Abstract
Dithiolopyrrolone (DTP) group antibiotics were first isolated in the early half of the 20th century, but only recently has research been reawakened by insights gained from the synthesis and biosynthesis of this structurally intriguing class of molecules. DTPs are characterized by an electronically unique bicyclic structure, which contains a compact disulfide bridge between two ene-thiols. Points of diversity within the compound class occur outside of the bicyclic core, at the two amide nitrogens. Such modifications distinguish three of the most well studied members of the class, holomycin, thiolutin, and aureothricin; the DTP core has also more recently been identified in the marine antibiotic thiomarinol, in which it is linked to a marinolic acid moiety, analog of the FDA-approved topical antibiotic Bactroban® (GlaxoSmithKline). Dithiolopyrrolones exhibit relatively broad-spectrum antibiotic activity against many Gram-positive and Gram-negative bacteria, as well as strains of Mycobacterium tuberculosis. Additionally, they have been shown to exhibit potent and selective anti-cancer activity. Despite this promising profile, there is still much unknown about the mechanisms of action for DTPs. Early reports suggested that they inhibit yeast growth at the level of transcription and that this effect is largely responsible for their distinctive microbial static properties; a similar mechanism is supported in bacteria. Elucidation of biosynthetic pathways for holomycin in Streptomyces clavuligerus and Yersinia ruckeri and thiomarinol in Alteromonas rava sp. nov. SANK 73390, have contributed evidence suggesting that multiple mechanisms may be operative in the activity of these compounds. This review will comprehensively cover the history and development of dithiolopyrrolones with particular emphasis on the biosynthesis, synthesis, biological activity and mechanism of action.
1 Introduction
During the past century, many successful antibiotic discovery efforts owed their starts to natural products. In particular, in the 1940’s-1960’s, in what might be considered the “golden era” of natural product isolation, more than 50% of the natural product scaffolds that are currently employed clinically, were first identified and structurally characterized. In subsequent years, as antibiotic resistance rose, these scaffolds provided some of the raw materials for medicinal chemists’ to fashion the next generations of antibiotic therapeutics. Many natural products that were too toxic for antibiotic use could still be modified or else find their way into the clinic for alternative therapeutic indications. Still other compounds remain untapped or underexplored because a lack in understanding of their basic mechanism prevented efforts to rationally optimize their therapeutic efficacy.
The dithiolopyrrolones, also referred to as pyrrolinonodithioles or DTPs, are a structurally and electronically unique group of natural products that remain to be therapeutically exploited. Holomycin and thiolutin are perhaps the best-known representatives of the dithiolopyrrolone class of compounds, which is comprised of more than 25 variants isolated from a variety of soil- and marine-derived bacteria, including both Gram-positives and Gram-negatives. The dithiolopyrrolones exhibit very broad-spectrum antibiotic activity and potent antiangiogenic effects. Despite this attractive biological activity, neither DTPs nor DTP analogs are currently employed in the clinic, due to attendant toxicity. Importantly, the mechanism of biological activity and moreover, the potential cause of such side effects are still not well understood. Recent genome mining efforts have led to the elucidation of several DTP biosynthetic pathways and these results have combined with advances in synthetic methods to contribute to renewed interest in this intriguing class of compound. The present review aims for a comprehensive survey of the literature, dating back to the first accessible reports of dithiolopyrrolone isolations in 1948. Critical assessment of opportunities for new inquiry is provided in areas of biosynthesis, mechanism of action and therapeutic development.
2 Structures and Isolation
2.1 Structural characteristics
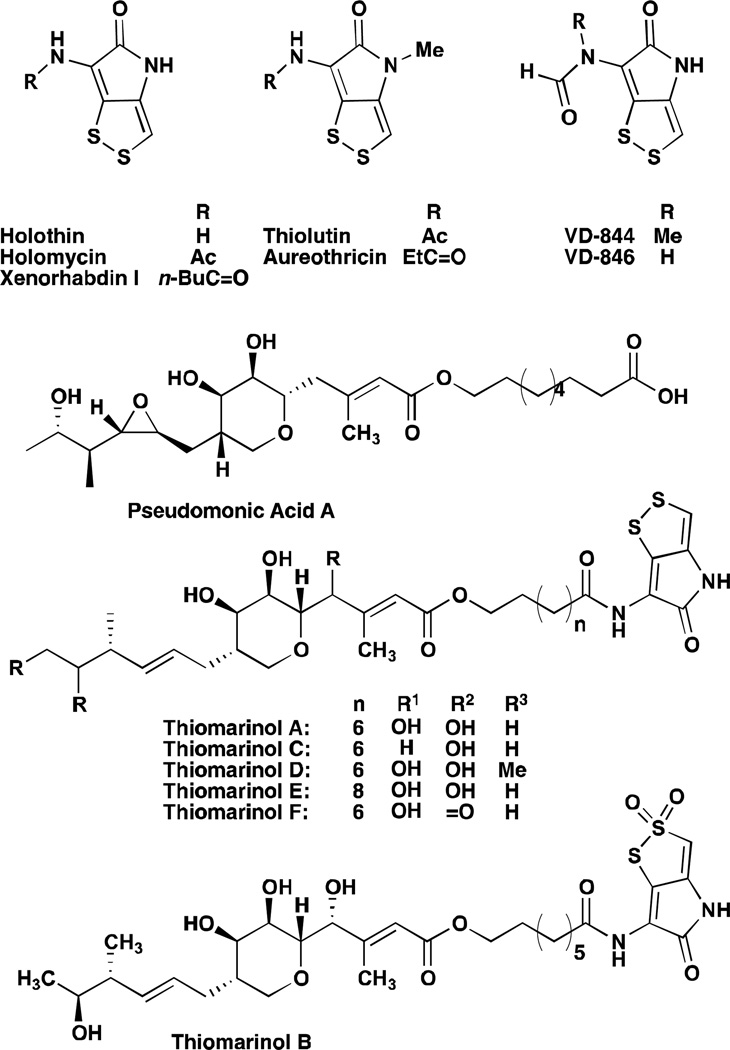
The strikingly simple architecture of dithiolopyrrolones (planar, no stereocenters) and low molecular weight (FW: 213.98 for holomycin, 228.0027 for thiolutin) belies the complexity of their electronic structure. Structures of several well-studied DTPs are illustrated in Figure 1. The Cys-Cys dipeptide-derived pyrrothine core contains a formally aromatic ene-dithiol-disulfide, which can be found in nature with substituent variation at either of the two nitrogen heteroatoms. In particular, the acyl chain of the exo-cyclic amide has been found in lengths varying from formyl to octyl, with several instances of branching or unsaturation also being reported. Meanwhile, the endo-cyclic pyrrolone nitrogen has been isolated in the secondary, unsubstituted, as well as the tertiary, N-methylated forms. The extended conjugation of the ene-dithiols gives rise to a distinctive yellow/gold color and strong UV absorbance at 388 nm. The infrared spectrum of thiolutin exhibits carbonyl bands at 1667 cm−1 and 1634 cm−1, NH stretching and bending absorption at 3205 cm−1 and 1550 cm−1, respectively, and strong C=C stretching absorption at 1600 cm−1 which are consistent with the highly conjugated aromatic structure of the compound; similar values have been reported for holomycin. In crystal structures of the naturally occurring DTP, VD-844, the S-C bond lengths, 1.73 A and 1.74 A, are significantly shorter than is typical for a single bond, (1.82 A) and are closer in length to the S-C bonds in thiophene, 1.718 A.1,2 The length of the N(4)-C(8) bond is reported at 1.33 A, a value intermediate between C-N bond lengths reported for pyrrole, 1.383 A and succinimide, 1.27 A, while the N(4)-C(5) distance of 1.37 A, is slightly longer than the typical 1.33 A for amide N-C bonds. While the exo-cyclic secondary amides of thiolutin and holomycin populate the favourable trans conformer, stabilized by a weak O-S interaction, the tertiary nitrogen of VD-844 exhibits two different conformations in the 100 MHz NMR; all protons in VD-844 at ordinary temperature give rise to two peaks. Coupling between the formyl and methyl protons is more pronounced in the cisoid conformation illustrated in Figure 1, 6.3 The existence of these rotamers presumably closely related in energy could be rigorously confirmed from the X-ray structure. Neither oxidation potentials nor explicit examples of reactivity or degradation reactions for the compact core have been reported in the literature.
Figure 1.
Structures of isolated dithiolopyrrolones.
2.2 DTPs from Actinomycetes
Aureothricin (5) is the earliest reported DTP in the literature; it was first isolated as a yellow crystalline solid from Streptomyces strain no. 26A related to Nocardia farcinica (referred to as A. farcinicus in the original isolation paper) in 1948, by Umezawa and co-workers.4 Notably, the compound was characterized by potent antibiotic activity against E. coli and a strong UV absorption max at 388 nm, distinctive of the pyrrothine core. The isolation of thiolutin was subsequently described from a strain of Streptomyces albus in 1950, but correct formulas of both aureothricin and thiolutin remained elusive until Celmer and co-workers reported their characterization via chemical degradation and derivatization.5–7 Further work by the same group demonstrated that thiolutin and aureothricin contain the same acidic degradation product, identified as the common core pyrrothine of the DTP antibiotics. This fragment could be used to generate either thiolutin or aureothricin via acetylation or propionylation, respectively. These same studies led to the definitive structural assignment of the two compounds. Holomycin was subsequently isolated from a strain of S. griseus and identified as desmethyl-thiolutin by researchers at ETH, Zurich.8 Holomycin was later identified among additional metabolites from the important industrial strain and clavulanic acid producer, Streptomyces clavuligerus.9
Several Actinomycete strains have been reported to produce multiple different DTPs. Aureothricin, thiolutin and a new compound, iso-butyropyrrothine were isolated as a mixture from another soil-derived Streptomyces of pimprina sp. in 1960, indicating for the first time that a single bacterium was capable of making multiple different acylation variants of the pyrrothine core.10 Similarly, holomycin and N-propionyl holothin were both isolated from a nitrosoguanidine mutant of a soil-derived Cephamycin C producer, Streptomyces sp. P6621 designated P6621-7N49.11 Variation in substitution on the exo-cyclic nitrogen of the pyrrothine could also be observed in isolates from an unidentified Streptomycete collected near Copenhagen in 1969.3 The two compounds purified in this study were named, separately, VD-844 and VD-846, and exhibited strong activity against Neisseria. Both NMR and elemental analysis were used to confirm the structures as 6 and 7. Most recently, thiolutin, iso-butyropyrrothine and three new DTPs were isolated from the culture broth of Saccharothrix SA 233, a rare Actinomycetales present in the soils of palm groves from southern Algeria.12,13 Among the new compounds from the Saccharothrix strain are two containing the unusual and potentially highly electrophilic α,β-unsaturated tigloyl and 3-methyl-2-butenoyl groups.
2.3 DTPs from Proteobacteria
Isolation years, structures, and primary references are listed in Table 1. While the earliest isolations of dithiolopyrrolones came predominantly from soil-derived Gram-positive species, such as Streptomyces, more recently they have also been found from Gram-negative heterotrophs, as well as a number of marine derived species. McInerney and co-workers isolated five new DTP variants from bacteria of Xenorhabdus sp., termed xenorhabdins.14 Xenorhabdus are Gram-negative enterobacteria that form mutualistic associations with entomopathogenic soil nematodes and are known to be pathogenic towards a variety of insects. These bacteria proliferate in the killed host and favour reproduction of the nematodes by providing nutrients and by producing antibiotics that inhibit the growth of other microbial flora in the insect cadavers. Therefore, there is reason to believe that DTP production plays a role in this mutualistic life cycle. Compounds were isolated exclusively from Xenorhabdus in so-called phase one growth and showed antimicrobial and insecticidal activity.
Table 1.
Historical list of isolated dithiolopyrrolones, producer strains, dates, and references.
| Compound | Endo-cylic N | Exo-cylic N | Producer Strains | Isolation Year | References |
|---|---|---|---|---|---|
| Aureothricin | Methyl | Propionyl | Streptomyces | 1948 | Japan. Med. J. 1948, 1, 512–517. J. Am. Chem. Soc. 1955, 77, 2861–2865. |
| Thiolutin | Methyl | Acetyl |
Streptomyces albus Streptomyces luteosporeus |
1950 | J. Am. Chem. Soc. 1952, 74, 6304–6305. J. Am. Chem. Soc. 1955, 77, 2861–2865. |
| Holomycin | H | Acetyl |
Streptomyces griseus Streptomyces clavuligerus Photobacterium halotorelans Yersinia ruckeri |
1959 1979 2010 2013 |
Helv. Chim. Acta 1959, 42, 563–569. J. Antibiot. 1979, 32, 549–554. Mar Drugs 2010, 8, 2946–2960. J. Biol. Chem. 2013, 288, 14688–14697. |
| iso-Butyropyrrothine | Methyl | Isobutyryl | Streptomyces pimprina | 1960 | Cell. Mol. Life Sci. 1960, 16, 504–505. |
| VD-844 | H | Formyl, Methyl | Streptomyces | 1969 | J. Antibiot. 1969, 22, 233. J. Antibiot. 1969, 22, 231–232. |
| VD-846 | H | Formyl | Streptomyces | 1969 | J. Antibiot. 1969, 22, 233. |
| Hexanoylholothin | H | Hexanoyl | Xenorhabdus bovienii | 1991 | J. Nat. Prod. 1991, 54, 774–784. |
| iso-Heptanoylholothin | H | Isoheptynoyl | Xenorhabdus bovienii | 1991 | J. Nat. Prod. 1991, 54, 774–784. |
| Octanoylpyrrothine | H | Octanoyl | Xenorhabdus bovienii | 1991 | J. Nat. Prod. 1991, 54, 774–784. |
| Hexanoylpyrrothine | Methyl | Hexanoyl | Xenorhabdus bovienii | 1991 | J. Nat. Prod. 1991, 54, 774–784. |
| Thiomarinol A | H | Marinolyl (from marinolic acid) |
Alteromonas rava | 1993 | J. Antibiot. 1993, 46, 1834–1842. |
| Thiomarinol B | H | Marinolyl (from marinolic acid) |
Alteromonas rava | 1995 | J. Antibiot. 1995, 48, 907–909. |
| Thiomarinol C | H | 4-Deoxymarinolyl | Alteromonas rava | 1995 | J. Antibiot. 1995, 48, 907–909. |
| Thiomarinol D | H | 14-methylmarinolyl | Alteromonas rava | 1997 | J. Antibiot. 1997, 50, 449–452. |
| Thiomarinol E | H | 10-(hydroxymonyl)decanoyl | Alteromonas rava | 1997 | J. Antibiot. 1997, 50, 449–452. |
| Thiomarinol F | H | 13-oxomarinolyl | Alteromonas rava | 1997 | J. Antibiot. 1997, 50, 449–452. |
| Thiomarinol G | H | 4,6-dideoxy-8-hydroxymarinolyl | Alteromonas rava | 1997 | J. Antibiot. 1997, 50, 449–452. |
| iso-Heptanoylpyrrothine | Methyl | Isoheptynoyl | Xenorhabdus bovienii | 1991 | J. Nat. Prod. 1991, 54, 774–784. |
| PSA | Methyl | 3-methyl-2-butenoyl | Saccharothrix SA 233 | 2002 | J. Antibiot. 2002, 55, 696–701. J. Antibiot. 2002, 55, 702–706. |
| PSB | Methyl | Tigloyl | Saccharothrix SA 233 | 2002 | J. Antibiot. 2002, 55, 696–701. J. Antibiot. 2002, 55, 702–706. |
| PSC2 | Methyl | n-butyryl | Saccharothrix SA 233 | 2002 | J. Antibiot. 2002, 55, 696–701. J. Antibiot. 2002, 55, 702–706. |
In the patent literature, there are also reports of isolation of S-oxidized variants of the DTP core from Xenorhabdus bovenii.15 These thiosulfonate derivatives were observed in trace amounts in isolates, in combination with larger quantities of the unadulterated DTP compounds. It is further reported that the so-called xenorxides could be harvested in greater yield if the cell-free filtrate was left open to the air for extended periods of time from one week up to one month. These compounds were reportedly active against a variety of bacteria and fungi.
