ABSTRACT
Asymptomatic cytomegalovirus (CMV) replication occurs frequently in the genital tract in untreated HIV-infected men and is associated with increased immune activation and HIV disease progression. To determine the connections between CMV-associated immune activation and the size of the viral reservoir, we evaluated the interactions between (i) asymptomatic seminal CMV replication, (ii) levels of T cell activation and proliferation in blood, and (iii) the size and transcriptional activity of the HIV DNA reservoir in blood from 53 HIV-infected men on long-term antiretroviral therapy (ART) with suppressed HIV RNA in blood plasma. We found that asymptomatic CMV shedding in semen was associated with significantly higher levels of proliferating and activated CD4+ T cells in blood (P < 0.01). Subjects with detectable CMV in semen had approximately five times higher average levels of HIV DNA in blood CD4+ T cells than subjects with no CMV. There was also a trend for CMV shedders to have increased cellular (multiply spliced) HIV RNA transcription (P = 0.068) compared to participants without CMV, but it is unclear if this transcription pattern is associated with residual HIV replication. In multivariate analysis, the presence of seminal plasma CMV (P = 0.04), detectable 2-long terminal repeat (2-LTR), and lower nadir CD4+ (P < 0.01) were independent predictors of higher levels of proviral HIV DNA in blood. Interventions aimed at reducing seminal CMV and associated immune activation may be important for HIV curative strategies. Future studies of anti-CMV therapeutics will help to establish causality and determine the mechanisms underlying these described associations.
IMPORTANCE Almost all individuals infected with HIV are also infected with cytomegalovirus (CMV), and the replication dynamics of the two viruses likely influence each other. This study investigated interactions between asymptomatic CMV replication within the male genital tract, levels of inflammation in blood, and the size of the HIV DNA reservoir in 53 HIV-infected men on long-term antiretroviral therapy (ART) with suppressed HIV RNA in blood plasma. In support of our primary hypothesis, shedding of CMV DNA in semen was associated with increased activation and proliferation of T cells in blood and also significantly higher levels of HIV DNA in blood cells. These results suggest that CMV reactivation might play a role in the maintenance of the HIV DNA reservoir during suppressive ART and that it could be a target of pharmacologic intervention in future studies.
INTRODUCTION
Although antiretroviral therapy (ART) can suppress HIV RNA levels to below the limit of detection in the blood of HIV-infected individuals (1, 2), the virus persists despite prolonged continuous treatment, and the viral load rebounds when therapy is interrupted (3). The reason for this rapid rebound is the presence of a long-lived reservoir of latent HIV proviruses, which is established early in the course of infection (4, 5).
Several non-mutually exclusive mechanisms underlie HIV persistence in a very small proportion of memory CD4+ T cells during suppressive ART (6–9). First, the existence of residual HIV replication could lead to de novo infection of memory CD4+ T cells (10, 11). Second, the HIV latent reservoir could be maintained by proliferation of latently infected CD4+ T cells, driven by antigenic stimulation (12) or cytokines (homeostatic proliferation) (13, 14). The relative contributions of antigen-driven and homeostatic proliferation to the size and maintenance of the HIV reservoir are unclear, but both mechanisms likely play a role in HIV persistence (13, 15). Both mechanisms of HIV persistence (i.e., residual replication and cell proliferation) are modulated by inflammation and immune activation, widely described in patients receiving ART (16). Similarly, inflammatory cytokines and general CD4+ T cell activation can modulate HIV expression in latently infected cells, sensitize target cells for new cycles of infection (17, 18), lead to dysfunctional HIV-specific T cell responses (19, 20), and impair the clearance of HIV-infected cells (13). It is therefore crucial to understand which factors contribute to persistent immune activation and if the same factors also modulate and maintain the HIV reservoir.
Cytomegalovirus (CMV) infection is highly prevalent among HIV-infected individuals and is associated with increased levels of T cell activation and HIV disease progression (21, 22). We have demonstrated that asymptomatic CMV replication in the male genital tract is associated with higher levels of total HIV DNA in the blood of untreated HIV-infected individuals (23). Since our previous study investigated participants not receiving ART, the measured HIV DNA levels included both integrated and unintegrated viral forms and did not represent solely the latent HIV reservoir (24). It is unclear if these observed associations still persist during suppressive ART, which is relevant to inform future eradication strategies.
In this study, we evaluated the associations between (i) asymptomatic CMV replication within the male genital tract, (ii) levels of T cell activation and proliferation in blood, and (iii) the size and transcriptional activity of the HIV DNA reservoir in paired semen and blood samples from 53 HIV-infected CMV-seropositive men on long-term ART and with suppressed HIV RNA in blood plasma. The results described here will help inform the design of future clinical studies to investigate the use of anti-CMV therapeutics to establish any causal links between CMV shedding, immune activation and the persistence of the HIV reservoir and to better understand the mechanisms underlying the described associations.
MATERIALS AND METHODS
Participants, samples, and clinical laboratory tests.
Paired semen and blood samples collected from asymptomatic, chronically HIV-infected, sexually active men who have sex with men (MSM) who were prospectively enrolled in the California Collaborative Treatment Group (CCTG) 592 trial were included in this study (25). CCTG 592 is a study of an Internet-based behavioral intervention with MSM that included baseline collection of paired blood and semen from a total of 180 HIV-infected MSM who were on or off ART. For this study, we included baseline semen samples from a subset of 53 CMV-seropositive subjects who were receiving effective ART with blood plasma HIV RNA at <200 copies/ml within 3 months before the seminal-sample collection and who had peripheral blood mononuclear cell (PBMC) samples stored for further analysis. The individuals started ART during chronic HIV infection, but no information is available about the durations of HIV infection. Fifty subjects had HIV RNA at <50 copies/ml, while three had HIV RNA levels between 50 and 200 copies/ml. All included participants were CMV seropositive (26). Semen was collected and processed as previously described (27, 28). Blood CD4+ T lymphocyte absolute counts were measured by flow cytometry, and HIV RNA levels in blood plasma were quantified by the Amplicor HIV Monitor Test (Roche Molecular Systems Inc.).
