In vivo deletion of USP3, a deubiquitinating enzyme involved in DNA damage repair, increases the incidence of spontaneous cancer and impairs the proliferation and repopulation ability of HSCs.
Abstract
Histone ubiquitination at DNA breaks is required for activation of the DNA damage response (DDR) and DNA repair. How the dynamic removal of this modification by deubiquitinating enzymes (DUBs) impacts genome maintenance in vivo is largely unknown. To address this question, we generated mice deficient for Ub-specific protease 3 (USP3; Usp3Δ/Δ), a histone H2A DUB which negatively regulates ubiquitin-dependent DDR signaling. Notably, USP3 deletion increased the levels of histone ubiquitination in adult tissues, reduced the hematopoietic stem cell (HSC) reserves over time, and shortened animal life span. Mechanistically, our data show that USP3 is important in HSC homeostasis, preserving HSC self-renewal, and repopulation potential in vivo and proliferation in vitro. A defective DDR and unresolved spontaneous DNA damage contribute to cell cycle restriction of Usp3Δ/Δ HSCs. Beyond the hematopoietic system, Usp3Δ/Δ animals spontaneously developed tumors, and primary Usp3Δ/Δ cells failed to preserve chromosomal integrity. These findings broadly support the regulation of chromatin ubiquitination as a key pathway in preserving tissue function through modulation of the response to genotoxic stress.
Maintenance of genome integrity is crucial for organism physiology, including stem cell and tissue homeostasis and tumor avoidance (Hoeijmakers, 2009; Jackson and Bartek, 2009). To protect the genome from the continuous challenge of exogenous as well as endogenous genotoxic agents, cells have evolved highly effective mechanisms, collectively termed the DNA damage response (DDR; Lukas et al., 2011; Polo and Jackson, 2011). Evidence of an essential role for the DDR in tissue homeostasis has been provided in the hierarchically organized process of hematopoiesis. In fact, mouse models with defective genome maintenance mechanisms suffer from severe hematopoietic phenotypes and hematopoietic stem cell (HSC) deficiencies (Ito et al., 2004; Nijnik et al., 2007; Rossi et al., 2007). In these animals, the accumulation of DNA damage correlates with a progressive decline of HSC proliferation and self-renewal capacity (Rossi et al., 2007; Niedernhofer, 2008). Age-dependent accrual of DNA damage occurs also in WT murine and in human HSCs, emphasizing the impact of unresolved DNA stress on the functional decline of adult stem cells, with direct consequences on tissue function (Rossi et al., 2007; Rübe et al., 2011).
DNA damage can elicit different cellular responses in HSCs, depending on the nature and extent of the lesions. For instance, unrepaired DNA double-strand breaks (DSBs) can drive cell cycle arrest and senescence or apoptosis, thus contributing to impairment in stem cell function during aging. When imprecisely repaired, DNA damage can persist as a fixed mutation in HSCs, which can be propagated to daughter cells through self-renewal and differentiation and may ultimately lead to oncogenic transformation (Rossi et al., 2008; Sperka et al., 2012). Specific properties distinguish HSCs from more restricted progenitors. Among these features is cell cycle regulation, as HSCs are mostly in a quiescent state, whereas restricted progenitors divide more frequently (Kiel et al., 2007a). Furthermore, HSCs reside in hypoxic niches in the BM. Both conditions limit the chance of HSCs to be exposed to physiological DNA damage by replication errors or reactive oxygen species, respectively (Morrison et al., 1995; Morrison and Spradling, 2008; Rossi et al., 2008). Finally, a different DDR and the preferential use of certain DNA repair pathways may also play a role in determining how DNA damage is sensed by stem cells, repaired, or propagated to their progeny (Milyavsky et al., 2010; Mohrin et al., 2010).
The functional consequence of aging on the HSC pool is a reduced capacity in replenishing the blood system with mature cells, as well as an altered lineage potential (Rossi et al., 2008; Pang et al., 2011; Geiger et al., 2013). Recent data point to a contribution of DDR to this phenomenon. In fact, genotoxic stress/telomere attrition can activate an HSC-specific differentiation checkpoint that drives premature lymphoid differentiation, limiting the self-renewal potential of damaged HSCs and concomitantly depleting lymphoid-competent HSCs (Wang et al., 2012). Despite the recent advances, much remains to be learnt about the molecular pathways that limit the function of aging HSCs.
Ubiquitin (Ub) modification plays a central role in the response to DSBs, ensuring efficient DNA damage recognition, signaling, and repair (Lukas et al., 2011; Jackson and Durocher, 2013). After phosphorylation of the histone variant H2AX (yielding γH2AX), the coordinated activity of the RNF8 (Huen et al., 2007; Kolas et al., 2007; Mailand et al., 2007; Wang et al., 2007) and RNF168 E3 ligases leads to histone H2A and H2AX ubiquitination (Doil et al., 2009; Pinato et al., 2009; Stewart et al., 2009), which is required for efficient and stable accumulation of DDR factors, such as 53BP1 and the tumor suppressor BRCA1 (Lukas et al., 2011; Jackson and Durocher, 2013). DSB-induced ubiquitination engages several E3 ligases (Lukas et al., 2011; Jackson and Durocher, 2013). Among these, the Polycomb Ring1b/Bmi1 mono-ubiquitinate histone H2A on lysine 119 (K119; Ismail et al., 2010; Ginjala et al., 2011; Vissers et al., 2012). Under physiological conditions, this is a prevalent modification that can constitute ∼10–15% of nuclear H2A and it acts as a repressive mark for gene transcription (Goldknopf et al., 1977; Simon and Kingston, 2013). RNF8-RNF168 ligases target H2A on a distinct site, lysine 13/15 (Gatti et al., 2012; Mattiroli et al., 2012), and this K15ub mark is read by 53BP1 (Fradet-Turcotte et al., 2013). The interplay between the two Ub marks on H2A is still poorly understood (Shanbhag et al., 2010; Ginjala et al., 2011; Mattiroli et al., 2012).
Ubiquitination is dynamic and reversible (Komander et al., 2009), and to date only a few deubiquitinating enzymes (DUBs) have been implicated in negative regulation of the RNF8–RNF168 pathway (Lukas et al., 2011; Jackson and Durocher, 2013; Mosbech et al., 2013; Panier and Durocher, 2013). Among these, our group has identified the Ub-specific protease 3 (USP3). USP3 is a chromatin-associated DUB strongly active in reverting histone H2A and H2B mono-ubiquitination (Nicassio et al., 2007). Several observations link USP3 activity to DDR. First, transient USP3 silencing led to spontaneous DNA breaks, enhanced 53BP1 nuclear foci, and DDR activation in human cancer cells. Second, upon ionizing radiation (IR), USP3-depleted cells were unable to efficiently resolve γH2AX and Ub conjugates at DNA damage foci, with a concomitant delay in cell cycle recovery (Nicassio et al., 2007). Finally, overexpression of USP3 opposes the accumulation of RNF168 and the downstream mediator 53BP1 at IR-induced DSBs in a manner dependent on its catalytic activity (Doil et al., 2009; Mosbech et al., 2013; EC, unpublished data). Altogether, these findings support a role for USP3 in counteracting RNF8-RNF168–mediated signaling by removal of Ub marks from chromatin. However, it remains unresolved whether USP3 is key in chromatin ubiquitination in vivo. Here, we investigate the consequences of USP3 loss of function in vivo, which enabled us to discover a critical role for de-ubiquitination in maintaining tissue homeostasis under physiological conditions and stress response. More generally, our findings indicate that deubiquitination events are as critical as ubiquitination for preventing chronic genotoxic stress in vivo.
RESULTS
USP3-deficient mice show normal postnatal development and genome-wide increase in mono-ubiquitinated H2A and H2B
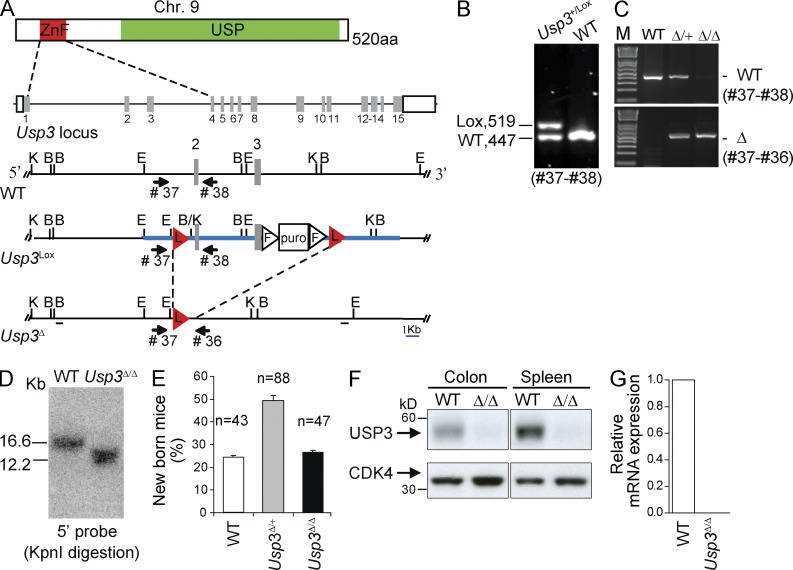
To investigate the biological role of USP3, we have generated conditional USP3-deficient mice (Fig. 1, A and B; Materials and methods). Germline disruption of the floxed Usp3 allele was achieved through the actin-cre deleter strain (Fig. 1, C and D). Usp3-deleted (Usp3Δ/Δ) pups were born at Mendelian ratio and with no overt phenotypes in the presence of effective USP3 deletion as measured by protein and mRNA levels (Fig. 1, E–G).
Figure 1.
Usp3Δ/Δ mice are viable. (A) Generation of conditional (Usp3Lox) and null (Usp3Δ) USP3 alleles. USP3 protein domains and gene locus are schematically represented. ZnF, zinc finger Ub binding domain (ZnF-UBP); USP, Ub-specific protease domain. The targeting construct for Usp3 (thick blue line) contains LoxP (L, red triangles) sites positioned in introns flanking exon 2 and 3. Numbered gray boxes: exons. Triangles: FRT (F) sites. Puro: puromycin Dtk selection cassette. Restriction enzymes used for screening: B, BamHI; E, EcoRI; K, KpnI. Thick black lines: DNA probes used in Southern blot analysis. (B) PCR analysis of genomic DNA isolated from targeted ES clones. (C–G) Actin-Cre deleter strain was used for germline deletion and intercrossing of Usp3Δ/+ mice produced Usp3Δ/Δ homozygous animals, confirmed by PCR analysis (C) and Southern blot (D) on tail tip DNA. (E) Genotype frequency per litter, on a total of 24 litters. n = number of born mice/genotype. Mean ± SD is shown. (F) Immunoblot of whole cell extract (WCE) from tissues from WT and Usp3Δ/Δ mice with anti-USP3 and anti-CDK4 antibody. (G) Reverse transcription qPCR analysis of the relative expression of USP3 transcript in WT and Usp3Δ/Δ MEFs (mouse embryonic fibroblasts).
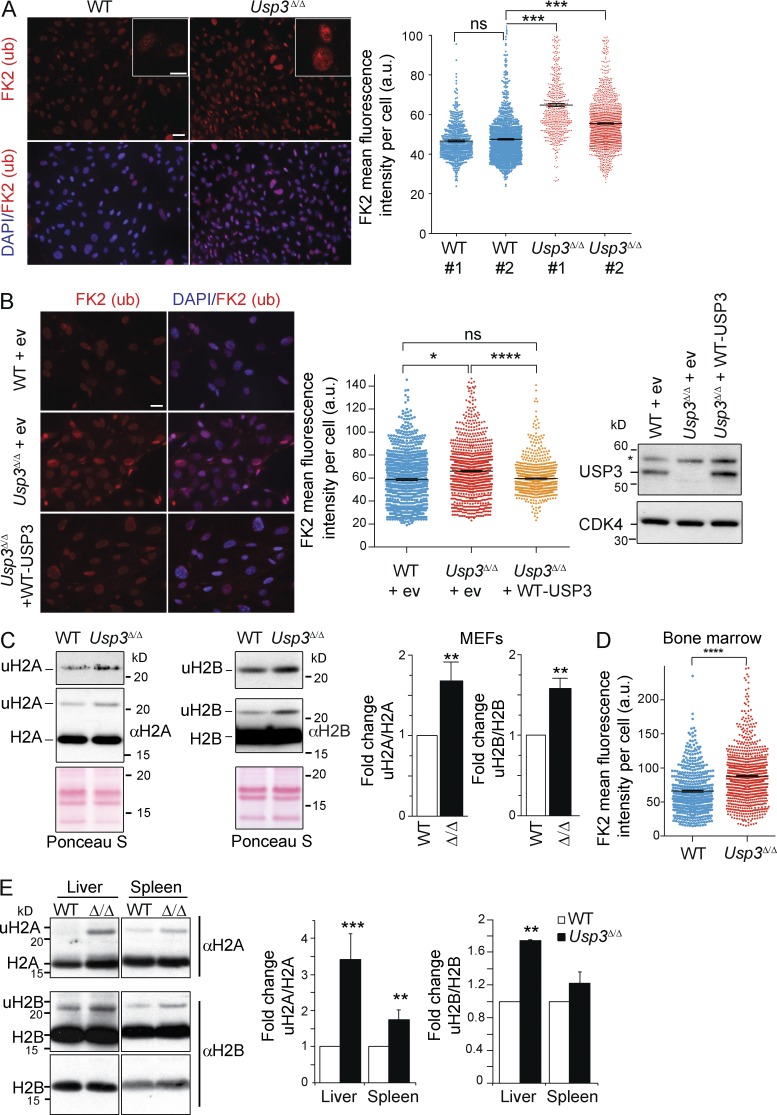
To address the effect of loss of USP3 on nuclear ubiquitination, we performed immunofluorescence analysis with the pan anti–mono- and –poly-ubiquitinated substrate FK2 antibody. USP3Δ/Δ primary mouse embryonic fibroblasts (MEFs) displayed a significantly higher FK2 signal and a pronounced enrichment in nuclear foci compared with WT cells (Fig. 2 A). Attributing increased ubiquitination to USP3 loss of function, ubiquitination levels were effectively rescued when an active USP3 protein was stably reexpressed in these cells (Fig. 2 B). To assess if histone ubiquitination contributed to the FK2 signal, we isolated core histones from MEFs. Immunoblotting revealed that the mono-ubiquitinated forms of H2A (uH2A) and H2B (uH2B) were reproducibly more abundant in Usp3Δ/Δ compared with WT MEFs (Fig. 2 C). Enhanced chromatin-bound FK2 Ub signal and uH2A/uH2B levels were also found in vivo, as exemplified by freshly isolated BM cells and by histone fractions purified from spleen and liver from Usp3Δ/Δ mice (Fig. 2, D and E). Together, our data indicate that USP3 is dispensable for normal embryogenesis and postnatal development but it is required to prevent accumulation of uH2A and uH2B in adult tissues, providing our previous in vitro data (Nicassio et al., 2007) with in vivo significance.
Figure 2.
