Abstract
Mediator is a key regulator of eukaryotic transcription1, connecting activators and repressors bound to regulatory DNA elements with RNA polymerase II (Pol II) 1-4. In the yeast S. cerevisiae, Mediator comprises 25 subunits with a total mass over 1 MDa 5,6, and is organized into three modules, termed Head, Middle/Arm and Tail 7-9. Our understanding of Mediator assembly and its role in regulating transcription has been impeded to date by limited structural information. Here, we report the crystal structure of the essential Mediator Head module (seven subunits, 223 kDa) at 4.3 Å resolution. Our structure reveals three distinct domains with the integrity of the complex centered on a bundle of ten helices from five different Head subunits. An intricate pattern of interactions within this helical bundle ensures stable assembly of the Head subunits, and provides the binding sites for general transcription factors (GTFs) and Pol II. Our structural and functional data suggest the Head module to juxtapose TFIIH and the carboxyl-terminal domain (CTD) of the largest subunit of Pol II, thereby facilitating CTD phosphorylation. Our results reveal architectural principles underlying the role of Mediator in the regulation of gene expression.
In the yeast S. cerevisiae, the Mediator Head module is composed of seven subunits (Med17, Med11, Med22, Med6, Med8, Med18, Med20) 10. Four subunits are encoded by SRB genes, first identified through a genetic screen for mutations suppressing the Pol II CTD truncation 11,12. The Head is essential for Mediator function since mutations in the Head module abolish mRNA synthesis in vivo 13,14 and in vitro 15, and eliminate Mediator interaction with promoters in vivo 10. The Head module is organized into three domains that can undergo significant conformational changes, and it interacts with the TATA-binding protein (TBP) subunit of general transcription factor TFIID and the Rpb4/Rpb7 Pol II subunits 16. The Head has also been shown to interact with TFIIH through the Med11 subunit 17. Determining the architecture of the Mediator Head module is therefore vital for insight into the mechanism by which Mediator controls gene expression.
We engineered the Head module to obtain the crystals of sufficient quality for structure determination (Supplementary Text 1). In our engineered Mediator Head, Med18 loop regions and the N-terminal 108 residues of Med17 were deleted, without apparent effect on the integrity of the complex (Supplementary Fig. 1). The modified Head module was labeled with seleno-methionine (SeMet), and purified as described16. Overcoming two major technical obstacles (Supplementary Text 2) resulted in SeMet crystals that diffract to 4.3 Å (Supplementary Table 1). The electron density map was calculated to 4.3 Å resolution (Supplementary Fig. 2) by SeMet Single Anomalous Dispersion (SAD) after inital phases had been obtained using Ta6Br14 and K3Ir(NO3)6 derivatives (Supplementary Text 3).
We started identification of the individual polypeptide constituents of the Mediator Head module by docking of the Med18-Med20-Med8C-terminal helix (CTH) complex structure (PDBID: 2HZS) 18 into the electron density map, followed by rigid-body refinement. The polypeptide chains of the other subunits were identified on the basis of the SeMet positions and their juxtaposition to large amino acid side chains within ordered regions of secondary structure (Methods). This approach permitted the unambiguous assignment of all discernible elements of secondary structure in the density map to individual Head module subunits (Figure 1b)(Supplementary Figs. 3-5).
Figure 1. Overall structure of the Mediator Head module.
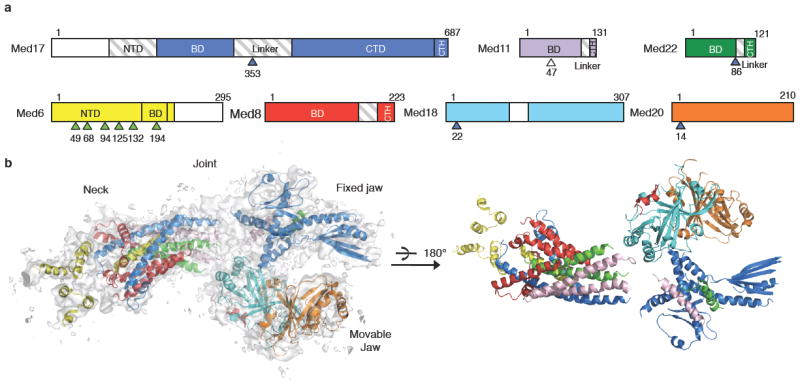
(a) Head module subunits domains. Med17 is shown in blue, Med11 in purple, Med22 in dark green, Med6 in yellow, Med8 in red, Med18 in cyan, and Med20 in orange. The region not modeled is shaded in gray, the region not present in the crystal is in white. Positions of med6 ts mutations are marked by arrows colored in green, srb suppressor mutations in blue, Med11 residue 47 (T) in white. NTD: N-terminal domain, BD: Bundle domain, CTD: C-terminal domain. (b) A ribbon model of the Mediator Head module is superimposed on the experimental electron density map contoured at 1.5 σ.
Our crystal structure is consistent with the 30-35 Å resolution molecular envelope of the Head module derived from single particle EM analysis (Supplementary Figs. 6, 7). Mediator Head can be described in terms of three major domains, a “Fixed jaw”, a “Movable jaw”, and a “Neck” (Figure 1b) (Supplementary Figs. 4, 5), with a “Central joint” between these domains. Our X-ray structure of the Head module reveals the overall architecture of the module and the domain boundaries. Each domain is connected though flexible loops and linkers at the Central joint (Figure 3c).
Figure 3. Structures of Fixed and Movable jaw domains.
