Abstract
N -methyl-D-aspartate (NMDA) receptors belong to the family of ionotropic glutamate receptors, which mediate most excitatory synaptic transmission in mammalian brains. Calcium permeation triggered by activation of NMDA receptors is the pivotal event for initiation of neuronal plasticity. Here we show the crystal structure of the intact heterotetrameric GluN1/GluN2B NMDA receptor ion channel at 4 Å. The NMDA receptors are arranged as a dimer of GluN1-GluN2B heterodimers with the two-fold symmetry axis running through the entire molecule composed of an amino terminal domain (ATD), a ligand-binding domain (LBD), and a transmembrane domain (TMD). The ATD and LBD are much more highly packed in the NMDA receptors than non-NMDA receptors, which may explain why ATD regulates ion channel activity in NMDA receptors but not in non-NMDA receptors.
Brain development and function rely on neuronal communication at a specialized junction called the synapse. In response to an action potential, neurotransmitters are released from the presynapse and activate ionotropic and metabotropic receptors at the postsynapse to generate a postsynaptic potential. Such synaptic transmission is a basis for experience dependent changes in neuronal circuits. The majority of excitatory neurotransmission in the human brain is mediated by transmission of a simple amino acid, L-glutamate (1), which activates metabotropic (mGluRs) and ionotropic glutamate receptors (iGluRs). iGluRs are ligand-gated ion channels that comprise three major families, AMPA (GluA1-4), kainate (GluK1-5), and NMDA receptors (GluN1, GluN2A-D, and GluN3A-B). Non-NMDA receptors can form functional homotetramers that respond to only L-glutamate. In contrast, NMDA receptors are obligatory heterotetramers mainly composed of two copies each of GluN1 and GluN2, which activate upon concurrent binding of glycine or D-serine to GluN1 and L-glutamate to GluN2 and relief of a magnesium block of the ion channel pore by membrane depolarization (2). Opening of NMDA receptor channels results in an influx of calcium ions that trigger signal transduction cascades that control strength of neural connectivity or neuroplasticity. Hyper or hypo activation of NMDA receptors is implicated in neurological disorders and diseases including Alzheimer’s disease, Parkinson’s disease, depression, schizophrenia, and ischemic injuries associated with stroke (3).
The NMDA receptor subunits, like other iGluR subunits, contain modular domains that are responsible for controlling distinct functions. In NMDA receptors, an amino terminal domain (ATD) contributes to control of ion channel open probability and deactivation speeds (4–6), and contains binding sites for subtype-specific allosteric modulator compounds including zinc (GluN2A and 2B), ifenprodil (GluN2B), and polyamines (GluN2B) (7–9). A ligand-binding domain (LBD) binds agonists and antagonists to control ion channel opening. A transmembrane domain (TMD) forms the heterotetrameric ion channel. A carboxyl terminal domain (CTD) associates with postsynaptic density proteins, which in turn facilitates intracellular signaling pivotal for neuroplasticity. In non-NMDA receptors, the ATD does not regulate ion channel activity, the LBD binds only one agonist, L-glutamate, and the TMD forms an ion channel pore with no voltage sensing capacity and with significantly less calcium permeability compared to NMDA receptors. The significantly shorter CTD interacts with postsynaptic proteins that are distinct from the NMDA receptor-associating proteins. Thus, despite being categorized in the same iGluR family, non-NMDA receptors and NMDA receptors have clear differences in basic ion channel physiology and pharmacology. The only crystal structure of an intact iGluR is the homotetrameric GluA2 AMPA receptor bound to an antagonist (10). In NMDA receptor families, structural information has been limited to that of isolated ATD (7, 8, 11) and LBD (12–15) extracellular domains. Thus, the modes of subunit and domain arrangement of intact heterotetrameric NMDA receptors have remained enigmatic. Moreover, the structure-function relationship of NMDA receptors has been difficult to dissect because functions such as ATD-mediated allosteric regulation, ligand-induced gating, and ion permeability occur in the context of heterotetramers and involve inter-subunit and domain interactions. Thus, to facilitate understanding of complex functions in NMDA receptors, we sought to capture the pattern of inter-subunit and -domain arrangement by crystallographic studies on the intact heterotetrameric GluN1a/GluN2B NMDA receptor ion channel.
