SUMMARY
The serine/threonine kinase LKB1 is a tumor suppressor whose loss is associated with increased metastatic potential. In an effort to define biochemical signatures of metastasis associated with LKB1 loss, we discovered that the EMT transcription factor Snail1 was uniquely upregulated upon LKB1-deficiency across cell types. The ability of LKB1 to suppress Snail1 levels was independent of AMPK, but required the related kinases MARK1 and MARK4. In a screen for substrates of these kinases involved in Snail regulation, we identified the scaffolding protein DIXDC1. Similar to loss of LKB1, DIXDC1 depletion results in upregulation of Snail1 in a FAK-dependent manner, leading to increased cell invasion. MARK1 phosphorylation of DIXDC1 is required for its localization to focal adhesions and ability to suppress metastasis in mice. DIXDC1 is frequently downregulated in human cancers, which correlates with poor survival. This study defines a novel AMPK-independent phosphorylation cascade essential for LKB1-dependent control of metastatic behavior.
INTRODUCTION
A critical question in cancer biology is the relationship between tumor initiating mutations, including oncogenes and tumor suppressor genes, and the propensity for tumors to metastasize (Hanahan and Weinberg, 2011). LKB1/STK11 is the causal gene inactivated in the inherited cancer disorder Peutz-Jeghers Syndrome and is also inactivated in ~25% of non-small cell lung cancers (Ding et al., 2008). Beyond effects on tumor initiation, loss of Lkb1 uniquely confers invasive and metastatic behavior in genetically engineered mouse models of cancer when directly compared to other tumor suppressors (e.g. Trp53, Rb, p16/Cdkn2a) (Carretero et al., 2010; Contreras et al., 2010; Ji et al., 2007; Liu et al., 2012). The enhanced metastatic potential of Lkb1-deficient tumors is also notable for occurring in cancers across multiple lineages, namely endometrium, lung, and melanoma (Contreras et al., 2008; Ji et al., 2007; Liu et al., 2012).
LKB1 is a protein kinase that through direct phosphorylation activates a family of 14 kinases related to the AMP-activated Protein Kinase (AMPK) (Hardie and Alessi, 2013). Through these downstream kinases, LKB1 has highly conserved functions in organismal metabolism and cell polarity, in addition to its tumor suppressor function (Jansen et al., 2009). Many of the best understood functions of LKB1 are attributable to its ability to activate AMPK, a central conserved regulator of metabolism and cell growth (Mihaylova and Shaw, 2011). However, LKB1 has been established as a highly conserved regulator of cell polarity as well, stemming from initial findings in Drosophila and C. elegans, where the LKB1 ortholog was identified as Partitioning-defective 4 (par-4) (Jansen et al., 2009). In comparison to AMPK, far less is known about the biological functions and molecular targets of the other kinases activated by LKB1, though one subfamily, the Microtubule Affinity-Regulating Kinase (MARKs) (or Partitioning-defective 1/Par1) genes, control apical-basal polarity and may be primary mediators of LKB1 action in that process (Ollila and Makela, 2011).
Numerous downstream substrates of AMPK and its related kinases have been identified that provide molecular mechanisms underlying how LKB1 coordinates cell growth and metabolism (Hardie and Alessi, 2013). For example, AMPK regulates cell growth and survival through substrates in the mTORC1 and autophagy pathways. AMPK and related LKB1-dependent kinases coordinate cellular and organismal metabolism through phosphorylation of metabolic enzymes, glucose transport regulators, and transcriptional regulators.
Despite this insight into how LKB1 loss leads to metabolic derangement and growth of primary tumors, there are no well-established molecular connections between AMPK or any of its related kinases with effectors known to be involved in the control of metastasis. Moreover, it is not known whether the potent metastasis suppressing activity of LKB1 is mediated by AMPK or by other kinases that it activates. We report here the identification of a direct substrate of the AMPK-related kinases MARK1 and MARK4 that appears to mediate effects of LKB1 on epithelial-mesenchymal transition, migration, and metastatic behavior in vivo.
RESULTS
Snail is upregulated from loss of LKB1 or its downstream kinases MARK1 and MARK4, independent of AMPK
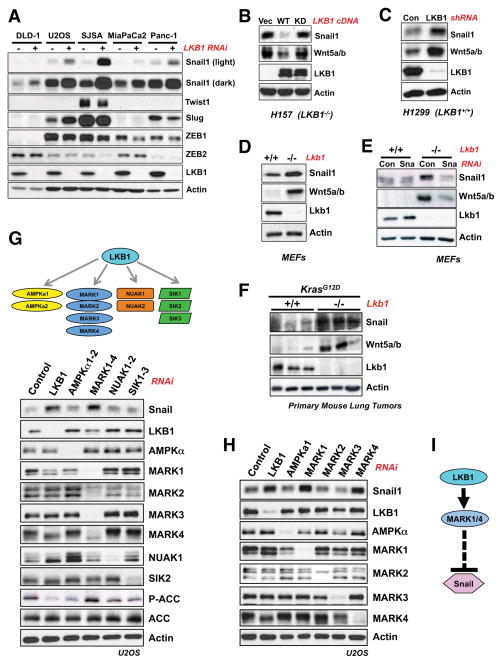
Given that LKB1-deficiency promotes aggressive metastatic behavior of diverse tumor types (Contreras et al., 2008; Ji et al., 2007; Liu et al., 2012), we sought to identify downstream effectors of LKB1 that control metastasis. Hallmarks of the metastatic cascade include spatially restrained epithelial cells acquiring the ability to invade local tissue, intravasate, and extravasate at distant sites. Experimental evidence suggests that this cascade can be triggered by the induction of an epithelial to mesenchymal transition (EMT), a transcriptional program orchestrated by re-expression of conserved transcription factors critical in embryonic development (Chaffer and Weinberg, 2011). We therefore investigated whether any EMT transcription factors were repressed by LKB1 by silencing LKB1 in a panel of LKB1-proficient tumor cell lines. Across cell lines from diverse tumor types, the transcription factor Snail1 was consistently upregulated when LKB1 was suppressed, in contrast to expression of Snail2 (Slug), ZEB1, ZEB2, or Twist (Figure 1A). In human NSCLC cell lines bearing LKB1 inactivation, expression of wild-type (WT) but not kinase-dead LKB1 reduced Snail1 levels (Figure 1B) and LKB1 knockdown in a LKB1-proficient NSCLC cell line increased Snail1 levels (Figure 1C). Surprisingly, the ability of LKB1 to suppress Snail1 was not confined to tumor cells or cells bearing oncogenic signals, as Lkb1−/− mouse embryonic fibroblasts (MEFs) exhibited Snail1 upregulation compared to littermate Lkb1+/+ MEFs (Figure 1D). In Lkb1-deficient NSCLC cells and MEFs, LKB1 re-expression suppressed Snail at the mRNA level (Figure S1A, S1B).
Figure 1. LKB1 regulates the EMT transcription factor Snail1 in a manner dependent on MARK1 and MARK4, but independent of AMPK.
(A) RNAi knockdown of LKB1 (20 μM) was performed across multiple human cancer cell lines of diverse tissue origin. DLD-1 cells are colorectal adenocarcinoma, U2OS and SJSA are osteosarcoma, and MiaPaCa2 and Panc-1 are pancreatic adenocarcinoma cell lines. Knockdown was performed for 72 hours and lysates were analyzed by western blotting with indicated antibodies.
(B) H157 human non-small cell lung cancer cells (LKB1-null) were reconstituted with a WT LKB1 (WT) or the kinase-dead K78I mutant (KD). Expression of Snail1 and its target gene Wnt5a were analyzed by western blotting. (Vec) denotes control reconstitution.
(C) RNAi knockdown of LKB1 (20 μM) for 72 hours in H1299 human lung cancer cells (LKB1-wt).
(D) Western blotting for Snail1 and its target gene Wnt5a in Lkb1+/+ and Lkb1fl/fl MEFs following infection with Ad-Cre.
(E) MEFs as described in (D) were transfected with siRNA oligos (50 μM) targeting mouse Snai1 (Sna). Expression of the Snail1 target Wnt5a was analyzed by western blotting.
(F) Primary mouse lung tumors from KrasG12D/+; Lkb1+/+ or KrasG12D/+; Lkb1fl/fl mice immunoblotted with indicated antibodies.
(G) LKB1 is the master upstream activating kinase of the AMPKR kinase family. siRNA oligos targeting AMPKR subfamilies were transfected into U2OS at a final concentration of 20 μM for 72 hours. Lysates were analyzed by western blotting with indicated antibodies.
(H) RNAi knockdown of individual MARK isoforms (20 μM) or AMPKα subunits (20 μM final) for 72 hours and immunoblotted with indicated antibodies.
(I) Pathway model. LKB1, functioning through the AMPK-related kinases MARK1 and MARK4, specifically represses the levels of the EMT transcription factor Snail1.
