Abstract
The most common risk factor for developing hepatocellular carcinoma (HCC) is chronic infection with hepatitis B virus (HBV). To better understand the evolutionary forces driving HCC we performed a near saturating transposon mutagenesis screen in a mouse HBV model of HCC. This screen identified 21 candidate early stage drivers, and a bewildering number (2860) of candidate later stage drivers, that were enriched for genes mutated, deregulated, or that function in signaling pathways important for human HCC, with a striking 1199 genes linked to cellular metabolic processes. Our study provides a comprehensive overview of the genetic landscape of HCC.
INTRODUCTION
Nearly 500,000 people are diagnosed with HCC each year and their overall 5-year survival rate is below 12%. The highest incidence of HCC is in regions where infection with HBV is endemic, and men are 2 to 4 times more likely to develop HCC than women. HCC related to infection with hepatitis B virus (HBV) has also become the fastest-rising cause of cancer-related death in the United States during the past two decades. While the use of emerging sequencing and genomics technologies has identified many mutated and/or differentially expressed genes in HCC, they have also uncovered a surprising amount of intra- and inter-tumor heterogeneity1-5. For these reasons, and because important DNA mutations can be concealed among the large number of passenger mutations present in these tumors, it has proven difficult to identify the complete complement of driver genes for HCC. Epigenetic silencing of tumor suppressor genes also frequently occurs in tumors6,7. This, combined with recent studies showing that there may be thousands of haploinsufficient tumor suppressor (TS) genes8, makes the identification of all driver genes for HCC additionally difficult. Published reports from the ENCODE project have also identified millions of functional elements, many of which are transcription factor binding sites that regulate the expression of genes often located hundreds of kilobases away9. Nearly 70% of disease-associated single nucleotide polymorphisms (SNPs) identified in genome-wide association studies (GWAS) map to these distal enhancers9, raising the possibility that non-coding mutations in these distal elements might also significantly contribute to cancer, and potentially explaining why some tumors have few or no mutated cancer genes10,11, even after extensive genome characterization. Tumors with a paucity of mutated genes might also have mutations in very infrequently mutated genes that we do not yet have the statistical power to detect.
One method for identifying these missing cancer genes, as well as to validate the hundreds of candidate cancer genes already described, is through comparative genomics involving transposon-based insertional mutagenesis12. Recently, it has become possible to mobilize the Sleeping Beauty (SB) transposon in basically any mouse tissue at high enough frequencies to induce virtually any kind of cancer13-15. Mutagenic SB transposons carry a strong promoter for activating proto-oncogenes and transcriptional stop cassettes for inactivating TS genes, and the transposons therefore tag cancer genes in tumor cells. Human tumor genomes are complex with multiple mutagenic processes operative. By contrast, transposons tag cancer genes directly, thus facilitating their identification.
Here, we sought to obtain a comprehensive list of genes functionally necessary to trigger HCC by performing a large-scale SB transposon mutagenesis screen13,14. Since the major etiology of human HCC is chronic infection with the HBV16 we mobilized SB in the livers 17 of transgenic mice predisposed to develop HCC due to expression of the toxic HBV surface antigen (HBsAg) in their livers 18. By using this mouse model we aimed to identify genes that could cooperate with HBV-induced liver inflammation in the induction of HCC, similar to that which occurs in most human HCC.
RESULTS
HBV-associated HCC mouse model for mutagenesis screening
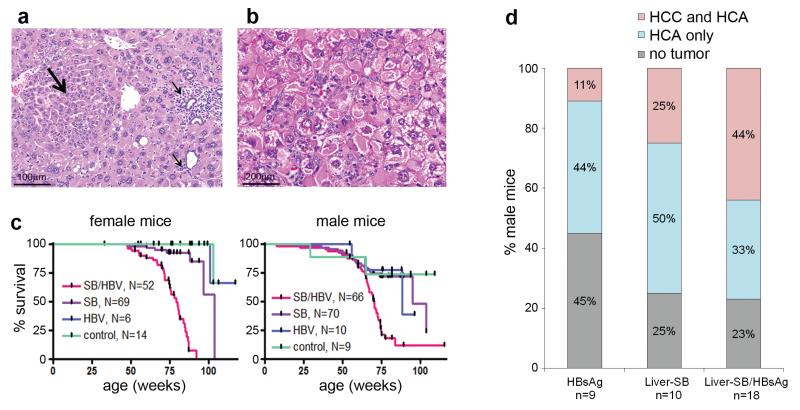
Liver-SB/HBsAg transgenic mice develop chronic liver inflammation with associated reactive hyperplasia (Figure 1a, Supplementary Figure 1), hepatocytomegaly, and ground glass hepatocytes (Figure 1b) similar to the human chronic hepatitis B and to the previously characterized HBsAg mice 18. Liver-SB/HBsAg modifications lead to the appearance of preneoplastic foci, visible from 19.7 weeks of age, followed by hepatocellular adenoma (HCA) and trabecular HCC (Supplementary Figure 2). SB induced a cooperative tumorigenic effect with HBsAg as Liver-SB/HBsAg mice displayed reduced survival (Figure 1c). We also noted a tendency to larger numbers of tumors (Supplementary Figure 2) and to more advanced stage disease compared to HBsAg mice alone (Figure 1d).
Figure 1. HBsAg/Liver-SB mice have chronic liver inflammation and accelerated formation of HCC.
a, Preneoplastic hepatocellular focus (large arrow) of hyperplastic hepatocytes, with inflammatory cells (small arrows) in a 22.8 week-old Liver-SB/HBsAg male mouse. b, Hepatocytes with endoplasmic reticulum inclusions, so called ground glass hepatocytes, as seen in human HBV-induced hepatitis. Sample from a 20.4 week-old Liver-SB/HBsAg male mouse. c, Kaplan-Meier survival curves for male and female mice of all four combinations of genotypes. Liver-SB/HBsAg (SB/HBV), Liver-SB (SB), HBsAg transgene (HBV) and littermate control mice carrying an inactive transposon (no transposase) and no HBsAg transgene (control). Female median survival was 79.1 weeks for Liver-SB/HBsAg and 103.7 weeks for Liver-SB (Logrank test p<10−4). Male median survival was 70.4 weeks for Liver-SB/HBsAg, 94.9 weeks for Liver-SB, and 88.4 for HBsAg mice (Logrank test p<10−4). d, Summary of histopathology performed on livers of moribund male mice of various phenotypes. We observe more of “HCC and HCA” in class “Liver-SB/HBsAg”, than in classes “Liver-SB” and “HBsAg” combined. Although a trend is visible, it was not significant (p=0.086, one–sided Fisher’s exact test on a contingency table). The overall penetrance for hepatic tumorigenesis in Liver-SB/HBV was 77%. HCC : hepatocellular carcinoma, HCA: hepatocellular adenoma.
To identify the genes mutated by SB that cooperate with HBsAg-associated inflammation in tumor induction, we PCR amplified and sequenced the transposon insertion sites from 250 tumors harvested from 34 mice19, which yielded 328,687 sequence reads and identified an average of 1315 unique transposon insertion sites per tumor. By comparing the location of the transposon insertions in all tumors we found a few tumors that were genetically related. These tumors were removed leaving 228 genetically unrelated tumors, which were subsequently used for downstream analysis (Supplementary Figures 3,4).
Large-scale screen identifies Common Insertion Sites (CISs)
We then screened for CISs, which are regions in the cancer genomes that contain a higher density of transposon insertions than predicted by chance and therefore are likely to contain a cancer gene (CISs on chromosome 1 could not be conclusively identified because of local transposon hopping and CISs on this chromosome were therefore excluded). A gene-centric CIS-calling method (gCIS) that looks for a higher density of transposon insertions within the coding regions of all RefSeq genes than predicted by chance20 identified 2525 gCIS genes (Supplementary Table 1a), while a non-gene-centric Gaussian Kernel Convolution (GKC) method21 that looks for a higher density of insertions within fixed kernel widths of 15Kb to 240Kb22 identified 2041 CISs containing 2103 genes (Supplementary Table 1b). There was a 83% overlap between the CISs identified by these two methods (p<2×10−16, Chi-square with Yates correction) and together, they identified 2871 CIS loci containing 2881 genes (Supplementary Table 1c, Supplementary Figure 5a). In vitro transposition cell culture assays have suggested that SB is a random insertional mutagen and the only requirement for insertion is a TA dinucleotide12. Although the GKC method scans the entire cancer genome for CISs, most CISs were located within or in close proximity to genes, providing additional evidence that SB targets the coding regions of genes that confer a selective advantage for tumor growth. gCISs not identified by GKC often contained very large (>300Kb) or small genes (<10Kb) and were probably missed in part because of the use of fixed kernel widths (Supplementary Figure 5b,c).
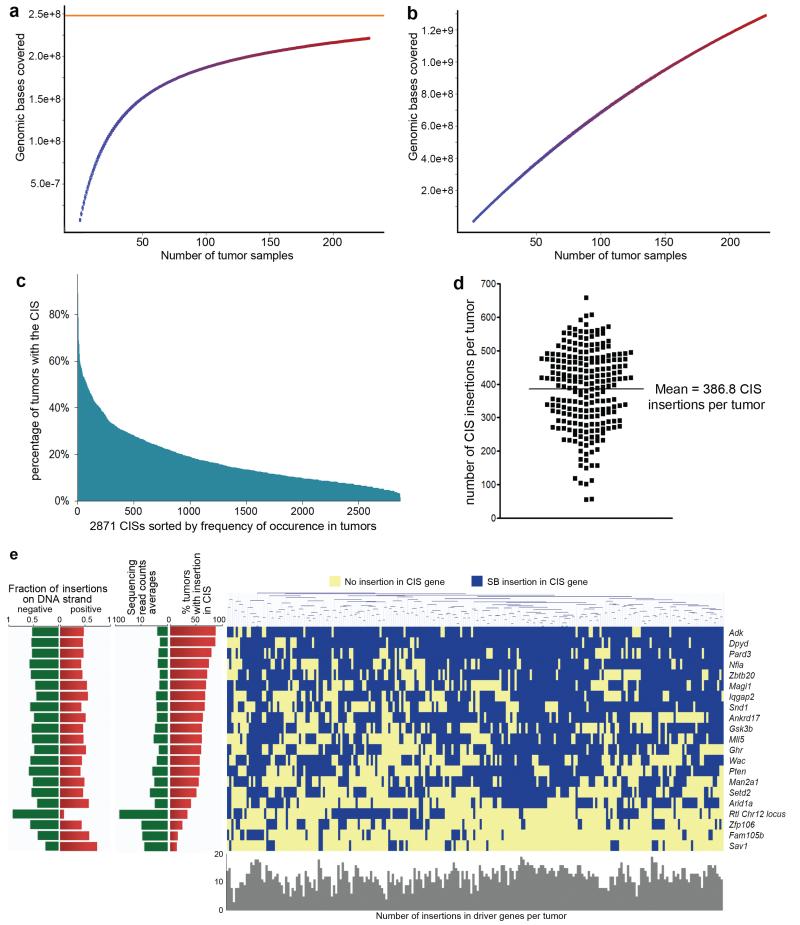
To estimate the genetic coverage of this screen we randomly selected subgroups of 10 to 220 tumors from the total of 228 tumors and then used GKC to identify the CISs for each subgroup. The number of tumors in each subgroup was then plotted against the genomic base pairs covered by all the CISs identified for each subgroup. The curve plateaued before reaching 100 tumors, indicating that the screen was approaching saturation with as few as a 100 tumors (Figure 2a,b). Adding more tumors merely identified additional CISs of lower frequency (Supplementary Figure 6,7). Consistent with this notion of saturation, this screen identified most of the CIS genes found previously in much smaller-scale HCC transposon screens performed in p53 mutant17 and Sav1-deficient mice (see below) (Supplementary Figure 8). To our knowledge this is the first saturating transposon screen for cancer genes reported for mice.