Production and structural study of heterotetrameric NMDA receptors
NMDA receptors are obligatory heterotetramers composed of two copies each of GluN1 and GluN2. Structural studies of heteromultimeric eukaryotic membrane proteins from a recombinant source have been hindered by difficulties in properly assembling multiple membrane proteins in recombinant expression host cells. After extensive exploration of expression methods, we succeeded in obtaining homogeneously assembled heterotetrameric NMDA receptors by expressing modified GluN1-4a and GluN2B subunits (GluN1a/GluN2Bcryst; fig. S1–4, see Supplementary Methods) in Sf9 insect cells using a recombinant baculovirus containing both of those subunits under the Hsp70 promoter from Drosophila melanogaster. Expression under conventional late promoters such as P10 and Polyhedrin promoters hampers proper heteromeric assembly of the NMDA receptor subunits (fig. S2). The GluN1a/GluN2Bcryst construct forms an ion channel that is opened upon glycine and L-glutamate application and allosterically regulated by ifenprodil and polyamines similarly to the wild-type receptor (fig. S5–6). GluN1a/GluN2Bcryst was crystallized in the presence of a GluN1 agonist, glycine, a GluN2 agonist, L-glutamate, and an ATD-binding allosteric inhibitor, ifenprodil. The structure was initially solved at 5.7 Å by molecular replacement using the extracellular domain structures as search probes (Supplementary Methods). Overall the extracellular domains of the receptor were well resolved and electron density for the TMD was of sufficient quality to conclude that TMD helices of GluN1a/GluN2Bcryst receptor are arranged similarly to those of GluA2 (fig. S7) (10). To improve the x-ray diffraction quality, we stabilized the heterotetramer by forming disulfide cross-links between subunits (fig. S1). Based on the 5.7 Å structure, we engineered a GluN2B Ser214Cys mutation to form a disulfide bond between the ATDs of two GluN2B subunits and pairs of mutations, the GluN1a Thr561Cys/Phe810Cys and GluN2B Asp557Cys/Ile815Cys, to tether the M1 helices to the M4 helices of the neighboring subunit at the TMD. The disulfide cross-linked mutant receptor (GluN1a/GluN2Bcrystx) is trapped in an inhibited state, which could be unlocked by application of reducing agents (fig. S6). The cross-linking improved the diffraction limit to better than 4 Å, which resulted in electron density sufficient to build most of the GluN1a/GluN2B NMDA receptor including the entire extracellular domains, TMD, linkers between ATD and LBD and between LBD and TMD except some residues in the cytoplasmic loops (GluN1a 583–604, 617–622 and 834–847, and GluN2B 570–601, 616–629 and 841–852), in the loop connecting the LBD to the TMD (GluN2B 541–548 and 803–806) and in the extracellular loops (GluN1a 95–104 and 442–444, and GluN2B 440–450) (Fig. 1, S3 and S4). The model for the TMD was built using GluA2 TMD as a guide and residue assignment was verified using selenomethionine labeling (fig. S8, table S1), and electron density for aromatic residues and cross-link sites. Even though structural refinement was conducted using the most advanced refinement methods for treating low resolution data (16, 17), we suggest cautious interpretation of our structural model at the TMD, since there is some level of positional uncertainty intrinsic to a 4 Å model. No significant difference in the architecture between GluN1a/GluN2Bcryst and GluN1a/GluN2Bcrystx was observed demonstrating that disulfide cross-linking of the subunits did not alter the overall structure.
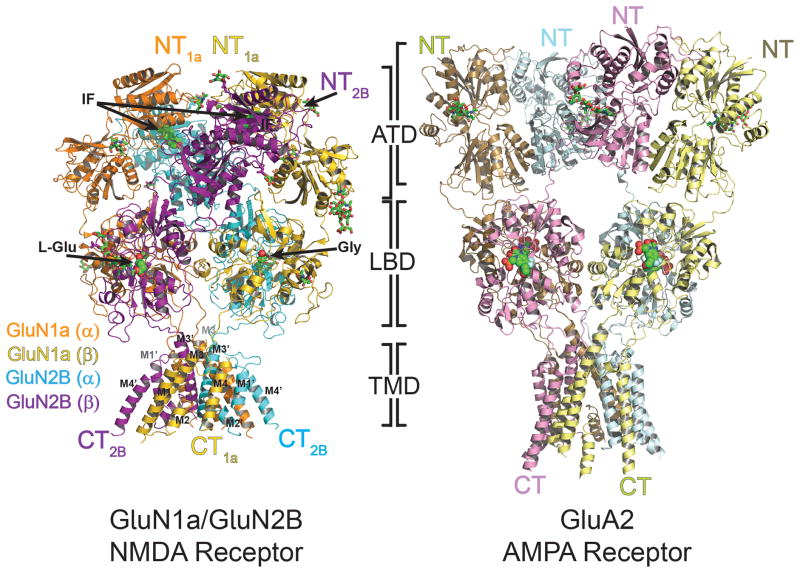
Figure 1. Overall structure of heterotetrameric GluN1a/GluN2B NMDA receptor and comparison with GluA2 AMPA receptor.
Overall structures of GluN1a/GluN2B NMDA receptor (left) and GluA2 AMPA receptor (right, PDB ID: 3KG2). Both structures are placed so that the tetramers of both receptors are in the similar orientation at the LBD layer. GluN1a and GluN2B subunits, labeled as GluN1a (α), GluN1a (β), GluN2B (α), GluN2B (β) are colored in orange, yellow, cyan and purple, respectively. The amino (NT) and carboxy (CT) termini are located on top and bottom, respectively. Ifenprodil (IF), located at the GluN1a/GluN2B ATD heterodimer interfaces, and agonists, glycine (Gly) and L-glutamate (L-Glu), lodged at the LBD clamshells, are shown in green spheres. N-glycosylation chains are shown in green sticks.
Overall Structure
The GluN1a/GluN2B NMDA receptor bound to glycine, L-glutamate, and ifenprodil is shaped like a hot-air balloon where the balloon and basket correspond to the extracellular domains and the TMD, respectively (Fig. 1). There is a clear boundary between the layers of LBD and TMD, while the ATD and LBD appear as a single unit. The GluN1a and GluN2B subunits assemble as the staggered GluN1-GluN2-GluN1-GluN2 (1-2-1-2) heterotetramer in every domain as previously predicted (10, 18, 19) (Fig. 1–2). The GluN1a/GluN2B ATD and LBD heterodimers are similar to isolated GluN1b/GluN2B ATD complexed to ifenprodil (rmsd 0.9 Å) (8) and isolated GluN1/GluN2A LBD complexed to glycine and L-glutamate (rmsd 1.1 Å) (12). Observed electron density for glycine, L-glutamate, and ifenprodil supports the view that the structure represents the allosterically inhibited state (fig. S9). The assembly of the NMDA receptor tetramer as a dimer-of-dimers at the extracellular region is similar to the organization of the GluA2 AMPA receptor (10, 20) and likely of other iGluR members. Despite the similarity in the pattern of tetrameric arrangement, the overall shape of GluN1a/GluN2B NMDA receptor is distinct from that of the “Y” shaped GluA2 AMPA receptor (Fig. 1) (10). This is attributed to tight packing of the ATD and LBD in GluN1a/GluN2B NMDA receptors. In contrast ATD and LBD interact minimally in GluA2 AMPA receptors.
