Summary
Broadly neutralizing antibodies (bNAbs) to HIV-1 envelope glycoprotein (Env) can prevent infection in animal models. Characterized bNAb targets, although key to vaccine and therapeutic strategies, are currently limited. We defined a new site of vulnerability by solving structures of bNAb 8ANC195 complexed with monomeric gp120 by X-ray crystallography and trimeric Env by electron microscopy. The site includes portions of gp41 and N-linked glycans adjacent to the CD4 binding site on gp120, making 8ANC195 the first donor-derived anti-HIV-1 bNAb with an epitope spanning both Env subunits. Rather than penetrating the glycan shield using a single variable region CDR loop, 8ANC195 inserted its entire heavy chain variable domain into a gap to form a large interface with gp120 glycans and regions of the gp120 inner domain not contacted by other bNAbs. By isolating additional 8ANC195 clonal variants, we identified a more potent variant, which may be valuable for therapeutic approaches using bNAb combinations with non-overlapping epitopes.
Introduction
The only target of neutralizing anti-HIV-1 antibodies is the envelope glycoprotein (Env) spike, a heterotrimer of gp120 and gp41 subunits. Single cell-based antibody cloning techniques have recently uncovered a large number of antibodies that can potently neutralize highly diverse HIV-1 variants by targeting Env (Burton et al., 2012; Klein et al., 2013b; Mascola and Haynes, 2013; Scheid et al., 2011; Wu et al., 2010). When transferred passively, broadly neutralizing antibodies (bNAbs) can prevent infection by HIV-1 or SHIV in humanized mice and macaques, respectively (Burton et al., 2012; Klein et al., 2013b; Mascola and Haynes, 2013). Moreover, combinations of bNAbs can also suppress established HIV-1 and SHIV infections (Barouch et al., 2013; Klein et al., 2012; Shingai et al., 2013). It is therefore believed that vaccines that elicit these antibodies would be protective against HIV-1.
Most of the bNAbs characterized to date target one of four major sites of vulnerability on HIV-1 Env: on gp120, the CD4 binding site, V1/V2, and the base of the V3 loop, and on gp41, the membrane proximal region (MPER) (Burton et al., 2012; Klein et al., 2013b; Mascola and Haynes, 2013). 8ANC195 is among a small group of bNAbs that does not appear to target any of these sites. Although only two of the B cells originally isolated from the 8ANC195 donor, an HIV-1 elite controller, belonged to the 8ANC195 clone, the antibodies produced by this clone complemented the neutralizing activity of antibodies produced by a more expanded B cell clone that targeted the CD4 binding site (Scheid et al., 2011).
8ANC195 is classified as a bNAb because it neutralized 66% of viruses in a diverse viral panel (Scheid et al., 2011). Like other anti-HIV-1 bNAbs (Klein et al., 2013a; Scheid et al., 2009a), 8ANC195 is highly somatically mutated, including insertions and deletions in the complementarity determining regions (CDRs) and framework regions (FWRs) of its heavy chain (HC) and light chain (LC). Although initial efforts to map the 8ANC195 epitope were unsuccessful (Scheid et al., 2011), computational analyses of neutralization data predicted that intact potential N-linked glycosylation sites (PNGSs) at positions 234gp120 and 276gp120 were essential for its activity (Chuang et al., 2013; West et al., 2013). These predictions were confirmed by evaluating the neutralization potency of 8ANC195 against mutant HIV-1 strains in vitro (Chuang et al., 2013; West et al., 2013) and in vivo (West et al., 2013). However, the precise 8ANC195 epitope on HIV-1 Env remained elusive.
Results
Crystal structures of 8ANC195 alone and in complex with gp120 and CD4
To determine the epitope recognized by 8ANC195 and investigate its neutralization mechanism, we solved the crystal structures of the Fab fragment of 8ANC195 alone and complexed with an HIV-1 clade A/E 93TH057 gp120 core and CD4 domains 1–2 (sCD4) at 2.1 Å and 3.0 Å resolution, respectively (Figure 1A,B; Table S1). To reduce glycan heterogeneity, five PNGSs on the core gp120 were removed by mutation (Asn88Glngp120, Asn289Glngp120, Asn334Glngp120, Asn392Glngp120, Asn448Glngp120), and the gp120 was expressed in HEK 293S GnTI −/− cells, which attach only high-mannose N-linked glycans to PNGSs, rather than a mixture of high-mannose, complex and hybrid-type N-linked glycans (Binley et al., 2010; Dunlop et al., 2010).
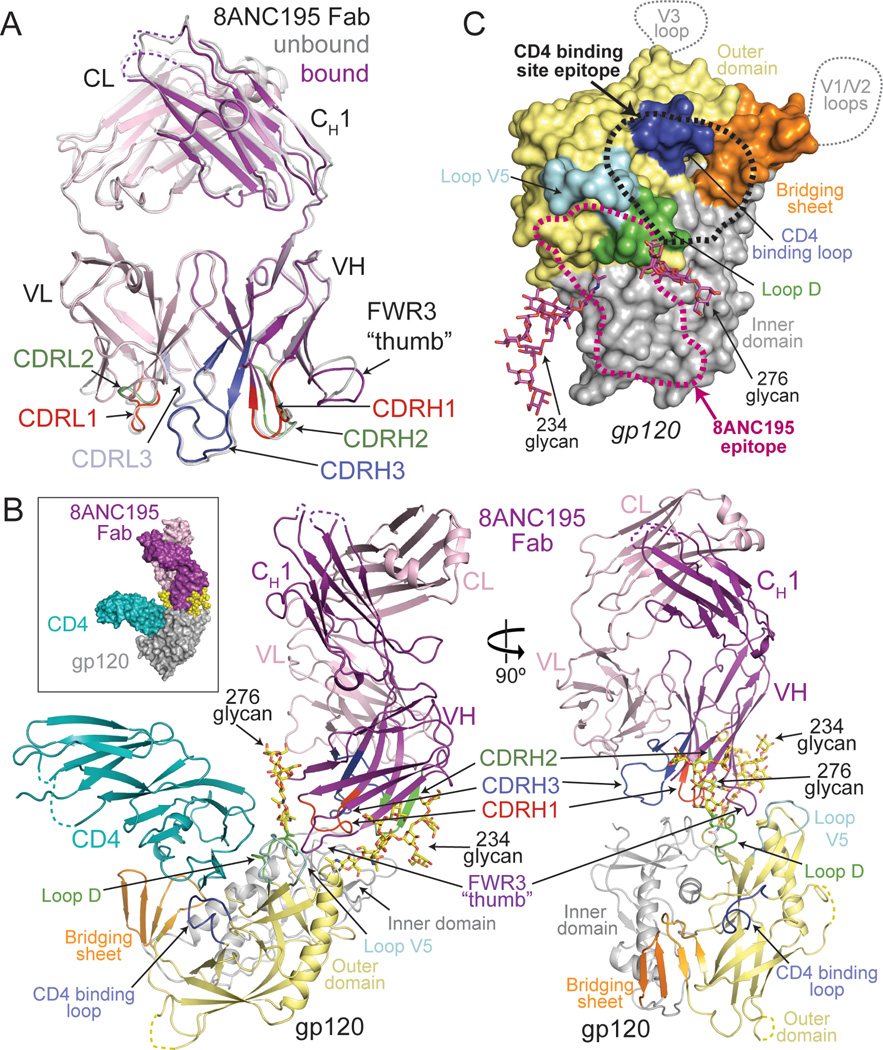
Figure 1. Crystal structures of 8ANC195 Fab and 8ANC195/gp120/sCD4 complex.
(A) Superimposition of unbound (grey) and bound (HC, purple; LC, light pink) structures of 8ANC195 Fab shown as ribbon diagrams. CDR loops are highlighted (CDRH1/CDRL1, red; CDRH2/CDRL2, green; CDRH3, dark blue; CDRL3, light blue) and a “thumb”-like loop formed by an insertion in FWR3 is indicated. Disordered loops are shown as dashed lines. (B) Space-filling model (inset) and ribbon diagram of ternary complex of 8ANC195 (HC, purple; LC, light pink), sCD4 (teal), and 93TH057 gp120 core (inner domain, grey; outer domain, light yellow; bridging sheet, orange; loop D, green; loop V5, cyan; CD4 binding loop, blue). Ordered glycans attached to Asn234gp120 and Asn276gp120 are shown as yellow sticks (oxygens, red; nitrogens, blue). Fab CDR loops are colored as in (A). sCD4 was omitted from the right panel for clarity. (C) Approximate locations of bNAb epitopes on a surface representation of the gp120 core. The epitopes of V3 and V1/V2 antibodies include regions of loops (dotted lines) not present in the gp120 core structure. CD4 binding site and 8ANC195 epitopes are outlined by black (CD4 binding site) and magenta (8ANC195) dots. Glycans included in the 8ANC195 epitope are shown in magenta. Subdomains of gp120 are colored as in (B). See also Figure S1.