Figure 2. Identification of driver genes for HCC in the Liver-SB/HBV screen.
a,b, CISs were called using the Gaussian Kernel Convolution (GKC) method from the indicated numbers of randomly chosen Liver-SB/HBV tumors. The resulting CIS genomic loci were overlapped with the CIS loci from the 228 tumors. The numbers of tumor samples used were plotted against the genomic bases covered. For each combination of tumors the median values and 25th and 75th percentiles were plotted. a, Near saturation of the screen is seen with a theoretical asymptote ( in orange) at 248,023,943 bases with an increasing number of samples. The percentage saturation obtained with 100 samples is 75.4%. b, This plot shows the data generated from randomized samples; the straightness of the line indicates no saturation. c, Distribution of all 2871 CISs according to the percent of time they are mutated in tumors. When a CIS gene was identified using both gCIS and GKC, the average percent was used. Only 79 CISs were mutated in more than 50% of the tumors, while 638 CISs were mutated in more than 25% of the tumors. d, Number of transposon insertions found in CIS loci in each tumor (represented as dots). The mean is 387 ± 118 CISs targeted per tumor. e, Clustering in all 228 tumors of the 21 HCC driver genes with highest sequencing read counts and frequency of occurrence in tumors. No statistically significant genetic correlations were found among the 21 driver genes.
Nearly 68% of the CIS genes were mutated in <20% of the tumors and on average each tumor contained 387 mutated CIS genes (Figure 2c,d). This is indicative of extensive intratumor heterogeneity, likely resulting from branching tumor evolution, similar to that reported for several human cancers23, combined with the large mutational load induced by SB. In this HCC model, every liver cell contains ~350 copies of the transposon13, which when induced to transpose, can transpose over and over again. In addition, because the transposon insertion sites are PCR amplified before being deep-sequenced, insertional mutations present in a small number of cells in the tumor can be identified. It is this ability to sample the extensive intratumor heterogeneity at such a deep level that we believe made it possible to approach saturation with only a 100 tumors.
General features of HCC CIS genes
The transposon13 contains a promoter used to deregulate oncogenes and stop cassettes in both transcriptional orientations to inactivate TS genes. Transposon insertions at oncogenes therefore tend to be located at the 5′ end of the gene in the same transcriptional orientation, while insertions in TS genes are usually located throughout the gene in either orientation. Insertion site patterns are thus often predictive of gene function. For the genes identified here and in other transposon screens performed in solid tumors13-15,17,19,22,24, the majority are predicted to function as TS genes. Most genes also have insertions only in one allele, suggesting they may function as haploinsufficient TS genes, although it remains possible that the other allele is silenced by an epigenetic or other mutational event. This is consistent with recent studies performed in human cells and tumors that identified hundreds if not thousands of haploinsufficient genes, which are able to drive cellular proliferation. These genes are postulated to operate in cooperative networks to maximize proliferative fitness25, and support other studies indicating that even partial inactivation of TS genes can contribute to tumorigenesis8.
Branching tumor evolution complicates efforts to implement personalized medicine and suggests that targeted therapies might be directed to genes mutated at the trunk of the evolutionary tree. Transposons provide powerful tools for identifying trunk genes since insertions in trunk genes are more likely to be associated with a higher number of sequencing reads. Despite the splinkerette PCR method preventing a perfect indication of read quantitation, previous studies show that CIS genes with high read depth are more likely to be highly penetrant, as seen with APC22. To identify potential trunk genes for HCC we again screened for CISs but this time using only the 25,197 SB insertions that were represented by at least four sequencing reads each. Among the 524 CISs identified (Supplementary Table 1d) were 21 outlier CISs that showed the highest sequencing read counts and frequency of occurrence (Figure 2e, Supplementary Figure 9a). Six genes including Arid1a10,11, Gsk3b26, Iqgap227, Magi128, Pten29 and Sav130,31 have known tumor suppressive roles in HCC, while Snd1 is described as an oncogene promoting HCC angiogenesis32,33. Two genes including Zbtb2034,35 and Ankrd1736,37 are involved in hepatocyte differentiation and maturation, while Zbtb20 is a transcription factor that represses the expression of α-fetoprotein, a widely used biomarker used for HCC surveillance. The Setd2 TS gene38 and the putative TS genes Adk39, Dpyd40,41, Mll542 and Nfia43,44 were also identified as potential driver genes for HCC, although there is no published evidence they are involved in HCC. Six driver genes also regulate hepatic metabolism with Pten45, Gsk3b46, Adk47,48, Zbtb2049 and Ghr50,51 regulating lipid and glucose metabolism, and Dpyd controling pyrimidine catabolism in hepatocytes. Seven driver genes including Mll5, Setd2, Wac, Arid1a, Nfia, Snd1 and Zbtb20 are transcription factors, cofactors or chromatin remodelers, highlighting the importance of the modulation of transcriptional programs in HCC. One driver CIS we named Rtl1 Chr12 locus (Figure 2e) is the merger of several CISs at the same imprinted locus52 (Rian, Rtl1, 6430526N21Rik genes and a microRNA cluster) and was targeted in 38.4% of the tumors. The majority (92%) of the transposon insertions at this CIS are located on the negative strand (Supplementary Figure 9b), indicating a potential oncogenic loss of imprinting and transcriptional activation of the Rtl1 gene, or one or more of the microRNAs located downstream. Interestingly, these microRNAs are among the most strongly up-regulated microRNAs in murine and human HCC53.
Comparisons of CIS genes with human data and other indications
We next asked whether the 2881 HCC CIS genes are specific for HCC by comparing them to the genes identified in transposon screens for colorectal cancer (CRC)22 and pancreatic adenocarcinoma (PDAC)19,24. These comparisons showed that >60% of the HCC CIS genes are also mutated in CRC and PDAC (p<2.10−16) (Supplementary Figure 10). This is not surprising since many cancer genes are known to function in more than one cancer cell type. HCC CIS genes are also significantly mutated in human HCC as shown by the very significant overlap with the genes somatically mutated in human HCC identified in two whole-genome HCC sequencing studies10,11, and in a genome-wide analysis of HBV integrations54 (Table 1). In addition, we found a significant enrichment among the genes listed in the Catalogue of Somatic Mutations in Cancer (COSMIC) database, which reports all of the somatic mutations identified in human HCC, and the genes listed in the Cancer Gene Census database, which aims to catalogue all of the driver genes in human cancers (Table 1). HCC CIS genes thus play important roles in human HCC and provide additional validation for the genes identified in human HCC.
Table 1. Enrichment of Liver-SB/HBV CIS genes in the genes mutated or misregulated in human cancer.
The enrichment p values are calculated using a 2×2 contingency table, Chi-Square test with Yates’ correction.
| Organism | Number of Genes in dataset (mouse chr1 excluded) | Number of overlapping CIS genes | % coverage of Human dataset | Enrichment p value | |
|---|---|---|---|---|---|
| Human HCC somatic mutations (ref 11) | Human | 858 | 177 | 20.6% | 9.53E-14 |
| Human HCC somatic mutations (ref 10) | Human | 189 | 52 | 27.5% | 4.31E-10 |
| COSMIC database for somatic mutations in human HCC | Human | 499 | 83 | 16.6% | 0.00423 |
|
| |||||
| Human HBV insertions (ref 54) | Human | 103 | 23 | 22.3% | 0.00340 |
|
| |||||
| CGC database (somatic mutations, all cancers) | Human | 468 | 120 | 25.6% | < 2.10−16 |
|
| |||||
| Genes mis-expressed in Human HCC (ref 4) | Human | 3784 | 987 | 26.1% | < 2.10−16 |
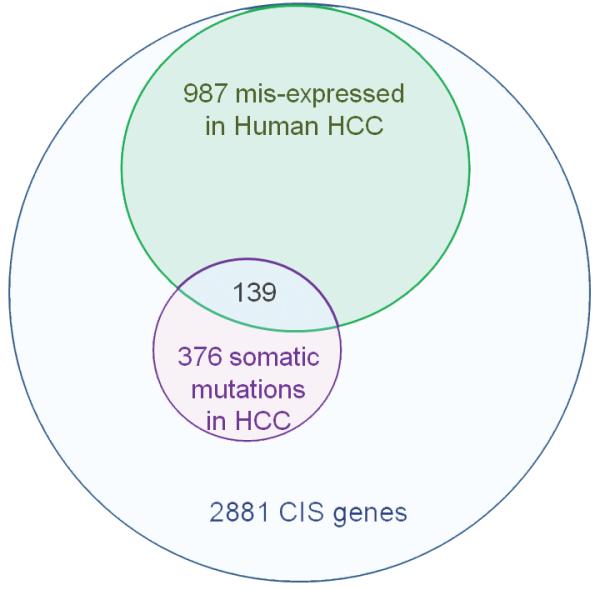
The expression of HCC CIS genes is also specifically deregulated in human HCC. Among 2189 HCC CIS genes contained in a large microarray dataset obtained from 223 HBV-positive HCC patients4 (Supplementary Figure 11a), 45% were significantly deregulated in human HCC (absolute fold change >1.5 and adjusted p-value <0.00001) (Supplementary Figure 11b; Supplementary Table 2), compared to 29% misregulated genes on the array that are not CIS genes (p <2.10−16). Differentially expressed HCC CIS genes also displayed significantly lower p-values and more of them were up-regulated than down-regulated, although this difference was not large (Supplementary Figure 11c-d). Comparison of the HCC microarray data with other CIS gene lists from PDAC19,24 and CRC22 screens showed the highest enrichment for HCC CIS genes (Supplementary Figure 11e), indicating better specificity when the data were from the same tumor type. Overall, 42.5% of the HCC CIS genes are mutated or mis-expressed in human HCC (Figure 3, Supplementary Table 3). The deregulation of 30 HCC CIS genes was subsequently confirmed by real time PCR using a microfluidic dynamic array containing 18 HBV-positive human tumors and 9 Liver-SB/HBsAg tumors (Supplementary Figure 11f; Supplementary Table 4). The direction of the expression change (up- or down-regulated) was positively correlated between human and mouse tumors (Supplementary Figure 11g).
Figure 3. Many Liver-SB/HBV CIS genes are deregulated or mutated in human cancer.