A core function of Snail family transcription factors is to create a cellular state favorable to cell migration and invasion (Thiery et al., 2009). Snail functions through repression of the E-cadherin promoter but also through transcriptional induction (Rembold et al., 2014) of mRNAs for extracellular matrix components, metalloproteinases, and numerous secreted growth factors, including the noncanonical Wnt ligands, Wnt5a and Wnt5b (Moreno-Bueno et al., 2006; Ren et al., 2011). As Wnt5a mRNA is highly upregulated in Lkb1-deficient gastrointestinal polyps (Lai et al., 2011), we examined the relationship between Wnt5a and Wnt5b levels and Snail1 levels across cell types. We found Snail1 was necessary (Figure S1C) and sufficient (Figure S1D) for induction of Wnt5a and Wnt5b in U2OS cells. Similarly, Wnt5a/Wnt5b levels paralleled Snail protein levels in various cell types when LKB1 was silenced (Figure 1B, 1C, 1D, S1B). Moreover, elevated Wnt5a/Wnt5b in Lkb1−/− MEFs was attenuated by knockdown of Snail1 (Figure 1E). Collectively these results indicate that Snail is necessary and sufficient for Wnt5a/5b expression in LKB1-deficient contexts, suggesting Wnt5a/5b levels may serve here as biomarkers of Snail activity.
Importantly, Snail expression was higher in lysates from lung tumors isolated from KrasLSL-G12D/+ Lkb1L/L mice than in tumors from KrasLSL-G12D/+ Lkb1+/+ mice, and Wnt5a/b levels paralleled elevated Snail levels here (Figure 1F). Since LKB1-deficiency enhances metastatic potential in KrasLSL-G12D/+ lung tumors (Ji et al., 2007), this is a specific context in which Snail could be mediating the bona fide metastasis-suppressing function of LKB1. We therefore sought to further elucidate the molecular mechanisms by which LKB1 controls Snail levels across cell types.
Because LKB1 can activate multiple AMPK-related kinases (“AMPKRs”), we first examined which downstream kinases controlled Snail levels. For screening purposes, we utilized U2OS cells as a human cell system in which LKB1 signaling is fully intact, but can be readily suppressed by RNAi-mediated silencing of LKB1. As previously observed (Figure 1A), LKB1 depletion in U2OS cells resulted in elevated Snail levels, yet surprisingly combined knockdown of the two genes encoding the AMPK catalytic subunits (AMPKa1 and AMPKa2) had no effect, even though phosphorylation of the AMPK substrate ACC was fully suppressed (Figure 1G). In contrast, knockdown of all four members of the MARK/Par-1 kinase subfamily resulted in robust Snail induction (Figure 1G). Knockdown of each of the MARK family members individually revealed that MARK1 (also known as Par1c) and MARK4 (Par1d) were most critical to suppression of Snail in these cells (Figure 1H), while the related kinases MARK2 and MARK3 had no effect on Snail levels. Deconvolution of the RNAi pools for LKB1 and MARK1 revealed that multiple independent siRNA duplexes against each target resulted in Snail induction, indicating that our observations are unlikely to be due to off-target silencing of unintended genes (Figure S1E).
Identification of DIXDC1 as a novel substrate of MARK1 and MARK that suppresses Snail levels
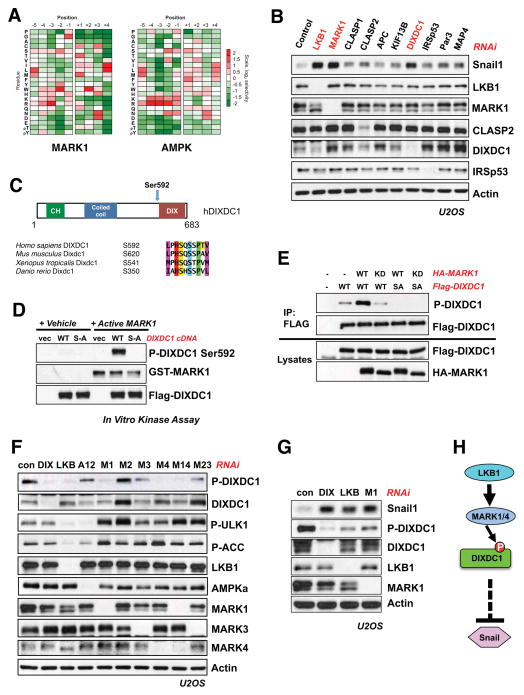
Next we sought to further dissect the mechanism by which MARK1 and MARK4 regulate Snail levels, as very little is known about these two kinases (Figure 1I). We have previously identified novel direct substrates of AMPK based on our determination of its phosphorylation site consensus motif using arrayed positional scanning peptide libraries (Figure S2A) (Egan et al., 2011; Gwinn et al., 2008; Mihaylova et al., 2011; Turk et al., 2006). To identify substrates of MARKs, we determined the substrate consensus motif for all four MARK kinases using the same method, and found them to have nearly identical profiles (Figure 2A, S2B). The optimal in vitro peptide phosphorylation sequence for the MARK kinases (“MARK motif”) was also quite similar to our previous described AMPK motif (Gwinn et al., 2008) (Figure 2A). This common consensus sequence includes strong preferences for aliphatic residues at positions −5 and +4 relative to the phosphorylation site, as well as for a basic residue at position −3. The similarity of these optimal sequences is consistent with previous studies demonstrating that phosphorylation sites induced by distinct members of the AMPKR family are similar in their primary sequence, and corresponds well with a survey of well-studied AMPK and MARK substrate phosphorylation sites (Shackelford and Shaw, 2009).
Figure 2. DIXDC1 is a novel substrate of MARK1 that suppresses Snail.
(A) Optimal phosphorylation consensus motifs were determined for all of the MARK kinases and compared to that of AMPK (Gwinn et al., 2008). The consensus motif for MARK1 is shown. In vitro kinase assays were performed using an arrayed set of peptide mixtures in which the indicated residue was fixed at the indicated position relative to a central phosphorylation site residue. The level of phosphorylation of each peptide mixture was quantified and normalized to an average value of 1 at each position to generate the displayed heat maps.
(B) siRNA knockdown (20 μM) of known and potential MARK substrates implicated in cytoskeletal signaling that were identified in the screen. Lysates were immunoblotted for induction of Snail compared to siRNA knockdown of LKB1 and MARK1 (20 μM).
(C) DIXDC1 domain structure and conservation of identified S592 residue across vertebrates.
(D) In vitro-IP kinase assay using recombinant Active MARK1. FLAG-tagged WT or S592A DIXDC1 was expressed and immunoprecipitated from HEK293T cells. Isolated DIXDC1 was then subjected to an in vitro kinase reaction with human MARK1. Reactions were immunoblotted with a phospho-specific antibody raised against the S592 residue. Vec = empty vector.
(E) MARK1 WT or KD (kinase-dead) cDNA constructs were expressed in HEK293T cells. DIXDC1 was immunoprecipitated and immunoblotted with P-DIXDC1 S592 antibody.
(F) siRNA knockdown for indicated genes (20 μM final) was performed in U2OS cells for 72 hrs. Lysates were immunoblotted with indicated antibodies. Key: Control RNAi (Con), AMPKα1 and AMPKα2 (A12), MARK1 (M1), MARK2 (M2), MARK3 (M3), MARK4 (M4), MARK1 and MARK4 (M14), MARK2 and MARK3 (M23).
(G) siRNA knockdown for indicated genes (20 μM) in U2OS cells for 72 hr. Lysates were immunoblotted with indicated antibodies.
(H) Pathway model. DIXDC1 is a novel LKB1-dependent substrate of MARK1 and MARK4 that represses Snail1 through an unknown mechanism..
We scanned the human proteome in silico to identify proteins containing sequences that matched the optimal MARK substrate motif. Repeating the search on other eukaryotic proteomes allowed us to narrow the initial list of ~1000 candidate sites in humans down to ~300 highly conserved sites (Figure S2B). Given that control of the cytoskeleton is a major conserved function of the MARK/Par1 kinase family, we focused our attention on 25 candidate MARK substrates bearing conserved phosphorylation motifs that either contained a Pfam domain or gene ontology (GO) designation related to the cytoskeleton. As a first test of whether these 25 candidates may be regulated by MARK kinases, we examined whether exogenous gain and loss of function of MARK1 kinase activity could regulate the phosphorylation of these proteins, as measured indirectly through their binding to the phospho-dependent binding scaffold protein 14-3-3 or recognition by a phospho-motif antibody generated against the optimal AMPK family substrate motif (Gwinn et al., 2008). Many of the known substrates of AMPK, the MARKs, and their related kinases inducibly bind to 14-3-3 upon their phosphorylation by these kinases (Shackelford and Shaw, 2009). This screening approach revealed a handful of previously reported MARK substrates - MAP4, Par3, KIF13B, IRSp53 (see Figure S2B, S2C), as well as four novel targets whose phosphorylation was controlled in a MARK-dependent manner (CLASP1, CLASP2, APC, DIXDC1) (Figure S2B, S2D).