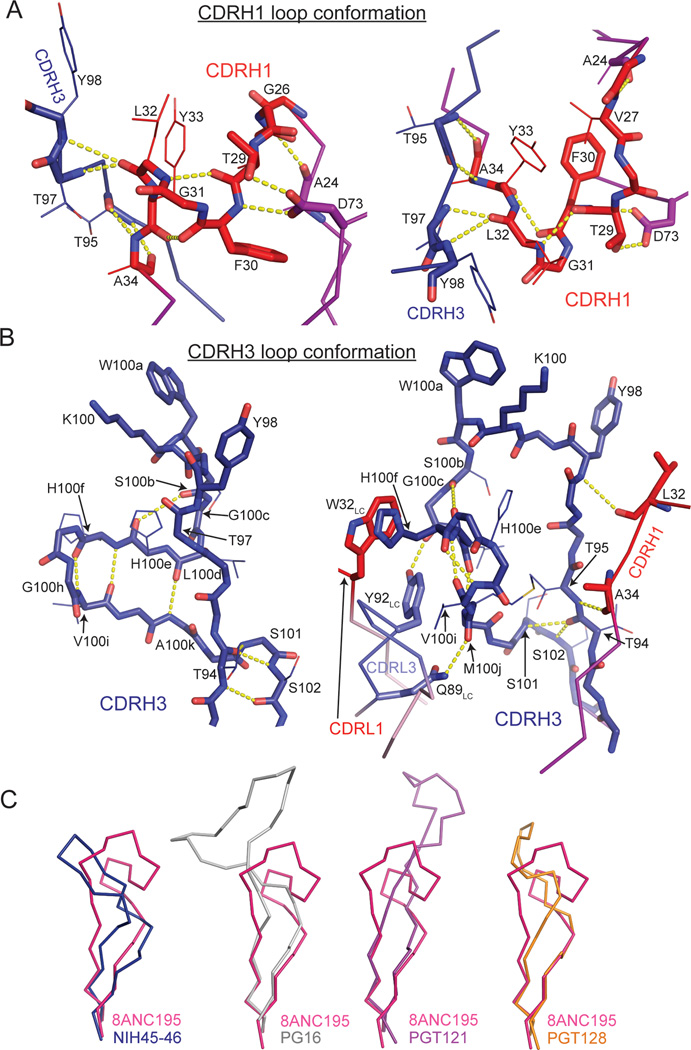
Comparison of 8ANC195 Fab in its free versus gp120-bound state revealed high structural similarity (RMSD = 0.7 Å for 236 Cα atoms of VH–VL) except for a 3.5 Å displacement of the loop connecting strands D and E in HC FWR3 (Figure 1A). The CDRH1 and CDRH3 loops were folded into hook-like tertiary structures in free and gp120-bound Fabs; therefore the conformations were not induced upon binding to gp120 (Figure 1A and Figure 2A,B). The CDRH3 architecture differed from CDRH3s in other antibodies including anti-HIV-1 antibodies with long CDR loops (Figure 2C). The CDRH1 loop conformation was stabilized by a hydrogen bond network among backbone atoms of CDRH1, burial of Phe30HC, and hydrogen bonds with Asp73HC and Thr97HC (Figure 2A). CDRH3 had a complex tertiary structure in which residues 95HC–100cHC formed a loop protruding ~10 Å from the antibody surface, and residues 100dHC–100kHC formed a β-sheet subdomain that was stabilized by hydrophobic stacking between His100fHC and Trp32LC and a hydrogen bond between Met100jHC and Gln89LC (Figure 2B). The side chain of Tyr91LC hydrogen bonded with the Gly100cHC carbonyl oxygen, stabilizing a kink in the loop that formed the transition between these secondary structure elements (Figure 2B).
Figure 2. Conformations of 8ANC195 CDRH1 and CDRH3 loops.
(A) The hook-like conformation of CDRH1 (red) is stabilized by burial of the hydrophobic Phe30HC side chain and hydrogen bonds within CDRH1 and with CDRH3 (blue) and FWR1 and FWR3 residues (Ala24HC and Asp73HC, respectively). (B) The complexed CDRH3 conformation (blue) consists of a protruding loop (residues 102HC–110HC) and a small β-sheet subdomain (residues 111HC–118HC) stabilized by multiple hydrogen bonds within CDRH3 as well as with CDRH1 and CDRL3. A hydrogen bond between Tyr92LC and Gly110HC stabilized the bifurcation of CDRH3 into its two subdomains. CDRH1 and CDRH3 loop backbone atoms are shown as sticks and side chains of residues important for stabilizing the loop conformations are shown as sticks (involved in direct contacts) or lines (backbone involved in contacts) (with exception of those of Tyr105HC, Lys107HC and Trp108HC, which are shown for clarity). (C) Comparison of CDRH3 loops in 8ANC195 and other anti-HIV-1 bNAbs. CDRH3 residues corresponding to 8ANC195HC residues 97–124 (pink) of NIH45–46 (blue, PDB 3U7Y), PG16 (grey, PDB 4DQO), PGT121 (purple, PDB 4FQC) and PGT128 (orange, PDB 3TYG) are shown as Cα traces.
The complex structure showed independent binding of sCD4 and 8ANC195 Fab to distinct sites on gp120 (Figure 1B). sCD4 interacted with the gp120 core as in other sCD4-gp120 structures (Kwong et al., 1998) (Figure S1A), thus its binding was not altered by the presence of the adjacent antibody, consistent with binding and neutralization experiments showing no effects of CD4 addition on 8ANC195 activity (Figure S1B,C). sCD4 did, however, contribute to crystal packing (Figure S1D), which may explain why diffraction-quality crystals failed to grow in its absence. In the ternary complex structure, 8ANC195 bound to a gp120 region adjacent to the CD4 binding site, contacting mainly the gp120 inner domain, loops D and V5, and a small patch of the gp120 outer domain (His352gp120–Asn354gp120) (Figure 1B,C).
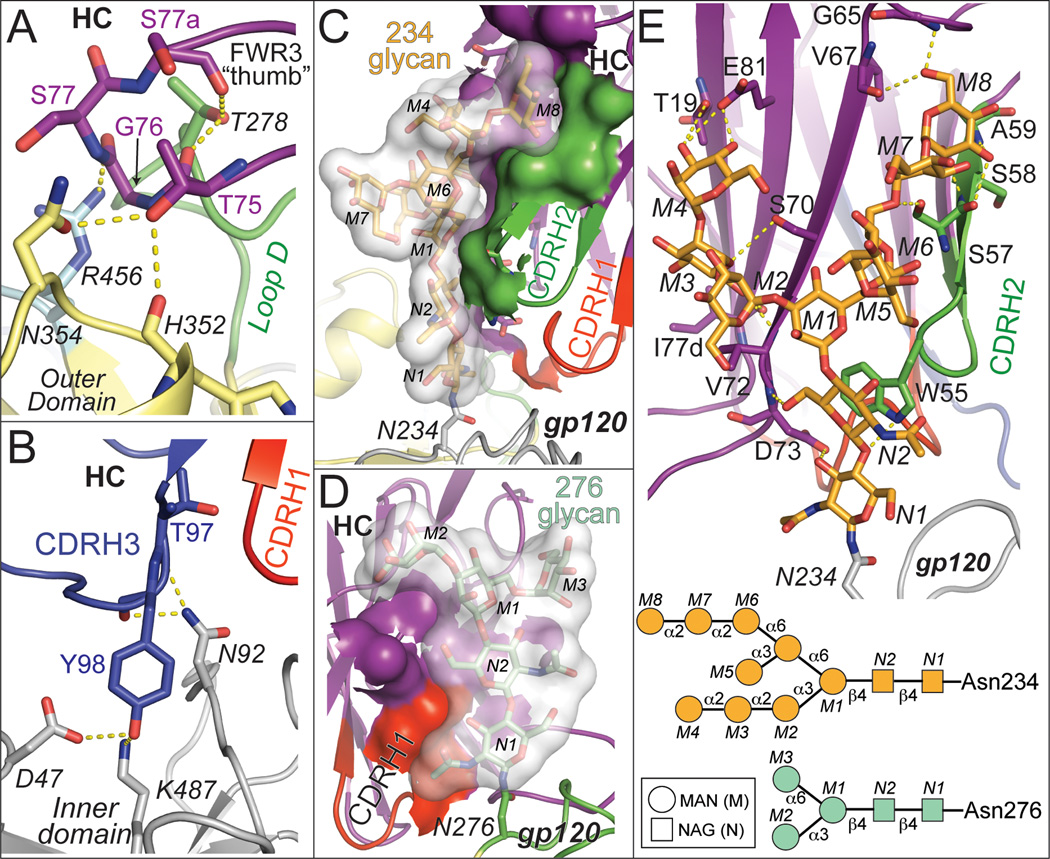
8ANC195 Fab bound gp120 core exclusively with its HC, using residues in FWRs and its three CDR loops to form an extensive interface (3,747 Å2 total buried surface area; 1,347 Å2 HC–gp120 protein contacts; 2,400 Å2 HC–gp120 glycan contacts) (Figure 1B, 3, Figure S2; Table S2). A loop in FWR3HC, consisting of somatically-mutated residues and extended by a four-residue insertion, reached like a thumb into the pocket formed by loops D, V5 and outer domain residues 352gp120–358gp120 (Figure 1A,B and 3A, Figure S2B). Deletion of the FWR3 insertion that results in the protruding “FWR3HC thumb” abrogated neutralization activity of 8ANC195 (Figure S1E). CDRH1 and CDRH3 contacted the gp120 inner domain (Figure 3B, Figure S2B), contributing to a 1,347 Å2 interface between the 8ANC195 HC and gp120 protein residues. The CDRH1 and CDRH3 loop conformations, conserved in the free Fab (Figure 1A, Figure 2A,B), were necessary for binding gp120 since extending these loops would result in clashes with gp120. The resulting antibody combining site was exquisitely suited to contacting portions of the inner domain of gp120 not targeted by other bNAbs (Figure 1C).
Figure 3. Contacts made by 8ANC195 HC with gp120 protein residues and glycans.
Labels for gp120 protein and glycan residues are italicized. Hydrogen bonds are shown as dashed yellow lines. (A) FWR3HC loop contacts with loop D (green), loop V5 (light blue), and outer domain loop (yellow). (B) 8ANC195 HC CDRH1 (red) and CDRH3 (blue) contacts with gp120 inner domain (grey). (C) Buried surface area between the Asn234gp120 glycan (transparent surface with glycan residues shown as sticks) and 8ANC195 (HC FWR residues in purple and CDRH2 in green). Antibody atoms buried by glycan interactions are shown as surfaces. (D) Buried surface area between the Asn276 glycangp120 (transparent surface with glycan residues shown as sticks) and 8ANC195 (HC FWR residues in purple and CDRH1 in red). Antibody atoms buried by glycan interactions are shown as surfaces. (E) Top: Contacts made by 8ANC195 HC FWR residues (purple) and CDRH2 (green) with Asn234gp120 glycan (orange). Glycan and protein residues involved in hydrogen bonds are shown as sticks. Bottom: schematic of ordered high mannose glycans on Asn234gp120 and Asn276 gp120 (bottom). See also Figure S2.