The studies were conducted with appropriate written subject consent and were approved by the Human Research Protections Program at the University of California, San Diego; the Los Angeles Biomedical Research Institute at Harbor-UCLA Medical Center; and the University of Southern California. Written informed consent was provided by all study participants.
Quantification of HIV DNA, 2-LTR circular HIV DNA, and herpesvirus DNA in PBMCs and semen.
DNA was extracted from 5 million PBMCs and 200 μl seminal plasma for each participant (QIAamp DNA minikit, Qiagen, CA). For PBMCs, total HIV DNA (Pol) and the 2-long-terminal-repeat (2-LTR) junction were quantified by droplet digital PCR (ddPCR) from extracted DNA (29). Briefly, 1,000 ng of DNA per replicate was digested with BSAJ1 enzyme (New England BioLabs) prior to ddPCR. Total HIV DNA (Pol) and 2-LTR PCRs were performed as a duplex assay with VIC (Pol)- and 6-carboxyfluorescein (FAM) (2-LTR)-labeled probes, respectively, with the following cycling conditions: 10 min at 95°C, 40 cycles consisting of a 30-s denaturation at 94°C followed by a 60°C extension for 60 s, and a final 10 min at 98°C. Total CMV and Epstein-Barr virus (EBV) DNA PCRs were performed as a duplex with FAM (CMV)- and HEX (EBV)-labeled probes, respectively, with the following cycling conditions: 10 min at 95°C, 40 cycles consisting of a 30-s denaturation at 94°C followed by a 54°C extension for 60 s, and a final 10 min at 98°C. A 1:10 dilution of the digested DNA was used for host cell ribonuclease P/MRP 30-kDa subunit (RPP30) PCR (probe VIC) and cycled with the same parameters as the Pol–2-LTR duplex. Copy numbers were calculated as the mean of replicate PCR measurements and normalized to 1 million CD4+ T cells as determined by RPP30 (total cell count) and flow cytometry (percentage of CD4 T cells). The levels of 7 herpesviruses (CMV; EBV; herpes simplex virus 1 [HSV-1] and HSV-2; and human herpesvirus 6 [HHV-6], HHV-7, and HHV-8) were also measured by real-time PCR in the DNA extracted from seminal plasma, as described previously (21).
Quantification of cellular HIV RNA in PBMCs.
Cellular HIV RNA was measured for the subset of 42 individuals with enough stored PBMCs to perform these analyses and completely suppressed HIV RNA levels in blood plasma (<50 copies/ml). The extracted RNA (500 ng) was reverse transcribed into 20 μl cDNA (iScript Advanced cDNA synthesis kit for RT-qPCR; Bio-Rad) using the manufacturer's protocol. The cDNA product (8 μl; approximately 200 ng) was added to the ddPCR reaction mixture. Unspliced HIV RNA (usGag) and multiply spliced (msTat-Rev) PCRs were performed as a duplex with HEX (usGag)- and FAM (msTat-Rev)-labeled probes, respectively, using primers and probes as previously described (30, 31). The cycling conditions for this PCR were as follows: 10 min at 95°C, 60 cycles consisting of a 30-s denaturation at 94°C followed by a 60°C extension for 60 s, and a final 10 min at 98°C. Copy numbers were calculated as the mean of replicate PCR measurements and normalized to total RNA as determined by the A260/A280 absorption ratio using a NanoDrop 2000 spectrophotometer (Thermo Scientific).
Anti-CMV IgG antibody levels.
As previously described (26), anti-CMV IgG antibody levels were measured in blood plasma. The number of units per ml were determined by interpolation from a standard curve of a known anti-CMV IgG solution (26, 32).
Multiparameter flow cytometry analysis.
For a subset of 49 patients, analysis of cellular activation and proliferation was performed by multicolor flow cytometry. Aliquots of 5 million PBMCs were quickly thawed at 37°C, resuspended in RPMI supplemented with 20% fetal bovine serum (FBS), incubated at room temperature for 20 min, centrifuged, and resuspended in 500 μl phosphate-buffered saline (PBS) staining buffer. Cell viability was assessed using the LIVE/DEAD Fixable Aqua Dead Cell Stain Kit (Life Technologies). Four subjects (out of the total 53 included subjects) were excluded because of <85% cell viability. Approximately 300,000 to 500,000 PBMCs per tube were stained for cell surface markers prior to fixation and, when required, followed with permeabilization for intracellular-protein detection (FOXP3/Transcription Factor Staining Buffer set; eBioscience). The stained cells were acquired on a FACSCanto (BD Biosciences). We used the following antibody combinations to evaluate immune activation and proliferation in T cells: (tube 1) HLA-DR–fluorescein isothiocyanate (FITC), CD45RA-phycoerythrin (PE), CD4–peridinin chlorophyll protein (PerCP)-Cy5.5, CD38–PE-Cy7, CD27-allophycocyanin (APC), CD3–APC-Cy7, and CD8-Pacific Blue; (tube 2) Ki67-FITC, CD45RA-PE, CD4–PerCP-Cy5.5, CCR7–PE-Cy7, CD27-APC, CD3–APC-Cy7, and CD8-Pacific Blue (BD Biosciences). The antibody combinations in tube 1 were used to assess naive (CD45RA+ CD27+), central memory (CD45RA− CD27+), effector memory (CD45RA− CD27−), and terminally differentiated (CD45RA+ CD27−) CD4 and CD8 T cell subsets. With tube 2, T cell subsets were defined as naive (CD45RA+ CD27+ CCR7+), central memory (CD45RA− CD27+ CCR7+), transitional memory (CD45RA− CD27+ CCR7−), and effector memory (CD45RA+/− CD27− CCR7−) in both the CD4+ and CD8+ subsets. Immune activation was defined by CD38+ HLA-DR+ expression and proliferation and by Ki67+ in total CD4+ and CD8+ T cell populations and subsets. To normalize the levels of HIV DNA and 2-LTR circles, the percentage of CD4+ T cells within total PBMCs was determined for each sample. All data analyses were performed using Flow Jo software (version 9.6.2).