USP3 deletion leads to a genome-wide increase in mono-ubiquitinated H2A (uH2A) and H2B (uH2B) in mouse cells and tissues. (A) Immunostaining of WT and Usp3Δ/Δ MEFs with anti-Ub (FK2) antibody (red) and DAPI (blue). The FK2 signal intensity per nucleus was quantified by ImageJ. A minimum of 1,000 cells/sample was analyzed. Data are means ± SEM of two independent MEF lines per genotype. Bars: 500 µm; (inset) 10 µm. (B) WT or Usp3Δ/Δ MEFs were infected with control retrovirus (empty vector, ev) or with retrovirus expressing WT USP3 (WT-USP3) and immunostained with FK2. Representative images and FK2 signal quantification as in A. Right panel: immunoblot of MEFs WCE for USP3 and CDK4 (*, nonspecific protein band). Data are means ± SEM of two independent experiments with a minimum of 800 cells/genotype. Bar, 500 µm. (C) Immunoblot of core histone fraction from WT and Usp3Δ/Δ MEFs. Quantification by ImageJ of the uH2A and uH2B signal normalized, respectively, to H2A or H2B, averaged from four (uH2A) or three (uH2B) independent MEF lines per genotype is shown. Data are means ± SD. (D) FK2 staining on freshly isolated BM cells from WT and Usp3Δ/Δ mice (n = 3 per genotype). Signal intensity was quantified as in A. WT, n = 555; Usp3Δ/Δ, n = 938. Data are means ± SEM. (E) Immunoblot of core histones fraction from liver and spleen of WT and Usp3Δ/Δ. uH2A and uH2B were quantified as in C. uH2A, n = 3 mice; uH2B, n = 2 mice per genotype. Data are means ± SD. For all panels: *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; ****, P ≤ 0.0001.
Loss of USP3 leads to shorter life span and increased cancer incidence
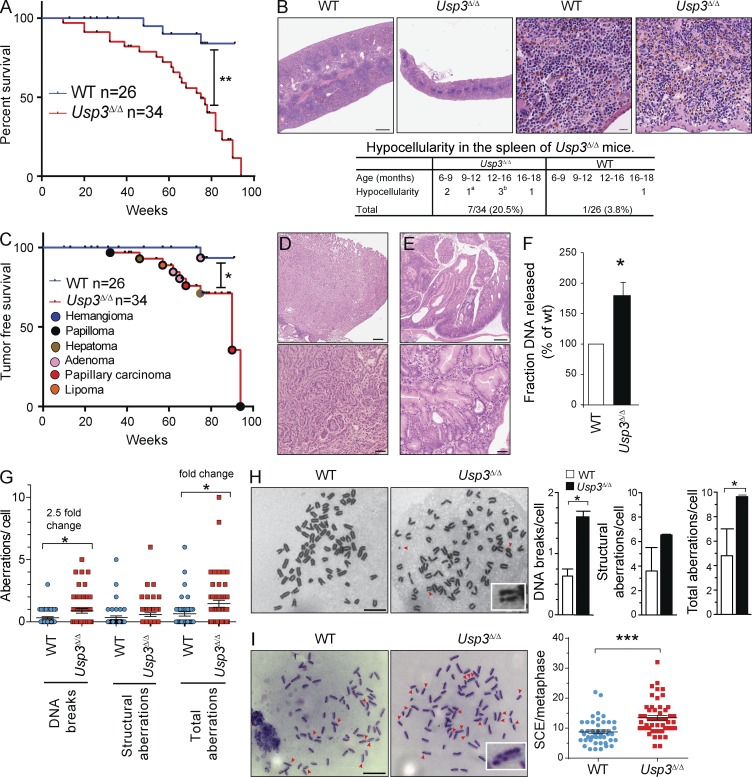
To examine the effect of USP3 deletion on animal life span, we monitored cohorts of Usp3Δ/Δ (n = 34) and WT (n = 26) mice up to 90 wk of age. Kaplan Meier survival analysis indicated that Usp3Δ/Δ mice have a significantly shorter lifespan compared with their WT littermates (Fig. 3 A).
Figure 3.
Usp3Δ/Δ mice exhibit shorter lifespan, increased tumorigenesis, and spontaneous genotoxic stress in MEFs. (A–E) Cohorts of WT (n = 26) and Usp3Δ/Δ (n = 34) mice were monitored for survival for 90 wk. (A) Kaplan Meier general survival analysis. (B) Histopathological analysis of spleens from WT and Usp3Δ/Δ mice and representative H&E-stained spleen sections from 5-mo-old animals. Bars: (left) 500 µm; (right) 20 µm. aLow myelopoiesis in one 10-mo-old Usp3Δ/Δ animal; blow lymphoid compartment in a 15-mo-old Usp3Δ/Δ mouse. (C) Kaplan Meier tumor-free survival analysis and distribution of tumor types in Usp3Δ/Δ mice. (D and E). H&E staining of histological sections of representative malignancies in Usp3Δ/Δ mice. (D) Moderately differentiated papillary carcinoma of the lung (17 mo). (E) Adenomatosis in the stomach (14 mo). Bars: (top) 500 µm; (bottom) 50 µm. (F) Constant field gel electrophoresis (CFGE) analysis of WT and Usp3Δ/Δ MEFs. Results are the mean ± SD of three independent experiments. (G) Quantification of chromosomal aberrations in metaphase preparations of WT and Usp3Δ/Δ MEF. A minimum of 42 cells/genotype was assessed. Mean ± SEM of one of two representative experiments is shown. (H) Metaphase analysis of WT and Usp3Δ/Δ MEFs immortalized with p53 knockdown (sh-p53). Arrowheads: chromatid break, chromosome fragment, ring chromosome. Inset, chromatid break. Results are the mean ± SD of three independent experiments with a minimum of 30 metaphase/genotype each counted. Bar, 10 µm. (I) SCEs analysis in WT and Usp3Δ/Δ sh-p53 MEFs. SCEs in representative metaphases are indicated by arrows. Inset, chromosome with double SCE. SCEs in a minimum of 48 cells/genotype were quantified. Mean ± SEM of one of two representative experiments is shown. Bar, 10 µm. For all panels: *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001. P-value was assessed by Log-rank test (A and C) or by Student’s t test (F–I).
We detected a hypocellular spleen in 7 out of 34 Usp3Δ/Δ mice (20.5%) between 6 and 18 mo, whereas only 1 out of 26 (3.8%) WT mice showed such phenotype at the age of 18 mo, suggesting a decline in adult tissue function (Fig. 3 B). Reinforcing this, clinicopathological analysis revealed that Usp3Δ/Δ mice had a significant decrease in tumor-free survival compared with their WT counterparts (Fig. 3 C). Spontaneous tumors occurred in 11 of 34 (32.3%) Usp3Δ/Δ mice compared with only 1 of 26 (3.8%) WT mice. (Fig. 3 C and Table S1). Pre-neoplastic lesions and a broad spectrum of tumor types in the lung, gastrointestinal tract, and brain were specifically associated with the Usp3Δ/Δ genotype (Table S1; Fig. 3, C–E). Collectively, our data show that Usp3Δ/Δ mice are susceptible to tissue function decline and neoplasia, a phenotype consistent with earlier studies on physiological ageing and experimentally enforced accumulation of DNA damage (Hoeijmakers, 2009; Jackson and Bartek, 2009).
Usp3Δ/Δ MEFs exhibit spontaneous chromosomal breaks and enhanced mitotic recombination
Tissue function decline may occur in the presence of sustained DNA damage (Hoeijmakers, 2009; Jackson and Bartek, 2009). We therefore sought to determine the impact of USP3 deletion on genome integrity. By comparing Usp3Δ/Δ to WT MEFs, we measured a significant increase in spontaneous DNA breaks by constant field gel electrophoresis as well as enhanced chromosomal breaks and structural abnormalities (including dicentric, ring chromosomes, and chromosome fusions) by metaphase spread analysis (Fig. 3, F and G).
Spontaneous DSBs upon USP3 loss may lead to gross genomic rearrangements by triggering DSB repair pathways. In cycling cells, DBS repair by homologous recombination results in crossover events at a low rate. These can be visualized cytologically as sister chromatid exchanges (SCEs), and their frequency is an indicator of chromosomal instability (Sonoda et al., 1999). To enable primary cell proliferation in the presence of active DDR, we immortalized WT and Usp3Δ/Δ MEFs with p53 knockdown (shp53-MEFs). The immortalization did not affect the chromosomal aberrations in Usp3Δ/Δ cells, which were comparable to those observed in p53-competent background (Fig. 3 H). In turn, we measured SCEs by 5′-bromodeoxyuridine labeling during DNA synthesis. Consistent with our hypothesis, the frequency of spontaneous SCEs was significantly higher in Usp3Δ/Δ shp53-MEFs compared with WT shp53-MEFs (Fig. 3 I). Collectively, our data support a role for USP3 in genome maintenance. Defects in this function are strongly associated with chronic genotoxic stress and are causal to spontaneous tumorigenesis, notably in mice lacking DDR components (Hoeijmakers, 2009; Jackson and Bartek, 2009).
USP3-deficient mice develop lymphopenia with age
Mouse models with a defective DDR display severe hematopoietic phenotypes and HSC deficiencies (Ito et al., 2004; Nijnik et al., 2007; Rossi et al., 2007). Consistently, Usp3Δ/Δ mice showed defects in hematopoietic tissues (Fig. 3 B). Therefore, we set out to determine whether USP3 deficiency functionally affected hematopoiesis.
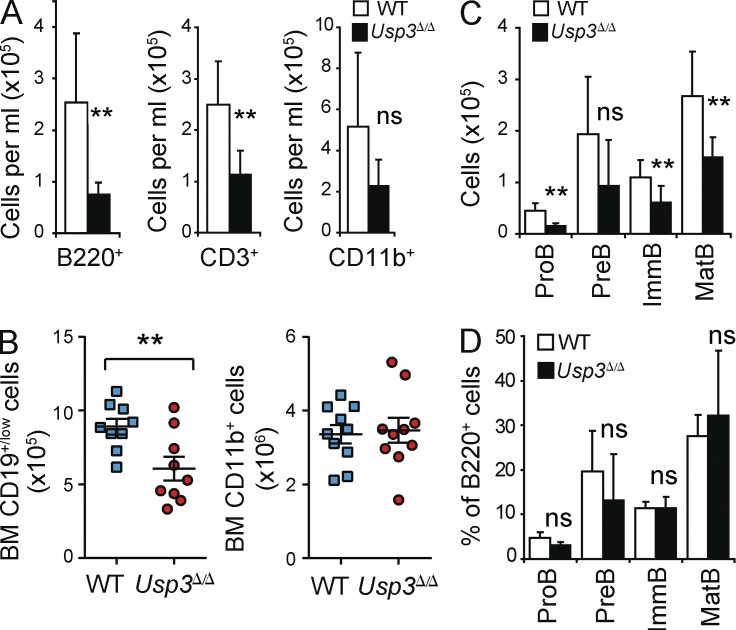
First, we analyzed peripheral blood in 10–17- and 44-wk-old (aged) animals. We found a considerable decrease in the numbers of white blood cells (WBCs) in Usp3Δ/Δ aged mice only compared with their WT littermates, suggesting an age-dependent cellular attrition in the hematopoietic system in Usp3Δ/Δ mice. Lymphocytes were more affected by USP3 deletion (Table 1).
Table 1.
Peripheral blood parameters of WT and Usp3Δ/Δ mice at 10–17 and 44 wk of age
| Cohort (n) | Age | WBC | Lymphocyte | RBC | Hgb | Plt |
| wk | ×109/liter | ×109/liter | ×1012/liter | mM/liter | ×109/liter | |
| WT (9) | 10-17 | 4.8 ± 1.3 | 4.4 ± 1.3 | 4.6 ± 0.3 | 4.5 ± 0.4 | 621 ± 184 |
| Usp3Δ/Δ (13) | 10-17 | 4.2 ± 1.4 | 3.8 ± 1.3 | 4.4 ± 0.2 | 4.3 ± 0.2 | 584 ± 192 |
| WT (10) | 44 | 5.7 ± 1.4 | 5.1 ± 1.0 | 4.6 ± 0.3 | 4.5 ± 0.3 | 628 ± 292 |
| Usp3Δ/Δ (13) | 44 | 3.6 ± 1.4*** | 3.2 ± 0.9*** | 4.3 ± 0.1* | 4.3 ± 0.2 | 506 ± 167 |
Hgb, hemoglobin; Plt, platelets. Significant difference between USP3Δ/Δ and WT mice as determined by Student’s t test: *, P ≤ 0.05; ***, P ≤ 0.001. Numbers indicate means ± SEM. Results are from three independent experiments.
Next, we quantified our results using flow cytometry. Significant absolute and relative loss of both B (B220+) and T (CD3+) lymphoid lineages was observed in the blood of aged mice, whereas alterations in the myeloid population (CD11b+) fell below the threshold for significance (Fig. 4 A and Fig. S1). Importantly, skewed hematopoiesis was mirrored in the BM of Usp3Δ/Δ mice. FACS analysis showed that the lymphoid compartment (CD19+) was substantially reduced in aged but not in 17-wk-old Usp3Δ/Δ mice, whereas the myeloid (CD11b+) lineage was not considerably affected (Fig. 4 B and not depicted). During B cell development, all B cell subpopulations were uniformly depleted in aged Usp3Δ/Δ mice only (Fig. 4, C and D). Thus, loss of USP3 significantly affected hematopoiesis, notably leading to age-dependent cell loss of B and T cell lineages.
Figure 4.
USP3-deficient mice develop lymphopenia with age. (A) Peripheral blood cell counts in aged (44 wk old) WT and Usp3Δ/Δ mice. B220+, B lymphocytes; CD3+, T lymphocytes; CD11b+, monocytes, granulocytes, and macrophages. Data are means ± SD. WT, n = 7; Usp3Δ/Δ, n = 7. Representative FACS profiles are shown in Fig. S1. (B) Flow cytometry analysis of BM of aged WT and Usp3Δ/Δ mice for lymphoid (CD19+/low) and myeloid (CD11b+) cell populations. Cell numbers per BM (2 femurs) are shown. Data are means ± SEM. WT, n = 10; Usp3Δ/Δ, n = 10. (C) Flow cytometry analysis of B cell differentiation in the BM of aged WT and Usp3Δ/Δ mice: Pre–B (B220low IgM− cKit− CD25+), Pro–B (B220low IgM− cKit+ CD25−), immature B (B220low IgM+), and mature B (B220high IgM+) cells. Cell numbers per BM (2 femurs) are shown. Data are means ± SD. WT, n = 8; Usp3Δ/Δ, n = 7. (D) Frequency (percentage of total B220+ B cell population) of the B cell subsets analyzed in C. Results are from two (A, C, and D) or three (B) independent experiments. For all panels: **, P ≤ 0.01; ns, not significant.
Qualitative and quantitative defects in the adult hematopoietic stem and progenitor cell compartment in Usp3Δ/Δ mice
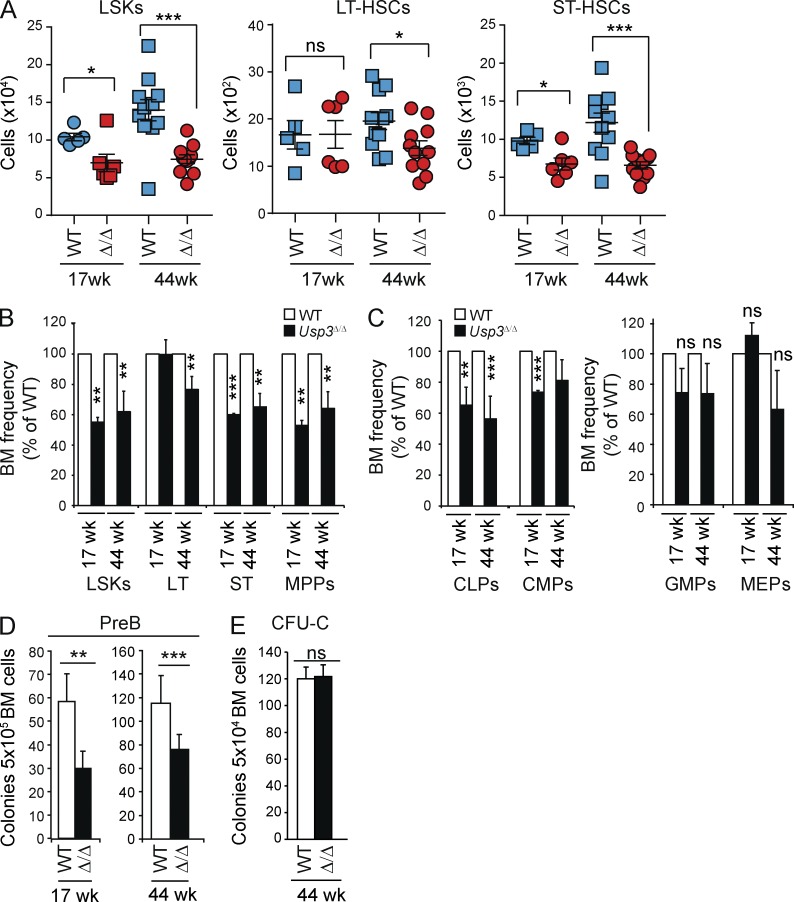
Because B cell defects occurred at early stages of commitment, the USP3 phenotype may arise in the hematopoietic stem and progenitor cell compartment rather than at later stages of development such as at B cell specification or during programmed rearrangements of the IgH locus (Alt et al., 2013). To test this, we performed multiparameter flow cytometry in 17- and 44-wk-old mice on the Lin−Sca1+c-kit+ (LSK) compartment, which is enriched for HSCs. This analysis revealed a profound decrease in the LSK numbers in Usp3Δ/Δ mice at both ages (Fig. 5, A and B; and Fig. S2 A). In addition, although the LSK population expanded in WT mice with age, consistent with previous results (Morrison et al., 1996), no increase in absolute numbers of LSKs was found in Usp3Δ/Δ 44-wk-old animals (Fig. 5 A). These results show that deficiency in the HSC-enriched LSK compartment precedes the reduction in mature cells observed in the blood of Usp3Δ/Δ mice, indicating that this compartment is dependent on USP3.
