to allow the infected cell to defend itself. Using the transferrin uptake model, the precise kinetics of ligand trafficking were defined, and it was demonstrated that without PLD function these events take approximately twice as long.
The recent development of isoform-preferring PLD inhibitors (12) presented an opportunity to disrupt the formation of PA by PLD and interrogate the contribution of each isoenzyme during influenza infection. Based on the results using small molecule inhibitors and RNAi, PLD2 is proposed as a host factor that is used for efficient influenza infection. Importantly, our data suggest that PLD1 also plays a role in influenza infection. PLD1 RNAi can inhibit influenza replication. The focus on PLD2 over PLD1 in this study stems from interests in viral entry and host factors that are exploited by influenza to gain entry. Because of their subcellular location, PLD1 associates with the Golgi complex, whereas PLD2 is distributed in the cell membrane. Hence, PLD2 regulation would be more relevant during entry, whereas PLD1 would have a role in viral egress, assembly, and budding (9) The role of PLD1 and its relationship to the signaling pathways in which PLD2 are also involved is an important area for future studies.
The PLD2 pathway is a potentially attractive target for development of therapeutic agents as minimal toxicity was observed from the administration of VU0364739 alone (any toxicity in vivo was associated with the vehicle, and no in vitro toxicity was observed), and knocking out PLD2 in a mouse model is not obviously deleterious. Additional research is necessary to decode how host factors are co-opted by viruses to aid their pathogenesis and the intervention necessary to prevent or minimize efficient viral infection. Because no current therapeutic alone is capable of completely protecting against influenza infection, future studies are required to evaluate the intriguing possibility that combinations of antiviral agents that work via distinct mechanisms could afford protection.
Acknowledgments
We thank Pavlina T. Ivanova, Stephen B. Milne, and David S. Myers for contributions to the mass spectrometry analysis of glycerophospholipid composition changes; Lana McClaren and Christine M. Oshansky for support in the lipid analysis and viral titer experiments; Michelle D. Armstrong for technical support and assistance with preparation of the manuscript; and the Cell and Tissue Imaging Center at St. Jude Children's Research Hospital, Paul Digard, and T. David K. Brown for assistance with microscopy and helpful conversations.
This work was supported, in whole or in part, by National Institutes of Health Grants U54MH084659, P01ESO013125, and HHSN2722008000058C and HHSN266200700005C from the NIAID. This work was also supported by Medical Research Council Grant RG53985.
- PLD
- phospholipase D
- Rig-I
- retinoic acid-inducible gene-1
- MxA
- myxovirus resistance gene A
- PC
- phosphatidylcholine
- PA
- phosphatidic acid
- NP
- nucleoprotein
- m.o.i.
- multiplicity of infection
- ANOVA
- analysis of variance
- TCID50
- median tissue culture infective dose
- LD50
- median lethal dose
- EID50
- 50% embryo infective dose.
REFERENCES
- 1. Rossman J. S., Jing X., Leser G. P., Lamb R. A. (2010) Influenza virus M2 protein mediates ESCRT-independent membrane scission. Cell 142, 902–913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Blanc M., Hsieh W. Y., Robertson K. A., Kropp K. A., Forster T., Shui G., Lacaze P., Watterson S., Griffiths S. J., Spann N. J., Meljon A., Talbot S., Krishnan K., Covey D. F., Wenk M. R., Craigon M., Ruzsics Z., Haas J., Angulo A., Griffiths W. J., Glass C. K., Wang Y., Ghazal P. (2013) The transcription factor STAT-1 couples macrophage synthesis of 25-hydroxycholesterol to the interferon antiviral response. Immunity 38, 106–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Morita M., Kuba K., Ichikawa A., Nakayama M., Katahira J., Iwamoto R., Watanebe T., Sakabe S., Daidoji T., Nakamura S., Kadowaki A., Ohto T., Nakanishi H., Taguchi R., Nakaya T., Murakami M., Yoneda Y., Arai H., Kawaoka Y., Penninger J. M., Arita M., Imai Y. (2013) The lipid mediator protectin D1 inhibits influenza virus replication and improves severe influenza. Cell 153, 112–125 [DOI] [PubMed] [Google Scholar]