Statistics.
Statistical analyses were performed using SAS (version 9.2). Each continuous variable was assessed for normal distribution. Viral-load variables were transformed to log10 values, and CMV shedding and the presence of the LTR were dichotomized (undetectable/detectable). Comparisons were performed using the Fisher exact text (for sparse data), t test (for continuous, normally distributed variables), or Mann-Whitney U test (for continuous, non-normally distributed variables). Correlation analyses were performed using parametric (Pearson) or nonparametric (Spearman) correlation coefficients. A multivariate linear regression for the association of detectable seminal CMV DNA with HIV DNA in blood was performed, adjusting for possible confounders that were found to be significant in the univariate analysis (P < 0.05). Analyses were performed on the complete data set of 53 subjects (including three subjects experiencing an HIV RNA blip of <200 copies/ml in the 3 months before sample collection) and on the reduced data set of 50 subjects with fully suppressed HIV RNA in blood plasma (<50 copies/ml). To estimate the average viral transcriptional activity per single HIV DNA copy (Pol), we calculated the ratio between cellular HIV RNA and total HIV DNA for each sample, as described previously (31, 33). Because of the exploratory character of the study, we decided not to correct our results for false detection (multiple comparisons).
RESULTS
Participants, samples, and clinical laboratory tests.
The study participants (n = 53) were HIV-infected men with a median age of 46 (95% interquartile range [IQR], 38 to 51) years who were receiving ART and had ≤200 copies of HIV RNA/ml of blood plasma. Fifty subjects had <50 copies of HIV RNA/ml, while three had levels between 50 and 200 copies of HIV RNA/ml. The median time on ART at the time of sample collection was 3.8 (IQR, 1.8 to 5.3) years, and the median CD4 T cell count was 625 (IQR, 535 to 739) cells/μl. Eighty-two percent were on a regimen including tenofovir, 41% were on a regimen including a nonnucleoside reverse transcriptase inhibitor (NNRTI), 61% were on a regimen including a protease inhibitor (PI), and 16% were on a regimen also including an integrase inhibitor. Self-reported levels of ART adherence during the preceding month were >90% of daily doses for 90.4% of the subjects. The subjects' characteristics are summarized in Table 1.
TABLE 1.
Subjects' characteristics
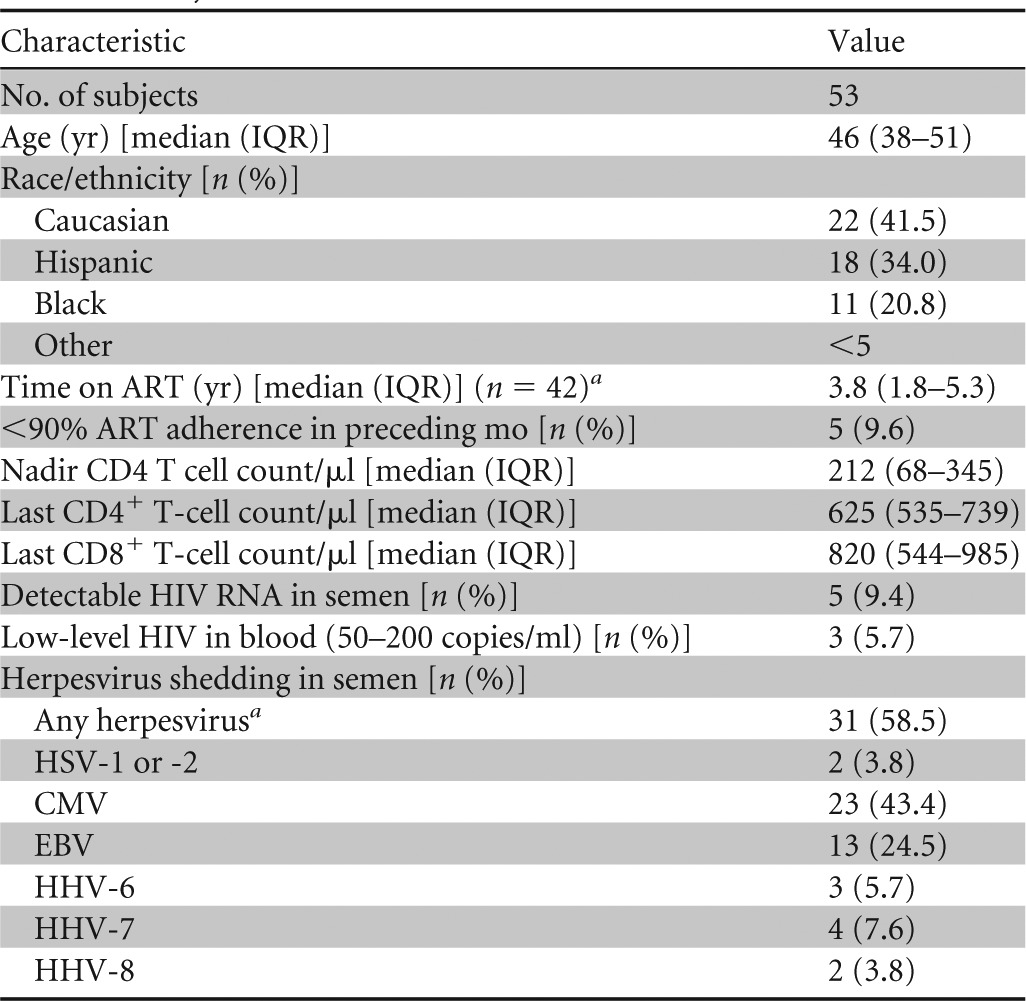
Time on ART was available for 42 subjects.