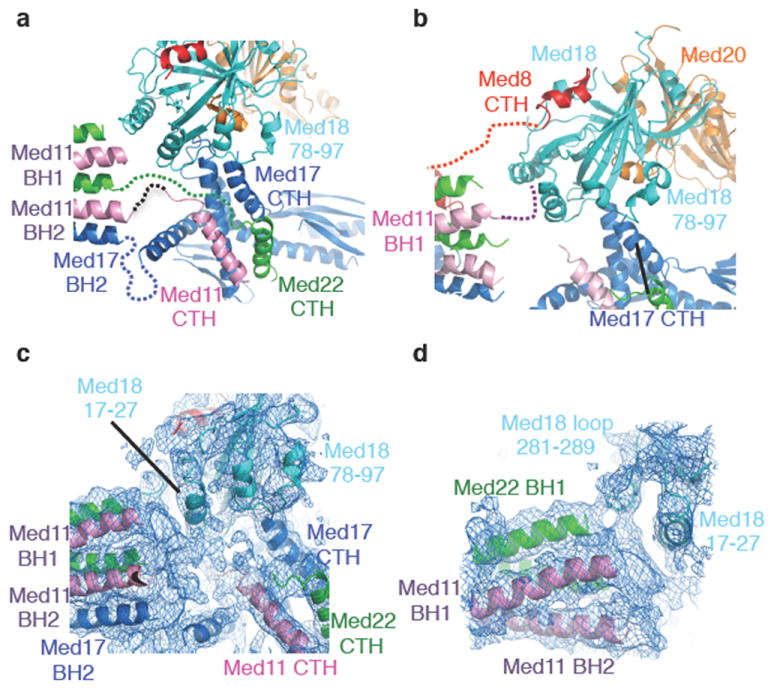
(a) Fixed jaw domain interactions. The linker region of Med17 (320-420), Med11 (94-110), and Med22 (87-100) are drawn as dotted lines. (b) Movable jaw domain interactions. Linker regions of Med8 and Med11 are drawn as dotted lines. (c) Electron density map at the central Joint region, displaying density corresponding to the linker regions of Med11, Med22, and Med17. (d) Electron density map at the junction of Med11, Med22, and Med18 subunits. The models of Med22 BH1, Med11 BH1, BH2 and Med18 loop (residues 17-27, 281-289) are superimposed.
Our previous work on expression and purification of the Head module pointed to a central role of Med17 in Head assembly 10. We extended this work through a comprehensive biochemical analysis combined with EM imaging for delineating the Med17 domain structure and elucidating its interactions with other Head components (Supplementary Text 4). The results support our model of the Mediator Head module architecture.
Assembly of the Head module starts with formation of the Mini-Head (Med17-Med11-Med22). Subsequently, Med8 and Med6 are added, followed by Med20-Med18 10. Our structure shows that a four helix-bundle, built by α-helices from Med11 (BH1, BH2), and Med22 (BH1, BH2) interact with BH2 of Med17 to form the larger helical bundle (Figures 2, 3)(Supplementary Fig. 4). This is consistent with the observation that omission of either Med11 or Med22 leads to disassembly of the Head 10. Med6 interacts with the Mini-Head through its single BH and Med8 serves to stabilize the central α-helical bundle by surrounding the central helices. Finally, the Med18-Med20 heterodimer binds to Core-Head primarily through the C-terminal helix (CTH) of Med8 (Figure 2).
Figure 2. Mechanism of Mediator Head module complex assembly.
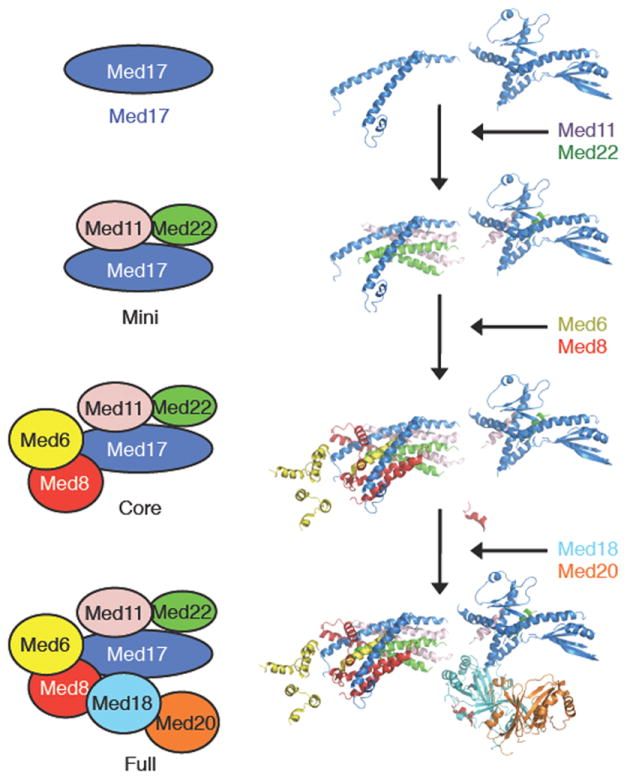
Models of the Mini (Med17, Med11 and Med22) and Core (Mini module with Med6 and Med8) Head module as derived from our crystal structure of the full Head module (Core module with Med18 and Med20) are depicted for comparison. Schematic diagrams of Head module components (panel left) and corresponding structures (right) are shown.
