Summary
During meiosis in C. elegans, unpaired chromosomes and chromosomal regions accumulate high levels of histone H3 lysine 9 dimethylation (H3K9me2), a modification associated with facultative heterochromatin assembly and the resulting transcriptional silencing [1, 2]. Meiotic silencing of unpaired DNA may be a widely conserved genome defense mechanism [3–5]. The mechanisms of meiotic silencing remain unclear, although both transcriptional and posttranscriptional processes are implicated [3–5]. Cellular RNA-dependent RNA polymerases (RdRPs) function in development and RNA-mediated silencing in many species [3, 6, 7] and in heterochromatin assembly in S. pombe [3, 8]. There are four C. elegans RdRPs, including two with known germline functions. EGO-1 is required for fertility and robust germline RNAi [9–11]. RRF-3 acts genetically to repress RNAi and is required for normal meiosis and spermatogenesis at elevated temperatures [12] (S. L’Hernault, personal communication). Among C. elegans RdRPs, we find that only EGO-1 is required for H3K9me2 enrichment on unpaired chromosomal regions during meiosis. This H3K9me2 enrichment does not require Dicer or Drosha nuclease or any of several other proteins required for RNAi. ego-1 interacts genetically with him-17, another regulator of chromatin and meiosis [13], to promote germ-line development. We conclude that EGO-1 is an essential component of meiotic silencing in C. elegans.
Results and Discussion
EGO-1 and RRF-3 Activity Regulate Meiotic H3K9me2 Accumulation
Since RdRP activity is required for meiosis and meiotic silencing in N. crassa and for heterochromatin assembly in S. pombe, we investigated the effect of RdRP mutations on meiotic heterochromatin assembly in C. elegans using anti-H3K9me2 antibodies. (Details of strains used throughout this report are described in the Supplemental Experimental Procedures.) We first investigated ego-1 mutants. We evaluated three types of unpaired chromosomes that become enriched for H3K9me2 during meiosis [1, 2]: (1) the single male X chromosome, (2) poorly paired X chromosomes that occur in him-8 (high incidence of males) mutants, and (3) an unpaired autosomal fragment, sDp3. Each type of unpaired DNA became enriched in H3K9me2 during meiosis in wild-type (ego-1(+)) animals and ego-1(0)/+ heterozygotes, but failed to do so in ego-1(0) null mutants (Figures 1A–1C). H3K9me2 accumulation was also defective in males carrying an ego-1 point mutation in a conserved residue in the RdRP catalytic domain in trans to a null allele (designated ego-1(RdRP/null)) (data not shown).
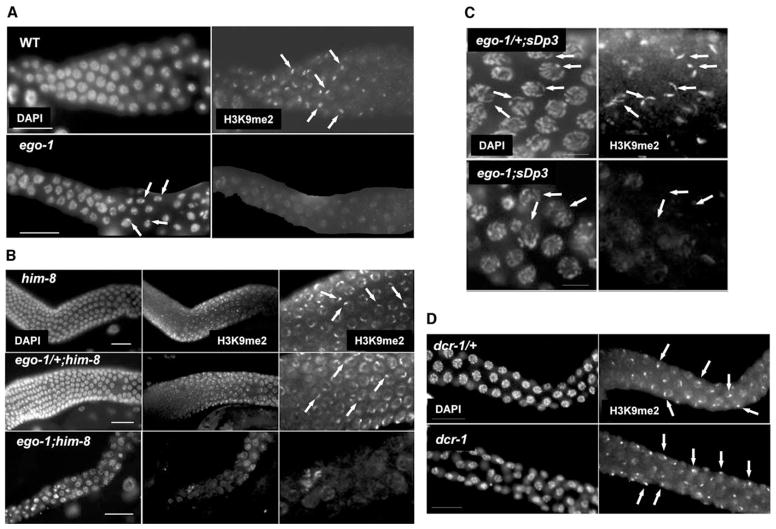
Figure 1. EGO-1 Activity Is Required for H3K9me2 Accumulation on Unpaired Chromosomes and Chromosomal Fragments during Meiosis.
Each panel shows meiotic nuclei stained with DAPI to visualize DNA or polyclonal anti-H3K9me2 antibody. ego-1 null alleles are used in all cases (see Supplemental Experimental Procedures). Photographs are oriented with distal tissue to the left.