Figure 5.
Reduced size of adult HSC and CLP compartments and impaired pre–B lymphoid colony-forming activity in vitro in Usp3Δ/Δ mice. (A–C) Multiparameter flow cytometry analysis of primitive hematopoietic populations. Gating strategies and representative FACS profiles are presented in Fig. S2. (A) Absolute cell numbers of primitive populations from BM (2 femurs and 2 hips bones) of WT and Usp3Δ/Δ mice: LSK (Lin− Sca1+ cKit+), LT-HSC (LSK, flk2/CD135−, CD150+, CD34−, LT-HSC), and ST-HSC (LSK, flk2/CD135−, CD150+, CD34+, ST-HSC). Mean ± SEM is shown. (B and C) Frequency of LSKs, LT-HSCs, ST-HSCs, and MPPs (B) or CLPs, CMPs, GMPs, and MEPs (C) in BM of Usp3Δ/Δ mice was calculated and normalized relative to WT animals. Mean ± SD is shown. (A–C) Results are from two (17 wk) or three (44 wk) independent experiments. 17 wk, n = 5 per genotype; 44 wk, n = 11 per genotype. (D and E) BM cells from WT or Usp3Δ/Δ mice were assayed for pre–B (D) or myeloid colony-forming (CFU-C; E) ability. Results are from at least two independent experiments, n = 3 per group per experiment. Mean ± SD is shown. For all panels: *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; ns, not significant.
The LSK compartment includes subpopulations of both long-term HSC (LT-HSC; cells capable of long-term reconstitution of the hematopoietic system) and short-term HSC (ST-HSC), as well as multipotent progenitors (MPPs; Osawa et al., 1996; Christensen and Weissman, 2001; Yeung and So, 2009). To discriminate between these, we used the CD150 SLAM HSC surface receptor in conjunction with the LSK, flt2/CD135, and CD34 markers (Kiel et al., 2007b; Fig. S2 A). In aged mice, USP3 loss resulted in significantly lower absolute numbers and frequencies of all three primitive populations: LT-HSCs (LSK, flk2/CD135−CD34−CD150+; 1.4-fold reduction), ST-HSCs (LSK, flk2/CD135−CD34+CD150+; 1.8-fold reduction), and MPPs (LSK, flk2/CD135+CD150−; twofold reduction). In contrast, at 17 wk of age, Usp3Δ/Δ animals only showed a reduction in ST-HSCs and MPPs, whereas the least proliferative LT-HSCs were less affected by USP3 deletion (Fig. 5, A and B).
In the more mature progenitor pools, USP3 loss resulted in a preferential decrease in common lymphoid progenitors (CLPs) already at 17 wk of age, with minimal changes in the numbers of common myeloid progenitors (CMPs), granulocyte-monocyte progenitors (GMPs), or megakaryocyte-erythrocyte progenitors (MEPs), indicating that the latter can functionally cope with a reduced pool of LSKs (Fig. 5 C and Fig. S2 B). Deficiency in CLP homeostasis was validated in vitro as colony formation capacity of Usp3Δ/Δ BM cells in methylcellulose specifically affected lymphoid progenitors (Fig. 5, D and E).
Thus, based on phenotypic profiling, loss of USP3 had a significant effect on the size of the HSC, LSK, and CLP compartments that was aggravated upon aging, and caused quantitative and qualitative defects in colony-forming ability in lymphoid progenitor populations. Together with the notion that early embryogenesis is not affected in Usp3Δ/Δ mice, the above results indicate that USP3 is functionally required for adult HSC maintenance.
USP3-deficient HSCs display cell-autonomous functional decline
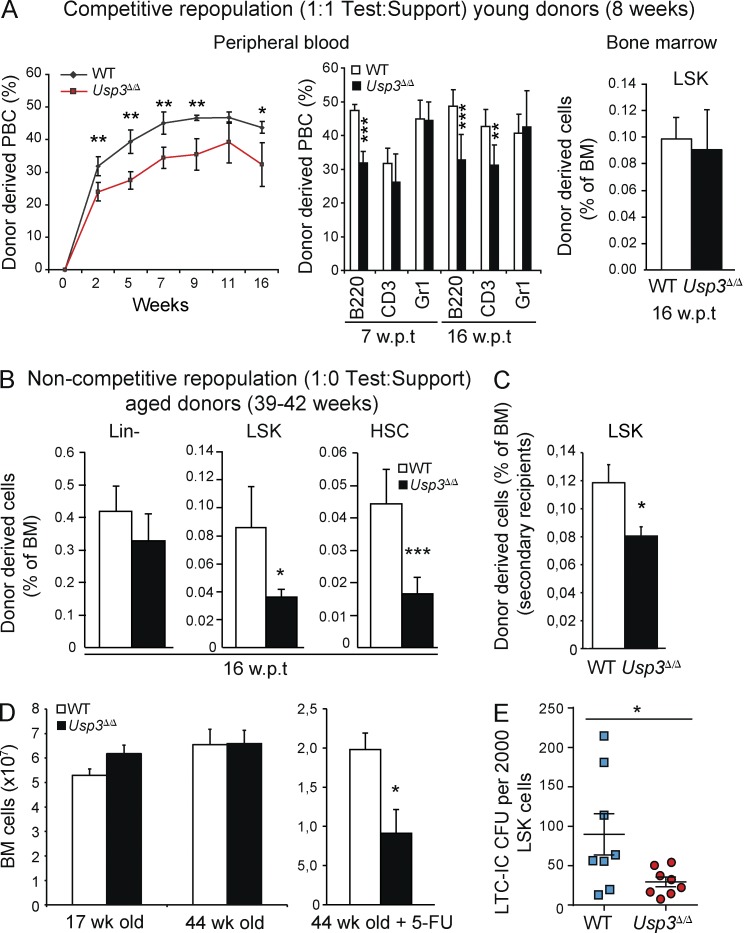
To investigate the basis of the Usp3Δ/Δ HSC phenotype, we performed in vivo and in vitro functional assays. To functionally test the HSC repopulation ability, we transplanted WT or Usp3Δ/Δ BM (CD45.2) cells in a 1:1 ratio with WT Bl6 competitor population (CD45.1) into lethally irradiated WT Bl6 recipients (CD45.1). In this competitive transplantation experiment, we chose to use young adult mice (8 wk old), before numerical defects would affect the Usp3Δ/Δ progenitor compartment. Consistent with our previous observation in Usp3Δ/Δ animals (Figs. 4 and 5), 2–16 wk post transplantation (wpt) peripheral blood analysis showed a moderate but significant decrease in the repopulating ability of Usp3Δ/Δ BM cells compared with WT donors (Fig. 6 A, left). In particular, Usp3Δ/Δ donor cells poorly contributed to the lymphoid B and T lineages (B220+ and CD3+, respectively). As expected, the myeloid (Gr1+) and the LSK compartments were unaffected (Fig. 6 A, middle and right, respectively). Next, we performed a noncompetitive repopulation assay, in which 100% of WT or Usp3Δ/Δ BM from 39–42-wk-old mice was transplanted. To cope with the reduced number of HSCs in Usp3Δ/Δ mice at this age, we normalized the BM dose to the actual HSCs present at the time of transplantation (stem cell equivalent BM). Nonetheless, Usp3Δ/Δ LSKs and primitive HSCs in recipient BM at 16 wpt showed significantly less engraftment (Fig. 6 B). Finally, we performed serial transplantations. Reasoning that sequential transplantation forces LT-HSC to self-renew and to differentiate to repeatedly reconstitute the recipient hematopoietic system (Purton et al., 2006), we chose to use BM cells from young (8 wk old) donor mice. Notably, at 12 wpt, secondary recipients of Usp3Δ/Δ BM displayed a significantly reduced donor-derived LSK population compared with WT (Fig. 6 C).
Figure 6.
USP3-deficient HSCs have a cell-autonomous defect in repopulating ability in vivo and in colony formation in vitro. (A) Competitive transplantation of BM cells from 8-wk-old WT or Usp3Δ/Δ (CD45.2; test) mice with WT (CD45.1; support) BM cells showing total reconstitution (left) and contribution of donor-derived cells to B cell (B220+), T cell (CD3+), and myeloid (Gr1+) lineages (middle) in the blood, or to LSKs in the BM (right) of irradiated recipients at the indicated wpt. Data are mean ± SD (n = 5 per genotype). One of two representative experiments is shown. PBC, peripheral blood cell. (B) Noncompetitive transplantation of BM cells from aged (39–42 wk old) WT or Usp3Δ/Δ mice. Donor-derived Lin−, LSKs, and HSCs in primary recipients at 16 wpt is shown. Data are mean ± SD (n = 5 per genotype). (C) WT or Usp3Δ/Δ BM cells from 8-wk-old mice were used in noncompetitive serial transplantations. Donor-derived LSKs in the BM of secondary recipients (separated by a 12 wk reconstitution period) are shown. Data are mean ± SEM (n = 5 per genotype). (D) Total BM cell numbers in WT or Usp3Δ/Δ mice at 17 and 44 wk of age (WT = 5, Usp3Δ/Δ = 6, in two independent experiments; 2 femurs and 2 hip bones) or in 44-wk-old mice (n = 3 per genotype) upon 5-FU treatment (2 femurs). Data are mean ± SEM. (E) LTC-IC assay using WT or Usp3Δ/Δ Lin− cells purified from 8–16-wk-old mice (three experiments, n = 4 mice/genotype/experiment). The number of LSKs in the Lin− populations was evaluated by phenotypic profiling before plating, and results are expressed as total number of CFU-C normalized to 2,000 LSK plated. Data are mean ± SEM. In all BM transplantations, BM cells corresponding to stem cell equivalents were transplanted. In B and C, BM cells from n = 3 donor mice per genotype were pooled before primary transplantation. For all panels: *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001.
To examine the impact of USP3 loss on HSC homeostasis in response to an external and temporally controlled source of hematopoietic stress, we exposed the mice to sequential injections of the pyrimidine analogue 5-Fluorouracil (5-FU). Although we did not detect BM hypocellularity in Usp3Δ/Δ mice at any age, a continuous regimen of 5-FU that induces severe myelosuppression exposed the defect in the BM reconstituting ability of aged Usp3Δ/Δ mice as estimated by the significant decrease in Usp3Δ/Δ BM cellularity compared with WT animals (Fig. 6 D). These data indicate that chronic stress exposure exacerbates the Usp3Δ/Δ phenotype in the early progenitor compartment, the LT-HSCs.
Finally, to corroborate our in vivo findings with an in vitro functional assay, we performed long-term culture–initiating cell (LTC-IC) assays, a retrospective assessment of the presence of functional HSCs via ex vivo propagation (van Os et al., 2008). To this end, we purified Lineage negative (Lin−) cells from 15–17-wk-old Usp3Δ/Δ and WT mice. After long-term culturing, Usp3Δ/Δ Lin− cells showed a significant decrease in colony-forming ability in methylcellulose, indicating a reduced activity of Usp3Δ/Δ primitive hematopoietic cells (Fig. 6 E). Collectively, these findings experimentally support a predominantly cell-autonomous function of USP3 in HSCs under both homeostatic conditions and stress-induced hematopoiesis.
USP3-deficient HSCs suffer from greater DNA damage in vivo and in vitro
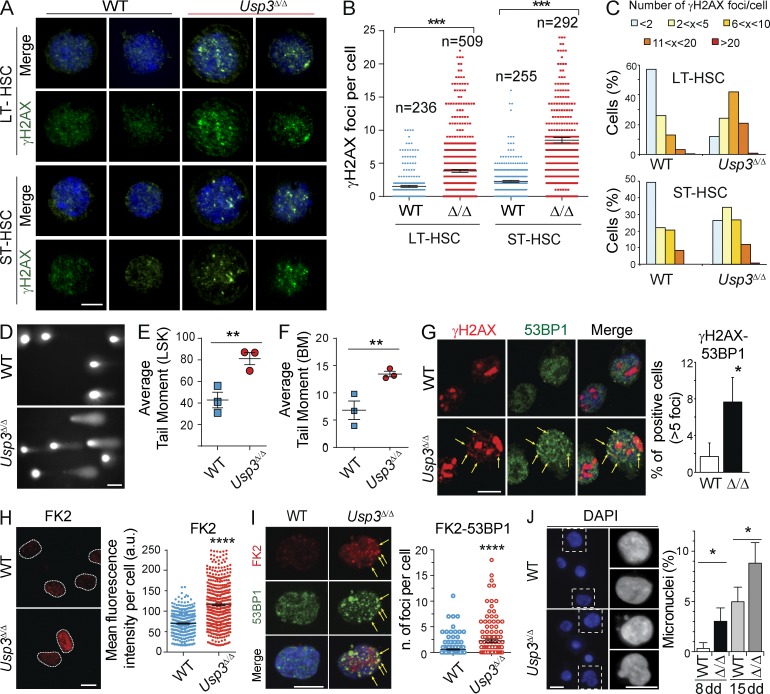
To test whether insufficient preservation of genome integrity underlies the Usp3Δ/Δ hematopoietic phenotype, we first performed immunostaining for γH2AX, a marker of DDR activation (Rogakou et al., 1998). We quantified the number of γH2AX foci in freshly isolated FACS-sorted (sorted) HSCs from aged (44 wk old) mice. Although only rare γH2AX foci were observed in WT HSCs (>80% of LT-HSCs and >60% of ST-HSCs showed no more than two foci per cell), a high percentage of the Usp3Δ/Δ LT-HSCs and ST-HSCs stained positive for γH2AX, with 45 and 85%, respectively, showing multiple foci (Fig. 7, A and B). Accumulation of γH2AX foci in Usp3Δ/Δ progenitors was a predominant, although not exclusive, feature of aged Usp3Δ/Δ HSCs (Fig. 7 C).
Figure 7.