Herpesvirus 1 to 8, excluding varicella virus.
Herpesviruses and viral reservoir characteristics.
Five participants (9.4%) were shedding HIV RNA, and 31 (58.5%) were shedding at least one herpesvirus in seminal plasma at the time of collection. Specifically, 23 (43.4%) were shedding CMV, 13 (24.5%) EBV, 2 (3.8%) HSV-2, 3 (5.7%) HHV-6, 4 (7.6%) HHV-7, and 2 (3.8%) HHV-8 (Table 1; see Fig. S1 in the supplemental material). None of the subjects were shedding HSV-1. Eleven subjects (21%) had detectable levels of CMV DNA in PBMCs (median, 4.2 copies/million cells among detectable samples; IQR, 2.6 to 5.8 copies), and 50 (94%) had detectable levels of EBV in PBMCs (median, 75 copies/million cells; IQR, 18.5 to 389 copies) (see Fig. S1 in the supplemental material).
Total levels of HIV DNA (Pol) as measured by ddPCR were detectable in 92% of PBMC samples (n = 49), with a median level of 2.54 log10 copies per million CD4+ T cells (IQR, 2.0 to 3.1 copies), while 2-LTR circles were detectable in 43% of all PBMC samples (n = 23), with an average level of 1.42 log10 copies per million CD4+ T cells among detectable samples (IQR, 1.19 to 1.89 copies). In the subset of 42 patients with completely undetectable HIV RNA levels and sufficient samples available, levels of unspliced HIV RNA were detectable in 95% of the samples (n = 40), while levels of multiply spliced HIV RNA encoding Tat and Rev were detectable in 64% of the samples (n = 27).
Associations between CMV replication, HIV DNA levels, immunological markers, and cellular HIV transcription. (i) Predictors of higher HIV DNA levels (n = 53).
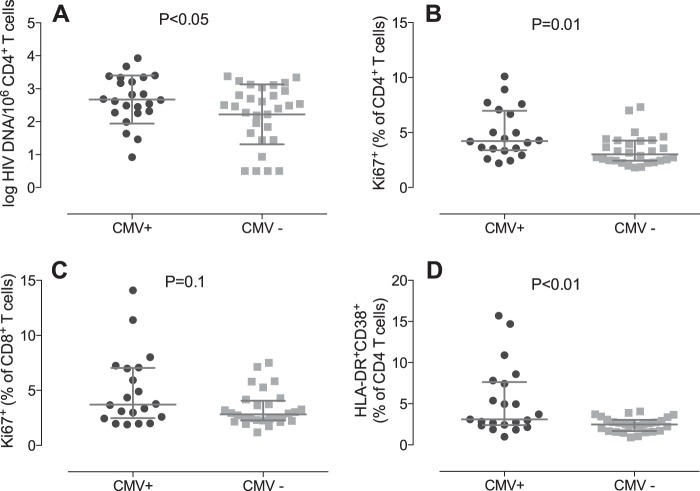
Univariate analysis revealed significantly higher levels of HIV DNA in the CD4+ T cells of participants with detectable seminal CMV than in those without detectable CMV in semen (mean, 2.84 versus 2.11 log10 copies per million CD4+ T cells; P < 0.05) (Fig. 1A). There was also a trend for increased levels of CMV IgG to be associated with higher levels of total log10 HIV DNA in CD4+ T cells (P = 0.08). As might be expected, the presence of detectable 2-LTR circles was associated with higher levels of total HIV DNA (P < 0.01), and we also found a significant negative correlation between the nadir CD4+ T cell count and HIV DNA Pol copies per CD4+ T cells (r = −0.29; P = 0.03), as previously described (34). The presence of seminal shedding of HIV and herpesviruses other than CMV, the CD4+ T cell count, age, time on ART, and self-reported adherence to ART were not associated with levels of total HIV DNA (Table 2).
FIG 1.
Levels of total HIV DNA per million CD4+ T cells (A) and percentages of proliferating (Ki67+) CD4+ T cells (B), proliferating (Ki67+) CD8+ T cells (C), and activated (HLA-DR+ CD38+) CD4+ T cells (D) in PBMCs of subjects with (CMV+) and without (CMV−) cytomegalovirus shedding in seminal plasma. P values were determined using a t test (A) and the Mann-Whitney Wilcoxon test (B, C, and D). The box plots display means and standard deviations (A) and medians and interquartile ranges (B, C, and D).
TABLE 2.