Repartition of the 42.5% Liver-SB/HBV CIS genes found to be either mutated or misexpressed in human HCC. A total of 1224 CISs from the Liver-SB/HBV screen were associated with a gene mutated or misexpressed in human HCC.
Transposon drive HCC via conserved pathways
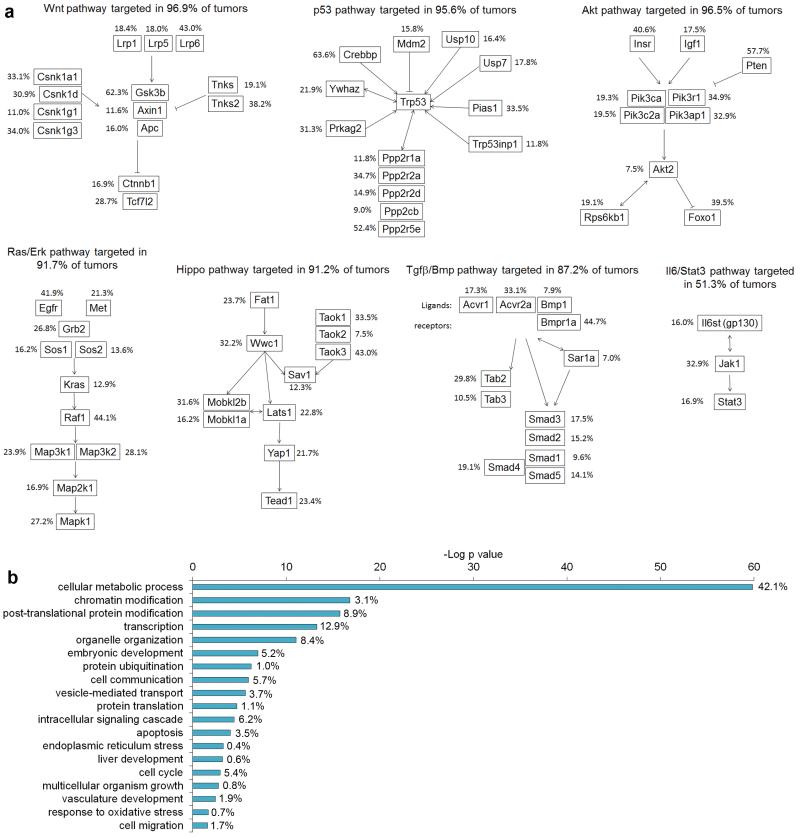
HCC CIS genes are also enriched in the major cancer signaling pathways known to be important for human HCC such as the Ras/Erk, p53, Akt, Wnt, and Tgfβ/Bmp pathways, together with the recently identified hepato-tumor suppressor Hippo pathway30 (Figure 4a, Supplementary Table 5, Supplementary Figure 12). Another known HCC signaling pathway, IL6/Stat3, was less enriched, possibly because the tumor microenvironement locally secretes IL6, thus activating this pathway in hepatocytes. A similar analysis for PDAC19,24 and CRC22 CIS genes showed that many of these signaling pathways are also enriched for mutations in these genes. Interestingly, the Hippo pathway was more highly mutated in HCC, whereas the Wnt pathway, which is critical for CRC22, was more highly mutated in CRC (Supplementary Figure 12a). Transposons also prominently targeted the Hippo pathway in tumors generated in a Sav1-deficient background (Methods and Supplementary Table 1e). Sav1 activates MST1, which results in the downstream activation of Hippo signalling55. Together, these results suggest that transposon-induced mutations in Hippo pathway genes cooperate with Sav1-deficiency to further deregulate Hippo signaling, and are consistent with the results from a transposon screen performed in Apc mutant mice, which identified a large number of CRC CIS genes that targeted the Wnt pathway22. Liver-SB/Sav1-deficient mice also developed multiple large liver tumors nearly two months earlier than Liver-SB/HBsAg mice, providing additional evidence for an oncogenic role for deregulated Hippo signaling in HCC.
Figure 4. Liver-SB/HBV CIS genes drive tumorigenesis through conserved cancer signaling pathways.
a, HCC CIS genes are enriched for genes in major oncogenic canonical signalling pathways. The percentages represent the fraction of tumors with a transposon insertion at the gene locus. Most tumors displayed insertions in genes in these canonical pathways. b, Gene ontology analysis performed with DAVID bioinformatics, biological processes. P values for enrichment are represented on a –log scale. The percentages indicate the proportion of HCC CIS genes found associated with a specific biological process.
HCC CIS genes target metabolic events
When we classified the HCC CIS genes according to their gene ontology we noticed a striking over-representation of genes linked to cellular metabolic processes (p=1.4 × 10−60, 1199 genes) (Figure 4b, Supplementary Table 6). A less striking enrichment for metabolic processes was detected for CRC and PDAC CIS genes (Supplementary Figure 13), while most other ontologies such as transcription or cell cycle displayed similar enrichments among all three tumor types. When we focused the gene classification on metabolic processes, we found 47 metabolic categories that were targeted by the transposon during HCC tumor development, against only 9 and 10 metabolic categories for CRC and PDAC CIS genes, respectively (Supplementary Figure 14). When we looked for enrichment for disease genes annotated in Ingenuity Pathway Analysis, we also found a highly significant enrichment for HCC CIS genes in metabolic diseases (265 genes, p = 4 × 10−6); genetic association to cancer was also highly evident (431 genes, p = 6 × 10−10). Overall, genes associated with protein, carbohydrate, lipid and nucleic acid metabolism were highly mutated by SB. Several other processes were also targeted including oxidoreduction and metabolism of ATP, organophosphate, hormones, isoprenoid and vitamins (Supplementary Table 6).
To determine whether similar genetic changes occur in human HCC we measured metabolic gene expression levels in 18 HBV-positive human HCC and 9 Liver-SB/HBsAg tumors using RT-qPCR. The deregulation of key metabolic genes was positively correlated in these two species, and similar gene expression signatures were found for most genes (Supplementary Figure 15). The classification of HCC CIS genes has thus highlighted the importance and specificity of targeting metabolic genes in HCC. The fact that normal hepatocytes display important general metabolic functions that most likely need to be reorganized during malignant transformation may explain the reprogramming by SB of many aspects of hepatocyte metabolism.
Metabolic profiling and mapping of mouse HCC tumors
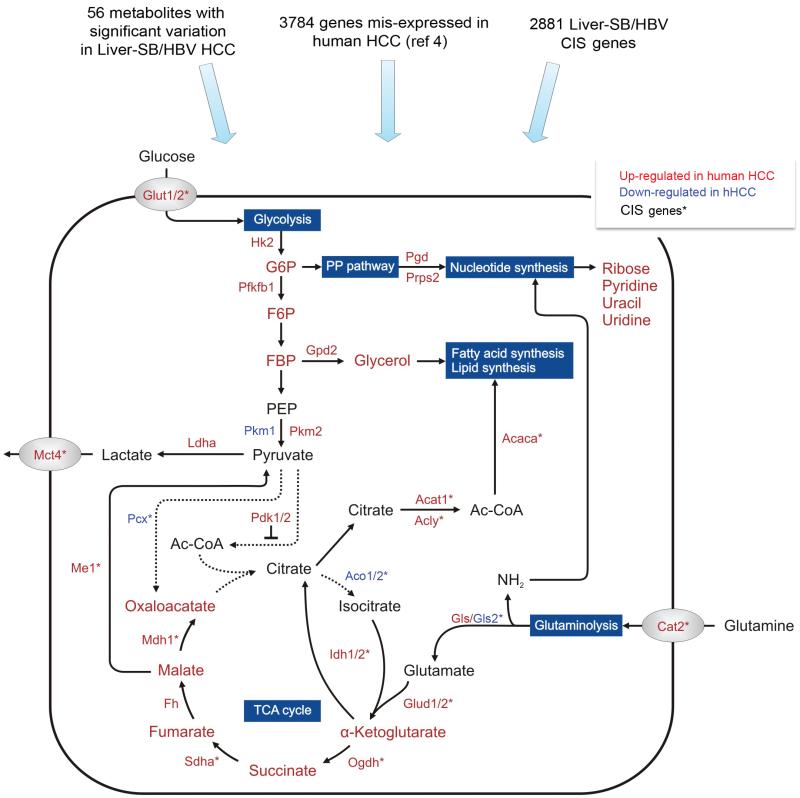
To better understand how genetic changes in metabolic genes affect hepatic tumor metabolism we performed non-targeted metabolic profiling on eight food-restricted Liver-SB/HBsAg animals. Chemical derivatisation and gas chromatography coupled to time-of-flight mass spectrometry (GC/TOFMS) were utilized to identify the differences in metabotypes between malignant (tumor masses > 3mm) and adjacent normal liver tissue. The quality of acquired metabolomics data was verified by principal component analysis (PCA) and partial least squares discriminant analysis (PLS-DA) (Supplementary Figure 16). Even with eight matched samples, statistical analyses using the t-test and Welch-test were able to identify 56 annotated metabolites significantly changed in tumors compared to normal adjacent tissues (Supplementary Figure 17). Consistent with the transposon data we noticed significant alterations corresponding to specific metabolic pathways. For instance, carbohydrate metabolism was affected through increased levels of glucose and fructose and reduced ribitol, ribonic acid and allonic acid. Levels of several amino acids including isoleucine, valine, aspargine, tyrosine, methionine, serine, leucine, phenylalanine, threonine and alanine were also significantly downregulated in tumors, consistent with data from human tumors56. Moreover, components of nucleic acid including ribose, pyridine, uracil and uridine were found at higher levels in tumors. We then integrated the genetic (Supplementary Table 1c), expression4, and metabolic results (Supplementary Table 7) in order to identify potential correlations. We noticed a marked consistency between genetic alterations, mRNA levels and actual amounts of metabolites, revealing a probable control of metabolic changes by genetic modifications at least to glycolysis, the tricarboxylic acid (TCA) cycle, glutaminolysis, the pentose phosphate (PP) pathway (Figure 5) and redox regulation (Supplementary Figure 18). Collectively, these metabolic alterations control bioenergetics, biosynthesis and redox regulation, which are known to optimize tumor cell metabolism for proliferation and survival.
Figure 5. Mapping of genomic and metabonomic data to metabolic pathways disrupted in HCC.