The Fixed jaw domain comprises the C-terminal helices (CTH) of Med11, Med22, and the C-terminal domain (CTD) of Med17. The Med11 CTH and Med22 CTH interact with the helical regions of the Med17 CTD. Med17 (residues 610-660) forms a β-sheet structure that lines the inner surface of the Fixed jaw and faces the Movable jaw (Figure 3a). The Med17 CTD interacts with the loop region of Med18. The functional importance of the Med17 CTD correlates with the biochemical activity of the Head module in vitro, as well as phenotypic analysis in vivo, as loss of the Med17 CTD abolishes the transcription activity of the Head module (Supplementary Fig. 12), and all Med17 CTD deletion mutants as well as internal deletion mutants result in lethality (Supplementary Fig. 13).
The Movable jaw is formed by the Med18-Med20-Med8 CTH complex. On the basis of previous EM studies 16, which demonstrated multiple orientations of this domain with respect to the rest of the Head module, we termed this domain the “Movable” jaw. Besides the interaction with the Med8 CTH 18, our complete Head module structure has revealed additional interactions with the Fixed jaw and Neck domains. First, the Med18 loop region formed by residues 78-97 interacts with the Med17 CTH of the Fixed jaw domain (Figure 3a, 3b). Second, the electron density corresponding to the N-terminus of the Med11 subunit (residues 1-20) indicates an interaction with Med18 (residues 17-27 and 281-289) (Figure 3c, 3d). The assignment of Med11 residues 1-20 was complicated by the substitution of Met17 to Ser17 (see Methods), and thus an unambiguous sequence marker is lacking. However, our biochemical data (Supplementary Text 5) support our architectural model in which a stable association of Med18-Med20 with the Head module requires binding to Med8 and at least one additional interaction (Med11 or Med17). The interactions with the CTH of Med17 and the NTD of Med11 are likely to be critical for the functional positioning and flexibility of the Movable jaw (Figure 3b-d)(Supplementary Fig. 6)16.
The Neck domain has an unusual, striking structure: a total of ten helices from five different subunits associate through the formation of a large helical bundle. The NTD of Med6 is located adjacent to the large helical bundle and consists of four α-helices (Figures 1b and 4a). The helical bundle of the Neck domain can be divided into two parts, a short bundle composed of 4 short α-helices, and a long bundle composed of 6 long α-helices. Three helices of the Med8 subunit (BH3, BH4, BH5) appear to stabilize the assembly of both short and long bundles, and thus, the entire Neck domain structure. TBP was reported to bind to the N-terminal 138 residues of Med8 18, which corresponds to BH1-BH5, all located on the surface of the Neck domain.
Figure 4. Structure of the Neck domain, and model of Head-Pol II-IIH.
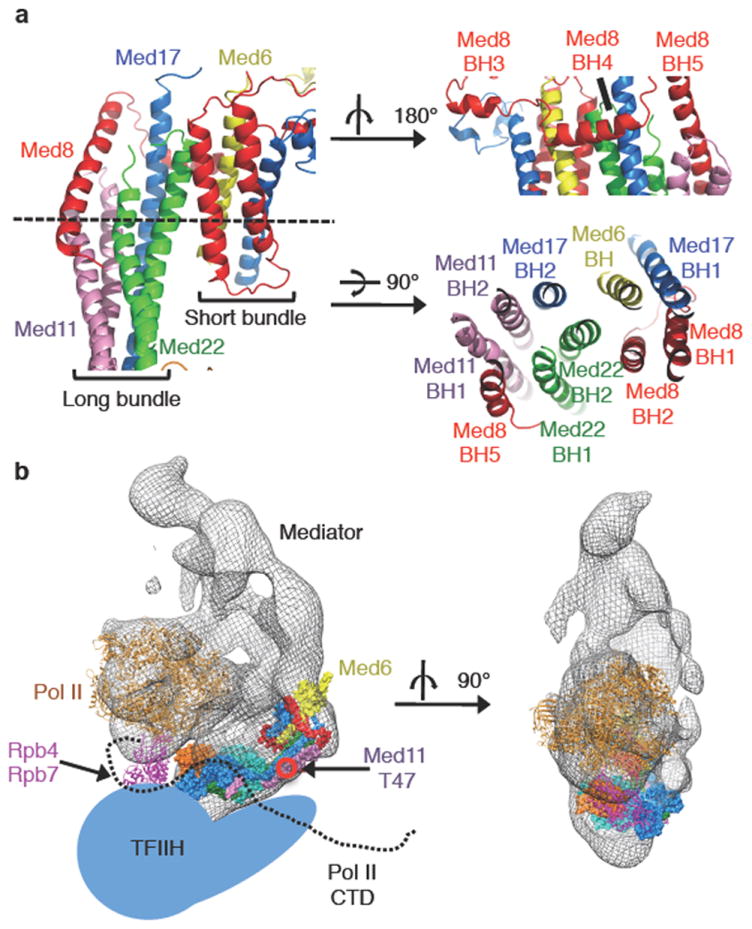
(a) The Neck domain is depicted from front, back (upper right), and top views (bottom right). (b) Model of Pol II-Mediator-TFIIH complex. Pol II and the Head structures were docked into the EM map of the Mediator-Pol II shown as a mesh 8. Head module is colored as in (Figure 1b). Core Pol II in brown, Rpb4/Rpb7 in purple, the Pol II CTD drawn as a black dotted line, TFIIH illustrated in a schematic representation (light blue). Location of Med11 residue 47 (T) is indicated.