Predictors of higher HIV DNA levels
| Factor | Mean log10 HIV DNA copies (95% CI)a | r value |
P valuea |
||
|---|---|---|---|---|---|
| Univariate (n = 53) | Multivariate |
||||
| n = 53 | Undetectable | ||||
| CMV seminal shedding | |||||
| Any | 2.84 (2.27–2.94) | 0.05 | 0.03 | 0.05 | |
| None | 2.11 (1.72–2.51) | ||||
| 2-LTR circlec | |||||
| Any | 2.84 (2.60–3.08) | <0.01 | <0.01 | <0.01 | |
| None | 2.03 (1.63–2.42) | ||||
| CD4+ T cell count | −0.20 | 0.16 | |||
| Nadir CD4+ T cell count | −0.34 | 0.03 | 0.01 | <0.01 | |
| Age | −0.04 | 0.82 | |||
| Days on ART (n = 42)d | −0.03 | 0.83 | |||
| CMV IgG (n = 52)e | 0.24 | 0.08 | |||
| EBV seminal shedding | |||||
| Any | 2.41 (2.03–2.70) | 0.86 | |||
| None | 2.37 (2.06–2.76) | ||||
| Detectable HIV in semen | |||||
| Any | 2.59 (1.63–3.55) | 0.6 | |||
| None | 2.36 (2.07–2.64) | ||||
| <90% ART adherence | |||||
| Yes | 1.81 (0.54–3.08) | 0.16 | |||
| No | 2.45 (2.17–2.73) | ||||
95% CI, 95% confidence interval.
Univariate P values were determined using a t test (for categorical variables) and Pearson correlation analysis (for continuous variables). Multivariate linear regression analysis was performed for variables with P values of ≤0.050 in univariate analysis and repeated including only the subset of 50 subjects with HIV RNA levels in blood of <50copies/ml. P values of ≤0.05 are in boldface.
Total HIV DNA log10 levels of 2-LTR were normalized per million CD4+ T cells.
Days on ART were available for 42 subjects.
CMV IgG was available for 52 subjects.
Multivariate analysis indicated that detectable seminal CMV was independently associated with higher levels of HIV DNA in CD4+ T cells (P = 0.04) after adjusting for the presence of 2-LTR circles and nadir CD4+ T cells. Of note, CMV IgG presented a trend toward an association with HIV DNA levels in univariate analysis (P = 0.08), but it became nonsignificant when added to the multivariate model (P = 0.69). Even after restricting the analysis to the 50 subjects with HIV RNA in blood plasma at <50 copies/ml, seminal CMV shedding remained independently associated with higher levels of HIV DNA (P = 0.04) (Table 2). Similarly, there was a significant but weak correlation between seminal CMV levels and CD4-associated HIV DNA levels in blood (P = 0.04; Pearson, r = 0.26), which remained significant after adjusting for the presence of 2-LTR circles and nadir CD4+ in a multivariate model. As previously reported (35), subjects with detectable CMV DNA showed a trend toward lower CD4+ T cell counts than non-CMV shedders (P = 0.039), but no difference was found for the nadir CD4+ T cell count (P = 0.24). The presence of CMV DNA in PBMCs was not associated with significantly higher HIV DNA levels (2.53 versus 2.26 log10; P = 0.43) or with higher CD4+ T cell immune activation or proliferation. Similarly, the amount of EBV DNA in PBMCs did not correlate with HIV DNA levels (P = 0.26).
(ii) Immunological markers (n = 49).
To investigate whether increased immune activation and proliferation of T cells could be a possible mechanism connecting CMV replication with higher levels of proviral HIV DNA, we grouped patients according to whether CMV was detectable in seminal plasma (Table 3). We observed that subjects with detectable CMV in semen (n = 21) had lower total CD4 T cell counts than those with undetectable seminal CMV (n = 28; P = 0.039). Moreover, the CD4 T cell compositions were different in the two groups: subjects with detectable CMV in semen had a significantly higher frequency of transitional memory CD4+ (median, 13.1% versus 10.23%; Mann-Whitney, P = 0.04) and effector memory CD4+ (12.6% versus 7.3%; P = 0.05) T cells in blood than those with undetectable seminal CMV. Subjects with detectable seminal CMV DNA also had higher levels of proliferating (Ki67+) total CD4+ T cells (P = 0.01) (Fig. 1B) and effector memory CD4+ T cells (P = 0.09) (Table 4). There was also a trend toward higher levels of proliferating CD8+ T cells (P = 0.1) (Fig. 1C) in those with detectable seminal CMV than in those whose levels were undetectable. Among subjects with CMV DNA in semen, there was also a significant correlation between HIV DNA levels and frequency of activated (P < 0.01; Spearman, r = 0.6) (Fig. 2A) and proliferating (P < 0.01; r = 0.64) (Fig. 2B) CD4+ T cells. However, we did not find similar correlations between the frequency of proliferating (r = −0.29; P = 0.15) and activated (r = −0.04; P = 0.85) CD4+ T cells and HIV DNA among CMV nonshedders. The trend, if any, was a negative correlation, opposite to what we observed for CMV shedders. After including the entire population (CMV shedders and nonshedders), there was no association between CD4+ proliferation and HIV DNA (r = 0.10; P = 0.51) and a diminished association between CD4+ activation and HIV DNA (r = 0.35; P = 0.02). Subjects with detectable seminal CMV DNA also had significantly increased levels of immune activation (HLA-DR+ CD38+) of total CD4+ T cells (P < 0.01) (Fig. 1D) and effector memory CD4+ T cells (P = 0.02) (Table 4). No difference was observed in CD8+ immune activation of T cells between the two groups. All the described associations remained significant after excluding the three subjects with HIV RNA levels in blood plasma at 50 to 200 copies/ml.
TABLE 3.