To understand the metabolic pathways disrupted in HCC at the genetic, mRNA, and metabolite levels, we used data from the Liver-SB/HBV screen (Supplementary Table 1c), human expression data4, and metabolomic results (Supplementary Table 7). Metabolites are written with larger font size, and genes with smaller font size. Red or green fonts mean the gene expression or metabolite amount is increased or decreased, respectively, in HCC tissue versus adjacent non-tumor tissue. CIS genes listed in Supplementary Table 1c are labeled with *. This map focuses on glycolysis, TCA cycle, glutaminolysis, and the pentose phosphate (PP) pathway. Despite the increased glucose uptake (Glut1/2) and consumption, the tumor cells predominantly expressed the pyruvate kinase M2 isoform (Pkm2), which converts phosphoenolpyruvate (PEP) to pyruvate less efficiently than Pkm1 70. This promotes the accumulation and shuttling of glycolytic intermediates such as glucose-6-phosphate (G6P), fructose-6-phosphate (F6P), and fructose-1,6-biphosphate 2 (FBP) to the pentose phosphate (PP) pathway and macromolecular synthesis69. Thus, this termination of the glycolysis metabolic pathway together with dysregulation of Pcx and Pdk1/2, would prevent pyruvate from entering the TCA cycle. This would also lead to a truncated TCA cycle and insufficient glucose-dependent citrate production. However, most of the TCA cycle genes (Idh1/2, Sdha, Fh, Odgh) and corresponding intermediates are up-regulated in tumor cells, except for the down-regulation of the aconitase gene family (Aco1/2), suggesting the TCA cycle is at least partially activated. Our results imply an activation of glutaminolysis (Cat2, Gls, Glud1/2) converting glutamine to α-ketoglutarate to replenish TCA cycle intermediates. Effective maintenance of citrate synthesis is possible through reductive carboxylation of glutamine-derived α-ketoglutarate by Idh1/273. These adaptations around glycolysis and the TCA cycle could allow rapid generation of both ATP for bioenergetics and important metabolites for biosynthesis. In addition, the breakdown of citrate by Acly could constitute a primary source of acetyl-CoA for fatty acid and lipid synthesis. Moreover, up-regulation of Ldha, could convert glutamine-derived pyruvate to lactate that is excreted by Mct4 transporter outside the cells68.
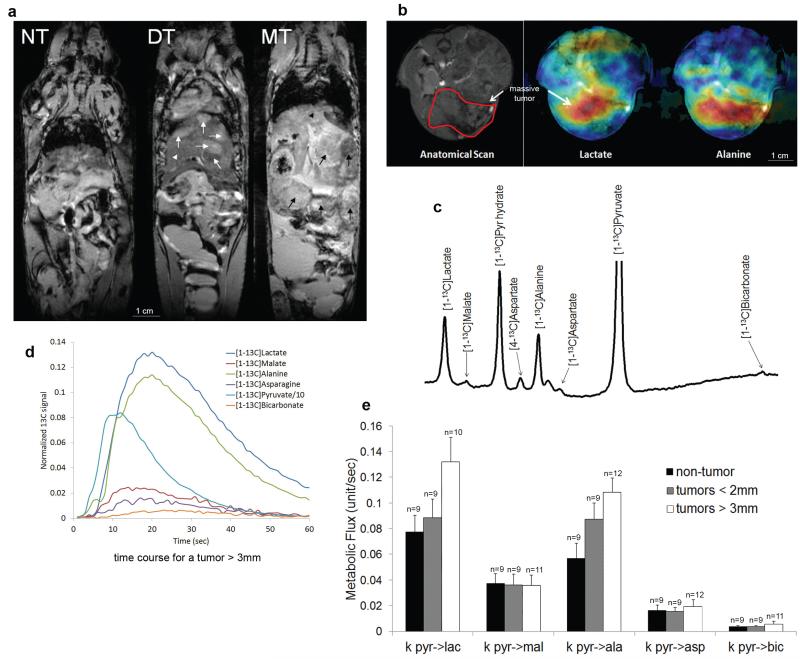
We also tested our interpretation of the integrated data using biochemical assays and in vivo live metabolic imaging 57. We found that intrahepatic lactate dehydrogenase (Ldh), alanine transaminase (Alt) and glutaminase (Gls) enzymatic activities were consistently more strongly activated in large hepatic tumors (Supplementary Figure 19), supporting the enhanced conversion of pyruvate to alanine and lactate, and the inclusion of glutamine possibly as an anaplerotic carbon source to supplement the increased metabolic fuel requirement in larger tumors58. Metabolic imaging with hyperpolarized pyruvate57 also showed a major conversion of pyruvate into alanine and lactate (Figure 6), while its conversion into bicarbonate and other TCA intermediates are unchanged, indicating a shift towards aerobic glycolysis and anabolic processes, relative to oxidative phosphorylation. It is to be noted however that pyruvate is only one of the fuel sources in HCC, and other processes generating ATP such as lipid catabolism have indeed been found to be significantly increased in HCC as well 56,59. Overall, these experimental findings were consistent with our predictions based on the integration of multiple datasets.
Figure 6. Quantitative changes in pyruvate metabolism can be detected by hyperpolarized carbon-13 magnetic resonance spectroscopy.
in vivo. a, T2-weighted MRI illustrates the development of HCC at different stages in Liver-SB/HBsAg animals. The anatomy appears normal in the pre-tumor stage (NT), at 2-5 months of age. However, at approximately 8 months of age, tumors (white arrows) began to appear within the normal liver (white arrow head). This is referred to as the developing tumor (DT) stage. Massive tumors (MT) were detected by MRI approximately two months later (black arrows). b, Live in vivo imaging provided the mapping of lactate and alanine production in HCC after hyperpolarized pyruvate injection (blue -> green -> yellow -> red represent amounts of molecules lowest to largest). c, Representation of the in vivo measured hyperpolarized carbon-13 spectra in the liver. Metabolites that were detected include [1-13C]lactate (183.0 ppm), [1-13C]malate (181.5 ppm), [1-13C]pyruvate hydrate (179.1 ppm), [4-13C]aspartate (177.8 ppm), [1-13C]alanine (176.4 ppm), [4-13C]oxaloacetate (175.6 ppm), [1-13C]aspartate (175.0 ppm), [1-13C]pyruvate (170.8 ppm) and [1-13C]bicarbonate (160.8 ppm). The pyruvate peak was truncated to better illustrate the downstream metabolite peaks. d, Representative time course depicting the simultaneous production of downstream metabolites upon infusion of hyperpolarized [1-13C]pyruvate in a mouse bearing a large liver tumor. e, Quantitative changes in pyruvate metabolism can be detected by hyperpolarized carbon-13 magnetic resonance spectroscopy in live mice in vivo. Rate of exchange of 13C label from [1-13C]pyruvate (pyr) to [[1-13C]lactate (lac), [1-13C]malate (mal), [1-13C]alanine (ala), [1-13C]aspartate (asp), and [1-13C]bicarbonate (bic) after [1-13C]pyruvate infusion into mice at different tumor development stages. ANOVA tests were performed for each metabolic flux result. At the FDR-corrected significance level of 0.05, no differences between group means were found. However, explicitly testing for a trend by computing Spearman correlation showed that the metabolic flux pyr->ala significantly increased with tumor size (rho = 0.51; FDR-corrected p=0.022). Although a strong trend was also observed for the metabolic flux pyr->lac, this did not reach statistical significance (rho = 0.43; FDR-corrected p = 0.052). The number of animals in groups is indicated above each bar. Error bars represent the standard error of the mean (SEM).
DISCUSSION
Here we report the first saturating transposon screen for any type of cancer. In total, we identified 2881 genes that were positively selected during HCC development, and 42.5% of these genes were also mutated or transcriptionally altered in human HCC. The human HCC genome contains a large spectrum of mutations including DNA amplifications, deletions, rearrangements, point mutations and loss of heterozygosity, in addition to epigenetic changes that together lead to tumor development. This has made it difficult to discriminate between the true driver and passenger mutations for HCC6. SB mutagenesis thus provides an important comparative genomics tool for identifying and validating the driver genes for HCC. We also found that many genes mutated by SB code for proteins that function in cancer signaling pathways known to be important for human HCC. In addition, we found that multiple genes from the same signaling pathway are often mutated in the same tumor. This is consistent with recent reports describing the Darwinian evolutionary processes operative in cancer cells60,61 and the recently recognized importance of convergent evolution to tumor development, in which the same gene or signaling pathway is mutated multiple times in different branches of the tumor evolutionary tree due to high selective pressure62. These results support the suggestions of others that it might be better to therapeutically target the signaling pathways themselves rather than the individually mutated genes.
HCC is thought to arise from the hepatocyte, a highly differentiated cell that has programed specialized functions. The transposon screen identified a surprisingly high enrichment for genes implicated in metabolic processes. This high enrichment seemed specific for HCC as it was not found in transposon screens for other types of cancer15,19,22,63. One of the major functions of hepatocytes is the control of metabolic homeostasis, including protein synthesis, transformation of carbohydrates, and synthesis of cholesterol, bile and phospholipids. Our metabolomics study is also consistent with assays performed using human samples of serum, urine64,65 or liver tissue56,59,66, demonstrating that these metabolic processes are indeed altered in transformed human hepatocytes. Using an integrative approach involving functional genomics and metabolomics, we further mapped the genetic, transcriptomic, and metabolomic results to core metabolic processes commonly affected in cancer67. This helped to uncover molecular mechanisms likely altered in HCC. This disrupted metabolism included the increased uptake of glucose and glutamine, which is implied by SB’s targeting of the membrane transporters Glut1/2 and Slc7a2 (Cat2), in addition to their up-regulated expression in human tumors. As human HCC tissues also show elevation of glucose and glutamine66,68 our data support that they may be genetically selected sources that fuel tumor HCC cell metabolism. We also identified a shift towards aerobic glycolysis and anabolic processes, relative to oxidative phosphorylation in liver tumors. Similar deregulation of metabolic genes was observed in human HCC, suggesting anabolic pathways, in conjunction with aerobic glycolysis (Warburg effect)69, contribute to the enhanced glucose metabolism in HCC. Notably, recent in vitro studies showed that heterogeneous nuclear ribonucleoproteins (hnRNP) could induce alternative splicing and thereby results in a high PKM2/PKM1 expression ratio. This change could be important for the promotion of cell proliferation and aerobic glycolysis in tumors70,71. In line with these studies we found that SB not only targets Hnrnpa1, but mouse and human HCCs also significantly overexpress this gene proportionally to PKM2 expression. This suggests that glucose is being shunted via the PP pathway towards the biosynthesis of molecules including nucleotides70, and supports the increased lactate production (aerobic glycolysis) that was observed in our in vivo metabolic imaging. Tumor cell proliferation diverts citrate towards fatty acid synthesis as the SB screen suggested. Since the TCA cycle is crucial for generating intermediate metabolites to ensure cell viability, anaplerotic pathways must be activated. Indeed, our results show glutaminolysis as such a possible carbon source with SB mutations in many genes regulating the glutaminolytic pathway72,73.
Metabolic adaptations associated with glucose and glutamine pathways in cancerous hepatocytes could also enhance the antioxidant defenses through NADPH-dependent glutathione and thioredoxin systems in order to maintain cellular redox balance. In this way, changes in glucose and glutamine pathways could support three basic needs for dividing tumor cells, namely bioenergetics, biosynthesis and redox regulation. Inhibiting glycolysis and/or glutaminolysis might therefore be efficacious in restricting liver tumor formation and progression, as suggested for other cancer types74. Genes identified by our transposon screen have thus provided a broader and deeper understanding of hepatocellular cancer genomics and metabolism as well as offering potential molecular and targets for cancer therapy for instance in signaling and metabolic pathways.