We tested whether knockdown of the four known and four novel MARK substrates could decrease or increase basal Snail expression levels in U2OS cells. Notably, loss of the novel target DIXDC1 demonstrated the greatest effect, triggering an upregulation of Snail levels similar to that induced by knockdown of MARK1 or LKB1 (Figure 2B). Three distinct siRNA oligos targeting DIXDC1 resulted in upregulation of Snail, ruling out an off-target effect of the siRNA pool used (Figure S2E).
DIXDC1 (also known as CCD1) is a poorly-studied scaffolding protein composed of three protein-protein interaction domains: an actin-binding calponin homology (CH) domain, a coiled-coil (CC) domain, and a dishevelled and axin (DIX) oligomerization domain (Figure 2C). Notably, the conserved candidate MARK phosphorylation site in DIXDC1, Ser592, lies just at the start of the DIX domain (Figure 2C). Given that WT but not kinase-dead MARK1 induced binding of DIXDC1 to 14-3-3, we examined whether the putative MARK1 phosphorylation site Ser592 was required for this association. Preventing phosphorylation by mutating Ser592 to Ala completely abolished the ability of wild-type MARK1 to induce DIXDC1 to bind 14-3-3 (Figure S2F). Recombinant, active MARK1 phosphorylated DIXDC1 in vitro, as judged by induced reactivity of WT, but not S529A mutant, DIXDC1 with a phospho-specific antibody raised against the site (Figure 2D). Similar results were observed with MARK4, demonstrating that both of these MARK kinases can phosphorylate DIXDC1 at Ser592 in vitro (Figure S2E). Using the phospho-specific antibody, we observed that overexpression of either WT MARK1, or LKB1 combined with its activating subunit STRADα, increased Ser592 phosphorylation of co-expressed DIXDC1 in HEK293T cells, while kinase inactive MARK1 was without effect (Figure S2H). These experiments indicate that exogenously expressed DIXDC1 can serve as a target for LKB1- and MARK1-dependent signals in cells.
To verify the endogenous kinase(s) responsible for phosphorylation of endogenous DIXDC1 at Ser592, we performed knockdown for LKB1, AMPK, and individual MARK family kinases. Silencing of LKB1, MARK1, or MARK4 resulted in loss of endogenous Ser592 DIXDC1 phosphorylation, while knockdown of AMPK or MARK2 or MARK3 had no effect (Figure 2F). The regulation of DIXDC1 Ser592 phosphorylation by MARK1 and MARK4 paralleled our observations that these kinases suppress Snail levels (Figure 1H). Furthermore, levels of phospho-Ser592 DIXDC1 were inversely correlated with the level of Snail expression (Figure 2G). These data suggest that the ability of DIXDC1 to suppress Snail levels may require LKB1- and MARK1/4- dependent phosphorylation of DIXDC1 Ser592. DIXDC1 knockdown also resulted in increased Wnt5a/b, an effect that required Snail (Figure S2I) as previously noted in the context of LKB1-deficiency (Figure 1E).
DIXDC1 localizes to focal adhesions in a Serine 592-dependent manner and regulates their maturation
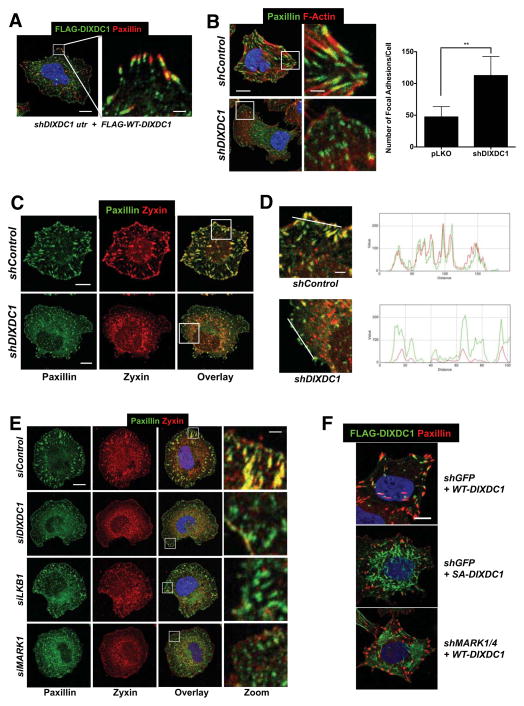
To investigate how DIXDC1 controls Snail levels, we first examined its subcellular localization. In U2OS cells, stably expressed epitope-tagged DIXDC1 localized primarily to focal adhesions, consistent the reported localization of endogenous DIXDC1 (Wang et al., 2006). DIXDC1 localized just distal to the focal adhesion protein paxillin, in a region where the focal adhesion complexes link to the intracellular actin cytoskeleton (Wehrle-Haller, 2012).
To determine if DIXDC1 affects focal adhesion dynamics, we examined the morphology of focal adhesions in U2OS knocked down for DIXDC1 using paxillin and zyxin as a markers for total and mature adhesions, respectively (Zaidel-Bar et al., 2003). At one hour post-plating, we noticed a striking contrast between mature, actin-linked adhesion plaques in control cells and an abundance of nascent, immature focal contacts in DIXDC1 shRNA cells, as judged by paxillin staining (Figure 3B, C). At this time, zyxin perfectly colocalized with paxillin in control cells, but was largely absent from focal contacts in DIXDC1 knockdown cells, indicating a defect in focal adhesion maturation (Figure 3C, D). Collectively, these experiments suggest for the first time that DIXDC1 controls the dynamics of focal adhesions. Importantly, knockdown for LKB1 or MARK1 in U2OS cells phenocopied the effect of DIXDC1 knockdown on focal adhesions as observed by zyxin and paxillin co-staining (Figure 3E).
Figure 3. DIXDC1 localizes to focal adhesions and regulates their maturation dependent on Ser592 phosphorylation.
(A) Immunolocalization of paxillin and stably expressed FLAG-DIXDC1 by lentiviral transduction in U2OS cells depleted for endogenous DIXDC1 by lentiviral shRNA transduction. Serum-starved cells were placed in suspension for 1 hr in serum-free media followed by plating onto collagen I coated coverslips in serum-free media for 1 hr to stimulate and synchronize focal adhesion formation. Scale bar = 10 μm (Left image). Scale bar = 2 μm (Zoom image).
(B) Immunolocalization of paxillin in U2OS cells stably depleted for DIXDC1 by lentiviral shRNA transduction and plated as described in (A). Representative image shown and total focal adhesion number/cell is quantified at right (n = 50 cells/condition). Scale bar = 10 μm. Scale bar = 2 μm (Zoom image). ** p < .0001 compared to pLKO control. Statistical analysis performed using an unpaired Student’s t-test.
(C) Colocalization of paxillin and zyxin in U2OS cells stably depleted for DIXDC1 by lentiviral shRNA transduction plated as described in (A). Scale bar = 10 μm.
(D) Magnified images of boxed regions in (C) showing individual spectra for paxillin and zyxin signal at peripheral adhesions. Scale bar = 2 μm.
(E) siRNA depletion of indicated genes (20 μM) in U2OS cells followed by paxillin and zyxin colocalization. Representative images are shown. Scale bar = 10 μm (Left images).
Scale bar = 1 μm (Zoom image).
(F) FLAG-DIXDC1 WT and S592A were transiently expressed by low level lentiviral transduction in U2OS cells. 24 hr post infection, cells were plated as described in (A). Focal adhesions were marked by immunostaining for paxillin. Scale bar = 10 μm.
To directly examine the effect of MARK1 phosphorylation of Ser592 on DIXDC1 action at focal adhesions, we compared stable cell lines expressing FLAG- DIXDC1WT or FLAG-DIXDC1S592A to cells with FLAG-DIXDC1WT but with shRNAs to MARK1 and MARK4. Notably, non-phosphorylatable FLAG-DIXDC1S592A was no longer capable of localizing to focal adhesions like FLAG-DIXDC1WT (Figure 3F). FLAG-DIXDC1WT was similarly mislocalized away from focal adhesions by knockdown for MARK1 and MARK4 (Figure 3F, right image). Collectively these results suggest that Ser592 phosphorylation is required for proper localization of DIXDC1 to focal adhesions.