USP3-deficient HSCs accumulate spontaneous DNA damage. (A–C) γH2AX immunostaining on sorted Lin− Sca1+ c-Kit+ CD150+ flk2/CD135− CD34− (LT-HSC) or CD34+ (ST-HSC) from 44-wk-old (A and B) or 17-wk-old (C) mice. Representative images (A) of LT-HSCs and ST-HSCs from 44-wk-old mice and quantification of the number of γH2AX foci/cell in HSCs from 44-wk-old (B) or 17-wk-old mice (minimum of 200 cells per genotype; C). Results are from two independent experiments. n = 3 mice/genotype/experiment. Bar, 5 µm. (D–F) Alkaline comet assay on sorted Usp3Δ/Δ LSKs (D and E) or total BM cells (F). Representative LSKs images (D) and the Average Tail Moment calculated by Comet Score on LSKs (E) or BM cells (F) are shown. A minimum of 150 comets was evaluated per sample. n = 3 per genotype, 44 wk old. Bar, 50 µm. (G–J) Sorted LT-HSCs from BM of 40–44 wk old mice were grown in liquid cultures and analyzed for DNA damage. (G) Immunostaining of γH2AX and 53BP1 on LT-HSCs after 8–11 d in culture. The percentage of cells containing >5 γH2AX and 53BP1 foci was evaluated in three independent experiments. n > 50 cells/genotype/experiment. Arrows: γH2AX-53BP1 colocalizing foci. Bar, 5 µm. (H) Immunostaining of FK2 LT-HSCs after 8–11 d in culture. Representative images and quantification by Image J of FK2 signal intensity from three independent experiments. Nuclei are outlined. n > 100 cells/genotype/experiment. Bar, 10 µm. (I) Co-immunostaining of FK2 and 53BP1 on LT-HSCs after 8–11 d in culture. The number of co-foci (arrows) per cell was quantified in 2 independent experiments, on a total of n = 145 (WT) or 80 (Usp3Δ/Δ) cells scored. Bar, 10 µm. (J) Percentage of micronuclei in LT-HSCs cultures after 8 or 15 d in culture. A minimum of 70 cells/sample was scored in three independent experiments. Bars, 10 µm. In all panels: *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; ****, P ≤ 0.0001. Results are mean ± SEM (B, E, F, H, and I) or mean ± SD (G and J).
To investigate the actual presence of DNA breaks, we performed an alkaline comet assay. A significantly higher proportion of increasingly damaged comet tails was detected in sorted LSKs and in total BM cells from aged Usp3Δ/Δ mice, reflecting accumulation of DNA breaks in these cells (Fig. 7, D–F). Positive detection of comet tails was not due to an increase in apoptosis (see below). Collectively, these results demonstrate that USP3 protects hematopoietic stem and progenitor cells from accumulation of spontaneous DNA damage in vivo.
To test whether DNA damage occurs in Usp3Δ/Δ HSCs owing to enforced proliferation upon stress-induced expansion, we sorted LT-HSCs from aged mice and investigated DNA damage accrual in actively proliferating HSCs in vitro. Analysis of γH2AX and 53BP1 foci indicated increased damage in cultured Usp3Δ/Δ LT-HSCs (Fig. 7 G). Consistent with our previous findings in MEFs, conjugated Ub at the chromatin was also enhanced in Usp3Δ/Δ LT-HSCs, as measured by FK2 immunofluorescence (Fig. 7 H). Because 53BP1 accumulation at DSB sites is dependent on (RNF168-mediated) ubiquitination (Doil et al., 2009; Stewart et al., 2009; Fradet-Turcotte et al., 2013), we asked whether enhanced FK2 would positively correlate with 53BP1 recruitment. Notably, double immunofluorescence staining revealed that colocalizing 53BP1-conjugated Ub foci were indeed enriched in Usp3Δ/Δ LT-HSCs, suggesting that Ub-dependent DNA damage signaling was enhanced in these cells (Fig. 7 I).
To determine possible functional consequences of DNA damage accumulation in the absence of USP3, we analyzed micronuclei formation as biomarkers of genotoxic stress (Fenech et al., 2011). DAPI analysis showed a higher frequency of micronuclei in Usp3Δ/Δ LT-HSCs cultures, which were exacerbated upon culturing (Fig. 7 J). Together, these findings are consistent with 17-wk-old USP3-deficient proliferating ST-HSCs being more vulnerable, and so more easily lost, than quiescent LT-HSCs (Fig. 5, A and B), and implicate an integral role for USP3 in genomic maintenance of HSCs through the control of chromatin ubiquitination.
Increased cellular aging underlies USP3-deficient BM defects
Persistent DNA damage may lead to either DDR-induced differentiation or cell cycle restriction, both of which may account for the phenotypes we discovered in USP3-deficient mice (Rossi et al., 2008; Sperka et al., 2012; Wang et al., 2012). To discriminate between the two possibilities, we first investigated the consequences of USP3 deletion on LSK homeostasis by performing genome-wide expression profiling. RNA-seq of two freshly isolated biological replicas of sorted LSKs from 8-wk-old Usp3Δ/Δ animals showed a very limited number of genes either slightly up- or down-regulated (<100 out of ∼25,000) in Usp3Δ/Δ LSKs, none of which are reported to be directly involved in HSC maintenance or to be linked to premature differentiation (Tables S2–S4). Thus, young adult LSKs perpetuated a stable gene expression program regardless of the homozygous deletion of USP3.
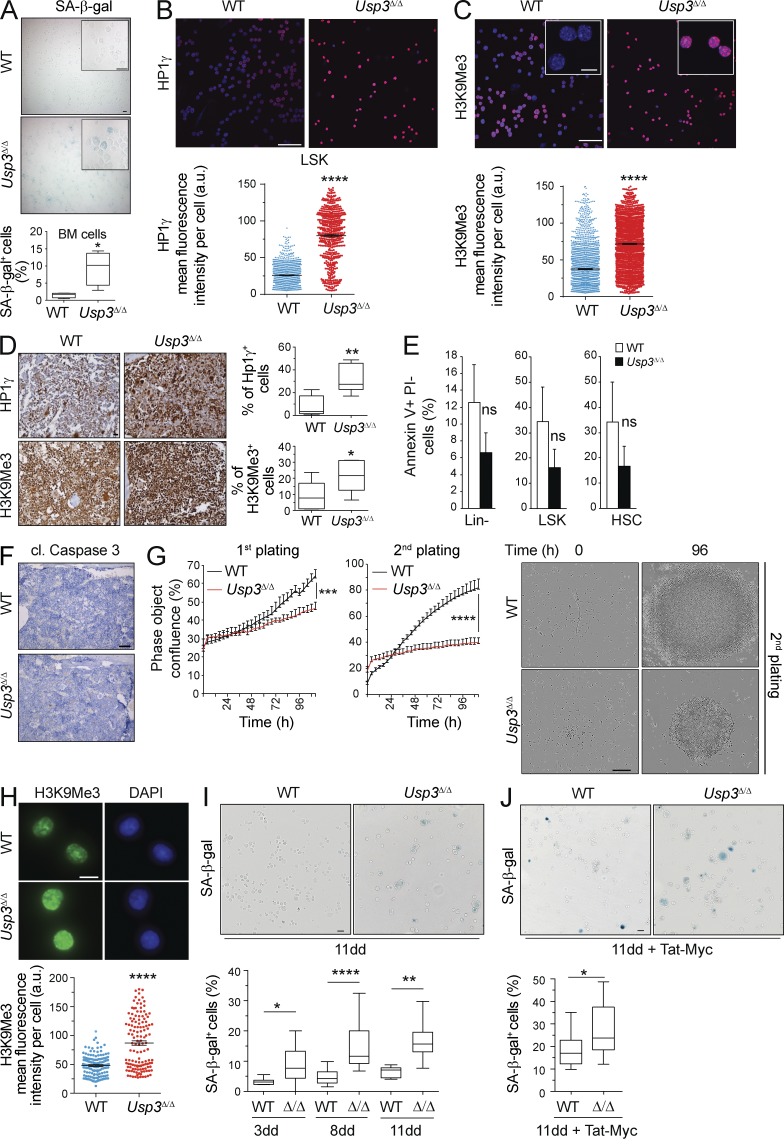
Next, we sought to understand whether enhanced DNA damage and persistent DDR signaling would affect cellular homeostasis in vivo. First, we asked whether USP3-deficient BM displayed senescence-associated β-galactosidase (SA-β-gal) activity directly on freshly isolated BM cells from 32-wk-old mice. Indeed, we measured a 6.5-fold increase in Usp3Δ/Δ SA-β-gal–positive senescent cells compared with WT (Fig. 8 A).
Figure 8.
Cellular senescence in Usp3Δ/Δ HSC compartment and BM. (A) Cytospins of BM cells from Usp3Δ/Δ and WT mice were assayed for SA-β-galactosidase activity. The percentage of SA-β-Gal–positive cells was quantified by counting 100 cells on three separate fields (n = 4 mice per genotype). Bar, 20 µm. (B and C) Cytospin preparations of sorted LSKs from BM of Usp3Δ/Δ or WT mice were immunostained for HP1γ (B) and H3K9Me3 (C). Focal nuclear staining is visible in insets. Signal intensity per nucleus was quantified by ImageJ. n = 3 per genotype. A minimum of 1,000 nuclei/sample was evaluated. Data are mean ± SEM of one of two representative experiments. Bars: 75 µm; (inset) 10 µm. (D) Immunostaining for HP1γ and H3K9Me3 on BM sections from Usp3Δ/Δ and WT mice. The percentage of positive cells was quantified in 3 fields on a minimum of 1,500 cells/field per sample. n = 7 per genotype. Bar, 20 µm. (E) Quantification of apoptotic (Annexin V positive and Propidium Iodide [PI] negative) freshly isolated hematopoietic subpopulations (mean ± SD) from WT or Usp3Δ/Δ. n = 3 per genotype. One of two representative experiments is shown. (F) Representative images of WT and Usp3Δ/Δ BM sections stained for apoptosis-indicating cleavage (cl.) of caspase 3. n = 6 mice per genotype. Bar, 500 µm. (G) Sorted LT-HSCs were plated after 8 d (first plating) or 11 d (second plating) in culture and monitored for growth. Kinetic measures the number of cells, recorded over time and plotted as phase contrast object confluence. n = 4 wells per data point. Mean ± SD of one of two representative experiments is shown. Representative images at time 0 and 96 h after plating are shown. Bar, 300 µm. (H) Immunostaining of in vitro expanded LT-HSCs for H3K9Me3. Signal quantification by ImageJ from two independent experiments is shown (mean ± SEM). n = 150 per genotype. Bar, 10 µm. (I) LT-HSCs cultures were assayed for SA-β-galactosidase activity after 3, 8, or 11 d (dd) in culture. A minimum of 350 (3dd), 2300 (8 dd), or 550 (11 dd) cells counted in 10 separate fields were evaluated. Bar, 20 µm. (J) LT-HSCs cultures were assayed for SA-β-galactosidase activity upon Tat-cMyc protein transduction. A minimum of 1,000 cells per genotype was evaluated in two replicate experiments. Bar, 20 µm. Mice were 32 wk old (A–D) or 40–44 wk old (E and F). G–J: LT-HSCs for in vitro expansion were isolated from 40–44-wk-old mice. For all panels: *, P ≤ 0.05; **, P ≤ 0.01; ****, P ≤ 0.0001; ns, not significant.
Second, we investigated whether senescence may be preceded by and associated with chromatin changes such as increased chromatin condensation and heterochromatin formation. These events were observed during replicative- and oncogene-induced senescence in human cells (Narita et al., 2003; Kuilman et al., 2010; Sulli et al., 2012) and in tumorigenesis in mice (Braig et al., 2005; Reimann et al., 2010). Notably, high expression and focal enrichment of the heterochromatin markers H3K9me3 and the heterochromatin protein 1γ (HP1γ) was apparent by immunostaining in the great majority of sorted Usp3Δ/Δ LSKs, compared with low levels of these markers in WT cells (68.7 ± 8.4% Usp3Δ/Δ vs. 31.7 ± 12.7% WT; and 74.6 ± 16.4% Usp3Δ/Δ vs. 7.8 ± 4.5 WT, respectively; Fig. 8, B and C). Immunohistochemistry on BM sections also revealed greater expression of HP1γ and of H3K9Me3 in Usp3Δ/Δ BM compared with WT littermates (Fig. 8 D). Thus, USP3-deficient LSK and BM cells display increased heterochromatinization and SA-β-gal activity in vivo, which are biomarkers for senescence.
In contrast, Annexin V analysis of early apoptotic cells in freshly isolated Lin−, LSKs, and HSCs revealed no significant differences between the groups, indicating that apoptosis is not the predominant factor underlying cell loss in these Usp3Δ/Δ populations (Fig. 8 E). Also, cleaved caspase 3–positive cells were not readily detected on BM tissue sections of Usp3Δ/Δ mice (Fig. 8 F).
We next tested the impact of cytokine-induced replication on LT-HSCs. Sorted LT-HSCs from 44-wk-old mice divided during the first 3 d of culture, regardless of the genotype. However, WT LT-HSCs subsequently expanded, whereas Usp3Δ/Δ cells exhibited limited division, most notably after passage (Fig. 8 G). This was accompanied by increased heterochromatinization, as indicated by H3K9Me3 immunofluorescence analysis (Fig. 8 H), and SA-β-gal activity (Fig. 8 I). Notably, the fraction of SA-β-gal–positive Usp3Δ/Δ cells progressively augmented upon longer culturing (Fig. 8 I).
To test the impact of replication stress on LT-HSC fitness, we exposed these cells to c-Myc using a cell-permeable protein transduction approach. Notably, a significantly higher percentage of Usp3Δ/Δ LT-HSCs displayed SA-β-gal activity upon Myc expression, suggesting that they are more sensitive to oncogene-induced senescence (Fig. 8 J). Our results indicate that Usp3Δ/Δ HSCs engage cell cycle restriction under the conditions of cytokine-induced and oncogene-induced replicative stress.
USP3 protects HSCs and mice from IR
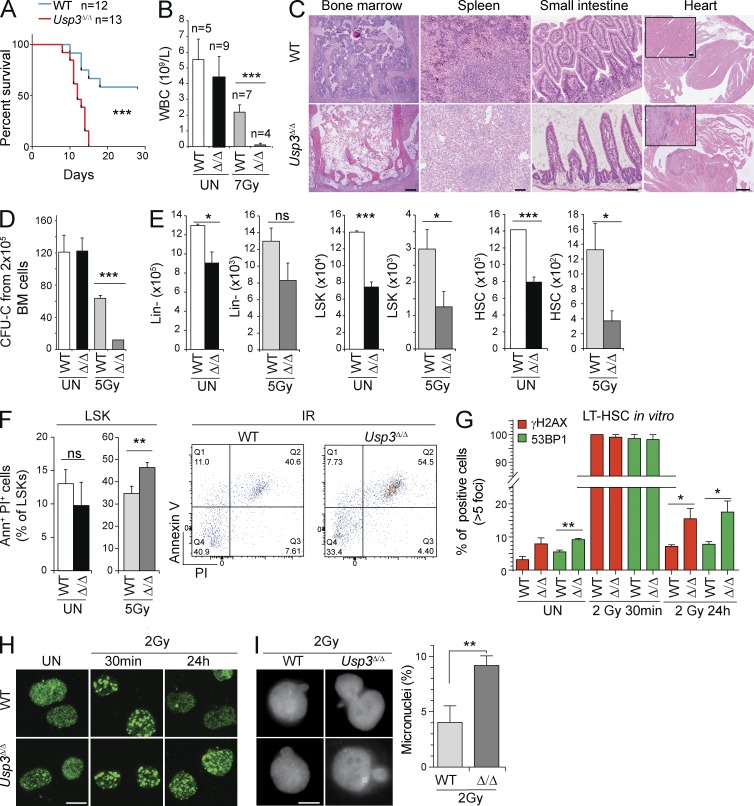
To test the importance of USP3 in preserving the hematopoietic progenitor compartment from the consequences of unscheduled DNA damage, we tested the ability of Usp3Δ/Δ mice to recover from an acute genotoxic stress after IR, which relies on the survival of HSCs to replenish the blood system. 8-wk-old WT and Usp3Δ/Δ animals were exposed to an experimentally determined sublethal dose of 7 Gy total body irradiation (TBI) and were monitored for 28 d. Usp3Δ/Δ mice started to lose weight around day 9 and all of them were sacrificed between days 10 and 16. In contrast, 60% of the WT mice survived (Fig. 9 A). Hematopoietic analysis revealed extreme BM failure (Fig. 9 B). Depletion of hematopoietic activity in BM and spleen was confirmed by histopathological analysis (Fig. 9 C).
