Summary
Specific protein-RNA interactions guide post-transcriptional gene regulation. Here we describe RNA Bind-n-Seq (RBNS), a method that comprehensively characterizes sequence and structural specificity of RNA binding proteins (RBPs), and its application to the developmental alternative splicing factors RBFOX2, CELF1/CUGBP1 and MBNL1. For each factor, we recovered both canonical motifs and additional near-optimal binding motifs. RNA secondary structure inhibits binding of RBFOX2 and CELF1, while MBNL1 favors unpaired Us but tolerates C/G pairing in motifs containing UGC and/or GCU. Dissociation constants calculated from RBNS data using a novel algorithm correlated highly with values measured by surface plasmon resonance. Motifs identified by RBNS were conserved, were bound and active in vivo, and distinguished the subset of motifs enriched by CLIP-Seq that had regulatory activity. Together, our data demonstrate that RBNS complements crosslinking-based methods and show that in vivo binding and activity of these splicing factors is driven largely by intrinsic RNA affinity.
Introduction
RBPs bind sequence and/or structural motifs in nuclear pre-mRNAs to direct their processing, and bind mature mRNAs to control their translation, localization, and stability. RBPs of the Rbfox, CUG-BP/Elav-like (CELF) and muscleblind-like (MBNL) families are important and highly conserved regulators of developmental and tissue-specific alternative splicing
Rbfox2, a close homolog of Rbfox1 (Underwood et al., 2005), is required for neural development (Gehman et al., 2012), regulates epithelial-mesenchymal transition (EMT) (Baraniak et al., 2006), and is required for human embryonic stem cell (ESC) survival (Yeo et al., 2009). The consensus binding motif for Rbfox proteins – UGCAUG or simply GCAUG – has been determined by systematic evolution of ligands by exponential enrichment (SELEX) and is conserved from nematodes through vertebrates (Jin et al., 2003; Ponthier et al., 2006). However, the iterative selection steps used in SELEX favor recovery of just the strongest binding motifs and may not detect moderate and lower affinity motifs. Only about ⅓ to ½ of Rbfox2 binding sites identified in vivo contain these canonical motifs (Jangi et al., 2014; Yeo et al., 2009), but it has remained unclear whether this RBP can recognize other sequence motifs. In general, motifs recognized by RBPs with lower affinity are more challenging to characterize, but such motifs may play biological roles that are as important as those played by higher affinity motifs. For RBPs that accumulate during development, like MBNLs, higher affinity motifs may be bound at earlier time points, while lower affinity motifs may specify regulation at later developmental time points or only in certain cell types where the RBP accumulates to high levels.
CELF1 and MBNL1 proteins are functionally linked by their roles in development and disease, often regulating the same splicing targets in an antagonistic fashion. In heart development, during which CELF protein levels decrease and MBNL proteins accumulate, this antagonism may sharpen developmental splicing transitions (Kalsotra et al., 2008). This developmental expression pattern reverses that seen in the muscle wasting disease myotonic dystrophy type 1 (DM1), in which expanded CUG repeats in the 3′ UTR of DMPK mRNAs reduce available cellular levels of MBNL proteins by sequestration (Mankodi et al., 2005; Taneja et al., 1995), and CELF1 proteins are stabilized by hyperphosphorylation (Kuyumcu-Martinez et al., 2007). CELF1 has three RNA recognition motifs (RRMs) that bind motifs with consensus UGU (Ladd et al., 2001; Marquis et al., 2006). MBNL1 has two pairs of zinc fingers that are reported to bind preferentially to YGCY (Y = C or U) motifs (Ho et al., 2004). To date, it has remained unclear whether MBNL1 primarily recognizes or single- or double-stranded RNA elements. CUG repeat RNA crystallizes as an A-form helix (Mooers et al., 2005), with C and G bases paired and Us unpaired, and additional biochemical studies have shown that a mismatched RNA hairpin structure is important for recognition by MBNL1 (Warf and Berglund, 2007). However, structures of MBNL1 zinc fingers co-crystallized with CGCUGU RNA suggested that MBNL1 recognizes single-stranded RNA (Teplova and Patel, 2008). Additionally, the role of motif spacing and of intervening sequences between tandem motifs remain largely uncharacterized.
Widely used methods for mapping protein-RNA interactions in vivo based on ultraviolet cross-linking and immunoprecipitation (CLIP) (Ule et al., 2003; Underwood et al., 2005) have contributed to understanding of post-transcriptional regulation. However, these techniques are laborious and require many selection steps that likely introduce various types of bias. Motif analysis from CLIP data is complicated by the fact that it does not distinguish binding by a single protein from binding of a protein complex, and it may preferentially detect uridine-rich sequences (Sugimoto et al., 2012). Iterative binding approaches like SELEX, including recent high-throughput versions (Campbell et al., 2012) identify consensus motifs, but are not quantitative and are biased towards the highest affinity motifs. A newer method, RNAcompete, uses in vitro RNA-protein binding followed by microarray analysis, enabling high-throughput identification of RNA binding motifs (Ray et al., 2009; Ray et al., 2013). However, the number of probes assayed and the low temperatures typically used make it difficult to analyze effects of RNA secondary structure on RNA binding, and RNAcompete does not yield Kd values. Quantitative biophysical measurements including Kd values can be obtained from methods such as electrophoretic mobility shift assays (EMSA) or surface plasmon resonance (SPR), but their throughput is quite low.
To better characterize the functions of biologically important RBPs, we sought to develop a method that would measure affinities to the full spectrum of bound RNAs in a quantitative and high-throughput manner. Methods for characterizing protein/DNA interactions that are both high-throughput and quantitative have been developed, including HT-SELEX and Bind-n-Seq, both of which use one-step binding to a pool of randomized DNA in vitro followed by deep sequencing (Jolma et al., 2010; Zykovich et al., 2009), and HiTS-FLIP, which directly measures protein bound to double-stranded DNA on a flow cell (Nutiu et al., 2011). We adapted the general approach used by HT-SELEX and Bind-n-Seq to the study of protein-RNA interactions in vitro in a method we call “RNA Bind-n-Seq” (RBNS). Our method adapts and extends these protein/DNA interaction assays in two important ways. First, we use multiple RBP concentrations to optimize analysis at different ranges of affinity. Second, we have expanded the analytical framework to more accurately estimate relative dissociation constants, and to assess the effects of RNA secondary structure on binding. RBNS analyses of RBFOX2, CELF1 and MBNL1 yielded comprehensive portraits of the sequence and RNA secondary structural determinants of RNA recognition by these factors. Analysis of data from systems in which these RBPs were depleted or inducibly over-expressed in mouse cells provided evidence of function for both non-canonical and canonical binding motifs identified in vitro. We observed good correlation between in vitro and in vivo binding overall, but found that motifs enriched by CLIP but not RBNS are not associated with regulatory activity. Therefore, RBNS aids in identification of high-confidence splicing-associated binding sites and is complementary to CLIP.
Results
Design considerations for RNA Bind-n-Seq experiments
RBNS is designed to dissect the sequence and RNA structural preferences of RBPs. A recombinantly expressed and purified RBP is incubated with a pool of randomized RNAs at several different protein concentrations, typically ranging from low nanomolar to low micromolar (Fig. 1A). The RNA pool typically consists of random RNAs of length λ = 40 nt flanked by short primers used to add the adapters needed for deep sequencing. This RNA pool design simplifies library preparation, avoids biases that can result from RNA ligation, and ensures that any bacterial RNA carried over from protein expression will not contaminate the sequenced library. (In the unusual case where the RBP has significant affinity to primer RNA, different primer sequences must be substituted.) In each experiment, the RBP is captured via a streptavidin binding peptide (SBP) tag. RBP-bound RNA is reverse-transcribed into cDNA and barcoded sequencing adapters are added by PCR to produce libraries for deep sequencing. Libraries corresponding to the input RNA pool and to 5 or more RBP concentrations (including zero RBP concentration as an additional control), are sequenced in a single Illumina HiSeq 2000 lane, typically yielding at least 15–20 million reads per library.
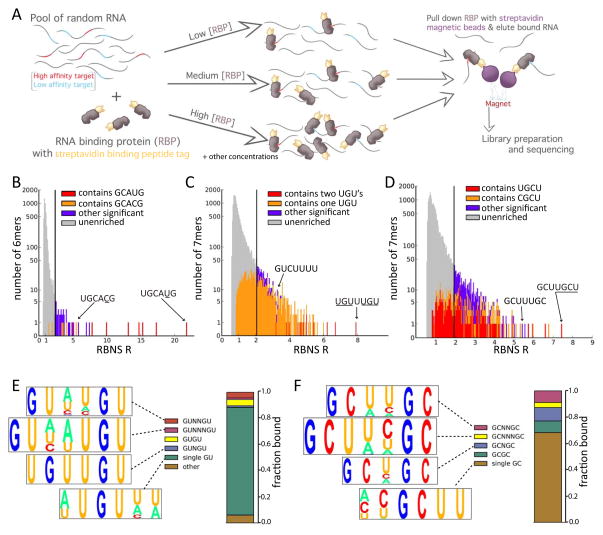
Figure 1. RNA Bind-n-Seq overview and motif enrichment analysis.
