Abstract
Alternative (M2) macrophage activation driven through interleukin 4 receptor α (IL-4Rα) is important for immunity to parasites, wound healing, the prevention of atherosclerosis and metabolic homeostasis. M2 polarization is dependent on fatty acid oxidation (FAO), but the source of fatty acids to support this metabolic program has not been clear. We show that the uptake of triacylglycerol substrates via CD36 and their subsequent lipolysis by lysosomal acid lipase (LAL) was important for the engagement of elevated oxidative phosphorylation (OXPHOS), enhanced spare respiratory capacity (SRC), prolonged survival and expression of genes that together define M2 activation. Inhibition of lipolysis suppressed M2 activation during infection with a parasitic helminth, and blocked protective responses against this pathogen. Our findings delineate a critical role for cell-intrinsic lysosomal lipolysis in M2 activation.
Macrophages exist throughout the body as resident components of most tissues. They are embryonically derived, seeded into tissues in utero, and maintained by in situ proliferation1,2. During inflammation additional macrophages develop from monocytes recruited from the bone marrow1, or from proliferation of resident cells3. Macrophages are crucial for immunity and can adopt different activation states depending on context. Interferon-γ(IFN-γ in combination with Toll-like receptor (TLR) agonists promotes M1 (or classical) activation, whereas the cytokines interleukin 4 (IL-4) and IL-13 promote M2 (or alternative) activation4,5. From the host defense standpoint, M1 macrophages are inflammatory and can play a positive role in immunity to microbial pathogens and tumors5. In contrast, M2 macrophages promote tissue repair and metabolic homeostasis, and play key roles in immunity to parasitic helminths5.
M1 and M2 macrophages have distinct metabolic phenotypes, which differ from those of resting macrophages6,7. M1 macrophages rely on aerobic glycolysis, while M2 macrophages use fatty acid oxidation (FAO, also known as β-oxidation) to fuel mitochondrial oxidative phosphorylation (OXPHOS). IL-4-induced changes in macrophage metabolism are dependent upon the transcription factor STAT6 and underpinned by the induction of expression of PPARγ-coactivator-1β (PGC1β) and associated mitochondrial biogenesis8. M2 activation is prevented by inhibition of FAO, whereas overexpression of PGC1β is sufficient to attenuate M1 activation in response to IFN-γ and lipopolysaccharide (LPS)8. Thus IL-4-induced FAO is critical for M2 activation. In other immune cells FAO is known to support cellular longevity9,10, but whether or not this is the case in macrophages is unclear.
Because of the health implications of M2 macrophage activation in various settings, there is considerable interest in understanding the cellular pathways that underpin the M2 phenotype. Despite the established importance of FAO for M2 activation, the source of fatty acids to support this process is unknown. The consensus view from work in other cell types is that fatty acids to meet metabolic and other requirements are released through a coordinated process of lipolysis from triacylglycerols stored in lipid droplets (LDs) that is initiated by the enzyme adipose triglyceride lipase (ATGL)11,12. However, the expression of CD36, which is a receptor for the endocytosis of triacylglycerol-rich lipoprotein particles, such as LDL and VLDL13,14, is induced in macrophages by IL-4 (ref. 15), and has been implicated in M2 activation16. Those findings suggested to us that the uptake and lipolysis of exogenous triacylglycerols may serve to generate fatty acids for FAO in M2 macrophages. Consistent with this view, we report here that lysosomal lipolysis mediated by lysosomal acid lipase (LAL), an enzyme that is expressed in macrophages as they differentiate from monocytes17, and which is further induced by stimulation with IL-4, plays an important role in M2 activation. Inhibition of this pathway suppressed changes in OXPHOS and macrophage longevity and the expression of key genes that mark M2 commitment. Many of the effects of lipolysis inhibition were recapitulated in CD36-deficient macrophages. Our data highlight a previously unappreciated role for LAL in macrophage cell-intrinsic lipolysis to support FAO, show that this enzyme is integral to the M2 activation pathway, and provide a framework for understanding the contribution of CD36 to these processes.
Results
M2 macrophage activation is dependent on fatty acid oxidation
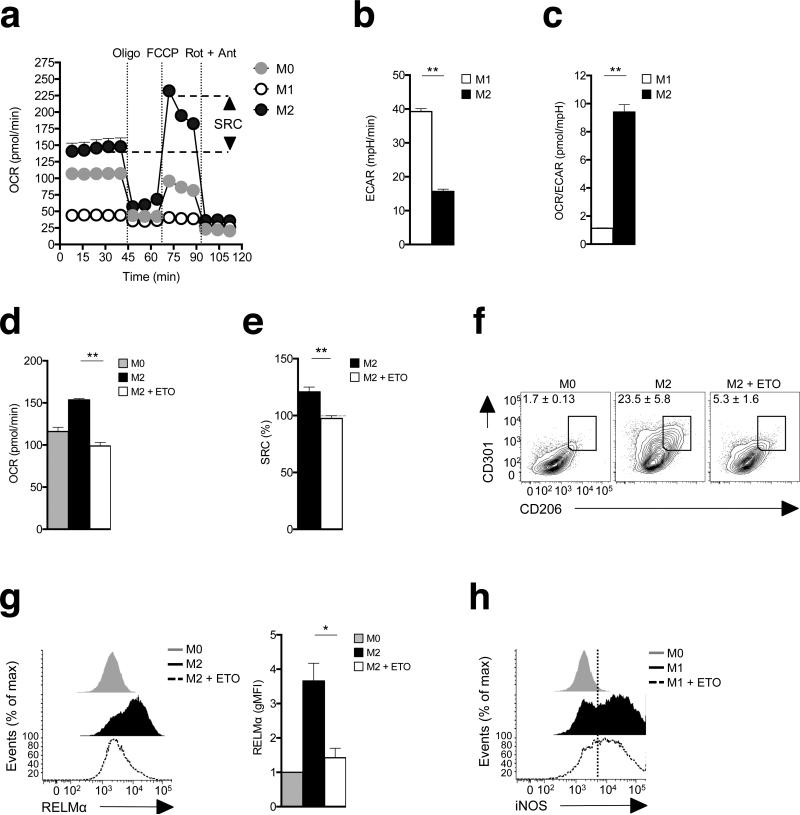
Using extracellular flux analysis, we compared oxygen consumption by M0 (unactivated), M1 and M2 bone marrow-derived macrophages. We found that M2 macrophages had enhanced mitochondrial oxygen consumption rates (OCR), and markedly increased spare respiratory capacity (SRC10,18 the quantitative difference between maximal uncontrolled OCR, and the initial basal OCR), indicative of increased commitment to OXPHOS (Fig. 1a). In contrast, we found no evidence of mitochondrial oxygen consumption in M1 macrophages (Fig. 1a), which rather rely on aerobic glycolysis, measured as the extracellular acidification rate, ECAR (Fig. 1b), to meet their bioenergetic needs19. The extent of the metabolic difference between M1 and M2 cells was apparent in the overall ratio of OXPHOS to aerobic glycolysis, which was 10-fold higher in M2 than M1 macrophages, reflecting polar opposite core metabolic programs (Fig. 1c). Consistent with previous reports, we found that commitment of M2 macrophages to FAO and OXPHOS was evident in the increased expression of genes in these pathways (Supplementary Fig. 1a,c)8,20. This pathway was also sensitive to the suppression of mitochondrial oxygen consumption by etomoxir, an inhibitor of Cpt1, a key FAO enzyme (Fig. 1d)8. We found that etomoxir also eradicated SRC in M2 macrophages (Fig. 1e), validating the link between FAO and heightened OXPHOS. Enhanced SRC was previously linked to longevity in memory CD8+ T cells10. We found this increased longevity to also be true in macrophages, where M2 cells exhibited a significant survival advantage compared to M0 and M1 cells in culture (Supplementary Fig. 2).
Figure 1.
M2 activation is marked by increased spare respiratory capacity and is dependent on fatty acid oxidation. (a) Bone marrow-derived macrophages were cultured in medium without (M0) or with IFN-γ + LPS (M1), or with IL-4 (M2) for 24 h and then oxygen consumption rates (OCR) was determined using an XF-96 Extracellular Flux Analyzer (EFA) during sequential treatments with oligomycin, FCCP, and rotenone/antimycin. Spare respiratory capacity (SRC), the quantitative difference between maximal uncontrolled OCR, and the initial basal OCR, is depicted in the plot. (b) M1 and M2 macrophage basal extracellular acidification rates (ECAR, changes in mpH per unit time), measured in real time using the EFA. (c) Ratios of OCR/ECAR for M1 and M2 macrophages. (d) The effect of etomoxir (ETO) on basal OCR in M2 macrophages. Basal OCR for M0 macrophages is shown as a reference. (e) The effect of ETO on M2 SRC as defined in (a). (f) The effect of ETO on CD206 and CD301 expression by M2 macrophages, as measured by flow cytometry. Expression on M0 macrophages is shown for reference. (g) Effect of ETO on RELMαexpression by M2 macrophages. (h) Effect of ETO on iNOS expression by M1 macrophages. In these experiments ETO was included in the culture from the time at which M2 or M1 polarizing conditions were initiated, and data were collected 24 h later. Data in a-e are means + SEM (a) or ± SEM of 5 or more technical replicates from one experiment, and are representative of more than three independent experiments.. In f, numbers represent % of macrophages falling within indicated gates; plots shown are from one representative experiment of 2, but numbers are mean ± SEM of data from both experiments. . In g histograms show plots from one experiment representative of 3, and the bar graph shows mean relative geometric mean fluorescence intensity (gMFI) values ± SEM from 3 independent experiments. In h, histograms show plots from one experiment representative of 2. The mean % of cells that were iNOS positive in these two experiments were 69.7% ± 1.3% (SEM) for M1 cells and 60.5% ± 9.7% for M1 plus ETO. P-values are from Student's t-test (*P < 0.05, **P < 0.0001).
M2 and M1 macrophage activation are marked by the differential expression of a large panel of genes, including Mrc1 (CD206), Clec10a (CD301), Retlna (RELMα), Pdcd1lg2
(PD-L2) and Nos2 (iNOS) (Supplementary Fig. 1b,c). Using flow cytometry, we found that etomoxir remarkedly inhibited the expression of CD301, CD206 and RELMα in macrophages cultured in M2 conditions (Fig. 1f,g), but had comparatively little effect on the expression of iNOS in cells cultured in M1 conditions (Fig. 1h). Thus, FAO, measureable as increased baseline OCR and increased SRC, is essential for M2 activation.