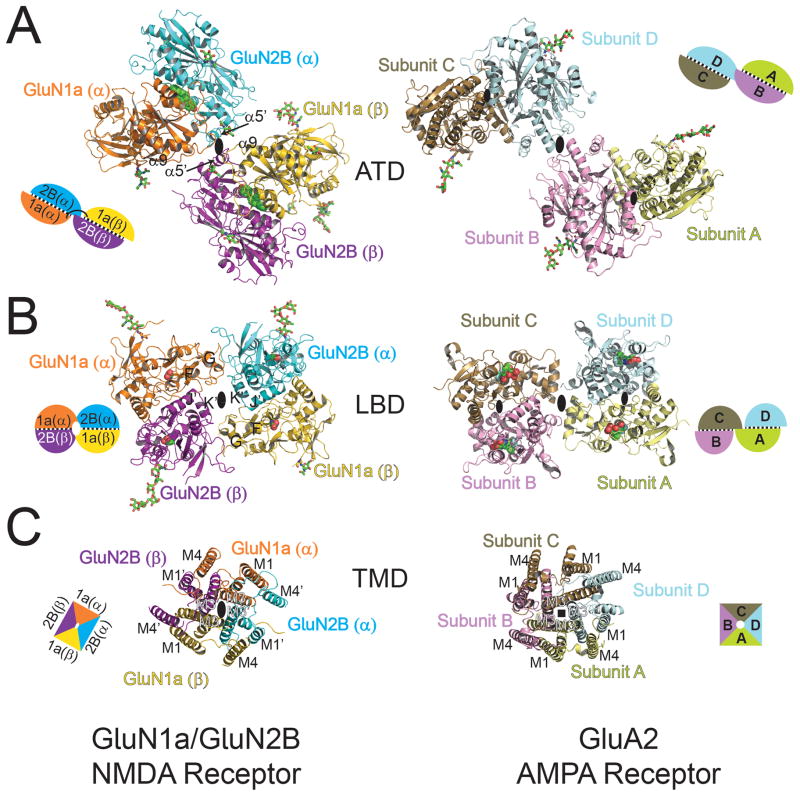
Figure 2. Domain-by-domain structural comparison of heteromeric GluN1a/GluN2B NMDA receptors and homomeric GluA2 AMPA receptor.
Structures of ATD (A), LBD (B), and TMD (C) viewed from the top of the receptors. All of the domains are assembled around the overall two-fold axis (large black oval) in GluN1a/GluN2B heterotetramers (left). In GluA2 homotetramers (right), the local two-fold axis (small black oval) runs within the ATD and LBD dimers, a two-fold axis (large black oval) runs between the ATD and LBD dimers, and the fourfold axis (black square) runs in the center of the TMD. Schematic figures next to the structures represent subunit organization at each domain, where subunits with black dots in between represent dimer pairs. Ifenprodil, glycine, L-glutamate, and ZK200775 are shown in spheres. N-glycosylation chains are shown in green sticks.
Organization of GluN1 and GluN2B subunits
There are two key features in the pattern of subunit arrangement. First, pseudo-symmetry mismatch is present between the extracellular region and the TMD. The GluN1a and GluN2B subunits are arranged in a 1-2-1-2 orientation with two-fold symmetry between the two GluN1a/GluN2B heterodimers in the ATD and LBD, but with pseudo-four-fold symmetry in the TMD (Fig. 2). A similar subunit arrangement is observed in the GluA2 homotetrameric structure that has two-fold symmetries within and between homodimers of the ATD and LBD and fourfold symmetry in the TMD (10) indicating that symmetry mismatch may be common to iGluR structures. The second important feature is swapping of dimer pairs between the ATD and LBD layers (Fig. 2A and B). In the ATD layer, heterodimer pairs assemble as GluN1a (α)/GluN2B (α) and GluN1 (β)/GluN2B (β) whereas in the LBD layer, they assemble as GluN1a (α)/GluN2B (β) and GluN1 (β)/GluN2B (α) (Fig. 2A and B). A similar pattern of domain swapping is also observed in the homotetrameric GluA2 AMPA receptor where subunits assemble as a dimer of A/B and C/D homodimers at the ATD and as a dimer of A/D and B/C dimers at the LBD (Fig. 2A and B) (10). Overall the conformations of GluN1a and GluN2B subunits approximately correspond to those of A/C and B/D subunits in the GluA2 AMPA receptor homotetramer. This assignment is based on the observation that the ATD-LBD linker and the M3-LBD linker are respectively more “distal” and “proximal” from the overall two-fold symmetry axis for GluN1a than GluN2B similar to the orientation of the A/C than B/D subunits in the GluA2 AMPA receptor (Fig. 2 and fig. S10).