A. Overview of the experimental method. Tagged protein is incubated with a diverse pool of RNA oligonucleotides of fixed concentration at each of several concentrations of protein. The RBP is pulled down using streptavidin-coated magnetic beads and the associated RNA is sequenced. The counts of sequences in this library are used to estimate proportions of bound RNA molecules, in comparison to input RNA, which is also sequenced.
B. Stacked histogram showing the distribution of RBNS R values of all RNA 6mers in the RBFOX2 experiment at a protein concentration of 365 nM. 6mers that contain specific 5mers, whether significant or not are shown in red or orange; other 6mers are colored based on whether their R value is at least 2 SD above the mean (purple), or not (gray). A log scale is used for the Y-axis.
C. As in B), but shows distribution of R values for all 7mers for CELF1 at a protein concentration of 64 nM.
D. As in C), but shows distribution of all 7mers for MBNL1 at a concentration of 250 nM.
E. Visualization of CELF1 binding preferences. The sequence content (displayed as a pictogram with letter height proportional to frequency), and estimated bound fraction of four groups of 7mer motifs are shown. The top 50 7mers were grouped and aligned based on their content and spacing of GU submotifs (Fig. S1D).
F. Visualization of the Mbnl1 binding preferences. As in E) but based on the top 50 7mer motifs for MBNL1, grouped by spacing of GC submotifs (alignments shown in Fig. S1E).
See also Figure S1.
Most RBPs bind single-stranded RNA sequence motifs 3–8 bases in length (Stefl et al., 2005). Here, we performed one experiment using the RBFOX2 RRM with short oligonucleotides (λ = 10 nt). However, we soon realized that use of longer sequences (λ = 40 nt) provided comparable affinity measurements to short linear motifs of size k (kmers) in the range of interest (about 3 to 10 nt, Fig. S1) while also enabling assessment of RNA secondary structural and other contextual effects on binding that cannot be assessed using 10mers. Size λ = 40 nt is closer to the in vivo situation where RBPs typically bind long RNAs, but is within the range where structure can be most accurately predicted by thermodynamic RNA folding algorithms (Hofacker, 2003).
RNA Bind-n-Seq comprehensively identifies known and novel motifs of RBPs
RBNS was performed using recombinant RBFOX2, MBNL1 and CELF1 proteins. For each protein, at each of several concentrations, motif read enrichment (“R”) values were calculated for each kmer (for k = 5, 6, 7) as the ratio of the frequency of the kmer in the selected pool to the frequency in the input RNA library. In our typical zero concentration experiment, 99.9% of 6mers had R values less than 1.19, and the highest value was 1.21, indicating little if any sequence bias from the apparatus. The false discovery rate (FDR) was 1.2% for CELF1 7mers as judged by the 0 nM RBP experiment, and was ~0 for the other proteins (Methods).
For RBFOX2, at all concentrations ≥14 nM the 6mer UGCAUG had the highest R value (Fig. 1B and below), confirming this well-known motif as the highest affinity 6mer. The enrichment of UGCAUG reached a maximum R of 22 at a protein concentration of 365 nM (Fig. 1B). We derived an equation relating the observed R value to the relative affinity (ratio of dissociation constants) between nonspecific and specific binding under idealized conditions (Supp. Methods, equation 18). With R = 22, k = 6 and λ = 40, this equation implies at least ~900-fold higher binding affinity to UGCAUG than to nonspecific 6mers. All 8 of the 6mers that contain GCAUG had significant R values (Fig. 1B), consistent with the known affinity of Rbfox proteins for this 5mer (Jin et al., 2003). Several 6mers containing GCACG were also significant, indicating that this 5mer represents an alternate RBFOX2 binding motif. Certain other 6mers not containing GCAUG or GCACG, but often containing GCAU, also had significant R values, suggesting that RBFOX2 has some affinity for other RNA motifs as well (Table S1).
Proteins of the CELF family preferentially bind to UG- and UGU-containing motifs (Marquis et al., 2006; Timchenko et al., 1996). For CELF1, a large number of 6mer and 7mer motifs had significant R values (7mer analysis shown in Fig. 1C). Inspection of these motifs showed that the highest R values were observed for 7mers containing two UGU triplets. In fact, all 7mers containing two UGUs were significantly enriched, suggesting that presence of two UGUs is sufficient for strong binding and that CELF1 tolerates presence or absence of a 1 nt spacer between UGUs (Fig. 1C). The highest 7mer R value observed for CELF1, R = ~8 for UGUUUGU, implies > ~250-fold binding affinity over background (Supp. Methods eq. 18), somewhat below that of RBFOX2 for UGCAUG. This observation and the fatter tail of the R value distribution emphasize that CELF1 binds a broader spectrum of motifs with lower affinity than RBFOX2. Of the top fifty 7mers, all contained at least one UGU. However, not every motif containing a single UGU was significant, and some 7mers lacking UGU were significantly enriched, indicating that RNA recognition by CELF1 is complex. Inspection of the top fifty CELF 7mers (Fig. S1D) suggested that they can be clustered into 4 classes matching GUNxGU for x= 0,1,2,3, and a fifth class containing a single GU (Fig. 1E).
MBNL1 is known to favor binding to YGCY motifs in vitro by SELEX (Goers et al., 2010), and GCUU and UGCU were the top 4mers by CLIP-Seq (Wang et al., 2012). The most enriched 7mers for MBNL1 contained either YGCU or GCUU, often supplemented by a second GC. The most enriched 7mer, GCUUGCU, contained both of these 4mers and had an R value near 9, slightly higher than the top value for CELF1 (Fig. 1D). Overall, 54% of 7mers containing YGCU, and 61% of those containing GCUU, but only 9% of those containing YGCC had significant R values, suggesting that MBNL’s specificity is better summarized as YGCU + GCUU rather than YGCY. MBNL1 7mers could be grouped into four classes matching GCNxGC for x= 0,1,2,3, and a fifth class matching YGCU (Fig. 1F). MBNL1’s observed preference for multiple GCs with variable spacing is consistent with previous studies (Cass et al., 2011).
Relative dissociation constants are accurately estimated from RBNS
To better understand the dependence of R values on RBP concentration and to assess the extent and effects of experimental noise, we modeled RBNS experiments and predicted the output under various assumptions. In an idealized setting in which an RBP binds a high affinity motif X with Kd= 5 nM and several moderate affinity motifs Y each with Kd= 30 nM (assuming binding with 1:1 stoichiometry and a Hill coefficient of 1), the fraction of each motif bound is expected to follow essentially a sigmoidal function of RBP concentration, with half maximal binding to the motif occurring at a free protein concentration near the Kd value (Fig. 2A). From the predicted binding fraction, assuming complete recovery of protein, the expected R value at each concentration can be determined under various assumptions about the affinity of the protein for non-specific RNA and the amount of non-specific RNA bound to the apparatus.
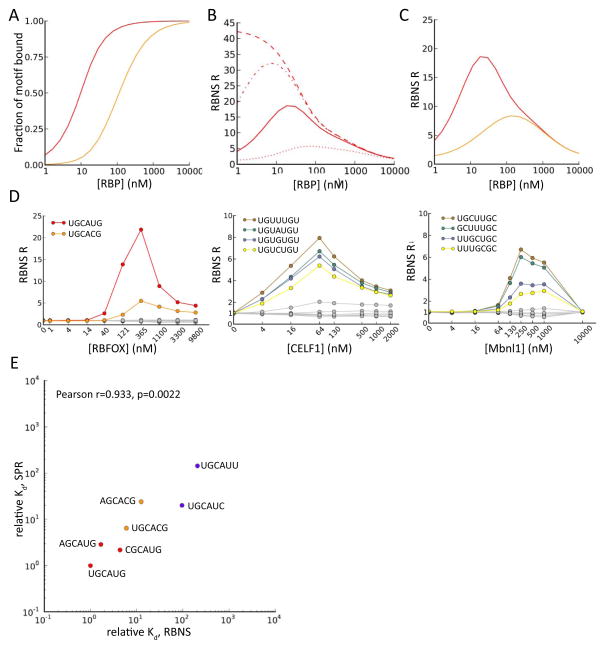
Figure 2. Modeling of RBNS data and estimation of dissociation constants.
A. Simulated output of RBNS under basic assumptions. Standard binding curves for two motifs of different binding affinities (see text).