The 8ANC195 Fab also made extensive interfaces with glycans attached to Asn234gp120 (buried surface area = 1,645 Å2) and Asn276gp120 (buried surface area = 755 Å2), consistent with its dependence on these PNGSs for neutralization (Chuang et al., 2013; West et al., 2013). Together with CDRH2, somatically-mutated FWR residues in strands B, C”, D and E contributed to an extensive interface with the Asn234gp120-associated N-glycan (usually high mannose in native HIV-1 Envs (Go et al., 2011)) that involved 10 sugar moieties, including specific interactions with terminal mannose residues (Figure 3C,E, Figure S2C–H). Although Man5GlcNAc2 glycans comprise the majority of N-glycans on gp120 proteins produced in GnTI −/− cells, Man6–9GlcNAc2 have also been found (Dunlop et al., 2010), and the D1,D3-Man8GlcNAc2 structure of the Asn234gp120-associated N-glycan was verified by simulated annealing omit maps (Figure S3E,H). A two-residue deletion at the CDRH2–FWR3HC boundary compared to the germline sequence permitted these interactions, since the longer loop would clash with inner domain residue Asn234gp120 and its neighbors. The Asn276gp120 glycan (a complex-type N-glycan in native HIV-1 Envs (Binley et al., 2010; Go et al., 2011), but high mannose in the crystallized gp120) was wedged between 8ANC195 and sCD4, where it contacted FWR residues in strands A and B and the N-terminal portion of CDRH1, forming an interface involving only the core pentasaccharide common to both high mannose and complex-type N-glycans (Figure 3D, Figure S2F,G). Recognition of the common core pentasaccharide has been suggested to be an adaptation evolved by glycan-dependent bNAbs such as PGT121 to recognize both complex-type and high mannose glycans at a particular PNGS on Env (Mouquet et al., 2012). In the case of the 8ANC195 interaction with the Asn276gp120 glycan, modeling indicates that the core fucose of a complex-type N-glycan could be accommodated by the antibody (Figure S3I), thus 8ANC195 may also exhibit promiscuous recognition of certain N-glycans.
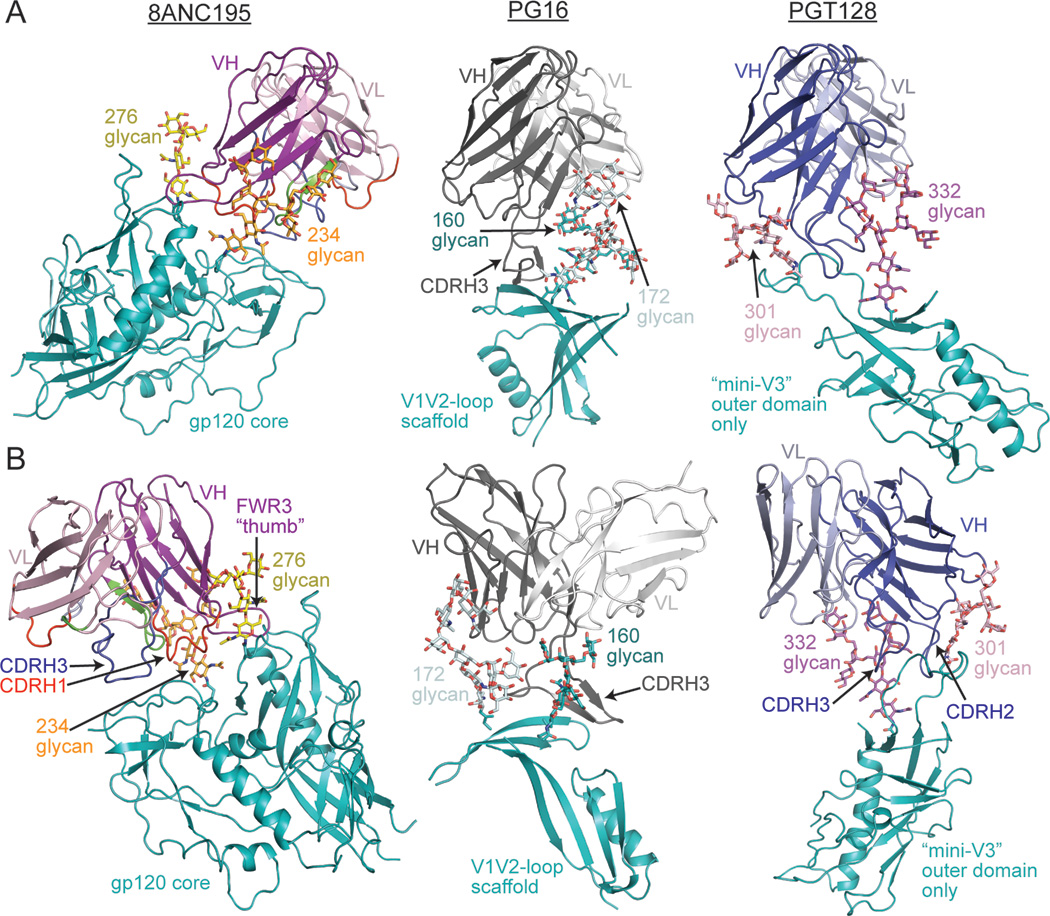
The 8ANC195 HC was bracketed by the Asn234gp120 and Asn276gp120 glycans in a manner analogous to interactions of HIV-1 antibodies that penetrate the Env glycan shield, such as PG16 (interactions with Asn160gp120 and Asn156gp120/Asn173gp120 glycans) (Pancera et al., 2013), PGT128 (with Asn301gp120 and Asn332gp120 glycans) (Pejchal et al., 2011) and PGT121 (with Asn332gp120 and Asn137gp120 glycans) (Julien et al., 2013a; Julien et al., 2013b; Mouquet et al., 2012) (Figure 4). However, in contrast to these antibodies, 8ANC195 contacts with gp120 were made exclusively by its HC; indeed, 33% of 8ANC195 VH domain residues not buried at the LC interface contacted gp120. In summary, the 8ANC195-gp120 structure demonstrated that 8ANC195 recognizes a novel epitope involving the Asn234gp120 and Asn276gp120 glycans, the gp120 inner domain, loop D and loop V5, which would be adjacent to gp41 in Env trimer (Julien et al., 2013a; Lyumkis et al., 2013).
Figure 4. Comparison of glycan-dependent bNAbs.
8ANC195 is “bracketed” by two glycans (Asn234gp120 glycan, orange; Asn276gp120 glycan, yellow) in the 8ANC195 Fab/gp120/sCD4 complex structure (left panels). For comparison, crystal structures of PG16 (grey, middle panels, PDB 4DQO) bound to a V1/V2 loop scaffold and PGT128 (blue, right panels, PDB 3TYG) bound to a V3 loop scaffold are shown with (A) the antibody HCs aligned to the 8ANC195 HC or (B) an alternative view showing their interactions with bracketing glycans (for PG16: Asn160gp120 glycan, teal/Asn172gp120 glycan, light cyan; for PGT128: Asn301gp120 glycan, pink/Asn332gp120 glycan, purple). The proteins are shown as ribbon diagrams and the glycans as stick representations.