Amongst other Gram-negative bacteria that have been shown to produce DTPs are P. halotolerans and Yersinia ruckeri, a marine-derived fish gut pathogen.16,17 The latter isolation was the result of a genome mining effort, which identified a DTP-biosynthetic cluster in the recently sequenced genome of Y. ruckeri and subsequently pursued grow-up and isolation of the putative “cryptic” metabolite. Holomycin was the structure isolated from both the Yersinia as well as P. halotolerans.
Intriguingly, several unique N-acyl derivatives have been isolated from a marine bacterium, Alteromonas rava sp. nov. SANK 73390. 18–22 These compounds, designated thiomarinols, contain the typical DTP pyrrothine connected to variants of the polyketide core of pseudomonic acid C (PA-C). Pseudomonic acid A (PA-A), a close structural homolog of PA-C, is the major constituent of mupirocin, the topical antibiotic developed by Smith-Kline-Beecham and presently marketed as Bactroban® in the US by the pharmaceutical company Glaxo-Smith-Kline. Therefore, the thiomarinols constitute highly novel examples of naturally occurring hybrid antibiotics. To date, six different DTP-containing thiomarinols have been reported in the literature and representative structures are illustrated in figure 1. Notably, the S-oxidized variant, thiomarinol B, which contains a thiosulfonate moiety was also isolated from SANK 73390 and reported to have near 4-fold greater antibiotic activity than the non-oxidized progenitor against a broad panel of Gram-negative and Gram-positive bacteria. Overall, the thiomarinols are more potent than the individual constituent antibiotics alone.
3 Biosynthesis and Regulation
3.1 Biosynthesis of holomycin
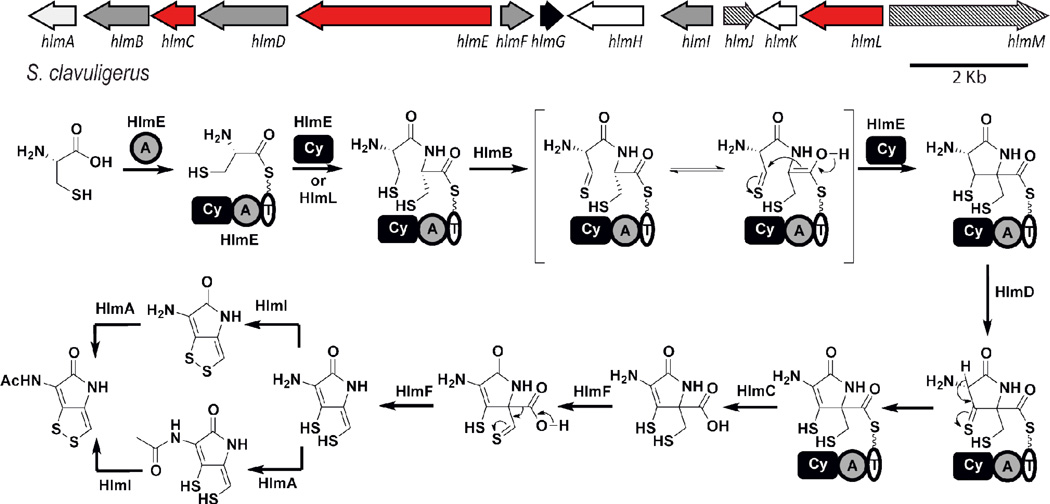
Although DTPs had been known for a long time, their biosynthesis remained poorly understood until work by Li and Walsh was reported in 2010.23 Whole genome sequencing of holomycin producer, S. clavuligerus, enabled identification and characterization of the holomycin biosynthetic pathway. The genome of S. clavuligerus was sequenced first by the Broad Institute and subsequently by two additional independent teams in the Netherlands and Korea. 24,25 The Broad Institute genome was assembled to four super contigs but not the whole genome. Takano and coworkers completed the full assembly and uncovered an interesting feature of the genome: it not only contains a 6.8 Mbp linear chromosome, which is characteristic of actinomycetes genomes, but also a large linear plasmid of 1.8 Mbp.24 Prediction of putative natural product clusters using bioinformatic software anti-SMASH revealed over 40 potential clusters, some of which are located on the linear chromosome and the rest on the linear plasmid.
In the first biosynthesis studies on holomycin, Li and Walsh mined the Broad Genome to map potential candidate clusters.23 Based on the structure of holomycin, two cysteine residues were hypothesized to serve as building blocks that might be activated by a non-ribosomal peptide synthetase (NRPS). Subsequent oxidations need to occur on the Cys-Cys dipeptide backbone in order to remove eight total electrons and a carboxylic acid yielding the unsaturated bicyclic DTP core (Figure 2). Therefore, a BLAST was carried out to search for a cluster of genes containing cysteine-specific NRPSs and relevant oxidoreductases in the S. clavuligerus genome. A putative holomycin gene cluster was discovered on the main chromosome. This gene cluster contains a single module NRPS with several non-canonical domains: a condensation/cyclization (Cy) domain, an adenylation (A) domain predicted to activate cysteine, and a thiolation (T) domain. Apart from the NRPS, four flavin-dependent oxidoreductases, an acyl transferase, and a standalone thioesterase (TE) were also found in the gene cluster. The gene cluster assignment was validated by gene replacement experiments focusing on the NRPS and the decarboxylase, which presumably removes the carboxylate from the C-terminal cysteine. Separate, in-frame deletions of these two genes resulted in complete abolishment of holomycin production in both the wild type and a holomycin-overproducing strain (discussed in Section 3.4). Additionally, in depth biochemical characterization was carried out on the NRPS and acyl transferase. Using a 32P radiolabeled pyrophosphate exchange assay, the adenylation domain was tested against Cys, Met, Ala and Ser and it was shown to be highly specific for Cys loading. Cysteine can be loaded on the NPRS as demonstrated by autoradiography using 14C-1C-Cys and later by phosphopantatheine (PPant) ejection assay (unpublished results). Activity of the acyl transferase was also reconstituted in vitro converting holothin to holomycin in the presence of acetyl CoA. This enzyme can also utilize propionyl and other CoA substrates generating holomycin analogs. It displays somewhat relaxed substrate specificity and can accommodate branched, unsaturated and even hydroxylated acyl CoAs with a preference for short chain CoAs. It does not seem to utilize any aromatic CoA variant such as benzoyl or sinapoyl CoA. This study combined genome-mining, genetic knockout and in vitro reconstitution to identify and validate an unprecedented pathway for holomycin biosynthesis. The genes involved in holomycin biosynthesis in S. clavuligerus were named hlm A through M.
Figure 2.
Holomycin gene cluster from S. clavuligerus and proposed biosynthetic pathway.
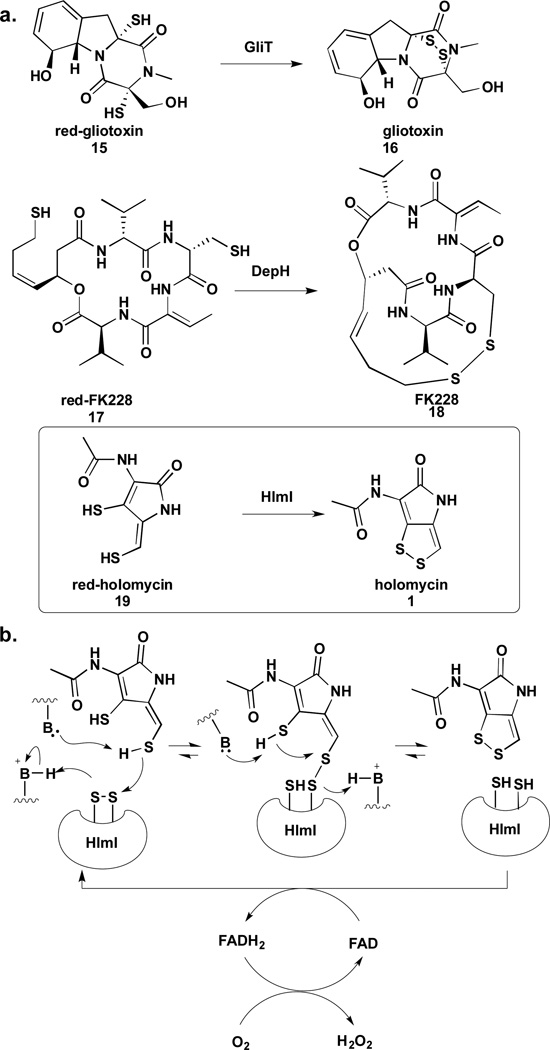
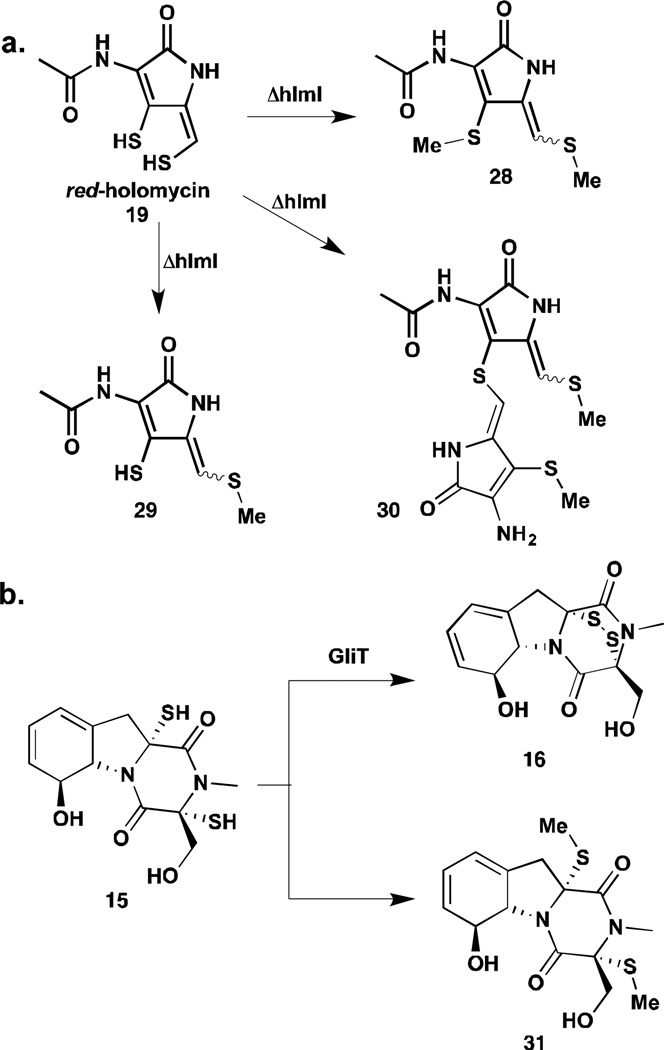
Li and Walsh also investigated the role of HlmI, a thioredoxin-oxidoreductase-like enzyme, in holomycin biosynthesis.26 HlmI shares 27–35% sequence homology with GliT and DepH from the biosynthetic pathways of disulfide-containing natural products gliotoxin (16) and FK228 (18) respectively. Both GliT and DepH have been shown to act as dithiol oxidases in late-stage biosynthetic modifications.27,28 HlmI was shown to catalyze an analogous reaction in holomycin biosynthesis, converting the holomycin dithiol to the corresponding disulfide (Figure 3a).26 Interestingly, the reduced form of holomycin (red-holomyin), substrate of HlmI, displays a strong UV absorption at 320 nm, which decreases dramatically when it is converted to the disulfide. Kinetic parameters for HlmI could be measured by following the changes in UV absorption at 320 nm in real-time. Although background oxidation was observed in vitro, the HlmI catalyzed-reaction exhibited a three-orders of magnitude rate enhancement over the background reaction. Phylogenetic analysis of HlmI, DepH, and GliT revealed that each enzyme belongs to a distinctive clade. These three clades constitute a small-molecule dithiol oxidase group that is evolutionarily distant from the protein thioredoxin oxidoreductases such as TrxB from E. coli. The small-molecule oxidases contain a conserved CXXC motif that is found in the active site of TrxB, suggesting a catalytic mechanism involving the formation of mixed disulfide intermediates between an active site cysteine and substrate thiol (Figure 3b). However, unlike TrxB and DepH, HlmI and GliT do were shown to utilize molecular oxygen instead of NADPH to drive the reaction equilibrium towards substrate oxidation; hydrogen peroxide is released as a by-product.26–29 The X-ray crystal structures of HlmI, DepH and GliT were published very recently.29 All three structures display similar heterodimeric fold as TrxB with a FAD molecule bound in each monomer. The differently sized substrate binding clefts of these enzymes were thought to account for their preference for different small molecule substrates. Interestingly, the HlmI structure has a glutathione (GSH) molecule covalently attached to one of the active site cysteines. It remains to be investigated whether the presence of the GSH molecule in HlmI is merely an artifact of crystallization or is indicative of a physiologically relevant phenomenon.
Figure 3.
Enzymatic oxidation of disulfide bonds in natural products and proposed mechanism of HlmI.