Lipolysis contributes to M2 activation
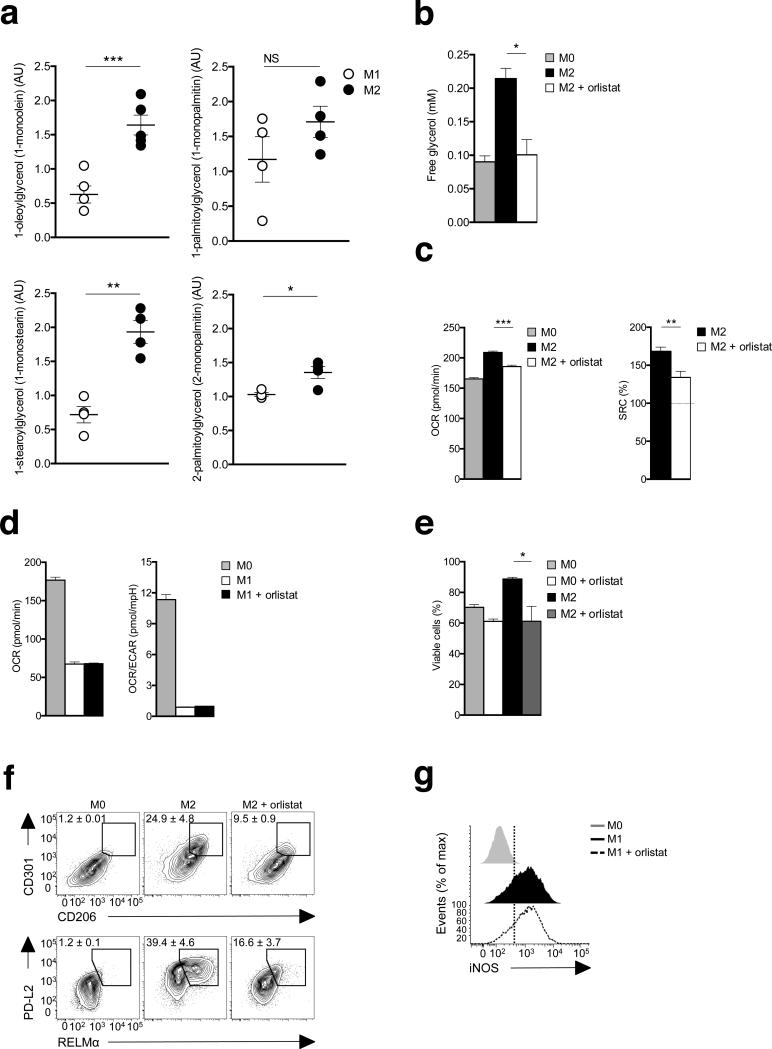
Cellular requirements for fatty acids can be met by enzymatically regulated lipolysis of triacylglycerols into diacylglycerols and monoacylglycerols, accompanied by fatty acid release11; an endpoint of this process is the release of glycerol from cells in which lipolysis is occurring11. We performed global metabolite profiling using mass spectrometry and found significantly increased amounts of 3 out of 4 monoacylglycerols measured in M2 cells, indicating that lipolysis is enhanced in these cells (Fig. 2a). Consistent with this conclusion, we found increased concentrations of extracellular glycerol were present in cultures of M2 compared with M0 macrophages (Fig. 2b). To test whether lipolysis is linked to FAO and OXPHOS, we stimulated cells with IL-4 in the presence or absence of tetrahydrolipistatin (Orlistat), a clinically utilized active site-directed lipase inhibitor21. We found that Orlistat inhibited IL-4-induced increases in lipolysis, as measured by glycerol production (Fig. 2b), and suppressed associated changes in OXPHOS (Fig. 2c), although it had no effect on the metabolic changes induced by M1 conditions (Fig. 2d). These reductions in OXPHOS and SRC in Orlistat-treated M2 cells were linked to significantly reduced survival, evident as in decreased viability by day 2 of culture (Fig. 2e). Orlistat had less marked effects on the survival of M0 cells (Fig. 2e). Within the short timeframe that M1 cells remained alive, we could detect no significant effect of inhibiting lipolysis on their survival (data not shown). Given the close link between FAO and M2 activation, we next tested the effects of inhibiting lipolysis on macrophages stimulated with IL-4. We found that Orlistat suppressed the IL-4 induced expression of CD206, CD301, PD-L2 and RELMα(Fig. 2f), but had no effect on iNOS expression in M1 macrophages (Fig. 2g). Together, these data indicate that lipolysis supports FAO and in this way is critical for M2 activation.
Figure. 2.
Lipolysis is essential for M2 activation and survival. (a) Abundance of the monoacylglycerols 1-oleoylglycerol, 1-palmitoylglycerol, 1stearoglycerol and 2-palmitoylglycerol in extracts of M2 compared to M1 macrophages were assessed by mass spectrometry (AU, arbitrary units). (b) Glycerol concentrations in 24 h culture supernatants of M0 macrophages, M2 macrophages, and M2 macrophages treated with the lipase inhibitor tetrahydrolipistatin (Orlistat, 100 μM). (c-g) The effect of inhibiting lipolysis with Orlistat on (c) basal OCR and SRC in M2 macrophages, (d) basal OCR, and on the basal OCR/ ECAR ratio in M1 macrophages, (e) M2 macrophage survival, (f) CD206, CD301, PD-L2 and RELMα expression by M2 macrophages, as measured by flow cytometry, (g) iNOS expression in M1 macrophages. In these experiments Orlistat was included in the culture from the time at which M2 or M1 polarizing conditions were initiated and data were collected 24 h (a-d, f, g) or 48 h (e) later. Data for M0 macrophages are shown in c,d,f, and g for reference. Data in a show means ± SEM from 4 independent experiments. Data in b-e are means ± SEM of 3 technical replicates from one experiment representative of 3. In f, numbers represent % of macrophages falling within indicated gates; plots shown are from one representative experiment of 2, but numbers are mean ± SEM of data from both experiments. In g histograms show plots from one experiment representative of 2. The mean % of cells that were iNOS positive in these two experiments were 68.2 ± 5.0% (SEM) for M1 cells and 70.2% ± 6.7% for M1 plus Orlistat. data are P values are from Student's t-test (*P < 0.05; **P < 0.01; ***P < 0.0001).
Lysosomal acid lipase is responsible for lipolysis upstream of FAO
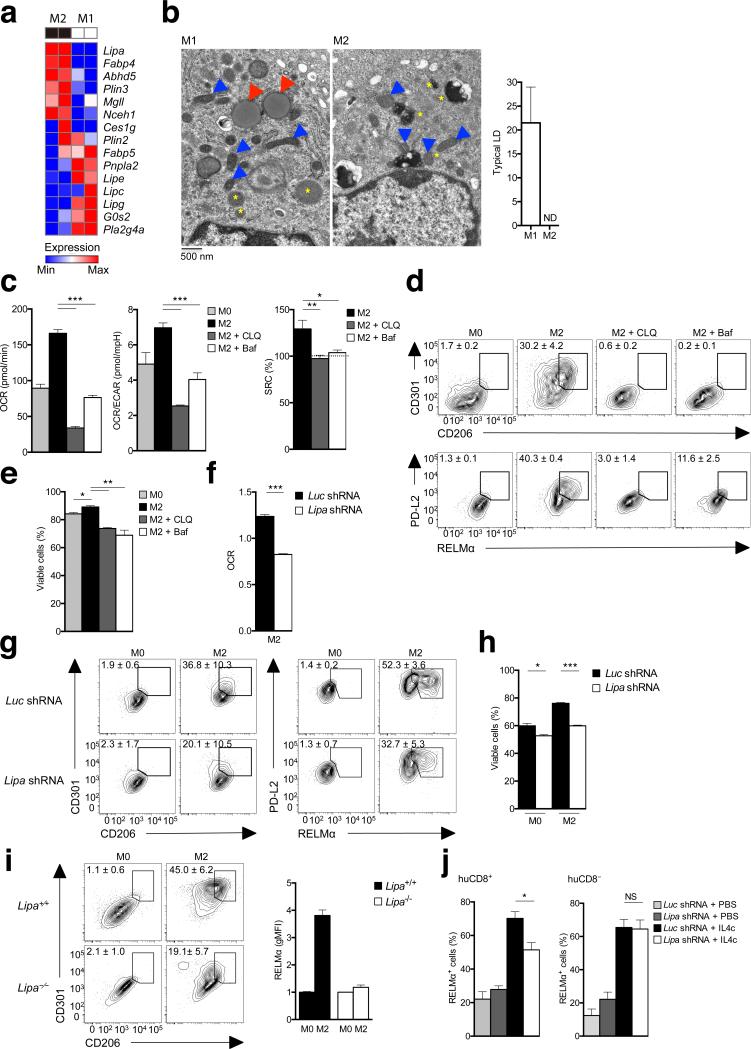
Our data implicated lipolysis as a critical mechanism for the generation of fatty acids for FAO and M2 activation in macrophages stimulated with IL-4. In other cells fatty acids for FAO are released from triacylglycerols stored with cholesterol esters in LDs by a process of lipolysis initiated by ATGL and continued by hormone-sensitive lipase (HSL), encoded by Pnpla2 and Lipe, respectively11,12,22. However, amongst genes encoding lipases, we found that Pnpla2 and Lipe were expressed more strongly in M1 cells than in M2 cells (Fig. 3a), suggesting that these genes might not play a role in M2 activation. To directly test this, we measured RELMα expression in peritoneal macrophages (PM) from Pnpla2–/– mice following the i.p. injection of IL-4–anti-IL-4 complexes (IL-4c), a procedure shown previously to induce peritoneal macrophage M2 activation23. We found that >90% of macrophages in Pnpla2–/– mice or in wild-type mice expressed RELMα following IL-4c injection, indicating that the loss of ATGL has no effect on M2 activation (Supplementary Fig. 3a). A substantial body of literature has associated M1 activation with the development of lipid droplets24, but whether or not lipid droplets are prevalent in M2 macrophages is unclear. Using transmission electron microscopy (TEM) we found that M1 macrophages contained easily identifiable lipid droplets, whereas M2 cells did not (Fig. 3b). Together with the lipase expression data and data from experiments using Pnpla2–/– macrophages, these findings suggest that M2 macrophages are not utilizing canonical lipid droplet-triacylglycerol hydrolysis pathways to generate fatty acids for FAO. Nevertheless, we did observe that neutral lipids as measured by flow cytometric detection of BODIPY stainin, the appearance of lipid droplets as visualized by electron microscopy, and the quantitation of triacylglycerols and cholesterol esters by mass spectrometry, accumulated in macrophages stimulated with IL-4 in the presence of Orlistat (Supplementary Fig. 3b-d). Mass spectrometry additionally confirmed that neutral lipid stores are greater in M1 macrophages than in M2 macrophages (Supplementary Fig. 3d). These findings indicate that M2 macrophages accumulate neutral lipid stores when lipolysis is inhibited.
Figure 3.