EM reconstruction of 8ANC195 bound to soluble Env trimer
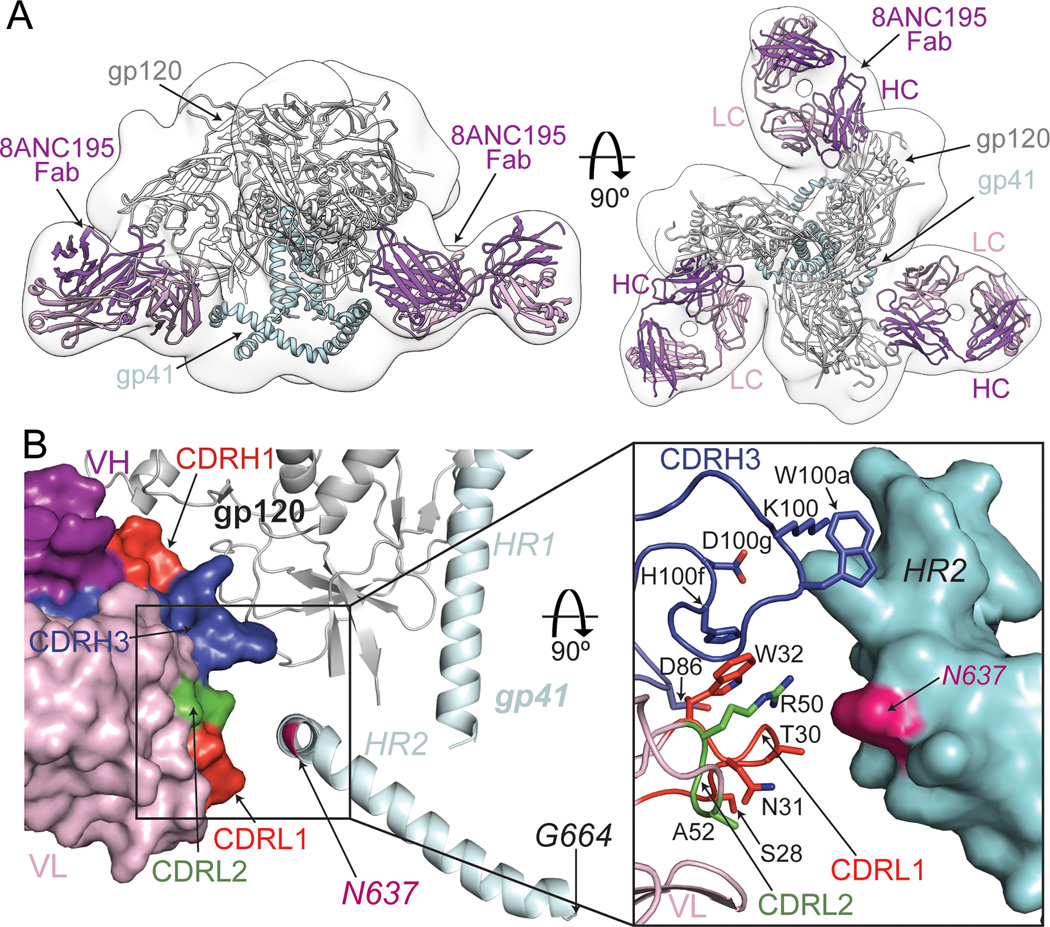
To investigate portions of the 8ANC195 epitope beyond the gp120 core, including potential contacts with gp41, we used negative stain single particle EM to determine the structure of 8ANC195 Fab bound to a soluble HIV-1 Env trimer with SOSIP mutations derived from strain BG505 (Figures 5A, S3) (Julien et al., 2013a; Lyumkis et al., 2013; Sanders et al., 2013). Independent docking of the BG505 Env trimer structure (PDB 4NCO) (Julien et al., 2013a) and 8ANC195 Fab resulted in a model wherein the Fab contacted both gp120 and gp41 within a single protomer (Figure 5A, Figure S3). As expected from the lack of gp41 in the complex crystal structure and the low resolution of the EM structure, a small difference in Fab placement was observed between the two structures (Figure S3D,E). The EM model placed the CDRL1, CDRL2, and portions of FWR3LC and CDRH3 in close proximity to the HR2 helix of gp41 (Figure 5B). Although gp41 residues were not definitively identified in the trimer crystal structure (Julien et al., 2013a), based on the assignment of the HR2 C-terminus as Gly664gp41 (Lyumkis et al., 2013), the kink in the HR2 helix was assigned as Asn637gp41 (Figure 5B, Figure S4), the asparagine of a highly conserved PNGS. This assignment was consistent with mapping of gp41 residues 651–664 to the end of a helix at the base of the Env trimer, as determined by difference mapping to determine the position of residue 650gp41 (Lyumkis et al., 2013). By counting backwards from residue 650gp41, the kink in the HR2 helix can be assigned as residue Asn637gp41. In addition, although the glycan at Asn637gp41 was not built into the model for the 4.7 Å crystal structure of the BG505.SOSIP trimer (Julien et al., 2013a), difference Fourier maps calculated using the PDB code 4NCO structure factors showed electron density for the first GlcNAc of this glycan (Figure S3G). The 8ANC195–Env trimer EM model predicted that the Asn637gp41-linked glycan and adjacent amino acid residues on HR2 interacted with 8ANC195 CDRH3, CDRL1 and CDRL2. Despite the predicted contacts with the Asn637gp41-linked glycan, a mutant YU2 pseudovirus lacking the glycan (Asn637gp41Gln) did not show decreased sensitivity to 8ANC195 (Table S3). However, the gp120 portion of the 8ANC195 epitope is so extensive that it could mask or compensate for contributions made by a smaller gp41 portion of the epitope. In addition, the VRC01-like bNAbs provide a precedent in which an Env glycan (Asn276gp120) is contacted by the antibody (Diskin et al., 2013), yet viral strains and mutants that do not contain the glycan can show increased, rather than decreased, sensitivity in neutralization assays (Li et al., 2011).
Figure 5. EM reconstruction of 8ANC195/Env trimer complex and model of 8ANC195 LC interactions with gp41 HR2.
(A) EM reconstruction of 8ANC195 Fab/BG505 SOSIP.664. Side (left) and top (right) views of EM density with the X-ray structures of BG505 SOSIP.664 (PDB ID 4NCO; gp120, grey; gp41, light blue) and 8ANC195 Fab fit independently of gp140 coordinates to the EM density (purple). See also Figure S3. (B) Close-up of 8ANC195 LC/HR2 region of EM complex structure (Fab placement is best fit/independently placed as in (A)). Left: Fab is shown as a surface representation with highlights (CDRL1, red; CDRL2, green; CDRH1, red; CDRH3, blue), and gp140 is shown as a ribbon diagram (gp120, grey; gp41, light blue). The position of Asn637gp41 (magenta) was deduced from the position of the C-terminus of the SOSIP.664 trimer (Gly664gp41). Right: 8ANC195 HC and LC residues (sticks) positioned to contact HR2, which is shown as a surface representation calculated from HR2 coordinates in PDB 4NCO with presumptive sidechains added to the polyalanine coordinates. See also Figure S4.
Light chain involvement in neutralization of HIV-1 by 8ANC195
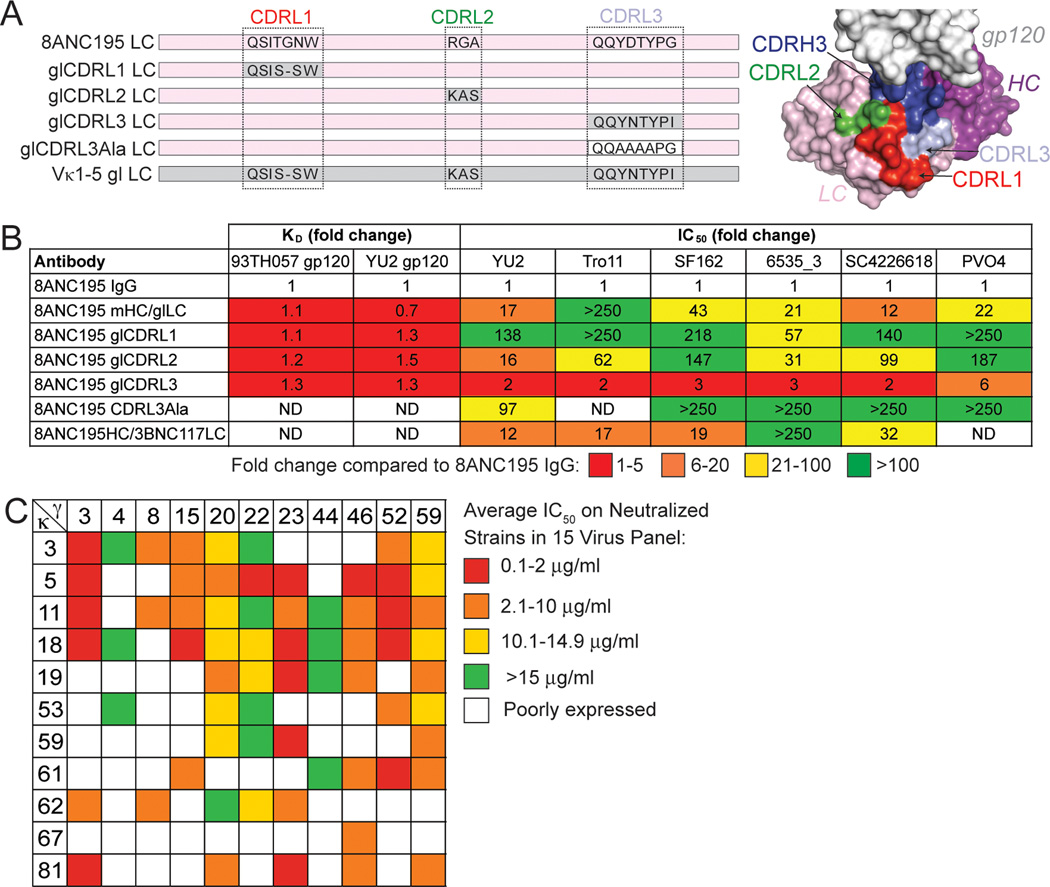
The EM reconstruction highlighted a potential role for 8ANC195 LC contacts to gp41. To assess LC contacts with trimeric Env, we tested chimeras consisting of the 8ANC195 HC paired with different LCs in neutralization and binding assays. The chimeras included a full germline LC, a mature LC with individual CDR loops reverted to their germline sequences or CDRL3 partially mutated to alanines, or the LC from the CD4 binding site antibody 3BNC117 (Figure 6A). As expected from the crystal structure in which all gp120 contacts were made by the 8ANC195 HC, the chimeras bound normally to gp120 core and to a full-length 93TH057 gp120 (Figure 6B, table S3), thus changes in the LC did not disrupt the HC portion of the antibody combining site.
Figure 6. Effects of LC sequence changes on 8ANC195 neutralization potency.
(A) Sequences of LC CDRs in constructs used with 8ANC195 HC to make chimeric IgGs (left) and location of CDRs on 8ANC195 structure (right). Sequences derived from the mature antibody are shown on a pink background, those derived from the germline precursor are shown on a grey background. The mutations introduced into CDRL3 in glCDRL3Ala are shown on a white background. (B) Effects of changes in 8ANC195 LC on binding to 93TH057 and YU2 gp120s and neutralization of viral strains, expressed as fold changes over results for 8ANC195 IgG. KD and IC50 values for these experiments are shown in table S3. (C) Heat map showing the expression and neutralization of randomly paired HCs and LCs from the bulk sort on a Tier 2 15-virus panel. Red squares represent average IC50 values on neutralized viral strains (arithmetic means) between 0.1 and 5 µg/ml; orange squares between 5.1 and 10 µg/ml, yellow squares between 10.1 and 14.9 µg/ml, and green squares above 15 µg/ml. White squares represent insufficient antibody expression. See also Figures S5 and S6.