(A) Wild-type (wt) male pachytene cells contain a bright focus of H3K9me2 staining associated with the condensed X chromosome. Weaker staining is visible on autosomes. H3K9me2 staining rapidly decreases as cells exit pachytene and start to condense as primary spermatocytes (also see Figure S1). In ego-1 mutant males, the condensed X chromosome is distinct in DAPI-stained preparations but fails to accumulate a bright focus of H3K9me2 staining (arrows). This result is consistent with previous work showing that condensation of the male X can occur in the absence of H3K9me2 enrichment [1]. Weak H3K9me2 staining is visible on all chromosomes. We note that occasional ego-1 nuclei had one or more brighter foci of staining that did not seem to localize to the X, suggesting that EGO-1 activity may also prevent inappropriate H3K9me2 accumulation on autosomes.
(B) him-8 and ego-1/+;him-8 pachytene cells have two bright foci of H3K9me2 staining, corresponding to the unpaired regions of the X chromosomes (arrows). In some nuclei, these regions are not adjacent. Paired autosomes have weaker staining, as in wild-type. In contrast, the bright foci of H3K9me2 staining are not evident in ego-1;him-8 pachytene cells.
(C) ego-1/+;sDp3 animals have 1–2 bright foci of H3K9me2 staining in pachytene cells (arrows). Two foci result when partial pairing occurs between the left end of sDp3 and the left end of an intact chromosome III, leaving a portion of sDp3 and a portion of the other chromosome III unpaired. In contrast, ego-1;sDp3 pachytene cells lack the bright H3K9me2 foci on both sDp3 and the unpaired region of the intact chromosome III.
(D) Both dcr-1/+ and dcr-1 males have a bright focus of H3K9me2 staining in pachytene cells that corresponds to the tightly condensed X chromosome (arrows). Weaker staining is associated with autosomes.
In wild-type hermaphrodites, H3K9me2 staining is briefly elevated on autosomes in mid-to-late pachytene and diplotene stages. This transient accumulation is excluded from paired X chromosomes, which undergo activation during oogenesis [1, 2]. In ego-1(0) hermaphrodites, we still observed a low level of H3K9me2 accumulation on autosomes (Figure 1B; data not shown). We conclude that EGO-1 activity is required for H3K9me2 enrichment on unpaired chromosomal regions during pachytene stage of meiosis, but not for accumulation on autosomes in later stages.
rrf-3(0) mutants have a temperature-sensitive germ-line phenotype [12]; therefore, we assayed H3K9me2 targeting in rrf-3(0) mutants raised at restrictive (25°C) temperature. We consistently observed H3K9me2 targeting to the male X in these animals (see Figure S1 in the Supplemental Data available with this article online). Strikingly, H3K9me2 staining persisted in rrf-3(0) mutants substantially longer than in wild-type, remaining strong in primary spermatocytes (Figure S1). H3K9me2 is normally depleted from chromatin by the time primary spermatocyte nuclei begin to visibly condense and before other modifications are extensively depleted from the autosomes (Figure 1A; Figure S1; [2]). The temperature-sensitive fertility defects in the rrf-3(0) mutants may correlate with the aberrant chromatin regulation that we observe in these animals. H3K9me2 accumulation during meiosis was normal in rrf-1(0) and rrf-2(0) males (data not shown).
Regulation of Repetitive DNA Arrays in the ego-1 Mutant Germline
C. elegans transgenes are often generated as multicopy, extrachromosomal arrays [14]. Expression of such trans-genes is typically detected in the soma, but not in the germline [2, 15]. Germline-silenced arrays accumulate a high density of H3K9me2 during pachytene stage (similar to the male X) and do not accumulate “active” chromatin modifications, e.g., H3 methylated on lysine 4 (H3K4me) [2]. We investigated whether EGO-1 is required for H3K9me2 accumulation on a repetitive extrachromosomal array, ccEx7291, and a repetitive array fused to a chromosomal fragment, svDp1. Strong H3K9me2 staining of both arrays was seen in wild-type and ego-1(0)/+ animals, but not in ego-1(0) animals (Figure 2A). Therefore, EGO-1 activity is critical for targeting H3K9me2 to unsynapsed, repetitive arrays. We note that these transgenes are not likely to be fully paired with any partner during meiosis.
Figure 2. H3K9me2 Accumulation on Extrachromosomal, Multicopy Transgenes Is Defective in ego-1 but Not in dcr-1 Mutants.
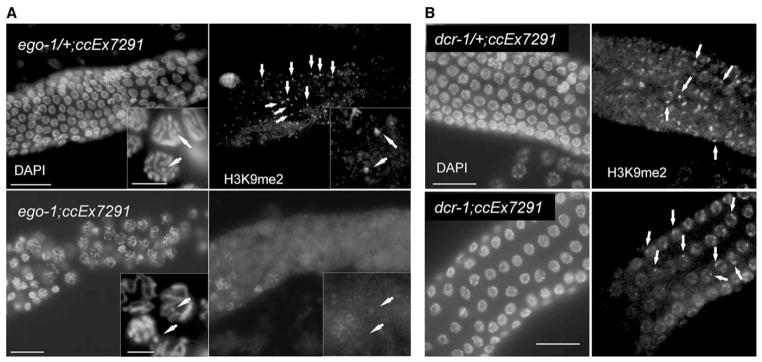
Each panel shows a portion of the meiotic germline from a strain carrying the repetitive extrachromosomal array, ex7291.