B. Simulated RBNS R values for a single high affinity 6mer motif as a function of protein concentration, under the assumption of different fixed amounts of nonspecific background (NSB) RNA recovery, independent of protein concentration (dashed: no NSB, dash/dot: low NSB, solid: moderate NSB, dotted: high NSB).
C. Simulated RBNS R values assuming presence of a single strong motif (red) and 10 weaker motifs (orange), including moderate background nonspecific binding.
D. RBNS R values for several top enriched 6mers or 7mers (colored) and several random 6mers/7mers (gray) are shown as a function of RBP concentration for each RBP studied. For RBFOX2, canonical UGCAUG and non-canonical UGCACG 6mers are shown. For CELF1, the four 7mers matching UGUNUGU are shown. For MBNL1, 7mers with two GCs at different spacings are shown, with flanking/intervening Us.
E. Comparison of relative Kd values for several RBFOX2 6mers as estimated by Bind-n-seq (at RBFOX concentration 121 nM), and as measured by SPR. Correlation is significant by Pearson test (R=0.933, P=2e-3). Motifs are colored as in Figure 1B.
See also Figure S4.
The modeled enrichment profiles (Fig. 2B) show that R values of high affinity motifs decrease as RBP concentrations become very high under all conditions tested. This effect is readily understood by considering that high RBP concentrations will tend to drive binding toward lower affinity RNAs (and high affinity motifs may become saturated), resulting in a lower fraction of high affinity motifs in RBP-bound RNA. These simulations also showed that even a small amount of nonspecific binding to the apparatus greatly reduces R values at very low RBP concentrations, because nonspecifically-recovered RNA dilutes the small amount of specifically-bound RNA. Together, these two effects produce a characteristic unimodal curve that peaks at intermediate RBP concentrations under a wide range of assumptions about affinities (Fig. 2C).
Unimodal enrichment profiles for highly enriched kmers were observed for RBFOX2, CELF1 and MBNL1, in general agreement with our model under the assumption of moderate levels of nonspecific background (Fig. 2D). In all cases, R values near 1 were observed at RBP concentrations of 0 nM and began to climb above 1 in the low (4 to 40) nM range, decreasing to near 1 at the highest (micromolar) protein concentrations. For each factor, the relative rankings of kmers obtained at different protein concentrations were highly correlated, supporting the assay’s robustness (Table S2).
Next, we sought to estimate Kd values from RBNS data. The initial quantity of each kmer present was estimated based on the input RNA concentration (1μM), and the concentration of bound RNA was then calculated from the total concentration of protein-RNA complex, measured by Bioanalyzer analysis (Methods). The fraction of bound RNA attributable to binding at each specific kmer was then estimated using a novel “streaming kmer assignment” (SKA) algorithm (Supp. Methods). SKA generalizes the analytical approach of Supp. Methods eq. 18 in that it accounts for arbitrarily complex combinations of affinities to different kmers. The SKA algorithm assigns binding to a specific kmer in each sequence probabilistically, based on continually updated estimates of relative binding preferences, using multiple passes through the sequence read data (Supp. Methods), somewhat analogous to the streaming assignment of ambiguously mapping sequence reads to a genome introduced in the recently described eXpress algorithm (Roberts and Pachter, 2013). Using simulated read data, we observed that assignments of binding locations within reads are more accurate when using SKA than when using raw R values or B values inferred using Supp. Methods eq. 18. In particular, SKA can distinguish bound motifs from motifs enriched through frequent overlap with bound motifs. For example, binding of RBFOX2 to GCAUG motifs will cause overlapping motifs of the form CAUGN (N = A, C, G or T) to be enriched in bound reads even if these motifs have no affinity for RBFOX2 except when preceded by a G. In these cases, the degree to which the bound motif is preferentially enriched enables the SKA algorithm to effectively “learn” to assign lower probabilities (typically near background levels) to overlapping motifs (Fig. S2, S3).
Using estimates of bound and free kmer concentrations, we define the “relative” Kd value of a kmer as the ratio of the kmer’s absolute dissociation constant to that of the highest affinity kmer (Methods). The kmers for which SKA predicts binding (those with absolute Kd< ~2000 nM) have relative Kd estimates spanning several orders of magnitude that are highly correlated to SPR measurements (r = 0.94, P < 0.001) (Fig. 2E). Similarly high correlations were observed relative to previously measured SPR data for RBFOX1, a close paralog of RBFOX2 with identical RNA binding domain (Fig. S4). Together, these observations demonstrate that RBNS yields quantitative measures of protein-RNA affinity.
Secondary structure inhibits binding of Rbfox and CELF proteins to RNA
RBNS can also be used to detect effects of RNA structure on binding of RBPs. We applied the thermodynamically-based Vienna RNAfold algorithm (Hofacker, 2003) to sequence reads in order to assess the contribution of RNA structure to RBP:RNA interactions. In a motif-centric analysis, we analyzed folding of all RNAs harboring high affinity UGCAUG, UGUUU, or UGCUGC motifs in RBFOX2, CELF1 or MBNL1 RBNS datasets, respectively (as well as other motifs), and in control libraries. The probability of intramolecular base pairing at each base in the motif was calculated from the energy-weighted ensemble of structures and averaged across the bases in the motif to give the “average base-pairing probability” (ABP). Sequence reads were then binned by their ABP, and R values were calculated separately for each combination of motif, protein concentration and ABP bin. In these analyses, the bin with lowest ABP (0.0–0.2) was invariably the most enriched for both RBFOX2 and CELF1 binding at all non-zero RBP concentrations (Fig. 3A), and R values decreased as ABP increased. Similar results were obtained when analyzing other top motifs for these two factors. Together, these data suggest that RBFOX2 and CELF1 preferentially recognize single-stranded RNA motifs and that intramolecular base-pairing directly competes with RBP recognition of these RNA motifs to a roughly similar extent for both proteins (Auweter et al., 2006; Edwards et al., 2013).
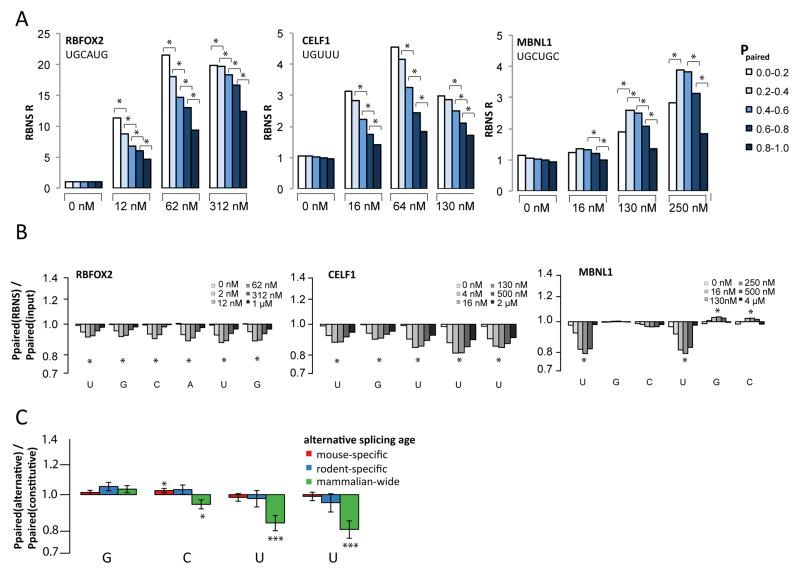
Figure 3. Impact of RNA secondary structure on recognition of RNA sequence motifs.
A. Using rnafold, the average Ppaired value across the bases in each instance of the indicated motif was used to assign each motif occurrence to one of the 5 Ppaired bins indicated, and an R value was calculated at each RBP concentration for each bin as the frequency in the selected library divided by that in the input library. R values are shown for several concentrations of the three proteins, with asterisks indicating statistical significance (Z score > 2, P < 0.05) between adjacent structure bins (Methods).
B. The ratio of the mean value of Ppaired in the bound library to that in the input control library is plotted on a log scale. Z scores were calculated for each selected library. Asterisks indicate bases where every selected library had |Z-score| > 2 (P < 0.05).
C. As in (B) for GCUU motifs located within 130 bases downstream of alternative exons of different evolutionary ages normalized to GCUU motifs in introns downstream of constitutive exons (Merkin et al., 2012). Error bars show SEM and asterisks indicate significance by Wilcoxon rank-sum test (* P < 0.05, *** P < 0.001).