Figure 9.
USP3 protects HSCs from genotoxic stress in vivo and in vitro. (A–C) Age-matched (8 wk old) WT (n = 12) and Usp3Δ/Δ (n = 13) mice were exposed to 7 Gy TBI and monitored for 28 d. (A) Kaplan Meier survival curve. P-value was determined by Log-rank test. (B) WBC counts of unirradiated or irradiated mice (WT, 28 d after TBI) and Usp3Δ/Δ (at time of sacrifice due to illness after TBI). Results are mean ± SD from two independent experiments. (C) Representative images of hematoxylin-eosin (H&E)–stained tissue sections of WT (28 d after TBI) and of Usp3Δ/Δ (at the time of sacrifice due to illness after 7 Gy TBI) mice. Bars: (BM) 100 µm; (spleen) 50 µm; (small intestine) 50 µm; (heart) 500 µm; (inset) 50 µm. (D) CFU-C from BM cells of 8-wk-old WT or Usp3Δ/Δ mice. Mice (n = 3 per genotype) were left untreated or subjected to TBI (5Gy). 7 d after IR, BM cells were isolated and plated on methylcellulose with cytokines. Results are means ± SD. (E and F) Absolute numbers (2 femurs and 2 hips bones) of Lin−, LSKs, and HSCs in 44-wk-old mice that were left untreated (UN) or exposed to 5 Gy TBI (5Gy) and sacrificed after 24 h (E). UN, n = 11 per genotype; IR, WT, n = 3; Usp3Δ/Δ, n = 4. (F) Increased cell death in Usp3Δ/Δ LSKs, as determined by Annexin V staining in mice as in E. UN, n = 3 per genotype; IR, WT, n = 3; Usp3Δ/Δ, n = 4. Results are means ± SEM. (G and H) Immunofluorescence of γH2AX and 53BP1 in WT or Usp3Δ/Δ LT-HSCs in culture for 8–10 d. IRIFs were scored in LT-HSCs after mock treatment or at 30 min and 24 h after 2Gy of IR. The percentage of cells containing >5 IRIFs are plotted ± SD. Representative images for 53BP1 staining are shown (H). A minimum of 50 cells/sample/experiment over two (γH2AX) or three (53BP1) independent experiments was evaluated. Bar, 10 µm. (I) Percentage of micronuclei in WT or Usp3Δ/Δ LT-HSCs in culture for 8–10 d. Cells were irradiated with 2Gy and micronuclei scored at 24 h after IR. Results are means of three independent experiments ± SD on a minimum of 70 cells/genotype. Bar, 5 µm. For all panels: *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ P ≤ 0.001; ns, not significant.
To investigate the role of USP3 in the response to IR outside the hematopoietic compartment, we analyzed the murine intestine, with homeostasis comparable to the hematopoietic system. Importantly, depletion of crypts in the mucosa of the small intestine was evident in Usp3Δ/Δ irradiated mice. Notably, multiple hemorrhagic lesions and severe degeneration also affected other organs such as the heart (Fig. 9 C).
Lack of BM reconstitution in irradiated USP3-deficient mice supports higher sensitivity of their hematopoietic stem and progenitor compartment. In vitro, we validated this observation by performing colony formation assays with BM cells from mice before and after IR treatment. Usp3Δ/Δ BM cells plated 7 d after exposure to 5 Gy showed significantly reduced colony formation. Consistent with our previous results (Fig. 5 F), no differences were observed with cells from nonirradiated mice (Fig. 9 D).
To investigate the consequences of IR on HSCs in vivo, mice were exposed to 5 Gy and sacrificed 24 h later. Notably, the Usp3Δ/Δ primitive progenitor compartment (i.e., LSKs and HSCs) was acutely decreased compared with WT controls (Fig. 9 E and Fig. S2 C). Reduction in LSK cellularity upon IR was accompanied by an increase in the fraction of Annexin V+/PI+ cells in Usp3Δ/Δ mice (Fig. 9 F).
To gain mechanistic insight into USP3-mediated protection of HSCs from genotoxic stress, we sorted LT-HSCs and analyzed their response to IR in vitro. Both WT and Usp3Δ/Δ HSCs showed a similar immediate induction of γH2AX and 53BP1 IR-induced foci (IRIF) upon 2 Gy. By 24 h after IR, however, although the great majority of IRIFs were resolved in WT cells, γH2AX and 53BP1 foci persisted in a significant fraction of Usp3Δ/Δ cells, suggesting a delay in DSB repair (Fig. 9, G and H). This was accompanied by an increase in the number of micronuclei, indicative of the actual presence of broken/lagging chromosome in Usp3Δ/Δ cells (Fig. 9 I).
Together, these data demonstrate that USP3-deficient HSCs are more sensitive to genotoxic stress in vivo and in vitro and support the notion that unresolved DNA damage affects the functional integrity of Usp3Δ/Δ HSCs in homeostasis, as well as upon aging and stress. Overall, the data point to USP3 as a critical mediator of DDRs in vivo.
DISCUSSION
We determine here the impact of negative regulation of DNA damage-induced Ub signaling on organism physiology, which was largely unexplored. USP3 is one of the few DUBs that have been linked to this aspect of DDR (Nicassio et al., 2007; Doil et al., 2009; Jackson and Durocher, 2013; Panier and Durocher, 2013; Sharma et al., 2014). Using a mouse genetics approach, we provide evidence that defective deubiquitination does have profound consequences in vivo, as USP3-deleted mice have a shortened lifespan, associated with a functional decline of the hematopoietic stem and progenitor compartment and with sporadic spontaneous neoplasia. Linked to these phenotypes is chronic genotoxic stress, most likely resulting from the altered histone ubiquitination upon USP3 loss, as measured in vivo and in vitro. In support of this, we find that USP3 protects HSCs from DNA damage through modulation of Ub-dependent DDR signaling. This role in HSC maintenance is, to our knowledge, a previously uncharacterized function of the RNF8-mediated ubiquitination pathway.
USP3 and cancer
Similarly to other components of the Ub-DDR network, such as 53BP1, RNF8, and RNF168 (Ward et al., 2003; Li et al., 2010; Bohgaki et al., 2011; Halaby et al., 2013), Usp3Δ/Δ mice are mildly tumor prone, with up to 9% of the animals showing neoplasia within 1 yr of age. This is consistent with the moderate, though measurable, magnitude of chromosomal aberrations in Usp3Δ/Δ cells. Because a broad spectrum of tumor types was observed, we propose this pathology to be related to a general tissue function decline. Progressive accumulation of mutations as a consequence of unresolved DNA damage in Usp3Δ/Δ cells may underlie this phenotype. Although a genome maintenance defect is a plausible explanation, our results do not rule out the possibility that USP3’s role in tumor suppression may also involve deubiquitination of still uncharacterized substrates, besides H2A and H2B, potentially regulating distinct cellular processes. The ability to deubiquitinate multiple substrates is in fact not unprecedented within the USPs (Komander et al., 2009), and a novel target for USP3, RIG-1, has been recently reported (Cui et al., 2014).
USP3 and Ub-mediated DDR in HSC maintenance
Pathology arising from Usp3 deficiency clearly manifests in hematopoiesis. Although hematopoietic homeostasis is achieved in young adult Usp3Δ/Δ mice, there is indeed a marked decline in HSC activity over time. In physiological conditions, loss of USP3 mainly exacerbate the decline of the lymphoid progenitor compartment, a phenotype reminiscent of an aged hematopoietic system, in which functional decline of aged HSCs is associated with immunodeficiency and skewed differentiation in the elderly (Rossi et al., 2008; Pang et al., 2011). USP3 is accountable for the age-dependent cellular attrition in HSCs, because the BM from young (8 wk old) Usp3Δ/Δ mice, although functionally compromised compared with WT of the same age, performed better than BM from aged (44 wk old) mice. In turn, a gradual loss of functional HSCs is further exacerbated under conditions of stress, as demonstrated by the reduced capacity of Usp3Δ/Δ HSCs for self-renewal and by the lineage skewing upon transplantation in vivo, as well as by the reduced proliferative potential in vitro. Again, these features are characteristic of an aged HSC phenotype (Geiger et al., 2013) and directly implicate USP3 in protecting HSCs from the consequences of physiological stress in vivo.
In agreement with the enhanced DNA breaks in MEFs, USP3 loss was further associated with spontaneous DNA damage in HSCs in vivo and in vitro. Our results support the DDR to be implicated in the functional defects of Usp3Δ/Δ HSCs, as they feature increased radiation sensitivity in vivo and prolonged DDR upon IR in vitro. These findings are in agreement with several studies demonstrating the dependence of the hematopoietic system on an intact DDR (Ito et al., 2004; Nijnik et al., 2007; Rossi et al., 2007; Niedernhofer, 2008). Normocellularity of the BM and a more pronounced skewing toward lymphoid lineages under homeostasis is shared with some DDR-deficient mouse models, such as the Ku80-deficient mice (Rossi et al., 2007). However, the Usp3Δ/Δ HSC phenotype as revealed upon BM transplantation appeared less severe compared with classical components of the HR, NHEJ, NER, or telomere maintenance pathways (Ito et al., 2004; Nijnik et al., 2007; Rossi et al., 2007; Niedernhofer, 2008). Because we observe enhanced ubiquitination in Usp3Δ/Δ mice, these data further support a role for USP3, and in turn for deubiquitination, as a modulator of DDR signaling, the alteration of which would have less severe consequences for genome integrity compared with mutations in core DSB repair factors. In fact, neither apoptosis nor DNA damage-induced premature lymphoid differentiation significantly occurred in Usp3Δ/Δ primitive hematopoietic progenitors. Rather, we observed features of senescence. Not surprisingly, these features, along with spontaneous DNA damage, were further enhanced when we challenged Usp3Δ/Δ primitive progenitors with replicative stress in vitro, thereby implicating USP3 in fine-tuning DDR during proliferation. In this, our data on USP3 provide a framework to further investigate how lack of control of the Ub-dependent DDR pathway contributes to the functional decline observed in aged HSCs.
Histone ubiquitination by RNF8-RNF168 is central in DDR activation (Lukas et al., 2011; Jackson and Durocher, 2013). This raised the hypothesis that deubiquitination by USP3 may in turn be important for restraining/silencing the DDR. Consistently, enhanced Ub conjugates were accompanied by spontaneous 53BP1 foci as well as a prolonged retention of 53BP1 upon IR in Usp3Δ/Δ HSCs. Notably, these data extend previous observations obtained with RNAi by us and others (Nicassio et al., 2007; Gudjonsson et al., 2012). The pathologies found in Usp3Δ/Δ mice further indicate that in vivo, this aspect of DDR may be equally relevant to DDR activation in genome maintenance. We can envision at least two deleterious scenarios that could result from increased association/stabilization of genome caretakers such as 53BP1 with chromatin as a consequence of USP3 depletion. First, enhanced binding of DDR mediators might initiate unnecessary repair events, thus contributing to genomic instability. Consistent with this possibility, the spreading of chromatin ubiquitination at DSBs due to amplification of the RNF168 pool has been linked to enhanced DSB repair (Gudjonsson et al., 2012). Second, hyperubiquitination may lead to prolonged association of DDR factors at damaged DNA sites, which could in turn delay silencing of DDR signaling and cell cycle recovery (Nicassio et al., 2007). If deubiquitination is delayed, we hypothesize that persistent uH2A and possibly uH2B conjugates, which have also been implicated in regulating DSB repair (Moyal et al., 2011; Nakamura et al., 2011), may become detrimental during subsequent genetic insults. In such a situation, repair factors may get targeted simultaneously to repaired and de novo broken DNA. Over time, this scenario is likely to destabilize the genome, leading to chromosomal aberrations, stem cell depletion, and cancer.
In summary, the USP3 knockout mouse model establishes the importance of USP3 in limiting chronic genotoxic stress and HSC aging. Also, it highlights the relevance of understanding the mechanistic basis of chromatin deubiquitination, a still poorly understood aspect of DDR, and how it contributes to diseases related to chromosomal metabolism and to the pathogenesis of cancer.
MATERIALS AND METHODS
Mouse strains, monitoring of the mice, and TBI.
Generation of conditional (Usp3Lox; MGI:5490049; FVB;129P2-Usp3<tm1.2Nki>) and null (Usp3Δ; MGI:5490048) Usp3 alleles was performed as described below. LoxP sites were positioned in introns flanking the ZnF-UBP domain coding region (exon 2 and 3) of Usp3. Actin-Cre was used for germline deletion. Mice in this study were in a mixed 129/Ola × FVB genetic background (MGI:5490048; FVB;129P2(B6)-Usp3<tm1.1Nki>). USP3-null lines generated from two independently targeted embryonic stem (ES) cell clones were analyzed in parallel for Mendelian offspring and for development and gave similar results. Mice used in BM reconstitution experiments were backcrossed into a C57BL/6J (The Jackson Laboratory) strain background (Ly5.2+) for 8 generations (MGI:5490048; B6.129P2(FVB)-Usp3<tm1.1Nki>). Mice were maintained in a specific pathogen-free environment. All animal experiments comply with local and international regulations and ethical guidelines and have been authorized by our local experimental animal committee at The Netherlands Cancer Institute (DEC-NKI). Mice were monitored for survival and tumor development for 22 mo. Time to death was defined as the latency between birth and unexpected death or a terminal disease stage indicated by >20% weight loss or other symptoms of severe sickness. Mice were sacrificed by CO2 asphyxiation and underwent necroscopy. Organs were fixed as described below for histopathological analysis. Hypocellularity in the spleen of Usp3Δ/Δ mice was evaluated by blind examination of H&E-stained spleen sections. Sensitivity to IR was assessed with the use of 2–4-mo-old littermates. Mice were irradiated with an experimentally defined sublethal dose of IR (7 Gy; HF320 Radiobiology Constant Potential X-ray Unit, Pantak, East Haven, CT) and monitored for 28 d.
Construction of USP3 targeting vector.
Targeting construct for USP3 was generated using the pFlexible targeting vector system (van der Weyden et al., 2005). The pFlexible vector (pAG14) contains the PGK-driven positive/negative selectable marker puroDtk and loxP (L) and FRT (F) recombination target sites. This allow for efficient conversion of a single targeting event into a null or conditional mouse line.
A BAC clone (bMQ-415O10; Geneservice) from 129S7/AB2.2 mouse DNA (indexed 129S7/SvEvBrd-Hprtb-m2 (AB2.2) library, displayed on the Ensembl Genome Browser [http://www.ensembl.org/Mus_musculus/] under the DAS source 129S7/AB2.2) containing the USP3 locus was used to generate the USP3 targeting construct, and DNA fragments were amplified from the BAC DNA by PCR using the Expand Long range dNTPack kit (Roche). Fragments were as follows: the 5′ homology arm (3.5 kb PCR fragment 5′ of exon 2 in the USP3 gene, flanked by AscI restriction site: forward primer, 5′-GGCGCGCCCTGCCTCTGCTTCCTTTAGT-3′; reverse primer, 5′-GGCGCGCCGAATTCGGTTAATAAACTGGTTTCCCA-3′), the conditional arm (4.3 kb PCR product containing exon 2 and 3, flanked by PacI sites; forward primer containing PacI-BamHI-KpnI sites, 5′-TTAATTAAGGATCCGGTACCTGTTGACTTCAAAAGAGAGTG-3′; reverse primer, 5′-TTAATTAACCTCTTAAAGTCTAATACACTTC-3′), and the 3′ homology arm (4.6 kb PCR fragment 3′ of exon 3, flanked by NotI sites; forward primer, 5′-GCGGCCGCAAGTTGGAGATAGCCATGCC-3′; reverse primer, 5′-GCGGCCGCCTGTTAAAAGCAGATAAGCAC-3′). PCR products were first cloned in pCR-XL-TOPO Vector (TOPO XL PRC Cloning kit; Invitrogen) using one shot Stbl3 chemically competent Escherichia coli (Invitrogen). The pCR-XL-TOPO–cloned homology arms were fully sequenced prior subcloning into the pFlexible vector using the AscI, PacI, and Not1 restriction sites incorporated by PCR. Primers used for sequencing are available upon request. The final pFlexible-USP3CKO-FRT (conditional knockout–Flpe recombinase target) construct was fully sequenced prior to electroporation.