Hyperactivation of FAK from loss of LKB1 or DIXDC1 function is responsible for elevated Snail
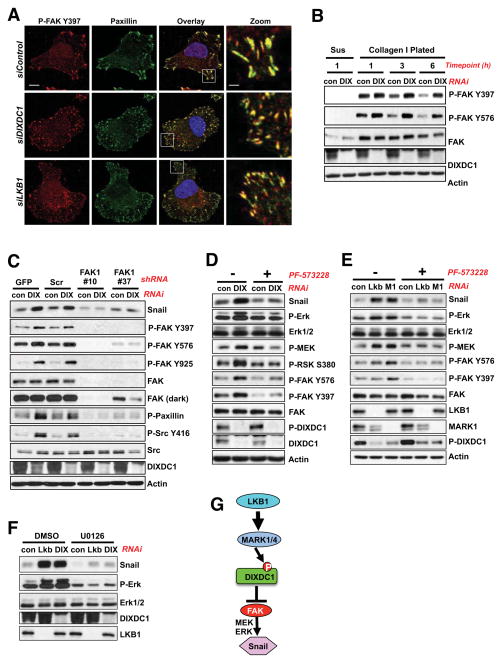
As FAK is normally recruited to nascent focal adhesions concurrently with paxillin (Wehrle-Haller, 2012), and FAK was reported to be hyperactivated in LKB1-deficient lung tumors (Carretero et al., 2010), we next examined the impact of DIXDC1 silencing on FAK. Activated Tyr397-phosphorylated FAK colocalized with paxillin 1 hour after plating, in both nascent adhesions in the DIXDC1 or LKB1 knockdown cells, as well as the larger more mature focal adhesions formed in the control cells (Figure 4A). Nascent adhesions are reportedly highly enriched for phospho-tyrosine (Zaidel-Bar et al., 2003). Consistent with this, cells bearing DIXDC1 knockdown exhibited a greater peak and prolonged activation of FAK upon plating (Figure 4B). The increase in FAK activation from DIXDC1 knockdown resulted in elevated Src signaling, as defined by Src Tyr416 phosphorylation or paxillin Tyr118 phosphorylation, which were both attenuated in cells expressing FAK shRNAs (Figure 4C) or by treatment with the small molecule FAK inhibitor PF-573228 (Slack-Davis et al., 2007) (Figure S4A). FAK silencing or inhibition also attenuated Snail1 induction from knockdown of DIXDC1 (Figure 4C, D), MARK1, or LKB1 (Figure 4E), consistent with a study reporting that FAK regulates Snail1 levels in MEFs (Li et al., 2011).
Figure 4. Hyperactivation of FAK upon loss of LKB1 or DIXDC1 is responsible for Snail induction.
(A) Immunolocalization of P-FAK Y397 and paxillin in cells transfected with siRNA oligos (20 μM) against DIXDC1 or LKB1 and plated as described in Figure 3A. Representative images are shown. Scale bar = 10 μm. Scale bar = 2 μm (Zoom image).
(B) Analysis of focal adhesion signaling upon plating on ECM. U2OS cells depleted of DIXDC1 by siRNA transfection (20 μM) were treated as described in Figure 3A and plated for the indicated timepoint. Lysates immunoblotted with indicated antibodies. Sus = Suspension 1 hr.
(C) U2OS cells were infected with lentiviral vectors expressing two separate shRNA duplexes to FAK1 for 24 hr. Immediately following infection, U2OS cells were depleted of DIXDC1 by siRNA transfection (20 μM) for 72 h. At 48 hours post siRNA transfection, cells were serum-starved for 24 hours before collection of cell lysates. GFP = shGFP control. Scr = Scramble control.
(D) U2OS cells were transfected with siRNA against DIXDC1 (20 μM) for 72 hr. 48 hr post RNAi transfection, cells were treated with 1 μM PF-573228 FAK inhibitor or DMSO vehicle control for 24 hr in the absence of serum. Lysates were analyzed by immunoblot with the indicated antibodies.
(E) siRNA (20 μM) knockdown of the indicated genes in U2OS cells for 72 hr. 48 hr post RNAi transfection, cells were treated with 1 μM PF-573228 FAK inhibitor or DMSO vehicle control for 24 hr in the absence of serum. Lysates were analyzed by immunoblot with the indicated antibodies.
(F) U2OS cells were transfected with siRNA against LKB1 or DIXDC1 (20 μM) for 72 hr. 48 hr post RNAi transfection, cells were treated with 20 μM U0126 (MEK inhibitor) or DMSO vehicle control for 24 hr in the absence of serum. Lysates were analyzed by immunoblot with the indicated antibodies.
(G) Model of proposed LKB1-MARK1-DIXDC1 signaling cascade.
Upon DIXDC1 knockdown, FAK also exhibited increased phosphorylation at Tyr925, which mediates binding to Grb2 and activation of the Ras-MEK-ERK pathway. Consistent with the increased P-Y925, endogenous GRB2 associated with more FAK and Src in the DIXDC1 knockdown cells (Figure S4B). Correspondingly, we observed higher levels of FAK-dependent activation of MEK, ERK1/2, and the ERK substrate RSK in DIXDC1 knockdown cells (Figure 4D).
To further examine what pathways downstream of FAK were involved in Snail upregulation, we treated cells with small molecule inhibitors of MEK, PI3K, Akt, or mTOR. Of these, only the MEK inhibitor U0126 caused complete loss of Snail protein (Figure 4F, S4C, S4D). This observation is consistent with previous studies reporting multiple mechanisms of ERK-dependent increases in Snail, including both mRNA induction (Barbera et al., 2004; Toettcher et al., 2013) and direct ERK phosphorylation sites in Snail suppressing protein turnover (Zhang et al., 2013). Collectively, our findings suggest that when LKB1, MARK1, or DIXDC1 function is compromised, FAK becomes hyperactivated resulting in MEK/ERK-dependent induction of Snail (Figure 4G).
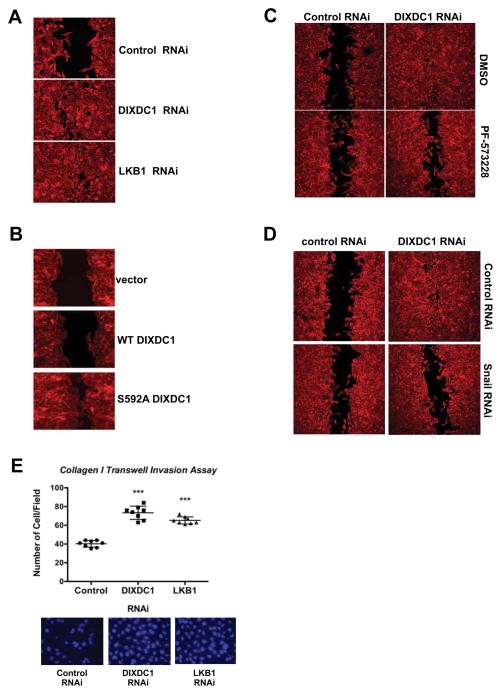
DIXDC1 suppresses cell migration and invasion in a Ser592-dependent manner
To examine how DIXDC1 loss impacts cell behavior, we examined cell migration and invasion. We first performed scratch assays to examine directed migration, in the presence of mitomycin C to prevent proliferation during the experiment. RNAi silencing of LKB1 or DIXDC1 resulted in increased wound healing, which was mimicked by stable overexpression of non-phosphorylatable DIXDC1S592A, but not DIXDC1WT (Figure 5A, 5B). This finding suggests that in this context, DIXDC1S592A behaves as a dominant negative allele to suppress the activity of the endogenous DIXDC1. Increased migration upon DIXDC1 knockdown was attenuated by FAK inhibition (Figure 5C, S5A) or Snail RNAi (Figure 5D). To determine if DIXDC1 similarly affected invasive behavior, cells were placed in a collagen-coated Boyden chamber and allowed to invade for 24 hours prior to fixation. Cells with stable LKB1 or DIXDC1 knockdown had a ~1.8-fold increase in cell invasion in that time window (Figure 5E). The enhanced collagen invasion of cells bearing DIXDC1 knockdown was blocked inhibition of FAK (Figure S5A).
Figure 5. DIXDC1 suppresses cell migration and invasion in a manner dependent on S592 phosphorylation.
(A) Scratch wound healing assay of U2OS cells depleted of indicated genes by siRNA transfection (20 μM) for 72 h. Migration media supplemented with 10 ug/ml Mitomycin C to block cell proliferation. Cells were stained 14 hr post-wounding with Rhodamine-conjugated Phalloidin to demarcate cell borders. Images representative of 3 independent experiments.
(B) Stable expression of indicated DIXDC1 cDNA by lentiviral transduction in U2OS cells. Scratch wound healing assays performed as in (A). Images from 2 independent experiments shown and representative of 3 experiments.
(C) siRNA transfection (20 μM) against indicated genes in U2OS cells for 72 h. Scratch wound healing assays performed as in (A). Media was supplemented with DMSO vehicle control or 1 μM PF-573228 at the time of wounding and incubated for the 14 hr migration period.
(D) siRNA knockdown of DIXDC1 (20 μM) and Snail1 (50 μM) in U2OS cells for 72 h. Scratch wound healing assays performed as in (A).