Lipolysis for M2 activation is mediated by lysosomal acid lipase. (a) RNA-Seq analysis of the relative expression in M2 and M1 macrophages of genes involved in lipolysis. (b) Transmission electron micrographs of bone marrow derived M1 and M2 macrophages. Blue arrowheads indicate mitochondria, yellow asterixes indicate lysosomes, and red arrowheads indicate lipid droplets. Graph shows numbers of lipid droplets (LD) per cell in M1 and M2 cells (150 cells per condition were examined, and data represent means ± SEM). (c) The effect of neutralizing lysosomal pH with chloroquine (CLQ) or Bafilomycin (Baf) on basal OCR, the ratio of OCR/ECAR, and SRC in M2 macrophages. Basal OCR and OCR/ECAR ratios for M0 macrophages are shown as a reference. (d) The effect of CLQ and Baf on CD206, CD301, PD-L2 and RELMα expression by M2 macrophages, as measured by flow cytometry. Expression for M0 macrophages is shown for reference. (e) Effects on M2 cell viability of culturing for 24 h in CLQ or Baf. (f) Basal OCR, relative to M0 macrophages, of M2 macrophages transduced with either control virus (Luc shRNA) or virus expressing shRNA against Lipa (Lipa shRNA). (g,h) Expression of CD206 and CD301 and of PD-L2 and of RELMα (g) and cellular viability (h) of macrophages transduced with Luc shRNA or Lipa shRNA prior to culture in M0 or M2 conditions. For (h), cells were cultured for 2 days before assessment. (i) Expression of CD206, CD301 and RELMα by wild-type (Lipa+/+) or Lipa-/-M0 or M2 macrophages. (j) %RELMα positive Lipa shRNA transduced vs Luc shRNA transduced huCD8+ peritoneal macrophages from bone marrow chimeric mice injected with PBS or IL-4c (left). Data from untransduced huCD8- peritoneal macrophages from the same mice are shown for comparison (right). Data in a are from 2 separate experiments. Images in b are from individual cells, representative of over 150 examined from 2 independent experiments. Data in the graph are mean ± SEM values of numbers of typical lipid droplets (LD) per cell, from 2 independent experiments Data in c, e and f are means ± SEM of 3 technical replicates from one experiment representative of 3. In d, g, and i, numbers represent % of macrophages falling within indicated gates; plots shown are from one representative experiment of 2, but numbers are mean ± SEM of data from both experiments. Data in bar graphs in h and i are of mean ± SEM values from 2-3 independent experiments. Data in j are from mean ± SEM values from a minimum of 2 independent mice. P values are from Student’s t-test (*P < 0.05; **P < 0.01; ***P < 0.0001).
We noted that, unlike genes encoding many other cellular lipases (including Pnpla2, Lipe, Lipc, Lipg), expression of Lipa was upregulated in M2 compared to M1 macrophages (Fig. 3a, Supplementary Fig. 1c), suggesting that it could be important for M2 activation. Lipa encodes LAL, a triacylglycerol lipase that is able to breakdown triacylglycerols delivered into the lysosomes by endocytosis of low density lipoproteins (LDLs and VLDLs)25. Since LAL is a known target of Orlistat26, we reasoned that the observed effects of Orlistat on macrophage FAO and M2 activation (Fig. 2) could reflect the inhibition of LAL function in this system. To investigate the role of LAL, which unlike the other cellular lipases has optimum activity at acidic rather than neutral pH, in M2 activation, we first neutralized lysosomal pH using chloroquine or Bafilomycin A1. We found that these conditions completely inhibited the IL-4–induced increases in OXPHOS, as measured by OCR, OCR/ECAR ratio and SRC (Fig. 3c). These effects were accompanied by the inhibition of IL-4–induced increases in CD206, CD301, PD-L2 and RELMα expression (Fig. 3d) and a decrease in longevity (Fig. 3e). These data support the view that an acidic compartment is essential for M2 activation and are consistent with a role for LAL in this process. To formally address this possibility, we targeted LAL using shRNAs, and found that increased OXPHOS, expression of CD206, CD301, RELMα and PD-L2, and increased longevity in response to IL-4 were significantly inhibited by the suppression of LAL expression (Fig. 3f-h). Suppression of LAL expression also resulted in the accumulation of lipid droplets in macrophages stimulated with IL-4 (Supplementary Fig. 3b). Additionally, we examined the ability of Lipa–/– macrophages to become M2 activated and found that increases in expression of CD206, CD301 and RELMα were significantly muted in Lipa–/– macrophages versus wild-type cells (Fig. 3i). Finally, we made bone marrow chimeras in which irradiated CD45.2 mice were reconstituted with CD45.1 bone marrow that had been bone transduced with retrovirus encoding shRNAs targeting Lipa or a control gene (luciferase). Progeny from transduced cells could be distinguished via expression of human CD8 encoded as a reporter by the retrovirus. We examined M2 activation of peritoneal macrophages derived from CD45.1+ transduced versus untransduced bone marrow cells following i.p. injection of IL-4c. We found that peritoneal macrophage RELMα expression in response to IL-4 was inhibited by the suppression of LAL expression (Fig. 3j). Together, these data indicate that cell-intrinsic lipolysis mediated by LAL plays a critical role in optimal macrophage M2 activation in response to IL-4.
Elevated OXPHOS and optimal M2 activation are dependent on CD36
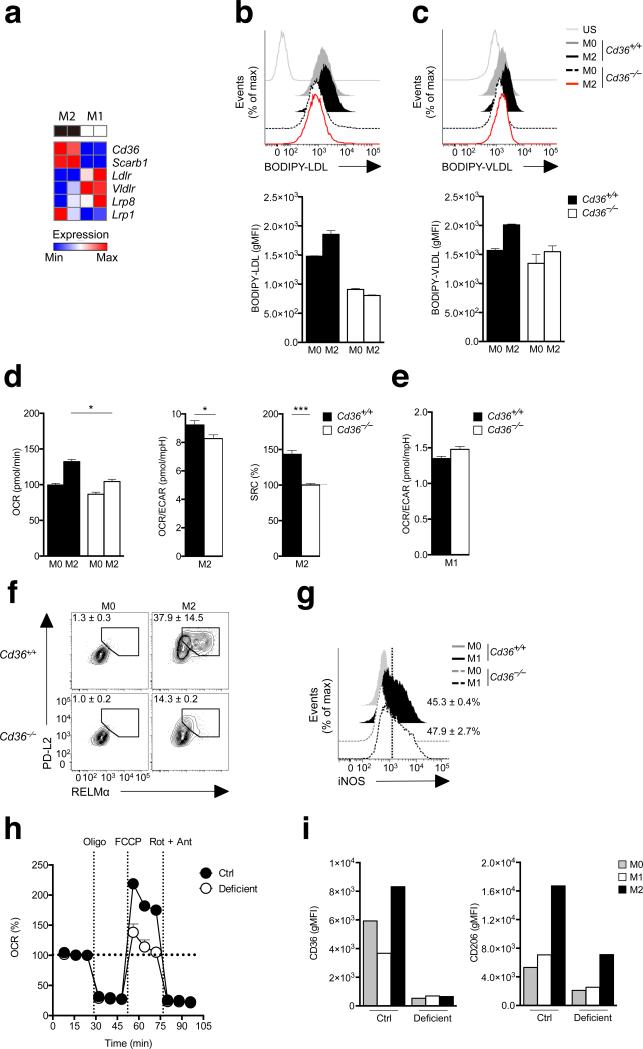
The important role for LAL in M2 macrophages suggested that triacylglycerol delivery to lysosomes is a key process in the generation of fatty acids to support FAO and M2 activation. We examined the expression of receptors that mediate the uptake of triacylglycerol-carrying serum lipoproteins in M2 macrophages and observed that CD36 was upregulated in M2 cells (Fig. 4a)15. The same was true for the related scavenger receptor Scarb1, but not for Ldlr1, Vldlr, or Lrp8 (Fig. 4a, Supplementary Fig. 1c). Based on these observations, and the fact that CD36 has been previously implicated in M2 activation16, we hypothesized that CD36 may play a role in increased FAO in M2 macrophages by facilitating the uptake of triacylglycerols. To examine this, we assessed responses to IL-4 of macrophages made from the bone marrow of Cd36–/– mice. We found that compared to M0 macrophages, wild-type M2 macrophages, but not Cd36–/– M2 macrophages, took up more LDL (Fig. 4b) and VLDL (Fig. 4c). In addition to being a receptor for LDL and VLDL, CD36 is a fatty acid translocase13,14. While we found M2 macrophages to be more capable than M0 cells of taking up palmitate, we observed no difference between wild-type and Cd36–/– cells in this regard (Supplementary Fig. 4a), suggesting that the fatty acid translocase activity of CD36 is not accounting for increased free fatty acid uptake by M2 cells. Compared to wild-type macrophages, Cd36–/– macrophages exhibited significantly lower IL-4 induced increases in OXPHOS as measured by OCR, the OCR/ECAR ratio and SRC (Fig. 4d), but were substantially similar in terms of the metabolic markers of M1 activation (Fig. 4e). Consistent with the critical role for FAO in M2 activation, Cd36–/– macrophages exhibited substantially reduced levels of expression of PD-L2 and RELMαin response to IL-4 (Fig. 4f). In comparison, iNOS expression was similar in wild-type and Cd36–/– macrophages stimulated in M1 conditions (Fig. 4g). We further addressed the role of CD36 by examining the effect on wild-type macrophage responses to IL-4 of inhibiting CD36 with sulfo-N-succinimidyl oleate (SSO)27. The findings from these experiments confirmed those from using Cd36–/– macrophages. We found that LDL uptake was substantially inhibited by SSO (Supplementary Fig. 4b). Moreover, SSO completely blocked IL-4-induced increases in OXPHOS as measured by OCR, the OCR/ECAR ratio and SRC (Supplementary Fig. 4c) but had no effect on metabolic markers of M1 activation (Supplementary Fig. 4d). SSO substantially inhibited expression of CD206, CD301, PD-L2 and RELMαin response to IL-4 (Supplementary Fig. 4e), but had no effect on iNOS expression in M1 macrophages (Supplementary Fig. 4f). Together, these data support the view that elevated CD36 expression in macrophages in response to IL-4 plays an important role in increased uptake of LDL and VLDL, and that this pathway supports FAO and M2 activation.
Figure 4.