Generation of Usp3CKO-FRT/+ ES cells.
The USP3 targeting vector was linearized with ClaI and electroporated into 129/Ola-derived ES cells of the E14 subclone IB10 (Robanus-Maandag et al., 1998). Cells were grown under 1.8 mg/ml puromycin selection on a lethally irradiated MEF feeder layer. After 5 d of drug selection, ∼300 puromycin resistant ES clones were picked and amplified in nonselective medium. High molecular weight DNA was isolated according to standard procedures (Jonkers et al., 2001), and PCR and Southern blot analysis were performed to identify homologous recombinants. Genomic DNA was amplified with a primer pair encompassing the LoxP1 site, primers 37 (forward, 5′-ATAATTGGCCTGATGACAGC-3′) and 38 (reverse, 5′-TCATCGTAGCTTGTGATTGC-3′), yielding a DNA fragment of 447 bp for the WT allele and of 519 bp for the targeted allele. For Southern blotting, the probes were generated and labeled by PCR. The 5′ probe (527 bp; forward primer, 5′-AAGAGGATTGAAGCATAGG-3′; reverse primer, 5′-TTTGAAATCCTAGGACACAGC-3′) hybridizes to a 10 kb BamHI restriction fragment in WT cells and a 7.2 kb fragment in homologous recombinants. The 3′ probe (600 bp; forward primer, 5′-ATAATGCTGTTAACAGGAACG-3′; reverse primer, 5′-AATTTCTCTTTCTAACTCCATG-3′) recognizes a 7.6 kb EcoRI fragment in WT cells and a 10 kb fragment in recombinants. The probe recognizing the puromycin cassette (470 bp; forward primer, 5′-GAGCTGCAAGAACTCTTCC-3′; reverse primer, 5′-ATATAAACCCGCAGTAGCG-3′) hybridizes to a 10 kb EcoRI fragment in recombinants only.
Generation of Usp3CKO-FRT conditional and Usp3-deleted (Δ) mice.
Three independent ES cell clones with homologous integration at the targeting site were injected into C57BL/6J blastocysts to generate chimeric mice. These chimeras were subsequently crossed with FVB/N females to produce heterozygous offspring. Heterozygous mice with successful germline transmission of the targeted allele were crossed with C57BL/6J mice expressing Flp recombinase (B6.Cg-Tg(ACTFLPe)9205Dym/A; Rodríguez et al., 2000) to remove the neomycin resistance cassette resulting in USP3lox allele. To obtain Cre-mediated deletion of the floxed allele in the germline, heterozygous USP3 conditional mice were crossed with Actin-Cre deleter mice (FVB/N), in which cre expression is under the human β-actin gene promoter and is expressed in all cells of the embryo by the blastocyst stage of development, and intercrossed to generate Usp3Δ/Δ mice.
Genotyping of Usp3 mice.
Genotyping of F1 tail DNA was performed by Southern blot using the same DNA probes as described above for ES cells. The 5′ probe recognizes a 16.6 kb KpnI DNA fragment in the WT allele, a 9.5 kb fragment in the USP3CKO-FRT allele, and a 12.2 fragment in the deleted allele, the 3′ probe hybridizes to a 7.6 kb EcoRI fragment in WT allele, a 10 kb fragment in the USP3CKO-FRT allele, and a 6 kb fragment in the deleted allele. After founders were established, genotyping of F2 DNA was performed by PCR. Tail DNA was extracted using the Direct PCR DNA Extraction Reagents (QIAGEN) and PCR performed using Platinum PCR Supermix (Invitrogen). The USP3 alleles were detected with the following primers: the LoxP1 site in intron 1, yielding a 447 and 519 bp product for the WT or USP3CKOFRT, respectively (forward primer 37, 5′-ATAATTGGCCTGATGACAGC-3′; reverse primer 38, 5′-TCATCGTAGCTTGTGATTGC-3′); the LoxP2 site in intron 3, yielding a 419 and a 519 bp product for the WT or the USP3lox, respectively (forward primer 35, 5′-GTAGCTACAGCACATACTGG-3′; reverse primer 36, 5′-ATAGACAGGACTTTACTACC-3′); and the USP3Δ allele, yielding a 544 bp product in the USP3Δ allele (forward primer 37 and reverse primer 36). The Cre transgene (a 200 bp PCR fragment) was detected with the following primers: forward primer, 5′-GTTTCACTGGTTATGCGG-3′; reverse, 5′-TGCCTTCTCTACACCTGC-3′. PCR for the FLP transgene yielded a 750 bp DNA fragment (forward primer, 5′-GGTCCAACTGCAGCCCAAGCTTCC-3′; reverse primer, 5′-GTGGATCGATCCTACCCCTTGCG-3′; Rodríguez et al., 2000).
Protein analysis.
Total protein lysate from MEFs and histone fractions from cells and tissues were prepared as previously described (Nicassio et al., 2007). A complete list of antibodies used in Western blotting is found in Table S5.
Hematology.
Peripheral blood was collected into EDTA-coated microtubes, and blood cell counts were analyzed on a COUNTER Ac·T dif (Beckman Coulter). For FACS analysis, blood was depleted from red blood cells by hypotonic lysis and staining was performed with fluorochrome-labeled antibodies specific for B220, CD3, CD11b. Samples were analyzed on a CyAn ADP Analyzer or on a LSR Fortessa (BD). A complete list of antibodies used in flow cytometry is found in Table S6.
Flow cytometric analyses and cell sorting of hematopoietic subpopulations.
BM mononuclear cells (MNCs) were stained for lymphoid and myeloid lineage markers. To analyze and isolate BM LSK, LT-HSC, ST-HSC, MPP, CLP, and CMP subpopulations (Osawa et al., 1996; Christensen and Weissman, 2001; Kiel et al., 2007b; Karsunky et al., 2008; Rodrigues et al., 2008; Yeung and So, 2009), MNCs were first stained with Lineage Cell Detection Cocktail-Biotinylated mouse antibody (Macs; Miltenyi Biotec). For FACS analysis, cells were then directly stained with fluorochrome-conjugated antibodies against Sca1 (Pacific blue), c-kit (APC), CD34 (FITC), CD135/Flk2 (PE), CD150 (PECy7), CD16/32 (PerCpCy5,5), and CD127 (PerCpCy5,5). For cell sorting of LSK, LT-HSC, and ST-HSC, depletion of lineage+ cells was performed using Biotin MicroBeads (Macs; Miltenyi Biotec) and magnetic columns (Macs; Miltenyi Biotec) before staining. Samples were analyzed with CyAn ADP Analyzer. Cell sorting was performed with a FACSAria (BD).
All FACS data were analyzed using FlowJo Software (Tree Star). Apoptosis in Lin−, LSKs, and HSCs (defined as above) was assessed in freshly isolated BM cells using the FITC Annexin V Apoptosis detection kit II (BD). Samples were analyzed with the LRS Fortessa (BD).
BM transplantation.
In competitive BM transplantation, 1.5 × 106 BM MNCs from 8-wk-old donor test animals (CD45.2, WT, or Usp3Δ/Δ) were mixed in a 1:1 ratio with WT competitor cells (CD45.1) and injected into lethally irradiated (2 × 5.5 Gy TBI) C57BL/6-CD45.1 recipient mice. Recipient mice peripheral blood was monitored by FACS analysis every 2 wk for 16 wk. Donor contribution and multilineage reconstitution were assessed based on the expression, respectively, of CD45.1/CD45.2 antigens or B220, CD3, and Gr1 markers in the CD45.2+ fraction. At 16 wpt, primary recipients were sacrificed and the frequency of donor-derived CD45.2 LSK assayed by phenotypic profiling. In noncompetitive transplantation, BM cells from 39–42-wk-old WT or Usp3Δ/Δ mice were transplanted. For each genotype, cells from 3 donor mice were pooled and 5 × 105 cells injected into lethally irradiated CD45.2 primary recipients. Flow cytometry staining for LSK and HSC of donors was performed to ensure that the HSC frequency in test and control BM would be comparable. Differences in HSC were corrected before transplantation to transplant stem cell equivalents. LSKs and HSCs analysis of primary recipient was performed at 16 wpt, as described above.
For serial transplantation, irradiated primary recipients were injected noncompetitively with 3 × 106 BM cells (stem cell equivalents) from 8-wk-old WT or Usp3Δ/Δ mice (for each genotype, BM cells from 3 mice were pooled). For secondary transplants, primary recipients were sacrificed at 12 wpt, BM cells from 3 recipients per genotype were pooled, and 3 × 106 of these cells were transplanted into lethally irradiated hosts. Secondary recipients were sacrificed at 12 wpt and analysis for donor-derived Lin−, LSKs, and HSCs was performed as described above. In each experiment, 5 recipient mice with test or control cells were assayed. Samples were analyzed with an LRS Fortessa.
5-FU treatment.
5 Fluorouracil (5-FU; Sigma-Aldrich) was administered to 44-wk-old mice weekly intraperitoneally at a dose of 150 mg/kg for 4 consecutive wk. BM cellularity (two femurs) was evaluated at 4 wk after the first treatment.
LTC-IC assay.
OP9 stromal cells were grown in Iscove’s modified Dulbecco’s medium supplemented with 20% FBS and 50 mM 2-mercaptoethanol. Confluent OP9 stromal layers in 6 wells were irradiated with 15 Gy and subsequently seeded with purified Lin− cells (15,000 cells per well) in triplicates. After 5 wk of culturing (MyeloCult M5350 [STEMCELL Technologies], supplemented with 276 ng/ml hydrocortisone; replacement of 1/2 medium volume every week), cells were harvested and seeded in methylcellulose for CFU-C colony assays (as below).
RNA-seq gene expression analysis.
For gene expression analysis, LSKs (FACS sorted from freshly isolated BM), naive splenic B cells (freshly FACS sorted), or FACS-sorted LPS-activated B cells were used. n = 4 Usp3Δ/Δ and n = 4 WT littermates (2 mo old). Cells from individual animals were pooled and total RNA was extracted. Samples were prepared using TruSeq protocols, and standard sample preparation protocols and RNA-seq was performed on a Hiseq2000 machine (Illumina) at the NKI Genomics Core Facility.
Naive splenic B cells were obtained by CD43 depletion using biotinylated anti CD43 (Clone S7; BD), and the IMag system (BD), as described by the manufacturer. Naive B cells were directly FACS sorted with fluorochrome-conjugated antibody specific for CD19 (APC), or cultured in vitro for 4 d in IMDM + 8% FBS and 50 µg/ml E. coli LPS (055:B5; Sigma-Aldrich) to obtain activated B cells, followed by sorting. Two independent experiments were performed. Cell sorting was performed by FACSAria (BD). The data discussed in this publication have been deposited in NCBI’s Gene Expression Omnibus (Edgar et al., 2002) and are accessible through GEO Series accession no. GSE58495.
HSC liquid culture, time-lapse imaging, and Tat-cMyc fusion protein delivery.
LT-HSCs were sorted from BM of 40–44-wk-old mice (WT n = 3, Usp3Δ/Δ n = 4). Cells from individual animals were pooled and grown in liquid cultures. Cells were grown in RHB-A Stem Cell Culture Medium (STEMCELL Technologies) containing SCF, Flt3-ligand, and TPO (50 ng/ml), and IL-3, IL-6, and IL-11 (25 ng/ml; PeproTech). Medium was replenished and cells were expanded in 3% oxygen to maintain optimal growth. To evaluate proliferation, 1,000 cells were plated in 96-well plates at day 8 or 11 of culture. 4 wells per conditions were imaged (phase-contrast) with a 4 h interval for 6 d using the IncuCyte FRL (Essen BioScience). Confluence was determined by the IncuCyte software, based on area (confluence) metrics. Plating of 500 cells/well gave similar results.
The cDNA of human transcription factor cMYC (NM_002467.3) was PCR amplified and subcloned in pGEX6P1 plasmid containing the membrane penetrating peptide YGRKKRRQRRR from HIV TAT protein. Expression of recombinant Tat-cMyc was induced in log phase growing bacteria E. coli BL21 DE3 strain (Invitrogen) upon treatment of 1 mM isopropyl-b-d-galactoside for 3 h at 30°C. Purification comprised a further step to remove GST tag from TAT-cMyc protein using PreScission Protease (GE Healthcare). Liquid cultures of HSCs were treated with 1 µg/ml of purified Tat-c-Myc for 5 d, with daily replenishing of the fusion protein.
Quantitative real-time (qRT) PCR.
Total RNA was extracted using TRIZOL reagent (Life Technologies) and cDNA was prepared using Superscript II RT and oligo(dT)n primers (Life technologies). qRT-PCR was performed on a StepOnePlus RT-PCR system (Applied Biosystems) using SYBR Green PCR Master Mix (Applied Biosystems). The amount of target, normalized to an endogenous reference (HPRT), was calculated by 2−ΔΔCT. Primer sequences were as follows: Hprt forward, 5′-CTGGTGAAAGGACCTCTCG-3′; Hprt reverse, 5′-TGAAGTACTCATTATAGTCAAGGGCA-3′; Usp3Δ forward 1, 5′-TCTGTTTGACTTGTTCAAGTGTCC-3′; and Usp3Δ reverse 1, 5′-ATGGTTGAGTAAGGGTATCTGTGC-3′.
Immunofluorescence and quantitative image analysis.
For FK2 immunofluorescence staining, MEFs were grown on coverslips. After washing in PBS, cells were permeabilized in 0.5% Triton X-100 in PBS on ice 10 min, followed by fixation in 4% paraformaldehyde in PBS at room temperature 10 min, and a second permeabilization. Coverslips were blocked in 2% BSA in PBS (PBS+) for 30 min at room temperature, and incubated with primary antibody in PBS+ for 1 h at room temperature. Next, they were washed 5 × 5 min in PBS+ and incubated with the secondary antibody for 45 min at room temperature. Coverslips were washed in PBS+ 2× and PBS 1×. Finally, nuclei were stained using 200 ng/ml DAPI for 1 min at room temperature. Coverslips were mounted using FluorSave reagent (EMD Millipore). Freshly isolated BM cells or in vitro cultured LT-HSCs were deposited on charged slides (Superfrost Plus; Menzel-Glaser) by cytospin and directly processed as above for staining with FK2 and FK2-anti-53BP1 coimmunostaining.
For immunostaining with antibodies against phosphorylated ser139 H2AX (γH2AX), 53BP1, HP1γ, and trimethylated H3K9 (H3K9Me3), FACS-sorted LSKs, LT-HSCs, and ST-HSCs, or in vitro cultured LT-HSCs deposited on coverslips by cytospin were directly fixed in 4% paraformaldehyde and subsequently processed as above. Counterstain was with Alexa Fluor 488– or Alexa Fluor 568–conjugated secondary antibodies and DAPI (Sigma-Aldrich). Micronuclei were scored on fixed cells stained with DAPI.