Though they share a dimer-of-dimers arrangement, the modes of subunit association in each domain differ significantly between NMDA and AMPA receptors. In the ATD, the two GluN1a/GluN2B heterodimers interact with each other at two interfaces involving upper lobes of the two GluN1a subunits (α and β) and lower lobes of the two GluN2B subunits (α and β). In contrast the GluA2 receptor has only one interface between Subunit B and D (Fig. 2A and fig. S11). The ATDs of NMDA receptor and AMPA receptor subunits have low sequence identity and this is reflected in the large differences in structures (7, 8). Consequently, the ATD layer is more compactly packed in GluN1a/GluN2B than in GluA2. At the LBD, the dimer-of-dimers arrangement is comparable between the GluN1a/GluN2B and GluA2 receptors. The GluN1a/GluN2B heterodimers interact between the lower lobe of GluN1a and the upper lobe of GluN2B at the two equivalent sites (Fig. 2B and S11). In GluA2, the equivalent regions are much more loosely packed, but instead, there is another closely packed region involving Subunit A and C (10). Whether these distinct modes of interactions at the LBD represent an architectural difference between NMDA and AMPA receptors or different functional states remains an open question. In the previous study, the GluA2 AMPA receptor was crystallized in the presence of an antagonist (ZK200775) whereas the GluN1a/GluN2B NMDA receptor was crystallized in the presence of the allosteric inhibitor, ifenprodil, and agonists, glycine and L-glutamate. Finally, at the TMD, the subunits form pseudo four-fold symmetrical interactions between the M3 helices at the center of the ion channel and between M4 helices of one subunit and M1 and M3 helices of the adjacent subunit, similar to the GluA2 AMPA receptor (Fig. 2C and fig. S11). Within the pore, the M2 helices are close to the M1 and M3 helices of the same subunit and the M3 helices of the adjacent subunit.
Inter-subunit interfaces and function
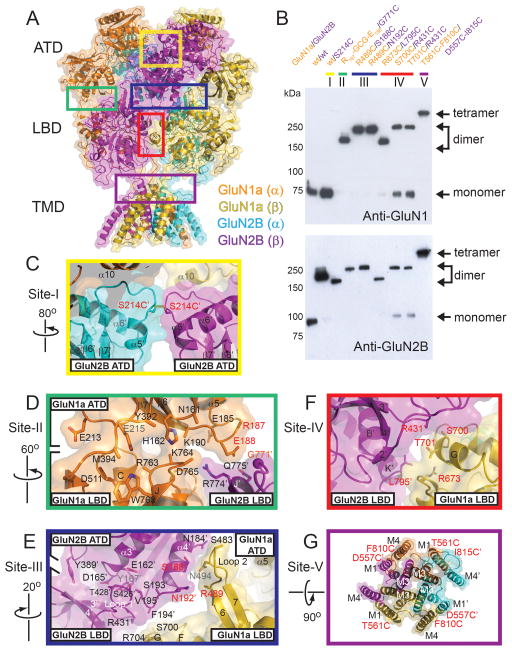
The crystal structure of the heterotetrameric GluN1a/GluN2B NMDA receptor shows inter-subunit interfaces that are distinct from those in the GluA2 AMPA receptor. Thus, we have tested whether the distinct interfaces observed in our crystal structure are present in intact NMDA receptors in the membrane environment. We engineered cysteine residues and tested for spontaneous disulfide bond formation at the following subunit interfaces: (I). GluN2B (ATD)-GluN2B (ATD); (II). GluN1a (ATD)-GluN2B (LBD); (III). GluN1a (LBD)-GluN2B (ATD); (IV). GluN1a (LBD)-GluN2B (LBD); and (V). GluN1a (TMD)-GluN2B (TMD) (Fig. 3). The Western blot experiments on the mutant GluN1a/GluN2B NMDA receptor proteins have shown that all of the single cysteine mutant pairs in the extracellular region form disulfide cross-links resulting in GluN2B–GluN2B homodimer formation (I) and GluN1a-GluN2B heterodimer formation (II, III and IV), whereas the double cysteine mutant pair in the TMD forms GluN1a-GluN2B-GluN1a-GluN2B disulfide cross-links resulting in heterotetramer formation (V) in the absence of a reducing agent (Fig. 3B). In the presence of reducing agents, or when a subunit with the cysteine mutation is expressed with a subunit with no mutation, bands representing monomers of GluN1a and GluN2B appear (fig. S12). This indicates that the disulfide cross-links are formed by engineered cysteine pairs, and validates the physiological relevance of the subunit arrangement observed in our crystal structure.
Figure 3. Inter- and intra-subunit interfaces in GluN1a/GluN2B NMDA receptors.
(A) Ribbon and surface representation of GluN1a/GluN2B NMDA receptor colored as in Fig. 1. Inter-subunit interfaces that are probed by disulfide cross-linking experiments are surrounded by colored boxes. (B) Western blot analysis of disulfide bond formation by cysteine substitutions at the subunit interfaces probed by anti-GluN1 (top) and anti-GluN2B (bottom) antibodies under non-reducing conditions. Arrows indicate positions of non-cross-linked monomers and cross-linked dimers and tetramers. (C–G) Close up views of the inter- and intra-subunit interfaces between GluN2B ATD (α) and GluN2B-ATD (β) (Site-I, yellow box) (C), between GluN1a ATD and GluN1a LBD, and GluN1a-ATD and GluN2B-LBD (Site-II, green box) (D), between GluN1a LBD and GluN2B ATD, and GluN2B ATD and GluN2B LBD (Site-III, blue box) (E), between GluN1a LBD and GluN2B LBD (Site-IV, red box) (F) and between GluN1a TMD and GluN2B TMD (Site-V, purple box) (G). Side chains without clear electron densities are modeled as alanine. Residues that are mutated to cysteine are labeled in red.