MBNL1 binding tolerates pairing of GCs but favors unpaired Us
The RNA structure analysis for MBNL1 yielded a different pattern, with the highest R values observed for motifs with moderate ABP in the range 0.2–0.6. To better understand the impact of RNA structure on MBNL1 binding, we calculated the base-pairing probability for each base in bound sequences containing UGCUGC, and normalized to that of UGCUGC-containing RNAs in the input library, matching for C+G% content (Methods). This analysis showed no preference for lower base-pairing probabilities at GC positions, but showed substantially reduced base-pairing of Us in bound sequences (Fig. 3B). A similar tolerance for pairing of the central GC dinucleotide and preference for unstructured flanking pyrimidine bases was observed for all high affinity MBNL1 motifs tested, including UGCUU, GCUUGC, CGCUU and GCUGCU, and remained when controlling for GpC dinucleotide content. Similar RNA folding analyses of data for RBFOX2 and CELF1 showed a relatively uniform preference for absence of structure at every position across the binding motif, again consistent with predominant binding to single-stranded RNA (Fig. 3B).
MBNL motifs with unpaired Us are associated with ancient alternative exons
In a recent comparative study, we classified conserved exons by their pattern of alternative or constitutive splicing across four mammals and one bird (Merkin et al., 2012), and observed that introns adjacent to exons alternatively spliced in all of the studied mammals (“ancient alternative exons”) are enriched for MBNL and Rbfox motifs, among others. Curiously, we found that MBNL1 binding to these introns (assayed by CLIP-Seq) exceeded that expected based on motif enrichment by several fold, implying that these introns possess contextual feature(s) that favor binding of MBNL proteins. Performing RNA folding analysis of introns adjacent to exons of different classes, we observed that Us occurring in MBNL motifs such as GCUU that occur near ancient alternative exons have lower base-pairing probability than similar motifs occurring near constitutive exons or more lineage-restricted alternative exons (which showed lower enrichment by CLIP) (Fig. 3C). These observations suggest that ancient alternative exons have been selected for presence of MBNL motifs in contexts where the Us are unpaired, likely to facilitate binding by MBNLs.
Motifs identified in vitro are predominantly bound in vivo
To assess the extent to which RBNS motifs are bound in vivo, we compared to CLIP-Seq data. For RBFOX, a modified version of the high-resolution iCLIP procedure (Konig et al., 2010) was performed using tagged RBFOX2 in mouse embryonic stem cells (mESCs) (Jangi et al., 2014), enabling mapping of sites of crosslinking at nucleotide resolution (Methods).
Sites of crosslinking corresponded in many cases to canonical UGCAUG motifs or to the alternate motif, GCACG, identified above. For example, an iCLIP cluster overlapping a GCACG motif was observed in intron 2 of the Dyrk1a gene (Fig. 4A). To systematically assess the in vivo binding specificity of RBFOX2, the number of crosslinking sites overlapping occurrences of UGCAUG and other motifs in introns and 3′ UTRs were compiled and visualized in a meta-motif representation (Fig. 4B). Sharp peaks of crosslinking density directly over UGCAUG sites were present in both introns and 3′ UTRs, illustrating the high specificity of RBFOX2 binding and the high precision of the iCLIP method (Fig. 4B; upper). We also observed distinct peaks of crosslink density overlapping occurrences of the alternate motif, GCACG, in both introns and 3′ UTRs (Fig. 4B; middle), despite the lack of Us in this motif and the lower abundance of GCACG in the transcriptome (which likely results from presence of a mutation-prone CpG dinucleotide). These peaks were RBFOX2-specific: CLIP-Seq data from an unrelated RBP showed no significant enrichment near canonical or alternate RBFOX2 motifs (Fig. 4B, bottom).
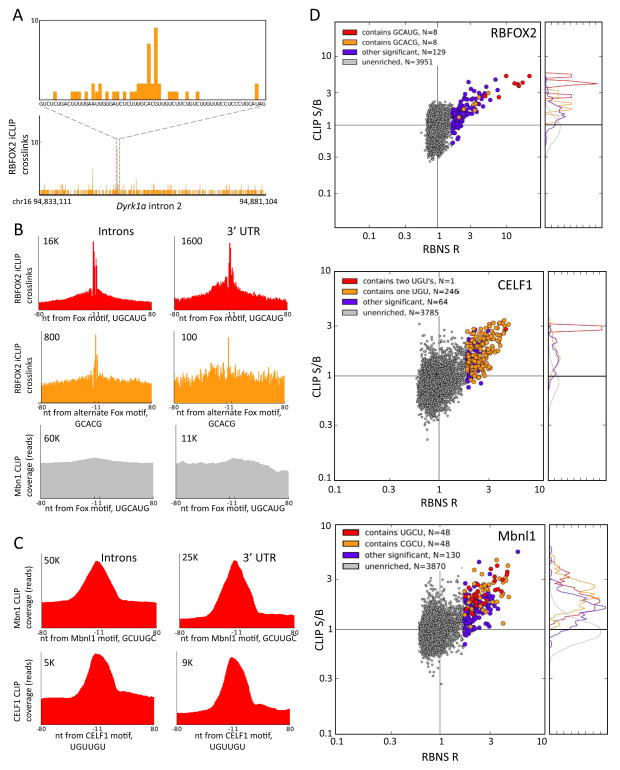
Figure 4. Preferential in vivo binding near RBNS motifs.
A. The distribution of RBFOX2 iCLIP crosslinking sites (mESCs) in intron 2 of the mouse Dyrk1a gene, showing a peak of crosslinks near the alternate motif, GCACG (orange box).
B. Meta-motif plots (cumulative number of crosslink sites) for RBFOX2 iCLIP data over all occurrences of UGCAUG (top row) in introns (left) and in 3′ UTRs (right), and similarly for the secondary motif GCACG (middle row). The bottom row shows a negative control: meta-motif plot of MBNL1 CLIP data (mouse myoblasts) in the vicinity of the RBFOX motif, UGCAUG. Numbers indicate y-axis scale.
C. Meta-motif plot of MBNL1 CLIP-seq coverage in the vicinity of the top MBNL 6mer, GCUUGC, in introns and 3′ UTRs (top row); similar data for CELF1 CLIP-Seq (mouse myoblasts) in the vicinity of the top CELF1 6mer, UUUUGU (bottom row).
D. Scatter plots of CLIP-Seq S/B (Methods) versus RBNS R values for each protein analyzed, using same concentrations as in Figure 1, but using 6mers rather than 7mers for CELF1 and MBNL1 to increase statistical power of CLIP S/B analysis. Top: RBFOX2 iCLIP data in introns. Middle: CELF1 CLIP data in introns. Bottom: MBNL1 CLIP data in 3′ UTRs. All significant 6mers containing the indicated submotifs are colored in red, orange, or purple; all non-significant 6mers are in gray. Histograms at right show the normalized distributions of CLIP S/B for the corresponding color-coded groups of 6mers.
See also Figure S5.
Similar analyses of MBNL1 motifs using Mbnl1 CLIP-Seq data from our previously published study with C2C12 mouse myoblasts (Wang et al., 2012) yielded a pronounced peak over MBNL motifs such as GCUUGC in introns and 3′ UTRs (Fig. 4C; upper). Analysis of CELF1 CLIP-Seq data from a study of this factor’s role in splicing and mRNA stability, also using mouse myoblasts (Wang et al., 2014), yielded a similar peak in the vicinity of canonical CELF motifs such as UGUUGU (Fig. 4C; lower). The peaks observed in the MBNL1 and CELF1 CLIP data were not as sharp as those observed for RBFOX2, likely reflecting the lower resolution of the standard CLIP-Seq protocol relative to the iCLIP protocol used for RBFOX2. Again, these peaks were RBP-specific (not shown).
We next compared in vitro and in vivo binding across a broader spectrum of motifs. We defined a CLIP “signal:background” (S/B) ratio for each motif as the total CLIP-Seq read coverage overlapping occurrences of the motif (“signal”) divided by the average of the CLIP coverage in 40 nt regions located at −80…−41 upstream and +41…+80 downstream of the motif, representing the background level of CLIP density in motif-containing transcripts. Comparing CLIP S/B values to RBFOX2 RBNS R values across all 6mers, we observed a strong correlation of these values for the set of motifs with significant R values, but not for other 6mers (Fig. 4D; left). In fact, 96% of 6mer motifs with significant R value had a CLIP-Seq S/B above the median value for all 6mers (Table S3), including not only all 6mers containing the canonical 5mer GCAUG but also all of those containing the alternate 5mer GCACG. Similar trends were observed for CELF1 and MBNL1, with CLIP-Seq S/B above the median observed for 96% of CELF1 and 99% of MBNL1 6mers with significant R values (Table S3; data for intronic sites in Fig. 4D; data for 3′ UTR sites in Fig. S5). These observations suggest that the intrinsic binding preferences identified by RBNS determine in vivo binding locations of these proteins to a surprisingly large extent. The observation that virtually all RBNS enriched motifs had CLIP signal above the median suggests that a substantial majority of motifs detected in vitro by RBNS are bound in vivo to at least some extent. However, this relationship was not reciprocal: many motifs with high CLIP S/B were bound in vitro, but many others lacked significant in vitro binding, a phenomenon that we explore below.