Digital images were acquired using a microscope (AxioObserver Z1; Carl Zeiss) with an ORCA-ER CCD (charge-coupled device) camera (C4742-80-12AG; Hamamatsu) and AxioVision software (20 and 40× magnification). Confocal sections were acquired with a confocal laser microscope system (SP2 AOBS; Leica) with LCS software (40 and 63× magnification) by sequential scanning. A complete list of antibodies used in IF and immunohistochemistry is found in Table S5.
We used ImageJ software (version 1.43; National Institutes of Health) for quantitative image analysis. The DAPI channel (Blue) was used to select the nuclei of the cells in the field, and pixel intensity in each nucleus was measured on the second channel, red channel for FK2, HP1γ, and H3K9Me3. Data are presented as fluorescence intensity per cell or percentage of positive cells. For HP1γ and H3K9Me3, cells with a pixel intensity >50 (on a 1–255 scale) were considered positive. For counting of γH2AX foci, foci in the green channel were defined as particles >0.2 mm2 with intensity >80 (on a 1–255 scale), and images were counted blind. Cells were isolated from 3 mice per genotype at 17 or 44 wk of age. Sorted cells from individual animals were pooled and deposited on cytospin for IF. Two independent experiments were performed and 200–500 cells were counted per genotype per experiment. For counting γH2AX/53BP1 foci, or FK2/53BP1 foci, in in vitro expanded LT-HSCs, foci were defined in each channel as above and foci positive for both signals were scored as colocalizing. For quantification of immunohistochemistry slides, cells with pixel intensity >50 (for HP1γ) or 45 (for H3K9Me3, on a 1–255 scale) were considered positive. Data were analyzed using Excel (Microsoft) and Prism 6 (GraphPad Software).
Histopathological analysis and immunohistochemistry.
For histopathological analysis, tissues and organs were collected and fixed in formalin (or ethanol-acetic acid-formalin [EAF]) for 48 h (or 24 h in EAF). Fixed tissues and organs were subsequently dehydrated, embedded in paraffin, and 2-µm sections were prepared and stained with hematoxylin and eosin (H&E) for routine examination. For immunohistochemistry, 4-µm-thick sections were prepared and staining for activated Caspase 3, H3K9Me3, or Hp1γ was performed according to the manufacturers’ recommendations. Secondary antibodies were coupled to Strep–AB complex, and immunodetection was performed using Liquid DAB Substrate Chromogen System (Dako). For H3K9Me3, slides were blocked with Powervision block (DPCB blocking) and binding was detected using the Powervision peroxidase kit (DPVR110 HRP). Counterstaining was performed with hematoxylin.
MEF isolation and retroviral transduction.
MEFs were cultured in 3% oxygen in DMEM (Life Technologies) supplemented with 10% FBS (Sigma-Aldrich) and 1% penicillin/streptomycin (Life Technologies). MEFs were isolated from embryonic day 14.5 embryos. In brief, fetal tissue was rinsed in PBS, minced, rinsed twice in PBS, and incubated with trypsin/EDTA (Life Technologies) for 10–15 min at 37°C and subsequently thoroughly dissociated in medium. After removal of large tissue clumps, the remaining cells were plated out in a 75-cm2 flask. After 48 h, confluent cultures were frozen down. Retrovirus carrying empty vector or short-hairpin (sh) RNA directed against p53 (pRetroSUPER sh-p53; Dirac and Bernards, 2003), pBABE-Puro empty vector, or expressing WT-USP3 was used to infect MEFs overnight. The next day, infected MEFs were split in medium containing puromycin to select the infected population. Efficient p53 knockdown was verified by Western blotting.
Chromosome analysis and SCE.
Isolated MEFs were grown in 3% O2. Analysis of chromosomal aberrations and SCE was performed on Giemsa-stained metaphase spreads as previously described (Neijenhuis et al., 2010). Images of metaphase spreads were captured using a microscope (Axiovert S100; Carl Zeiss) equipped with a camera (AxioCam Color; Carl Zeiss) and AxioVision software. For SCE analysis, 3 × 105 cells were plated in 10-cm dishes. After 24 h, 10 µM BrdU was added, incubated 40 h, the last 4 h of which in the presence of 0.2 µg/ml Colcemid. Metaphase spreads were prepared as above with the following exceptions. Metaphase spreads were stained with Hoechst 5 µg/ml for 12 min, rinsed with water, and washed with water for 5 min. Next, the slides were covered with water and coverslips and exposed to UV light (366 nm) for 1 h. Coverslips were removed and slides were rinsed using 2× SSC (Sodium Citrate buffer; 300 mM NaCl and 30 mM Trisodium Citrate) at 65°C 15 min. Finally, coverslips were washed with water 5 min and stained with Giemsa. Imaging and analysis was as described above.
Constant field gel electrophoresis, comet assay, and β-galactosidase activity assay.
DSBs in primary MEFs were measured by Constant field gel electrophoresis (CFGE) as previously described (Neijenhuis et al., 2010). Alkaline comet assay was performed on total BM or LSK cells freshly sorted from 10-mo-old mice using the Comet assay kit (Trevigen). Images were taken using a microscope (AxioObserver Z1; Carl Zeiss) equipped with a CCD camera and AxioVision software; comets were scored using CometScore software acquiring comet metrics as described (CometScore Tutorial) and analyzed using Excel. Senescence-associated β-galactosidase activity was assessed in cytospin preparation of freshly isolated BM cells from 32-wk-old littermates or sorted and in vitro cultured LT-HSCs using the Senescence-associated β-galactosidase staining kit (Cell Signaling Technology). Phase contrast pictures were captured using a microscope (Axiovert S100; Carl Zeiss) equipped with a camera (AxioCam Color) and AxioVision software.
Pre-B and CFU-C colony-forming assay.
BM MNCs cells were seeded on 35-mm culture dishes in triplicate in methylcellulose medium supplemented with cytokines: MethoCult GF M3434 (for CFU-C, 5 × 104 cells or 2 × 105 cells, as indicated) and MethoCult M3630 (for CFU–Pre-B, 5 × 105 cell; STEMCELL Technologies). CFU-Cs (colony forming units in culture) include CFU-GEMM (granulocyte, erythroid, macrophage, megakaryocyte), multipotential progenitors and lineage-restricted progenitors of the erythroid (BFU-E, burst-forming unit–erythroid), and granulocytic, monocyte-macrophage (CFU-GM). Cultures were incubated at 37°C under 5% CO2. Colonies were quantified at day 8.
Statistics.
Statistical analysis was performed using an unpaired two-tailed Student’s t test (Prism 6.0). Animal survival experiments were analyzed with a Log-rank nonparametric test and expressed as Kaplan-Meier survival curves.
Online supplemental material.
Fig. S1 shows representative FACS profiles for lymphoid and myeloid populations in the blood of 44-wk-old WT and Usp3Δ/Δ mice (relative to Fig. 4 B). Fig. S2 shows gating strategy and representative FACS profiles for hematopoietic stem and progenitor subpopulations (relative to Fig. 6 and Fig. 9 E). Table S1 shows neoplastic lesions and hyperplasia found in Usp3Δ/Δ mice. Table S2 lists genes validated by qRT-PCR in Usp3Δ/Δ and WT LSKs. Tables S3 and S4 contain RNA-seq analysis showing transcripts up-regulated (Table S3) or down-regulated (Table S4) in Usp3Δ/Δ LSKs compared with WT LSKs. Table S5 lists antibodies used for Western blot, immunofluorescence, or immunohistochemistry of murine samples. Table S6 lists fluorochrome-conjugated antibodies used for surface staining of murine samples. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20131436/DC1.
Supplementary Material
Acknowledgments
We thank J. Jonkers and K. Nacerddine for advise on USP3 targeting strategy; F. van Diepen and A. Pfauth for FACS sorting; L. Ommen and L. Brooks for assistance in digital microscopy; R. Kerkhoven, I. de Rink, and Arno Velds from the NKI Genomic Core Facility for RNA-seq; R. Bin Ali, M. Snoek, and P. Steiger for technical support; K. De Lint for assistance with the IncuCyte; and the personnel of the Animal Pathology Department and NKI animal facility for tissue management and animal husbandry. We thank A. Sparmann and M. Innocenti for helpful discussion and critical reading of the manuscript.
This work was supported by the Dutch Cancer Society (KWF) to E. Citterio and M. van Lohuizen (NKI-2007-3877 and NKI-2012-5665), to G. Gargiulo and M. van Lohuizen (NKI 2013-6030), and to H. Jacobs (NKI-2008-4112 and NKI-2012-5243), and by the Dutch Organization for Scientific Research NWO Vidi to E. Citterio (864.08.011).
The authors declare no competing financial interests.
Footnotes
Abbreviations used:
- CLP
- common lymphoid progenitor
- CMP
- common myeloid progenitor
- DDR
- DNA damage response
- DSB
- DNA double-strand break
- DUB
- deubiquitinating enzyme
- ES
- embryonic stem
- GMP
- granulocyte-monocyte progenitor
- HSC
- hematopoietic stem cell
- IR
- ionizing radiation
- IRIF
- IR-induced foci
- LSK
- Lin−Sca1+c-Kit+
- LT-HSC
- long-term HSC
- LTC-IC
- long-term culture–initiating cell
- MEP
- megakaryocyte-erythrocyte progenitor
- MNC
- mononuclear cell
- MPP
- multipotent progenitor
- SCE
- sister chromatid exchange
- ST-HSC
- short-term HSC
- Ub
- ubiquitin
- USP3
- Ub-specific protease 3
- WBC
- white blood cell
- wpt
- weeks post transplantation
References
- Alt, F.W., Zhang Y., Meng F.-L., Guo C., and Schwer B.. 2013. Mechanisms of programmed DNA lesions and genomic instability in the immune system. Cell. 152:417–429 10.1016/j.cell.2013.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohgaki, T., Bohgaki M., Cardoso R., Panier S., Zeegers D., Li L., Stewart G.S., Sanchez O., Hande M.P., Durocher D., et al. 2011. Genomic instability, defective spermatogenesis, immunodeficiency, and cancer in a mouse model of the RIDDLE syndrome. PLoS Genet. 7:e1001381 10.1371/journal.pgen.1001381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braig, M., Lee S., Loddenkemper C., Rudolph C., Peters A.H., Schlegelberger B., Stein H., Dörken B., Jenuwein T., and Schmitt C.A.. 2005. Oncogene-induced senescence as an initial barrier in lymphoma development. Nature. 436:660–665 10.1038/nature03841 [DOI] [PubMed] [Google Scholar]
- Christensen, J.L., and Weissman I.L.. 2001. Flk-2 is a marker in hematopoietic stem cell differentiation: a simple method to isolate long-term stem cells. Proc. Natl. Acad. Sci. USA. 98:14541–14546 10.1073/pnas.261562798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui, J., Song Y., Li Y., Zhu Q., Tan P., Qin Y., Wang H.Y., and Wang R.F.. 2014. USP3 inhibits type I interferon signaling by deubiquitinating RIG-I-like receptors. Cell Res. 24:400–416 10.1038/cr.2013.170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dirac, A.M., and Bernards R.. 2003. Reversal of senescence in mouse fibroblasts through lentiviral suppression of p53. J. Biol. Chem. 278:11731–11734 10.1074/jbc.C300023200 [DOI] [PubMed] [Google Scholar]
- Doil, C., Mailand N., Bekker-Jensen S., Menard P., Larsen D.H., Pepperkok R., Ellenberg J., Panier S., Durocher D., Bartek J., et al. 2009. RNF168 binds and amplifies ubiquitin conjugates on damaged chromosomes to allow accumulation of repair proteins. Cell. 136:435–446 10.1016/j.cell.2008.12.041 [DOI] [PubMed] [Google Scholar]
- Edgar, R., Domrachev M., and Lash A.E.. 2002. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 30:207–210 10.1093/nar/30.1.207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenech, M., Kirsch-Volders M., Natarajan A.T., Surralles J., Crott J.W., Parry J., Norppa H., Eastmond D.A., Tucker J.D., and Thomas P.. 2011. Molecular mechanisms of micronucleus, nucleoplasmic bridge and nuclear bud formation in mammalian and human cells. Mutagenesis. 26:125–132 10.1093/mutage/geq052 [DOI] [PubMed] [Google Scholar]
- Fradet-Turcotte, A., Canny M.D., Escribano-Díaz C., Orthwein A., Leung C.C., Huang H., Landry M.C., Kitevski-LeBlanc J., Noordermeer S.M., Sicheri F., and Durocher D.. 2013. 53BP1 is a reader of the DNA-damage-induced H2A Lys 15 ubiquitin mark. Nature. 499:50–54 10.1038/nature12318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatti, M., Pinato S., Maspero E., Soffientini P., Polo S., and Penengo L.. 2012. A novel ubiquitin mark at the N-terminal tail of histone H2As targeted by RNF168 ubiquitin ligase. Cell Cycle. 11:2538–2544 10.4161/cc.20919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiger, H., de Haan G., and Florian M.C.. 2013. The ageing haematopoietic stem cell compartment. Nat. Rev. Immunol. 13:376–389 10.1038/nri3433 [DOI] [PubMed] [Google Scholar]
- Ginjala, V., Nacerddine K., Kulkarni A., Oza J., Hill S.J., Yao M., Citterio E., van Lohuizen M., and Ganesan S.. 2011. BMI1 is recruited to DNA breaks and contributes to DNA damage-induced H2A ubiquitination and repair. Mol. Cell. Biol. 31:1972–1982 10.1128/MCB.00981-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldknopf, I.L., French M.F., Musso R., and Busch H.. 1977. Presence of protein A24 in rat liver nucleosomes. Proc. Natl. Acad. Sci. USA. 74:5492–5495 10.1073/pnas.74.12.5492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudjonsson, T., Altmeyer M., Savic V., Toledo L., Dinant C., Grøfte M., Bartkova J., Poulsen M., Oka Y., Bekker-Jensen S., et al. 2012. TRIP12 and UBR5 suppress spreading of chromatin ubiquitylation at damaged chromosomes. Cell. 150:697–709 10.1016/j.cell.2012.06.039 [DOI] [PubMed] [Google Scholar]
- Halaby, M.J., Hakem A., Li L., El Ghamrasni S., Venkatesan S., Hande P.M., Sanchez O., and Hakem R.. 2013. Synergistic interaction of Rnf8 and p53 in the protection against genomic instability and tumorigenesis. PLoS Genet. 9:e1003259 10.1371/journal.pgen.1003259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeijmakers, J.H.J.2009. DNA damage, aging, and cancer. N. Engl. J. Med. 361:1475–1485 10.1056/NEJMra0804615 [DOI] [PubMed] [Google Scholar]
- Huen, M.S., Grant R., Manke I., Minn K., Yu X., Yaffe M.B., and Chen J.. 2007. RNF8 transduces the DNA-damage signal via histone ubiquitylation and checkpoint protein assembly. Cell. 131:901–914 10.1016/j.cell.2007.09.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ismail, I.H., Andrin C., McDonald D., and Hendzel M.J.. 2010. BMI1-mediated histone ubiquitylation promotes DNA double-strand break repair. J. Cell Biol. 191:45–60 10.1083/jcb.201003034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito, K., Hirao A., Arai F., Matsuoka S., Takubo K., Hamaguchi I., Nomiyama K., Hosokawa K., Sakurada K., Nakagata N., et al. 2004. Regulation of oxidative stress by ATM is required for self-renewal of haematopoietic stem cells. Nature. 431:997–1002 10.1038/nature02989 [DOI] [PubMed] [Google Scholar]