The organization of the helical bundle in the Neck domain may produce a relatively rigid structure that could mechanically convey regulatory signals. Several observations suggest that Med6 may function as an interface between the Mediator Head and Middle modules, and transduce a mechanical signal from the Tail/Middle to the Head and onto Pol II (Supplementary Text 6).
Mediator stimulates the phosphorylation of Ser5 in the Pol II CTD by TFIIH 19, which promotes dissociation of Mediator from Pol II 20,21, an important step in the transition from initiation to elongation of transcription22. Our structural and biochemical data along with relevant previous observations12,17,23,24 suggests an interaction of the Pol II CTD, the Mediator Head module and TFIIH: First, mutation of Thr47 to Ala in Med11 affects the interaction of TFIIH with the Head module in vivo, resulting in a reduction of Pol II CTD Ser5 phosphorylation 17. Thr47 of Med11 is located near the center of the two symmetrical long helical bundles of the Neck, which thus could constitute the docking surface for TFIIH (Figure 4b)(Supplementary Fig. 16c). Second, three out of four suppressor mutations of Pol II CTD truncation, Med17 (Gly353 to Cys), Med22 (Asn86 to Lys), and Med18 (Thr22 to Ile), map to the Central joint region 12,24 (Supplementary Fig. 16a), suggesting a functional interaction between the CTD and this portion of the Head, consistent with previous observations 23. Third, the Head module within the Mediator-Pol II structure (Figure 4b) (Supplementary Fig. 17) is located near the base of the CTD. Finally, our biochemical data show that the Head module stimulates phosphorylation of the Pol II CTD by TFIIH (Supplementary Text 7)(Supplementary Fig. 18). Therefore, we suggest that the Head module may function as a scaffold that juxtaposes TFIIH and the Pol II CTD, thereby facilitating CTD phosphorylation (Figure 4b). Our Mediator Head structure reveals intricate interaction networks, notably the striking multi-helical bundle in the Neck domain, engaging five Mediator subunits in a single structure unit. Such interactions could not have been determined from structures of individual subunits alone, or from analyzing pairwise small domain-domain interactions, but only by study of the multi-protein complex in its entirety.
Methods
Construction of the vectors for complex expression, and for yeast genetics
All the vectors used in this study are summarized in Supplementary Tables 3 and 4. For expression of the modified Head module for crystallization, DNA sequences corresponding to residues 1-108 of Med17 were removed from vector pFL-10xHis-Med17 (pYT49) 16 by the SLIC method26, yielding pFL-10xHis-Med17 (109-687) (pYT171). DNA sequences corresponding to residues 109-140 and 71-156 of Med18, respectively, were removed from vector pSPL-Med18-Med20 (pYT75) 16, yielding pSPL-Med20-Med18 (Δ109-140) (pYT115), and pSPL-Med20-Med18 (Δ71-156) (pYT114). To eliminate an alternative translation start site, the methionine (residue 17) of Med11 was mutated to serine (pYT311). Finally, the transfer vector for the modified Head module was generated by fusing three vectors, pYT171, pYT114, and pUCDM-Med6-Med22-Med11-Med8 (pYT120) by Cre-LoxP recombination as described 25.
DNA sequences corresponding to residues 1-16 of Med11, were removed from the vector pYT111 by SLIC, yielding the vector pUCDM-Med22-Med11(Δ1-16) (pYT147). Fusion of pYT171, pYT147 and pYT120 with either pYT114 or, alternatively, pYT115 generated the expression vectors for a series of double Med18-Med11 partial Head module deletion mutants.
The constructs for Med17 mutagenesis were generated as follows. BamHI and HindIII fragments corresponding to the C-terminal deletion mutants of Med17 were generated by first introducing a stop codon and a HindIII site (TAAAAGCTT) into pBacPAK9-10His-SRB4 (MED17) vector 10 adjacent to the sequence corresponding to residues 108, 200, 300, 400, 500, and 600 of Med17, respectively, by using the QuickChange mutagenesis kit (Stratagene), followed by BamHI and HindIII digestion and gel purification. The purified fragments were cloned into the BamHI and HindIII sites of pFL vector 25, yielding vectors pYT165-170 (Supplementary Table 3). The N-terminal deletion, as well as the internal deletion mutant constructs of Med17, were generated by removing DNA sequences corresponding to residues 1-108, 1-201, 1-302, 1-400, 101-200, 201-300, and 301-400, respectively, from pFL-10xHis-Med17 by the SLIC method, yielding the vectors pYT183-186, and pYT289-291 (Supplementary Table 3). These vectors were fused with pUCDM-Med6-Med22-Med18-Med20-Med11-Med8 (pYT151), yielding vectors encoding for Head modules comprising Med17 mutant forms. The vector pYT151 was created by two rounds of sequential cloning of PmeI and the AvrII fragments containing Med18-Med20, and Med22-Med11 into SpeI and NruI sites of pUCDM-Med6-Med8 (pYT110).
Introduction of the deletion mutations into the yeast shuttle vector pCT127, carrying the wild-type MED17 gene, was also carried out by SLIC. The yeast shuttle vectors used in this study are listed at Supplementary Table 4.
Expression and purification of the Head module and mutants
Expression and purification of the recombinant Head module, the mutant forms and sub-complex was carried out in insect cells using the MultiBac system25. Production of high-titer viruses in Sf9 cells, and expression and purification of recombinant Head modules of Mediator, and its mutant forms were carried out as described previously16.