Alternate and canonical motifs are associated with alternative splicing regulation
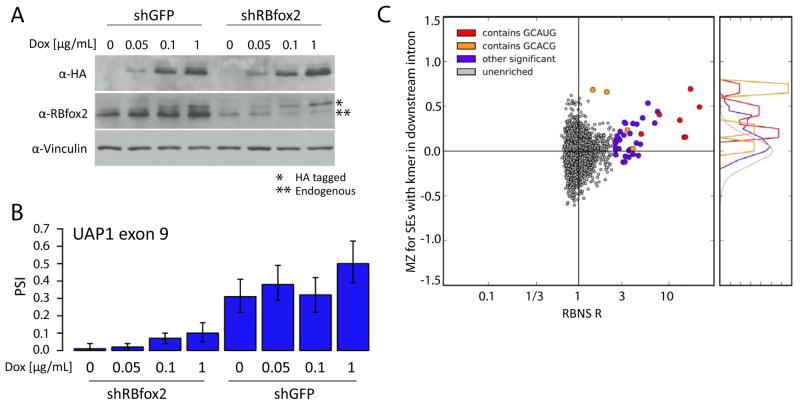
To explore the splicing regulatory activity of the RBFOX2 motifs identified by RBNS, mESCs with a range of RBFOX2 expression levels were generated. Over-expression of RBFOX2 to different extents was achieved by administration of various concentrations of doxycycline to an mESC line containing a tetracycline-inducible version of RBFOX2 (Jangi et al., 2014). Inhibition of RBFOX2 expression was achieved by stably introducing vectors expressing short hairpin RNAs (shRNAs) targeting the 3′ UTR of the endogenous gene (or shRNAs targeting GFP as a control). RNA-Seq analysis of cell lines expressing 8 different levels of RBFOX2 proteins was then performed to assess changes in alternative splicing.
Expression of Rbfox2 increased from 12 FPKM (fragments per kilobase of exon per million mapped fragments) in the lowest condition (shFOX2, 0 μg/mL DOX) to an FPKM of 32 at the highest induced level (shGFP, 1 μg/mL Dox), ranging from 40% to 123% of endogenous levels, still lower than occurs in certain mouse tissues (Fig. S6). Protein levels were confirmed by Western analysis (Fig. 5A). To systematically assess the consistency of changes in splicing, we defined a “monotonicity Z-score” (MZ) for each exon whose “percent spliced in” (PSI) value changed significantly (Wang et al., 2014). MZ captures the extent to which the exon’s PSI consistently increases (MZ > 0) or consistently decreases (MZ < 0) in a set of conditions with increasing levels of a regulatory factor, as is expected to occur for direct regulatory targets.
Figure 5. Splicing regulatory activity of RNA motifs from analysis of splicing factor perturbation data.
A. Western analysis of Rbfox2 in tet-inducible Rbfox2 mESC lines. Cells were treated with either a control hairpin targeting GFP (left lanes) or a hairpin targeting endogenous Rbfox2 mRNAs (right lanes). Cells were treated with 0, 0.05, 0.1 or 1 μg/mL of Dox to induce exogenous FLAG-tagged Rbfox2. Western shows endogenous and tagged Rbfox2 as well as a loading control (Vinculin).
B. The percent spliced in (PSI) values shown for a highly Rbfox2-sensitive alternative exon in pyrophosphorylase Uap1 in mESCs at each of the 8 different Rbfox2 levels shown above (2 hairpins × 4 levels of Dox). Error bars show 95% confidence intervals.
C. Distribution of RBFOX2 monotonicity Z-scores (Methods) versus RBFOX2 RBNS R values for all 6mers. MZ scores were calculated for 1442 skipped exons in mESC-expressed genes using the Rbfox2 perturbation system shown in A). For each 6mer, the average MZ score of all exons which had the 6mer in the first 200 bases of the downstream intron was calculated. Coloring as in Figure 5. RBNS-enriched 6mers had significantly higher MZ scores than unenriched 6mers (KS test, p=2e-7).
See also Figure S6.
Applying this approach to a set of mouse alternative exons, the exons with the highest MZ scores were exon 9 of the UAP1 gene (MZ = 2.98) and the EIIIB exon of Fibronectin1 (MZ = 2.81). The latter is a well-established Rbfox2 target whose downstream intron contains six canonical UGCAUG motifs (Huh and Hynes, 1993; Jin et al., 2003; Lim and Sharp, 1998). RNA-Seq data for the regulated UAP1 exon are displayed in Figure 5B, showing that the PSI value increases from below 10% in conditions where Rbfox2 is depleted, to 61% in the highest over-expression condition. To assess the extent to which particular sequence motifs were associated with splicing regulation, we defined an MZ score for each 6mer as the average MZ value of alternative exons which have the 6mer present in the first 200 bases of the downstream intron, a region in which RBFOX2 binding is associated with activation of exon inclusion (Ponthier et al., 2006; Yeo et al., 2009). Comparing motif MZ scores with RBNS R values of 6mers, we observed that >80% of 6mers with significant R values had positive MZ scores, consistent with a role in enhancement of splicing in response to increased RBFOX2 levels (Fig. 5C). Positive MZ scores were observed not only for all 6mers containing the canonical GCAUG 5mer, but also for all 6mers containing the GCACG alternate motif, supporting that this motif confers RBFOX-dependent splicing regulation.
RBNS detects sequence bias in CLIP data
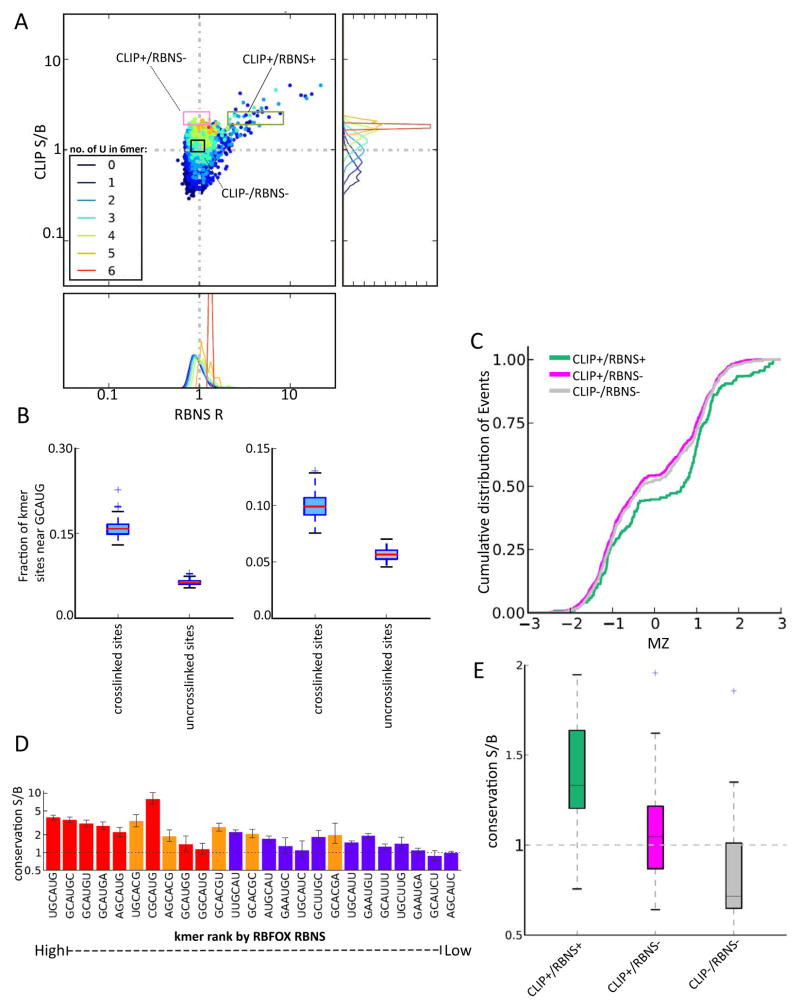
CLIP-Seq is a widely used and effective technique for mapping RBP binding sites in vivo {Licatalosi, 2008 #270;(Sugimoto et al., 2012). However, the absence of alternative comprehensive high-resolution methods for measuring in vivo binding has made it difficult to critically assess CLIP data for systematic biases or sources of false positives and false negatives. Previous studies have shown that CLIP favors U-rich sequences, because uridines form RNA-protein crosslinks more readily than other bases (Sugimoto et al., 2012). Coloring 6mers according to the number of Us that they contained in the plot of RBFOX2 CLIP S/B against RBNS R values revealed a group of 6mers with high U content (≥ 4 U out of 6) at the top center of the distribution with high CLIP S/B but no significant RBNS enrichment (Fig. 6A). By contrast, the remainder of 6mers with high CLIP S/B also had significant positive RBNS R values and contained moderate numbers of Us (usually 1 or 2). This observation and the systematic trend for higher iCLIP S/B values to be associated with higher U content (Fig. 6A; right) suggested that U-richness systematically and substantially enhances detection by CLIP, to an extent that essentially nonspecific (low specificity) protein-RNA interactions may be detected in contexts that are sufficiently U-rich.