Characteristics of CMV shedders versus nonshedders
| Characteristic | Value |
P value between groupsa | |
|---|---|---|---|
| Any CMV (n = 23) | No CMV (n = 30) | ||
| Age (yr) [median (IQR)] | 45 (36–49) | 46 (39–52) | 0.286 |
| Time on ARTb (yr) [median (IQR)] (n = 42) | 3.7 (1.6–6.4) | 3.8 (1.8–5.3) | 0.895 |
| Nadir CD4 T cell count/μl [median (IQR)] | 200 (39–329) | 233 (94–408) | 0.266 |
| Last CD4+ T cell count/μl [median (IQR)] | 542 (453–734) | 660 (577–752) | 0.039 |
| Last CD8+ T cell count/μl [median (IQR)] | 848 (544–1023) | 634 (507–985) | 0.253 |
| Detectable HIV RNA in semen [n (%)] | 3 (13) | 2 (7) | 0.420 |
| Low-level HIV in blood (50–200 copies/ml) [n (%)] | 2 (9) | 1 (3) | 0.573 |
| CMV IgG (U/ml) [median (IQR)] | 34.0 (27.2–44.0) | 34.0 (23.9–42.4) | 0.451 |
| Herpesvirus shedding in semen [n (%)] | |||
| Any herpesvirus except CMV | 9 (39) | 8 (27) | 0.384 |
| HSV-1 or -2 | 1 (4) | 1 (3) | 1.000 |
| EBV | 8 (35) | 5 (17) | 0.198 |
| HHV-6 | 0 (0) | 3 (10) | 0.249 |
| HHV-7 | 2 (9) | 2 (7) | 1.000 |
| HHV-8 | 1 (4) | 1 (3) | 1.000 |
P values were determined using the Mann-Whitney Wilcoxon test. P values of ≤0.1 are in boldface.
Time on ART information was available only for a subset of 42 participants.
TABLE 4.
Differences in T cell proliferation and activation between seminal CMV shedders and nonshedders
| Characteristica | Value |
Univariate P valueb | |
|---|---|---|---|
| Any CMV (n = 21) | No CMV (n = 28) | ||
| T cell proliferation | |||
| Ki67+ (% of CD4+ T cells) [median (IQR)] | |||
| Total | 4.2 (3.4–6.9) | 3.0 (2.4–4.3) | 0.01 |
| Naive | 1.9 (1.3–2.7) | 1.5 (1.1–2.6) | 0.44 |
| Central memory | 3.9 (3.1–5.1) | 3.0 (2.5–4.8) | 0.1 |
| Transitional memory | 4.0 (3.4–5.4) | 3.5 (2.3–4.9) | 0.14 |
| Effector memory | 5.8 (3.8–10.2) | 4.2 (3.3–5.1) | 0.09 |
| Ki67+ (% of CD8+ T cells) [median (IQR)] | |||
| Total | 3.7 (2.5–7.1) | 2.82 (2.3–3.9) | 0.1 |
| Naive | 2.4 (1.5–5.1) | 1.73 (1.0–2.6) | 0.13 |
| Central memory | 4.2 (2.8–8.9) | 3.1 (2.5–5.2) | 0.13 |
| Transitional memory | 2.7 (2.2–6.8) | 2.7 (1.9–4.4) | 0.2 |
| Effector memory | 4.5 (2.2–5.9) | 2.9 (2.2–4.0) | 0.35 |
| T cell activation | |||
| HLA-DR+ CD38+ (% of CD4+ T cells) [median (IQR)] | |||
| Total | 3.1 (2.4–7.4) | 2.5 (1.7–3.0) | <0.01 |
| Naive | 2.2 (1.2–3.5) | 1.7 (1.0–2.3) | 0.27 |
| Central memory | 2.4 (1.6–2.7) | 1.9 (1.1–2.7) | 0.21 |
| Effector memory | 5.8 (4.0–14.8) | 3.8 (2.5–5.6) | 0.02 |
| Terminally differentiated | 25.5 (9.2–33.1) | 13.2 (6.8–28.3) | 0.18 |
| HLA-DR+ CD38+ (% of CD8+ T cells) [median (IQR)] | |||
| Total | 20.3 (13.2–23.9) | 15.6 (8.3–21.8) | 0.25 |
| Naive | 10.3 (6.8–13.0) | 11.4 (4.0–17.1) | 0.71 |
| Central memory | 15.6 (11.0–21.0) | 13.3 (6.62–17.5) | 0.31 |
| Effector memory | 21.5 (12.1–24.5) | 15.7 (7.2–22.4) | 0.17 |
| Terminally differentiated | 25.1 (15.8–33.7) | 24.3 (10.8–31.6) | 0.57 |
Subsets were defined as naive (CD45RA+ CD27+ CCR7+), central memory (CD45RA− CD27+ CCR7+), transitional memory (CD45RA− CD27+ CCR7− for proliferation only), effector memory (CD45RA+/− CD27− CCR7−), and terminally differentiated (CD45RA+ CD27− for activation only) in CD4 and CD8 T cell subsets. Immunological data were available for a subset of 49 subjects.
Univariate P values were determined using the Mann-Whitney Wilcoxon test. P values of ≤0.1 are in boldface.
FIG 2.
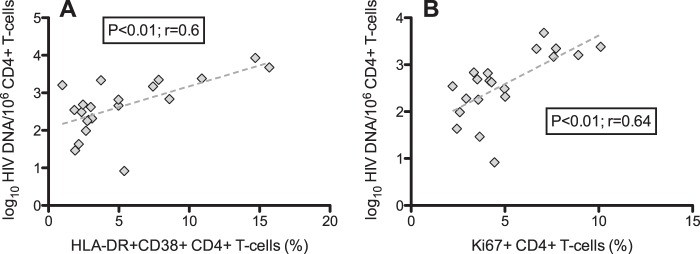
Correlative analysis between levels of log10 total HIV DNA per million CD4+ T cells and percentages of activated (HLA-DR+ CD38+) (A) and proliferating (Ki67+) (B) CD4+ T cells among patients with detectable CMV DNA in seminal plasma (n = 20). The P values were calculated using Pearson correlation analysis. Dashed line indicates the regression line.