Preparation of SeMet labeled Head module will be described elsewhere (Imasaki et al, manuscript in preparation). Briefly, the insect cells were cultured in methionine-free medium (Expression Systems, Davis, CA) overnight prior to baculovirus infection. L-seleno-methione (20 mg/L) (Sigma-Aldrich) was added at sequential 24-hour intervals. Cells were harvested 96 hours after infection. SeMet labeled complex was purified as described above.
Limited proteolysis and identification of the peptides fragments
A total of 135 μg of the recombinant Head module was incubated at 37 °C with Chymotrypsin (Sigma-Aldrich) at a final concentration of 0.01 mg/ml in a volume of 150 μl in Buffer A (20 mM Tris-HCl (pH 8.0), 100 mM NaCl, and 1 mM DTT). 20 μl aliquots were taken at 0, 5, 10, 30 and 60 min, and 15 μl of PMSF stock solution (100 mg/ml) was added to stop the reaction by inhibiting the protease. Aliquots were applied to 12.5 % SDS-PAGE, and transferred onto a Sequi-Blot™ PVDF membrane (Bio-Rad). Protein bands were stained by Coomassie Brilliant Blue (R-250). Protein bands resulting from proteolysis during the time course were identified, excised, and subjected to Edman degradation using a Procise 494 instrument from Applied Biosystems (AB) as described27. Stepwise-liberated PTH-amino acids were identified using an “on-line” HPLC system (AB) equipped with a PTH C18 (2.1×220 mm; 5 micron particle size) column (AB).
Crystallization and data collection
Crystals were obtained at 293 K by hanging-drop vapor diffusion against a reservoir solution of 0.1 M Tris-HCl (pH 7.6) containing 10-12.5% (w/v) PEG-6K, and 0.4 M (NH4)2SO4. Crystals were transferred into the reservoir solution containing 25% triethylene glycol (TEG). The crystals were flash frozen for data collection at 100 K. SDS-PAGE analysis of the dissolved crystals confirmed the presence of all 7 subunits. However, in situ proteolysis resulted in about 10% of the Med17 subunits being shortened at the N-terminus by 76 residues and almost 100% of the Med6 subunits shortened at the C-terminus by 80 residues (Supplementary Fig. 1). Diffraction data were collected at beamline 23ID at the Advanced Photon Source (APS) at Argonne National Laboratory. All diffraction data were processed with HKL2000 28. Twinning rates of the datasets were analyzed by using program Phenix Xtriage29.
Structure determination of the Mediator Head module
Initial phases were determined by two approaches: (i) Ta6Br14 Single Isomorphous Replacement with Anomalous Scattering (SIRAS) and (ii) Iridium Single Anomalous Dispersion (SAD). Ta6Br14 derivative crystals were prepared by soaking the native Head module crystals into reservoir solution containing 1 mM Ta6Br14. Initial phase was determined by SIRAS at 7.5 Å resolution. Density modification by the program Parrot extended the phase to 4.3 Å using the SeMet dataset. Iridium (Ir) derivatives of the crystal of Mediator Head module were prepared by soaking the crystals into crystallization reservoir solution containing 10 mM of K3Ir(NO3)6. Initial Ir phase was obtained by SAD using the programs, ShelxD and Phaser30,31. The phase was extended followed by density modification by program Parrot 32 with the SeMet dataset. However, the maps obtained at this stage were not yet interpretable.
To improve the maps, we used these maps together and applied the following methods: (i) locating SeMet sites in the crystal (ii) non-crystallographic symmetry (NCS) averaging between three molecules in the NCS by program DM 33, (iii) partial model building into the clearly discernible rod-like electron density from α-helices followed by rigid body refinement by using the programs COOT and Refmac534, and (iv) re-calculating phases by model and SeMet SAD phasing with program Phaser, utilizing the partial model and SeMet positions. Iterative rounds combining these procedures were performed until the model covered all interpretable secondary structure elements. Eventually, we could identify 98 SeMet sites. To minimize model bias, phases were re-calculated by SeMet SAD with program Phaser, using only the positions of these 98 selenium sites, and these improved SAD phases guided the final model building steps.
Model building and refinement
Assignment of polypeptide identities was carried out as follows: The published structure, Med18-Med20-Med8 (CTH) (PDBID: 2HZS) 18 was manually docked into the electron density map followed by rigid body refined by using the program COOT. Then the α-helices for the further polypeptides of Head module were manually built, and connected. Next, β sheets were manually built into the unassigned structured regions in the electron density, which corresponded to the Neck and Fixed jaw domains. Subsequently, we began assigning the polypeptide identities at the Neck domain. We tracked specific SeMet labeling patterns dictated by the presence of Met in the primary sequence of the polypeptides, and the presence of bulky regions corresponding to aromatic residue positions, as markers. We also utilized secondary structure predictions for additional guidance. First, Med8 BH1, Med17 BH1, and Med22 BH2 were found in α-helical bundle region in the Neck domain based on their primary-sequence specific, unique SeMet labeling pattern: these regions all contain more than two SeMet peaks and the spacing of SeMet peaks were consistent with the corresponding amino acid sequences in the subunits. This assignment was consistent with the secondary structure predictions indicating α-helical structure. The remaining Med8 residues 60-170, as well as Med22 BH1, were easily assigned by tracing from the Med8 BH1 helix back to the Med8 C-terminus, and the Med22 BH2 helix. This assignment was validated by the fact that their Met locations nicely aligned to anomalous peaks on the experimental map. Next, we identified the NTD of Med6 based on its unique SeMet positions, and also identified Med6 BH based on a specific location of SeMet (Met48), a bulky aromatic ring (Phe 52) (Supplementary Fig. 19a), and continuity from the NTD of Med6, consistent with secondary structure predictions. The C-terminal 80 residues of Med6 were proteolyzed in the crystals (Supplementary Fig.1). Consequently, no density was found corresponding to the C-terminus of Med6. We traced Med17 BH1, and identified the longest helix in the Neck domain as the BH2 helix of the Med17 based on a single SeMet (Met 313) and the aromatic side chain of Tyr 269 (Supplementary Fig. 19b); this assignment was consistent with secondary structure predictions. Finally, the two remaining continuous α-helices in the Neck domain were identified as Med11 BH1 and Med11 BH2 because of one unique SeMet position of the Med11. This assignment matches perfectly to the secondary structure prediction as well.