- 4. Tam V. C., Quehenberger O., Oshansky C. M., Suen R., Armando A. M., Treuting P. M., Thomas P. G., Dennis E. A., Aderem A. (2013) Lipidomic profiling of influenza infection identifies mediators that induce and resolve inflammation. Cell 154, 213–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ehrhardt C., Marjuki H., Wolff T., Nürnberg B., Planz O., Pleschka S., Ludwig S. (2006) Bivalent role of the phosphatidylinositol-3-kinase (PI3K) during influenza virus infection and host cell defence. Cell. Microbiol. 8, 1336–1348 [DOI] [PubMed] [Google Scholar]
- 6. Chen C., Zhuang X. (2008) Epsin 1 is a cargo-specific adaptor for the clathrin-mediated endocytosis of the influenza virus. Proc. Natl. Acad. Sci. U.S.A. 105, 11790–11795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Carrasco M., Amorim M. J., Digard P. (2004) Lipid raft-dependent targeting of the influenza A virus nucleoprotein to the apical plasma membrane. Traffic 5, 979–992 [DOI] [PubMed] [Google Scholar]
- 8. Campbell S. M., Crowe S. M., Mak J. (2001) Lipid rafts and HIV-1: from viral entry to assembly of progeny virions. J. Clin. Virol. 22, 217–227 [DOI] [PubMed] [Google Scholar]
- 9. Roth M. G. (2008) Molecular mechanisms of PLD function in membrane traffic. Traffic 9, 1233–1239 [DOI] [PubMed] [Google Scholar]
- 10. Selvy P. E., Lavieri R. R., Lindsley C. W., Brown H. A. (2011) Phospholipase D: enzymology, functionality, and chemical modulation. Chem. Rev. 111, 6064–6119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sieczkarski S. B., Brown H. A., Whittaker G. R. (2003) Role of protein kinase C βII in influenza virus entry via late endosomes. J. Virol. 77, 460–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Scott S. A., Selvy P. E., Buck J. R., Cho H. P., Criswell T. L., Thomas A. L., Armstrong M. D., Arteaga C. L., Lindsley C. W., Brown H. A. (2009) Design of isoform-selective phospholipase D inhibitors that modulate cancer cell invasiveness. Nat. Chem. Biol. 5, 108–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lavieri R. R., Scott S. A., Selvy P. E., Kim K., Jadhav S., Morrison R. D., Daniels J. S., Brown H. A., Lindsley C. W. (2010) Design, synthesis, and biological evaluation of halogenated N-(2-(4-Oxo-1-phenyl-1,3,8-triazaspiro[4.5]decan-8-yl)ethyl)benzamides: discovery of an isoform-selective small molecule phospholipase D2 inhibitor. J. Med. Chem. 53, 6706–6719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Scott S. A., Mathews T. P., Ivanova P. T., Lindsley C. W., Brown H. A. (2014) Chemical modulation of glycerolipid signaling and metabolic pathways. Biochim. Biophys. Acta 1841, 1060–1084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bligh E. G., Dyer W. J. (1959) A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 37, 911–917 [DOI] [PubMed] [Google Scholar]
- 16. Liu S. T., Sharon-Friling R., Ivanova P., Milne S. B., Myers D. S., Rabinowitz J. D., Brown H. A., Shenk T. (2011) Synaptic vesicle-like lipidome of human cytomegalovirus virions reveals a role for SNARE machinery in virion egress. Proc. Natl. Acad. Sci. U.S.A. 108, 12869–12874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Brown H. A., Henage L. G., Preininger A. M., Xiang Y., Exton J. H. (2007) Biochemical analysis of phospholipase D. Methods Enzymol. 434, 49–87 [DOI] [PubMed] [Google Scholar]
- 18. Ivanova P. T., Milne S. B., Byrne M. O., Xiang Y., Brown H. A. (2007) Glycerophospholipid identification and quantitation by electrospray ionization mass spectrometry. Methods Enzymol. 432, 21–57 [DOI] [PubMed] [Google Scholar]
- 19. Myers D. S., Ivanova P. T., Milne S. B., Brown H. A. (2011) Quantitative analysis of glycerophospholipids by LC-MS: acquisition, data handling, and interpretation. Biochim. Biophys. Acta 1811, 748–757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lam V., Duca K. A., Yin J. (2005) Arrested spread of vesicular stomatitis virus infections in vitro depends on interferon-mediated antiviral activity. Biotechnol. Bioeng. 90, 793–804 [DOI] [PubMed] [Google Scholar]