(A) ex7291 is visible in DAPI-stained preparations of both ego-1/+ and ego-1 tissue (see insets; arrows indicate extrachromosomal array). A bright focus of H3K9me2 staining, corresponding to ex7291, is visible in ego-1/+ (arrows) but not ego-1 tissue.
(B) A bright focus of H3K9me2 staining, corresponding to ex7291, is visible in both dcr-1/+ and dcr-1 tissue. Scale bar equals 20 μm; inset scale bar equals 5 μm.
ccEx7291 carries a GFP reporter, let-858::gfp. let-858:: gfp has been used to detect genes that participate in germline silencing, including components of both transcriptional and posttranscriptional mechanisms (TGS and PTGS, respectively) [15–18]. Although ccEx7291 did not accumulate H3K9me2 in the ego-1(0) mutant, we did not detect germline expression of LET-858::GFP (data not shown). We tested other GFP-tagged transgenes and also found that they remained silenced in ego-1(0) germ cells (data not shown).
Our findings may reflect the regulation of multicopy arrays by multiple mechanisms. We examined H3K9me2 accumulation on ccEx7291 in rde-2 (RNAi defective) mutants, which are defective in germline PTGS and transgene silencing [19]. Although LET-858::GFP was expressed (at a low level), ccEx7291 accumulated normal H3K9me2 levels (Figure S2). H3K9me2 accumulation is likely just one of several mechanisms that limit expression of repetitive DNA in the germline. Weak germline expression of extrachromosomal arrays is seen in certain PTGS mutants, including dcr-1 and rde-2, and also in mes mutants, which are defective in regulation of the silencing mark, H3K27me [15, 19–23; this report]. We may fail to see accumulation of LET-858::GFP in ego-1 mutant germlines because these other repressive mechanisms remain active.
H3K9me2 Accumulation in dcr-1/Dicer and drsh-1/Drosha Mutants
Dicer endonuclease functions in heterochromatin assembly in many species and may act in the biogenesis of short RNAs that recruit the chromatin-modifying machinery to target loci [3, 8, 24, 25]. We investigated whether C. elegans DCR-1/Dicer activity is required for H3K9me2 accumulation on unpaired DNA during meiosis. H3K9me2 accumulation appeared normal on the X chromosome in dcr-1(0) adult males, on partially paired X chromosomes in dcr-1(0);him-8 adult hermaphrodites, and on autosomes in late pachytene-diplotene stage in dcr-1(0) hermaphrodites (Figure 1D; data not shown). Moreover, H3K9me2 accumulated normally on ccEx7291 in a dcr-1(0) background (Figure 2B). Therefore, DCR-1 does not seem to be required for H3K9me2 accumulation during meiosis. One caveat is that these dcr-1(0) mutants were derived from dcr-1(0)/+ heterozygous mothers and therefore had inherited maternal product. Embryos lacking both maternal and zygotic DCR-1 arrest during early development and cannot be assayed for defects in germline chromatin (data not shown).
We evaluated the importance of another RNase III-type nuclease, Drosha [26]. H3K9me2 staining was normal in drsh-1(0) males and drsh-1(0);him-8 hermaphrodites derived from drsh-1/+ mothers, and in animals where maternal and zygotic Drosha was knocked down by RNAi (data not shown). Therefore, Drosha activity does not appear to be to be required for meiotic H3K9me2 accumulation.
We analyzed H3K9me2 accumulation in males carrying mutations in several other RNAi pathway components. RDE-1 (an Argonaute/Piwi/PAZ protein), RDE-2 (a novel protein), and RDE-4 (a dsRNA binding protein) are required for RNAi upon introduction of exogenous dsRNA [19, 27]. RDE-2 is also required for normal germ-line development under conditions of stress [19, 28]. We observed normal H3K9me2 staining in male rde-1, rde-2, and rde-4 mutants raised at both normal and elevated temperatures (data not shown). Hence, proteins required for RNAi per se are not required for the normal pattern of H3K9me2 accumulation during meiosis. C. elegans contains another 26 predicted Piwi/PAZ proteins (see Wormbase, http://www.wormbase.org/; [19]), several of which are known to function in RNAi-related phenomena. We observed normal H3K9me2 staining in alg-1 (Argonaute-like gene required for microRNA function) [29] mutant males and upon RNAi knockdown of several other Piwi/PAZ candidates assayed thus far (data not shown). Systematic analysis of single and multiple Piwi/PAZ mutants is required to fully evaluate whether this protein family participates in meiotic heterochromatin assembly. Interestingly, RRF-1, DCR-1, and several RDE proteins have recently been reported to function in transcriptional silencing of a multicopy transgene in the C. elegans soma [30], although the H3K9 methylation status of repetitive transgenes in somatic cells is not known.