(E) Collagen transwell migration assay. Transwell filters were coated with collagen I and U2OS cells depleted for the indicated genes by siRNA transfection were plated onto filters in serum-free media supplemented with 10 ug/ml Mitomycin C for 24 h. Filters were excised and migrated cells were stained with DAPI and counted. *** = p<.0001 relative to control. Statistical analysis performed using an unpaired Student’s t-test.
Dixdc1 dictates metastatic behavior in non-small cell lung cancer lines
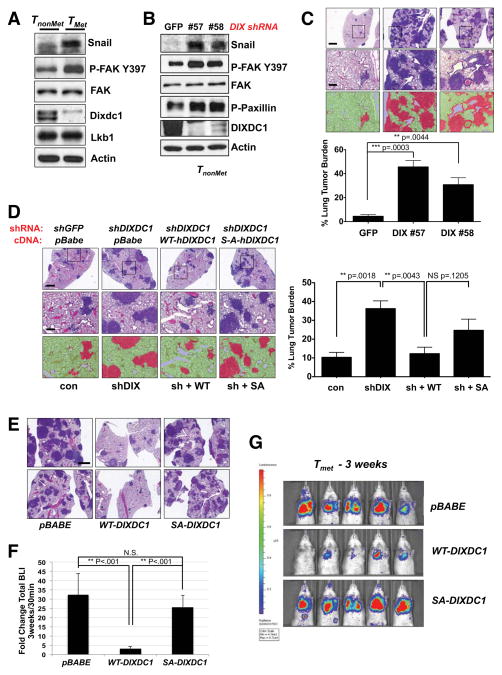
Given the role of Lkb1 in the suppression of lung tumor metastasis in mouse Kras-dependent NSCLC models (Carretero et al., 2010; Ji et al., 2007), we wanted to explore the function of Dixdc1 in lung tumor cell lines with defined metastatic characteristics. We utilized a panel of KrasG12D, p53−/− mutant lung adenocarcinoma cell lines derived from nonmetastatic and metastatic primary mouse lung tumors (TnonMet and TMet respectively) that retain the metastatic capacity of their tumor of origin (Winslow et al., 2011). Notably, while Lkb1 protein levels were unaffected, Dixdc1 protein and mRNA levels were lower in the TMet lines compared to the TnonMet lines (Figure 6A, S6A), whereas Snail and FAK Phospho-Y397 levels were higher in the metastatic setting. Thus Dixdc1 reduction and the previously observed associated biochemical signature of DIXDC1 suppression correlated with metastatic potential in these cell lines. Furthermore, stable knockdown of Dixdc1 in TnonMet cells resulted in hyperactivation of FAK signaling and increased Snail levels to an extent similar to that found in the TMet cells (Figure 6B).
Figure 6. DIXDC1 suppresses lung colonization of primary mouse lung cancer cell lines in a manner dependent on S592 phosphorylation.
(A) Lysates from non-metastatic (TnonMet) 368T1 and metastatic (TMet) 393T5 Kras G12D; p53−/− mouse primary lung tumor cell lines immunoblotted with the indicated antibodies.
(B) Stable depletion of Dixdc1 by lentiviral shRNA transduction with two independent shRNA sequences in TnonMet 368T1 cells. Lysates were immunoblotted with the indicated antibodies.
(C) Lung colonization assay. TnonMet 368T1 cells were stably depleted of endogenous Dixdc1 by lentiviral transduction with two independent shRNA sequences. Cells were then injected intravenously in the lateral tail vein of syngeneic 129/Bl6 F1 mice. Lungs were harvested 3 weeks post transplantation and representative hematoxylin & eosin (H&E) -stained sections are shown. Total lung tumor burden was quantified using morphometric Inform software. Scale bar (top image) = 1mm. Scale bar (zoom image) = 100 μm. Statistical analysis performed using an unpaired Student’s t-test.
(D) Rescue of enhanced colonization efficiency by enforced expression of WT, but not S592A mutant hDIXDC1 in TnonMet cells. Cells were stably depleted for endogenous Dixdc1 or GFP control by lentiviral shRNA transduction. Knockdown cells were then reconstituted with a WT or S592A hDIXDC1 cDNA using retroviral transduction. Stably selected cells were injected intravenously and colonization efficiency was assessed by H&E staining 3 weeks post transplantation. Total lung tumor burden was quantified using morphometric Inform software. Scale bar (top image) = 1mm. Scale bar (zoom image) = 100 μm. Statistical analysis performed using an unpaired Student’s t-test.
(E) Suppression of lung colonization by highly metastatic TMet 393T5 cells through enforced expression of WT, but not S592A mutant hDIXDC1. TMet 393T5 cells, which express low levels of Dixdc1 (A), were transduced with a retroviral vector encoding the cDNA for WT or S592A mutant hDIXDC1. Cells were transduced with a lentiviral vector expressing firefly luciferase and subsequently injected intravenously in the lateral tail vein of syngeneic 129/Bl6 F1 mice. Representative H&E sections of mouse lungs at 3 weeks post-transplantation are shown. Scale = 1mm.
(F) Quantification of total bioluminescence at 3 weeks (E) compared to 30 min post tumor cell intravenous transplantation. ** p < .001 unpaired Student’s t-test. N.S. = not significant.
(G) At 3 weeks post-transplantation, mice from (E) (n = 5/group) were imaged using IVIS bioluminescence imaging. Signal intensity representing lung tumor burden in shown. All data are represented as the mean ± SEM.
To further investigate Dixdc1 function in these murine lung adenocarcinoma cell lines, we first confirmed that Lkb1, Mark1, and Mark4 regulate endogenous Dixdc1 Ser592 phosphorylation in the TnonMet cells (Figure S6B). As observed in U2OS cells, acute RNAi to Mark1 or Dixdc1 also resulted in hyperactivation of FAK upon plating (Figure S6C). Conversely, stable enforced expression of Dixdc1 in the TMet cells suppressed FAK signaling (Figure S6D). Consistent with regulation of the FAK-Snail signature, Dixdc1 knockdown in TnonMet cells resulted in increased invasion (Figure S6E).
To directly examine the impact of loss of Dixdc1 on metastatic potential in vivo, we stably knocked down Dixdc1 with two independent shRNAs in TnonMet cells and performed a lung colonization assay in syngeneic mice (Winslow et al., 2011). Dixdc1 knockdown dramatically increases the lesion number and total tumor burden (Figure 6C), without affecting the growth rate of these cells in culture (Figure S6F). The increase in tumor burden in TnonMet cells observed from Dixdc1 silencing was comparable to that induced by shRNA to Lkb1 (Figure S6G). The increased tumor burden in the TnonMet cell lines bearing Dixdc1 shRNA was reversed by stable re-expression of a wild-type, but not S592A mutant, hDIXDC1 (Figure 6D). Moreover, stable expression of WT, but not S592A mutant, hDIXDC1 in TMet cells, which lack appreciable Dixdc1 (Figure 6A, S6H), caused potent suppression of experimental metastasis (Figure 6E–G), providing strong evidence for the importance of this single phosphorylation site in governing metastasis in vivo. Quantification by luciferase imaging confirmed the decrease in total tumor burden (Figure 6F, G). The suppression of metastasis by overexpression of WT Dixdc1 also demonstrates that the TMet cells are capable of receiving Lkb1-dependent signals to Dixdc1, but simply lack sufficient Dixdc1 expression as compared to the non-metastatic cells.
DIXDC1 is frequently lost in human cancers
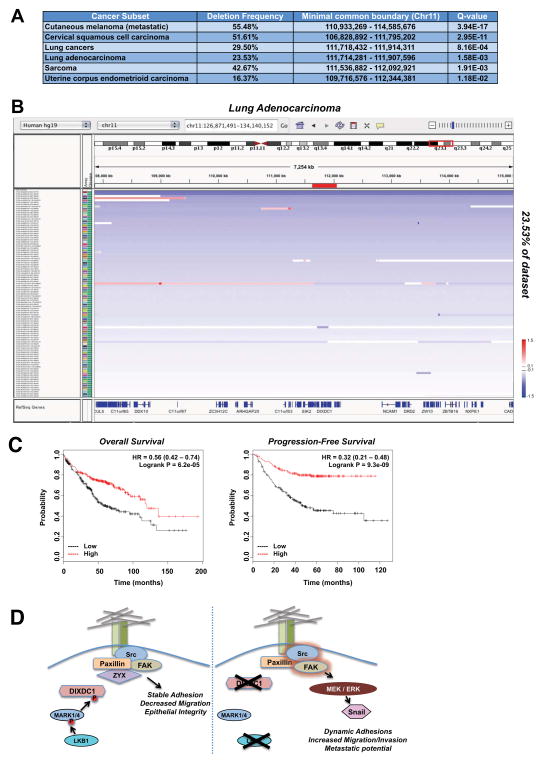
Though the TMet cell lines retain Lkb1, given their loss of Dixdc1 mRNA expression and potent effects of Dixdc1 on tumor burden in this setting, we wondered whether DIXDC1 may be deleted, mutated, or downregulated in human tumors. We examined copy number variations for human DIXDC1, which lies in a broad tumor suppressor region on chromosome 11q23.1. GISTIC analysis reveals that DIXDC1 is significantly deleted in 11 out of 27 cancer subsets across the entire Tumorscape/TCGA dataset of 9041 tumors. Notably, DIXDC1 is most significantly deleted independent of a peak region in metastatic cutaneous melanoma, lung cancer, and cervical squamous cell carcinoma (Figure 7A). Although infrequent, we were able to observe patient samples that displayed a focal, highly specific deletion at the DIXDC1 locus (Figure 7B). Interestingly, melanoma, lung cancer, and cervical carcinoma represent well-defined settings where loss of Lkb1 in GEMM models modulates metastasis and FAK/Src signaling (Liu et al., 2012). In support of this analysis, using a new algorithm (CONEXIC) that combines gene expression and copy number alterations to identify driver events in melanoma, suppression of DIXDC1 was identified as one of the top 30 drivers in this tumor type (Akavia et al., 2010). Importantly, DIXDC1 mRNA expression correlated better with global gene expression changes that any of the other 16 genes in a focal deletion region of 11q23.1 or the focal deletion of 11q23.1 itself (Akavia et al., 2010), reinforcing that alterations in DIXDC1 mRNA levels are a critical measure of its functionality beyond the subset of tumors bearing LKB1 mutations where DIXDC1 is inactivated from loss of Ser592 phosphorylation.