CD36 is critical for M2 activation. (a) RNA-Seq analysis of the relative expression in M2 vs. M1 macrophages of genes involved in the uptake of triacylglycerol-carrying proteins. (b, c) The uptake of BODIPY labeled LDL (a) and VLDL (b) by M0 and M2 WT (Cd36+/+) and Cd36-/- macrophages, as measured by flow cytometry. Top panels show histograms from one experiment, bottom panels show gMFI from duplicate experiments. Fluoresence staining was compared to unstained cells (US). (d) basal OCR, the ratio of basal OCR to basal ECAR, and SRC in M2 WT and Cd36-/- M2 macrophages, (e) basal OCR and the ratio of basal OCR to basal ECAR in WT and Cd36-/- M1 macrophages, (f) PD-L2 and RELMα expression by WT and Cd36-/- M2 macrophages, (g) iNOS expression by WT and Cd36-/- M1 macrophages. Data in b-g from M0 macrophages are shown for comparison. (h) OCR readings, in real time, for human control and CD36-deficient M0 and M2 macrophages (derived from monocytes that were cultured in M-CSF for 8 days prior to treatment with cytokine), at basal and following sequential treatments with oligomycin, FCCP, and rotenone/antimycin A. Data have been baselined relative to each other. (i) Expression of CD36 and CD206 in control and CD36 deficient human M0, M1 and M2 monocyte-derived macrophages. Data in a are from 2 separate experiments. In b-c data in histograms are from one experiment representative of 2, and data in bar graphs represent mean ± SEM values from 2 independent experiments. In d and e data are means ± SEM of 3 technical replicates from one experiment representative of 2 independent experiments. Numbers in f and g represent % of macrophages falling within the indicated gates. In f, plots are from 1 experiment representative of 2, but numbers represent mean ± SEM values from 2 independent experiments. In g graphs are from one experiment representative of 2, but numbers represent mean ± SEM values from 2 independent experiments. In h data are mean + SEM values from a minimum of 4 technical replicates from cells from one subject. P values are from Student's t-test (*P < 0.01; **P < 0.0001).
Next we tested the ability of monocyte-derived macrophages from a CD36-deficient patient to become alternatively activated in response to IL-4. We found that SRC was substantially diminished in human CD36-deficient M2 macrophages compared to CD36-sufficient human M2 macrophages (Fig. 4h). Expression of CD206, the mannose receptor C type 1 (MRC1), is a marker of human M2 activation, as is expression of CD3628. Thus we used flow cytometry to examine CD36 and CD206 expression in M0, M1 and M2 macrophages from CD36-sufficient and -deficient subjects. We found that CD36 and CD206 expression were elevated in M2 versus M0 and M1 macrophages, and that expression of CD206 was substantially lower in CD36-deficient macrophages stimulated with IL-4 compared to CD36-sufficient macrophages (Fig. 4i). Together with our findings from mouse Cd36–/– macrophages and the SSO inhibitor studies, these data from human CD36-deficient macrophages strongly support a significant role for CD36 in M2 activation.
Our data suggested that the lysosomal lipolysis of exogenous triacylglycerols acquired through a CD36-dependent mechanism is important for optimal M2 activation. In this case, we reasoned that depriving macrophages of exogenous triacylglycerols should impact M2 activation. We cultured macrophages in complete medium containing or lacking serum for 24 h and then measured the expression of key M2 marker genes following an additional 24 h of stimulation with IL-4. In these experiments, M2 activation was substantially inhibited, but not prevented, by the absence of serum (Supplementary Fig. 5a). Consistent with our data implicating CD36-mediated uptake of LDL and VLDL in M2 macrophages, we found that under serum-free conditions, M2 activation could be promoted by the addition of LDL and VLDL (Supplementary Fig. 5a). The finding that M2 activation could proceed to some extent in the absence of serum, suggested that macrophages have an alternative pathway to fuel FAO. A candidate for such an alternative pathway is fatty acid synthesis. To explore this possibility, we examined how M2 activation was affected by (5-(Tetradecyloxy)-2-furoic Acid (TOFA), an inhibitor of acetyl-CoA carboxylase, which catalyzes an early step in the synthesis of fatty acid from acetyl-CoA. TOFA completely blocked M2 activation in serum-free conditions and, in the presence of serum, reduced activation to levels observed in inhibitor-free, serum-free conditions (Supplementary Fig. 5b). These data suggest that macrophages possess an underlying pathway through which, in the presence of IL-4, they synthesize fatty acids to support M2 activation, and that optimal M2 activation requires contributions of exogenous triacylglycerols and de novo synthesized fatty acids.
These findings raised the question of whether M2 macrophages first incorporate endogenously synthesized fatty acids into triacylglycerols prior to liberating them by lipolysis for FAO, as has been suggested to be the dominant pathway in other cell types11. We examined this by first testing the effect of Orlistat on M2 activation in serum-free conditions. We found that the inhibition of lipolysis under these conditions did inhibit M2 activation (Supplementary Fig. 5b), indicating that endogenously synthesized fatty acids are incorporated into triacylglycerols in M2 macrophages. Triacylglycerols synthesized in this way are stored as lipid droplets, but our ultrastructural analysis failed to identify lipid droplets in M2 cells (Fig. 3b). However, we did observe lipid droplets in macrophages stimulated with IL-4 in the presence of Orlistat and in macrophages transduced with shRNA targeting Lipa (Supplementary Fig. 3b). Since the inhibition of lipolysis precludes the synthesis of these neutral lipid stores from fatty acids liberated from acquired triacylglycerols, we assume that they are produced from de novo synthesized fatty acids. Taken together, our data raise the possibility that in addition to a primary pathway in which exogenous triacylglycerols are broken down in the lysosomal compartment to support FAO, M2 macrophages are also able to synthesize triacylglycerols from de novo synthesized fatty acids and hydrolyze these via the same pathway.
M2 activation during helminth infection is LAL-dependent
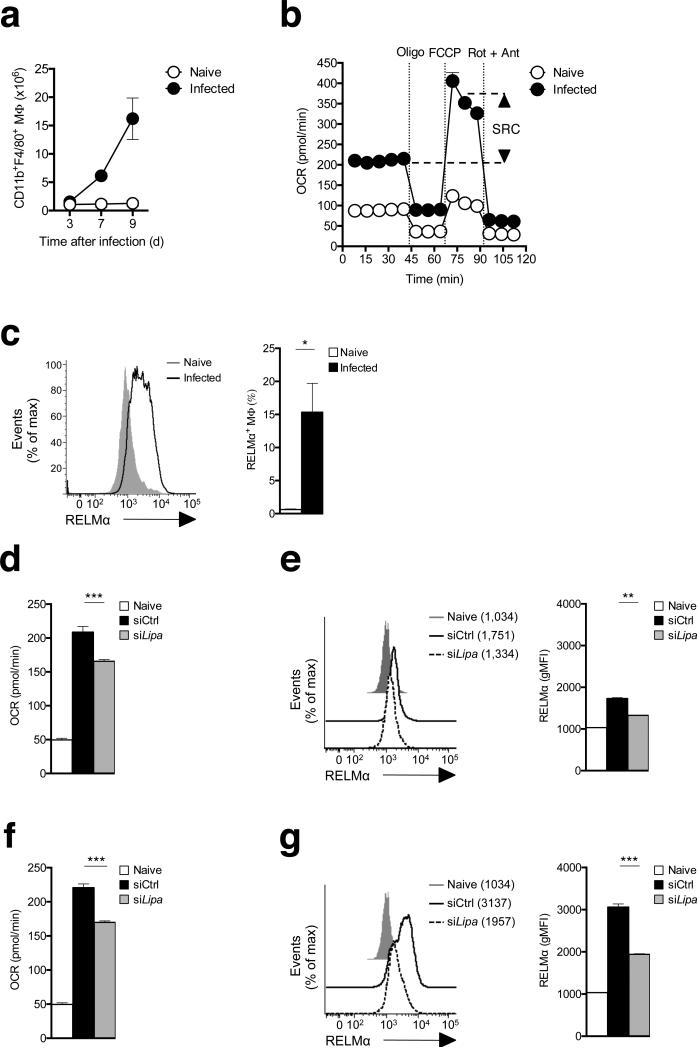
Our data indicated that lipolysis is essential for the metabolic switch and associated changes in the expression of key genes that occur as macrophages become alternatively activated in response to IL-4. To assess whether lipolysis is critical for M2 activation in vivo, we used the Heligmosomoides polygyrus bakeri (H. polygyrus) mouse model of human intestinal helminth infection, in which M2 macrophages play an essential role in immunity29. We found that peritoneal macrophage numbers increased significantly over time during this infection (Fig. 5a), and that peritoneal macrophages from infected mice exhibited the metabolic profile of IL-4-activated bone marrow macrophages, with heightened baseline OCR and marked SRC (Fig. 5b). Consistent with previous observations, a substantial percentage of peritoneal macrophages from H. polygyrus-infected mice expressed the M2 marker RELMα (Fig. 5c)30. We found that maintenance of the peritoneal M2 macrophage activation phenotype is LAL-dependent in this setting since increases in OCR and RELMα expression were significantly inhibited when peritoneal macrophages were treated ex-vivo with siRNAs targeting Lipa (Fig. 5d,e). The dependence of M2 activation on LAL was also apparent when peritoneal cells were maintained in a type 2 environment through the addition of IL-4 in vitro (Fig. 5f,g). These data indicate that macrophages in a type 2 cytokine environment in vivo exhibit lipolysis-dependent changes in metabolism and M2 marker gene expression that are equivalent to those induced by IL-4 in bone marrow derived macrophages in vitro.
Figure 5.
The peritoneal cavity is a site of M2 macrophage activation during infection with H. polygrus, and M2 macrophage activation is LAL-dependent. (a) Total numbers of CD11b+F4/80+ peritoneal macrophages in naïve mice and in infected mice at various days post-infection with H. polygyrus. (b) OCR readings, in real time, for macrophages from uninfected (Naïve) or infected mice (day 9) at basal and following sequential treatments with oligomycin, FCCP, and rotenone/antimycin A. SRC is depicted in the plot. (c) RELMα expression in peritoneal macrophages from naïve mice and from mice infected with H. polygyrus for 9 days. The graph to the right shows the % of all peritoneal cells that are RELMα+ macrophages. (d,f) Baseline OCR of ex vivo peritoneal macrophages from naïve mice, or from infected mice following treatment with control (siCtrl) or Lipa (siLipa) siRNAs and culture for 12 h in the absence (d) or presence (f) of added IL-4. (e,g) RELMα expression by the macrophage populations described in d and f. In a data are means ± SEM for cells taken from 2 - 4 individual mice from one experiment representative of 3. In b, data are mean + SEM values for 5 technical replicates for cells from 2 – 4 mice per group from 1 experiment representative of 3. In c the histogram shows data from cells pooled from 2 individual mice and the bar graph shows mean ± SEM values from 2 – 3 individual mice per group from 1 experiment representative of 3. In d, and f, bar graphs show mean ± SEM values for 3 technical replicates for cells pooled from 2 mice per group from 1 experiment representative of 2. In e data in histograms are from 1 mouse, representative of 2 mice per group, representative of 2 experiments. In g, data are mean ± SEM values for data from 2 mice per group from one experiment representative of 2. P values are from Student's t-test (*P < 0.05; **P < 0.0001).