ego-1 Interacts Genetically with him-17
ego-1 loss-of-function mutants have meiotic defects; leptotene-zygotene is protracted, and univalents are present in some oocytes at diakinesis (Figure 3C) [9, 10]. Univalents can result from defective chromosomal pairing, synaptonemal complex formation, or recombination [31]. Recent findings in yeast and C. elegans suggest that recombination relies on proper chromatin structure [13, 32]. Mutations in C. elegans him-17 disrupt formation of double-strand breaks and lead to meiotic chromosome segregation defects [13]. They also severely reduce H3K9me2 accumulation on the male X chromosome and delay the transient H3K9me2 accumulation on hermaphrodite autosomes [13]. HIM-17 is a novel protein with several copies of the THAP domain implicated in DNA binding [13]. Since mutations in ego-1 and him-17 disrupt H3K9me2 enrichment of the male X chromosome and cause defects in early meiosis, we looked for a genetic interaction between them.
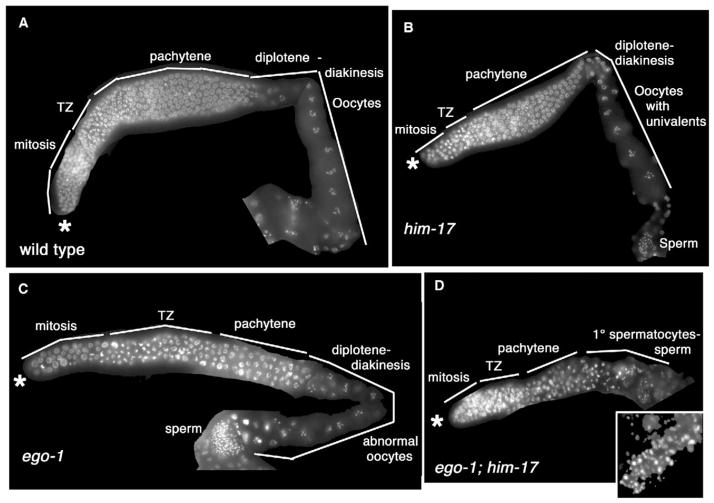
Figure 3. ego-1 Interacts Genetically with him-17.
Each panel shows one hermaphrodite gonad arm at 24 hr post-L4 stage. Asterisk indicates distal end of each arm. TZ, “transition zone” containing germ cells in early meiotic prophase (leptotene-zygotene stages). ego-1 and him-17 null alleles are shown (see Supplemental Experimental Procedures). Animals were raised at 20°C.
(A) The wild-type germline includes distal mitotic cells, meiotic cells, and oocytes in diakinesis stage of meiosis. Sperm are present at the proximal end (although most are out of the plane of focus).
(B) Organization of the him-17 germline is normal.
(C) Overall organization of the ego-1 germline is normal, although some nuclei are enlarged, the leptotene-zygotene region is expanded, and oocytes are small.
(D) The ego-1;him-17 germline is small and lacks oocytes. Sperm nuclei are irregular in size (inset).
Wild-type C. elegans hermaphrodites produce sperm during the L4 larval stage and switch to oogenesis at the time of the adult molt. At 24 hr post-L4 stage, the wild-type hermaphrodite gonad contained 589 (±21; n = 2) mitotic/early meiotic germ cells (including oocytes) and a reserve of >100 sperm (Figure 3A). In him-17(0) mutants raised at 20°C, germline function and organization are grossly normal, except for the appearance of univalent chromosomes in oocytes (Figure 3B) [13]. At 24 hr post-L4 stage, him-17 mutants had 418 (±2; n = 2) and ego-1 mutants had 265 (±40; n = 8) mitotic/early meiotic germ cells; >100 sperm were present in each case.
Adult ego-1;him-17 hermaphrodites raised at 20°C have more severe germline defects compared with each single mutant (Figure 3D). At 24 hr post-L4 stage, the gonad arm contained only 50 (±26) sperm and 154 (±34) mitotic/early meiotic germ cells (n = 6); oocytes were absent. Sperm nuclei were irregular in size and larger than normal, perhaps reflecting a combination of aneuploidy caused by chromosome segregation defects and improper chromosome condensation caused by a chromatin defect (Figure 3D, inset). By 48 hr post-L4 stage, only ~2% of ego-1;him-17 germ lines contained any oocytes (n = 54). The absence of oocytes may reflect either a sex determination defect (failure to make the sperm-to-oocyte switch) or the small germ-line (which does not produce sperm on schedule). ego-1;him-17(0) males also had small germlines and irregular sperm nuclei (data not shown).