ONLINE METHODS
Mice and histology
We utilized the following alleles to generate a mouse model of HBV-associated hepatocellular carcinoma: HBsAg transgenic18, Alb-Cre75, T2/Onc2 (6113)13 or T2Onc3 (12740) and Rosa26-lsl-SB1114. The resulting cohorts were on a mixed B6.129 genetic background. Alb-Cre/+; T2Onc2/+; Rosa26-lsl-SB11/+; HBsAg/+ mice were used for the SB screen. Alb-Cre/+; T2Onc3/+; Rosa26-lsl-SB11/+; HBsAg/+ were used for metabolic imaging. All animals were monitored on a biweekly basis in accordance with IACUC guidelines. Full necropsies were performed. For DNA extraction, liver tumors were measured and snap-frozen. For histology, livers were 10% formalin fixed and paraffin-embedded. 5μm sections were processed for haematoxylin and eosin staining, or orcein staining. All slides were reviewed by our veterinary pathologist, Dr. Jerrold M. Ward. Histological classification of hepatic lesions and tumors were as previously reported18,76.
Identification of transposon insertions sites
Identification of transposon insertion sites was performed using splinkerette PCR to produce barcoded PCR products that were pooled and sequenced on the 454 GS-Titanium (Roche) platform. Reads from sequenced tumors were mapped to the mouse genome assembly NCBI m37 and merged together to identify non-redundant Sleeping Beauty (SB) insertion sites. Cloning and mapping of the transposon insertion sites were performed as previously described19,22. 328,687 non-redundant insertion sites (Supplementary Dataset 1) were used to identify CISs using a gene centric method20 and a Gaussian Kernel Convolution (GKC) statistical framework method21.
Gaussian Kernel Convolution (GKC) method for common insertion site (CIS) identification
Any insertions sites on the transposon donor mouse chromosome 1 for tumors derived from T2/Onc2 were excluded from common insertion site (CIS) analysis. The likelihood of ‘local hopping’ of the transposon is increased where the transposon array is located77. This phenomenon can significantly increase the background level of transposon insertion events, thereby complicating CIS analysis. 328,687 non-redundant insertion sites were used to identify CISs using a Gaussian kernel convolution (GKC) statistical framework21. The previous GKC analysis approach22 was enhanced by utilizing multiple kernel scales (widths of 15K, 30K, 50K, 75K, 120K and 240K nucleotides). CISs predicted across multiple scales and overlapping in their genomic locations were clustered together, such that the CIS with the smallest genomic “footprint” was reported as the representative CISs. For highly significant CISs with narrow spatial distributions of insertion sites, the 15K kernel is typically the scale on which CISs are identified. The P value for each CIS was adjusted by chromosome and a cut-off of p<0.05 was used.
Pruning genetically similar tumors from the same animals
We initially sequenced 250 tumors from 34 mice and called CISs using the GKC method on all tumors (with a non-stringent p-value significance threshold to capture many loci). Using the boundaries of the CIS loci, we obtained genomic positions that defined 2110 bins on the genome. We scored every tumor for every bin as having an insertion in the bin or not. We computed the Spearman correlation between all tumors and performed hierarchical clustering (complete linkage). When tumors from the same animal clustered together as pairs in the dendrogram, we retained only the tumor, which had the largest total number of insertions. The aim of the pruning is to ensure that there is at least a single tumor from another animal that is most similar to a tumor from a given animal. In theory, pairs of clustered tumors from the same animal can remain after the pruning step if, for example, three tumors originally clustered together and only one was removed. To resolve this problem the pruning step can be repeated iteratively. After the pruning step, we tested whether the median Spearman correlation between the 228 remaining same-animal-tumors and the median Spearman correlation between the non-same-animal tumors was statistically significant. If this was not the case, we terminated the pruning process. On our dataset a single pruning round was sufficient. We used the remaining 228 tumor samples further to call CISs with both GKC and gCIS methods.
Analysis of screen saturation
In order to determine whether the SB screen had reached saturation, and if so, what number of samples was sufficient to reach saturation, the Gaussian Kernel Convolution (GKC) CIS calls from all 228 samples were analyzed using the ACT software package78. ACT considers genomic locations generated by multiple samples for specific biological phenomenon under study (e.g.ChIP-seq peaks) to determine the saturation of a screen. The program considers the various combinations in which samples can be added so that the increase in base pair coverage is a range of values based on all the samples. The results can be depicted as a series of boxplots showing the increase in base pair coverage, where the boxplot at each position n on the x-axis shows the coverage values of all combinations of n samples. Boxplots that approach a horizontal asymptote indicate that the coverage has reached saturation.
For the GKC CISs generated by all 228 samples, the insertion sites that contributed to CISs were extracted, resulting in a set of approximately 131,000 sites. The insertion sites were then selected per sample and pseudo-kernels of 7.5k nucleotides either side of each insertion were applied to mimic GKC kernels of 15k nucleotides. Overlapping kernels within each sample were merged into continuous genomic regions. These 228 modified insertion files were then analyzed using ACT. For each combination of samples the median values, and 25th and 75th percentiles were plotted using the ggplot2 visualization package for the R statistical analysis platform. As a control, the 228 samples were reanalyzed where the same number of insertion sites per sample was selected at random across the mouse genome (excluding chromosome 1, the donor chromosome). The pseudo-15k nucleotide kernels were applied. Figure 2a shows the saturation plot for all 228 samples, clearly indicating a ‘knee’ in the profile as the graph asymptotes with an increasing number of samples. Visual inspection of the graph indicates that below 50 samples, the screen does not appear to have reached saturation. From 100 samples upwards there appears to be sufficient coverage to report saturation. Conversely, Supplementary Figure 4a for the randomized samples is virtually a straight line, indicating no saturation. While the analysis does not produce a clear-cut asymptote this is to be expected due to the type of data under consideration. ACT was designed to analyze such data as ChIP-seq arrays for predicting transcription factor binding sites. In these scenarios ChIP-seq replicates should ideally report the same key binding sites/genomic locations. Hence across multiple samples the same locations should be reported. For SB screens however, while insertions in the same gene will be found from different samples, the locations of the insertion sites will not overlap perfectly, even with the addition of the 15k nucleotide pseudo kernels. Hence each sample will introduce novel regions, such that the overall coverage will continue to increase even if the screen has truly reached a ‘saturation’ point. Also, not all samples will contribute to all CISs. Different combinations of samples will thereby result in varying coverage’s, causing the coverage profile not to asymptote perfectly.
Cross-species oncogenomics analysis
Gene expression assays, conducted on 223 HBV-positive human patients diagnosed with HCC, was used to compare tumors and paired non-tumor tissues. The microarray data are publicly available and downloadable at the Gene Expression Omnibus database with accession number GSE145204. Human hepatocellular carcinoma (HHC) expression profiles were downloaded from the GEO database (GSE14520). Only the 223-paired samples hybridized to the HG-U133A platform were selected. Then, CEL files were Robust Multi-array Average (RMA) normalized and differential gene expression between normal and tumor cells were computed using Affy and limma package, implemented in Bioconductor. As a result, adjusted P-value (FDR-based p-value adjustment according to Benjamini and Hochberg) and fold-change were obtained for each probset. Our mouse candidate cancer genes were converted to human genes using three different databases of homologous genes (MGI, HomoloGene and Ensembl)79-81. The obtained human genes were then mapped to the list of Affymetrix platform probe sets. For genes detected with more than one probe set, the probe set with best-adjusted p-value was considered as gene representative. The threshold of significance used was the absolute value of fold change more than 1.5, and adjusted p value less than 0.00001.
Smaller scale liver mutagenesis screen in the Sav1 deficient background
This study was performed in a Sav1 deficient background with Alb-Cre/+, T2Onc2/+, Rosa26-lsl-SB11/+, Sav1fl/fl animals. The Sav1 conditional allele was previously described30. A total of 136 large tumors from 31 animals were utilized for analysis. Most animals were euthanized at 8 to 9 months of age, when they had multiple large tumors. The protocol for genomic DNA extraction, barcoded amplification, and sequencing on Illumina Hi-Seq2000 was previously described20. 6,251,915 reads were mapped to the mouse genome (mm9) and filtered when they were represented by very few sequence reads20. Finally, the gene-centric CIS identification method (gCIS) identified 73 common insertion sites (Supplementary Table 1e).
Patient Samples
Resected tumor samples were obtained from patients undergoing curative resection for HCC (n=172). Operations took place between 1991-2009 at the National Cancer Centre, Singapore. All samples were collected in accordance with the requirements of the local Singapore Ethical Committees and informed consent was obtained from all subjects. The patient demographics and clinical characteristics have already been described in a previous study82. Total RNA was isolated using TRIzol (Invitrogen) following the manufacturer’s instructions, and the RNA concentration was quantified by NanoDrop (Thermo Scientific). RNA samples were reverse transcribed to cDNA using the SuperScript III cDNA Synthesis Kit (Invitrogen) in 10 μl reactions containing 1 μg total RNA and oligo(dT) primers according to manufacturer’s instructions.
Gene expression analysis from patient samples
Specific primers targeting genes of interest were designed using Primer Express software version 3.0. Primer sequences used for specific target amplification (STA pre-amplification reaction) as well as qRTPCR are listed in Supplementary Table 4. For the STA reaction, each cDNA sample was pre-amplified with 200nM pooled STA primer mix and Tagman PreAmp Master Mix (Applied Biosystems) in a 5ul reaction, which was run for 14 cycles according to the manufacturer’s protocol. To remove unincorporated primers, each sample was treated with Exonuclease I (Fermentas) following incubation at 37°C for 30 minutes. For inactivation, the mix was in a second step, incubated at 80°C for 15 minutes. At the end of the Exonuclease I treatment, the reactions were diluted 1:5 in TE buffer (pH 8.0) prior to use for qRT-PCR. The Fluidigm BioMark™ real-time PCR system and 48.48 Microfluidic Dynamic Arrays were employed for high-throughput qRT-PCR analysis83. As volume per inlet is 5 μl, the 6 μl volume per inlet with overage was prepared. For the samples, 2.7 μl of each STA and ExoI-treated sample were mixed with 20X DNA Binding Dye Sample Loading Reagent (Fluidigm) and 2X SsoFast EvaGreen SuperMix with Low ROX (Bio-Rad). For the gene expression assays, 0.3 μl of mix primer pairs (100 uM) was added with 2X Assay Loading Reagent (Fluidigm) following the addition of 1X TE buffer to 6 ul volume. Prior to loading the samples and assays into the inlets, the chip was primed in the NanoFlex 4-IFC Controller. The samples and assays were then loaded into the inlets of the dynamic array. Following loading and mixing of the samples and assays into the chip by the IFC Controller, PCR was run with the following reactions conditions: 50°C for 2 minutes, 95°C for 10 minutes, followed by 40 cycles of 95°C for 15 seconds and 60°C for 60 seconds. Global threshold and linear baseline correction were automatically calculated for the entire chip. The melting curve analysis and cycle threshold (Ct) were provided by Fluidigm Real-Time PCR Analysis software version 3. Fold change in expression of genes of interest between liver tumor and adjacent non-tumor samples were determined using the comparative Ct method following the formula: 2−ΔCt(tumor)/2−ΔCt(non-tumor) 84. GUSB and Gusb were used as the internal control gene in human and mouse samples, respectively. The -ΔCt data obtained from this calculation was then used to generate the heatmap as well as supervised hierarchical clustering of tumor and adjacent non-tumor samples by dChip software. In addition, statistical analysis was performed by a standard t-test to identify significant differentially expressed genes (p ≤ 0.05) between two studied groups for each species.