Figure 7. DIXDC1 is frequently downregulated in human cancer.
(A) Characterization of deletion of the DIXDC1 locus. Summary of deletion frequency and boundary of alteration of the DIXDC1 locus in the indicated tumor types. Data was obtained through query of the TCGA TumorScape database (Broad Institute).
B) Lung adenocarcinoma patients exhibiting a deletion at the DIXDC1 locus visualized using the Integrative Genomic Viewer (IGVv2.3; Broad Institute). Blue bars represent degree of deletion, white bars represent no alteration, and red bars represent an amplification. DNA copy number ratio is relative to a reference somatic DNA sample. Note the single patient representing a focal deletion at the DIXDC1 locus.
(C) Correlation of DIXDC1 expression with patient survival in lung adenocarcinoma. DIXDC1 expression was stratified as high vs. low against median expression. Overall survival and
Analysis of DIXDC1 mRNA expression in human NSCLC cancer datasets revealed its frequent downregulation (Figure S7A–D). Interestingly, Dixdc1 was previously identified as one of only 97 genes genome-wide that was cooperatively suppressed at the mRNA level by Kras and p53 mutation in a pattern correlating with the malignant phenotype (McMurray et al., 2008) (Fig. S7E). The 97 genes identified in this dataset proved relevant across a broad panel of human tumor cell lines, suggesting they may be relevant for tumorigenesis across cell contexts. To further explore DIXDC1 expression in human cancer, we next examined DIXDC1 levels as a function of patient outcome in an annotated set of NSCLC tumors. Mining a recent meta-analysis of published lung cancer microarray datasets that identified biomarkers related to survival (Gyorffy et al., 2013), we found that low DIXDC1 expression in lung adenocarcinoma patients is significantly correlated with decreased progression-free and overall survival (Figure 7C). As survival in such cohorts is largely due to metastasis, this suggests in these contexts low DIXDC1 mRNA may correlate with increased metastatic frequency.
DISCUSSION
In this study, we identified the poorly studied scaffold protein DIXDC1 as the first direct substrate of LKB1-dependent kinases connected to metastatic progression. Phenotypes observed from gain and loss of DIXDC1 function also help provide a direct molecular basis for the only previously noted biomarkers of Lkb1-deficient metastasis: 1) hyperactivation of FAK/Src family kinase signaling (Carretero et al. 2010; Liu et al., 2012) and 2) increased expression of some EMT target genes (Carretero et al., 2010). This effect is mediated by MARK1/4 dependent activation of DIXDC1, which suppresses the FAK-ERK-Snail1 signaling axis (Figure 7D). While MARK1 and MARK4 do not appear redundant in the regulation of DIXDC1, it remains unclear whether this reflects their formation of a heterodimeric complex or alternative possibilities to be pursued in future studies. Phosphorylation of DIXDC1 at Ser592 by MARK1/4 is required for its proper targeting to focal adhesions and required for DIXDC1 to suppress metastatic growth in vivo. In addition to the functional inactivation of DIXDC1 in LKB1−/− tumors from loss of Ser592 phosphorylation, we found that deletions at the DIXDC1 locus were highly selected for in the exact tumor subsets where LKB1 has been characterized as a dominant regulator of metastasis. It is also worth noting that few endogenous suppressors of EMT are known, which may explain why DIXDC1 downregulation would be selected for independent of LKB1 mutation, as its loss promotes Snail-dependent gene expression, stimulating invasion and tumor microenvironment remodeling. Analysis of DIXDC1 and its regulation by other pathways that modulate EMT will be of great interest, both in the context of different tumor types and developmental processes.
Molecularly, our data suggests that DIXDC1 is involved in focal adhesion maturation. We observed that loss of DIXDC1 results in an accumulation of nascent focal adhesions, which were largely absent for zyxin staining. Zyxin-null cells display increased rates of cell motility as well as hyper-activation of the FAK/Src signaling module (Hoffman et al., 2006; Mise et al., 2012), consistent with the phenotypes we observe with zyxin mislocalization in DIXDC1-depleted cells. In a broad context, downregulation of factors that regulate the maturation of focal adhesions such as DIXDC1 and zyxin might be selected for in those cells gaining increased invasion potential. How does this role for DIXDC1 in suppressing cell migration and invasion fit with the few previously reported functions for DIXDC1? Previous studies identified Nudel and Dvl proteins as the predominant in vivo DIXDC1 interaction partners (Shiomi et al., 2003; Singh et al., 2010; Wong et al., 2004), which interestingly have both been reported to regulate cell motility and focal adhesion dynamics via effects on FAK and paxillin (Matsumoto et al., 2010; Shan et al., 2009). We have confirmed DIXDC1 association with endogenous Nudel and Dvl, though we have not been able to detect their association with FAK or paxillin thus far. Future studies will be aimed at identifying a DIXDC1 interaction network during adhesion and focal adhesion maturation to better define how it suppresses FAK signaling. The central involvement of FAK in LKB1-dependent metastatic potential fits well with emerging evidence that hyperactivation of FAK and Src may represent a central biochemical pathway broadly required for metastasis in tumors from many different tissues of origin (Shibue and Weinberg, 2009; Zhang et al., 2009). This study suggests that certain initiating genetic alterations may provide robust metastatic fitness by activating FAK/SRC from even early stages of tumorigenesis, alleviating the need for additional genetic lesions to enhance metastatic potential.
Notably, phosphorylation of DIXDC1 downstream of LKB1 is mediated by the poorly studied kinases MARK1 and MARK4, and independent of AMPK. DIXDC1 represents one of the first AMPK-independent effectors of LKB1 in human cancer and indicates that in some contexts the tumor suppressor activity of LKB1 is unlikely to be mediated by AMPK alone. The finding that LKB1 actively suppresses the Snail1 EMT transcription factor through a novel signaling pathway downstream of MARK1 and MARK4 suggests much remains to be decoded for this ancient pathway connecting metabolism to cell polarity and cytoskeletal control. This study also provides further molecular insight into how initiating mutations in the LKB1 tumor suppressor leads simultaneously to immediate alterations in mTOR signaling, autophagy, metabolism, as well as FAK signaling, here via loss of phosphorylation of a single serine residue in a single downstream effector. Regulation of such seemingly diverse cell processes by LKB1 may underlie its unique potency for tumor initiation and metastatic progression in some tissues, and warrants further investigation into identifying the full repertoire of conserved substrates of the LKB1-dependent kinases to reveal additional rate-limiting regulators of cell biology and disease.
EXPERIMENTAL PROCEDURES
Plasmid Constructs
The cDNA for human DIXDC1 was cloned from a cDNA library prepared from U2OS cells and sequence verified to match the sequence of the long isoform of human DIXDC1 (NM_001037954.2). DIXDC1 cDNA was subcloned into pENTR4-FLAG (Addgene #17423) or pENTR4-myc (edited from pENTR4-FLAG) to create FLAG or myc tagged ENTR clones, respectively. ENTR clones for human LKB1 and LYK5 (STRADalpha) were obtained from Invitrogen (IOH21169, IOH45129 respectively). Human Snail cDNA (Addgene #16218) and Slug cDNA (Addgene #31698) were PCR amplified and subcloned to pDONR221 with BP Clonase (Invitrogen). Site-directed mutagenesis was performed using QuikChange II XL (Stratagene) according to the manufacturers instructions. All ENTR/DONR clones were sequence verified to ensure no additional mutations. To create mammalian expression vectors, ENTR clones were recombined into DEST vectors using LR clonase (Invitrogen). DEST vectors used in this study include: pQCXIB (Addgene #17400), pQCXIN (Addgene #17395), pLentiCMV/TO DEST (Addgene #17293), pcDNA3 N-term myc DEST, pcDNA3 N-term FLAG DEST, pBabe-Hygro DEST. pEBG2T-hMARK1 WT and T215A(KD) were a gift from Dario Alessi (Dundee, UK).