Several subsequent studies provided additional evidence that the gene cluster of hlmA-M is responsible for holomycin biosynthesis. Deng H. and coworkers reported heterologous expression of this cluster in S. albus.30 A genomic library was constructed from S. clavuligerus and a cosmid containing holomycin operons was located by screening for the dithiol oxidase HlmI. Expression of this cosmid in S. albus led to production of holomycin. Ikeda and coworkers cloned and introduced a minimal sequence containing 13 genes hlm A through M into an engineered heteorologous expression host S. avermitilis SUKA 22, which resulted in a strain that stably produces holomycin and holothin in higher yield than wild type S. clavuligerus.31 In addition, a putative dithiolopyrrolone cluster was identified using a genome-mining approach in Yersinia ruckeri ATCC29473. This cluster contains genes that are homologous to holomycin biosynthetic genes from S. clavuligerus and was anticipated to make dithiolopyrrolone derivatives. Indeed holomycin production was confirmed in Y. ruckeri by high-resolution mass spectrometry and NMR. Holomycin was also isolated as an antimicrobial compound from marine bacterium P. halotolerans on a global ocean expedition, expanding the current known holomycin producers from soil actinomycetes to fish pathogen and marine Vibrionaceae.16
3.2 Biosynthesis of thiomarinol
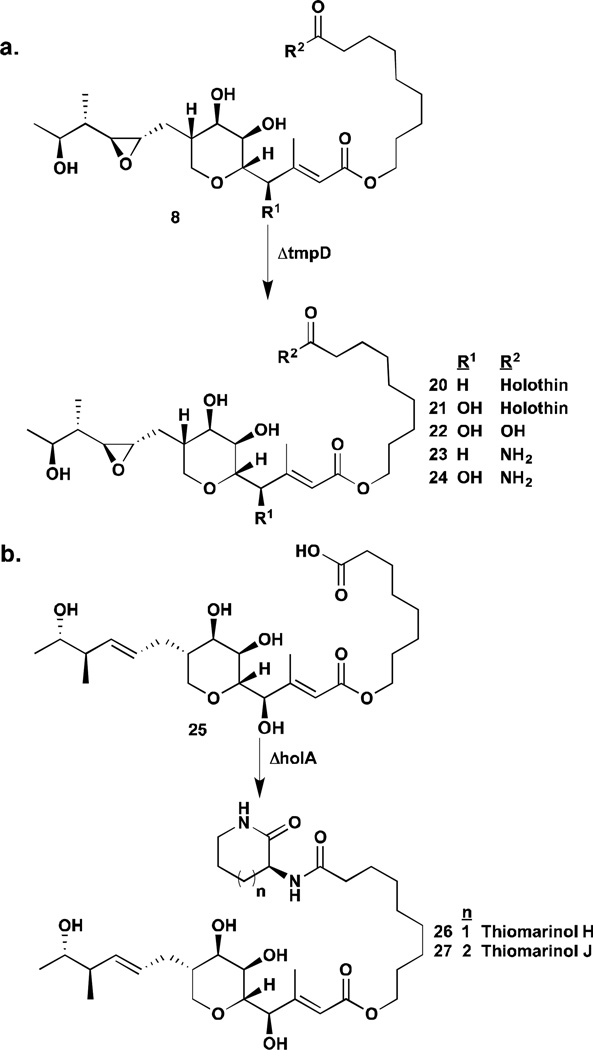
Thiomarinol contains a DTP core linked to a marinolic acid moiety, similar in structure to the antibiotic mupirocin. Mupirocin was isolated from soil and plant associated bacterium Pseudomonas fluorescens and its biosynthesis has been extensively studied in the last decade.32 The gene cluster for thiomarinol, however, was not uncovered until 2011 when Thomas and co-workers carried out whole-genome sequencing of the thiomarinol producer Pseudoalteromonas sp SANK 73390.33 A 97 kb plasmid was found to encode many genes similar to those of the mupirocin gene cluster including type I polyketide synthetase likely responsible for the biosynthesis of the marinolic acid fragment. In addition, a set of genes, unrelated to mupirocin biosynthesis, was discovered downstream of the mup cluster. These genes, hom A through H, share 35–64% sequence homology with members of the hlm cluster from S. clavuligerus with some genomic rearrangement. The gene cluster assignment was validated by targeted gene knockouts in the PKS gene tmpD and NRPS gene holA (homologue of hlmE), thought to be key biosynthetic genes for marinolic acid and DTP respectively. Both mutations resulted in loss of production of thiomarinol. Interestingly, the ΔtmpD mutant still retains the ability to generate DTP compounds and ΔholA the ability to generate marinolic acid, demonstrating that the two pathways are independent of each other. The thiomarinol cluster additionally contains an amide ligase-like gene tmlU, absent from the mupirocin or holomycin pathway. Deletion of tmlU led to production of both marinolic acid and DTPs but not full-length thiomarinol, suggesting that this gene is involved in linking the two moieties together. These mutants, ΔtmpD and ΔholA, were then utilized in a “mutasynthesis” study, which gave rise to several novel DTP molecules; the mutasynthetic approach is discussed in further detail in the next section.
3.3 Precursor-directed biosynthesis of dithiolopyrrolones
Before the discovery of the full DTP biosynthetic pathway, engineered biosynthetic efforts at DTP compounds were accomplished mainly by means of precursor-directed feeding studies in the producing bacteria. Actinomycete strain Saccharothrix algeriensis NRRL B-24137 was used perhaps most extensively in these studies; this strain naturally produces five different DTPs, including thiolutin, senecioyl-, tigloyl-, isobutyryl- and butanoyl- DTP. In one study, addition of benzoic, valeric or cinnamic acids into the growth media led to the discovery of four new DTP compounds, benzoyl-, valeryl-, demethyl-benzoyl-, and formoyl-DTP.34 Another study using the same bacterial strain, but supplemented with sorbic acid allowed formation of crotonyl-, sorbyl-, 2-hexonyl-, and 2-methyl-3- pentenoyl- DTP.35 Some of these new compounds were likely generated through direct incorporation of the organic acids, such as benzoic-, valeryl- and sorbyl- DTP, whereas others came from rerouting of the exogenously supplied acids. The effects of sulfur-containing amino acids on DTP production were also investigated. L-Cysteine and L-cystine at 5–10 mM induced the highest level of production of DTPs in S. algeriensis, while L-methionine and DL-ethionine, a methionine antimetabolite and antagonist, strongly inhibited the production of DTPs at all concentrations.36 These results can be explained by the specificity of NRPS HlmE towards L-cysteine over L-methionine (L-cystine is likely reduced to L-cysteine in the cellular environment) and suggest that methionine and ethionine are inhibitors of DTP biosynthetic enzymes.
The identification of DTP gene clusters turned a new leaf in the generation of new DTP analogs. The activity of acyltransferase HlmA in the holomycin pathway could be reconstituted in vitro. This enzyme utilizes CoA substrates of different chain lengths to generate novel DTPs with various acyl groups including isovaleryl-, hexanoyl, octanoyl-, palmitoyl-, oleoyl-, and 12-hydroxy-stearoyl DTPs demonstrating the power of engineered biosynthesis with purified enzymes (also described in Section 3.1).23 Because the PKS and DTP pathways appeared to be independent of each other in making hybrid-antibiotic thiomarinol, mutants disrupted in PKS gene tmpD, essential for making marinolic acids, and NRPS gene holA, a homolog of holomycin biosynthetic gene hlmE, were used in a “mutasynthesis” study.37 The ΔtmpD and ΔholA mutant bacteria were fed with alternative acids and amines respectively. The ΔtmpD strain utilized pseudomonic acid A, which differs from marinolic acid A by an extra carbon and a 4,5-oxo group, to generate a new pseudomonic acid-DTP hybrid molecule (Scheme 1a). Mutant ΔholA incorporated anhydroornithine and anhydrolysine in place of DTP to synthesize thiomarinol H and J (Scheme 1b). 100% incorporation of these “unnatural” substrates could not be achieved, potentially due to the selectivity of the biosynthetic enzymes. This study demonstrated the flexibility of the thiomarinol biosynthetic pathway to generate new compounds. However, the activity of most of the compounds generated in this study is comparable or less than that of the native compound against B. subtilis or MRSA, suggesting that a wider chemical space needs to be explored.
Scheme 1.
Mutasynthetic preparation of thiomarinol derivatives.
3.4 Regulation
Variable production of holomycin in wild type S. clavuligerus has been mentioned in several reports.23,38,39 In general the production level appears to be very low, and sometimes below detection limit. Liras and co-workers discovered that a mutant disrupted in ccaR, encoding for an autoregulator protein that activates β-lactam antibiotic clavulanate production, generated a small amount of a yellow antimicrobial compound, subsequently identified as holomycin.38 The level of holomycin production was measured in other mutants that are blocked in clavulanate biosynthesis. Mutants in ceaS (carboyxethylarginine synthease), bls (β-lactam synthetase), or oat2 (ORF6, ornithine acetyltransferase), all of which encode for enzymes responsible for chemical transformations in clavulanate biosynthesis, did not produce clavulanate or holomycin. However genetic inactivation of cyp (putative cytochrome P450), ORF12 (putative β-lactamase protein), or oppA2 (previously designated as ORF15 encoding for a putative polypeptide permease) resulted in a complete loss of clavulanate production and a dramatic increase in holomycin level. The roles of these proteins in clavulanate biosynthesis have not been fully elucidated.40 One potential explanation is that the clavulanate pathway shares common intermediates with the holomycin pathway. However, the recent discoveries on holomycin biosynthesis suggest that it is unlikely that common intermediates exist between the two pathways. Furthermore, ORF12 and oppA2 may be involved in resistance or regulation rather than catalysis: ORF12 protein contains a conserved motif present in β-lactamases, and OppA2 is proposed to transport arginine, a necessary substrate for clavulanate biosynthesis, and/or sensing signaling peptides.41 Cyp and ORF11 constitute a cytochrome P450 and ferrodoxin pair that may serve as an oxidative complex or sensor. Therefore, the more likely scenario is that the clavulanate and holomycin pathways are cross-regulated in the producing bacterium, the mechanisms of which remain to be investigated. Additionally, although disruption in arginine regulator ArgR was reported to negatively impact clavulanate biosynthesis and positively affect holomycin production, the role of ArgR in either pathway is likely indirect.42
Transcriptional analysis of wild type S. clavuligerus and holomycin overproducers yielded initial insights into the regulatory mechanisms of the holomycin pathway.39 This pathway was shown to contain eight distinct transcriptional units, some of which are initiated in the coding regions of the previous gene. The mutants oppA2::aph, claR::aph (deletion mutant in clavulanate regulator claR, another holomycin overproducer) and ccaR::tsr demonstrated higher expression levels of genes in the holomycin pathway than the wild type. These results delineate the core genes that are involved in holomycin biosynthesis and suggest that holomycin production is controlled at the transcript level. A significant increase in transcription was observed for genes hlm A through I and L. The gene hlmK was expressed at a lower level in the overproducer than the wild type, suggesting that this gene is not involved in holomycin biosynthesis. This result is consistent with the bioinformatics prediction that hlmK encodes for an inactive thioesterase, which no longer contains an active site triad. Interestingly, the expression level of putative transcriptional regulators hlm J and M remained the same, indicating that the overproduction of holomycin does not result from overexpression of either regulator. In the same study, a proteomics level analysis revealed large protein spots for HlmD, F and G in the overproducer, whereas the signals of these proteins were barely detectable in the wild type. This observation agrees with transcript data for the genes hlmD, F, and G, expression of which was greatly elevated in the overproducer. It is possible that these three enzymes catalyze the rate-limiting steps in holomycin biosynthesis and therefore need to be expressed at a higher level than the rest of the proteins in the pathway. Interestingly, a rhodanese-like enzyme RhlA was also found highly expressed in the proteome of holomycin overproducer strain oppA2::aph.43 The rhodanese superfamily of enzymes typically contains one or more rhodanese structural modules and is thought to utilize persulfides to transfer sulfurs to other biosynthetic enzymes.44,45 The role of RhlA in holomycin biosynthesis remains to be elucidated. RhlA may be involved in trafficking sulfur to cysteine or holomycin biosynthesis enzymes.
3.5 Outlook on biosynthesis
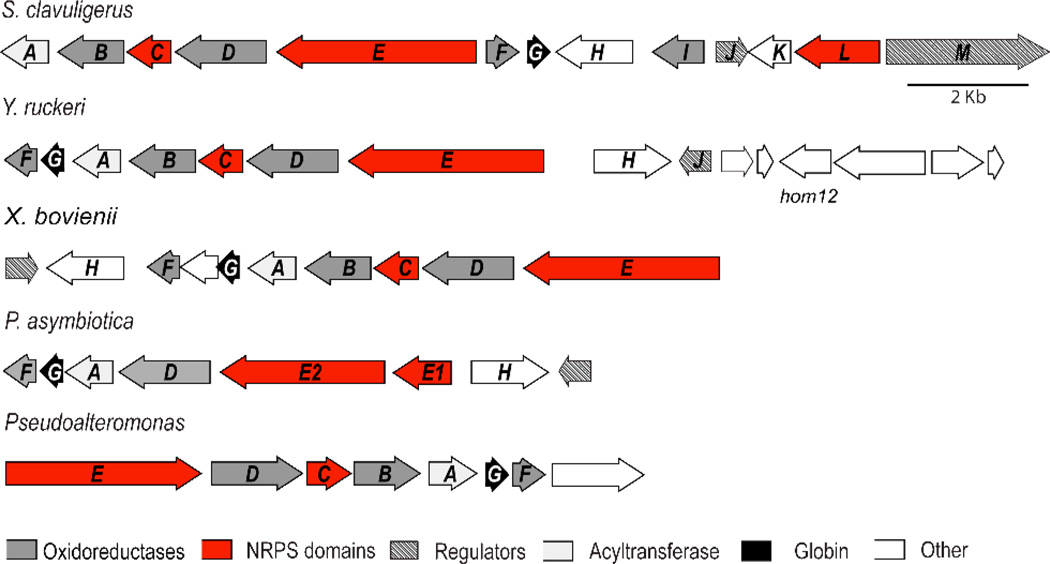
Currently, three DTP biosynthetic clusters have been sequenced and confirmed: two for holomycin from S. clavuligerus, Y. ruckeri and one for thiomarinol from Pseudoalteromonas. Mining of sequenced genomes revealed a fourth putative DTP cluster from Xenorhabdus bovienii SS-2004. Although this particular strain has not been reported to produce DTP-like molecules, DTP compounds have been isolated from closely related strains of X. bovienii whose genomes were not yet sequenced. Therefore, it is likely that this sequenced X. bovienii strain produces xenorhabdin-like molecules and its putative DTP cluster is included in the comparative gene cluster analysis (Figure 4). The comparative analysis demonstrates that seven genes hlm A through F are shared among all four clusters and are likely to be essential for the synthesis of the dithiolopyrrolone core. These genes also displayed similar fold of increased expression in a transcriptional profiling assay of holomycin overproducers, consistent with the proposal.39 The activities of hlm A, E and I gene products have been well characterized both biochemically and genetically. The order and mechanisms of the remainder of enzymatic transformations in DTP biosynthesis remain to be further investigated. The oxidation reactions catalyzed by flavoenzymes Hlm B, D and F likely involve new chemistry and are of particular interest for future research.
Figure 4.
Comparison of characterized and cryptic DTP biosynthetic clusters.
4 Mode of action and self-resistance
Dithiolopyrrolones were first isolated as antibiotics and subsequently tested against fungi and mammalian cells. They display broad-spectrum activities against both Gram-positive and Gram-negative bacteria, fungi, and a number of cancer cell lines. Strains and antibacterial activities of several hallmark DTPs are summarized in Table 2. Studies aimed at elucidating a common mechanism of action for dithiolopyrrolones have primarily employed thiolutin and/or holomycin.
Table 2.