- 21. Scott S. A., Xiang Y., Mathews T. P., Cho H. P., Myers D. S., Armstrong M. D., Tallman K. A., O'Reilly M. C., Lindsley C. W., Brown H. A. (2013) Regulation of phospholipase D activity and phosphatidic acid production after purinergic (P2Y6) receptor stimulation. J. Biol. Chem. 288, 20477–20487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Everitt A. R., Clare S., Pertel T., John S. P., Wash R. S., Smith S. E., Chin C. R., Feeley E. M., Sims J. S., Adams D. J., Wise H. M., Kane L., Goulding D., Digard P., Anttila V., Baillie J. K., Walsh T. S., Hume D. A., Palotie A., Xue Y., Colonna V., Tyler-Smith C., Dunning J., Gordon S. B., GenISIS Investigators, MOSAIC Investigators, Smyth R. L., Openshaw P. J., Dougan G., Brass A. L., Kellam P. (2012) IFITM3 restricts the morbidity and mortality associated with influenza. Nature 484, 519–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Antonescu C. N., Danuser G., Schmid S. L. (2010) Phosphatidic acid plays a regulatory role in clathrin-mediated endocytosis. Mol. Biol. Cell 21, 2944–2952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Corrotte M., Chasserot-Golaz S., Huang P., Du G., Ktistakis N. T., Frohman M. A., Vitale N., Bader M.-F., Grant N. J. (2006) Dynamics and function of phospholipase D and phosphatidic acid during phagocytosis. Traffic 7, 365–377 [DOI] [PubMed] [Google Scholar]
- 25. Lakadamyali M., Rust M. J., Zhuang X. (2004) Endocytosis of influenza viruses. Microbes Infect. 6, 929–936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Skehel J. J., Cross K., Steinhauer D., Wiley D. C. (2001) Influenza fusion peptides. Biochem. Soc. Trans. 29, 623–626 [DOI] [PubMed] [Google Scholar]
- 27. Verhelst J., Parthoens E., Schepens B., Fiers W., Saelens X. (2012) Interferon-inducible Mx1 protein inhibits influenza virus by interfering with functional viral ribonucleoprotein complex assembly. J. Virol. 86, 13445–13455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Elvers M., Stegner D., Hagedorn I., Kleinschnitz C., Braun A., Kuijpers M. E., Boesl M., Chen Q., Heemskerk J. W., Stoll G., Frohman M. A., Nieswandt B. (2010) Impaired αIIbβ3 integrin activation and shear-dependent thrombus formation in mice lacking phospholipase D1. Sci. Signal. 3, ra1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Oliveira T. G., Chan R. B., Tian H., Laredo M., Shui G., Staniszewski A., Zhang H., Wang L., Kim T.-W., Duff K. E., Wenk M. R., Arancio O., Di Paolo G. (2010) Phospholipase D2 ablation ameliorates Alzheimer's disease-linked synaptic dysfunction and cognitive deficits. J. Neurosci. 30, 16419–16428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Yang L., Seifert A., Wu D., Wang X., Rankovic V., Schröder H., Brandenburg L. O., Höllt V., Koch T. (2010) Role of phospholipase D2/phosphatidic acid signal transduction in μ- and δ-opioid receptor endocytosis. Mol. Pharmacol. 78, 105–113 [DOI] [PubMed] [Google Scholar]
- 31. Cruchaga C., Karch C. M., Jin S. C., Benitez B. A., Cai Y., Guerreiro R., Harari O., Norton J., Budde J., Bertelsen S., Jeng A. T., Cooper B., Skorupa T., Carrell D., Levitch D., Hsu S., Choi J., Ryten M., UK Brain Expression Consortium, Sassi C., Bras J., Gibbs J. R., Hernandez D. G., Lupton M. K., Powell J., Forabosco P., Ridge P. G., Corcoran C. D., Tschanz J. T., Norton M. C., Munger R. G., Schmutz C., Leary M., Demirci F. Y., Bamne M. N., Wang X., Lopez O. L., Ganguli M., Medway C., Turton J., Lord J., Braae A., Barber I., Brown K., Alzheimer's Research UK Consortium, Pastor P., Lorenzo-Betancor O., Brkanac Z., Scott E., Topol E., Morgan K., Rogaeva E., Singleton A. B., Hardy J., Kamboh M. I., St George-Hyslop P., Cairns N., Morris J. C., Kauwe J. S., Goate A. M. (2014) Rare coding variants in the phospholipase D3 gene confer risk for Alzheimer's disease. Nature 505, 550–554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Munck A., Böhm C., Seibel N.M., Hashemol Hosseini Z., Hampe W. (2005) Hu-K4 is a ubiquitously expressed type 2 transmembrane protein associated with the endoplasmic reticulum. FEBS J. 272, 1718–1726 [DOI] [PubMed] [Google Scholar]
- 33. Padrón D., Tall R. D., Roth M. G. (2006) Phospholipase D2 is required for efficient endocytic recycling of transferrin receptors. Mol. Biol. Cell 17, 598–606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Czarny M., Fiucci G., Lavie Y., Banno Y., Nozawa Y., Liscovitch M. (2000) Phospholipase D2: functional interaction with caveolin in low-density membrane microdomains. FEBS Lett. 467, 326–332 [DOI] [PubMed] [Google Scholar]
- 35. Mahankali M., Peng H. J., Cox D., Gomez-Cambronero J. (2011) The mechanism of cell membrane ruffling relies on a phospholipase D2 (PLD2), Grb2 and Rac2 association. Cell. Signal. 23, 1291–1298 [DOI] [PMC free article] [PubMed] [Google Scholar]