Next, we focused on the Fixed jaw domain. By subtracting the polypeptides already assigned to the Neck and Movable jaw (see above), the Fixed jaw should only contain the C-terminal regions of subunits Med11, Med17 and Med22. First, based on continuity, SeMet position (Met 422), aromatic ring position (Tyr 423) (Supplementary Fig. 19c), and secondary structure prediction, helix 420-455, β-sheet 456-480, helix 496-523 and 540-570 of Med17were identified. The remainder of the electron density in this region was continuous, and thus enabled us to trace Med17 completely to its C-terminus. We identified Med17 CTH and β-sheet with α-helix 600-608, based on the SeMet positions and α-helix length from secondary structure prediction. Finally, we assigned two remaining helices: Med11 CTH was identified based on the presence of one SeMet peak, and we assigned the last helix to Med22 CTH, which entirely lacks SeMet.
Initially, all models were refined by program CNS DEN35, refinement with strong NCS restraints between the three independent complexes in the asymmetric unit and twinning refinement. Then, the model was refined by using program Phenix with NCS restraints with single refined group isotropic temperature factor for each subunit, Ramachandran restraint, TLS refinement, and twinning refinement. The final model exhibits good geometry with 91.3 %, 8.0 %, 0.7 % of the amino acid residues in the most favored, allowed, and disallowed regions of the Ramachandran plot, respectively. All structural illustrations and electron density maps were prepared with PYMOL (www.pymol.org) and COOT. PSIPRED was used for secondary structure prediction36.
Docking of the X-ray structure into the EM map
The model of 12-subunit Pol II was docked into Mediator-RNA polymerase II holoenzyme structure 8 followed by docking the X-ray model of the Head module into the density corresponding to the Mediator Head module, by using the program Chimera 37.
EM sample preparation, data collection, and image analysis
We diluted purified Head module deletion mutants in buffer containing 25 mM KCl, 25 mM Tris-HCl (pH 7.8), and 5 mM DTT. For preparation of all EM samples, about 3μl of protein solution were applied to a carbon-coated Maxtaform, 300-mesh Cu/Rh EM specimen grid (Ted Pella, Inc., Redding, CA) freshly glow discharged in the presence of amyl amine. The particles were then preserved by staining with a 2.0% (w/w) uranyl acetate solution using the sandwich carbon layer technique 38,39The images were recorded under low-dose conditions using a Tecnai Spirit (Philips/FEI) microscope equipped with a LaB6 filament and operating at an accelerating voltage of 120 kV. Images were recorded on a Tietz (TVIPS GmbH) CCD camera at 42,000X magnification and approximately 1 μm underfocus, resulting in a final pixel size corresponding to 5.06 Å.
The images were initially analyzed using the ml_align2d program, a multi-reference 2D alignment routine with a maximum-likelihood target function 40 implemented in the Xmipp package 41. Averages derived from the ml_align2d program were utilized to run iterative alternating rounds of supervised multi-reference alignment/classification and reference-free alignment as described 42 to improve the homogeneity of the image classes.
In vitro transcription and the C-terminal domain (CTD) phosphorylation assays
In vitro transcription assay to assess activity of the recombinant Head module and its mutant form using srb4ts mutant crude extract was described previously 10. Quantification of transcripts on an absolute scale was performed using a FLA-5100 FUJIFILM fluorescent image analyzer and the MultiGauge software package after addition of 1 nCi of-32P UTP to the gel 5 min before the end of the run. The CTD phosphorylation assay was performed as described 15.
Yeast phenotypic analysis
The shuttle vectors carrying the MED17 mutations are described in Supplementary Table 4. The shuttle vectors were introduced into yeast strain Z572 by plasmid shuffling, and grown on SC medium containing 5-FOA at 30 °C as described 15.
Supplementary Material
Acknowledgments
We thank Lou Messerle for Ta6Br14 metal cluster, Tom Hurley for critical reading of the manuscript, Millie Georgiadis for discussions, Craig Kaplan for his advice for yeast genetics, Thomas Earnest for sharing the beamtime, and Laura Fabrizio for assisting the N-terminal sequence analysis. We thank the CCP4 summer school funded by NCI (Y1-CO-1020), and NIGMS (Y1-GM-1104) for their assistance with the twinning data analysis. Y.T. thanks the instructors at “The X-Ray Methods Course” at Cold Spring Harbor Laboratory. This work was supported by US National Science Foundation grant MCB 0843026 (Y.T.), an American Heart Association 0735395N (Y.T.) and a Human Frontier Science Program long-term fellowship (T.I.), NIH grants R01 GM67167 (to F.J.A.), GM36659 (to R.D.K) NCI Cancer Center Support Grant P30 CA08748 (to MSKCC Microchemistry and Proteomics Core Laboratory), and the European Commission Framework Program 7 projects INSTRUCT and P-CUBE (to I.B.). X-ray data were collected at the GM/CA-CAT at the Advanced Photon Source (APS), Argonne National Laboratory, Argonne, IL. GM/CA-CAT is funded by NIH (Y1-CO-1020, Y1-GM-1104), and APS was supported by DOE (DE-AC02-06CH11357). Portions of this research were carried out at the Stanford Synchrotron Radiation Lightsource (SSRL) supported by DOE, and NIH, and The Advanced Light Source (ALS) supported by DOE (DE-AC02-05CH11231).