What are the molecular determinants that favor the 1-2-1-2 arrangement over 1-1-2-2 arrangement? Previous studies have shown that the ATD is important for allosteric modulation and for controlling open probability and deactivation speeds (4, 5), but not required for formation of functional ion channels (21). Furthermore, interactions of helices in the TMD are similar to those in the homotetrameric GluA2 AMPA receptor structure, thus, the structural determinant for the 1-2-1-2 subunit arrangement may reside in the LBDs. In silico construction of GluN1a-GluN2B heterotetramers in the 1-1-2-2 format by superposing the GluN1 LBD structure onto the GluN2B LBD structure and vice versa revealed steric clashes in both the GluN1a-GluN1a and the GluN2B-GluN2B interfaces (fig. S13). The GluN1a-GluN1a interaction is prevented by a collision between Loop1 of one GluN1a and Helix G of the other, whereas the GluN2B-GluN2B interaction is disfavored by a steric hindrance between Helix K′ of one subunit and Helix E′ and F′ of the other (fig. S13). Thus, while the TMD is essential for tetramerization, the structural features in the LBD appear to favor the 1-2-1-2 arrangement.
The GluN1a/GluN2B heterotetrameric structure shows that some residues and motifs previously shown to play important roles in function are located at the interface between GluN1-GluN2B heterodimers (fig. S14). One example is Loop 1′ of GluN2B located within the LBD. The equivalent motif in GluN2A has been suggested to play a major role in negative cooperativity between the glycine and L-glutamate binding sites (22). Another example is a point mutation on Helix E of the GluN1 LBD lobe (Asp669Asn), which has previously been shown to affect gating properties by altering sensitivity to pH, spermine and ifenprodil (23). Our cross-linking results also showed “trapping” GluN1a-GluN2B ATD attenuates the ion channel activity. Taken together these results suggest that rearrangement between the two GluN1a-GluN2B dimers may be important for activity. Consistent with this, a recent study on GluA2 AMPA receptor showed that rearrangement of two GluA2 homodimers at the LBD regulates ion channel gating activity (24).
Inter-domain interaction between ATD and LBD and function
One of the major functional differences between NMDA receptors and non-NMDA receptors is the ATD-mediated regulation of ion channel activity present in the former and absent in the latter. In NMDA receptors, binding of allosteric modulators at the ATD alters agonist potency at the LBD indicating tight functional coupling between the ATD and the LBD (25). This coupling is structurally well represented by the more extensive ATD-LBD interaction in NMDA receptors (3,107 Å2) than in AMPA receptors (1,470 Å2). The crystal structure shows the two major sites, Site-II and –III, mediating the tight ATD-LBD association mainly through hydrophilic interactions even though the exact mode of residue-by-residue contacts cannot be pinpointed due to limitation in resolution (Fig. 3D and E). In Site-II, GluN1a ATD and the GluN1a/GluN2B LBD heterodimer are packed together through interaction between the loop extending from Helix α5 to Strand β7 at GluN1a ATD and GluN2B Helix J′ at the GluN1a-GluN2B interface at LBD (Fig. 3D). The region around GluN1a Helix α5 is where the twenty one residue long loop that contains numbers of basic amino acids would be present in the GluN1b splice variant. This loop, encoded by exon 5, has been shown to accelerate the deactivation time course (26), and to influence allosteric modulation by decreasing potency of protons, polyamines, and Zn2+ (27). In Site-III, the GluN1a LBD and the GluN1a/GluN2B ATD heterodimer are packed together by GluN1a Loop 2 “wedging” into the interface between GluN1a ATD and GluN2B ATD. Furthermore, GluN1a Helix F and G, GluN2B Loop 1′, and the loop extending from GluN2B Helix α4′ stack onto each other to further stabilize the ATD-LBD interaction (Fig. 3E). Helix F in GluN1 has been implicated in gating control, thus, this may be a key locus where ATD may have an impact on gating properties (28). Overall, the ATD and the LBD are in a tight arrangement that is suited to transmit structural changes between domains (fig. S15).