Reported antibacterial activities of select dithiolopyrrolones. MICs reported in mg/mL from disc diffusion assays for holomycin (2), thiolutin (4), and thiomarinols A (9) and B(14)2.
| Strains | Holomycin (2) | Strains | Thiolutin (4) | Thiomarinols | A (9) | B (14) |
|---|---|---|---|---|---|---|
| Enterobacter cloacae N1 | 64 | Agrobacterium tumefaciens No 2410 | 100 | AT. pneumoniae 846 (R) | 0.8 | 0.2 |
| Enterococcus faecalis 1 | 4 | Alternaria sp. | 20 | Enterobacter cloacae 963 | 3.1 | 0.8 |
| Escherichia coli DC2 | 0.5 | Bacillus coagulans CIP 6625 | <0.2 | Enterococcus faecalis 68 1 | 0.8 | 0.05 |
| Escherichia coli MG1655 | 0.2 | Bacillus subtilis ATCC 6633 | 2 | Escherichia coli 609 | 1.5 | 0.8 |
| Haemophilus influence Q1 | 0.3 | Candida albicans IPA 200 | 20 | Escherichia coli NIHJ JC-2 | 3.1 | 0.8 |
| Klebsiella pneumoniae E70 | 8 | Escherichia coli ATCC 10536 | >100 | Klebsiella pneumoniae 806 | 1.5 | 0.8 |
| Moraxella catarrhalis 1502 | 0.3 | Fusarium culmorum | 10 | Morganella morganii 1 5 10 | 12.5 | 6.2 |
| Morganella morganii 1 | 64 | Fusarium equiseti | 10 | Proteus vulgaris 1420 | 0.2 | 0.05 |
| Proteus mirabilis C889 | 4 | Fusarium oxysporum f.sp. lini Foln 3–5 | 40 | Pseudomonas aeruginosa 1001 | 0.8 | 0.2 |
| Pseudomonas aeruginosa Dalgleish | 64 | Klebsiella pneumoniae CIP 82.91 | 1 | Pseudomonas aeruginosa No. 7 | 0.8 | 0.4 |
| Staphylococcus aureus 8325-4 | 8 | Micrococcus luteus ATCC 9314 | <0.2 | Pseudomonas aeruginosa PA01 | 0.4 | 0.8 |
| Staphylococcus aureus Carter37 | 2 | Pseudomonas aeruginosa CIP A22 | >100 | Salmonella en teritidis | 1.5 | 0.4 |
| Staphylococcus epidermidis PHLN20 | 1 | Pseudomonas syringae No 1882 | 100 | Serratia marcescens IAM1 184 | 6.2 | 3.1 |
| Streptococcus agalactiae Hester | 2 | Saccharomyces cerevisiae ATCC 4226 | 10 | Staphylococcus aureus 209P JC-1 | <0.01 | <0.01 |
| Streptococcus pneumoniae ERY2 | 0.1 | Salmonella enterica CIP 81.3 | >100 | Staphylococcus aureus 535 (MRSA) | <0.01 | <0.01 |
| Streptococcus pyogenes CN10 | 16 | Staphylococcus aureus CIP 7625 | 20 | Staphylococcus aureus 56R | <0.01 | <0.01 |
4.1 DTPs as inhibitors of transcription
Studies in the 1970s by Tipper and co-workers first revealed the ability of thiolutin to inhibit macromolecular synthesis in Saccharomyces cerevisiae.46 Thiolutin at a concentration of 2–4 µg/mL completely shut down both RNA and protein synthesis in yeast whole cells and spheroplasts.46 The effect on RNA synthesis was immediate, however inhibition of protein synthesis appeared more gradual. The activity of thiolutin was directly tested in a subsequent study against partially purified yeast RNA polymerase I, II and III in vitro and a strong inhibitory effect was observed against all three polymerases.47 These results are consistent with the hypothesis that thiolutin inhibits DNA-dependent RNA polymerases in yeast. Similar studies were carried out in E. coli. As observed in yeast, thiolutin at a concentration of 5 ug/mL rapidly arrested incorporation of radioactive precursors for DNA, RNA and protein synthesis in E. coli.48 However, no activity was detected for thiolutin against E. coli RNA polymerase in vitro. 49 Furthermore, it was also unclear which step of RNA synthesis thiolutin inhibits; results of experiments on beta-galactosidase induction in E. coli have suggested both transcription initiation and chain elongation as potential targets of thiolutin mediated transcription inhibition.49,50 These contrasting results cast doubt on the proposal that RNA polymerase is the primary target of thiolutin. Nonetheless, these early studies provided valuable insights into thiolutin mode of action, demonstrating for the first time that thiolutin is bacteriostatic and that this effect is reversible. Furthermore, the activity appears to be independent of the DNA template or the presence of reducing agents such as DTT.47,48
The mode of action of thiolutin was further investigated by several efforts that screened for spontaneous resistant mutants. Two thiolutin-resistant mutants in E. coli were mapped to two different loci on the genome.51 Spontaneous mutants of Salmonella typhimurium were only conditionally resistant to holomycin in enriched media.52 Whole genome sequencing technologies were not available at the time and therefore the sites of mutations could not be determined unambiguously. The study in S. typhimurium again showed the in vivo activity of thiolutin to inhibit RNA synthesis and the lack of in vitro activities against bacterial RNA polymerases.52 The activity of thiolutin was also tested against five actinomycete strains that each natively produce a RNA polymerase inhibiting antibiotic including tolypomycin Y, geldanamycin, rifamycin, streptovaricin and streptolydigin.53 The results are surprising in that thiolutin displayed strong inhibitory activity against all five strains, which are naturally resistant to their own antibiotics. However, thiolutin again failed to inhibit partially purified RNA polymerase, this time isolated from each of the five strains. This result, together with the lack of cross-resistance of thiolutin with other RNA polymerase inhibitors, seems to suggest that thiolutin has a different mode of action against prokaryotes versus eukaryotes.
Two decades after the original study, Chopra and coworkers revisited DTP mode of action focusing on holomycin. Interestingly, in contrast to the potent activity observed for thiolutin in yeast, holomycin was reported to be completely inactive against S. cerevisiae or Candida kefyr in this study.54 However, holomycin still displayed activity in E. coli, similar to thiolutin. It readily inhibited RNA and protein synthesis in vivo, but only weakly inhibited E. coli RNA polymerase in vitro. The authors considered the possibility that holomycin inhibits the aminoacylation of tRNAs and thereby induces a stringent response. They subsequently ruled out this possibility because holomycin demonstrated similar activity against the ΔrelA mutant, which does not have a stringent response system, as the wild type E. coli strain. To explain these data, the authors suggested that holomycin and thiolutin are pro-drugs that need to be activated in E. coli cells to inhibit transcription. Mechanisms responsible for the difference in activity against eukaryotes and prokaryotes, however, remain elusive. It is also unclear why holomycin and thiolutin display such dramatically different antifungal activities when their structures differ so minimally.
Despite the ambiguities in DTP mechanism of action in bacteria, the inhibitory effects of thiolutin against eukaryotic RNA polymerases have been exploited in measurements of mRNA stability in budding yeast, fungi and dinoflagellate.55–59 In these studies, thiolutin is used to stop transcription and prevent the formation of new mRNA. The levels of existing mRNAs can be determined at different time points after thiolutin treatment to calculate the half lives of these mRNAs. For instance, thiolutin was used as one of the five chemical inhibitors of transcription in a genome-wide analysis of mRNA stability in Saccharomyces cerevisiae.56 The global transcript stability profiles of wild type yeast following chemical treatment were compared to that of a temperature sensitive yeast RNA polymerase mutant under heat shock conditions. While the stability profiles are different for the chemical vs. heat shock stress conditions, two of the chemicals tested, thiolutin and 1,10-phenanthroline, displayed similar global effects. 1,10-Phenanthroline is a metal chelator that is thought to inhibit RNA polymerase activity by sequestering magnesium. The authors suggest that it is possible that thiolutin utilizes a similar metal chelation mechanism of action, but no further experimental evidence is provided. Interestingly, thiolutin was also reported to inhibit mRNA degradation, which activity potentially conflicts with its use in RNA stability studies.60 Thus, the use of thiolutin in a handful of such RNA stability remains suggestive of a polymerase mediated mechanism of action, at least when it comes to yeast and fungi.
4.2 DTP self-resistance
Resistance mechanisms that ride along side of the natural product biosynthetic pathway in producer cells are also important to hypotheses on the mechanism. Additional insights into the mode of action of DTPs have been garnered from studies on their biosynthesis and self-resistance. Thioredoxin oxidoreductase-like enzyme, HlmI, was characterized as a dithiol oxidase that efficiently converts the dithiol to a disulfide in holomycin biosynthesis.26 Deletion of the hlmI gene in the producer abolished the production of holomycin and also resulted in a phenotype that is hypersensitive to exogenous holomycin. It appeared that HlmI is not only involved in holomycin biosynthesis but also self-resistance. One hypothesis is that holomycin acts as a pro-drug and the disulfide becomes reduced in the reducing environment inside the target cells to generate red-holomycin, the active form of the antibiotic and also the penultimate metabolite in the holomycin pathway. HlmI keeps holomycin disulfide in the oxidized form in the producer S. clavuligerus and protects the producer from the action of the antibiotic. To test this hypothesis and investigate how hlmI deletion mutants cope with the toxicity of red-holomycin, differential metabolomic profiling was carried on hlmI deletion mutants along with wild type S. clavuligerus and a holomycin-overproducer by high-resolution mass spectrometry and NMR.61 These profiling experiments did not detect any red-holomycin, instead uncovering a number of novel metabolites unique to the ΔhlmI mutants including mono- and di-S-methylated species at the reduced enethiols (Figure 5a). An intriguing thioether adduct was also observed in significantly higher quantity in ΔhlmI strains than in the wild type or holomycin overproducer. These results revealed the reactivity of the endo- and exo-cyclic ene-thiols in red-holomycin, which can be incapacitated by S-methylation or dimerization in the bacteria as a back-up plan for self-protection; these data further support a role of the reduced ene-thiols in the antimicrobial mechanism of action of holomycin.
Figure 5.
Mechanisms of bacterial resistance to dithiols. a) Shunt products isolated from hlmI knock-outs and b) methylated gliotoxins isolated from producer strains.
An analogous role for the gliotoxin dithiol oxidase, GliT, was previously demonstrated. Deletion of GliT in a gliotoxin producer renders the fungus extremely sensitive to gliotoxin and gliotoxin non-producers gains resistance to this toxin by expression of GliT.28,62 Additionally, the isolation of bismethyl thiol forms of gliotoxin biosynthetic intermediates that are substantially less toxic than gliotoxin presents another example of thiol methylation as a detoxification strategy (Figure 5b).63,64 Besides disulfide reduction, it is also possible that DTPs are activated in some target cells through sulfur oxidation to thiosulfonate, which have been isolated as variants for xenorhabdins and thiomarinols. An E. coli mutant with a heightened sulfur oxidase activity was reported as hypersensitive to thiolutin. 65 Whether activated by reduction or oxidation, the disulfides in DTPs are clearly important for activities. These studies, though not identifying specific targets, provide an important chemical perspective to the reactivity of DTPs and potential mechanisms of action.
An interesting yet somewhat unexpected self-resistance mechanism was revealed for holomycin in Gram-negative bacterial producer Y. ruckeri and suggested an alternative mode of action for this antibiotic.17 This particular holomycin gene cluster encodes an rRNA (Guanine-N1)methytransferase-like enzyme Hom12, which appeared to be involved in self-resistance, as opposed to the dithiol oxidase-like resistance protein found in the S. clavuligerus holomycin pathway. The ability of Y. ruckeri to survive treatment of high concentration of holomycin was abolished by deletion of hom12 and restored by complementation with hom12 on a plasmid. Overexpression of this gene in E. coli provides protection against holomycin treatment to a previously holomycin-sensitive strain. The authors concluded based on these results that Hom12 methylates RNA as a resistance mechanism against the action of holomycin to inhibit transcription. However, rRNA methyltransferases are often found as immunity proteins for antibiotics that exert their mechanisms of action through inhibition of the ribosome. For example, chloramphenicol/florfenicol resistance gene (cfr) and erythromycin resistance gene (erm) both encode for SAM-dependent methyltransferases that modify the 23S rRNA of the 50S ribosome and prevent the antibiotics from binding to their ribosomal targets.66 The discovery of an rRNA methyltransferase as immunity gene in the holomycin pathway suggests that holomycin acts as an inhibitor of translation rather than transcription. Interestingly, the holomycin gene cluster from S. clavuligerus does not contain any hom12 homolog, whereas a dithiol oxidase-like enzyme homologous to hlmI could not be located in the genome of Y. ruckeri. It appears that the Yersinia and Streptomyces producers utilize two alternative and perhaps complementary resistance mechanisms.
Of particular interest is the mode of action and resistance mechanism of thiomarinol, a DTP-polyketide hybrid antibiotic. The marinolic acid portion of thiomarinol is very similar in structure to mupirocin, which has been shown to target isoleucyl tRNA synthetase. It is curious whether the addition of a DTP moiety alters the target or specificity of this antibiotic. The thiomarinol cluster in Pseudoalteromonas encodes a putative Ile-tRNA synthetase TmlM, homologous to the mupirocin resistance protein MupM.33 Expression of tmlM in E. coli confers high level of resistance to thiomarinol, but mupM expression only led to minimal resistance. Similar finding was reported against methicillin-resistant Staphylococcus aureus (MRSA) that contains a copy of Ile-tRNA synthetase highly resistant to mupirocin. While mupirocin was completely ineffective towards this strain, thiomarinol displayed some level of activity, which was clearly lower than its activity against mupirocin-sensitive S. aureus strain. The cross resistance between thiomarinol and mupirocin suggests that the former shares the same target as the latter but is much more active. The DTP portion of thiomarinol may provide additional contact to Ile-tRNA synthetase or serve as a mechanism-based inhibitor to irreversibly inactivate this enzyme.
4.3 Activity and Mode of Action Against Cancer
Cytotoxicity is described in some of the early reports on DTP isolation, but little regarding their anticancer effects can be found prior to a report by Ishizuka and co-workers at the Microbial Chemistry Research Foundation.67 This group identified thiolutin, aureothricin, and thioaurin in a screen of low-molecular-weight natural products able to inhibit adhesion of human umbilical vein endothelial cells (HUVECs) to vitronectin. Of these compounds, thiolutin inhibited adhesion of HUVECs to vitronectin most effectively, with an IC50 of 0.83 µM. In vivo experiments showed that thiolutin significantly suppressed angiogenesis induced by tumor cells in a mouse dorsal air sac model. To explore the mechanism of inhibition the researchers examined the effect of thiolutin on intracellular cell adhesion signaling. They found that the amount of paxillin in HUVECs was significantly reduced by thiolutin treatment, while those of other focal adhesion proteins including vinculin and focal adhesion kinase (FAK) were not. Metabolic labeling experiments further showed that thiolutin enhanced degradation of paxillin in HUVECs and protease inhibitors (MG115 and E64-D) were capable of partially decreasing the rate of paxillin degradation, restoring thiolutin-induced inhibition of HUVEC adhesion to vitronectin.
Following up on the antiangiogenic activity of thiolutin, Karger and Roberts found that thiolutin inhibits endothelial cell adhesion by perturbing interactions with components of the actin and intermediate filament cytoskeleton.68,69 Using a series of immunoprecipitation and LC/MS experiments they demonstrated first that Hsp27 phosphorylation in endothelial cells is stimulated by treatment with thiolutin and subsequently that this effect specifically ablates interactions with nestin and collapses nestin filaments. Increased Hsp27 phosphorylation is a conserved response of endothelial cells to at least seven other angiogenesis inhibitors, but thiolutin proved exceptionally active, stimulating robust phosphorylation even at concentrations of < 1 µM. The inhibitory effects of thiolutin on endothelial cell adhesion were shown to be transient, potentially due to a compensatory increase in Hsp27 protein levels. Additionally, thiolutin-mediated Hsp27 phosphorylation was shown to require p38 kinase, but is itself independent of increased p38 phosphorylation.
The antimitotic activity of VD844 (6) was uncovered in a study that aimed to isolate the cytotoxic component from an ascidian-associated Streptomyces strain.70 Bioactivity-guided fractionation of this strain yielded an orange crystalline compound, whose structure corresponds to that of VD844. As described in Section 2.2, VD844 was first isolated in 1969 as a potent antimicrobial.3 In the 2014 study, VD844 was shown to inhibit the growth of metastic prostate cancer cells at low µM concentration. Further analyses revealed that VD844 treatment results in aberrant cell cycle and accumulation of polynucleated cells. Therefore, VD844 was thought to modulate cytokinesis and induce apoptosis in cancer cells. The expression level of several proteins involved in cytokinesis, including polo-like kinase 1 (Plk1) and protein regulator of cytokinesis 1 (Prc1), appeared to be reduced after VD844 treatment.