Figure 6. RBNS distinguishes subsets of CLIP-seq motifs with and without regulatory activity.
A. RBFOX2 iCLIP S/B in 3′ UTRs is plotted against RBFOX2 RBNS R value for all 6mers (as in Fig. 4D), with points colored by the number of U bases present in the 6mer as indicated. The distribution of iCLIP S/B values is shown at right, and the distribution of RBNS R values are shown below, for each group of 6mers binned by U content. Log scale is used on both axes.
B. RBFOX primary motifs have increased frequency near crosslinked CLIP+/RBNS− sequences. For each CLIP+/RBNS− motif in either introns (left) or 3′ UTRs (right), the fraction of motifs that had a GCAUG within 40 nt was calculated for all motif occurrences that were crosslinked in iCLIP or uncrosslinked.
C. Cumulative distribution of MZ scores for sets of alternative exons grouped by presence of specific 6mer motifs in first 200 nt of downstream intron. Groups of 6mers are colored as in A).
D. Conservation S/B of the top RBFOX2 6mer motifs by RBNS in mammalian 3′ UTRs. Motifs are listed in descending order of R value and colored as in previous figures.
E. Box plots of the distributions of conservation S/B for 6mers grouped as in A).
To determine the extent to which CLIP+/RBNS− motifs result from binding to U-rich sequences near authentic RBFOX motifs, we analyzed the sequences surrounding crosslinked CLIP+/RBNS− motifs (Fig. 6B). While we observed a ~2-fold increase in GCAUG motifs near these sites (within 40 nt) relative to uncrosslinked occurrences of these motifs, presence of a nearby GCAUG motif was observed for only ~15% of crosslinked sites associated with CLIP+/RBNS− motifs (Fig. 6B). These data suggest that some CLIP signal for such motifs comes from binding to nearby canonical motifs, but that most such binding derives from crosslinking of protein that is associated with RNA non-specifically or via interaction with other RBPs.
To assess the splicing activity of motifs detected exclusively by CLIP, we compared the splicing regulatory activity of three sets of motifs: (i) 6mers with high CLIP S/B, but low RBNS R values (the CLIP+/RBNS− set); (ii) 6mers with significant RBNS R values and CLIP S/B values in the same range as the previous set (CLIP+/RBNS+); and (iii) a negative control group of sequences that lacked enrichment by CLIP or RBNS (CLIP−/RBNS−) (Fig. 6A). Comparing the splicing regulation of cassette exons whose downstream introns contain 6mers from each set revealed a clear pattern: exons associated with the CLIP+/RBNS+ set had significantly higher MZ scores than those associated with either control 6mers, or with CLIP+/RBNS− 6mers. Furthermore, the CLIP+/RBNS set was no more likely to be associated with high MZ values than the control set (Fig. 6C). Thus, no evidence was found that the CLIP+/RBNS− set of motifs has regulatory activity. Instead, the simplest explanation is that these motifs result from transient nonspecific interactions of protein with RNA, with U-rich sequences preferentially captured relative to other nonspecifically bound RNAs. This analysis shows that RBNS can provide information useful for interpretation of CLIP-Seq data. On the other hand, the observation that essentially all significant RBNS 6mers also had high CLIP S/B values argues against the existence of a class of CLIP-invisible (.g., uncrosslinkable) RNA motifs, at least for RBFOX2.
RBNS motifs are conserved across mammals
Motifs that contribute to regulation of conserved alternative splicing events should often be evolutionarily conserved, and the canonical binding motifs of RBFOX2, MBNL1 and CELF1 are highly conserved in introns flanking alternative exons and in 3′ UTRs (Daughters et al., 2009; Merkin et al., 2012; Sugnet et al., 2006; Wang et al., 2012; Wang et al., 2008). Adapting a method previously developed to assess conservation of microRNA target sites in mRNAs (Friedman et al., 2009), we assessed the conservation of significant RBFOX2 RBNS motifs in orthologous UTRs of 23 mammalian species. UTRs were chosen over introns because they can be more reliably aligned in most cases. For this analysis, we calculated for each 6mer the fraction of its occurrences in conserved introns that were evolutionarily conserved over at least a minimum evolutionary branch length (the “signal”). We measured a similar fraction for a cohort of control 6mers matched for genomic abundance, C+G% and CpG dinucleotide content, defining the mean conserved fraction over these control 6mers as the “background”. For RBFOX motifs, almost all 6mers containing the canonical GCAUG 5mer had conservation signal:background (S:B) ratio significantly above 1, indicating preferential conservation (Fig. 6D). Furthermore, 6mers containing the alternative motif GCACG had S:B values nearly as high, further supporting the in vivo regulatory function of this motif. Some but not all of the remaining RBNS motifs also had significant S:B values, supporting function. No significant conservation was detected for the set of CLIP+/RBNS− 6mers (Fig. 6E), consistent with lack of regulatory activity. By contrast, the set of CLIP+/RBNS+ motifs matched for CLIP density showed significant conservation (Fig. 6E).
Discussion
Here we have described a method, RBNS, and associated analytical approaches, that provides comprehensive and quantitative information about the spectrum of RNA motifs bound by an RBP. As affinities for all kmers are assessed simultaneously, this approach may prove attractive as an alternative to traditional low-throughput quantitative methods. To address more targeted questions related to specific RBPs, various details of the RBNS experimental setup could be varied, including the length or composition of the input RNA or the presence of additional protein factors that is hypothesized to cooperate or compete with the protein being pulled down. Instead of random RNA, total cellular RNA, mRNA or RNA immunoprecipitaed with an RBP could be used to limit sampled sequences to potential in vivo binding sites. This approach could enable detection of binding to sites with complex architecture engineered by evolution, but would substantially reduce sequence diversity, limiting the power to analyze binding to longer motifs or effects of RNA structure. Current sequencing technologies limit motif size to about 10 bases, but there are strategies to circumvent these limits (Supp. Methods).
Complexity of RNA binding affinity spectra
The depth of data generated in this approach yields information across a broad range of binding affinities, particularly when several RBP concentrations are used, enabling detection of weaker but significant motifs, such as GCACG for RBFOX2. For this particular example, the structure of the RBFOX1 RRM domain (which is identical to that of RBFOX2) has been solved by NMR, in complex with RNA representing canonical motif, UGCAUG (Auweter et al., 2006). The substitution of U for C in the fifth position of the 6mer would not introduce a steric clash, and one of the two hydrogen bonds that RBFOX1 makes with U5 would be preserved with a C in this position (Auweter et al., 2006). Together, these observations suggest that RBFOX proteins can bind GCACG in a manner similar to their binding of GCAUG, albeit with somewhat lower affinity. These observations, and similar results for a variety of variants of classical CELF1 and MBNL1 motifs, lead us to conclude that RBPs often have rather complex RNA binding affinity spectra, often centered on core dinucleotides, such as the GUs and GCs present in CELF1 and MBNL1 motifs, respectively. We also found that GCACG motifs are bound in vivo, and are associated with sequence conservation and splicing regulatory activity to an extent similar to canonical motifs. These and similar observations for a variety of variant CELF1 and MBNL1 motifs argue that secondary motifs with affinities within an order of magnitude or so of the optimal motif often play conserved roles in splicing regulation.
We envision several types of applications for RBNS and the resulting data. These applications include modeling and predicting changes in RBP occupancy and regulatory activity in response to changes in RBP abundance or activity occurring during development, between cell types, or in different cell states (e.g., EMT, disease versus normal), and predicting the regulatory consequences of genetic variation (e.g., disease gene mutations or polymorphisms) on RBP binding and regulatory activity. For these applications, the quantitative precision of the Fi and Kd values from the SKA algorithm may prove useful. Other potential applications include understanding the influence of RNA secondary structure on RBP binding and function, and interpreting CLIP-Seq data. These last two applications are discussed below.