In contrast to gp120 binding, neutralization potencies assayed against native Env spike trimers were decreased by changes in the 8ANC195 LC. For example, reverting CDRL1 and CDRL2 sequences to germline precursor sequences (changing 3 of 7 and 3 of 3 residues, respectively) almost completely abrogated neutralization of YU2, an 8ANC195-sensitive strain. Changes to CDRL3 led to a moderate reduction in neutralization potency, as did substituting the 3BNC117 LC for the cognate LC (Figure 6B, table S3). A chimeric IgG with one of the most conservatively-substituted LCs (Thr–Gly–Asn, mature CDRL1 containing a one-residue insertion, reverted to Ser–Ser, germline CDRL1) displayed unchanged binding to gp120, yet showed reductions in neutralization potency of up to 250-fold. Similarly, conservative changes in CDRL2 (Arg–Gly–Ala, the mature CDRL2, reverted to the germline Lys–Ala–Ser sequence) caused large reductions in neutralization potencies but had little effect on gp120 binding. Overall the data showed differential sensitivities of the binding and neutralization assays to changes in the 8ANC195 LC that were distant from the gp120 surface, which supported the EM results suggesting that LC, and CDRL1 and CDRL2 in particular, contacted gp41.
Isolation of clonal members of the 8ANC195 family
To further investigate Env recognition by 8ANC195, we isolated additional members of this antibody clone from the original donor by single cell sorting using gp120 stabilized in the CD4-bound conformation (2CC core) as bait (Figure S5). From 1536 single 2CC core-binding B cells, 10 (0.7%) were clonally related to 8ANC195, and of these, only four differed slightly from the two previously-described members (1 to 3 and 1 to 7 residue differences in the HCs and LCs, respectively) (Figure S5). Consistent with the limited sequence diversity, these antibodies exhibited similar potencies to 8ANC195 in neutralization assays against a panel of 15 Tier 2 viruses (Figure S5C and Table S4).
Reasoning that the 2CC core bait might fail to capture some 8ANC195 family members, we used clone-specific primers to amplify 8ANC195 variants from purified populations of CD19+ IgG+ memory B cells (Figure S6). We obtained 128 HC and 100 LC sequences that were clonally related to 8ANC195 and displayed greater sequence diversity than antibodies obtained using antigen-specific selection (Figure S6). Combinations of the 11 HC and 11 LC genes exhibiting greatest diversity were co-transfected in order to evaluate their neutralizing activity against a 15-member Tier 2 virus panel. Three of 67 (4.5%) new antibodies were at least as broad and potent as 8ANC195 (Figure 6C and Table S5). One of these, γ52HCκ5LC, was 5-fold more potent than 8ANC195 (neutralized 12 of 15 viruses with a mean IC50 of 0.45 µg/ml as compared to 2.3 µg/ml for 8ANC195) (Table S5E), a potency and breadth against this virus panel that was comparable to bNAbs that target non-overlapping sites (Mouquet et al., 2012; Wu et al., 2010) such as VRC01 (neutralized 12 of 15 viruses with a 0.56 µg/ml mean IC50) and more broad but less potent than 10–1074 (neutralized 6 of 15 viruses with a mean of 0.09 µg/ml).
The LC was critical to the activity of the more potent γ52HCκ5LC variant, as demonstrated by diminished neutralization potencies when κ5LC was swapped for either κ3LC or κ11LC (Figure 6C). The weaker neutralization could be explained by differences between κ5LC and κ3LC at solvent-exposed residues in CDRL2 (53LC and 54LC) and FWRL3 (64LC), and a nearby buried residue (34LC) that may affect the structural integrity of CDRL1. Modeling of YU2 gp41 residues into the Env trimer structure (Julien et al., 2013a) suggested that 8ANC195 positions 53LC and 54LC were adjacent to the Asn637gp41 PNGS (Figures S4D, S7). The improved neutralizing activity of κ5LC compared with the other newly-isolated LCs was associated with small side chains at positions 34LC (Val), 53LC (Ala) and 54LC (Ala), whereas κ3LC or κ11LC, which were less broadly neutralizing when paired with identical HCs, included bulkier and/or charged side chains that would clash with the nearby gp41 glycan. κ5LC was the only LC containing an S64RLC substitution and this single change compared to the 8ANC195 LC may account for the 5-fold improved potency of γ52HCκ5LC. Residues in the immediate vicinity of Asn637gp41 might also modify neutralization; a computational analysis of neutralization panel data using the Antibody Database program (West et al., 2013) suggested that Glu632gp41 was associated with stronger neutralization.
Discussion
8ANC195 defines a novel site of HIV-1 Env vulnerability that contacts gp120 glycans and portions of the gp120 inner domain not targeted by other bNAbs. A single particle EM reconstruction, supported by site-directed mutagenesis, suggested that the epitope spans gp120 and gp41 (Figure 7). As such, 8ANC195 represents the first patient-derived anti-HIV-1 antibody that spans both subunits of the Env trimer. An antibody generated by phage display was reported to exhibit binding to gp120 as well as to a gp41-Fc fusion protein, but it was selected in vitro for these independent binding activities and there was no evidence that it contacts both Env subunits simultaneously on a native Env trimer (Zhang et al., 2012). By contrast, our structural, mutagenesis and neutralization results suggest that the 8ANC195 epitope spans both subunits in a native trimer. Rather than penetrating the glycan shield using only a single CDR loop, a strategy employed by antibodies such as PG9 and PGT128 (McLellan et al., 2011; Pejchal et al., 2011), 8ANC195 inserted its entire HC variable region into a gap in the shield to form a large interface, of which >50% involved contacts to gp120 glycans. The recognition of both Env subunits by 8ANC195 may facilitate neutralization by preventing conformational changes required for gp41-mediated fusion of the host cell and viral membranes.
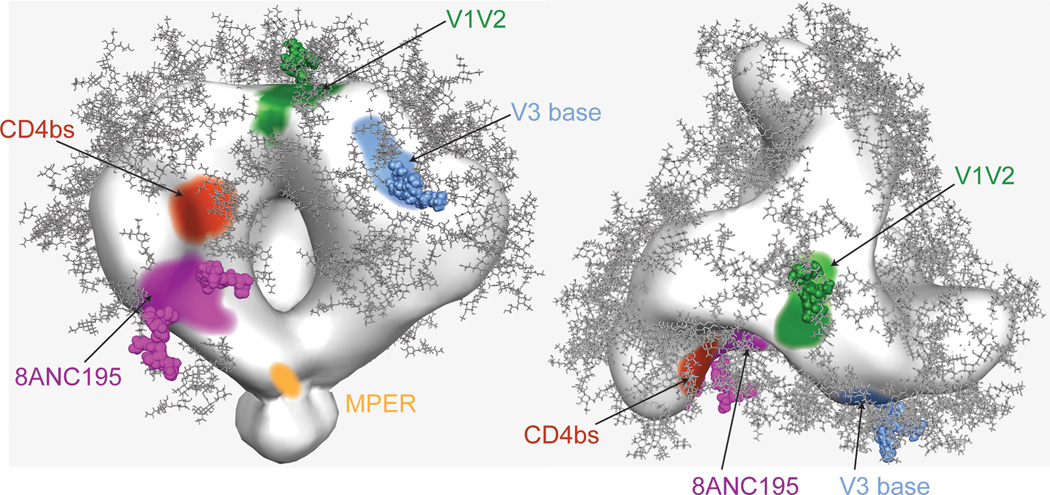
Figure 7. Locations of bNAb epitopes on HIV-1 Env trimer.
EM density map of Env trimer including MPER region (trimer map EMD-5019 from (Liu et al., 2008)) showing approximate epitope locations for antibodies targeting the 8ANC195 epitope (purple with Asn234gp120 and Asn276gp120 glycans shown as spheres), CD4 binding site (red), V3 loop/Asn332gp120 glycan (blue with Asn332gp120 glycan shown as spheres), V1/V2 loop/Asn160gp120 glycan (green with Asn160gp120 glycan shown as spheres), and MPER (yellow).
8ANC195 was one of two family members of a rare B cell clone and the original antibody was less potent than some bNAbs targeting the CD4 binding site (Scheid et al., 2011; Wu et al., 2010) or V3 loop (Mouquet et al., 2012; Walker et al., 2011). Although it was not possible to obtain large numbers of 8ANC195 variants by standard single cell cloning techniques (Scheid et al., 2009b), randomly combining HCs and LCs obtained from memory B cells without antigen-specific sorting demonstrated that the target of this antibody supported neutralization activity comparable to that against the most vulnerable sites on Env. Since 8ANC195 arose in an HIV-1–infected donor, it is possible for the human immune system to generate antibodies against this new epitope, making it a valuable site to include on potential immunogens. Indeed, the footprint of this newly-identified bNAb on gp120 primarily involves the inner domain, contrary to suggestions that efforts be made to focus the antibody response on the gp120 outer domain only (Nabel et al., 2011). Characterization of the potential gp120-gp41 bridging epitope highlights the importance of using properly-folded gp120-gp41 Env trimers (Julien et al., 2013a; Lyumkis et al., 2013; Sanders et al., 2013) or building epitope scaffolds containing the gp120 outer and inner domains plus a portion of gp41 for immunogen design.
In addition to identifying vaccine targets, bNAbs could potentially be used therapeutically in established HIV-1 infections, especially bNAb combinations that limit escape by recognizing non-overlapping epitopes (Barouch et al., 2013; Klein et al., 2012; Shingai et al., 2013). Potent variants of 8ANC195 may be particularly valuable in this respect since the epitope does not overlap with the targets of CD4 binding site, V2 loop, V3 loop or MPER antibodies.
Experimental Procedures
Protein expression and purification
The antibodies used in this study were produced and purified as in previously-described studies (Diskin et al., 2011). Briefly, 8ANC195 and its clonal variants, 3BNC60, and chimeric antibody (mature HC / various LC; γ/κ combinations of newly isolated 8ANC195 variants) IgGs were expressed by transiently transfecting HEK293-6E cells with vectors containing the appropriate heavy and light chain genes. Secreted IgGs were purified from cell supernatants using protein A affinity chromatography (GE Healthcare). For neutralization assays, IgGs were diluted to 1 mg/mL stocks in 20 mM Tris pH 8.0, 150 mM sodium chloride (TBS buffer). 8ANC195 Fab was expressed with a 6x-His tag on the C-terminus of CH1 as described for IgGs and purified using Ni2+-NTA affinity chromatography (GE Healthcare) and Superdex 200 16/60 size exclusion chromatography (GE Healthcare).