Antibodies
Cell Signaling Technology antibodies used: Wnt5a/b (#2530), Snail (#3879), Slug (#9585), ZEB1 (#3396), LKB1 (#3047), P-ACC S79 (#3661), Total ACC (#4190), Axin2 (#2151), MARK2 (#9118), MARK3 (#9311), AMPKalpha (#2532), P-ULK1 S555 (#5869), Nuak1 (#4458), SIK2 (#6919), GST (#2622), myc-tag (#2272), P-Src family Y416 (#2113), Src (#2109), P-Paxillin Y118 (#2541), Pathscan I for P-ERK1/2 and P-Akt S473 (#5301), Total ERK1/2 (#4695), P-S6K (#9234), HA-tag (#3724), P-MEK1/2 (#9154), P-p90RSK S380 (#11989), P-FAK Y925 (#3284). Epitomics antibodies used: Phospho-FAK Y397 (#2211-1), Phospho-FAK Y576/577 (#2183-1), Total FAK1 (#2146-1), Zyxin (#3586-1). BD Transduction Labs antibodies used: Paxillin (P13520). Sigma antibodies used: Actin (A5441), Flag polyclonal (F7425). Protein Tech antibodies used: MARK1 (21552-1-AP), MARK2 (15492-1-AP). Millipore antibodies used: ZEB2 (ABT332), MARK4 (07-699). Abcam antibodies used Twist (ab50887), IRSp53 (ab15697). CLASP2 antibody was from Santa Cruz Biotechnology (sc-98440). DIXDC1 total antibody was from R&D Systems (AF5599). Phospho-DIXDC1 S592 was developed in collaboration with Antony Wood at Cell Signaling Technologies (CST, Danvers, MA).
Cell lines
U2OS, H157, H1299, SJSA, Panc-1, MiaPaca-2, and 293T were purchased from ATCC. Littermate-derived Lkb1+/+ and Lkb1−/− MEFs were described previously (Shaw et al., 2004). TnonMet and TMet primary mouse lung cancer cell have been described previously (Winslow et al., 2011). Specific lines used in this study include 368T1, 394T4, 802T4, 565T2, and 393T5. All cells were maintained in DMEM (Mediatech, Manassas, VA) supplemented with 10% fetal bovine serum (Hyclone, Thermo Scientific) and cultured at 37°C in 10% CO2.
Animal Studies
Tumor lysates from KrasG12D;Lkb1+/+ and KrasG12D;Lkb1−/− primary lung tumors were generated from Lox-Stop-Lox KrasG12D and Lkb1flox/flox mice as previously described (Shackelford et al., 2013). Mice used for syngeneic mouse transplantation assays were 129/Bl6 F1 hybrid mice (Jackson Laboratories) as described previously (Winslow et al., 2011).. Mice were 7–9 weeks old at time of injections. For lung colonization assays, 7 × 104 cells resuspended in 200 μl PBS were injected through the lateral tail vein. Mice were sacrificed 3 weeks after injection and lungs were harvested for tissue processing.
Supplementary Material
HIGHLIGHTS.
LKB1 suppresses Snail1 levels across diverse normal and cancer cell types
MARK1 and MARK4 but not AMPK mediate the suppression of Snail by LKB1
MARK1/4 phosphorylation of DIXDC1 is required to suppress Snail via effects on FAK
DIXDC1 suppress metastasis in lung adenocarcinoma models in Ser592-dependent manner
Acknowledgments
We thank David Shackelford for the Kras, Lkb1+/+ and Kras, Lkb1L/L tumor lysates used in Figure 1F. We thank Drs. Anthony Wood and Gary Kasof at Cell Signaling Technology for developing the Phospho-Ser592 DIXDC1 and total DIXDC1 antibodies in conjunction with J.M.G. and R.J.S. J.M.G. performed all experiments in Figures 1–7 except Figure 2A, Figure S2B, Figure 6C–G, and Figure S6G. J.M.G. performed mouse experiments for Figure 6 with assistance from R.U.S. Murine lung cancer TMet and TnonMet cell lines utilized in Figure 6 were generated and characterized by M.M.W who advised on their use. Figure 2A and S2A were performed by H.J.L and B.E.T. The study was designed by J.M.G. and R.J.S., who analyzed the data and wrote the paper with feedback from all authors. J.M.G. was supported by the predoctoral T32 CMG training grant to UCSD/Salk. M.M.W. is the recipient of a V Foundation for Cancer Research Scholar Award and this work is supported in part by National Institutes of Health grants R01CA17533601 to M.M.W. and R01GM104047 to B.E.T. This work in the laboratory of R.J.S. was funded by the Howard Hughes Medical Institute, R01CA172229, P01CA120964, and also supported in part through the Salk CCSG P30 CA014195, the Samuel Waxman Cancer Research Foundation, and The Leona M. and Harry B. Helmsley Charitable Trust grant #2012-PG-MED002.
Footnotes
Supplemental Information contains seven supplemental figures and figure legends, Extended Experimental Procedures.
The authors declare no competing financial interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Akavia UD, Litvin O, Kim J, Sanchez-Garcia F, Kotliar D, Causton HC, Pochanard P, Mozes E, Garraway LA, Pe’er D. An integrated approach to uncover drivers of cancer. Cell. 2010;143:1005–1017. doi: 10.1016/j.cell.2010.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbera MJ, Puig I, Dominguez D, Julien-Grille S, Guaita-Esteruelas S, Peiro S, Baulida J, Franci C, Dedhar S, Larue L, et al. Regulation of Snail transcription during epithelial to mesenchymal transition of tumor cells. Oncogene. 2004;23:7345–7354. doi: 10.1038/sj.onc.1207990. [DOI] [PubMed] [Google Scholar]
- Carretero J, Shimamura T, Rikova K, Jackson AL, Wilkerson MD, Borgman CL, Buttarazzi MS, Sanofsky BA, McNamara KL, Brandstetter KA, et al. Integrative genomic and proteomic analyses identify targets for Lkb1-deficient metastatic lung tumors. Cancer Cell. 2010;17:547–559. doi: 10.1016/j.ccr.2010.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaffer CL, Weinberg RA. A perspective on cancer cell metastasis. Science. 2011;331:1559–1564. doi: 10.1126/science.1203543. [DOI] [PubMed] [Google Scholar]
- Contreras CM, Akbay EA, Gallardo TD, Haynie JM, Sharma S, Tagao O, Bardeesy N, Takahashi M, Settleman J, Wong KK, et al. Lkb1 inactivation is sufficient to drive endometrial cancers that are aggressive yet highly responsive to mTOR inhibitor monotherapy. Dis Model Mech. 2010;3:181–193. doi: 10.1242/dmm.004440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding L, Getz G, Wheeler DA, Mardis ER, McLellan MD, Cibulskis K, Sougnez C, Greulich H, Muzny DM, Morgan MB, et al. Somatic mutations affect key pathways in lung adenocarcinoma. Nature. 2008;455:1069–1075. doi: 10.1038/nature07423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan DF, Shackelford DB, Mihaylova MM, Gelino S, Kohnz RA, Mair W, Vasquez DS, Joshi A, Gwinn DM, Taylor R, et al. Phosphorylation of ULK1 (hATG1) by AMP-activated protein kinase connects energy sensing to mitophagy. Science. 2011;331:456–461. doi: 10.1126/science.1196371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gwinn DM, Shackelford DB, Egan DF, Mihaylova MM, Mery A, Vasquez DS, Turk BE, Shaw RJ. AMPK phosphorylation of raptor mediates a metabolic checkpoint. Mol Cell. 2008;30:214–226. doi: 10.1016/j.molcel.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- Hardie DG, Alessi DR. LKB1 and AMPK and the cancer-metabolism link - ten years after. BMC biology. 2013;11:36. doi: 10.1186/1741-7007-11-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen M, Ten Klooster JP, Offerhaus GJ, Clevers H. LKB1 and AMPK family signaling: the intimate link between cell polarity and energy metabolism. Physiol Rev. 2009;89:777–798. doi: 10.1152/physrev.00026.2008. [DOI] [PubMed] [Google Scholar]
- Ji H, Ramsey MR, Hayes DN, Fan C, McNamara K, Kozlowski P, Torrice C, Wu MC, Shimamura T, Perera SA, et al. LKB1 modulates lung cancer differentiation and metastasis. Nature. 2007;448:807–810. doi: 10.1038/nature06030. [DOI] [PubMed] [Google Scholar]
- Lai C, Robinson J, Clark S, Stamp G, Poulsom R, Silver A. Elevation of WNT5A expression in polyp formation in Lkb1+/− mice and Peutz-Jeghers syndrome. J Pathol. 2011;223:584–592. doi: 10.1002/path.2835. [DOI] [PubMed] [Google Scholar]