(iii) Cellular HIV transcription (n = 42).
For a subset of 42 subjects with available PBMC aliquots and completely undetectable HIV RNA in blood plasma, we investigated if the presence of seminal CMV replication was associated with the frequency and levels of 2-LTR circles, cell-associated HIV RNA (unspliced and multiply spliced, encoding Tat-Rev), and increased average transcriptional activity (estimated as the cellular HIV RNA/HIV DNA ratio). Subjects with detectable CMV DNA in semen (n = 15) had higher levels of multiply spliced cell-associated HIV RNA (P = 0.017) and a trend toward increased average transcriptional activity (P = 0.068) compared to those with undetectable seminal CMV (n = 27). No association was observed between CMV shedding and unspliced HIV RNA (frequency or levels) or 2-LTR circles (Table 5).
TABLE 5.
Differences in cellular transcription between seminal CMV shedders and non-shedders
| Characteristica | Valueb |
Univariate P valuec | |
|---|---|---|---|
| Any CMV (n = 15) | No CMV (n = 27) | ||
| HIV RNA copies (us)/ng RNA/CD4 [median (IQR)] | 0.24 (0.09–0.83) | 0.14 (0.08–0.32) | 0.247 |
| HIV RNA copies (ms)/ng RNA/CD4 [median (IQR)] | 0.024 (0.0067–0.103) | 0.003 (0.0–0.03) | 0.017 |
| 2-LTR (copies/106 CD4+ T cells) [median (IQR)] | 0 (0–29.6) | 0 (0–19.7) | 0.983 |
| Any detectable (us) HIV RNA [n (%)] | 15 (100) | 25 (92.6) | 0.530 |
| Any detectable (ms) HIV RNA [n (%)] | 12 (80) | 15 (55.6) | 0.180 |
| Any detectable 2-LTR [n (%)] (total, 50) | 8 (38.1) | 13 (44.8) | 1.000 |
| HIV RNA (us)/HIV DNA ratio [median (IQR)] | 2.17 (1.16–5.63) | 1.57 (0.71–3.13) | 0.183 |
| HIV RNA (ms)/HIV DNA ratio [median (IQR)] | 0.31 (0.01–0.63) | 0 (0–0.42) | 0.068 |
Levels of unspliced (us) cellular HIV RNA normalized per ng CD4 T cells, levels of multiply spliced (ms) cellular HIV RNA encoding Tat-Rev normalized per ng CD4 T cells, and total log10 levels of 2-LTR normalized per million CD4+ T cells. The HIV RNA/HIV DNA ratio is the average number of HIV RNA transcripts (i.e., transcriptional activity) per HIV DNA copy. Cellular HIV RNA data were available for a subset of 42 subjects with undetectable HIV RNA levels in blood (<50 copies/ml).
Patients with HIV blips in blood were excluded from the analysis.
Univariate P values were determined using the Mann-Whitney Wilcoxon test; P values of <0.1 are in boldface.
DISCUSSION
The long-lived latent HIV reservoir that persists during suppressive ART is a major obstacle in achieving a cure (9). We previously demonstrated that asymptomatic CMV shedding in the genital tract by untreated HIV-infected men is associated with increased local and systemic T cell immune activation and higher levels of total HIV DNA in blood (21, 23). Understanding these relationships in the setting of ART is most relevant to the HIV cure research agenda. The present study in HIV-infected men whose infections were well suppressed with ART demonstrated that asymptomatic shedding of CMV in the male genital tract is associated with more than 5-fold higher mean levels of HIV provirus in peripheral CD4+ lymphocytes. We also found higher levels of multiply spliced HIV RNA transcripts in the cells of study participants with detectable CMV in semen. Although the current study design does not allow causality to be inferred, it does support the theory that CMV shedding in the male genital tract drives local and systemic immune activation with a subsequent increase in the HIV reservoir. Confirming this hypothesis will require a clinical trial of antiviral therapy aimed at stopping CMV shedding in the male genital tract as a way to reduce the HIV reservoir. The study presented here provides a rationale for such an interventional study.
The size of the latent HIV reservoir is relatively maintained throughout the life of an HIV-infected individual (9). There are two possible explanations for this persistence. First, residual or intermittent viral replication replenishes the pool of infected cells over time. Second, HIV-infected cells undergo homeostatic or antigen-driven proliferation. Of course, both these mechanisms could occur and interact to different degrees across individuals. This study demonstrated higher levels of multiply spliced HIV RNA transcripts in the cells of study participants with detectable CMV in semen but no difference in unspliced HIV RNA or 2-LTR circles. This discordant HIV RNA expression pattern (i.e., higher levels of multiply spliced than of unspliced HIV RNA) likely represents “blocked early-stage latency” as a form of latently HIV-infected cells (36, 37). We also found a trend for CMV shedders to have increased multiply spliced HIV RNA/HIV DNA ratios but no difference in unspliced HIV RNA/HIV DNA ratios. Altogether, these results suggest that the presence of asymptomatic CMV replication might be associated with some degree of cellular HIV transcription, but it remains unclear if this isolated increase in msRNA transcription is associated with ongoing infection.