A truncated gp120 from the HIV-1 strain 93TH057 containing mutations Asn88Glngp120, Asn289Glngp120, Asn334Glngp120, Asn392Glngp120, Asn448Glngp120 was produced by transiently transfecting HEK293-S (GnTI−/−) cells adapted for growth in suspension by the Caltech Protein Expression Center with a pTT5 vector encoding His-tagged gp120. Secreted gp120 was captured on Ni2+-NTA resin (GE Healthcare) and further purified using Superdex 200 16/60 size exclusion chromatography (GE Healthcare).
Soluble CD4 domains 1 and 2 (sCD4) and sCD4K75T were produced as described previously (Diskin et al., 2010). Briefly, the pACgp67b vector encoding 6x-His-tagged sCD4 or sCD4K75T (residues 1–186 of mature CD4) was used to make infectious baculovirus particles using BaculoGold (BD Biosynthesis). Protein was expressed in Hi5 cells, captured on a Ni2+-NTA column (GE Healthcare) and further purified using Superdex 200 16/60 size exclusion chromatography (GE Healthcare). To remove an N-linked glycan introduced by mutation in sCD4K75T, the protein was treated with Endoglycosidase H (New England Biolabs) for 16 hours at 25° C and then purified by Superdex 200 16/60 size exclusion chromatography (GE Healthcare).
For complex crystallization trials, purified 8ANC195 Fab, 93TH057 gp120 and EndoH-treated sCD4K75T were incubated at a 1:1:1 molar ratio for 2 hours at 25 °C. The complex was purified by Superdex 200 10/300 size exclusion chromatography (GE Healthcare) and the peak corresponding to 8ANC195 Fab/gp120/sCD4K75T complex concentrated to 16 mg/mL in TBS buffer. For crystallization of 8ANC195 Fab alone, the protein was concentrated to 20 mg/mL in TBS buffer.
Purified BG505 SOSIP trimers (Julien et al., 2013a; Julien et al., 2013b; Sanders et al., 2013) for EM studies were the gift of Dr. John P. Moore (Weill Cornell Medical College).
Crystallization
Crystals of 8ANC195 Fab (space group P41212, a = 66.5 Å, b = 66.5 Å, c = 219.0 Å; one molecule per asymmetric unit) were obtained upon mixing a protein solution at 11 mg/mL with 0.1M HEPES pH 7, 20% PEG 6,000, 10 mM zinc chloride at 20°C. Crystals were briefly soaked in mother liquor solution supplemented with 20% ethylene glycol before flash cooling in liquid nitrogen. Crystals of the 8ANC195 Fab/93TH057 gp120/sCD4K75T complex (space group P212121, a = 66.5 Å, b = 132.5 Å, c = 142.8 Å; one molecule per asymmetric unit) were obtained upon mixing a protein solution at 16 mg/mL with 14% polyethylene glycol 3,350, 0.1 M HEPES pH 7.3, 2% benzamidine HCl at 20°C. Crystals were briefly soaked in mother liquor solution supplemented with 30% ethylene glycol before flash cooling in liquid nitrogen.
Crystallographic data collection, structure solution and refinement
X-ray diffraction data for 8ANC195 Fab crystals were collected at the Argonne National Laboratory Advanced Photon Source (APS) beamline 23-ID-D using a MAR 300 CCD detector. X-ray diffraction data for 8ANC195 Fab/93TH057 gp120/sCD4K75T complex crystals were collected at the Stanford Synchrotron Radiation Lightsource (SSRL) beamline 12-2 using a Pilatus 6M pixel detector (Dectris). The data were indexed, integrated and scaled using XDS (Kabsch, 2010). The 8ANC195 Fab structure was solved by molecular replacement and refined to 2.13 Å resolution using an iterative approach involving refinement and verification of model accuracy with simulated annealing composite omit maps using the Phenix crystallography package (Adams et al., 2010), and manually fitting models into electron density maps using Coot (Emsley and Cowtan, 2004). The final model (Rwork = 20.2%; Rfree = 24.2%) includes 3,321 protein atoms, 15 ligand atoms and 178 water molecules (Table S1). 99.54%, 0.46% and 0.0% of the residues were in the favored, allowed and disallowed regions, respectively, of the Ramachandran plot. Disordered residues that were not included in the model include residues 127–134, 214–219 and the 6x-His tag of the 8ANC195 heavy chain, and residues 213–214 of the light chain. The 8ANC195 Fab/93TH057 gp120/sCD4K75T complex structure was solved by molecular replacement and refined to 3.0 Å resolution as described for the Fab structure. In addition to considering I/σI and completeness of the highest resolution shell (2.1% and 99.9%, respectively), we used the CC1/2 statistic (Karplus and Diederichs, 2012) (correlation coefficient between two random halves of the data set where CC1/2 > 10%) to determine the high-resolution cutoff for our data. Phenix (Adams et al., 2010) was used to compute CC1/2 (85.4% for the highest resolution shell and 99.8% for the entire data set), supporting our high-resolution cutoff determination. To prevent phase bias, no glycan residues were present during initial stages of refinement. Glycans were built manually in Coot (Emsley and Cowtan, 2004) into simulated annealing composite omit maps calculated using Phenix (Adams et al., 2010) throughout the refinement process. The final model (Rwork = 23.5%; Rfree = 27.2%) includes 7195 protein atoms and 408 atoms of carbohydrates and ligands (Table S1). 96.92%, 3.08% and 0.0% of the residues were in the favored, allowed and disallowed regions, respectively, of the Ramachandran plot. Disordered residues that were not included in the model include residues 126–135, 185–194, 214–219 and the 6x-His tag of the 8ANC195 heavy chain, residues 212–214 of the light chain, residues 125–197 (V1/V2 substitution), 302–324 (V3 substitution), residues 396–408 (a total of 6 residues from V4), residues 492–494 and the 6x-His tag of 93TH057 gp120 and residues 106–111, 150–152, 178–186 and the 6x-His tag of sCD4K75T.
Buried surface areas were calculated using PDBePISA (Krissinel and Henrick, 2007) and a 1.4 Å probe. Superimposition calculations were done and molecular representations were generated using PyMOL (Schrödinger, 2011). Pairwise Cα alignments were performed using PDBeFold (Krissinel and Henrick, 2004).
ELISAs
High-binding 96-well ELISA plates (Costar) were coated overnight with 5 µg/well of purified gp120 in 100 mM sodium carbonate pH 9.6. After washing with TBS containing 0.05% Tween 20, the plates were blocked for 2 h with 1% BSA, 0.05% Tween-TBS (blocking buffer) and then incubated for 2 h with 8ANC195 IgG (1 µg/mL) mixed with 1:2 serially diluted solutions of potential antibody competitors (sCD4, J3 VHH, 3BNC60 Fab, NIH45–46 Fab) in blocking buffer (competitor concentration range from 5 to 320 µg/mL). After washing with TBS containing 0.05% Tween 20, the plates were incubated with HRP-conjugated goat anti-human IgG antibodies (Jackson ImmunoReseach) (at 0.8 µg/ml in blocking buffer) for 1 hour. The ELISAs were developed by addition of HRP chromogenic substrate (TMB solution, BioLegend) and the color development stopped by addition of 10% sulfuric acid. Experiments were performed in duplicate.
Surface plasmon resonance
Experiments were performed using a Biacore T100 (Biacore) using a standard single-cycle kinetics method. YU-2 and 93TH057 gp120 proteins were primary amine-coupled on CM5 chips (Biacore) at a coupling density of 1,000 RUs and one flow cell was mock coupled using HBS-EP+ buffer. 8ANC195 and chimeric IgGs were injected over flow cells at increasing concentrations (62.5 to 1,000 nM), at flow rates of 20 µl/min with 5 consecutive cycles of 2 min association/1 min dissociation and a final 10 min dissociation phase. Flow cells were regenerated with 3 pulses of 10 mM glycine pH 2.5. Apparent binding constants (KD (M)) were calculated from single-cycle kinetic analyses after subtraction of backgrounds using a 1:1 binding model without a bulk reflective index (RI) correction (Biacore T100 Evaluation software). Binding constants for bivalent IgGs are referred to as “apparent” affinities to emphasize that the KD values include potential avidity effects.
Neutralization assays
A TZM-bl/pseudovirus neutralization assay was used to evaluate the neutralization potencies of the antibodies as described (Montefiori, 2005). Pseudoviruses were generated by cotransfection of HEK 293T cells with an Env expression plasmid and a replication-defective backbone plasmid. Neutralization assays were preformed in-house for evaluating 8ANC195 LC chimeras (Table S3 and Figure 4B) and by the Collaboration for AIDS Vaccine Discovery (CAVD) core neutralization facility for testing the newly isolated 8ANC195 clonal variants (Tables S4, S5; Figures S5, S6). Some of the in-house data were derived from neutralization assays that were dispensed automatically by a Freedom EVO® (Tecan) liquid handler (IC50 values derived from manual and robotic assays agreed to within 2–4 fold). In all cases, neutralization was monitored by the reduction of a Tat-induced reporter gene (luciferase) in the presence of a three-or five-fold antibody dilution series (each concentration run in duplicate or triplicate) after a single round of pseudovirus infection in the TZM-bl cell line (Montefiori, 2005). Nonlinear regression analysis was used to calculate the concentrations at which half-maximal inhibition was observed (IC50 values).
Negative-stain EM
The BG505 SOSIP.664/8ANC195 Fab complex and grids were prepared as described previously (Kong et al., 2013). The data were collected on an FEI Tecnai T12 electron microscope coupled with a Tietz TemCam-F416 4k × 4k CMOS camera using the LEGINON interface (Suloway et al., 2005). Images were collected in 10° increments from 0° to −40° using a defocus range of 0.6 – 0.9 µm at a magnification of 52,000x, resulting in a pixel size of 2.05 Å at the specimen plane. Particles were selected using DogPicker (Voss et al., 2009) within the Appion software package (Lander et al., 2009), and sorted from reference-free 2D class averages using the SPARX package (Penczek et al., 1992). An initial model was generated by common lines from class averages using the EMAN2 package (Tang et al., 2007) and was refined using 11,637 unbinned particles. The refinement was carried out using the SPARX package (Penczek et al., 1994) with C3 symmetry applied. The resulting resolution at a 0.5 Fourier Shell Correlation (FSC) cut-off was 18.7 Å (Figure S3B,C). Molecular representations were generated using Chimera (Pettersen et al., 2004).