Effects of structure on RNA binding
The impact of RNA structure on protein-RNA interactions can be inferred using RBNS. For RBFOX2 and CELF1, both of which bind RNA through RRM domains, our RNA folding analyses suggested strong preferences for binding of single-stranded RNA. Analysis of MBNL1, which binds RNA through zinc fingers, revealed a strong preference for unpaired Us but no significant bias for or against unpaired G and C bases in UGC-containing motifs, suggesting either that MBNL can melt paired GC dinucleotides or that it can recognize them even when they are base-paired. CUG repeat RNA, which is tightly bound by MBNL proteins both in vitro and in vivo (Teplova and Patel, 2008)Kino, 2004 #191;Fardaei, 2001 #317}, crystallizes as a hairpin with paired GCs separated by unpaired U-U bulges (Mooers et al., 2005), consistent with the pattern of MBNL binding preferences observed here. Intron 4 of cardiac troponin T (cTNT), a well-characterized MBNL binding and regulatory target, also contains multiple paired GCs flanked by unpaired pyrimidine bulges (Warf and Berglund, 2007). Consistently, biochemical evidence has shown that MBNL binds with high affinity to pairs of GC dinucleotides with a wide range (~1–15 bases) of intervening pyrimidine bases (Goers et al., 2010) Cass, 2011 #96}. This structural signature is consistent with RNA looping around MBNL proteins such that different zinc fingers interact with different GCs. RNA looping as a mechanism of RNA recognition has been proposed for PTB (Oberstrass et al., 2005; Perez et al., 1997) and is also consistent with the crystal structure of MBNL1 zinc fingers 3 and 4 (Teplova and Patel, 2008).
RBNS enhances interpretation of CLIP data
RBNS appears to yield a less biased portrait of the spectrum of RNA motifs bound by an RBP than do methods based on UV crosslinking, making it a useful complement to CLIP-based methods (including iCLIP and PAR-CLIP). The subset of CLIP-enriched motifs that were not detected by RBNS lacked evidence of regulatory activity or sequence conservation, arguing that they do not reflect biologically relevant binding. In practice, when crosslinking to a CLIP+/RBNS− motif that is located in close proximity to a CLIP+/RBNS+ motif is observed, our analyses imply that in most cases this binding should be attributed to the CLIP+/RBNS+ motif. Applying this sort of correction automatically might improve inference of regulatory elements. When comparing the extent of binding to two or more different regions, we expect that RBNS affinities could be used to correct for the crosslinking bias inherent in CLIP and improve the accuracy of quantitation.
Experimental Procedures
Cloning, expression and purification of proteins
Full length CELF1, MBNL1 (1-260), and RBFOX2 (100-194) were cloned downstream of a GST-SBP tandem affinity tag. Both truncated MBNL1 and RBFOX2 constructs contain all RNA binding domains, including all four MBNL1 Zinc finger domains and RBFOX2’s single RNA recognition motif (RRM). The proteins were recombinantly expressed, purified via the GST tag and the GST tag cleaved off with Prescission protease (GE).
RBNS
RBNS was performed after purifying a given RBP and in vitro transcribing RBNS input RNA, experimental details can be found in supplemental methods. 7–10 concentrations of RBP, including a no RBP condition was equilibrated in Binding buffer for 30 minutes at room temperature or 37 degrees in the case of RBFOX RBNS. RBNS input random RNA was then added to a final concentration of 1uM with 40 U of Superasin (Ambion) and incubated for 1 hour at room temperature or 37 degrees. To pull down tagged RBP and interacting RNA each RNA/protein solution was then added to 1 mg of washed streptavidin magnetic beads and incubated for one hour. Unbound RNA was removed from the beads and the beads were washed once with 1 mL of wash buffer. The beads were incubated at 70 degrees for 10 minutes in 100 uL of elution buffer (10mM tris pH 7.0, 1mM EDTA, 1%SDS) and the eluted material collected. Bound RNA was extracted, reverse transcribed into cDNA, and then amplified by PCR. See Supplemental methods for a more detailed description of the RBNS protocol
R values
Motif R values were calculated as the motif frequency in the RBP-selected pool over the frequency in the input RNA library. Frequencies were controlled for respective library read depth. R values were considered significant if greater than 2 standard deviations from the mean. The rate of kmer enrichment in the no protein condition, relative to the input library was defined as the FDR.
SKA analysis
The streaming kmer assignment algorithm is described in Supplemental Methods. See also Figures S2 and S3.
Monotonicity Z-scores
Each of the eight RNA-seq libraries was mapped to the mouse genome (mm9) with Tophat and the alternative splicing of skipped exon (SE) events was analyzed with MISO (Katz et al., 2010) as follows. Significantly changing (Bayes factor ≥ 5.0) events were identified from all pairwise comparisons between the libraries. The difference between the number of comparisons where the higher RBFOX concentration showed significantly more inclusion and the number where the lower RBFOX concentration showed more inclusion was calculated for all events. For each skipped exon event the monotonicity score was defined to be the Z score of this difference out of a control set of differences generated by shuffling the order of the RBFOX concentration datasets.
Supplementary Material
Highlights.
RBNS is a method for comprehensive, quantitative mapping of RNA binding specificity
RBNS identifies motifs recognized by RBFOX2, CELF1 and MBNL1 proteins
RNA structure inhibits binding, except MBNL1 tolerates G/C pairing in UGC motifs
RBNS distinguishes subsets of functional and non-functional CLIP-seq motifs
Acknowledgments
We thank Andy Berglund for advice on expression of RNA binding proteins, Eric Wang for helpful discussions, and Tom Cooper, Wendy Gilbert and A. B. for helpful suggestions on the text. We also thank Vincent Butty for his help with conservation analyses and Albert Tai for performing SPR analysis. This work was funded by an NIH NRSA Postdoctoral Fellowship (N.L.) and by grants from the NIH (C.B.B.).
Footnotes
Software
All software described here will be made freely available for academic use on github.
Accession numbers
(Will be added after acceptance)
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Auweter SD, Fasan R, Reymond L, Underwood JG, Black DL, Pitsch S, Allain FH. Molecular basis of RNA recognition by the human alternative splicing factor Fox-1. EMBO J. 2006;25:163–173. doi: 10.1038/sj.emboj.7600918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baraniak AP, Chen JR, Garcia-Blanco MA. Fox-2 mediates epithelial cell-specific fibroblast growth factor receptor 2 exon choice. Mol Cell Biol. 2006;26:1209–1222. doi: 10.1128/MCB.26.4.1209-1222.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell ZT, Bhimsaria D, Valley CT, Rodriguez-Martinez JA, Menichelli E, Williamson JR, Ansari AZ, Wickens M. Cooperativity in RNA-protein interactions: global analysis of RNA binding specificity. Cell reports. 2012;1:570–581. doi: 10.1016/j.celrep.2012.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cass D, Hotchko R, Barber P, Jones K, Gates DP, Berglund JA. The four Zn fingers of MBNL1 provide a flexible platform for recognition of its RNA binding elements. BMC Mol Biol. 2011;12:20. doi: 10.1186/1471-2199-12-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daughters RS, Tuttle DL, Gao W, Ikeda Y, Moseley ML, Ebner TJ, Swanson MS, Ranum LP. RNA gain-of-function in spinocerebellar ataxia type 8. PLoS genetics. 2009;5:e1000600. doi: 10.1371/journal.pgen.1000600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards JM, Long J, de Moor CH, Emsley J, Searle MS. Structural insights into the targeting of mRNA GU-rich elements by the three RRMs of CELF1. Nucleic acids research. 2013;41:7153–7166. doi: 10.1093/nar/gkt470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehman LT, Meera P, Stoilov P, Shiue L, O’Brien JE, Meisler MH, Ares M, Jr, Otis TS, Black DL. The splicing regulator Rbfox2 is required for both cerebellar development and mature motor function. Genes Dev. 2012;26:445–460. doi: 10.1101/gad.182477.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goers ES, Purcell J, Voelker RB, Gates DP, Berglund JA. MBNL1 binds GC motifs embedded in pyrimidines to regulate alternative splicing. Nucleic Acids Res. 2010;38:2467–2484. doi: 10.1093/nar/gkp1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho TH, Charlet BN, Poulos MG, Singh G, Swanson MS, Cooper TA. Muscleblind proteins regulate alternative splicing. EMBO J. 2004;23:3103–3112. doi: 10.1038/sj.emboj.7600300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofacker IL. Vienna RNA secondary structure server. Nucleic Acids Res. 2003;31:3429–3431. doi: 10.1093/nar/gkg599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh GS, Hynes RO. Elements regulating an alternatively spliced exon of the rat fibronectin gene. Molecular and cellular biology. 