Work on anticancer activities of DTPs demonstrated the therapeutic potential of this class of natural products in cancer treatment. Although these studies were carried out in different cancer cell lines and disease models and different DTP analogues were utilized, it is clear that DTPs have complex pharmacological effects in cancer. The underlying molecular mechanisms responsible for the observed antiangiogenic and antimitotic effects of DTPs remain to be elucidated.
5 Total synthesis of naturally occurring dithiolopyrrolones and analogues
Since the definitive structural assignments of thiolutin and aureothricin in 1955 and the reported crystal structure of VD-844 in 1960, there have been eight reported total syntheses of DTPs in the literature, giving rise to several reports of substantially modified analogues and a number of patents. The majority of syntheses build the top half pyrrolone by variations on either Dieckman cyclizations or else Knorr-type condensations with further elaboration by installation of the lower half sulfur moieties or exocyclic amide at varying stages of the synthesis. Variations on this theme have allowed differential substitution patterns at the three “non-quaternary" carbons in thiolutin (C3, N4, and the exocyclic acyl chain), as well as substitutions at many of the heteroatoms.
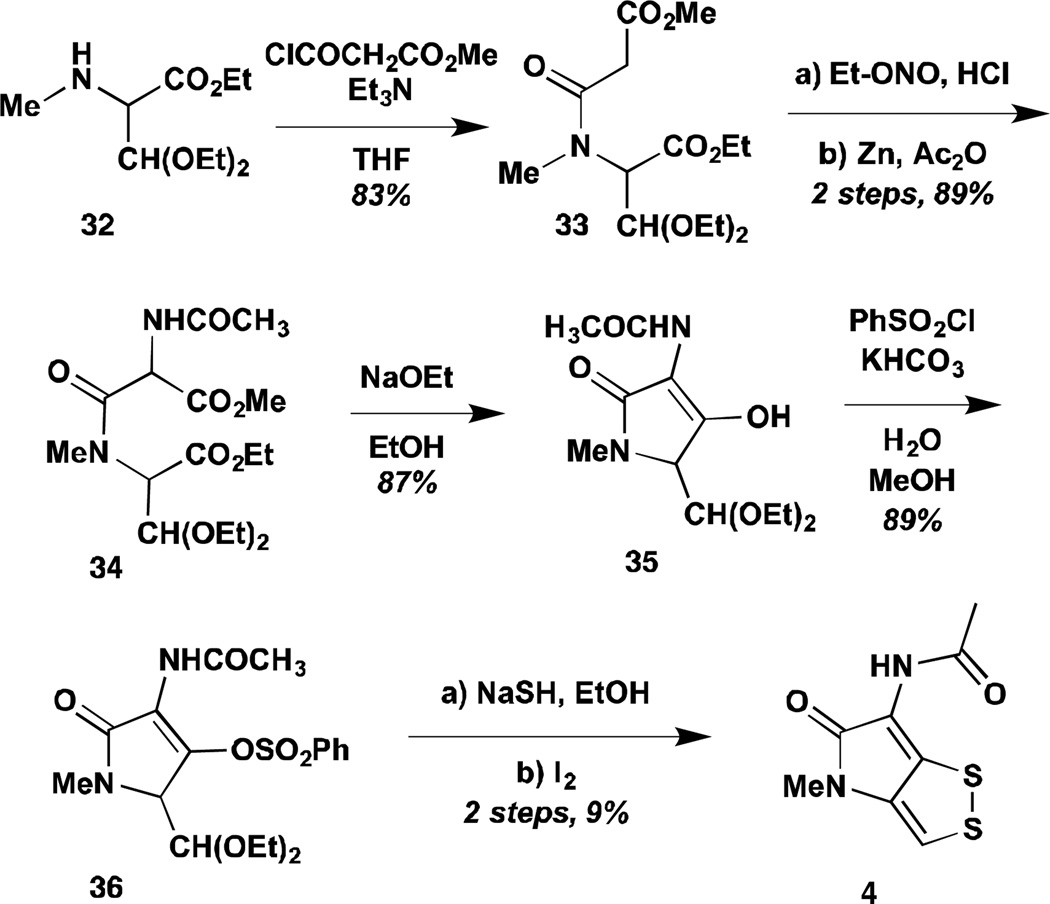
Schmidt and Geiger reported the first total synthesis of a dithiolopyrrolone in 1962.71 This route was deployed to access thiolutin, holomycin, and aureothricin (Scheme 2) via the previously reported deacylation protocol used to determine the structures of several of these compounds. Thus, N-methyl-1-ethoxycarbonyl-2-diethoxyethylamine could be condensed with methoxycarbonylacetyl chloride to give acetal 33. Nitrosation and acetylative reduction led to the pre-Dieckman substrate, which cyclized in the presence of sodium ethoxide. Subsequent tosyl protection and treatment with sodium sulfide installed the two thiols, which could at last be oxidized with catalytic iodine. The authors later reported improved yields by early stage thiolysis of acetal 33 with thioacetic acid to give the bis-acetado-dithioacetal.72
Scheme 2.
Total synthesis of thiolutin by Schmidt & Geiger.
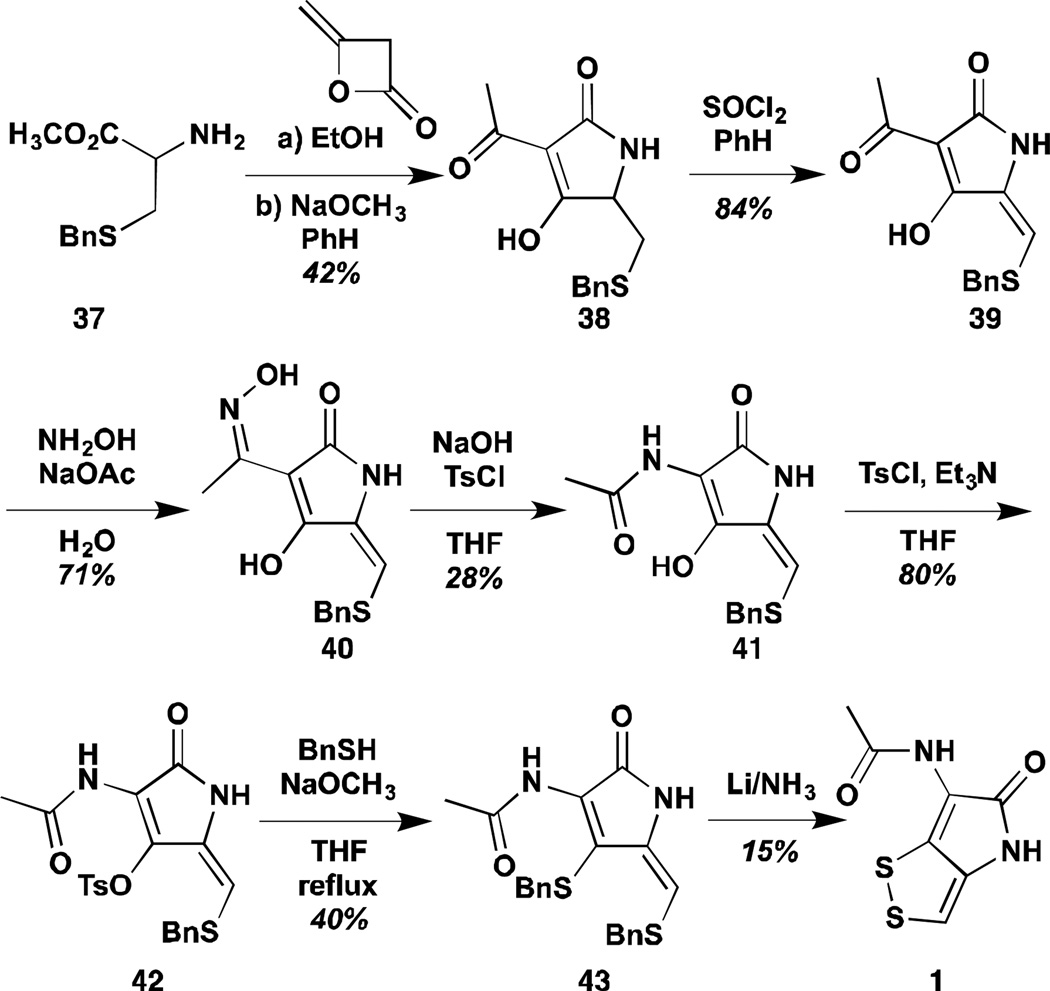
Shortly after the Schmidt synthesis, Buchi and Lukas reported preparation of holomycin by a seven-step process proceeding from benzyl protected cysteine (Scheme 3).73,74 Amidation of the cysteine nitrogen with diketene was followed in one-pot by cyclization/condensation. The exo-cyclic ketone of the thus constructed tetramic acid 38 could then be transformed to the requisite amide via an elegant Beckman rearrangement. The remaining steps closely resemble the Schmidt synthesis with exception of the use of benzyl protecting groups for the thiols. The latter could be removed with dissolving metal to provide 18 mg of holomycin in 0.3% overall yield without any chromatographic purification steps.
Scheme 3.
Total synthesis of holomycin by Buchi & Lukas.
A group led by Ian Harrison at Syntex reported a synthesis of holomycin in 1977 (Scheme 4).75,76 Theirs is a hybrid between the Schmidt pyrrolone strategy and the aminolysis chemistry developed by Hagio. Additionally, the Syntex synthesis uses an early stage dithioketal protection of the disulfide after the method developed by Kishi. Thus, the dithioketal is formed by condensation of methythioglycolate with p-methoxyacetophenone and subsequent Dieckman cyclization. Aminolysis in liquid ammonia provided the enamine 47. This could be acylated with methoxyalyl chloride and cyclized to the pyrrolinone in sodium amide. Harrison and co-workers report that direct replacement by the amino group did not occur on treatment with ammonia. Instead, preparation of the mesylate allowed reaction with higher amines, such as p-methoxybenzylamine. Oxidative deprotection of the dithiane and exo-cyclic amine, followed by acylation gave the carboxy-analogue of holomycin 52. Lastly, decarboxylation under Krapcho conditions yielded holomycin.
Scheme 4.
Syntex synthesis of holomycin.
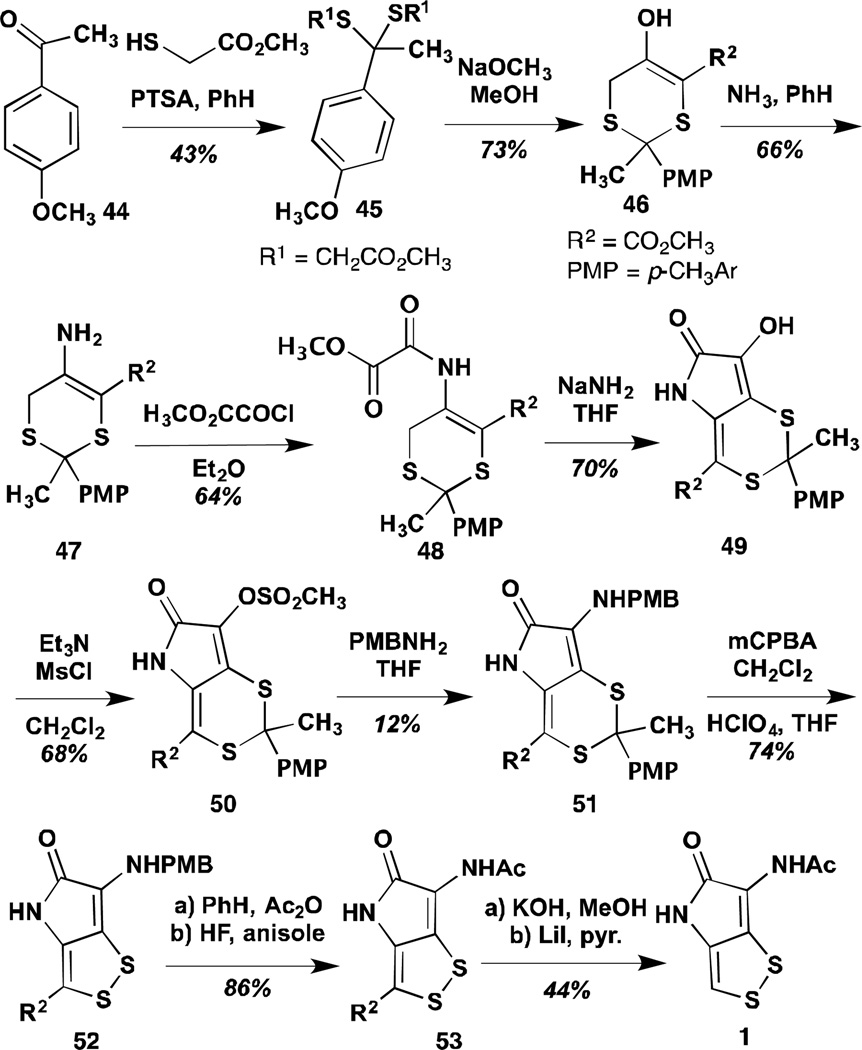
Stachel and co-workers accessed thiolutin by two distinct routes, reported concurrently (Scheme 5).77 Although brutal in terms of their step-count as well as many of the yields of key steps, the Stachel syntheses allowed this group to access the most diverse array of analogues, structurally and electronically, reported to date. Analogues were generated as intermediates in many steps of the synthesis. The shorter of the two Stachel syntheses begins with 1,2-dithiole-3-thione 54, prepared by an undisclosed method, and proceeds by homologation to give a mixture of cis- and trans-conjugate esters. Acid catalyzed lactam formation gave thiolutin analogue 56, which underwent smooth nitration to yield analogue 57. The nitro group could be further reduced and acetylated to yield the N-methyl analogue of Harrison intermediate 58, which could again be decarboxylated to give, in this case, thiolutin. The second route reported by the Stachel group initiates from a dithiane, similar to the Syntex work. A sequence of alkylation, aminal formation and cyclization set up the substrate 61 with two tertiary alcohols ready to undergo elimination to the dithiane protected analogue, 63. Acetylation to give the β-keto amide again set up a late stage Beckman rearrangement, as in the Buchi synthesis and final deprotection of the dithiane yielded thiolutin. Stachel and co-workers deprotected many of the intermediates from this synthesis to employ in biological testing of analogues. These data are discussed in a later section on analogues.
Scheme 5.