Transmembrane domain and the extracellular vestibule
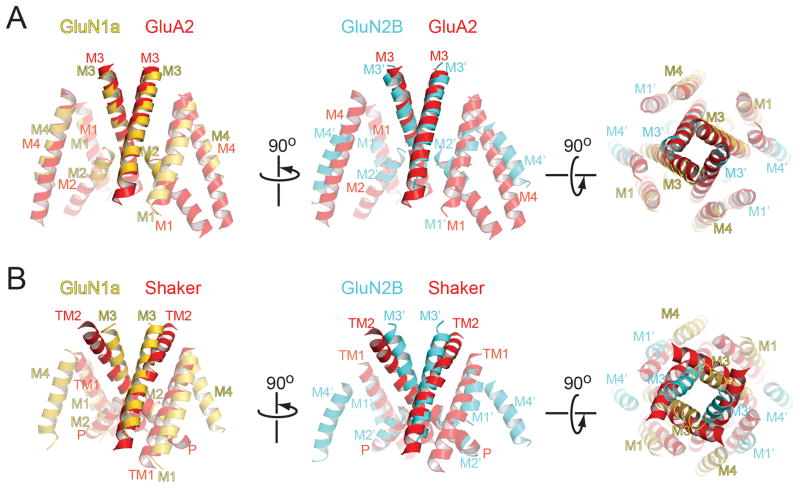
The TMD of the GluN1a/GluN2B NMDA receptor forms a heterotetrameric ion channel with pseudo four-fold symmetry similar in overall shape to the homotetrameric ion channel of the GluA2 AMPA receptors (rmsd 2.2 Å) except the M4 helix in GluN2B (Fig. 4A). The tetrameric crossing of the M3 helices occludes the ion penetration pathway to a similar degree in the presumed allosterically inhibited GluN1a/GluN2B NMDA receptor to the closed GluA2 AMPA receptor (Fig. 4A). This crossing of the M3 helices occurs around the highly conserved SYTANLAAF motifs in iGluRs, mutations of which are known to modify gating properties (29). The ion channel pore (M1 to M3) of the GluN1a/GluN2B NMDA receptor shows high structural similarity to that of KcsA potassium channels in a closed conformation (30) (rmsd 2.4 Å) despite the low sequence identity (19%) (fig. S16). In contrast, shaker (31) and MthK (32) potassium channels in open conformation do not superpose well, mainly due to bending of their TM2 helices compared to the M3 helices in GluN1a/GluN2B NMDA receptor (Fig. 4B and fig. S16). On the basis of structural similarities with the potassium channel, we speculate that gating of the GluN1a/GluN2B NMDA receptor may involve rearrangement of M3 helices.
Figure 4. Structural comparison of NMDA receptor ion channel with GluA2 AMPA receptor and Shaker potassium channels.
TMDs of GluN1a subunits (yellow, left panel) and GluN2B subunits (cyan, middle panel) are superposed onto the ion channel regions (red) of the closed conformation of GluA2 AMPA receptor (PDB ID: 3KG2) (A), open conformation of Shaker potassium channel (PDB ID: 2R9R) (B). The superposed structures are viewed from the side (left and middle panels) or from the extracellular side (right panel). Superposition is performed using Secondary-structure matching (SSM) tool in the program Coot. Loops are excluded from the figure for clarity.
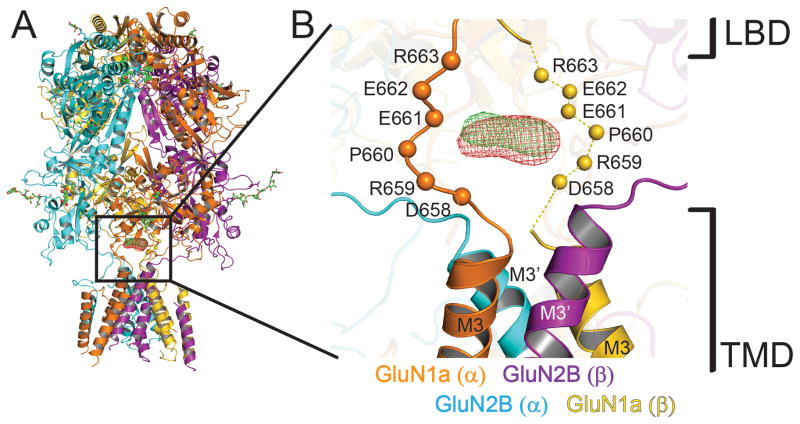
One of the hallmarks of NMDA receptor function is the high permeation of calcium ions, which plays a major role in neuronal plasticity as well as excitotoxicity. The crystal structure complexed with holmium or gadolinium, lanthanides known to occupy calcium binding sites in many biological macromolecules (33, 34), show binding between the LBD-TMD linkers from the two GluN1 subunits around the center of the ion channel (Fig. 5). A set of acidic residues in GluN1 (DRPEER motif) located in this region is critical for the high calcium flux characteristic of NMDA receptors (35). Thus, the lanthanide binding site along with the previous electrophysiological study further confirms the presence of the calcium pool located right outside of the ion channel. A similar charge-based ion pooling mechanism has been suggested for cation conductance in P2X4 receptors and nicotinic acetylcholine receptors (36, 37). Despite extensive efforts, the regions of the TMD that determine voltage-dependent Mg2+ block and Ca2+ permeation (38) are not resolved in this crystal structure. Structure-based understanding of cation selectivity and voltage-dependent Mg2+ block is thus a question that remains to be addressed.
Figure 5. Putative calcium binding site at the extracellular vestibule.
(A) Overall structure of GluN1a/GluN2B NMDA receptors with the anomalous Fourier difference maps for holmium (green mesh; from the 7.5 Å dataset) and gadolinium (red mesh; from the 7.8 Å dataset) countered at 4.5 σ. (B) Close up view of the boxed region in panel A. Holmium and gadolinium binding sites are located at the extracellular vestibule over the bundle of M3 helices. Cα atoms of the residues on GluN1a DRPEER motif from the GluN1a/GluN2Bcrystx structure are shown as spheres. Residues for the disordered DRPEER motif (shown as dashed lines) on the GluN1a (β) protomer (yellow) are positioned based on the structural alignment of the GluN1a (α) protomer (orange).
Conclusion
The crystal structure of GluN1a/GluN2B NMDA receptors in the current study reveals the patterns of inter-subunit and –domain interactions, which are different from those observed in AMPA receptors. These differences widely reflect functional differences. The structure will serve as a template for designing experiments that addresses functional questions specific to NMDA receptors. Finally, the defined subunit interfaces should serve as an important blueprint for design of therapeutic compounds.