Inhibition of lipolysis suppresses parasite elimination
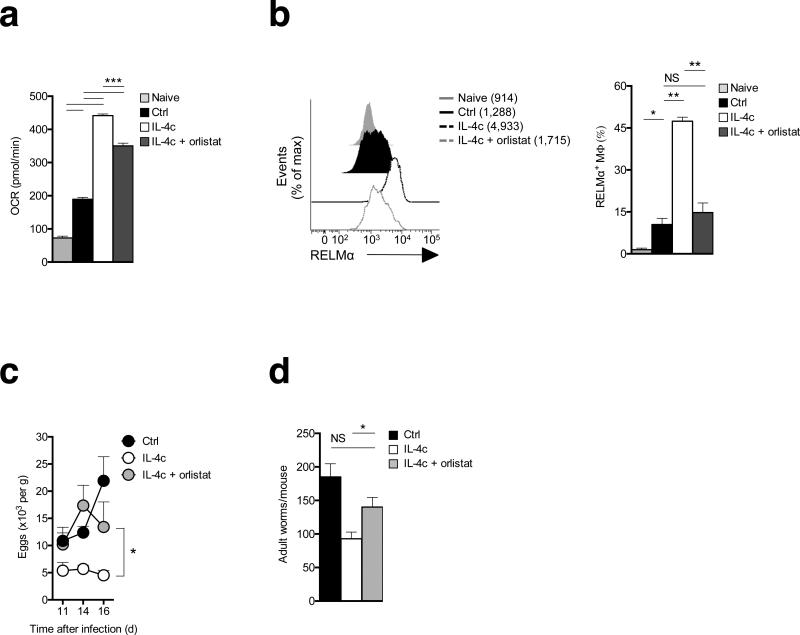
Primary infection with H. polygyrus in B6 mice is chronic despite the development of a type 2 cytokine response. However, injection of IL-4c into infected mice results in reductions in adult parasite numbers, and M2 macrophages are implicated in the protective mechanism induced by this cytokine3,29,31. Given our findings, we reasoned that lipolysis should be crucial for the development of M2 macrophages in these settings, and therefore that inhibition of this pathway would limit the expression of M2 macrophage-dependent protective immune responses. We found that IL-4c injected after primary infection markedly enhanced peritoneal macrophage OXPHOS (Fig. 6a) and increased RELMα expression (Fig. 6b), while simultaneously significantly reducing parasite burden, as measured by egg and adult worm counts (Fig. 6c,d). Consistent with a major role for lipolysis in M2 polarization, we found that enhanced M2 activation and concomitant protective effects associated with IL-4c injection were significantly inhibited by parenteral Orlistat treatment (Fig. 6a-e).
Figure 6.
Inhibition of lipolysis suppresses IL-4 driven M2 macrophage activation in vivo and the elimination of a primary H. polygyrus infection. Mice were infected and at day 9 groups of infected mice were injected with PBS (Ctrl), IL-4c, or IL-4c plus Orlistat. IL-4c injections were repeated on day 14 post infection. Orlistat treatment was repeated on days 11, 14 and 16 post infection. The experiment was terminated at day 17 after infection. (a) Basal OCR readings of peritoneal macrophages isolated from naïve mice, and from infected mice treated as shown. (b) RELMα expression by peritoneal macrophages from the various treatment groups. Left panel shows mean fluorescence intensity (MFI) and right panel shows % of macrophages staining positive for RELMα. (c) Fecal egg counts over time in mice treated as indicated. (d) Adult worm counts at day 17 in mice treated as indicated. In a, data are means ± SEM from 5 technical replicates of cells from 3 – 6 mice per group from one experiment representative of 2. In b (bar graphs) c and d, data are means ± SEM from 3 - 6 mice per group from one experiment representative of 2. In b, the histograms are from cells from one mouse per group, representative of 3 – 6 mice per group from one experiment representative of 2. P values are from Student's t-test, NS = not significant. Data are from individual experiments representative of two independent experiments. (*P < 0.05; **P < 0.0001)
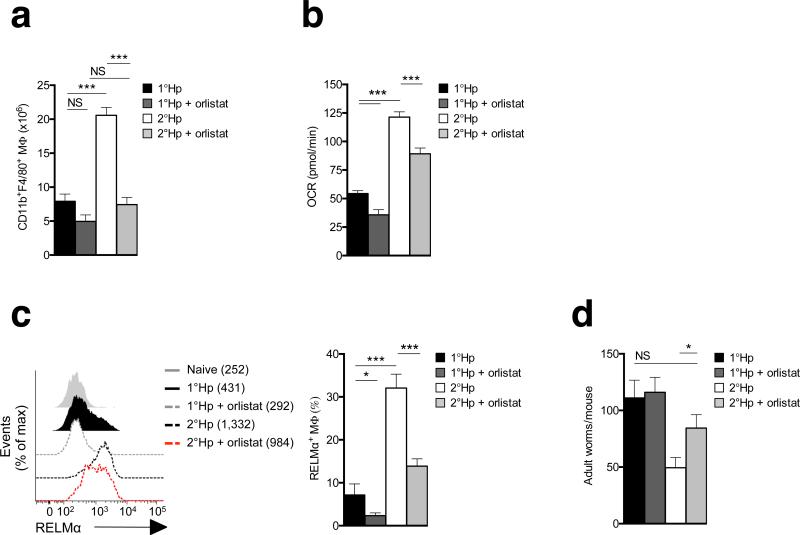
Mice in which primary H. polygyrus is cleared by chemotherapy exhibit a degree of immunity to secondary infection29. We compared the peritoneal macrophage response during primary H. polygyrus infection to that of a secondary re-infection following parasite clearance by administration of pyrantel pamoate and resting the animals for five weeks. We found that peritoneal macrophage numbers (Fig. 7a) and the magnitude of the increase in OXPHOS and RELMαexpression (Fig. 7b,c) were all significantly greater in mice responding to secondary infection. This correlated with resistance to secondary infection, apparent as a >50% drop in adult worm burdens arising from secondary vs. primary infections (Fig. 7d). We then asked whether inhibition of lipolysis following secondary infection affected enhanced M2 activation, or resistance to secondary infection. We found that Orlistat treatment prevented the increase in peritoneal macrophage numbers that occurred during secondary infection (Fig. 7a), inhibited all measured parameters of enhanced M2 activation (Fig. 7b,c) and suppressed resistance to reinfection in immune mice (Fig. 7d).
Figure. 7. Inhibition of lipolysis suppresses resistance to reinfection with H.polygyrus.
Mice were infected with H. polygyrus and treated with the anti-helminthic pyrantel pamoate prior to receiving a secondary infection (2°Hp). At the time of secondary infection, additional groups of mice were infected for the first time to provide primary H. polygyrus infection controls (1°Hp). Infected mice were treated (or not) with Orlistat on the day of infection and on days 2, 4, 6 and 8 post infection. The experiment was terminated on day 9 post 1° or 2° infection. (a) Numbers of CD11b+F4/80+ peritoneal macrophages in mice treated as indicated.(b) Basal OCR readings of peritoneal macrophages isolated from mice treated as indicated. (c) RELMα expression by peritoneal macrophages from the various treatment groups. Left panel shows mean fluorescence intensity (MFI) and right panel shows % of macrophages staining positive for RELMα. (d) Adult worm counts in mice treated as indicated. In a, c,(bar graph) and d, data are means ± SEM from 4 – 10 independently assessed mice per group from 1 experiment representative of 2. In b, data represent mean + SEM values for 5 technical replicates from cells from 4 – 8 or more mice per group from one experiment representative of 2. In c, data in the histogram are from 1 mouse representative of 3 – 8 mice per group from 1 experiment representative of 2. The numbers in the key are MFI values. In d data are from 5 – 8 individually assessed mice per experimental group. Data are from individual experiments representative of two independent experiments. P values are from Student's t-test, NS = not significant (*P < 0.05; **P < 0.0001).
Given the experimental approach used here, there was a possibility that Orlistat was affecting not only macrophage function, but also adaptive immunity. To address this, we used 4get/KN2 IL-4 reporter mice to enumerate CD4+ type 2 helper T cells (TH2) cells in mesenteric lymph nodes following primary or secondary infections with or without Orlistat treatment. We found that inhibition of lipolysis during the development of the effector or secondary effector response had no measurable effect on the adaptive response, as measured by the overall cellularity of the MLN (Supplementary Fig. 6a), or the numbers of IL-4–secreting TH2 cells in the MLN (Supplementary Fig. 6b). Furthermore, examining IL-4 reporter expression in all live cells from mice treated with Orlistat failed to reveal any effects on non-TH2 sources of IL-4 (data not shown). In summary, our data show that the inhibition of lipolysis is able to block macrophage M2 activation associated with resistance to the helminth parasite H. polygyrus, and in so doing suppress immunity to this pathogen
Discussion
We have found that accentuated FAO and related SRC in macrophages stimulated with IL-4 are dependent on a pathway of cell-intrinsic lipolysis mediated by the lysosomal enzyme LAL. Consistent with the necessity to generate fatty acids to fuel FAO and the importance of FAO for M2 activation, we found that LAL is expressed to a greater extent in M2 than in M1 macrophages, and is critical for full macrophage M2 activation in response to IL-4.
Mutations in LAL cause Wolman's disease and cholesterol ester storage disease (CESD), which are characterized by varying degrees of accumulation of triacylglycerols and cholesterol esters in the liver, intestine and adrenal glands, but also in blood vessels and spleen. Notably, triacylglycerol accumulation in macrophages is associated with deficiencies in LAL activity, and atherosclerosis, a disease ameliorated by M2 activation32,33 develops prematurely in CESD (discussed in 25). Moreover, genome-wide association studies (GWAS) have revealed that polymorphisms in Lipa are associated with premature human coronary artery disease at least 3 distinct genetic backgrounds34, 35. Thus a genetic link exists between LAL and a disease in which M2 macrophages are considered to play an important protective role. The clinical evidence provides support for the possibility that LAL plays an important role in M2 macrophage biology. Lipa–/– mice provide a model for human LAL deficiencies, and have provided insight into the role played by LAL in the immune system. In these mice, which die prematurely, lymphopoiesis is reduced and accompanied by a myeloproliferative disorder and multi-organ sterile inflammation36, which can be reversed by the macrophage-specific expression of human LAL37, supporting the view that macrophage LAL plays a critical role in the homeostatic control of inflammation. Those data are consistent with our finding that LAL is essential for M2 activation and the known ability of M2 activation to ameliorate inflammation in many settings. Interestingly, bone marrow transplantation has been used to treat Wolman's disease38, supporting the view that lack of LAL expression in cells of hematopoietic origin is critical for the pathogenesis of this disease.