We examined H3K9me2 accumulation in ego-1;him-17 germlines to determine whether H3K9me2 levels were further reduced beyond either single mutant, but the abnormal chromosome morphology made it difficult to determine how the staining pattern was affected. The overall pattern appeared to be aberrant. A low level of H3K9me2 staining was seen, particularly in the middle and proximal end of the germline where meiotic nuclei are typically present, and some highly condensed nuclei were more brightly stained (data not shown).
EGO-1 Protein Is Enriched in the Nuclear Fraction of Cell Lysates
We used affinity-purified anti-EGO-1 polyclonal antibodies to assay for EGO-1 in nuclear extracts (see Supplemental Experimental Procedures). These antibodies do not work for immunofluorescence studies with a variety of fixation conditions, but they performed well in protein blots [11]. We prepared nuclear-enriched and total cellular protein extracts and performed Western blots with anti-EGO-1, anti-lamin (a nuclear control), and anti-myosin (a cytoplasmic control) antibodies, and we quantified the results. Most of the total cellular EGO-1 protein was detected in the nuclear extract, as would be expected if EGO-1 directly interacts with chromatin (Figure S3).
Role of EGO-1 in Meiotic Silencing
We find that EGO-1 activity is required for H3K9me2 enrichment on unsynapsed chromosomal regions during meiosis in C. elegans. This process appears to be dependent on an RdRP but independent of Dicer, and thus to be distinct from the mechanism of heterochromatin assembly described in S. pombe in which both RdRP and Dicer activities are required [3, 8]. Recognition/repression of unpaired DNA in meiosis occurs in diverse species [3, 4] and may protect against spurious recombination and/or significant changes in the ordered sequences along homologs. Meiotic silencing may also protect the germline from expression/movement of foreign DNA that has inserted into the genome and transposable genetic elements that have been mobilized. As such, meiotic silencing may complement RNA silencing mechanisms that repress the movement of transposable genetic elements via a posttranscriptional process [33].
It is unclear how unpaired DNA is recognized and repressed, and evidence suggests that different mechanisms may act in different species [3, 4]. Mammals use a chromatin-based mechanism that includes H3K9me2 and other histone modifications [4]. In contrast, N. crassa uses a mechanism that is thought to be posttranscriptional [3, 5]. Intriguingly, the N. crassa mechanism requires an RdRP, SAD-1, whose function is essential for meiosis. Perhaps EGO-1 participates in an ancient chromatin-based genome defense mechanism that has been adapted for essential germ cell processes. Indeed, gene expression profiling suggests that EGO-1 activity ultimately represses expression of genes whose activity may be detrimental to germline function and upregulates expression of genes whose activity is critical for germline function (V.E.V., V. Reinke, and E.M.M., unpublished data). It will be interesting to determine whether EGO-1 activity regulates the pattern of chromatin assembly at the repressed genes.
Although we detected only the gross level changes in H3K9me2 accumulation in ego-1 germ cells, we expect that changes also occur at autosomal sites too scattered for detection by our methods. ego-1 and him-17 single mutants have subtly different defects in H3K9me2 accumulation (this study; [13]); therefore, EGO-1 and HIM-17 may regulate heterochromatin assembly at overlapping, but nonidentical, sets of chromosomal sites. The ego-1;him-17 phenotype may arise from the presence of a larger number of misregulated sites in the double mutant and/or additive defects from their nonoverlapping roles in other processes.
We propose two alternative models for how EGO-1 might target H3K9me2 to unpaired DNA (Figure 4). In both models, an RNA polymerase transcribes unpaired DNA, perhaps through unregulated initiation. EGO-1, in a complex with an RNA helicase (RHA-1) analogous to the RdRC complex in S. pombe [34], recognizes and binds the RNA as it is synthesized. RHA-1 is also required for H3K9me2 accumulation in C. elegans germ cells, but only at elevated temperatures, perhaps reflecting a purely structural role in this process [35]. In one model, EGO-1 converts ssRNA to dsRNA, which then triggers assembly of a RITS-type complex that promotes formation of H3K9me2 modifications, as proposed in other systems (see [8]). This model is supported by our finding that H3K9me2 accumulation is disrupted in ego-1(RdRP/null) putative catalytic mutants. The dsRNA may not require cleavage by a Dicer-type activity, perhaps reflecting a nonprocessive RdRP activity similar to that described for N. crassa QDE-1 in vitro [36]. In an alternative model, EGO-1 acts as a scaffold protein, bringing the chromatin-modifying machinery into proximity with unpaired DNA through its interaction with the transcript. There is ample precedent for the involvement of noncoding transcripts in facultative heterochromatin assembly, from X-inactivation to epigenetic imprinting [3, 4, 8, 37, 38], as well as recent evidence that transcription is required for targeting H3K9me2 to heterochromatic loci [39].