Supplementary Material
Acknowledgments
We thank staffs at the 23-ID-B and D beamlines at the Advanced Photon System in the Argonne National Laboratory and the BL41XU beamline at the Spring 8 for their excellent beamline supports. Noriko Simorowski is thanked for her technical support. We thank Hongjie Yuan and Stephen Traynelis for sharing unpublished data with us and for critical comments on this manuscript. We also thank Stephen Harrison for making important comments on this work. This work was supported by National Institute of Health (MH085926 to HF), Mirus Research Award (to HF), and a Robertson Research Fund of Cold Spring Harbor Laboratory (to HF). Coordinates and structure factors have been deposited in the Protein Data Bank under accession code 4PE5.
References and Notes
- 1.Hayashi T. Effects of sodium glutamate on the nervous system. Keio J Med. 1954;3:192. [Google Scholar]
- 2.Mayer ML, Westbrook GL, Guthrie PB. Voltage-dependent block by Mg2+ of NMDA responses in spinal cord neurones. Nature. 1984 May 17–23;309:261. doi: 10.1038/309261a0. [DOI] [PubMed] [Google Scholar]
- 3.Traynelis SF, et al. Glutamate receptor ion channels: structure, regulation, and function. Pharmacol Rev. 2010 Sep;62:405. doi: 10.1124/pr.109.002451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gielen M, Siegler Retchless B, Mony L, Johnson JW, Paoletti P. Mechanism of differential control of NMDA receptor activity by NR2 subunits. Nature. 2009 Jun 4;459:703. doi: 10.1038/nature07993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yuan H, Hansen KB, Vance KM, Ogden KK, Traynelis SF. Control of NMDA receptor function by the NR2 subunit amino-terminal domain. The Journal of neuroscience. 2009 Sep 30;29:12045. doi: 10.1523/JNEUROSCI.1365-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hansen KB, Furukawa H, Traynelis SF. Control of assembly and function of glutamate receptors by the amino-terminal domain. Mol Pharmacol. 2010 Oct;78:535. doi: 10.1124/mol.110.067157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Karakas E, Simorowski N, Furukawa H. Structure of the zinc-bound amino-terminal domain of the NMDA receptor NR2B subunit. Embo J. 2009 Dec 16;28:3910. doi: 10.1038/emboj.2009.338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Karakas E, Simorowski N, Furukawa H. Subunit arrangement and phenylethanolamine binding in GluN1/GluN2B NMDA receptors. Nature. 2011 Jul 14;475:249. doi: 10.1038/nature10180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mony L, Zhu S, Carvalho S, Paoletti P. Molecular basis of positive allosteric modulation of GluN2B NMDA receptors by polyamines. Embo J. 2011 Aug 3;30:3134. doi: 10.1038/emboj.2011.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sobolevsky AI, Rosconi MP, Gouaux E. X-ray structure, symmetry and mechanism of an AMPA-subtype glutamate receptor. Nature. 2009 Dec 10;462:745. doi: 10.1038/nature08624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Farina AN, et al. Separation of domain contacts is required for heterotetrameric assembly of functional NMDA receptors. The Journal of neuroscience. 2011 Mar 9;31:3565. doi: 10.1523/JNEUROSCI.6041-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Furukawa H, Singh SK, Mancusso R, Gouaux E. Subunit arrangement and function in NMDA receptors. Nature. 2005 Nov 10;438:185. doi: 10.1038/nature04089. [DOI] [PubMed] [Google Scholar]
- 13.Vance KM, Simorowski N, Traynelis SF, Furukawa H. Ligand-specific deactivation time course of GluN1/GluN2D NMDA receptors. Nat Commun. 2011;2:294. doi: 10.1038/ncomms1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yao Y, Harrison CB, Freddolino PL, Schulten K, Mayer ML. Molecular mechanism of ligand recognition by NR3 subtype glutamate receptors. Embo J. 2008 Aug 6;27:2158. doi: 10.1038/emboj.2008.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jespersen A, Tajima N, Fernandez-Cuervo G, Garnier-Amblard EC, Furukawa H. Structural Insights into Competitive Antagonism in NMDA Receptors. Neuron. 2014 Jan 22;81:366. doi: 10.1016/j.neuron.2013.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Murshudov GN, et al. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr D Biol Crystallogr. 2011 Apr;67:355. doi: 10.1107/S0907444911001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schroder GF, Levitt M, Brunger AT. Super-resolution biomolecular crystallography with low-resolution data. Nature. 2010 Apr 22;464:1218. doi: 10.1038/nature08892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Salussolia CL, Prodromou ML, Borker P, Wollmuth LP. Arrangement of subunits in functional NMDA receptors. J Neurosci. 2011 Aug 3;31:11295. doi: 10.1523/JNEUROSCI.5612-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Riou M, Stroebel D, Edwardson JM, Paoletti P. An alternating GluN1-2-1-2 subunit arrangement in mature NMDA receptors. PLoS One. 2012;7:e35134. doi: 10.1371/journal.pone.0035134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Das U, Kumar J, Mayer ML, Plested AJ. Domain organization and function in GluK2 subtype kainate receptors. Proc Natl Acad Sci U S A. 2010 May 4;107:8463. doi: 10.1073/pnas.1000838107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rachline J, Perin-Dureau F, Le Goff A, Neyton J, Paoletti P. The micromolar zinc-binding domain on the NMDA receptor subunit NR2B. J Neurosci. 2005 Jan 12;25:308. doi: 10.1523/JNEUROSCI.3967-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Regalado MP, Villarroel A, Lerma J. Intersubunit cooperativity in the NMDA receptor. Neuron. 2001 Dec 20;32:1085. doi: 10.1016/s0896-6273(01)00539-6. [DOI] [PubMed] [Google Scholar]