Pathway and gene ontology analyses
Gene-annotation enrichment analyses were completed with DAVID Bioinformatics v6.785. The list of all human genes was used as default background. For gene ontology analyses GOTERM and PANTHER were used. For pathway analyses, KEGG was selected. We obtained enrichment of genes that display a genetic association with cancer through DAVID using GENETIC_ASSOCIATION_DB_DISEASE and OMIM.
Metabolomics experiments
Chemicals for metabolomics experiments
HPLC-grade chloroform and methanol were purchased from Tedia Company Inc. (Fairfield, OH, USA). HPLC-grade toluene was purchased from JT Baker (Center Valley, PA, USA). MSTFA (N-methyl-N-(trimethylsilyl)trifluoroacetamide) with 1% TMCS (trimethylchlorosilane) and methoxyamine hydrochloride in pyridine (MOX) were purchased from Pierce (Rockford, IL, USA). Urease of Sigma type III, alkane standard mixture (C10 to C40), and sodium sulfate (anhydrous) were obtained from Sigma Aldrich (St. Louis, MO). Milli Q water (Millipore, Bedford, MA, USA). All other chemicals were of analytical grade.
Mouse Liver Tissue Sample Preparation
Each control and tumor liver tissue were weighed accurately and transferred to labeled 15 mL glass centrifuge tubes. Monophasic extraction solvent consisting of chloroform-methanol-water (2:5:2, v/v/v) was added to each sample such that the liver tissue concentration of each sample was 20 mg/mL. The tissue-solvent mixtures were homogenized by ultrasonicating at ambient temperature (24-28°C) for 90 min followed by mill-mixing using stainless steel beads for another 15-20 min. The homogenates were centrifuged at 3000 rpm at ambient temperature for 3 min and 800 μL of each supernatant was transferred to a clean 15 mL glass tube. Quality control (QC) samples (n=3) were prepared by pooling equal amounts of supernatant from 4 control and 4 tumor mouse liver tissues. These samples were dried under a gentle flow of nitrogen at 50°C for 60 min in the Turbovap LV (Caliper Life Sciences, Hopkinton, MA, USA). 100 μL of toluene (kept anhydrous with sodium sulfate) was added to each of the tissue extracts, vortexed for 1 min and dried at 50°C for 45 min using Turbovap LV in order to eliminate any trace of water which might interfere with the GC/MS analysis. The dried metabolic extract was derivatized first with 40 μL of MOX (20 mg/mL) at ambient temperature for 16 h. Subsequently, 60 μL of MSTFA with 1% TCMS was added to the mixture and incubated for 30 min at 70°C to form the trimethylsilyl (TMS) derivatives. The mixtures were cooled and 90 μL of each derivatized sample was transferred into a vial and subjected to GC/TOFMS analysis.
GC/TOFMS Analysis
The gas chromatography time-of-flight mass spectrometry analysis (GC/TOFMS) of the derivatized liver samples was performed using Pegasus 4D GC×GC/TOFMS (LECO Corporation, St. Joseph, MI, USA. The chromatography separation was conducted in one dimension (1D) GC mode where a 30 m DB-1 GC column with internal diameter of 250 μm and film thickness of 0.25 μm (Agilent Technologies, Santa Clara, CA) was used. Helium was used as the carrier gas at a flow rate of 1.5 mL/min. An injection volume of 1 μL was used and the injector split ratio was set to 1:5. The injector, transfer line and ion source temperatures were maintained at 220, 200, and 250 °C, respectively, throughout each analysis. Oven temperature was programmed at 70 °C for 0.2 min, increased at 10°C/min to 270°C where it was held for 10 min and further increased at 40°C/min to 310°C where it was held for 9 min. The MS was operated in EI mode (70 eV) and the detector voltage was set at 1800 V. Data acquisition was performed in the full scan mode from m/z 40 to 600 with an acquisition rate of 20 spectra/s.
Metabolic Data Preprocessing
Each chromatogram obtained from GC/TOFMS analysis was processed for baseline correction, noise reduction, smoothing, peak deconvolution, analyte alignment, preliminary analyte identification and area calculation using a data processing method created using ChromaTOF software version 4.21 (LECO Corporation). Only peaks with a signal-to-noise ratio (S/N) greater than 210 were utilized for further analysis. The area of each peak was calculated using unique mass. Preliminary metabolite identifications were performed for peaks with a similarity index of more than 60% and these were assigned putative metabolite identities based on the National Institute of Standards and Technology (NIST) library, LECO/Feihn Metabolomics library (LECO Corporation) and internally compiled spectral libraries. The resulting data table subsequent to data preprocessing was then exported to an Excel spreadsheet. For certain samples, missing values in their respective data tables were replaced by either integrating the baseline at retention times where peaks were confirmed to be missing or manually integrating peaks which were confirmed to have their retention times significantly shifted. The total area normalization for each sample was performed by dividing the integrated area of each analyte by the sum of all peak areas of analytes present in the sample. The normalization aided in correcting variations due to the amount of liver collected, sample preparation and analysis.
Chemometric Data Analysis
Normalized data were exported to SIMCA-P+ (Version 12.0, Umetrics, Umeå, Sweden) to perform principal component analysis (PCA) to identify clustering trends, as well as to detect and exclude outliers. Prior to PCA analysis, GC/TOFMS data was mean-centered and unit variance scaled. A DModX plot was calculated to check for any outliers. QC samples were also analyzed in the PCA analysis to ensure that the data acquisition for GC/TOFMS metabolic profiling was reproducible for all samples. After initial overview of the GC/TOFMS data using PCA analysis, the data were subjected to supervised data analysis using partial least-squares discriminant analysis (PLS-DA) where a model was built and utilized to identify marker metabolites that accounted for the differentiation in the two groups consisting of liver tumor tissues and normal-matched liver tissues. To validate and investigate overfitting of the data in the PLS-DA model, permutation tests with 100 iterations were carried out using SIMCA-P+. This permutation test compared the goodness of fit of the original model with the goodness of fit of several models based on data where the order of the Y-observations were randomly permuted, while the X-matrix was kept intact.
Marker Metabolite Screening
The criteria for the selection of marker metabolites were namely, variable importance values (VIP) greater than 1.0 and t-test (Welch’s correction) p value less than 0.05. The fold change of each marker metabolite detected within the control and tumor groups was calculated. Fold change value of less than one indicated a higher level of marker metabolite in the normal tissue. Hierarchical clustering was performed using algorithms within dChip software according to established methods86. Pearson correlation subtracted from unity was used as the distance metric, using the centroid linkage method, which provides bounded distances in the range (−2, 2). P value threshold for function enrichment was < 0.01.
Ex vivo biochemical enzyme activity assays
For each enzyme activity assay, 100 mg of liver tissue was homogenized in 200 μL of ice-cold 100 mM tris HCl buffer, then centrifuged for 10 min at 13,000 × g to remove insoluble material. All assays were based on continuous spectrophotometric rate determination. Hepatic ALT, AST and GLS enzyme activities were measured with commercially available enzymatic assay kits (Biovision, catalog #752-100, #753-100; Sigma-Aldrich, EC 3.5.1.2).
Magnetic resonance spectroscopy (MRS) and metabolic imaging
This technology is partially described in a recent publication 57.
Pyruvate polarization and dissolution (metabolic imaging)
40 mg of [1-13C]pyruvic acid (99% Carbon-13 labeled, Part # 677175, Sigma Aldrich), doped with 15 mM trityl-radical (OXO63, GE Healthcare) and 3 μl of gadoterate meglumine (10 mM, Dotarem®, Guerbet), was hyperpolarized in a polarizer, with 45 min of microwave irradiation. The sample was subsequently dissolved in a pressurized and heated alkaline solution, containing 100 mg/liter ethylenediaminetetraacetic acid (EDTA) to yield a solution of 80 mM hyperpolarized sodium [1-13C]pyruvate with a polarization of 45% and physiological temperature and pH87.
Metabolic flux in the liver measured by hyperpolarized carbon-13 MRS
Mice were positioned in a 9.4 T horizontal bore MR scanner interfaced to a DD2 console (Varian Medical Systems), and inserted into a dual-tuned (1H/13C) mouse volume coil (diameter, 39 mm). Correct positioning was confirmed by the acquisition of an axial proton gradient-echo image: echo time/repetition time (TE/TR), 8.0/100.0 ms; matrix size, 128 × 128; field-of-view (FOV), 30 × 30 mm; slice thickness, 1.0 mm; excitation flip angle, 30°. A respiratory-gated shim was used to reduce the proton linewidth to approximately 160 Hz. Immediately before injection, a respiratory-gated 13C MR pulse-acquire spectroscopy sequence was initiated. 250-350 μL (0.5 mmol/kg body weight) of hyperpolarized pyruvate was intravenously injected over 3 s into the anesthetized mouse. To localize signal over the liver, a 4 to 6 mm slice excitation was applied. Sixty individual liver spectra were acquired over 1 min after injection (TR, 1 s; excitation flip angle, 25°; sweep width, 4000 Hz; acquired points, 2048; frequency centered on the pyruvate resonance).