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature/nature.
Author Contributions T.I., I.B., and Y.T. implemented the MultiBac system; T.I. was mainly responsible for protein complex preparation, crystallization, data collection, data analysis and model building in collaboration with Y.T.; T.I., H.E-B., and P.T. carried out mass spectroscopy analysis; G.Calero, G.L.K. and Y.T. carried out the initial crystallization and data collection supervised by R.D.K.; Y.T., T.I. and F.C. designed and carried out expression of the mutant Head modules and their biochemical characterization; Y.T. and K. Y. designed and carried out yeast genetic experiment; Y.T. carried out in vitro transcription assay and the CTD kinase assay; G. Cai, K-L.T. and F.A. carried out EM imaging study on the Head module and its mutants; T.I., F.J.A. and Y.T. discussed and interpreted all results; Y.T. supervised the X-ray, biochemical and yeast genetic work, and wrote the manuscript in collaboration with T.I., I.B., F.J.A., and R.D.K.
Coordinates and structure factors have been deposited at the Protein Data Bank under accession code 3RJ1.
The authors declare no competing financial interests.
References
- 1.Kornberg RD. Mediator and the mechanism of transcriptional activation. Trends Biochem Sci. 2005;30:235–239. doi: 10.1016/j.tibs.2005.03.011. [DOI] [PubMed] [Google Scholar]
- 2.Conaway RC, Sato S, Tomomori-Sato C, Yao T, Conaway JW. The mammalian Mediator complex and its role in transcriptional regulation. Trends Biochem Sci. 2005;30:250–255. doi: 10.1016/j.tibs.2005.03.002. [DOI] [PubMed] [Google Scholar]
- 3.Boube M, Joulia L, Cribbs DL, Bourbon HM. Evidence for a mediator of RNA polymerase II transcriptional regulation conserved from yeast to man. Cell. 2002;110:143–151. doi: 10.1016/s0092-8674(02)00830-9. [DOI] [PubMed] [Google Scholar]
- 4.Malik S, Roeder RG. The metazoan Mediator co-activator complex as an integrative hub for transcriptional regulation. Nat Rev Genet. 2010;11:761–772. doi: 10.1038/nrg2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bjorklund S, Gustafsson CM. The yeast Mediator complex and its regulation. Trends Biochem Sci. 2005;30:240–244. doi: 10.1016/j.tibs.2005.03.008. [DOI] [PubMed] [Google Scholar]
- 6.Guglielmi B, et al. A high resolution protein interaction map of the yeast Mediator complex. Nucleic Acids Res. 2004;32:5379–5391. doi: 10.1093/nar/gkh878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Asturias FJ, Jiang YW, Myers LC, Gustafsson CM, Kornberg RD. Conserved structures of mediator and RNA polymerase II holoenzyme. Science. 1999;283:985–987. doi: 10.1126/science.283.5404.985. [DOI] [PubMed] [Google Scholar]
- 8.Davis JA, Takagi Y, Kornberg RD, Asturias FA. Structure of the yeast RNA polymerase II holoenzyme: Mediator conformation and polymerase interaction. Mol Cell. 2002;10:409–415. doi: 10.1016/s1097-2765(02)00598-1. [DOI] [PubMed] [Google Scholar]
- 9.Cai G, Imasaki T, Takagi Y, Asturias FJ. Mediator structural conservation and implications for the regulation mechanism. Structure. 2009;17:559–567. doi: 10.1016/j.str.2009.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Takagi Y, et al. Head module control of mediator interactions. Mol Cell. 2006;23:355–364. doi: 10.1016/j.molcel.2006.06.007. [DOI] [PubMed] [Google Scholar]
- 11.Nonet ML, Young RA. Intragenic and extragenic suppressors of mutations in the heptapeptide repeat domain of Saccharomyces cerevisiae RNA polymerase II. Genetics. 1989;123:715–724. doi: 10.1093/genetics/123.4.715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thompson CM, Koleske AJ, Chao DM, Young RA. A multisubunit complex associated with the RNA polymerase II CTD and TATA-binding protein in yeast. Cell. 1993;73:1361–1375. doi: 10.1016/0092-8674(93)90362-t. [DOI] [PubMed] [Google Scholar]
- 13.Thompson CM, Young RA. General requirement for RNA polymerase II holoenzymes in vivo. Proc Natl Acad Sci USA. 1995;92:4587–4590. doi: 10.1073/pnas.92.10.4587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Holstege FC, et al. Dissecting the regulatory circuitry of a eukaryotic genome. Cell. 1998;95:717–728. doi: 10.1016/s0092-8674(00)81641-4. [DOI] [PubMed] [Google Scholar]
- 15.Takagi Y, Kornberg RD. Mediator as a general transcription factor. J Biol Chem. 2006;281:80–89. doi: 10.1074/jbc.M508253200. [DOI] [PubMed] [Google Scholar]
- 16.Cai G, et al. Mediator Head module structure and functional interactions. Nat Struct Mol Biol. 2010;17:273–279. doi: 10.1038/nsmb.1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Esnault C, et al. Mediator-dependent recruitment of TFIIH modules in preinitiation complex. Mol Cell. 2008;31:337–346. doi: 10.1016/j.molcel.2008.06.021. [DOI] [PubMed] [Google Scholar]