- Jackson, S.P., and Bartek J.. 2009. The DNA-damage response in human biology and disease. Nature. 461:1071–1078 10.1038/nature08467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson, S.P., and Durocher D.. 2013. Regulation of DNA damage responses by ubiquitin and SUMO. Mol. Cell. 49:795–807 10.1016/j.molcel.2013.01.017 [DOI] [PubMed] [Google Scholar]
- Jonkers, J., Meuwissen R., van der Gulden H., Peterse H., van der Valk M., and Berns A.. 2001. Synergistic tumor suppressor activity of BRCA2 and p53 in a conditional mouse model for breast cancer. Nat. Genet. 29:418–425 10.1038/ng747 [DOI] [PubMed] [Google Scholar]
- Karsunky, H., Inlay M.A., Serwold T., Bhattacharya D., and Weissman I.L.. 2008. Flk2+ common lymphoid progenitors possess equivalent differentiation potential for the B and T lineages. Blood. 111:5562–5570 10.1182/blood-2007-11-126219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiel, M.J., He S., Ashkenazi R., Gentry S.N., Teta M., Kushner J.A., Jackson T.L., and Morrison S.J.. 2007a. Haematopoietic stem cells do not asymmetrically segregate chromosomes or retain BrdU. Nature. 449:238–242 10.1038/nature06115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiel, M.J., Radice G.L., and Morrison S.J.. 2007b. Lack of evidence that hematopoietic stem cells depend on N-cadherin-mediated adhesion to osteoblasts for their maintenance. Cell Stem Cell. 1:204–217 10.1016/j.stem.2007.06.001 [DOI] [PubMed] [Google Scholar]
- Kolas, N.K., Chapman J.R., Nakada S., Ylanko J., Chahwan R., Sweeney F.D., Panier S., Mendez M., Wildenhain J., Thomson T.M., et al. 2007. Orchestration of the DNA-damage response by the RNF8 ubiquitin ligase. Science. 318:1637–1640 10.1126/science.1150034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komander, D., Clague M.J., and Urbé S.. 2009. Breaking the chains: structure and function of the deubiquitinases. Nat. Rev. Mol. Cell Biol. 10:550–563 10.1038/nrm2731 [DOI] [PubMed] [Google Scholar]
- Kuilman, T., Michaloglou C., Mooi W.J., and Peeper D.S.. 2010. The essence of senescence. Genes Dev. 24:2463–2479 10.1101/gad.1971610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, L., Halaby M.J., Hakem A., Cardoso R., El Ghamrasni S., Harding S., Chan N., Bristow R., Sanchez O., Durocher D., and Hakem R.. 2010. Rnf8 deficiency impairs class switch recombination, spermatogenesis, and genomic integrity and predisposes for cancer. J. Exp. Med. 207:983–997 10.1084/jem.20092437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukas, J., Lukas C., and Bartek J.. 2011. More than just a focus: The chromatin response to DNA damage and its role in genome integrity maintenance. Nat. Cell Biol. 13:1161–1169 10.1038/ncb2344 [DOI] [PubMed] [Google Scholar]
- Mailand, N., Bekker-Jensen S., Faustrup H., Melander F., Bartek J., Lukas C., and Lukas J.. 2007. RNF8 ubiquitylates histones at DNA double-strand breaks and promotes assembly of repair proteins. Cell. 131:887–900 10.1016/j.cell.2007.09.040 [DOI] [PubMed] [Google Scholar]
- Mattiroli, F., Vissers J.H.A., van Dijk W.J., Ikpa P., Citterio E., Vermeulen W., Marteijn J.A., and Sixma T.K.. 2012. RNF168 ubiquitinates K13-15 on H2A/H2AX to drive DNA damage signaling. Cell. 150:1182–1195 10.1016/j.cell.2012.08.005 [DOI] [PubMed] [Google Scholar]
- Milyavsky, M., Gan O.I., Trottier M., Komosa M., Tabach O., Notta F., Lechman E., Hermans K.G., Eppert K., Konovalova Z., et al. 2010. A distinctive DNA damage response in human hematopoietic stem cells reveals an apoptosis-independent role for p53 in self-renewal. Cell Stem Cell. 7:186–197 10.1016/j.stem.2010.05.016 [DOI] [PubMed] [Google Scholar]
- Mohrin, M., Bourke E., Alexander D., Warr M.R., Barry-Holson K., Le Beau M.M., Morrison C.G., and Passegué E.. 2010. Hematopoietic stem cell quiescence promotes error-prone DNA repair and mutagenesis. Cell Stem Cell. 7:174–185 10.1016/j.stem.2010.06.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison, S.J., and Spradling A.C.. 2008. Stem cells and niches: mechanisms that promote stem cell maintenance throughout life. Cell. 132:598–611 10.1016/j.cell.2008.01.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison, S.J., Uchida N., and Weissman I.L.. 1995. The biology of hematopoietic stem cells. Annu. Rev. Cell Dev. Biol. 11:35–71 10.1146/annurev.cb.11.110195.000343 [DOI] [PubMed] [Google Scholar]
- Morrison, S.J., Wandycz A.M., Akashi K., Globerson A., and Weissman I.L.. 1996. The aging of hematopoietic stem cells. Nat. Med. 2:1011–1016 10.1038/nm0996-1011 [DOI] [PubMed] [Google Scholar]
- Mosbech, A., Lukas C., Bekker-Jensen S., and Mailand N.. 2013. The deubiquitylating enzyme USP44 counteracts the DNA double-strand break response mediated by the RNF8 and RNF168 ubiquitin ligases. J. Biol. Chem. 288:16579–16587 10.1074/jbc.M113.459917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyal, L., Lerenthal Y., Gana-Weisz M., Mass G., So S., Wang S.-Y., Eppink B., Chung Y.M., Shalev G., Shema E., et al. 2011. Requirement of ATM-dependent monoubiquitylation of histone H2B for timely repair of DNA double-strand breaks. Mol. Cell. 41:529–542 10.1016/j.molcel.2011.02.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura, K., Kato A., Kobayashi J., Yanagihara H., Sakamoto S., Oliveira D.V., Shimada M., Tauchi H., Suzuki H., Tashiro S., et al. 2011. Regulation of homologous recombination by RNF20-dependent H2B ubiquitination. Mol. Cell. 41:515–528 10.1016/j.molcel.2011.02.002 [DOI] [PubMed] [Google Scholar]
- Narita, M., Nuñez S., Heard E., Narita M., Lin A.W., Hearn S.A., Spector D.L., Hannon G.J., and Lowe S.W.. 2003. Rb-mediated heterochromatin formation and silencing of E2F target genes during cellular senescence. Cell. 113:703–716 10.1016/S0092-8674(03)00401-X [DOI] [PubMed] [Google Scholar]
- Neijenhuis, S., Verwijs-Janssen M., van den Broek L.J., Begg A.C., and Vens C.. 2010. Targeted radiosensitization of cells expressing truncated DNA polymerase beta. Cancer Res. 70:8706–8714 10.1158/0008-5472.CAN-09-3901 [DOI] [PubMed] [Google Scholar]
- Nicassio, F., Corrado N., Vissers J.H., Areces L.B., Bergink S., Marteijn J.A., Geverts B., Houtsmuller A.B., Vermeulen W., Di Fiore P.P., and Citterio E.. 2007. Human USP3 is a chromatin modifier required for S phase progression and genome stability. Curr. Biol. 17:1972–1977 10.1016/j.cub.2007.10.034 [DOI] [PubMed] [Google Scholar]
- Niedernhofer, L.J.2008. DNA repair is crucial for maintaining hematopoietic stem cell function. DNA Repair (Amst.). 7:523–529 10.1016/j.dnarep.2007.11.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nijnik, A., Woodbine L., Marchetti C., Dawson S., Lambe T., Liu C., Rodrigues N.P., Crockford T.L., Cabuy E., Vindigni A., et al. 2007. DNA repair is limiting for haematopoietic stem cells during ageing. Nature. 447:686–690 10.1038/nature05875 [DOI] [PubMed] [Google Scholar]
- Osawa, M., Hanada K., Hamada H., and Nakauchi H.. 1996. Long-term lymphohematopoietic reconstitution by a single CD34-low/negative hematopoietic stem cell. Science. 273:242–245 10.1126/science.273.5272.242 [DOI] [PubMed] [Google Scholar]
- Pang, W.W., Price E.A., Sahoo D., Beerman I., Maloney W.J., Rossi D.J., Schrier S.L., and Weissman I.L.. 2011. Human bone marrow hematopoietic stem cells are increased in frequency and myeloid-biased with age. Proc. Natl. Acad. Sci. USA. 108:20012–20017 10.1073/pnas.1116110108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panier, S., and Durocher D.. 2013. Push back to respond better: regulatory inhibition of the DNA double-strand break response. Nat. Rev. Mol. Cell Biol. 14:661–672 10.1038/nrm3659 [DOI] [PubMed] [Google Scholar]
- Pinato, S., Scandiuzzi C., Arnaudo N., Citterio E., Gaudino G., and Penengo L.. 2009. RNF168, a new RING finger, MIU-containing protein that modifies chromatin by ubiquitination of histones H2A and H2AX. BMC Mol. Biol. 10:55 10.1186/1471-2199-10-55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polo, S.E., and Jackson S.P.. 2011. Dynamics of DNA damage response proteins at DNA breaks: a focus on protein modifications. Genes Dev. 25:409–433 10.1101/gad.2021311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purton, L.E., Dworkin S., Olsen G.H., Walkley C.R., Fabb S.A., Collins S.J., and Chambon P.. 2006. RARγ is critical for maintaining a balance between hematopoietic stem cell self-renewal and differentiation. J. Exp. Med. 203:1283–1293 10.1084/jem.20052105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reimann, M., Lee S., Loddenkemper C., Dörr J.R., Tabor V., Aichele P., Stein H., Dörken B., Jenuwein T., and Schmitt C.A.. 2010. Tumor stroma-derived TGF-β limits myc-driven lymphomagenesis via Suv39h1-dependent senescence. Cancer Cell. 17:262–272 10.1016/j.ccr.2009.12.043 [DOI] [PubMed] [Google Scholar]
- Robanus-Maandag, E., Dekker M., van der Valk M., Carrozza M.L., Jeanny J.C., Dannenberg J.H., Berns A., and te Riele H.. 1998. p107 is a suppressor of retinoblastoma development in pRb-deficient mice. Genes Dev. 12:1599–1609 10.1101/gad.12.11.1599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues, N.P., Boyd A.S., Fugazza C., May G.E., Guo Y., Tipping A.J., Scadden D.T., Vyas P., and Enver T.. 2008. GATA-2 regulates granulocyte-macrophage progenitor cell function. Blood. 112:4862–4873 10.1182/blood-2008-01-136564 [DOI] [PubMed] [Google Scholar]
- Rodríguez, C.I., Buchholz F., Galloway J., Sequerra R., Kasper J., Ayala R., Stewart A.F., and Dymecki S.M.. 2000. High-efficiency deleter mice show that FLPe is an alternative to Cre-loxP. Nat. Genet. 25:139–140 10.1038/75973 [DOI] [PubMed] [Google Scholar]
- Rogakou, E.P., Pilch D.R., Orr A.H., Ivanova V.S., and Bonner W.M.. 1998. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J. Biol. Chem. 273:5858–5868 10.1074/jbc.273.10.5858 [DOI] [PubMed] [Google Scholar]
- Rossi, D.J., Bryder D., Seita J., Nussenzweig A., Hoeijmakers J., and Weissman I.L.. 2007. Deficiencies in DNA damage repair limit the function of haematopoietic stem cells with age. Nature. 447:725–729 10.1038/nature05862 [DOI] [PubMed] [Google Scholar]
- Rossi, D.J., Jamieson C.H., and Weissman I.L.. 2008. Stems cells and the pathways to aging and cancer. Cell. 132:681–696 10.1016/j.cell.2008.01.036 [DOI] [PubMed] [Google Scholar]
- Rübe, C.E., Fricke A., Widmann T.A., Fürst T., Madry H., Pfreundschuh M., and Rübe C.. 2011. Accumulation of DNA damage in hematopoietic stem and progenitor cells during human aging. PLoS ONE. 6:e17487 10.1371/journal.pone.0017487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanbhag, N.M., Rafalska-Metcalf I.U., Balane-Bolivar C., Janicki S.M., and Greenberg R.A.. 2010. ATM-dependent chromatin changes silence transcription in cis to DNA double-strand breaks. Cell. 141:970–981 10.1016/j.cell.2010.04.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma, N., Zhu Q., Wani G., He J., Wang Q.E., and Wani A.A.. 2014. USP3 counteracts RNF168 via deubiquitinating H2A and γH2AX at lysine 13 and 15. Cell Cycle. 13:106–114 10.4161/cc.26814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon, J.A., and Kingston R.E.. 2013. Occupying chromatin: Polycomb mechanisms for getting to genomic targets, stopping transcriptional traffic, and staying put. Mol. Cell. 49:808–824 10.1016/j.molcel.2013.02.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonoda, E., Sasaki M.S., Morrison C., Yamaguchi-Iwai Y., Takata M., and Takeda S.. 1999. Sister chromatid exchanges are mediated by homologous recombination in vertebrate cells. Mol. Cell. Biol. 19:5166–5169PubMed [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperka, T., Wang J., and Rudolph K.L.. 2012. DNA damage checkpoints in stem cells, ageing and cancer. Nat. Rev. Mol. Cell Biol. 13:579–590 10.1038/nrm3420 [DOI] [PubMed] [Google Scholar]
- Stewart, G.S., Panier S., Townsend K., Al-Hakim A.K., Kolas N.K., Miller E.S., Nakada S., Ylanko J., Olivarius S., Mendez M., et al. 2009. The RIDDLE syndrome protein mediates a ubiquitin-dependent signaling cascade at sites of DNA damage. Cell. 136:420–434 10.1016/j.cell.2008.12.042 [DOI] [PubMed] [Google Scholar]
- Sulli, G., Di Micco R., and d’Adda di Fagagna F.. 2012. Crosstalk between chromatin state and DNA damage response in cellular senescence and cancer. Nat. Rev. Cancer. 12:709–720 10.1038/nrc3344 [DOI] [PubMed] [Google Scholar]
- van der Weyden, L., Adams D.J., Harris L.W., Tannahill D., Arends M.J., and Bradley A.. 2005. Null and conditional semaphorin 3B alleles using a flexible puroΔtk loxP/FRT vector. Genesis. 41:171–178 10.1002/gene.20111 [DOI] [PubMed] [Google Scholar]
- van Os, R.P., Dethmers-Ausema B., and de Haan G.. 2008. In vitro assays for cobblestone area-forming cells, LTC-IC, and CFU-C. Methods Mol. Biol. 430:143–157 10.1007/978-1-59745-182-6_10 [DOI] [PubMed] [Google Scholar]
- Vissers, J.H., van Lohuizen M., and Citterio E.. 2012. The emerging role of Polycomb repressors in the response to DNA damage. J. Cell Sci. 125:3939–3948 10.1242/jcs.107375 [DOI] [PubMed] [Google Scholar]
- Wang, B., Matsuoka S., Ballif B.A., Zhang D., Smogorzewska A., Gygi S.P., and Elledge S.J.. 2007. Abraxas and RAP80 form a BRCA1 protein complex required for the DNA damage response. Science. 316:1194–1198 10.1126/science.1139476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, J., Sun Q., Morita Y., Jiang H., Gross A., Lechel A., Hildner K., Guachalla L.M., Gompf A., Hartmann D., et al. 2012. A differentiation checkpoint limits hematopoietic stem cell self-renewal in response to DNA damage. Cell. 148:1001–1014 10.1016/j.cell.2012.01.040 [DOI] [PubMed] [Google Scholar]
- Ward, I.M., Minn K., van Deursen J., and Chen J.. 2003. p53 Binding protein 53BP1 is required for DNA damage responses and tumor suppression in mice. Mol. Cell. Biol. 23:2556–2563 10.1128/MCB.23.7.2556-2563.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeung, J., and So C.W.. 2009. Identification and characterization of hematopoietic stem and progenitor cell populations in mouse bone marrow by flow cytometry. Methods Mol. Biol. 538:301–315 10.1007/978-1-59745-418-6_15 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.