MRS data analysis
Liver 13C MR spectra were analyzed using the AMARES algorithm as implemented in the jMRUI software package. Spectra were baseline and direct-current offset-corrected based on the last half of acquired points. Peaks corresponding to [1-13C]pyruvate and its metabolic derivatives [1-13C]lactate, [1-13C]malate, [1-13C]pyruvate hydrate, [1-13C]alanine, [4-13C]oxaloacetate, [1-13C]aspartate, [1-13C]bicarbonate were fitted with prior knowledge assuming a Lorentzian line shape, peak frequencies, relative phases, and linewidths. Quantified peak areas were plotted against time in Excel (Microsoft). The rate of exchange of the 13C label from the hyperpolarized pyruvate to each of its downstream metabolites was calculated with a kinetic model specifically designed to assess hyperpolarized pyruvate metabolism, using the fitted peak areas as input data. The model accounts for many of the variables in the hyperpolarized experiment, including the rate of injection, the initial polarization level of the hyperpolarized pyruvate, and the rate of decay of each of the hyperpolarized compounds. Firstly the change in [1-13C]pyruvate signal over the 60s acquisition time was fitted to the integrated [1-13C]pyruvate peak area data using equation [1]:
| [1] |
In this equation, Mpyr(t) represents the [1-13C]pyruvate peak area as a function of time. This equation fits the parameters kpyr, the rate constant for pyruvate signal decay (s−1), rateinj, the pyruvate arrival rate (a.u. s−1), tarrival, the pyruvate arrival time (s) and tend, the time correlating with the end of the injection (s). These parameters were then used in equation [2] along with the dynamic [1-13C]malate and [1-13C]aspartate to calculate kpyr→X, the rate constant for exchange of pyruvate to each of its metabolites (s−1) and kX, the rate constant for signal decay of each metabolite (s−1) which was assumed to consist of metabolite T1 decay and signal loss from the low flip angle radiofrequency pulses. In equation [2], t’ = t – tdelay, where tdelay represents the delay between pyruvate arrival and metabolite appearance caused by the traversal of the pyruvate through the cardiopulmonary circulation before arrival at the hepatic arteries.
| [2] |
Metabolic imaging
A 2D gradient-echo chemical shift imaging (CSI) pulse sequence was implemented in a 9.4 T MRI (magnetic resonance imaging) scanner. Imaging begins about 16 seconds after infusion of hyperpolarized pyruvate to ensure phase encoding starts only when all metabolites began to form. They were then axially imaged over a time-course of 2 mins with repetition time (TR) = 120 ms, echo-time (TE) = 0.6 ms, flip angle (FA) = 5°, spectral width (SW) = 80 ppm, spectral points = 256, field-of-view (FOV) = 25.6 × 25.6 mm2, matrix size = 16×16, slice thickness = 5 mm. Post-processing with Matlab picked up the peak of each metabolite in the spectra, filled up its k-space, zero-filled to 32 × 32, followed by 2d fourier transform. A reference proton image was acquired with matrix size = 128 × 128, TR = 100 ms, TE = 1.28 ms and slice thickness = 1 mm.
Supplementary Material
ACKNOWLEDGMENTS
We acknowledge Kurt Reifenberg at Johannes Gutenberg University, Germany for giving us the HBsAg mouse strain (originated from Frank Chisari). We also thank Keith Rogers and Susan Rogers at the IMCB Histopathology core for their necropsy and histotechnology assistance. We thank Pearlyn Cheok, Nicole Lim, and Dorothy Chen for their help with mouse breeding and monitoring. This work was supported by the Biomedical Research Council, Agency for Science, Technology and Research, Singapore and the Cancer Prevention Research Institute of Texas (CPRIT). A.G.R. and D.J.A. are supported by the Wellcome Trust and Cancer-Research UK. N.A.J. and N .G.C. are both CPRIT Scholars in Cancer Research.
Footnotes
COMPETING FINANCIAL INTERESTS: The authors declare no competing financial interests.
REFERENCES
- 1.Chen X, et al. Gene expression patterns in human liver cancers. Mol Biol Cell. 2002;13:1929–1939. doi: 10.1091/mbc.02-02-0023.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Imbeaud S, Ladeiro Y, Zucman-Rossi J. Identification of novel oncogenes and tumor suppressors in hepatocellular carcinoma. Semin Liver Dis. 2010;30:75–86. doi: 10.1055/s-0030-1247134. [DOI] [PubMed] [Google Scholar]
- 3.Mas VR, et al. Genes involved in viral carcinogenesis and tumor initiation in hepatitis C virus-induced hepatocellular carcinoma. Mol Med. 2009;15:85–94. doi: 10.2119/molmed.2008.00110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Roessler S, et al. A unique metastasis gene signature enables prediction of tumor relapse in early-stage hepatocellular carcinoma patients. Cancer Res. 2010;70:10202–10212. doi: 10.1158/0008-5472.CAN-10-2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wurmbach E, et al. Genome-wide molecular profiles of HCV-induced dysplasia and hepatocellular carcinoma. Hepatology. 2007;45:938–947. doi: 10.1002/hep.21622. [DOI] [PubMed] [Google Scholar]
- 6.Herath NI, Leggett BA, MacDonald GA. Review of genetic and epigenetic alterations in hepatocarcinogenesis. J Gastroenterol Hepatol. 2006;21:15–21. doi: 10.1111/j.1440-1746.2005.04043.x. [DOI] [PubMed] [Google Scholar]
- 7.Herceg Z, Paliwal A. Epigenetic mechanisms in hepatocellular carcinoma: how environmental factors influence the epigenome. Mutat Res. 2011;727:55–61. doi: 10.1016/j.mrrev.2011.04.001. [DOI] [PubMed] [Google Scholar]
- 8.Berger AH, Knudson AG, Pandolfi PP. A continuum model for tumour suppression. Nature. 2011;476:163–169. doi: 10.1038/nature10275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.ENCODE Project Consortium et al. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fujimoto A, et al. Whole-genome sequencing of liver cancers identifies etiological influences on mutation patterns and recurrent mutations in chromatin regulators. Nat Genet. 2012;44:760–764. doi: 10.1038/ng.2291. [DOI] [PubMed] [Google Scholar]
- 11.Guichard C, et al. Integrated analysis of somatic mutations and focal copy-number changes identifies key genes and pathways in hepatocellular carcinoma. Nat Genet. 2012;44:694–698. doi: 10.1038/ng.2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Copeland NG, Jenkins NA. Harnessing transposons for cancer gene discovery. Nat Rev Cancer. 2010;10:696–706. doi: 10.1038/nrc2916. [DOI] [PubMed] [Google Scholar]
- 13.Dupuy AJ, Akagi K, Largaespada DA, Copeland NG, Jenkins NA. Mammalian mutagenesis using a highly mobile somatic Sleeping Beauty transposon system. Nature. 2005;436:221–226. doi: 10.1038/nature03691. [DOI] [PubMed] [Google Scholar]
- 14.Dupuy AJ, et al. A modified sleeping beauty transposon system that can be used to model a wide variety of human cancers in mice. Cancer Res. 2009;69:8150–8156. doi: 10.1158/0008-5472.CAN-09-1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rad R, et al. PiggyBac transposon mutagenesis: a tool for cancer gene discovery in mice. Science. 2010;330:1104–1107. doi: 10.1126/science.1193004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang JD, Roberts LR. Hepatocellular carcinoma: A global view. Nat Rev Gastroenterol Hepatol. 2010;7:448–458. doi: 10.1038/nrgastro.2010.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Keng VW, et al. A conditional transposon-based insertional mutagenesis screen for genes associated with mouse hepatocellular carcinoma. Nat Biotechnol. 2009;27:264–274. doi: 10.1038/nbt.1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chisari FV, et al. Molecular pathogenesis of hepatocellular carcinoma in hepatitis B virus transgenic mice. Cell. 1989;59:1145–1156. doi: 10.1016/0092-8674(89)90770-8. [DOI] [PubMed] [Google Scholar]
- 19.Mann KM, et al. Sleeping Beauty mutagenesis reveals cooperating mutations and pathways in pancreatic adenocarcinoma. Proc Natl Acad Sci USA. 2012;109:5934–5941. doi: 10.1073/pnas.1202490109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brett BT, et al. Novel molecular and computational methods improve the accuracy of insertion site analysis in Sleeping Beauty-induced tumors. PLoS One. 2011;6:e24668. doi: 10.1371/journal.pone.0024668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.de Ridder J, Uren A, Kool J, Reinders M, Wessels L. Detecting statistically significant common insertion sites in retroviral insertional mutagenesis screens. PLoS Comput Biol. 2006;2:e166. doi: 10.1371/journal.pcbi.0020166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.March HN, et al. Insertional mutagenesis identifies multiple networks of cooperating genes driving intestinal tumorigenesis. Nat Genet. 2011;43:1202–1209. doi: 10.1038/ng.990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Swanton C. Intratumor Heterogeneity: Evolution through Space and Time. Cancer Res. 2012;72:4875–4882. doi: 10.1158/0008-5472.CAN-12-2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Perez-Mancera PA, et al. The deubiquitinase USP9X suppresses pancreatic ductal adenocarcinoma. Nature. 2012;486:266–270. doi: 10.1038/nature11114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Solimini NL, et al. Recurrent hemizygous deletions in cancers may optimize proliferative potential. Science. 2012;337:104–109. doi: 10.1126/science.1219580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Whittaker S, Marais R, Zhu AX. The role of signaling pathways in the development and treatment of hepatocellular carcinoma. Oncogene. 2010;29:4989–5005. doi: 10.1038/onc.2010.236. [DOI] [PubMed] [Google Scholar]
- 27.Schmidt VA, Chiariello CS, Capilla E, Miller F, Bahou WF. Development of hepatocellular carcinoma in Iqgap2-deficient mice is IQGAP1 dependent. Mol Cell Biol. 2008;28:1489–1502. doi: 10.1128/MCB.01090-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang G, Liu T, Wang Z. Downregulation of MAGI1 associates with poor prognosis of hepatocellular carcinoma. J Invest Surg. 2012;25:93–99. doi: 10.3109/08941939.2011.606875. [DOI] [PubMed] [Google Scholar]
- 29.Horie Y, et al. Hepatocyte-specific Pten deficiency results in steatohepatitis and hepatocellular carcinomas. J Clin Invest. 2004;113:1774–1783. doi: 10.1172/JCI20513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lu L, et al. Hippo signaling is a potent in vivo growth and tumor suppressor pathway in the mammalian liver. Proc Natl Acad Sci USA. 2010;107:1437–1442. doi: 10.1073/pnas.0911427107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zheng T, Wang J, Jiang H, Liu L. Hippo signaling in oval cells and hepatocarcinogenesis. Cancer Lett. 2011;302:91–99. doi: 10.1016/j.canlet.2010.12.008. [DOI] [PubMed] [Google Scholar]
- 32.Santhekadur PK, et al. Multifunction protein staphylococcal nuclease domain containing 1 (SND1) promotes tumor angiogenesis in human hepatocellular carcinoma through novel pathway that involves nuclear factor kappaB and miR-221. J Biol Chem. 2012;287:13952–13958. doi: 10.1074/jbc.M111.321646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yoo BK, et al. Increased RNA-induced silencing complex (RISC) activity contributes to hepatocellular carcinoma. Hepatology. 2011;53:1538–1548. doi: 10.1002/hep.24216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kojima K, et al. MicroRNA122 is a key regulator of alpha-fetoprotein expression and influences the aggressiveness of hepatocellular carcinoma. Nat Commun. 2011;2:338. doi: 10.1038/ncomms1345. [DOI] [PubMed] [Google Scholar]