Stachel syntheses of thiolutin
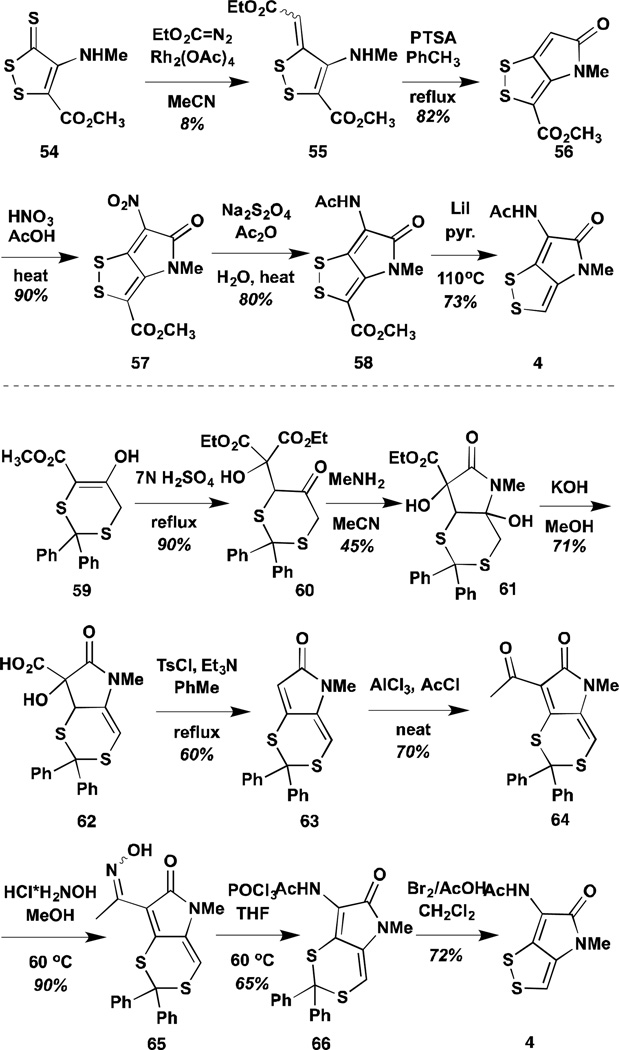
Concurrent with the work of Stachel and co-workers, a group from Ciba-Geigy reported a short and versatile synthesis that could access thiolutin in five steps and 18% overall yield from 1,3-dichloroacetone, a relatively inexpensive starting material.78 There are two key advances over prior art in the Ciba-Geigy synthesis: 1) early stage introduction of the sulfur heteroatoms and 2) a one-pot imine formation, acylation, and cyclization, promoted by titanium tetrachloride (Scheme 6a). Similar to the Hagio synthesis, the Ciba-Geigy synthesis uses molten ammonium acetate to convert the 3-hydroxy lactam to a 3-amino lactam. This product could be acylated and deprotected under mild conditions to furnish thiolutin and analogues. In addition to the N-methyl thiolutin substitution pattern the researchers sought to incorporate an aromatic substituent on the pyrrolone nitrogen. However, under the harsh conditions necessary to affect alcohol to amine conversion, the pyrrolone underwent transamidation to yield the unsubstituted pyrrolone core of holomycin (bottom of panel in Scheme 6a).
Scheme 6.
Modern syntheses of dithioyrrolones. a) Ciba-Geigy synthesis, b) Nielsen synthesis, and c) Welichem synthesis.
The Ciba-Geigy sequence has proven the most robust to date and has been adapted and improved upon by several groups in recent years to generate analogues. Nielsen and co-workers replaced the tert-butyl protecting groups used in the Ciba-Geigy synthesis with para-methoxybenzyl groups and, similarly, employed para-methoxybenzylamine in the pyrrolone formation reaction to yield substrates that could be fully deprotected in refluxing trifluoroacetic acid (Scheme 6b).79 The latter deprotection has the benefits of installing the disulfide and trifluoro-acetylating the exo-cyclic amine, which protecting group can be further removed under milder conditions than standard acetate to allow variation of the acyl chain substituents
A group of researchers from Welichem, Burnaby, BC, also employed the trifluoro-acetylation strategy, but installed it with trifluoroacetic acid in advance of thiol deprotection with mercury(II)acetate and iodine (Scheme 6c).80 This provides a cumulative increase in yield over the two steps for deprotection and acylation. Another significant modification developed by the Welichem researchers is aminolysis of the 3-hydroxy lactam in molten ammonium butyrate. Presumably the lower melting point of the ammonium butyrate and its greater hydrophobicity dissolve the substrate and effect the reaction at a lower temperature, which prevents the transamidation reaction with anilide substituents that had previously been reported by the Ciba-Geigy group. Thus, the Welichem group has been able to report a variety of N-aryl analogues.
6 Analogue Synthesis and Activity
As described above, the exact mechanism of action of DTPs is not well understood, although several targets have been advanced in the literature. Nonetheless, the somewhat eccentric electronic architecture of the DTPs has led a number of groups to try and accentuate or modify the measureable phenotypic activity by altering the DTP core structure. Unfortunately, there has been relatively no conservation of assay criteria amongst the studies and there is little basis to compare many of the different analogues.
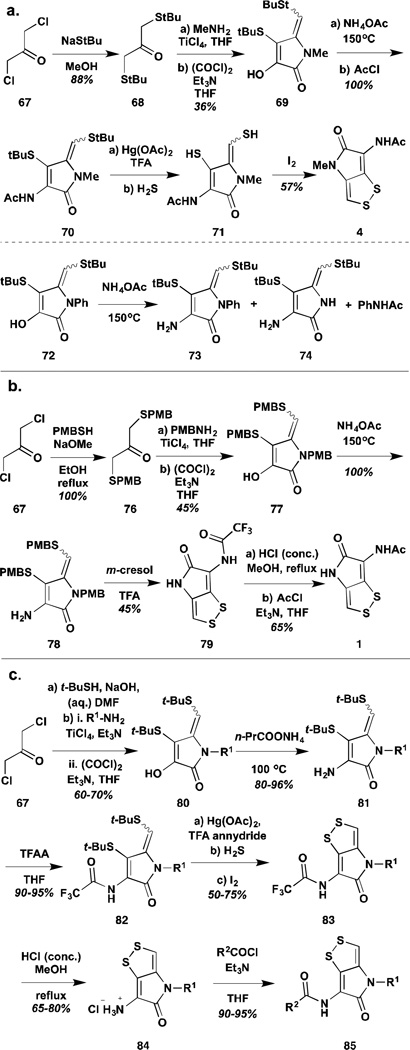
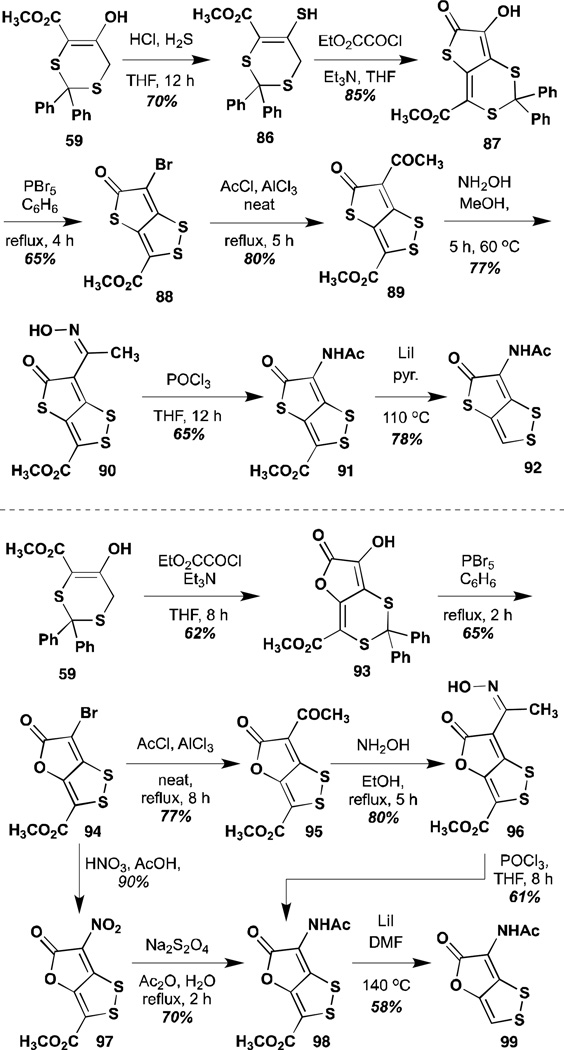
The most electronically diverse set of analogues has come from the efforts of Stachel and co-workers. In their series of six papers, proceeding from their initial synthesis of thiolutin, Stachel and co-workers published close to three-dozen analogues, including substitutions of the exo-cyclic nitrogen, the pyrrolone amide, and portions of the disulfide itself.77,81–84 With respect to the pyrrolone amide, they published two major classes of analogues: the thieno-1,3-dithienes, substituting the ring nitrogen with a sulfur atom, and the related oxygen substitution in the 1,3-dithiino-furans (Scheme 7).81,82 Synthesis of these thia- and oxa- analogues proceeded in similar fashion, from the protected intermediate 59. Substitution, in the case of the thia- analogue subsequently gave a substrate capable of undergoing cyclization to the variant pyrrolone core. Group transfer/replacement reactions, similar to those used in their thiolutin synthesis gave the final products.
Scheme 7.
Syntheses of thia- and oxa-analogues of holomycin.
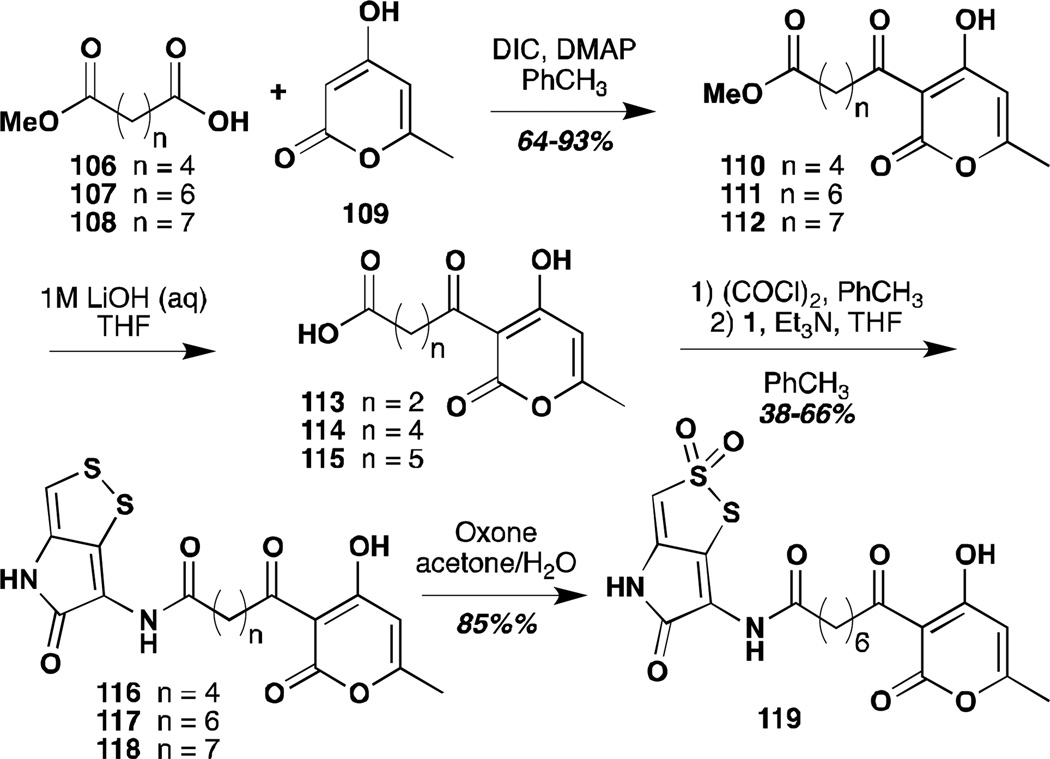
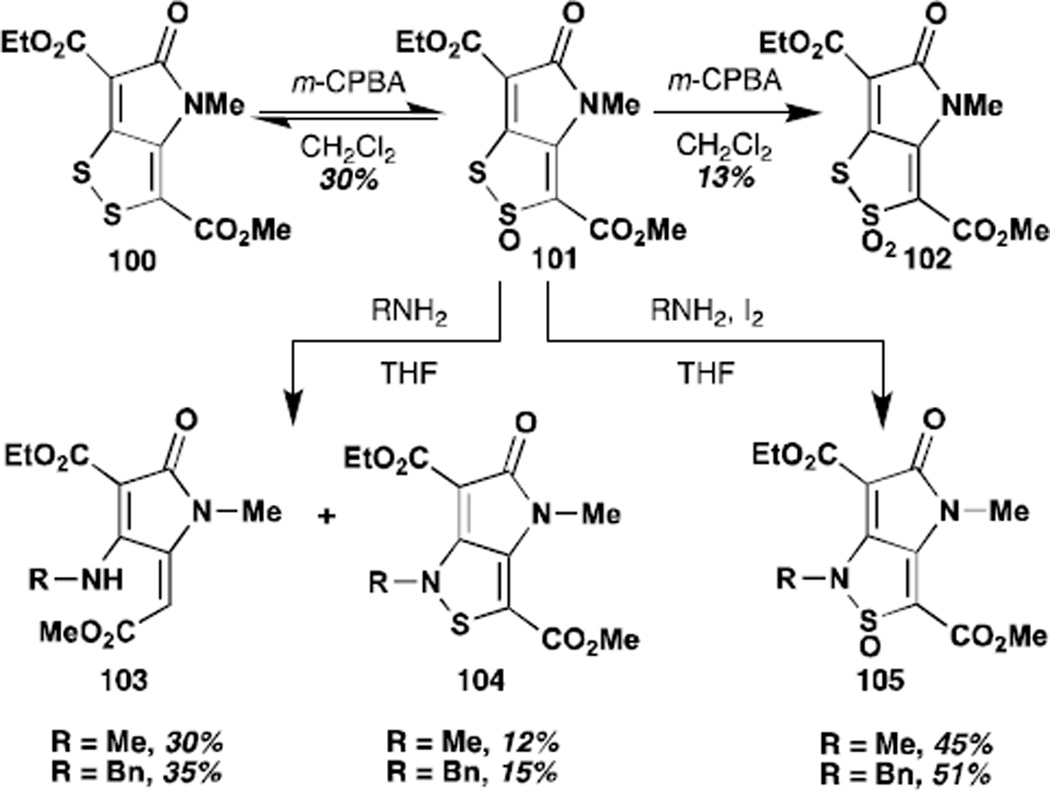
The Stachel group also took up the question of the activity of S-oxides of their DTP analogues.83 Only very few experiments were reported with holomycin or thiolutin themselves. Holomycin underwent sluggish oxidation with m-CPBA to give the S(2)-dioxide in low to moderate yield. The subsequent reactivity of this compound is not presented. In contrast, several of the more electron deficient analogues underwent facile oxidation to both the S(2)-monoxides and the S(2) and S(1)-dioxides. In particular, diester 100 underwent oxidation to the monoxide with stoichiometric m-CPBA and further oxidation to the thiosulfonate in presence of generous excess. In both cases, the regioselectivity of oxidation was rigorously demonstrated by x-ray crystallography. Both of these compounds were reported to undergo facile disproportionation. The monoxide 101 could be readily reduced with 1,2-dimethylhydrazine or thiophenol. Whereas the diester itself and the dioxide were impervious to treatment with amines under most conditions, the monoxide reacted to give the isothiazole, a process that could be further accelerated by addition of iodine. A mechanism is proposed, but no further evidence is provided. Similar reactivity was reported for donor substituted N-alkyl pyrrothines and their thia-analogues. The authors suggest that this reactivity profile could be relevant to the biological mechanism of action of the DTPs, but the corresponding experiments with unsubstituted DTPs are not reported. Thus the biological relevance of this reaction manifold remains speculative at best.