- 18.Lariviere L, et al. Structure and TBP binding of the Mediator head subcomplex Med8-Med18-Med20. Nat Struct Mol Biol. 2006;13:895–901. doi: 10.1038/nsmb1143. [DOI] [PubMed] [Google Scholar]
- 19.Kim YJ, Bjorklund S, Li Y, Sayre MH, Kornberg RD. A multiprotein mediator of transcriptional activation and its interaction with the C-terminal repeat domain of RNA polymerase II. Cell. 1994;77:599–608. doi: 10.1016/0092-8674(94)90221-6. [DOI] [PubMed] [Google Scholar]
- 20.Svejstrup JQ, et al. Evidence for a mediator cycle at the initiation of transcription. Proc Natl Acad Sci USA. 1997;94:6075–6078. doi: 10.1073/pnas.94.12.6075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Max T, Sogaard M, Svejstrup JQ. Hyperphosphorylation of the C-terminal repeat domain of RNA polymerase II facilitates dissociation of its complex with mediator. J Biol Chem. 2007;282:14113–14120. doi: 10.1074/jbc.M701345200. [DOI] [PubMed] [Google Scholar]
- 22.Payne JM, Laybourn PJ, Dahmus ME. The transition of RNA polymerase II from initiation to elongation is associated with phosphorylation of the carboxyl-terminal domain of subunit IIa. J Biol Chem. 1989;264:19621–19629. [PubMed] [Google Scholar]
- 23.Kang JS, et al. The structural and functional organization of the yeast mediator complex. J Biol Chem. 2001;276:42003–42010. doi: 10.1074/jbc.M105961200. [DOI] [PubMed] [Google Scholar]
- 24.Koleske AJ, Buratowski S, Nonet M, Young RA. A novel transcription factor reveals a functional link between the RNA polymerase II CTD and TFIID. Cell. 1992;69:883–894. doi: 10.1016/0092-8674(92)90298-q. [DOI] [PubMed] [Google Scholar]
- 25.Fitzgerald DJ, et al. Protein complex expression by using multigene baculoviral vectors. Nat Methods. 2006;3:1021–1032. doi: 10.1038/nmeth983. [DOI] [PubMed] [Google Scholar]
- 26.Li MZ, Elledge SJ. Harnessing homologous recombination in vitro to generate recombinant DNA via SLIC. Nat Methods. 2007;4:251–256. doi: 10.1038/nmeth1010. [DOI] [PubMed] [Google Scholar]
- 27.Tempst P, Geromanos S, Elicone C, Erdjument-Bromage H. Improvements in Microsequencer Performance for Low Picomole Sequence Analysis. Methods. 1994;6:248–261. doi: 10.1006/meth.1994.1027. [DOI] [Google Scholar]
- 28.Otwinowski Z, Minor W. Processing of X-ray Diffraction Data Collected in Oscillation Mode. Methods in Enzymology. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 29.Adams P, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sheldrick G. A short history of SHELX. Acta Crystallogr A. 2008;64:112–122. doi: 10.1107/S0108767307043930. [DOI] [PubMed] [Google Scholar]
- 31.McCoy A, et al. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cowtan K. Recent developments in classical density modification. Acta Crystallogr D Biol Crystallogr. 2010;66:470–478. doi: 10.1107/S090744490903947X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cowtan K. Modified phased translation functions and their application to molecular-fragment location. Acta Crystallogr D Biol Crystallogr. 1998;54:750–756. doi: 10.1107/s0907444997016247. [DOI] [PubMed] [Google Scholar]
- 34.Emsley P, Lohkamp B, Scott W, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schroder GF, Levitt M, Brunger AT. Super-resolution biomolecular crystallography with low-resolution data. Nature. 2010;464:1218–1222. doi: 10.1038/nature08892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bryson K, et al. Protein structure prediction servers at University College London. Nucleic Acids Res. 2005;33:W36–38. doi: 10.1093/nar/gki410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pettersen E, et al. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 38.Stoffler G, Stoffler-Meilicke M. The ultrastructure of macromolecular complexes studied with antibodies. De Gruyter; 1983. pp. 409–455. [Google Scholar]
- 39.Tischendorf GW, Zeichhardt H, Stoffler G. Determination of the location of proteins L14, L17, L18, L19, L22, L23 on the surface of the 5oS ribosomal subunit of Escherichia coli by immune electron microscopy. Mol Gen Genet. 1974;134:187–208. doi: 10.1007/BF00267715. [DOI] [PubMed] [Google Scholar]
- 40.Scheres SH, et al. Maximum-likelihood multi-reference refinement for electron microscopy images. J Mol Biol. 2005;348:139–149. doi: 10.1016/j.jmb.2005.02.031. [DOI] [PubMed] [Google Scholar]
- 41.Sorzano CO, et al. XMIPP: a new generation of an open-source image processing package for electron microscopy. J Struct Biol. 2004;148:194–204. doi: 10.1016/j.jsb.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 42.Brignole EJ, Smith S, Asturias FJ. Conformational flexibility of metazoan fatty acid synthase enables catalysis. Nat Struct Mol Biol. 2009;16:190–197. doi: 10.1038/nsmb.1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