- Li XY, Zhou X, Rowe RG, Hu Y, Schlaepfer DD, Ilic D, Dressler G, Park A, Guan JL, Weiss SJ. Snail1 controls epithelial-mesenchymal lineage commitment in focal adhesion kinase-null embryonic cells. J Cell Biol. 2011;195:729–738. doi: 10.1083/jcb.201105103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Monahan KB, Pfefferle AD, Shimamura T, Sorrentino J, Chan KT, Roadcap DW, Ollila DW, Thomas NE, Castrillon DH, et al. LKB1/STK11 inactivation leads to expansion of a prometastatic tumor subpopulation in melanoma. Cancer Cell. 2012;21:751–764. doi: 10.1016/j.ccr.2012.03.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto S, Fumoto K, Okamoto T, Kaibuchi K, Kikuchi A. Binding of APC and dishevelled mediates Wnt5a-regulated focal adhesion dynamics in migrating cells. EMBO J. 2010;29:1192–1204. doi: 10.1038/emboj.2010.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMurray HR, Sampson ER, Compitello G, Kinsey C, Newman L, Smith B, Chen SR, Klebanov L, Salzman P, Yakovlev A, et al. Synergistic response to oncogenic mutations defines gene class critical to cancer phenotype. Nature. 2008;453:1112–1116. doi: 10.1038/nature06973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mihaylova MM, Shaw RJ. The AMPK signalling pathway coordinates cell growth, autophagy and metabolism. Nat Cell Biol. 2011;13:1016–1023. doi: 10.1038/ncb2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mihaylova MM, Vasquez DS, Ravnskjaer K, Denechaud PD, Yu RT, Alvarez JG, Downes M, Evans RM, Montminy M, Shaw RJ. Class IIa Histone Deacetylases Are Hormone-Activated Regulators of FOXO and Mammalian Glucose Homeostasis. Cell. 2011;145:607–621. doi: 10.1016/j.cell.2011.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno-Bueno G, Cubillo E, Sarrio D, Peinado H, Rodriguez-Pinilla SM, Villa S, Bolos V, Jorda M, Fabra A, Portillo F, et al. Genetic profiling of epithelial cells expressing E-cadherin repressors reveals a distinct role for Snail, Slug, and E47 factors in epithelial-mesenchymal transition. Cancer Res. 2006;66:9543–9556. doi: 10.1158/0008-5472.CAN-06-0479. [DOI] [PubMed] [Google Scholar]
- Ollila S, Makela TP. The tumor suppressor kinase LKB1: lessons from mouse models. J Mol Cell Biol. 2011;3:330–340. doi: 10.1093/jmcb/mjr016. [DOI] [PubMed] [Google Scholar]
- Rembold M, Ciglar L, Yanez-Cuna JO, Zinzen RP, Girardot C, Jain A, Welte MA, Stark A, Leptin M, Furlong EE. A conserved role for Snail as a potentiator of active transcription. Genes Dev. 2014;28:167–181. doi: 10.1101/gad.230953.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren D, Minami Y, Nishita M. Critical role of Wnt5a-Ror2 signaling in motility and invasiveness of carcinoma cells following Snail-mediated epithelial-mesenchymal transition. Genes Cells. 2011;16:304–315. doi: 10.1111/j.1365-2443.2011.01487.x. [DOI] [PubMed] [Google Scholar]
- Shackelford DB, Abt E, Gerken L, Vasquez DS, Seki A, Leblanc M, Wei L, Fishbein MC, Czernin J, Mischel PS, et al. LKB1 inactivation dictates therapeutic response of non-small cell lung cancer to the metabolism drug phenformin. Cancer Cell. 2013;23:143–158. doi: 10.1016/j.ccr.2012.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shackelford DB, Shaw RJ. The LKB1-AMPK pathway: metabolism and growth control in tumour suppression. Nat Rev Cancer. 2009;9:563–575. doi: 10.1038/nrc2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan Y, Yu L, Li Y, Pan Y, Zhang Q, Wang F, Chen J, Zhu X. Nudel and FAK as antagonizing strength modulators of nascent adhesions through paxillin. PLoS Biol. 2009;7:e1000116. doi: 10.1371/journal.pbio.1000116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw RJ, Kosmatka M, Bardeesy N, Hurley RL, Witters LA, DePinho RA, Cantley LC. The tumor suppressor LKB1 kinase directly activates AMP-activated kinase and regulates apoptosis in response to energy stress. Proc Natl Acad Sci U S A. 2004;101:3329–3335. doi: 10.1073/pnas.0308061100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibue T, Weinberg RA. Integrin beta1-focal adhesion kinase signaling directs the proliferation of metastatic cancer cells disseminated in the lungs. Proc Natl Acad Sci U S A. 2009;106:10290–10295. doi: 10.1073/pnas.0904227106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiomi K, Uchida H, Keino-Masu K, Masu M. Ccd1, a novel protein with a DIX domain, is a positive regulator in the Wnt signaling during zebrafish neural patterning. Curr Biol. 2003;13:73–77. doi: 10.1016/s0960-9822(02)01398-2. [DOI] [PubMed] [Google Scholar]
- Singh KK, Ge X, Mao Y, Drane L, Meletis K, Samuels BA, Tsai LH. Dixdc1 is a critical regulator of DISC1 and embryonic cortical development. Neuron. 2010;67:33–48. doi: 10.1016/j.neuron.2010.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slack-Davis JK, Martin KH, Tilghman RW, Iwanicki M, Ung EJ, Autry C, Luzzio MJ, Cooper B, Kath JC, Roberts WG, et al. Cellular characterization of a novel focal adhesion kinase inhibitor. J Biol Chem. 2007;282:14845–14852. doi: 10.1074/jbc.M606695200. [DOI] [PubMed] [Google Scholar]
- Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139:871–890. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- Toettcher JE, Weiner OD, Lim WA. Using optogenetics to interrogate the dynamic control of signal transmission by the Ras/Erk module. Cell. 2013;155:1422–1434. doi: 10.1016/j.cell.2013.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turk BE, Hutti JE, Cantley LC. Determining protein kinase substrate specificity by parallel solution-phase assay of large numbers of peptide substrates. Nat Protoc. 2006;1:375–379. doi: 10.1038/nprot.2006.57. [DOI] [PubMed] [Google Scholar]
- Wang X, Zheng L, Zeng Z, Zhou G, Chien J, Qian C, Vasmatzis G, Shridhar V, Chen L, Liu W. DIXDC1 isoform, l-DIXDC1, is a novel filamentous actin-binding protein. Biochem Biophys Res Commun. 2006;347:22–30. doi: 10.1016/j.bbrc.2006.06.050. [DOI] [PubMed] [Google Scholar]
- Wehrle-Haller B. Structure and function of focal adhesions. Curr Opin Cell Biol. 2012;24:116–124. doi: 10.1016/j.ceb.2011.11.001. [DOI] [PubMed] [Google Scholar]
- Winslow MM, Dayton TL, Verhaak RG, Kim-Kiselak C, Snyder EL, Feldser DM, Hubbard DD, DuPage MJ, Whittaker CA, Hoersch S, et al. Suppression of lung adenocarcinoma progression by Nkx2-1. Nature. 2011;473:101–104. doi: 10.1038/nature09881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong CK, Luo W, Deng Y, Zou H, Ye Z, Lin SC. The DIX domain protein coiled-coil-DIX1 inhibits c-Jun N-terminal kinase activation by Axin and dishevelled through distinct mechanisms. J Biol Chem. 2004;279:39366–39373. doi: 10.1074/jbc.M404598200. [DOI] [PubMed] [Google Scholar]
- Zaidel-Bar R, Ballestrem C, Kam Z, Geiger B. Early molecular events in the assembly of matrix adhesions at the leading edge of migrating cells. J Cell Sci. 2003;116:4605–4613. doi: 10.1242/jcs.00792. [DOI] [PubMed] [Google Scholar]
- Zhang K, Corsa CA, Ponik SM, Prior JL, Piwnica-Worms D, Eliceiri KW, Keely PJ, Longmore GD. The collagen receptor discoidin domain receptor 2 stabilizes SNAIL1 to facilitate breast cancer metastasis. Nat Cell Biol. 2013;15:677–687. doi: 10.1038/ncb2743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang XH, Wang Q, Gerald W, Hudis CA, Norton L, Smid M, Foekens JA, Massague J. Latent bone metastasis in breast cancer tied to Src-dependent survival signals. Cancer Cell. 2009;16:67–78. doi: 10.1016/j.ccr.2009.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.