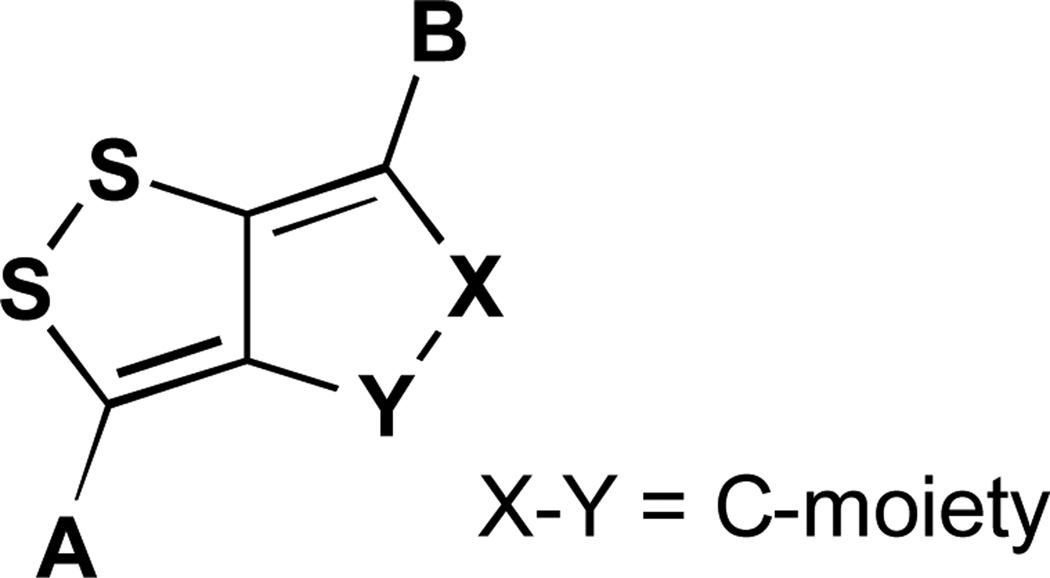
Unfortunately, these efforts have led to only moderate insights into structure-activity relationships as compounds have been deployed in only one assay to date.85 Nonetheless, working with this compound set, Stachel has demonstrated the importance of the disulfide moiety in activity against M. tuberculosis (Mtb). The results of these structure activity relationships are listed in Table 3. Several intermediates from the syntheses described above are also included in the table. Many of the analogues tested approach isoniazid in activity against Mtb and are much more effective against M. kansasii and M. avium.
Table 3.
Antimycobacterial activity of dithiolopyrrolone analogues (minimum inhibitory concentrations, MIC, µmol 1−1)
| Compound | Molecular Fragment |
MIC, µM |
|||||
|---|---|---|---|---|---|---|---|
| A | B | C | M. tuber. | M. kans. | M. avium. | M. fortuit. | |
| 88 | COOCH3 | Br | –SCO– | 60 | 60 | 60 | 250 |
| 91 | COOCH3 | NHCOCH3 | –SCO– | 60 | 30 | 500 | – |
| 92 | H | NHCOCH3 | –SCO– | 125 | 250 | 250 | 500 |
| 94 | COOCH3 | Br | –OCO– | 125 | 125 | 500 | >500 |
| 98 | COOCH3 | NHCOCH3 | –OCO– | 250 | 250 | 250 | >500 |
| 101 | COOCH3 | COOC2H5 | –SCO– | 30 | 30 | 30 | – |
| 102 | COOCH3 | OC2H5 | –SCO– | 30 | 30 | 30 | – |
| 103 | COOCH3 | COOCH3 | –N(CH3)SO2– | 60 | 60 | 250 | >500 |
| 104 | H | COOCH3 | –N(CH3)SO2– | 60 | 60 | 250 | 500 |
| 105 | COOCH3 | CONHNH2 | –N(CH3)SO2– | 500 | 500 | 500 | >500 |
| 106 | COOCH3 | NHCOCH3 | –N(CH3)SO2– | >500 | >500 | >500 | >500 |
| 107 | H | NHCOCH3 | –N(CH3)SO2– | 250 | 500 | 500 | 250 |
| 108 | H | OH | –SCO– | 60 | 60 | 60 | 250 |
| 109 | COOCH3 | H | –N(CH3)CO– | 30 | 30 | 60 | 60 |
| 110 | COOCH3 | Br | –N(CH3)CO– | 30 | 30 | 125 | 125 |
| 111 | COOCH3 | NO2 | –N(CH3)CO– | 250 | 250 | 500 | 500 |
| 112 | COOCH3 | COOCH3 | –N(CH3)CO– | 125 | 125 | 125 | >500 |
| 113 | COOCH3 | OC2H5 | –N(CH3)CO– | 125 | 125 | 60 | 500 |
| 114 | COOCH3 | OCH3 | –NHCO– | 60 | 60 | 500 | >500 |
| 115 | COOCH3 | SO2NH2 | –NHCO– | >500 | >500 | >500 | >500 |
| 116 | COOCH3 | Br | –NHCO– | 30 | 30 | 125 | 125 |
| 117 | COOCH3 | SCOOC2H5 | –NHCO– | 60 | 60 | 500 | 500 |
| 120 | COOCH3 | COOC2H5 | –OCO– | 60 | 60 | 125 | 125 |
| 121 | COOCH3 | OC2H5 | –OCO– | 125 | 250 | 125 | 500 |
| 122 | COOCH3 | NO2 | –OCO– | 250 | >500 | >500 | >500 |
| 123 | H | NO2 | –SCO– | 30 | 60 | 125 | 125 |
| 124 | COOCH3 | NHCH2C6H5 | –SCO– | 500 | >500 | >500 | >500 |
| 125 | COOCH3 | H | –SCO– | 7 | 7 | 60 | 125 |
| 126 | COOCH3 | OH | –SCO– | 125 | 125 | 125 | >500 |
| 128 | COOCH3 | NO2 | –SCO– | 60 | 60 | 30 | 250 |
| 130 | COOCH3 | COOCH3 | –N(CH3)SO2– | 500 | 500 | >500 | >500 |
Welichem deployed their improved synthesis to generate a number of N-aryl derivatives of holomycin.80,86 They additionally varied the N-acyl substituent, incorporating a number of aromatics in this position as well. Compounds were examined for antiproliferative activity in two actively growing human cancer cell lines, H460 (lung) and LCC6 (breast). While the natural dithiolopyrrolone from Xenorhabdus sp. used as a standard in the studies, exhibited high nanomolar activity, several analogues exhibited much more potent efficacy. The whole set of N-aryl analogues was submitted for biological profiling in the National Cancer Institute (NCI) panel of 60 cancer cell lines. Compounds were selective for solid cancer cells over haematological cancer cells, but NCI COMPARE algorithm yielded no significant activity correlations with compounds in the NCI database, suggesting that the dithiolopyrrolones act through a unique mechanism of action.
Wang and co-workers expanded on the Welichem results with N-aryl substituted analogues by incorporating N-carbonyl groups on the exo-cyclic nitrogen, presumably to improve solubility of the otherwise lipophilic moiety.87 The original compound, ZL-004, was initially disclosed in the patent literature, with subsequent analogues reported in the primary literature in 2013. While the investigators did not report a naturally occurring DTP, such as holomycin or thiolutin in their assays, they did demonstrate potent elevation of leukocyte counts in normal mice peripheral blood relative to recombinant human granulocyte colony stimulating factor (rhG-CSF) as a positive control. Their phenyl-carbamoyl analogue further demonstrated antileukopeania activity in a cyclophosphamide (CTX)-treated mouse model of leukopenia.
Given the pronounced activity of thiomarinol, a hybrid antibiotic comprised of holomycin coupled to marinolic acid, it seems likely that the DTP core could be incorporated into other such hybrids by combination with other novel antibiotics.88 Thus, Hayashi and co-workers have constructed a number of new DTP-based antibiotics by coupling holothin to myxopyronine analogues (Scheme 9). Hybrid compound 115 exhibited substantially improved antimicrobial activity over the relevant myxopyrrone subcomponent, but diminished activity relative to the DTP fragment. Importantly, compound 115 effected micromolar activity (14 µM) in vitro against purified E. coli RNA polymerase, the natural target of myxopyronins.
Scheme 9.
Synthesis of myxopyrrin/holothin hybrids.
7 Summary and Outlook
Dithiolopyrrolones (DTPs) have a long history, though they have failed to induce the kind of protracted medicinal campaigns that have followed so many other natural products. They exhibit reasonably potent and broad-spectrum antibiotic activity, which is mitigated by moderate to strong toxicity. DTPs have an extremely compact molecular architecture, the majority of which is taken up by the electronically unique bridged disulfide ring that is believed to be their primary pharmacophore. One can speculate that the synthetic challenge of this core prevented intensive efforts on improvement of their pharmacology during the early years, and that the lack of a readily definable molecular target dampened enthusiasm to further explore these molecules in the modern days. Yet strong incremental advances have been reported since the DTPs were very first isolated and, in recent years, several key discoveries have contributed to renewed interest in this intriguing class of molecules. First and perhaps foremost, the discovery of the thiomarinols has inspired imagination of the potential for a viable DTP-based antibiotic: thiomarinols represent not only an extremely rare example of a naturally occurring hybrid antibiotic, but a hybrid which is primarily comprised of an already FDA-approved fragment analogue, pseudomonic acid. Second, the more recent elucidations by several groups, of several dithiolopyrrolone biosynthetic pathways have yielded substantive insights into the mechanisms of resistance and, thereby, mechanisms of action of DTP group antibiotics. Such insights will undoubtedly help in further considerations of the precise molecular targets of DTP action. Third, as the result of numerous, compounded efforts, rapid, scalable, and manipulatable syntheses of the dithiolopyrrolones and many of their substantially modified analogues have been developed and deployed to garner initial structure-activity-relationships.
One important question that has not yet been fully addressed in the DTP literature is that of the nature of the toxic form of the DTP core and what chemical reactivity, if any, might attend the relevant metabolites. Studies with the holomycin dithiol oxidase HlmI have suggested that the availability of the potentially nucleophilic ene-dithiol of reduced DTPs is relevant to their inhibitory activity. This hypothesis is further strengthened by the observation of methylated variants in isolates from an HlmI knockout strain; methylation could be a strategy to blunt the free thiols of reduced DTPs. Even so, there is the further question of the electrophilicity of the conjugated eneamide. In its oxidized form, conjugate addition to the eneamide would invoke the penalty of disrupting DTP aromaticity. Under strongly reducing conditions, however, in the ene-dithiol form, the eneamide would likely be available for conjugate addition. Such reactivity could potentially explain the observation of dimer by-product 30 in HlmI isolates. Alternatively, the reduced disulfide has at least the potential to engage in metal chelation, although the close proximity of the two sulfur atoms in the constrained five membered ring suggests that this would be a tight fit. Lastly, a different kind of redox activity is also available to sulfur atoms, especially within the sulfur economy of the cell, that is conversion to oxides of higher state by reactive oxygen species or other molecular oxidants. This potential pathway is exemplified by the activity of the thiosulfonate variant of thiomarinol, thiomarinol B (14). Although the compound was isolated in the thiosulfonate oxidation state, its very existence suggests that other levels are possible, such as the thiosulfinate and the sulfinic or sulfonic acid. Stachel and co-workers demonstrated the extreme electrophilicity of thiosulfonates of the tetramate analogues of holomycin and thiolutin. However, the reactivity of the DTP thiosulfonate itself was not examined and it is not easily predictable. Metabolism of dithioles by mitochondrial cytochrome P450s is well known and it can be imagined, in the case of a DTP, that oxidation, either chemical or enzymatic, could provide an alternate route to disrupting the aromatic stabilization of the core. Such oxidation is important to the action of other dithiole containing compounds, such as leinamycin.
These multiple manifolds of potential reactivity highlight the large gaps in our current knowledge of this group of natural products and some of the problems that must be considered in any future endeavours to understand their biological activity. Still, substantial synthetic advances have put materials in the hands of researchers and undoubtedly, the tools have been fashioned and the stage is set to trade in on the promise of compounds like thiomarinol or variants thereof. It can equally be imagined that the elucidation of DTP biosynthetic pathways will give rise to identification of ever more profoundly modified variants, like thiomarinol, from genome mining and heterologous expression efforts.
Figure 6.
Generic structure of compounds from Table 3.
Scheme 8.
Reactivity of S-oxidized analogues of holomycin.
Acknowledgements
We thank Dr. Scott E. Allen (UNC) for aid with figures and careful reading of the manuscript. We also thank Christopher Neumann (Seattle Genetics) for helpful discussions. Research in the laboratory of B. L. is supported by University of North Carolina at Chapel Hill and NIH grant 4R00GM099904-02. We sincerely acknowledge all of our colleagues for their work cited in this review.
Contributor Information
Bo Li, Email: boli@email.unc.edu.
Christopher T. Walsh, Email: christopher_walsh@hms.harvard.edu.
Albert A. Bowers, Email: abower2@email.unc.edu.