Human Samples
Human samples were collected after signed informed consent in accordance with Institutional Review Board (IRB)-reviewed protocols by all participating institutions. Patient 8 was selected from a cohort of elite controllers that were followed at the Ragon Institute in Boston.
Isolation of 8ANC195 variants
Single Cell clonal variants of 8ANC195 were isolated by 2CC core-specific single cell sorting, followed by reverse transcription and immunoglobulin gene amplification as described previously (Scheid et al., 2011). Immunoglobulin genes were cloned into heavy and light chain expression vectors and co-transfected for IgG production as described previously (Tiller et al., 2008). IgG+ CD19+ memory B cells were bulk sorted on a FACS AriaIII cell sorter. Bulk mRNA was extracted using TRIzol (Invitrogen) and reverse transcribed as previously described (Scheid et al., 2011). 8ANC195-related heavy and light chain genes were PCR amplified using clone-specific primers. Amplification products were gel purified and cloned into TOPO TA sequencing vectors (Invitrogen) and expression vectors as described previously (Tiller et al., 2008).
Phylogenetic tree and Alignment assembly
Phylogenetic trees were assembled using Geneious Neighbor-Joining Tree Software. Sequence Alignments were performed using DNA Star Clustal W alignment software.
Computational analysis
The program AntibodyDatabase (West et al., 2013) was used to analyze 8ANC195 neutralization panel data from refs. (Scheid et al., 2011) and (Chuang et al., 2013). This method attempts to model the variation in neutralization potency across strains based on a sum of terms (“rules”) corresponding to specific residues or potential N-linked glycosylation site (PNGS) positions. With the free residual option deselected, the analysis finds a rule corresponding to ~3-fold better 8ANC195 neutralization for strains with Glu632gp41. This correlation appears to hold across clades based on neutralization data for strains having the most favorable glycosylation pattern (PNGS at 234gp120 and 276gp120, and not at 230gp120) (West et al., 2013). For all clades, the residue at 632gp41 versus geometric mean IC50s for 8ANC195 on strains with the most favorable glycosylation pattern was a follows: Glu, 0.43 µg/mL (n=53) versus Asp, 1.31 µg/mL (n=51). For separate clades, the correlations were Clade A: Glu, 0.47 µg/mL (n=3); Asp, 1.30 µg/mL (n=24); Clade B: Glu, 0.18 µg/mL (n=15); Asp, 0.72 µg/mL (n=6); Clade C: Glu, 0.32 µg/mL (n=2); Asp, 1.31 µg/mL (n=20).
Supplementary Material
Highlights.
Broadly neutralizing antibody 8ANC195 recognizes a new epitope on HIV-1 Env
8ANC195 epitope bridges gp120 and gp41 subunits of HIV-1 Env
8ANC195 inserts heavy chain variable domain into gap in Env glycan shield
8ANC195 epitope involves gp120 glycans and protein residues of gp120 inner domain
Acknowledgements
This research was supported by the National Institute Of Allergy And Infectious Diseases of the National Institutes of Health Grant HIVRAD P01 AI100148 (P.J.B. and M.C.N.) and HIVRAD P01 AI082362 (A.B.W); (the content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health), the Bill and Melinda Gates Foundation [Collaboration for AIDS Vaccine Discovery Grants 1040753 (P.J.B.) and 38619s (M.C.N.) Comprehensive Antibody-Vaccine Immune Monitoring Consortium Grant 1032144 (M.S.S.)], NIH Center for HIV/AID Vaccine Immunology and Immunogen Discovery (CHAVI-ID) 1UM1 AI100663-01 (M.C.N.), American Cancer Society Grant PF-13-076-01-MPC (to L.S.), a Dissertation Fellowship from the California HIV/AIDS Research Program (J.H.L), and the Molecular Observatory at Caltech supported by the Gordon and Betty Moore Foundation. The EM work was conducted at the National Resource for Automated Molecular Microscopy at The Scripps Research Institute, which is supported by the Biomedical Technology Research Center program (GM103310) of the National Institute of General Medical Sciences. We thank John P. Moore and Albert Cupo (Weill Cornell Medical College) for purified SOSIP trimers, Anna Gazumyan and Cassie Liu (Rockefeller University) for help with antibody expression, and the Caltech Protein Expression Center for producing antibody and gp120 proteins, generation of suspension-adapted HEK293-S cells and use of the Biacore T100. Operations at the Stanford Synchrotron Radiation Lightsource are supported by the US Department of Energy and the National Institutes of Health. We thank the beamline staff at the Advanced Photon Source GM/CA-CAT for use and support for beamline 23ID-D. M.C.N. and P.J.B. are Howard Hughes Medical Institute investigators.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author contributions
L.S. solved and analyzed crystal structures, performed and analyzed binding studies and analyzed antibody mutant data; J.F.S. isolated 8ANC195 variants and analyzed neutralization assays; JH.L. solved and analyzed EM structure; A.P.W.Jr. performed computational analysis; C.C. performed binding assays; H.G. expressed and purified proteins; P.N.P.G., R.M. and M.S.S. performed and analyzed neutralization assays; L.S., J.F.S., JH.L., A.B.W., M.C.N. and P.J.B. analyzed data and wrote the manuscript.
Data Deposition
Crystallographic atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org, for the 8ANC195 Fab (4P9M) and 8ANC195 Fab-gp120-CD4 complex (4P9H) structures. The EM reconstruction have been deposited in the Electron Microscopy Data Bank, www.ebi.ac.uk/pdbe/emdb (XXXX).
References
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barouch DH, Whitney JB, Moldt B, Klein F, Oliveira TY, Liu J, Stephenson KE, Chang HW, Shekhar K, Gupta S, et al. Therapeutic efficacy of potent neutralizing HIV-1-specific monoclonal antibodies in SHIV-infected rhesus monkeys. Nature. 2013;503:224–228. doi: 10.1038/nature12744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binley JM, Ban YE, Crooks ET, Eggink D, Osawa K, Schief WR, Sanders RW. Role of complex carbohydrates in human immunodeficiency virus type 1 infection and resistance to antibody neutralization. Journal of virology. 2010;84:5637–5655. doi: 10.1128/JVI.00105-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton DR, Poignard P, Stanfield RL, Wilson IA. Broadly neutralizing antibodies present new prospects to counter highly antigenically diverse viruses. Science. 2012;337:183–186. doi: 10.1126/science.1225416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang GY, Acharya P, Schmidt SD, Yang Y, Louder MK, Zhou T, Kwon YD, Pancera M, Bailer RT, Doria-Rose NA, et al. Residue-level prediction of HIV-1 antibody epitopes based on neutralization of diverse viral strains. Journal of virology. 2013;87:10047–10058. doi: 10.1128/JVI.00984-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diskin R, Klein F, Horwitz JA, Halper-Stromberg A, Sather DN, Marcovecchio PM, Lee T, West AP, Jr, Gao H, Seaman MS, et al. Restricting HIV-1 pathways for escape using rationally designed anti-HIV-1 antibodies. The Journal of experimental medicine. 2013;210:1235–1249. doi: 10.1084/jem.20130221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diskin R, Marcovecchio PM, Bjorkman PJ. Structure of a clade C HIV-1 gp120 bound to CD4 and CD4-induced antibody reveals anti-CD4 polyreactivity. Nature structural & molecular biology. 2010;17:608–613. doi: 10.1038/nsmb.1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diskin R, Scheid JF, Marcovecchio PM, West AP, Jr, Klein F, Gao H, Gnanapragasam PN, Abadir A, Seaman MS, Nussenzweig MC, et al. Increasing the potency and breadth of an HIV antibody by using structure-based rational design. Science. 2011;334:1289–1293. doi: 10.1126/science.1213782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunlop DC, Bonomelli C, Mansab F, Vasiljevic S, Doores KJ, Wormald MR, Palma AS, Feizi T, Harvey DJ, Dwek RA, et al. Polysaccharide mimicry of the epitope of the broadly neutralizing anti-HIV antibody, 2G12, induces enhanced antibody responses to self oligomannose glycans. Glycobiology. 2010;20:812–823. doi: 10.1093/glycob/cwq020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Go EP, Hewawasam G, Liao HX, Chen H, Ping LH, Anderson JA, Hua DC, Haynes BF, Desaire H. Characterization of glycosylation profiles of HIV-1 transmitted/founder envelopes by mass spectrometry. Journal of virology. 2011;85:8270–8284. doi: 10.1128/JVI.05053-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julien JP, Cupo A, Sok D, Stanfield RL, Lyumkis D, Deller MC, Klasse PJ, Burton DR, Sanders RW, Moore JP, et al. Crystal Structure of a Soluble Cleaved HIV-1 Envelope Trimer. Science. 2013a;342:1477–1483. doi: 10.1126/science.1245625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julien JP, Sok D, Khayat R, Lee JH, Doores KJ, Walker LM, Ramos A, Diwanji DC, Pejchal R, Cupo A, et al. Broadly neutralizing antibody PGT121 allosterically modulates CD4 binding via recognition of the HIV-1 gp120 V3 base and multiple surrounding glycans. PLoS pathogens. 2013b;9:e1003342. doi: 10.1371/journal.ppat.1003342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch W. Integration, scaling, space-group assignment and post-refinement. Acta Crystallogr D Biol Crystallogr. 2010;66:133–144. doi: 10.1107/S0907444909047374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karplus PA, Diederichs K. Linking crystallographic model and data quality. Science. 2012;336:1030–1033. doi: 10.1126/science.1218231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein F, Diskin R, Scheid JF, Gaebler C, Mouquet H, Georgiev IS, Pancera M, Zhou T, Incesu RB, Fu BZ, et al. Somatic mutations of the immunoglobulin framework are generally required for broad and potent HIV-1 neutralization. Cell. 2013a;153:126–138. doi: 10.1016/j.cell.2013.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein F, Halper-Stromberg A, Horwitz JA, Gruell H, Scheid JF, Bournazos S, Mouquet H, Spatz LA, Diskin R, Abadir A, et al. HIV therapy by a combination of broadly neutralizing antibodies in humanized mice. Nature. 2012;492:118–122. doi: 10.1038/nature11604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein F, Mouquet H, Dosenovic P, Scheid JF, Scharf L, Nussenzweig MC. Antibodies in HIV-1 vaccine development and therapy. Science. 2013b;341:1199–1204. doi: 10.1126/science.1241144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong L, Lee JH, Doores KJ, Murin CD, Julien JP, McBride R, Liu Y, Marozsan A, Cupo A, Klasse PJ, et al. Supersite of immune vulnerability on the glycosylated face of HIV-1 envelope glycoprotein gp120. Nature structural & molecular biology. 2013;20:796–803. doi: 10.1038/nsmb.2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krissinel E, Henrick K. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr D Biol Crystallogr. 2004;60:2256–2268. doi: 10.1107/S0907444904026460. [DOI] [PubMed] [Google Scholar]
- Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. Journal of molecular biology. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- Kwong PD, Wyatt R, Robinson J, Sweet RW, Sodroski J, Hendrickson WA. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature. 1998;393:648–659. doi: 10.1038/31405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander GC, Stagg SM, Voss NR, Cheng A, Fellmann D, Pulokas J, Yoshioka C, Irving C, Mulder A, Lau PW, et al. Appion: an integrated, database-driven pipeline to facilitate EM image processing. Journal of structural biology. 2009;166:95–102. doi: 10.1016/j.jsb.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, O'Dell S, Walker LM, Wu X, Guenaga J, Feng Y, Schmidt SD, McKee K, Louder MK, Ledgerwood JE, et al. Mechanism of neutralization by the broadly neutralizing HIV-1 monoclonal antibody VRC01. Journal of virology. 2011;85:8954–8967. doi: 10.1128/JVI.00754-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Bartesaghi A, Borgnia MJ, Sapiro G, Subramaniam S. Molecular architecture of native HIV-1 gp120 trimers. Nature. 2008;455:109–113. doi: 10.1038/nature07159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyumkis D, Julien JP, de Val N, Cupo A, Potter CS, Klasse PJ, Burton DR, Sanders RW, Moore JP, Carragher B, et al. Cryo-EM Structure of a Fully Glycosylated Soluble Cleaved HIV-1 Envelope Trimer. Science. 2013;342:1484–1490. doi: 10.1126/science.1245627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mascola JR, Haynes BF. HIV-1 neutralizing antibodies: understanding nature's pathways. Immunological reviews. 2013;254:225–244. doi: 10.1111/imr.12075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLellan JS, Pancera M, Carrico C, Gorman J, Julien JP, Khayat R, Louder R, Pejchal R, Sastry M, Dai K, et al. Structure of HIV-1 gp120 V1/V2 domain with broadly neutralizing antibody PG9. Nature. 2011;480:336–343. doi: 10.1038/nature10696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montefiori DC. Coligan John E, et al., editors. Evaluating neutralizing antibodies against HIV, SIV, and SHIV in luciferase reporter gene assays. Current protocols in immunology. 2005;Chapter 12(Unit 12):11. doi: 10.1002/0471142735.im1211s64. [DOI] [PubMed] [Google Scholar]
- Mouquet H, Scharf L, Euler Z, Liu Y, Eden C, Scheid JF, Halper-Stromberg A, Gnanapragasam PN, Spencer DI, Seaman MS, et al. Complex-type N-glycan recognition by potent broadly neutralizing HIV antibodies. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:E3268–E3277. doi: 10.1073/pnas.1217207109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabel G, Kwong P, Mascola J. Progress in the rational design of an AIDS vaccine. Philosophical transactions - Royal Society Biological sciences. 2011;366:2759–2765. doi: 10.1098/rstb.2011.0096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pancera M, Shahzad-Ul-Hussan S, Doria-Rose NA, McLellan JS, Bailer RT, Dai K, Loesgen S, Louder MK, Staupe RP, Yang Y, et al. Structural basis for diverse N-glycan recognition by HIV-1-neutralizing V1-V2-directed antibody PG16. Nature structural & molecular biology. 2013;20:804–813. doi: 10.1038/nsmb.2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pejchal R, Doores KJ, Walker LM, Khayat R, Huang PS, Wang SK, Stanfield RL, Julien JP, Ramos A, Crispin M, et al. A potent and broad neutralizing antibody recognizes and penetrates the HIV glycan shield. Science. 2011;334:1097–1103. doi: 10.1126/science.1213256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penczek P, Radermacher M, Frank J. Three-dimensional reconstruction of single particles embedded in ice. Ultramicroscopy. 1992;40:33–53. [PubMed] [Google Scholar]
- Penczek PA, Grassucci RA, Frank J. The ribosome at improved resolution: new techniques for merging and orientation refinement in 3D cryo-electron microscopy of biological particles. Ultramicroscopy. 1994;53:251–270. doi: 10.1016/0304-3991(94)90038-8. [DOI] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera--a visualization system for exploratory research and analysis. Journal of computational chemistry. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Sanders RW, Derking R, Cupo A, Julien JP, Yasmeen A, de Val N, Kim HJ, Blattner C, de la Pena AT, Korzun J, et al. A next-generation cleaved, soluble HIV-1 Env Trimer, BG505 SOSIP.664 gp140, expresses multiple epitopes for broadly neutralizing but not non-neutralizing antibodies. PLoS pathogens. 2013;9:e1003618. doi: 10.1371/journal.ppat.1003618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheid JF, Mouquet H, Feldhahn N, Seaman MS, Velinzon K, Pietzsch J, Ott RG, Anthony RM, Zebroski H, Hurley A, et al. Broad diversity of neutralizing antibodies isolated from memory B cells in HIV-infected individuals. Nature. 2009a;458:636–640. doi: 10.1038/nature07930. [DOI] [PubMed] [Google Scholar]
- Scheid JF, Mouquet H, Feldhahn N, Walker BD, Pereyra F, Cutrell E, Seaman MS, Mascola JR, Wyatt RT, Wardemann H, et al. A method for identification of HIV gp140 binding memory B cells in human blood. J Immunol Methods. 2009b;343:65–67. doi: 10.1016/j.jim.2008.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheid JF, Mouquet H, Ueberheide B, Diskin R, Klein F, Olivera TY, Pietzsch J, Fenyo D, Abadir A, Velinzon K, et al. Sequence and Structural Convergence of Broad and Potent HIV Antibodies That Mimic CD4 Binding. Science. 2011;333:1633–1637. doi: 10.1126/science.1207227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrödinger L. The PyMOL Molecular Graphics System (The PyMOL Molecular Graphics System) 2011 [Google Scholar]
- Shingai M, Nishimura Y, Klein F, Mouquet H, Donau OK, Plishka R, Buckler-White A, Seaman M, Piatak M, Jr, Lifson JD, et al. Antibody-mediated immunotherapy of macaques chronically infected with SHIV suppresses viraemia. Nature. 2013;503:277–280. doi: 10.1038/nature12746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suloway C, Pulokas J, Fellmann D, Cheng A, Guerra F, Quispe J, Stagg S, Potter CS, Carragher B. Automated molecular microscopy: the new Leginon system. Journal of structural biology. 2005;151:41–60. doi: 10.1016/j.jsb.2005.03.010. [DOI] [PubMed] [Google Scholar]
- Tang G, Peng L, Baldwin PR, Mann DS, Jiang W, Rees I, Ludtke SJ. EMAN2: an extensible image processing suite for electron microscopy. Journal of structural biology. 2007;157:38–46. doi: 10.1016/j.jsb.2006.05.009. [DOI] [PubMed] [Google Scholar]
- Tiller T, Meffre E, Yurasov S, Tsuiji M, Nussenzweig MC, Wardemann H. Efficient generation of monoclonal antibodies from single human B cells by single cell RT-PCR and expression vector cloning. J Immunol Methods. 2008;329:112–124. doi: 10.1016/j.jim.2007.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voss NR, Yoshioka CK, Radermacher M, Potter CS, Carragher B. DoG Picker and TiltPicker: software tools to facilitate particle selection in single particle electron microscopy. Journal of structural biology. 2009;166:205–213. doi: 10.1016/j.jsb.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker LM, Huber M, Doores KJ, Falkowska E, Pejchal R, Julien JP, Wang SK, Ramos A, Chan-Hui PY, Moyle M, et al. Broad neutralization coverage of HIV by multiple highly potent antibodies. Nature. 2011;477:466–470. doi: 10.1038/nature10373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West AP, Jr, Scharf L, Horwitz J, Klein F, Nussenzweig MC, Bjorkman PJ. Computational analysis of anti-HIV-1 antibody neutralization panel data to identify potential functional epitope residues. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:10598–10603. doi: 10.1073/pnas.1309215110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Yang ZY, Li Y, Hogerkorp CM, Schief WR, Seaman MS, Zhou T, Schmidt SD, Wu L, Xu L, et al. Rational design of envelope identifies broadly neutralizing human monoclonal antibodies to HIV-1. Science. 2010;329:856–861. doi: 10.1126/science.1187659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang MY, Yuan T, Li J, Rosa Borges A, Watkins JD, Guenaga J, Yang Z, Wang Y, Wilson R, Li Y, et al. Identification and characterization of a broadly cross-reactive HIV-1 human monoclonal antibody that binds to both gp120 and gp41. PloS one. 2012;7:e44241. doi: 10.1371/journal.pone.0044241. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.