Figure 4. Two Models for the Role of EGO-1 in Silencing Unpaired DNA during Meiosis.
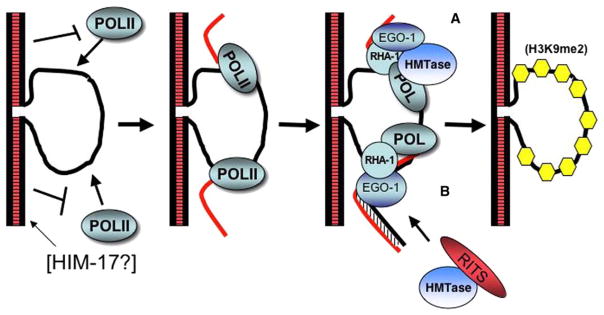
(A and B) Unpaired DNA (A) associates with and (B) is transcribed by a DNA-dependent RNA polymerase that is not active on paired DNA. HIM-17 may participate in this process.
(C) EGO-1 binds to the RNA and recruits chromatin modifiers, e.g., histone methyltransferase (HMTase), to the region. RHA-1 may function in an RdRC-like complex with EGO-1 at this step. HMTase may directly bind to EGO-1 for its recruitment. Alternatively, EGO-1 generates dsRNA, which then triggers assembly of a RITS-like complex. RITS-like activity then recruits HMTase.
(D) The end result of the EGO-1-RNA association is accumulation of H3K9me2 modifications preferentially on the unpaired DNA.
Supplementary Material
Acknowledgments
We thank C. David Allis, Jun Kelly Liu, and Henry Epstein for antibodies, Tim Schedl and Rodolfo Aramayo for discussion, Katherine Walstrom, Rob Martienssen, John Belote, and anonymous reviewers for comments on the manuscript, Claudia Zraly and Larry Smart for advice on methods, and Meera Sundaram, Shohei Mitani, and Geraldine Seydoux for strains. Some strains used in this study were provided by the Caenorhabditis Genetics Center, which is funded by the National Institutes of Health National Center for Research Resources. Funds for this study were provided by the National Science Foundation and Syracuse University (to E.M.M.) and the National Institutes of Health and Emory University Research Council (to W.G.K.).
Footnotes
Supplemental Data include three figures and Supplemental Experimental Procedures and can be found with this article online at http://www.current-biology.com/cgi/content/full/15/21/▪▪▪/DC1/.
References
- 1.Bean CJ, Schaner CE, Kelly WG. Meiotic pairing and imprinted X chromatin assembly in Caenorhabditis elegans. Nat Genet. 2004;36:100–105. doi: 10.1038/ng1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kelly WG, Schaner CE, Dernberg AF, Lee MH, Kim SK, Villeneuve AM, Reinke V. X-chromosome silencing in the germline of C. elegans. Development. 2002;129:479–492. doi: 10.1242/dev.129.2.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Matzke MA, Birchler JA. RNAi-mediated pathways in the nucleus. Nat Genet. 2005;6:24–35. doi: 10.1038/nrg1500. [DOI] [PubMed] [Google Scholar]
- 4.Lee JT. Sex chromosome inactivation: the importance of pairing. Curr Biol. 2005;15:R249–R252. doi: 10.1016/j.cub.2005.03.024. [DOI] [PubMed] [Google Scholar]
- 5.Hynes MJ, Todd RB. Detection of unpaired DNA at meiosis results in RNA-mediated silencing. Bioessays. 2003;25:99–103. doi: 10.1002/bies.10241. [DOI] [PubMed] [Google Scholar]
- 6.Ahlquist P. RNA-dependent RNA polymerases, viruses, and RNA silencing. Science. 2002;296:1268–1273. doi: 10.1126/science.1069132. [DOI] [PubMed] [Google Scholar]
- 7.Tijsterman M, Ketting RF, Plasterk RHA. The genetics of RNA silencing. Annu Rev Genet. 2002;36:489–519. doi: 10.1146/annurev.genet.36.043002.091619. [DOI] [PubMed] [Google Scholar]
- 8.Reddy KC, Villeneuve AM. C. elegans HIM-17 links chromatin modification and competence for initiation of meiotic recombination. Cell. 2004;118:439–452. doi: 10.1016/j.cell.2004.07.026. [DOI] [PubMed] [Google Scholar]
- 9.Grewal SIS, Rice JC. Regulation of heterochromatin by histone methylation and small RNAs. Curr Opin Cell Biol. 2004;16:230–238. doi: 10.1016/j.ceb.2004.04.002. [DOI] [PubMed] [Google Scholar]
- 10.Smardon A, Spoerke JM, Stacey SC, Klein ME, Mackin N, Maine EM. EGO-1 is related to RNA-directed RNA polymerase and functions in germline development and RNA interference in C. elegans. Curr Biol. 2000;10:169–178. doi: 10.1016/s0960-9822(00)00323-7. [DOI] [PubMed] [Google Scholar]