It has been suggested previously that compared to M1 macrophages, which exert their functions over short time periods, M2 macrophages are engaged in activities which are more prolonged, and that the relative efficiency of FAO versus glycolysis is well suited to meet the metabolic requirements of these roles7. Consistent with this view we found that M2 macrophages survive longer than M1 macrophages in culture and possess marked SRC, a characteristic which in CD8+ T lymphocytes is linked to the enhanced FAO and cellular longevity of memory cells10. In parallel studies LAL was also shown to be responsible for the generation of fatty acids for FAO in memory CD8+ T cells, a process that is driven by IL-1539, suggesting that LAL has broadly assumed the role of coordinating cytokine (IL-4 or IL-15)-induced increases in FAO to support longevity in immune cells. Interestingly, the expansion of M2 peritoneal macrophages that occurs in immune mice following challenge infection with H. polygyrus did not occur when lipolysis was inhibited. This finding may reflect reduced survival of these cells due to reduced FAO, as was apparent in tissue-cultured Orlistat-treated M2 macrophages. An alternative explanation is that this failure is the result of reduced proliferation within the M2 macrophage population. Recent reports have indicated that helminth infection-induced M2 macrophages proliferate in situ, and that population expansion in these cases is driven by proliferation rather than monocyte recruitment and differentiation23,30. Thus, in addition to blocking the expression of genes associated with M2 activation, inhibition of lipolysis may also effectively block IL-4-induced macrophage proliferation, especially since fatty acids derived from the lipolysis of triacylglycerols can serve as building blocks for the synthesis of membrane lipids, which is critical in cells that are undergoing division. Lipolysis of triacylglycerols may also generate fatty acids that can serve as ligands for nuclear receptors such a PPARs, which regulate the expression of FAO genes and are critical for M2 activation40. It is feasible that inhibition of lipolysis influences macrophage fate following exposure to IL-4 by blocking these processes.
A recent report showed that IL-4 induces increased lipolysis in adipocytes by promoting HSL activity41. Nevertheless, we found that M2 activation associated with increased OXPHOS occurred normally in both ATGL-deficient and HSL-deficient (data not shown) macrophages, indicating that these cells use alternative pathways of lipolysis to liberate fatty acids from triacylglycerols. Our data show that this alternative pathway is mediated by the lysosomal lipase LAL, the expression of which we found to be increased in M2 macrophages, and which has also been reported to be expressed more in human M2 than M1 macrophages28. Interestingly, genes encoding lysosomal proteins are enriched by chromatin immunoprecipitation for STAT6 binding42, indicating that IL-4 generally induces lysosomal function. Our data provide strong support for the lysosomal lipolysis of triacylglycerols as being critical for optimal M2 activation, and implicate the uptake of triacylglycerol-carrying proteins such as VLDL and LDL through CD36 as playing a role in this process. Interestingly, we found that IL-4-induced increases in OXPHOS and expression of CD206, CD301, PD-L2 and RELMα were unaffected by deletion of Ldlr itself (data not shown). Recent reports have indicated that the uptake of oxidized LDL via CD36 and platelet-activating factor receptor43, and the expression of the LDLR-related protein 1 (Lrp1)44 are sufficient to promote IL-4-independent expression of certain genes linked to the M2 phenotype, although the mechanisms underlying these effects are unclear. Moreover, the neutral cholesterol esterase hydrolases Nceh1 and Ces1g have been implicated in the ability of macrophages to play an anti-inflammatory role in atherosclerosis45,46, and expression of both of these, as well as of Lrp1 was somewhat elevated after stimulation with IL-4.
Our findings suggest that in addition to fueling FAO through the lipolysis of exogenous triacylglycerols, M2 macrophages can utilize endogenous triacylglycerols, generated from de novo fatty acid synthesis, for the same purpose. We believe that these endogenously synthesized triacylglycerols contribute to lipid droplet formation in M2 cells in which lipolysis has been inhibited. On balance, the data indicate that the lipolysis of endogenously synthesized triacylglycerols is also mediated by LAL, since suppression of LAL expression in M2 cells also results in lipid droplet accumulation. It is unclear at present how LAL gains access to lipid droplets in M2 macrophages. In other nonadipocytes, lysosome-dependent autophagy has been reported to be important for the lipolysis of triacylglycerols in lipid droplets38,47. We have not found evidence to support this pathway in M2 macrophages cultured in complete medium (data not shown), but it is feasible that autophagy programs are engaged when access to fatty acids from exogenous triacylglycerols is restricted. Regardless, autophagy-independent lysosomal lipolysis of lipid droplets by macrophages is not unprecedented48, so it remains feasible that the lipolysis of endogenously synthesized triacylglycerols occurs through novel mechanisms. These possibilities are under investigation.
M2 macrophages play important roles in immune responses to helminth parasites, acting to directly affect parasite health and persistence in sites of infection, and/or to regulate T cell responses and associated immunopathology. Using two models of immune elimination of the intestinal helminth parasite H. polygyrus, we found that inhibition of lipolysis significantly diminished anti-parasite immunity promoted by the injection of IL-4 into mice carrying a primary infection, and inhibited resistance to secondary infection in mice that were challenged following drug-cure of a primary infection. In both cases reductions in immunity were tightly correlated with reductions in OXPHOS and M2 marker expression in peritoneal macrophages. We found no evidence for an effect of inhibiting lipolysis on TH2 cell responses, indicating that observed differences in immunity were unlikely to be linked to reductions in the production of type 2 effector cytokines by the adaptive response, although we have not formally ruled out an effect on other sources of IL-4 or IL-13. We infer from these findings that lipolysis plays a role in the M2 activation in vivo in the setting of helminth infections. There have been no reports of a role for LAL in immunity to parasitic helminths in mice or humans, but the latter may reflect the relative lack of GWAS to identify quantitative trait loci affecting human resistance to helminth parasites.
Collectively, our data indicate that macrophages utilize the lysosomal pathway to mobilize fatty acids to facilitate FAO and the M2 activation program. Our findings suggest that approaches for gain or loss of function of LAL may offer therapeutic potential in diseases where M2 macrophages play protective or detrimental effects.
Methods
Animals and in vivo experiments
C57BL/6J mice, 4get/KN2 mice, B6 Cd36–/– mice and B6.129P2-Pnpla2tm1Rze/J (Pnpla2–/–) mice were bred and maintained in specific pathogen free conditions under protocols approved by the institutional animal care committee at Washington University School of Medicine, and were used at 8-12 weeks of age. Lipa+/+ and Lipa–/– mice were maintained at Indiana State University under SPF conditions. In some experiments, mice were injected i.p. with tetrahydrolipistatin (Orlistat; 200 mg/kg, Cayman Chemical) or with 300 μl of 3% thioglycollate (Sigma) immediately prior to IL-4 complexed to mAb anti-IL-4 (IL-4c; containing 5 μg of IL-4, PeproTech, and 25 μg of anti-IL-4 clone 11B11, BioXcell)49. For infection with H. polygyrus bakeri, we followed protocols described in detail previously50. Mice were infected orally by gavage with 200 infective L3 stage larvae. For secondary infection experiments, adult H. polygyrus were eliminated from infected mice by oral administration of pyrantel pamoate (1 mg/mouse; Columbia Laboratory) at day 14 post-primary infection, and > 5 weeks later the mice were challenge-infected with 200 L3 stage larvae by gavage. At day 9 post challenge infection, mice were sacrificed and parasite burdens were measured. To count worm, intestines were removed, opened longitudinally, and placed into a metal strainer on top of a 50 ml tube filled with phosphate buffered saline (PBS; Corning Inc.) for 4 h at 37 °C. Parasites dropped through the filter into the tube and were recovered for counting on a dissecting microscope. Parasite eggs were enumerated by floating eggs in feces collected from individual mice on saturated sodium chloride prior to collection, and counting under a microscope.
Isolation of cells from mice
Peritoneal exudate cells (PECs) were harvested from sacrificed naïve or infected mice by peritoneal lavage with 10 ml of sterile PBS containing 5% FBS (HyClone) per mouse. Total numbers of PECs and peritoneal macrophages were determined by cell counting in combination with multi-color flow cytometry. For analysis of lymphocyte activation by flow cytometry, single cell suspensions were prepared from lymphoid organs by forcing them through 70 μm strainers, and washing with RPMI containing 5% FBS (Corning Inc). Red blood cells were lysed with ACK lysis buffer (0.15 M NH4Cl, 1 mM KHCO3, 0.1 mM EDTA). Bone marrow was flushed from the femur and tibia using RPMI containing 5% FBS and dissociated into a single cell suspension by repeated pipetting. Cells were maintained on ice until use or analysis.
Preparation of macrophages from bone marrow and macrophage activation
Bone marrow cells were differentiated in the presence of recombinant mouse M-CSF (20 ng/ml; R&D Systems) in complete medium (RPMI 1640 containing 10 mM glucose, 2 mM L-glutamine, 100 U/mL of penicillin/streptomycin and 10% FBS) for 7 days. Day 7 macrophages were washed and variously stimulated with IL-4 (20 ng/ml; PeproTech) or lipopolysaccharide (LPS, 20 ng/ml; Sigma) plus IFN-γ (50 ng/ml; R&D Systems) in the presence or absence of 200 μM etomoxir (ETO; Sigma), 20 μM sulfo-n-succinimidyl oleate (SSO), 30 μM chloroquine (CLQ; Sigma), 0.1 μM bafilomycin A1 (Baf; Tocris Bioscience), 100 μM Orlistat (Cayman), 20 μM 5-(Tetradecyloxy)-2-furoic Acid (TOFA, Sigma), 5 or 50 μg/ml of LDL (Kalen Biomedical, LLC) and VLDL (Kalen Biomedical, LLC) for 24 h. Macrophages were then harvested and analyzed by flow cytometry for expression of markers of M1 or M2 activation. In some experiments the survival of macrophages was monitored following M2 or M1 activation. For these experiments, bone marrow-derived macrophages (0.5 × 106 cells per well of a 48-well plate) were cultured in complete medium with mouse M-CSF (20 ng/ml) and stimulated with IL-4 or LPS plus IFN-γ. Cell viability was monitored by flow cytometric analysis of 7-amino-actinomycin D (7-AAD; BioLegend) staining of F4/80+ cells over time. Culture medium and stimulation signals were replenished every 1 or 2 days.
Identification of human CD36-sufficient and -deficient subjects
Peripheral blood mononuclear cells (PBMC) were isolated from venous blood samples obtained from a CD36-deficient adult subject and an adult subject with normal CD36 expression using Ficoll-Paque (GE Health Care) and density gradient centrifugation. The CD36-deficient subject is homozygous for the G allele (allele frequency ~10%) of coding SNP rs3111938 (T/G), which introduces a stop codon and results in a truncated protein that is degraded. Homozygosity at the G allele results in complete CD36 deficiency51. The study was approved by the Washington University School of Medicine Institutional Review Board with written informed consent obtained from each subject.
Isolation of mononuclear cells from human peripheral blood and human macrophage activation
Monocytes were purified from PBMCs using human CD14 MicroBeads (MACS) and then differentiated into macrophages in complete RPMI medium containing 20 ng/ml recombinant human M-CSF (R&D Systems) for 8 days. Human macrophages were harvested on day 8, and stimulated with 20 ng/ml human IL-4 (R&D Systems) or 50 ng/ml human IFN-γ (R&D Systems) plus 20 ng/ml LPS for 24 h.