- 23.Kashiwagi K, Fukuchi J, Chao J, Igarashi K, Williams K. An aspartate residue in the extracellular loop of the N-methyl-D-aspartate receptor controls sensitivity to spermine and protons. Mol Pharmacol. 1996 Jun;49:1131. [PubMed] [Google Scholar]
- 24.Lau AY, et al. A conformational intermediate in glutamate receptor activation. Neuron. 2013 Aug 7;79:492. doi: 10.1016/j.neuron.2013.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zheng F, et al. Allosteric interaction between the amino terminal domain and the ligand binding domain of NR2A. Nat Neurosci. 2001 Sep;4:894. doi: 10.1038/nn0901-894. [DOI] [PubMed] [Google Scholar]
- 26.Vance KM, Hansen KB, Traynelis SF. GluN1 splice variant control of GluN1/GluN2D NMDA receptors. J Physiol. 2012 Aug 15;590:3857. doi: 10.1113/jphysiol.2012.234062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Traynelis SF, Hartley M, Heinemann SF. Control of proton sensitivity of the NMDA receptor by RNA splicing and polyamines. Science. 1995 May 12;268:873. doi: 10.1126/science.7754371. [DOI] [PubMed] [Google Scholar]
- 28.Inanobe A, Furukawa H, Gouaux E. Mechanism of partial agonist action at the NR1 subunit of NMDA receptors. Neuron. 2005 Jul 7;47:71. doi: 10.1016/j.neuron.2005.05.022. [DOI] [PubMed] [Google Scholar]
- 29.Zuo J, et al. Neurodegeneration in Lurcher mice caused by mutation in delta2 glutamate receptor gene. Nature. 1997 Aug 21;388:769. doi: 10.1038/42009. [DOI] [PubMed] [Google Scholar]
- 30.Zhou Y, Morais-Cabral JH, Kaufman A, MacKinnon R. Chemistry of ion coordination and hydration revealed by a K+ channel-Fab complex at 2.0 A resolution. Nature. 2001 Nov 1;414:43. doi: 10.1038/35102009. [DOI] [PubMed] [Google Scholar]
- 31.Long SB, Tao X, Campbell EB, MacKinnon R. Atomic structure of a voltage-dependent K+ channel in a lipid membrane-like environment. Nature. 2007 Nov 15;450:376. doi: 10.1038/nature06265. [DOI] [PubMed] [Google Scholar]
- 32.Ye S, Li Y, Jiang Y. Novel insights into K+ selectivity from high-resolution structures of an open K+ channel pore. Nat Struct Mol Biol. 2010 Aug;17:1019. doi: 10.1038/nsmb.1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li W, Aldrich RW. Activation of the SK potassium channel-calmodulin complex by nanomolar concentrations of terbium. Proc Natl Acad Sci U S A. 2009 Jan 27;106:1075. doi: 10.1073/pnas.0812008106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liddington RC, et al. Structure of simian virus 40 at 3.8-A resolution. Nature. 1991 Nov 28;354:278. doi: 10.1038/354278a0. [DOI] [PubMed] [Google Scholar]
- 35.Watanabe J, Beck C, Kuner T, Premkumar LS, Wollmuth LP. DRPEER: a motif in the extracellular vestibule conferring high Ca2+ flux rates in NMDA receptor channels. J Neurosci. 2002 Dec 1;22:10209. doi: 10.1523/JNEUROSCI.22-23-10209.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Imoto K, et al. Rings of negatively charged amino acids determine the acetylcholine receptor channel conductance. Nature. 1988 Oct 13;335:645. doi: 10.1038/335645a0. [DOI] [PubMed] [Google Scholar]
- 37.Kawate T, Michel JC, Birdsong WT, Gouaux E. Crystal structure of the ATP-gated P2X(4) ion channel in the closed state. Nature. 2009 Jul 30;460:592. doi: 10.1038/nature08198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Siegler Retchless B, Gao W, Johnson JW. A single GluN2 subunit residue controls NMDA receptor channel properties via intersubunit interaction. Nat Neurosci. 2012 Mar;15:406. doi: 10.1038/nn.3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fitzgerald DJ, et al. Protein complex expression by using multigene baculoviral vectors. Nat Methods. 2006 Dec;3:1021. doi: 10.1038/nmeth983. [DOI] [PubMed] [Google Scholar]
- 40.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 41.McCoy AJ, et al. Phaser crystallographic software. J Appl Crystallogr. 2007 Aug 1;40:658. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004 Dec;60:2126. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 43.Nicholls RA, Long F, Murshudov GN. Low-resolution refinement tools in REFMAC5. Acta Crystallogr D Biol Crystallogr. 2012 Apr;68:404. doi: 10.1107/S090744491105606X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010 Feb;66:213. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Davis IW, Murray LW, Richardson JS, Richardson DC. MOLPROBITY: structure validation and all-atom contact analysis for nucleic acids and their complexes. Nucleic Acids Res. 2004 Jul 1;32:W615. doi: 10.1093/nar/gkh398. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.