- 11.Qiao L, Lissemore JL, Shu P, Smardon A, Gelber M, Maine EM. Enhancers of glp-1, a gene required for cell-signaling in C. elegans, define a set of genes required for germline development. Genetics. 1995;141:551–569. doi: 10.1093/genetics/141.2.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vought VE, Ohmachi M, Lee MH, Maine EM. EGO-1, a putative RNA-directed RNA polymerase, promotes germline proliferation in parallel with GLP-1/Notch signaling and regulates the spatial organization of nuclear pore complexes and germline P granules in C. elegans. Genetics. 2005;170:1121–1132. doi: 10.1534/genetics.105.042135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Simmer F, Tijsterman M, Parrish S, Koushika SP, Nonet ML, Fire A, Ahringer J, Plasterk RHA. Loss of the putative RNA-directed RNA polymerase RRF-3 makes C. elegans hypersensitive to RNAi. Curr Biol. 2002;12:1317–1319. doi: 10.1016/s0960-9822(02)01041-2. [DOI] [PubMed] [Google Scholar]
- 14.Stinchcomb DT, Shaw JE, Carr SH, Hirsh D. Extrachromosomal DNA transformation of Caenorhabditis elegans. Mol Cell Biol. 1985;5:3484–3496. doi: 10.1128/mcb.5.12.3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kelly WG, Xu S, Montgomery MM, Fire A. Distinct requirements for somatic and germline expression of a generally expressed Caenorhabditis elegans gene. Development. 1997;146:227–238. doi: 10.1093/genetics/146.1.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jedrusik MA, Schulze E. A single histone H1 iso-form (H1.1) is essential for chromatin silencing and germline development in Caenorhabditis elegans. Development. 2001;128:1069–1080. doi: 10.1242/dev.128.7.1069. [DOI] [PubMed] [Google Scholar]
- 17.Couteau F, Guerry F, Muller F, Palladino F. A heterochromatin protein 1 homologue in Caenorhabditis elegans acts in germline and vulval development. EMBO Rep. 2002;3:235–241. doi: 10.1093/embo-reports/kvf051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim JK, Gabel HW, Kamath RS, Tewari M, Pasquinelli A, Rual JF, Kennedy S, Dybbs M, Bertin N, Kaplan JM, et al. Functional genomic analysis of RNA interference in C. elegans. Science. 2005;308:1164–1167. doi: 10.1126/science.1109267. [DOI] [PubMed] [Google Scholar]
- 19.Tabara H, Sarkissian M, Kelly WG, Fleenor J, Grishok A, Timmons L, Fire A, Mello CC. The rde-1 gene, RNA interference, and transposon silencing in C. elegans. Cell. 1999;99:123–132. doi: 10.1016/s0092-8674(00)81644-x. [DOI] [PubMed] [Google Scholar]
- 20.Holdeman R, Nehrt S, Strome S. MES-2, a maternal protein essential for viability of the germline in Caenorhabditis elegans, is homologous to a Drosophila Polycomb group protein. Development. 1998;125:2457–2467. doi: 10.1242/dev.125.13.2457. [DOI] [PubMed] [Google Scholar]
- 21.Bender LB, Cao R, Zhang Y, Strome S. THE MES-2/MES-3/MES-6 complex and regulation of histone H3 methylation in C. elegans. Curr Biol. 2004;14:1639–1643. doi: 10.1016/j.cub.2004.08.062. [DOI] [PubMed] [Google Scholar]
- 22.Ketting RF, Haverkamp THA, van Leunen HGAM, Plasterk RHA. mut-7 of C. elegans, required for transposon silencing and RNA interference, is a homolog of Werner Syndrome helicase and RNAseD. Cell. 1999;99:133–141. doi: 10.1016/s0092-8674(00)81645-1. [DOI] [PubMed] [Google Scholar]