Retroviral transduction
Retroviral transduction of macrophages was accomplished using a protocol that we have used previously to transduce bone marrow derived dendritic cells52. Sequences for luciferase and for Lipa short hairpin RNAs (shRNAs) were obtained from Open Biosystems and cloned into the MSCV-LTRmir30-PI8 retroviral vector, encoding huCD8 as a reporter. Two independent sequences were used to target Lipa: TCAAGTCCAGCATTCACTG, and TGTGCTTCAGAGACCAGGT. Recombinant retroviruses were used for spin infection (800 x g, 2 h) of day 3 bone marrow macrophage cultures. After 7 days in culture with M-CSF, macrophages were harvested and transduced macrophages were gated based on huCD8 expression. Both shRNAs were used for experiments, with similar results, but only data from experiments that utilized TGTGCTTCAGAGACCAGGT are shown. For the bone marrow chimera, bone marrow cells from donor mice were collected and cultured in complete RPMI medium containing 10 ng/ml IL-3, 10 ng/ml IL-6 and 50 ng/ml murine c-kit ligand for 24 h. Cells were then transduced with retroviral shRNAs targeting Lipa or luciferase, and 5 × 105 transduced cells were intravenously injected into irradiated recipients. Recipients were used 6 – 10 weeks later.
Treatment of peritoneal macrophages with small interfering RNA (siRNA)
An siRNA targeting mouse LAL (Lipa; AAAGCUAUGAAACCAUGGTg) was purchased from Applied Biosystems, and Silencer Negative control siRNA#1, which is not matched to any sequence in the mouse genome, was also provided by the manufacturer and used as a control. siRNAs (50 nM) were complexed with Lipofectamine 2000 and delivered into peritoneal macrophages as per the manufacturer's instructions. siRNA-treated peritoneal macrophages were then stimulated with 20 ng/ml of IL-4 for 48 h prior to analysis.
Metabolism assays
For real-time analysis of extracellular acidification rates (ECAR) and oxygen consumption rates (OCR), macrophages were analyzed using an XF-96 Extracellular Flux Analyzer (EFA, Seahorse Bioscience) as described in detail for bone marrow-derived dendritic cells previously52. Three or more consecutive measurements were taken under basal conditions and following the sequential addition of 1 μM oligomycin to inhibit the mitochondrial ATP synthase, 1.5 μM fluoro-carbonyl cyanide phenylhydrazone (FCCP), a protonophore, which uncouples ATP synthesis from oxygen consumption by the electron transport chain, and 100 nM rotenone plus 1 μM antimycin A (Rot/Ant; all drugs for this assay were purchased from Sigma), which inhibit the electron transport chain. In this assay, basal oxygen consumption can be established by measuring OCR in the absence of drugs. Declines in OCR following the addition of oligomycin and rotenone and antimycin are expected and indicate that cells are consuming oxygen for mitochondrial OXPHOS. Spare respiratory capacity (SRC) is calculated as the difference between basal OCR and maximal OCR following the addition of FCCP. Maximal OCR occurs following the addition of FCCP since cells attempt to maintain a proton gradient across the inner mitochondrial membrane by increasing the consumption of oxygen by the electron transport chain10,53. Global metabolite profiling of stimulated macrophages was performed using mass spectrometry by Metabolon, Durham, NC. Glycerol in macrophage supernatants was measured using absorbance at 570 nm after incubation with Free Glycerol Reagent (Sigma) and by comparison to a glycerol standard (Sigma). For the lipidomic analysis, washed cell pellets were frozen and thawed and homogenized in 500 μl of PBS using an Omni Bead Ruptor 24 (Omni International, Inc.). 50 μl of each homogenate was reserved for protein measurement. 450 μl of each homogenate was used for lipidomic analysis. A modified Bligh-Dyer extraction method was used to extract lipids in the presence of an internal standard mixture. Mass spectrometric measurement was performed with a Shimadzu 10A HPLC system coupled to a TSQ Quantum Ultra triple quadrupole mass spectrometer operated in SRM mode under ESI(+). Different analytical HPLC columns and mobile phases were employed for different analytes to achieve the optimal sensitivity and separation. A pooled lipid extract from study samples was used as a quality control (QC), and was injected between every sample to verify the instrumental consistency, which is described as %CV of QC. Only the species with (%CV of QC) < 15% were reported. Data processing was conducted with Xcalibur software (Thermo). The concentration of an analyte was calculated as the concentration of its corresponding internal standard multiplied by the peak area ratio of the analyte to the internal standard, based on the assumption that the MS responses of the analyte and internal standard are the same.
Flow Cytometry
Cells were kept at 4 °C and blocked with 5 μg/ml of anti-CD16/32 (clone 93, eBiosciences) before surface staining with antibodies to CD11b (clone M1/70, eBiosciences), F4/80 (clone BM8, eBiosciences), CD206 (clone C068C2, Biolegend), CD301 (clone ER-MP23, AbD Serotec), PD-L2 (clone TY25, eBiosciences), CD62L (clone MEL-14, BD Biosciences), CD44 (clone IM7, BD Biosciences), CD4 (clone RM4-5, BD Biosciences), and/or human CD2 (clone RPA-2.10, BD Biosciences). For intracellular staining of RELMα and iNOS, cells were fixed in 4% ultrapure paraformaldehyde, and stained with rabbit anti-RELMα (PeproTech) and mouse anti-Nos2 (clone C-11; Santa Cruz Biotechnology) for 1 h at 22°C in 0.2% saponin buffer, followed by incubation with appropriate fluorochrome-conjugated anti-rabbit or anti-mouse IgG (both Jackson Immunoresearch) in the same buffer. BODIPY labeled LDL (10 μg/ml, Invitrogen), BODIPY labeled VLDL (10 μg/ml, Kalen Biomedical, LLC) and BODIPY labeled palmitate (1 μM, Invitrogen) were used in conjunction with flow cytometry for uptake experiments. Intracellular neutral lipids were stained with 500 ng/ml BODIPY 493/503 (Invitrogen) and measured by flow cytometry. All macrophage staining data shown represent cells that were gated as CD11b+F4/80+, and living based on staining with LIVE/DEAD (Invitrogen) or 7-amino-actinomycin D (eBiosciences). Human macrophages were stained with antibodies to human CD14 (clone MφPg, BD Biosciences), human CD36 (clone CB38, BD Biosciences) and human CD206 (clone 15-2, Biolegend). Staining results are shown for CD14+ cells. Data were acquired on a FACSCanto II flow cytometer (BD Biosciences), and analyzed with FlowJo v.9.5.2 (Tree Star Inc.).
RNA-sequencing
mRNA was extracted from lysates of cells that had been stimulated for 24 h, using oligodT beads (Invitrogen). cDNA synthesis, sequencing and sequence analysis were performed as described53. Raw and processed sequencing data were deposited to Pubmed.
Transmission electron microscopy
For ultrastructural analysis, cells were fixed in 2% paraformaldehyde, 2.5% glutaraldehyde (Polysciences Inc.), 0.05% malachite green (Sigma) in 100 mM sodium cacodylate buffer, pH 7.2 for 1 h at 22°C. The malachite green was incorporated into the fixative to stabilize lipid constituents soluble in aqueous glutaraldehyde. Samples were washed in cacodylate buffer and post-fixed in 1% osmium tetroxide (Polysciences Inc.) for 1 h. Samples were then rinsed extensively in distilled H2O prior to en bloc staining with 1% aqueous uranyl acetate (Ted Pella Inc.) for 1 h. Following several rinses in dH2O, samples were dehydrated in a graded series of ethanol and embedded in Eponate 12 resin (Ted Pella Inc.). Sections of 95 nm were cut with a Leica Ultracut UC7 ultramicrotome (Leica Microsystems Inc.), stained with uranyl acetate and lead citrate, and viewed on a JEOL 1200 EX transmission electron microscope (JEOL USA Inc.) equipped with an AMT 8 megapixel digital camera (Advanced Microscopy Techniques).
Statistical analysis
Data were analyzed using Graphpad Prism (v5). Comparisons for two groups were calculated using One-way ANOVA and, where indicated, unpaired two-tailed Student's t-tests. Differences were considered significant when P-values were below 0.05. No randomization or exclusion of data points was used. Pilot in vivo studies were used for estimation of the sample size required to ensure adequate power.
Supplementary Material
Acknowledgements
We would like to thank G. Haemmerle and R. Zechner (University of Graz) for permission to use the Pnpla2–/– mice, and C. Semenkovich and R. Gross for providing us with these animals from their breeding programs. We thank H. Virgin, E.l Gautier (Pierre & Marie Curie University) and S. Ivanov for many helpful discussions, and the staff of the Department of Pathology & Immunology Flow Cytometry Core and the Metabolomics Core of the Diabetic Cardiovascular Disease Center for expert technical assistance. The work was supported by the National Institutes of Health grants to E.J.P. (AI32573, CA164062), E.L.P (AI091965 and CA158823), N.A.A. (DK060022), H. D. (HL087001) and C.Y. (CA138759, CA152099).
Footnotes
Contributions
S.C-C.H., B.E., Y.I, D.O'S., M.N., J.F.U., N.A.A., M.N.A., E.L.P. and E.J.P, designed experiments. S.C-C.H., B.E., Y.I., D.O'S., M.N., A.M.S., W.B., L.L-G., W.Y.L., C.M.O'N., C.Y., and H.D. performed experiments. S.C-C.H., B.E., M.N., D.O'S., W.Y.L, C.M.O'N, N.A.A., M.N.A., E.L.P. and E.J.P. analyzed data. S.C-C.H. and E.J.P. wrote the paper.
Accession codes:
GEO, RNA-seq dataset GSE53053
Competing financial interests
The authors declare no competing financial interests.