1993;13:5301–5314. doi: 10.1128/mcb.13.9.5301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jangi M, Boutz PL, Paul P, Sharp P. Rbfox2 controls autoregulation and gene expression in RNA binding protein networks. Genes & Dev. 2014;28:637–651. doi: 10.1101/gad.235770.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Y, Suzuki H, Maegawa S, Endo H, Sugano S, Hashimoto K, Yasuda K, Inoue K. A vertebrate RNA-binding protein Fox-1 regulates tissue-specific splicing via the pentanucleotide GCAUG. EMBO J. 2003;22:905–912. doi: 10.1093/emboj/cdg089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolma A, Kivioja T, Toivonen J, Cheng L, Wei G, Enge M, Taipale M, Vaquerizas JM, Yan J, Sillanpaa MJ, et al. Multiplexed massively parallel SELEX for characterization of human transcription factor binding specificities. Genome Res. 2010;20:861–873. doi: 10.1101/gr.100552.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalsotra A, Xiao X, Ward AJ, Castle JC, Johnson JM, Burge CB, Cooper TA. A postnatal switch of CELF and MBNL proteins reprograms alternative splicing in the developing heart. Proc Natl Acad Sci U S A. 2008;105:20333–20338. doi: 10.1073/pnas.0809045105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz Y, Wang ET, Airoldi EM, Burge CB. Analysis and design of RNA sequencing experiments for identifying isoform regulation. Nat Methods. 2010;7:1009–1015. doi: 10.1038/nmeth.1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konig J, Zarnack K, Rot G, Curk T, Kayikci M, Zupan B, Turner DJ, Luscombe NM, Ule J. iCLIP reveals the function of hnRNP particles in splicing at individual nucleotide resolution. Nature structural & molecular biology. 2010;17:909–915. doi: 10.1038/nsmb.1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuyumcu-Martinez NM, Wang GS, Cooper TA. Increased steady-state levels of CUGBP1 in myotonic dystrophy 1 are due to PKC-mediated hyperphosphorylation. Mol Cell. 2007;28:68–78. doi: 10.1016/j.molcel.2007.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladd AN, Charlet N, Cooper TA. The CELF family of RNA binding proteins is implicated in cell-specific and developmentally regulated alternative splicing. Mol Cell Biol. 2001;21:1285–1296. doi: 10.1128/MCB.21.4.1285-1296.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim LP, Sharp PA. Alternative splicing of the fibronectin EIIIB exon depends on specific TGCATG repeats. Molecular and cellular biology. 1998;18:3900–3906. doi: 10.1128/mcb.18.7.3900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mankodi A, Lin X, Blaxall BC, Swanson MS, Thornton CA. Nuclear RNA foci in the heart in myotonic dystrophy. Circ Res. 2005;97:1152–1155. doi: 10.1161/01.RES.0000193598.89753.e3. [DOI] [PubMed] [Google Scholar]
- Marquis J, Paillard L, Audic Y, Cosson B, Danos O, Le Bec C, Osborne HB. CUG-BP1/CELF1 requires UGU-rich sequences for high-affinity binding. Biochem J. 2006;400:291–301. doi: 10.1042/BJ20060490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkin J, Russell C, Chen P, Burge CB. Evolutionary dynamics of gene and isoform regulation in Mammalian tissues. Science. 2012;338:1593–1599. doi: 10.1126/science.1228186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mooers BH, Logue JS, Berglund JA. The structural basis of myotonic dystrophy from the crystal structure of CUG repeats. Proc Natl Acad Sci U S A. 2005;102:16626–16631. doi: 10.1073/pnas.0505873102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nutiu R, Friedman RC, Luo S, Khrebtukova I, Silva D, Li R, Zhang L, Schroth GP, Burge CB. Direct measurement of DNA affinity landscapes on a high-throughput sequencing instrument. Nat Biotechnol. 2011;29:659–664. doi: 10.1038/nbt.1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberstrass FC, Auweter SD, Erat M, Hargous Y, Henning A, Wenter P, Reymond L, Amir-Ahmady B, Pitsch S, Black DL, et al. Structure of PTB bound to RNA: specific binding and implications for splicing regulation. Science. 2005;309:2054–2057. doi: 10.1126/science.1114066. [DOI] [PubMed] [Google Scholar]
- Perez I, McAfee JG, Patton JG. Multiple RRMs contribute to RNA binding specificity and affinity for polypyrimidine tract binding protein. Biochemistry. 1997;36:11881–11890. doi: 10.1021/bi9711745. [DOI] [PubMed] [Google Scholar]
- Ponthier JL, Schluepen C, Chen W, Lersch RA, Gee SL, Hou VC, Lo AJ, Short SA, Chasis JA, Winkelmann JC, et al. Fox-2 splicing factor binds to a conserved intron motif to promote inclusion of protein 4.1R alternative exon 16. The Journal of biological chemistry. 2006;281:12468–12474. doi: 10.1074/jbc.M511556200. [DOI] [PubMed] [Google Scholar]
- Ray D, Kazan H, Chan ET, Pena Castillo L, Chaudhry S, Talukder S, Blencowe BJ, Morris Q, Hughes TR. Rapid and systematic analysis of the RNA recognition specificities of RNA-binding proteins. Nat Biotechnol. 2009;27:667–670. doi: 10.1038/nbt.1550. [DOI] [PubMed] [Google Scholar]
- Ray D, Kazan H, Cook KB, Weirauch MT, Najafabadi HS, Li X, Gueroussov S, Albu M, Zheng H, Yang A, et al. A compendium of RNA-binding motifs for decoding gene regulation. Nature. 2013;499:172–177. doi: 10.1038/nature12311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts A, Pachter L. Streaming fragment assignment for real-time analysis of sequencing experiments. Nature methods. 2013;10:71–73. doi: 10.1038/nmeth.2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefl R, Skrisovska L, Allain FH. RNA sequence- and shape-dependent recognition by proteins in the ribonucleoprotein particle. EMBO reports. 2005;6:33–38. doi: 10.1038/sj.embor.7400325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto Y, Konig J, Hussain S, Zupan B, Curk T, Frye M, Ule J. Analysis of CLIP and iCLIP methods for nucleotide-resolution studies of protein-RNA interactions. Genome Biol. 2012;13:R67. doi: 10.1186/gb-2012-13-8-r67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugnet CW, Srinivasan K, Clark TA, O’Brien G, Cline MS, Wang H, Williams A, Kulp D, Blume JE, Haussler D, et al. Unusual intron conservation near tissue-regulated exons found by splicing microarrays. PLoS computational biology. 2006;2:e4. doi: 10.1371/journal.pcbi.0020004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taneja KL, McCurrach M, Schalling M, Housman D, Singer RH. Foci of trinucleotide repeat transcripts in nuclei of myotonic dystrophy cells and tissues. J Cell Biol. 1995;128:995–1002. doi: 10.1083/jcb.128.6.995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teplova M, Patel DJ. Structural insights into RNA recognition by the alternative-splicing regulator muscleblind-like MBNL1. Nat Struct Mol Biol. 2008;15:1343–1351. doi: 10.1038/nsmb.1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timchenko LT, Miller JW, Timchenko NA, DeVore DR, Datar KV, Lin L, Roberts R, Caskey CT, Swanson MS. Identification of a (CUG)n triplet repeat RNA-binding protein and its expression in myotonic dystrophy. Nucleic acids research. 1996;24:4407–4414. doi: 10.1093/nar/24.22.4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ule J, Jensen KB, Ruggiu M, Mele A, Ule A, Darnell RB. CLIP identifies Nova-regulated RNA networks in the brain. Science. 2003;302:1212–1215. doi: 10.1126/science.1090095. [DOI] [PubMed] [Google Scholar]
- Underwood JG, Boutz PL, Dougherty JD, Stoilov P, Black DL. Homologues of the Caenorhabditis elegans Fox-1 protein are neuronal splicing regulators in mammals. Mol Cell Biol. 2005;25:10005–10016. doi: 10.1128/MCB.25.22.10005-10016.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ET, Cherone J, Cooper TA, Burge CB. Functional antagonism between CELF and Muscleblind proteins in the nucleus and cytoplasm. 2014 in preparation. [Google Scholar]
- Wang ET, Cody NA, Jog S, Biancolella M, Wang TT, Treacy DJ, Luo S, Schroth GP, Housman DE, Reddy S, et al. Transcriptome-wide regulation of pre-mRNA splicing and mRNA localization by muscleblind proteins. Cell. 2012;150:710–724. doi: 10.1016/j.cell.2012.06.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warf MB, Berglund JA. MBNL binds similar RNA structures in the CUG repeats of myotonic dystrophy and its pre-mRNA substrate cardiac troponin T. RNA. 2007;13:2238–2251. doi: 10.1261/rna.610607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo GW, Coufal NG, Liang TY, Peng GE, Fu XD, Gage FH. An RNA code for the FOX2 splicing regulator revealed by mapping RNA-protein interactions in stem cells. Nat Struct Mol Biol. 2009;16:130–137. doi: 10.1038/nsmb.1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zykovich A, Korf I, Segal DJ. Bind-n-Seq: high-throughput analysis of in vitro protein-DNA interactions using massively parallel sequencing. Nucleic Acids Res. 2009;37:e151. doi: 10.1093/nar/gkp802. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.