- 23.Robert VJP, Sijen T, van Wolfwinkel J, Plasterk RHA. Chromatin and RNAi factors protect the C. elegans germline against repetitive sequences. Genes Dev. 2005;19:782–787. doi: 10.1101/gad.332305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Petrie VJ, Wuitschick JD, Givens CD, Kosinski AM, Partridge JF. RNA interference (RNAi)-dependent and RNAi-independent association of the Chp1 chromo-domain protein with distinct heterochromatic loci in fission yeast. Mol Cell Biol. 2005;25:2331–2346. doi: 10.1128/MCB.25.6.2331-2346.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kanellopoulou C, Muljo SA, Kung AL, Ganesan S, Drapkin R, Jenuwein T, Livingston DM, Rajewsky K. Dicer-deficient mouse embryonic stem cells are defective in differentiation and centromeric silencing. Genes Dev. 2005;19:489–501. doi: 10.1101/gad.1248505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Denli AM, Tops BB, Plasterk RH, Ketting RF, Hannon GJ. Processing of primary microRNAs by the Microprocessor complex. Nature. 2004;432:231–235. doi: 10.1038/nature03049. [DOI] [PubMed] [Google Scholar]
- 27.Tabara H, Yigit E, Siomi H, Mello CC. The dsRNA binding protein RDE-4 interacts with RDE-1, DCR-1, and a DExH-box helicase to direct RNAi in C. elegans. Cell. 2002;109:861–871. doi: 10.1016/s0092-8674(02)00793-6. [DOI] [PubMed] [Google Scholar]
- 28.Tops BB, Tabara H, Sijen T, Simmer F, Mello CC, Plasterk RH, Ketting RF. RDE-2 interacts with MUT-7 to mediate RNA interference in Caenorhabditis elegans. Nucleic Acids Res. 2005;33:347–355. doi: 10.1093/nar/gki183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Grishok A, Pasquinelli AE, Conte D, Li N, Parrish S, Ha I, Baillie DL, Fire A, Ruvkun G, Mello CC. Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell. 2001;106:23–34. doi: 10.1016/s0092-8674(01)00431-7. [DOI] [PubMed] [Google Scholar]
- 30.Grishok A, Sinskey JL, Sharp PA. Transcriptional silencing of a transgene by RNAi in the soma of C. elegans. Genes Dev. 2005;19:683–696. doi: 10.1101/gad.1247705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Page SL, Hawley RS. Chromosome choreography: the meiotic ballet. Science. 2003;301:785–789. doi: 10.1126/science.1086605. [DOI] [PubMed] [Google Scholar]
- 32.Maleki S, Keeney S. Modifying histones and initiating meiotic recombination; new answers to an old question. Cell. 2004;118:404–406. doi: 10.1016/j.cell.2004.08.008. [DOI] [PubMed] [Google Scholar]
- 33.Vastenhouw NL, Plasterk RH. RNAi protects the Caenorhabditis elegans germline against transposition. Trends Genet. 2004;20:314–319. doi: 10.1016/j.tig.2004.04.011. [DOI] [PubMed] [Google Scholar]
- 34.Motamedi MR, Verdel A, Colmenares SU, Gerber SA, Gygi SP, Moazed D. Two RNAi complexes, RITS and RDRC, physically interact and localize to noncoding centromeric RNAs. Cell. 2004;119:789–802. doi: 10.1016/j.cell.2004.11.034. [DOI] [PubMed] [Google Scholar]
- 35.Walstrom KM, Schmidt D, Bean CJ, Kelly WG. RNA helicase A is important for germline transcriptional control, proliferation, and meiosis in C. elegans. Mech Dev. 2005;122:707–720. doi: 10.1016/j.mod.2004.12.002. [DOI] [PubMed] [Google Scholar]
- 36.Makeyev EV, Bamford DH. Cellular RNA-dependent RNA polymerase involved in posttranscriptional gene silencing has two distinct activity modes. Mol Cell. 2002;10:1417–1427. doi: 10.1016/s1097-2765(02)00780-3. [DOI] [PubMed] [Google Scholar]
- 37.Craig J. Heterochromatin—many flavours, common themes. Bioessays. 2004;27:17–28. doi: 10.1002/bies.20145. [DOI] [PubMed] [Google Scholar]
- 38.Anderson AA, Panning B. Epigenetic gene regulation by noncoding RNAs. Curr Opin Cell Biol. 2003;15:281–289. doi: 10.1016/s0955-0674(03)00041-3. [DOI] [PubMed] [Google Scholar]
- 39.Schramke V, Sheedy DM, Denli AM, Bonila C, Ekwall K, Hannon GJ, Allshire RC. RNA-inference-directed chromatin modification coupled to RNA polymerase II transcription. Nature. 2005;435:1275–1279. doi: 10.1038/nature03652. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.