- 35.Xie Z, et al. Zinc finger protein ZBTB20 is a key repressor of alpha-fetoprotein gene transcription in liver. Proc Natl Acad Sci USA. 2008;105:10859–10864. doi: 10.1073/pnas.0800647105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Flaim CJ, Chien S, Bhatia SN. An extracellular matrix microarray for probing cellular differentiation. Nat Methods. 2005;2:119–125. doi: 10.1038/nmeth736. [DOI] [PubMed] [Google Scholar]
- 37.Watt AJ, et al. A gene trap integration provides an early in situ marker for hepatic specification of the foregut endoderm. Mech Dev. 2001;100:205–215. doi: 10.1016/s0925-4773(00)00530-x. [DOI] [PubMed] [Google Scholar]
- 38.Duns G, et al. Histone methyltransferase gene SETD2 is a novel tumor suppressor gene in clear cell renal cell carcinoma. Cancer Res. 2010;70:4287–4291. doi: 10.1158/0008-5472.CAN-10-0120. [DOI] [PubMed] [Google Scholar]
- 39.Jackson RC, Morris HP, Weber G. Adenosine deaminase and adenosine kinase in rat hepatomas and kidney tumours. Br J Cancer. 1978;37:701–713. doi: 10.1038/bjc.1978.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nii A, et al. Significance of dihydropyrimidine dehydrogenase and thymidylate synthase mRNA expressions in hepatocellular carcinoma. Hepatol Res. 2009;39:274–281. doi: 10.1111/j.1872-034X.2008.00457.x. [DOI] [PubMed] [Google Scholar]
- 41.Queener SF, Morris HP, Weber G. Dihydrouracil dehydrogenase activity in normal, differentiating and regnerating liver and in hepatomas. Cancer Res. 1971;31:1004–1009. [PubMed] [Google Scholar]
- 42.Cheng F, et al. Camptothecin-induced downregulation of MLL5 contributes to the activation of tumor suppressor p53. Oncogene. 2011;30:3599–3611. doi: 10.1038/onc.2011.71. [DOI] [PubMed] [Google Scholar]
- 43.Bernard F, et al. Alterations of NFIA in chronic malignant myeloid diseases. Leukemia. 2009;23:583–585. doi: 10.1038/leu.2008.228. [DOI] [PubMed] [Google Scholar]
- 44.Johnson MR, Look AT, DeClue JE, Valentine MB, Lowy DR. Inactivation of the NF1 gene in human melanoma and neuroblastoma cell lines without impaired regulation of GTP.Ras. Proc Natl Acad Sci USA. 1993;90:5539–5543. doi: 10.1073/pnas.90.12.5539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vinciguerra M, Foti M. PTEN at the crossroad of metabolic diseases and cancer in the liver. Ann Hepatol. 2008;7:192–199. [PubMed] [Google Scholar]
- 46.Cohen P, Frame S. The renaissance of GSK3. Nat Rev Mol Cell Biol. 2001;2:769–776. doi: 10.1038/35096075. [DOI] [PubMed] [Google Scholar]
- 47.Bjursell MK, et al. Adenosine kinase deficiency disrupts the methionine cycle and causes hypermethioninemia, encephalopathy, and abnormal liver function. Am J Hum Genet. 2011;89:507–515. doi: 10.1016/j.ajhg.2011.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Boison D, et al. Neonatal hepatic steatosis by disruption of the adenosine kinase gene. Proc Natl Acad Sci USA. 2002;99:6985–6990. doi: 10.1073/pnas.092642899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sutherland AP, et al. Zinc finger protein Zbtb20 is essential for postnatal survival and glucose homeostasis. Mol Cell Biol. 2009;29:2804–2815. doi: 10.1128/MCB.01667-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fan Y, et al. Liver-specific deletion of the growth hormone receptor reveals essential role of growth hormone signaling in hepatic lipid metabolism. J Biol Chem. 2009;284:19937–19944. doi: 10.1074/jbc.M109.014308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mavalli MD, et al. Distinct growth hormone receptor signaling modes regulate skeletal muscle development and insulin sensitivity in mice. J Clin Invest. 2010;120:4007–4020. doi: 10.1172/JCI42447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.da Rocha ST, Edwards CA, Ito M, Ogata T, Ferguson-Smith AC. Genomic imprinting at the mammalian Dlk1-Dio3 domain. Trends Genet. 2008;24:306–316. doi: 10.1016/j.tig.2008.03.011. [DOI] [PubMed] [Google Scholar]
- 53.Luk JM, et al. DLK1-DIO3 genomic imprinted microRNA cluster at 14q32.2 defines a stemlike subtype of hepatocellular carcinoma associated with poor survival. J Biol Chem. 2011;286:30706–30713. doi: 10.1074/jbc.M111.229831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sung WK, et al. Genome-wide survey of recurrent HBV integration in hepatocellular carcinoma. Nat Genet. 2012;44:765–769. doi: 10.1038/ng.2295. [DOI] [PubMed] [Google Scholar]
- 55.Edgar BA. From cell structure to transcription: Hippo forges a new path. Cell. 2006;124:267–273. doi: 10.1016/j.cell.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 56.Budhu A, et al. Integrated metabolite and gene expression profiles identify lipid biomarkers associated with progression of hepatocellular carcinoma and patient outcomes. Gastroenterology. 2013;144:1066–1075. e1061. doi: 10.1053/j.gastro.2013.01.054. doi:10.1053/j.gastro.2013.01.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lee P, et al. In vivo hyperpolarized carbon-13 magnetic resonance spectroscopy reveals increased pyruvate carboxylase flux in an insulin resistant mouse model. Hepatology. 2013;57:515–524. doi: 10.1002/hep.26028. [DOI] [PubMed] [Google Scholar]
- 58.DeBerardinis RJ, Cheng T. Q’s next: the diverse functions of glutamine in metabolism, cell biology and cancer. Oncogene. 2010;29:313–324. doi: 10.1038/onc.2009.358. doi:10.1038/onc.2009.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Beyoglu D, et al. Tissue metabolomics of hepatocellular carcinoma: Tumor energy metabolism and the role of transcriptomic classification. Hepatology. 2013;58:229–238. doi: 10.1002/hep.26350. doi:10.1002/hep.26350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Anderson K, et al. Genetic variegation of clonal architecture and propagating cells in leukaemia. Nature. 2011;469:356–361. doi: 10.1038/nature09650. [DOI] [PubMed] [Google Scholar]
- 61.Hou Y, et al. Single-cell exome sequencing and monoclonal evolution of a JAK2-negative myeloproliferative neoplasm. Cell. 2012;148:873–885. doi: 10.1016/j.cell.2012.02.028. [DOI] [PubMed] [Google Scholar]
- 62.Gerlinger M, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med. 2012;366:883–892. doi: 10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Berquam-Vrieze KE, et al. Cell of origin strongly influences genetic selection in a mouse model of T-ALL. Blood. 2011;118:4646–4656. doi: 10.1182/blood-2011-03-343947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gao H, et al. Application of 1H NMR-based metabonomics in the study of metabolic profiling of human hepatocellular carcinoma and liver cirrhosis. Cancer Sci. 2009;100:782–785. doi: 10.1111/j.1349-7006.2009.01086.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang B, et al. Metabonomic profiles discriminate hepatocellular carcinoma from liver cirrhosis by ultraperformance liquid chromatography-mass spectrometry. J Proteome Res. 2012;11:1217–1227. doi: 10.1021/pr2009252. [DOI] [PubMed] [Google Scholar]
- 66.Yang Y, et al. Metabonomic studies of human hepatocellular carcinoma using high-resolution magic-angle spinning 1H NMR spectroscopy in conjunction with multivariate data analysis. J Proteome Res. 2007;6:2605–2614. doi: 10.1021/pr070063h. [DOI] [PubMed] [Google Scholar]
- 67.Ward PS, Thompson CB. Metabolic reprogramming: a cancer hallmark even warburg did not anticipate. Cancer Cell. 2012;21:297–308. doi: 10.1016/j.ccr.2012.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chen T, et al. Serum and urine metabolite profiling reveals potential biomarkers of human hepatocellular carcinoma. Mol Cell Proteomics. 2011;10:M110.004945. doi: 10.1074/mcp.M110.004945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cairns RA, Harris IS, Mak TW. Regulation of cancer cell metabolism. Nat Rev Cancer. 2011;11:85–95. doi: 10.1038/nrc2981. [DOI] [PubMed] [Google Scholar]
- 70.Christofk HR, et al. The M2 splice isoform of pyruvate kinase is important for cancer metabolism and tumour growth. Nature. 2008;452:230–233. doi: 10.1038/nature06734. [DOI] [PubMed] [Google Scholar]
- 71.David CJ, Chen M, Assanah M, Canoll P, Manley JL. HnRNP proteins controlled by c-Myc deregulate pyruvate kinase mRNA splicing in cancer. Nature. 2010;463:364–368. doi: 10.1038/nature08697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.DeBerardinis RJ, et al. Beyond aerobic glycolysis: transformed cells can engage in glutamine metabolism that exceeds the requirement for protein and nucleotide synthesis. Proc Natl Acad Sci USA. 2007;104:19345–19350. doi: 10.1073/pnas.0709747104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wise DR, et al. Hypoxia promotes isocitrate dehydrogenase-dependent carboxylation of alpha-ketoglutarate to citrate to support cell growth and viability. Proc Natl Acad Sci USA. 2011;108:19611–19616. doi: 10.1073/pnas.1117773108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Vander Heiden MG. Targeting cancer metabolism: a therapeutic window opens. Nat Rev Drug Discov. 2011;10:671–684. doi: 10.1038/nrd3504. [DOI] [PubMed] [Google Scholar]
- 75.Postic C, Magnuson MA. DNA excision in liver by an albumin-Cre transgene occurs progressively with age. Genesis. 2000;26:149–150. doi: 10.1002/(sici)1526-968x(200002)26:2<149::aid-gene16>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- 76.Thoolen B, et al. Proliferative and nonproliferative lesions of the rat and mouse hepatobiliary system. Toxicol Pathol. 2010;38:5S–81S. doi: 10.1177/0192623310386499. [DOI] [PubMed] [Google Scholar]
- 77.Morrow M, Samanta A, Kioussis D, Brady HJ, Williams O. TEL-AML1 preleukemic activity requires the DNA binding domain of AML1 and the dimerization and corepressor binding domains of TEL. Oncogene. 2007;26:4404–4414. doi: 10.1038/sj.onc.1210227. [DOI] [PubMed] [Google Scholar]
- 78.Jee J, et al. ACT: aggregation and correlation toolbox for analyses of genome tracks. Bioinformatics. 2011;27:1152–1154. doi: 10.1093/bioinformatics/btr092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Eppig JT, et al. Mouse genome informatics (MGI) resources for pathology and toxicology. Toxicol Pathol. 2007;35:456–457. doi: 10.1080/01926230701310536. [DOI] [PubMed] [Google Scholar]
- 80.Kasprzyk A. BioMart: driving a paradigm change in biological data management. Database. 2011;2011:bar049. doi: 10.1093/database/bar049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sayers EW, et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2012;40:D13–25. doi: 10.1093/nar/gkr1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Chew V, et al. Chemokine-driven lymphocyte infiltration: an early intratumoural event determining long-term survival in resectable hepatocellular carcinoma. Gut. 2012;61:427–438. doi: 10.1136/gutjnl-2011-300509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Spurgeon SL, Jones RC, Ramakrishnan R. High throughput gene expression measurement with real time PCR in a microfluidic dynamic array. PLoS One. 2008;3:e1662. doi: 10.1371/journal.pone.0001662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 85.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 86.Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Schroeder MA, et al. Measuring intracellular pH in the heart using hyperpolarized carbon dioxide and bicarbonate: a 13C and 31P magnetic resonance spectroscopy study. Cardiovasc Res. 2010;86:82–91. doi: 10.1093/cvr/cvp396. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.