References
- 1.Geissmann F, et al. Development of monocytes, macrophages, and dendritic cells. Science. 2010;327:656–661. doi: 10.1126/science.1178331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schulz C, et al. A lineage of myeloid cells independent of Myb and hematopoietic stem cells. Science. 2012;336:86–90. doi: 10.1126/science.1219179. [DOI] [PubMed] [Google Scholar]
- 3.Gause WC, Wynn TA, Allen JE. Type 2 immunity and wound healing: evolutionary refinement of adaptive immunity by helminths. Nat Rev Immunol. 2013;13:607–614. doi: 10.1038/nri3476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gordon S. Alternative activation of macrophages. Nat Rev Immunol. 2003;3:23–35. doi: 10.1038/nri978. [DOI] [PubMed] [Google Scholar]
- 5.Wynn TA, Chawla A, Pollard JW. Macrophage biology in development, homeostasis and disease. Nature. 2013;496:445–455. doi: 10.1038/nature12034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rodriguez-Prados JC, et al. Substrate fate in activated macrophages: a comparison between innate, classic, and alternative activation. J Immunol. 2010;185:605–614. doi: 10.4049/jimmunol.0901698. [DOI] [PubMed] [Google Scholar]
- 7.Odegaard JI, Chawla A. Alternative macrophage activation and metabolism. Annu Rev Pathol. 2011;6:275–297. doi: 10.1146/annurev-pathol-011110-130138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vats D, et al. Oxidative metabolism and PGC-1beta attenuate macrophage-mediated inflammation. Cell Metab. 2006;4:13–24. doi: 10.1016/j.cmet.2006.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pearce EL. Metabolism in T cell activation and differentiation. Curr Opin Immunol. 2010;22:314–320. doi: 10.1016/j.coi.2010.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van der Windt GJ, et al. Mitochondrial respiratory capacity is a critical regulator of CD8+ T cell memory development. Immunity. 2012;36:68–78. doi: 10.1016/j.immuni.2011.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zechner R, et al. FAT SIGNALS--lipases and lipolysis in lipid metabolism and signaling. Cell Metab. 2012;15:279–291. doi: 10.1016/j.cmet.2011.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kienesberger PC, Pulinilkunnil T, Nagendran J, Dyck JR. Myocardial triacylglycerol metabolism. J Mol Cell Cardiol. 2013;55:101–110. doi: 10.1016/j.yjmcc.2012.06.018. [DOI] [PubMed] [Google Scholar]
- 13.Su X, Abumrad NA. Cellular fatty acid uptake: a pathway under construction. Trends Endocrinol Metab. 2009;20:72–77. doi: 10.1016/j.tem.2008.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bharadwaj KG, et al. Chylomicron- and VLDL-derived lipids enter the heart through different pathways: in vivo evidence for receptor- and non-receptor-mediated fatty acid uptake. J Biol Chem. 2010;285:37976–37986. doi: 10.1074/jbc.M110.174458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Feng J, et al. Induction of CD36 expression by oxidized LDL and IL-4 by a common signaling pathway dependent on protein kinase C and PPAR-gamma. J Lipid Res. 2000;41:688–696. [PubMed] [Google Scholar]
- 16.Oh J, et al. Endoplasmic reticulum stress controls M2 macrophage differentiation and foam cell formation. J Biol Chem. 2012;287:11629–11641. doi: 10.1074/jbc.M111.338673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ries S, et al. Transcriptional regulation of lysosomal acid lipase in differentiating monocytes is mediated by transcription factors Sp1 and AP-2. J Lipid Res. 1998;39:2125–2134. [PubMed] [Google Scholar]
- 18.Nicholls DG, et al. Bioenergetic profile experiment using C2C12 myoblast cells. J Vis Exp. 2010 doi: 10.3791/2511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.O'Neill LA, Hardie DG. Metabolism of inflammation limited by AMPK and pseudo-starvation. Nature. 2013;493:346–355. doi: 10.1038/nature11862. [DOI] [PubMed] [Google Scholar]
- 20.Thomas GD, et al. The biology of nematode- and IL4Ralpha-dependent murine macrophage polarization in vivo as defined by RNA-Seq and targeted lipidomics. Blood. 2012;120:e93–e104. doi: 10.1182/blood-2012-07-442640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heck AM, Yanovski JA, Calis KA. Orlistat, a new lipase inhibitor for the management of obesity. Pharmacotherapy. 2000;20:270–279. doi: 10.1592/phco.20.4.270.34882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schweiger M, et al. Adipose triglyceride lipase and hormone-sensitive lipase are the major enzymes in adipose tissue triacylglycerol catabolism. J Biol Chem. 2006;281:40236–40241. doi: 10.1074/jbc.M608048200. [DOI] [PubMed] [Google Scholar]
- 23.Jenkins SJ, et al. Local macrophage proliferation, rather than recruitment from the blood, is a signature of TH2 inflammation. Science. 2011;332:1284–1288. doi: 10.1126/science.1204351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McLaren JE, Michael DR, Ashlin TG, Ramji DP. Cytokines, macrophage lipid metabolism and foam cells: implications for cardiovascular disease therapy. Prog Lipid Res. 2011;50:331–347. doi: 10.1016/j.plipres.2011.04.002. [DOI] [PubMed] [Google Scholar]
- 25.Sheriff S, Du H, Grabowski GA. Characterization of lysosomal acid lipase by site-directed mutagenesis and heterologous expression. J Biol Chem. 1995;270:27766–27772. doi: 10.1074/jbc.270.46.27766. [DOI] [PubMed] [Google Scholar]
- 26.Hadvary P, Sidler W, Meister W, Vetter W, Wolfer H. The lipase inhibitor tetrahydrolipstatin binds covalently to the putative active site serine of pancreatic lipase. J Biol Chem. 1991;266:2021–2027. [PubMed] [Google Scholar]
- 27.Harmon CM, Luce P, Beth AH, Abumrad NA. Labeling of adipocyte membranes by sulfo-N-succinimidyl derivatives of long-chain fatty acids: inhibition of fatty acid transport. J Membr Biol. 1991;121:261–268. doi: 10.1007/BF01951559. [DOI] [PubMed] [Google Scholar]
- 28.Martinez FO, Gordon S, Locati M, Mantovani A. Transcriptional profiling of the human monocyte-to-macrophage differentiation and polarization: new molecules and patterns of gene expression. J Immunol. 2006;177:7303–7311. doi: 10.4049/jimmunol.177.10.7303. [DOI] [PubMed] [Google Scholar]
- 29.Reynolds LA, Filbey KJ, Maizels RM. Immunity to the model intestinal helminth parasite Heligmosomoides polygyrus. Semin Immunopathol. 2012;34:829–846. doi: 10.1007/s00281-012-0347-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jenkins SJ, et al. IL-4 directly signals tissue-resident macrophages to proliferate beyond homeostatic levels controlled by CSF-1. J Exp Med. 2013;210:2477–2491. doi: 10.1084/jem.20121999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Anthony RM, et al. Memory T(H)2 cells induce alternatively activated macrophages to mediate protection against nematode parasites. Nat Med. 2006;12:955–960. doi: 10.1038/nm1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cardilo-Reis L, et al. Interleukin-13 protects from atherosclerosis and modulates plaque composition by skewing the macrophage phenotype. EMBO Mol Med. 2012;4:1072–1086. doi: 10.1002/emmm.201201374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stohr R, Federici M. Insulin resistance and atherosclerosis: convergence between metabolic pathways and inflammatory nodes. Biochem J. 2013;454:1–11. doi: 10.1042/BJ20130121. [DOI] [PubMed] [Google Scholar]
- 34.Wild PS, et al. A genome-wide association study identifies LIPA as a susceptibility gene for coronary artery disease. Circ Cardiovasc Genet. 2011;4:403–412. doi: 10.1161/CIRCGENETICS.110.958728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vargas-Alarcon G, et al. Single Nucleotide Polymorphisms within LIPA (Lysosomal Acid Lipase A) Gene Are Associated with Susceptibility to Premature Coronary Artery Disease. A Replication in the Genetic of Atherosclerotic Disease (GEA) Mexican Study. PLoS One. 2013;8:e74703. doi: 10.1371/journal.pone.0074703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Du H, et al. Lysosomal acid lipase-deficient mice: depletion of white and brown fat, severe hepatosplenomegaly, and shortened life span. J Lipid Res. 2001;42:489–500. [PubMed] [Google Scholar]
- 37.Yan C, et al. Macrophage-specific expression of human lysosomal acid lipase corrects inflammation and pathogenic phenotypes in lal−/− mice. Am J Pathol. 2006;169:916–926. doi: 10.2353/ajpath.2006.051327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Singh R, et al. Autophagy regulates lipid metabolism. Nature. 2009;458:1131–1135. doi: 10.1038/nature07976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.O'Sullivan D, et al. Memory CD8+ T cells use cell intrinsic lipolysis to support the metabolic programming necessary for development. Immunity. doi: 10.1016/j.immuni.2018.07.018. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chawla A. Control of macrophage activation and function by PPARs. Circ Res. 2010;106:1559–1569. doi: 10.1161/CIRCRESAHA.110.216523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tsao CH, Shiau MY, Chuang PH, Chang YH, Hwang J. Interleukin-4 Regulates Lipid Metabolism by Inhibiting Adipogenesis and Promoting Lipolysis. J Lipid Res. 2013 doi: 10.1194/jlr.M041392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brignull LM, et al. Reprogramming of lysosomal gene expression by interleukin-4 and Stat6. Bmc Genomics. 2013;14 doi: 10.1186/1471-2164-14-853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rios FJ, et al. Oxidized LDL induces alternative macrophage phenotype through activation of CD36 and PAFR. Mediators Inflamm. 2013;2013:198193. doi: 10.1155/2013/198193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.May P, Bock HH, Nofer JR. Low density receptor-related protein 1 (LRP1) promotes anti-inflammatory phenotype in murine macrophages. Cell Tissue Res. 2013;354:887–889. doi: 10.1007/s00441-013-1699-2. [DOI] [PubMed] [Google Scholar]
- 45.Bie J, Zhao B, Ghosh S. Atherosclerotic lesion progression is attenuated by reconstitution with bone marrow from macrophage-specific cholesteryl ester hydrolase transgenic mice. Am J Physiol Regul Integr Comp Physiol. 2011;301:R967–974. doi: 10.1152/ajpregu.00277.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sekiya M, et al. Ablation of neutral cholesterol ester hydrolase 1 accelerates atherosclerosis. Cell Metab. 2009;10:219–228. doi: 10.1016/j.cmet.2009.08.004. [DOI] [PubMed] [Google Scholar]
- 47.Ouimet M, et al. Autophagy regulates cholesterol efflux from macrophage foam cells via lysosomal acid lipase. Cell Metab. 2011;13:655–667. doi: 10.1016/j.cmet.2011.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xu XY, et al. Obesity Activates a Program of Lysosomal-Dependent Lipid Metabolism in Adipose Tissue Macrophages Independently of Classic Activation. Cell Metab. 2013;18:816–830. doi: 10.1016/j.cmet.2013.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Finkelman FD, et al. Anti-cytokine antibodies as carrier proteins. Prolongation of in vivo effects of exogenous cytokines by injection of cytokine-anti-cytokine antibody complexes. J Immunol. 1993;151:1235–1244. [PubMed] [Google Scholar]
- 50.Camberis M, Le Gros G, Urban J., Jr. Animal model of Nippostrongylus brasiliensis and Heligmosomoides polygyrus. Curr Protoc Immunol. 2003 doi: 10.1002/0471142735.im1912s55. Chapter 19, Unit 19 12. [DOI] [PubMed] [Google Scholar]
- 51.Love-Gregory L, et al. Common CD36 SNPs reduce protein expression and may contribute to a protective atherogenic profile. Hum Mol Genet. 2011;20:193–201. doi: 10.1093/hmg/ddq449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Krawczyk CM, et al. Toll-like receptor-induced changes in glycolytic metabolism regulate dendritic cell activation. Blood. 2010;115:4742–4749. doi: 10.1182/blood-2009-10-249540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Everts B, et al. Commitment to glycolysis sustains survival of NO-producing inflammatory dendritic cells. Blood. 2012;120:1422–1431. doi: 10.1182/blood-2012-03-419747. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.