Notes and references
- 1.Jensen BB. The Journal of Antibiotics. 1969;22:231–232. doi: 10.7164/antibiotics.22.231. [DOI] [PubMed] [Google Scholar]
- 2.Jensen B. Acta Crystalographica. 1971;27:392–400. [Google Scholar]
- 3.Von Daehne W, Godtfredsen WO, Tybring L, Schaumburg K. The Journal of Antibiotics. 1969;22:233. doi: 10.7164/antibiotics.22.233. [DOI] [PubMed] [Google Scholar]
- 4.Umezawa H, Maeda K, Kosaka H. Japan. Med. J. 1948;1:512–517. [Google Scholar]
- 5.Celmer WD, Tanner FW, Harfenist M, Lees TM, Solomons IA. J. Am. Chem. Soc. 1952;74:6304–6305. [Google Scholar]
- 6.Celmer WD, Solomons IA. Antibiot. Annu. 1953:622–625. [Google Scholar]
- 7.Celmer WD, Solomons IA. J. Am. Chem. Soc. 1955;77:2861–2865. [Google Scholar]
- 8.Ettlinger L, Gaumann E, Hutter R, Keller-Schierlein W, Kradolfer F, Neipp L, Prelog V, Zahner H. Helv. Chim. Acta. 1959;42:563–569. [Google Scholar]
- 9.Kenig M, Reading C. The Journal of Antibiotics. 1979;32:549–554. doi: 10.7164/antibiotics.32.549. [DOI] [PubMed] [Google Scholar]
- 10.Bhate DS, Hulyalkar RK, Menon SK. Cell. Mol. Life Sci. 1960;16:504–505. [Google Scholar]
- 11.Okamura K, Soga K, Shimauchi Y, Ishikura M, Lein J. The Journal of Antibiotics. 1977;30:334–336. doi: 10.7164/antibiotics.30.334. [DOI] [PubMed] [Google Scholar]
- 12.Lamari L, Zitouni A, Boudjella H, Badji B, Sabaou N, Lebrihi A, Lefebvre G, Seguin E, Tillequin F. The Journal of Antibiotics. 2002;55:696–701. doi: 10.7164/antibiotics.55.696. [DOI] [PubMed] [Google Scholar]
- 13.Lamari L, Zitouni A, Dob T, Sabaou N, Lebrihi A, Germain P, Seguin E, Tillequin F. The Journal of Antibiotics. 2002;55:702–706. doi: 10.7164/antibiotics.55.702. [DOI] [PubMed] [Google Scholar]
- 14.McInerney BV, Gregson RP, Lacey MJ, Akhurst RJ, Lyons GR, Rhodes SH, Smith DRJ, Engelhardt LM, White AH. J. Nat. Prod. 1991;54:774–784. doi: 10.1021/np50075a005. [DOI] [PubMed] [Google Scholar]
- 15.Webster JM, Li J, Chen G. PCT Int. Appl. 1999;27 [Google Scholar]
- 16.Wietz M, Mansson M, Gotfredsen CH, Larsen TO, Gram L. Marine Drugs. 2010;8:2946–2960. doi: 10.3390/md8122946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Qin Z, Baker AT, Raab A, Huang S, Wang T, Yu Y, Jaspars M, Secombes CJ, Deng H. Journal of Biological Chemistry. 2013;288:14688–14697. doi: 10.1074/jbc.M112.448415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shiozawa H, Kagasaki T, Kinoshita T, Haruyama H, Domon H, Utsui Y, Kodama K, Takahashi S. The Journal of Antibiotics. 1993;46:1834–1842. doi: 10.7164/antibiotics.46.1834. [DOI] [PubMed] [Google Scholar]
- 19.Shiozawa H, Kagasaki T, Torikata A, TANAKA N, Fujimoto K, Hata T, Furukawa Y, Takahashi S. The Journal of Antibiotics. 1995;48:907–909. doi: 10.7164/antibiotics.48.907. [DOI] [PubMed] [Google Scholar]
- 20.Shiozawa H, Shimada A, Takahashi S. The Journal of Antibiotics. 1997;50:449–452. doi: 10.7164/antibiotics.50.449. [DOI] [PubMed] [Google Scholar]
- 21.Shiozawa H, Takahashi H, Kagasaki T. Jpn. Kokai Tokkyo Koho. 1996:8. [Google Scholar]
- 22.Baggaley KH, Broom NJP, Brown P, O'Hanlon PJ, Osborne NF. PCT Int. Appl. 1994:44. [Google Scholar]
- 23.Li B, Walsh CT. Proceedings of the National Academy of Sciences. 2010;107:19731–19735. doi: 10.1073/pnas.1014140107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Medema MH, Trefzer A, Kovalchuk A, van den Berg M, Muller U, Heijne W, Wu L, Alam MT, Ronning CM, Nierman WC, Bovenberg RAL, Breitling R, Takano E. Genome Biology and Evolution. 2010;2:212–224. doi: 10.1093/gbe/evq013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Song JY, Jeong H, Yu DS, Fischbach MA, Park HS, Kim JJ, Seo JS, Jensen SE, Oh TK, Lee KJ, Kim JF. Journal of Bacteriology. 2010;192:6317–6318. doi: 10.1128/JB.00859-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li B, Walsh CT. Biochemistry. 2011;50:4615–4622. doi: 10.1021/bi200321c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang C, Wesener SR, Zhang H, Cheng Y-Q. Chemistry & Biology. 2009;16:585–593. doi: 10.1016/j.chembiol.2009.05.005. [DOI] [PubMed] [Google Scholar]
- 28.Scharf DH, Remme N, Heinekamp T, Hortschansky P, Brakhage AA, Hertweck C. J. Am. Chem. Soc. 2010;132:10136–10141. doi: 10.1021/ja103262m. [DOI] [PubMed] [Google Scholar]
- 29.Scharf DH, Groll M, Habel A, Heinekamp T, Hertweck C, Brakhage AA, Huber EM. Angew Chem Int Ed Engl. 2014 doi: 10.1002/anie.201309302. [DOI] [PubMed] [Google Scholar]
- 30.Huang S, Zhao Y, Qin Z, Wang X, Onega M, Chen L, He J, Yu Y, Deng H. Process Biochemistry. 2011;46:811–816. [Google Scholar]
- 31.Komatsu M, Komatsu K, Koiwai H, Yamada Y, Kozone I, Izumikawa M, Hashimoto J, Takagi M, Omura S, Shin-Ya K, Cane DE, Ikeda H. ACS Synth Biol. 2013;2:384–396. doi: 10.1021/sb3001003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thomas CCM, Hothersall JJ, Willis CLC, Simpson TJT. Nature Publishing Group. 2010;8:281–289. [Google Scholar]
- 33.Fukuda D, Haines AS, Song Z, Murphy AC, Hothersall J, Stephens ER, Gurney R, Cox RJ, Crosby J, Willis CL, Simpson TJ, Thomas CM. PLoS ONE. 2011;6:e18031–e18031. doi: 10.1371/journal.pone.0018031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bouras N, Merrouche R, Lamari L, Mathieu F, Sabaou N, Lebrihi A. Process Biochemistry. 2008;43:1244–1252. [Google Scholar]
- 35.Merrouche R, Bouras N, Coppel Y, Mathieu F, Sabaou N, Lebrihi A. FEMS Microbiology Letters. 2011;318:41–46. doi: 10.1111/j.1574-6968.2011.02246.x. [DOI] [PubMed] [Google Scholar]
- 36.Bouras N, Mathieu F, Sabaou N, Lebrihi A. Journal of Applied Microbiology. 2006;100:390–397. doi: 10.1111/j.1365-2672.2005.02762.x. [DOI] [PubMed] [Google Scholar]
- 37.Murphy AC, Fukuda D, Song Z, Hothersall J, Cox RJ, Willis CL, Thomas CM, Simpson TJ. Angew Chem Int Ed Engl. 2011;50:3271–3274. doi: 10.1002/anie.201007029. [DOI] [PubMed] [Google Scholar]
- 38.de la Fuente A, Lorenzana LM, Martín JF, Liras P. Journal of Bacteriology. 2002;184:6559–6565. doi: 10.1128/JB.184.23.6559-6565.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Robles-Reglero V, Santamarta I, Álvarez-Álvarez R, Martin JF, Liras P. J Biotechnol. 2013;163:69–76. doi: 10.1016/j.jbiotec.2012.09.017. [DOI] [PubMed] [Google Scholar]
- 40.Song JY, Jensen SE, Lee KJ. Appl Microbiol Biotechnol. 2010;88:659. doi: 10.1007/s00253-010-2801-2. [DOI] [PubMed] [Google Scholar]
- 41.Mackenzie AK, Valegård K, Iqbal A, Caines MEC, Kershaw NJ, Jensen SE, Schofield CJ, Andersson I. Journal of Molecular Biology. 2010;396:332. doi: 10.1016/j.jmb.2009.11.045. [DOI] [PubMed] [Google Scholar]
- 42.Yin H, Xiang S, Zheng J, Fan K, Yu T, Yang X, Peng Y, Wang H, Feng D, Luo Y, Bai H, Yang K. Applied and Environmental Microbiology. 2012;78:3431–3441. doi: 10.1128/AEM.07699-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nárdiz N, Santamarta I, Lorenzana LM, Martín JF, Liras P. Microbial Biotechnology. 2010;4:216–225. doi: 10.1111/j.1751-7915.2010.00222.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mueller EGE. Nat Chem Biol. 2006;2:185–194. doi: 10.1038/nchembio779. [DOI] [PubMed] [Google Scholar]
- 45.Cipollone R, Ascenzi P, Visca P. IUBMB Life. 2007;59:51. doi: 10.1080/15216540701206859. [DOI] [PubMed] [Google Scholar]
- 46.Jimenez A, Tipper DJ, Davies J. Antimicrobial Agents and Chemotherapy. 1973;3:729–738. doi: 10.1128/aac.3.6.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tipper DJ. Journal of Bacteriology. 1973;116:245–256. doi: 10.1128/jb.116.1.245-256.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Khachatourians GG, Tipper DJ. Antimicrobial Agents and Chemotherapy. 1974;6:304–310. doi: 10.1128/aac.6.3.304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sivasubramanian N, Jayaraman R. Molec. Gen. Genet. 1976;145:89–96. doi: 10.1007/BF00331562. [DOI] [PubMed] [Google Scholar]
- 50.Khachatourians GG, Tipper DJ. Journal of Bacteriology. 1974;119:795–804. doi: 10.1128/jb.119.3.795-804.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sivasubramanian N, Jayaraman R. Molec. Gen. Genet. 1980;180:609–615. doi: 10.1007/BF00268068. [DOI] [PubMed] [Google Scholar]
- 52.Joshi A, Verma M, Chakravorty M. Antimicrobial Agents and Chemotherapy. 1982;22:541–547. doi: 10.1128/aac.22.4.541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Roza J, Blanco MG, Hardisson C, Salas JA. The Journal of Antibiotics. 1986;39:609–612. doi: 10.7164/antibiotics.39.609. [DOI] [PubMed] [Google Scholar]
- 54.Oliva B, O'Neill A, Wilson JM, O'Hanlon PJ, Chopra I. Antimicrobial Agents and Chemotherapy. 2001;45:532–539. doi: 10.1128/AAC.45.2.532-539.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Das B, Butler JS, Sherman F. Molecular and Cellular Biology. 2003;23:5502–5515. doi: 10.1128/MCB.23.16.5502-5515.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Grigull J, Mnaimneh S, Pootoolal J, Robinson MD, Hughes TR. Molecular and Cellular Biology. 2004;24:5534–5547. doi: 10.1128/MCB.24.12.5534-5547.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kebaara BW, Nielsen LE, Nickerson KW, Atkin AL. Génome. 2006;49:894–899. doi: 10.1139/g06-046. [DOI] [PubMed] [Google Scholar]
- 58.Guan QQ, Zheng WW, Tang SS, Liu XX, Zinkel RAR, Tsui K-WK, Yandell BSB, Culbertson MRM. PLoS Genet. 2006;2:e203–e203. doi: 10.1371/journal.pgen.0020203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rossini C, Taylor W, Fagan T, Hastings JW. Chronobiol Int. 2003;20:963–976. doi: 10.1081/cbi-120025248. [DOI] [PubMed] [Google Scholar]
- 60.Pelechano V, Pérez-Ortín JE. Yeast. 2008;25:85–92. doi: 10.1002/yea.1548. [DOI] [PubMed] [Google Scholar]
- 61.Li B, Forseth RR, Bowers AA, Schroeder FC, Walsh CT. ChemBioChem. 2012;13:2521–2526. doi: 10.1002/cbic.201200536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schrettl MM, Carberry SS, Kavanagh KK, Haas HH, Jones GWG, O'Brien JJ, Nolan AA, Stephens JJ, Fenelon OO, Doyle SS. PLoS Pathog. 2010;6:e1000952–e1000952. doi: 10.1371/journal.ppat.1000952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Forseth RR, Fox EM, Chung D, Howlett BJ, Keller NP, Schroeder FC. J. Am. Chem. Soc. 2011;133:9678–9681. doi: 10.1021/ja2029987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kirby GW, Robins DJ, Sefton MA, Talekar RR. J. Chem. Soc., Perkin Trans. 1980;1:119. [Google Scholar]
- 65.Juhl MJ, Clark DP. Applied and Environmental Microbiology. 1990;56:3179–3185. doi: 10.1128/aem.56.10.3179-3185.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Leclercq R. Clin. Infect. Dis. 2002;34:482–492. doi: 10.1086/324626. [DOI] [PubMed] [Google Scholar]
- 67.Minamiguchi K, Kumagai H, Masuda T, Kawada M, Ishizuka M, Takeuchi T. Int. J. Cancer. 2001;93:307–316. doi: 10.1002/ijc.1321. [DOI] [PubMed] [Google Scholar]
- 68.Jia Y, Wu S-L, Isenberg JS, Dai S, Sipes JM, Field L, Zeng B, Bandle RW, Ridnour LA, Wink DA, Ramchandran R, Karger BL, Roberts DD. Cell Stress and Chaperones. 2009;15:165–181. doi: 10.1007/s12192-009-0130-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dai S, Jia Y, Wu S-L, Isenberg JS, Ridnour LA, Bandle RW, Wink DA, Roberts DD, Karger BL. J. Proteome Res. 2008;7:4384–4395. doi: 10.1021/pr800376w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Abreu PA, Sousa TS, Jimenez PC, Wilke DV, Rocha DD, Freitas HPS, Pessoa ODL, La Clair JJ, Costa-Lotufo LV. ChemBioChem. 2014 doi: 10.1002/cbic.201300717. [DOI] [PubMed] [Google Scholar]
- 71.Schmidt U, Geiger F. Angew Chem Int Ed Engl. 1962;1:265–265. [Google Scholar]
- 72.Schmidt U, Geiger F. Liebigs Ann. Chem. 1963;664:168–188. [Google Scholar]
- 73.Buchi G, Lukas G. J. Am. Chem. Soc. 1963;85:647–648. [Google Scholar]
- 74.Buchi G, Lukas G. J. Am. Chem. Soc. 1964;86:5654–5658. [Google Scholar]
- 75.Ellis JE, Fried JH, Harrison IT, Rapp E, Ross CH. J. Org. Chem. 2001;42:2891–2893. doi: 10.1021/jo00437a023. [DOI] [PubMed] [Google Scholar]
- 76.Harrison IT, Niem T, Ross CH. Chem. Ind. (London) 1980:157–158. [Google Scholar]
- 77.Stachel H-D, Nienaber J, Zoukas T. Liebigs Ann. Chem. 1992:473–480. [Google Scholar]
- 78.Dell I, Godfrey CR, Wadsworth DJ. 1992;504:384–384. [Google Scholar]
- 79.Hjelmgaard T, Givskov M, Nielsen J. Org. Biomol. Chem. 2007;5:344. doi: 10.1039/b616411k. [DOI] [PubMed] [Google Scholar]
- 80.Li B, Lyle MPA, Chen G, Li J, Hu K, Tang L, Alaoui-Jamali MA, Webster J. Bioorganic & Medicinal Chemistry. 2007;15:4601–4608. doi: 10.1016/j.bmc.2007.04.017. [DOI] [PubMed] [Google Scholar]
- 81.Stachel H-D, Schorp M, Zoukas T. Liebigs Ann. Chem. 1992;1992:1039–1044. [Google Scholar]
- 82.Stachel H-D, Zoukas T, Huβlein M, Nienaber J, Schorp M. Liebigs Ann. Chem. 1993;1993:305–311. [Google Scholar]
- 83.Schachtner JE, Zoukas H-D, Stachel T, Polborn K, Nöth H. Journal of Heterocyclic Chemistry. 1999;36:161–175. [Google Scholar]
- 84.Stachel H-D, Eckl E, Immerz-Winkler E, Kreiner C, Weigand W, Robl C, Wünsch R, Dick S, Drescher N. Helv. Chim. Acta. 2002;85:4453–4467. [Google Scholar]
- 85.Stachel H-D, Odlerová Ž, Waisser K. Collect. Czech. Chem. Commun. 1997;62:510–515. [Google Scholar]
- 86.Webster JM, Li J, Chen G. U.S. 2000:4. [Google Scholar]
- 87.Tan X, Li C, Yu Z, Wang P, Nian S, Deng Y, Wu W, Wang G. Chem. Pharm. Bull. 2013;61:351–357. doi: 10.1248/cpb.c12-00791. [DOI] [PubMed] [Google Scholar]
- 88.Yakushiji F, Miyamoto Y, Kunoh Y, Okamoto R, Nakaminami H, Yamazaki Y, Noguchi N, Hayashi Y. ACS Med. Chem. Lett. 2013;4:220–224. doi: 10.1021/ml300350p. [DOI] [PMC free article] [PubMed] [Google Scholar]