Persistent immune activation is associated with multiple manifestations of end organ damage during HIV infection, including neurocognitive impairment, cancer, and cardiovascular disease (16, 22, 38, 39). This chronic activation may also be an important factor in maintaining the HIV reservoir (16). Along with HIV itself, CMV could be an important mediator of immune activation, especially because a large fraction of an infected person's immune repertoire is targeted toward CMV (40, 41), and CMV reactivation in the genital tract is common, even in subjects with high CD4+ T cell counts during ART (42). Similar to our previous reports on ART-naive HIV-infected men, we found that asymptomatic CMV replication in the male genital tract was associated with higher levels of systemic CD4+ T cell activation and proliferation and also with higher levels of systemic CD4+ T cell-associated provirus, even during suppressive ART. Also, we found a strong association between CD4+ proliferation and activation with HIV DNA in the CMV shedders but no association among CMV nonshedders. This observation supports a hypothesis that CMV shedding induces an expansion of CD4 T cells, including HIV-infected cells.
Several nonexclusive mechanisms could explain these observations. First, chronic CMV replication could cause antigen-driven proliferation of CMV-specific CD4 T cells in blood. However, this form of cellular activation would result in HIV RNA expression rather than increased latency (15), and previous studies have suggested that CMV-specific T cells may be less likely to be infected by HIV in untreated individuals (43, 44). Future studies should investigate the dynamics of CMV-specific T cells in relation to CMV replication during suppressive ART, as well as determine the proportion of integrated HIV DNA in the large population of CMV-specific T cells. Second, CMV-associated immune activation could induce a non-T cell receptor (TCR)-mediated homeostatic proliferation of (HIV-infected) T cells and consequently lead to an increase in the HIV latent reservoir. This scenario seems less likely, since CMV replication tends to occur in effector tissues rather than in inductive lymphoid tissues, where CD4+ T cells typically undergo homeostatic proliferation (13).
Third, we observed that CMV shedding was associated with increased levels of multiply spliced HIV RNA in blood as a possible correlate of ongoing residual HIV replication. Along this line, recent reports suggest that there may be low-level reinfection of new CD4+ T cells despite apparently suppressive ART in many individuals, particularly those on PI-based regimens (which comprised the majority of subjects in the current study) (10, 45). On the other hand, the presence of a significant association between CMV shedding and HIV DNA levels after controlling for 2-LTR circles in our multivariate analysis suggests that CMV is driving HIV persistence independently of its effect on supporting active HIV replication, perhaps through proliferation of infected CD4+ T cells. Another possibility is that increased immune activation in HIV-infected individuals causes both homeostatic proliferation of HIV-infected cells and CMV replication in semen. However, a recent randomized study demonstrated a reduction in immune activation in HIV-infected individuals treated with the anti-CMV drug valganciclovir (46), suggesting that CMV replication is a driver of immune activation during HIV infection, though the study was limited by the fact that only 70% of the subjects had HIV RNA suppression with ART and the sample size was small (n = 30). Finally, the observed difference in HIV DNA levels between CMV shedders versus nonshedders might have been present before ART initiation, but to evaluate this possibility, we would need a sample collected before the initiation of ART.
The results from this study differ from observations made in subjects who were not suppressed on ART. Unlike our previous report (23), we did not find any association between the presence of detectable CMV in PBMCs and levels of the HIV DNA reservoir. However, subjects with detectable CMV in PBMCs had higher levels of proviral HIV DNA than those without CMV DNA (2.53 versus 2.26 log10 per million CD4+ T cells), but this difference was not statistically significant, likely as a consequence of the low frequency of detectable CMV in PBMCs (22%) compared to semen (44%). Also, the biological significance of detectable CMV DNA at such low levels in PBMCs is unclear and might not be associated with active CMV replication during ART. Additionally, in this study, we found an association between higher CMV IgG levels and proviral HIV DNA in univariate analysis, which was not observed in our previous, larger cohort of ART-naive individuals (26). However, the association between CMV IgG levels and proviral HIV DNA lost significance in a multivariate analysis, after adjusting for the presence of seminal CMV DNA. The reasons for this observation are unclear and may be a result of the use of ART in the study cohort. The dynamics of CMV replication and immune response before and after suppressive ART are complex and deserve further investigation. It is also unclear why CMV replication has less effect on CD8 T cells than on CD4 T cells. Immune activation and T cell proliferation are driven by homeostatic and viral factors that differentially affect the CD4 and CD8 T cell pools (47). CD4 T cell proliferation is driven by the homeostatic response to CD4 T cell depletion and by antigen stimulation (such as HIV or CMV), whereas CD8 T cell proliferation is mainly a result of antigen stimulation (47). The lower CD4 T cell counts in CMV shedders might drive homeostatic forces (i.e., increased homeostatic cytokines, such as interleukin 7 [IL-7]) to replenish this pool and might explain the more profound effects observed in the CD4 T cell compartment than in CD8 T cells. However, we cannot exclude a direct effect of CMV replication in both CD4 and CD8 T cell immune activation. These relationships deserve further investigation.
This study had a number of limitations. First, it used a cross-sectional study design of a cohort of chronically infected individuals in whom the duration of infection was not known; however, age and time on ART were not associated with the size of the proviral HIV DNA reservoir. Second, the study assessed CMV reactivation only in men, and it is not clear if similar mechanisms are at play in women. Third, it is not clear if the observed increase in HIV DNA in CD4+ T cells represents replication-competent latent provirus. In addition, the relatively small sample size limited our power to observe significant associations in subsets of CD4+ and CD8+ T cells. Most importantly, as mentioned above, because this was a cross-sectional, observational study, we cannot establish a causal relationship between CMV reactivation and HIV DNA levels, and the extent of immune activation during treated HIV infection could be a determinant of CMV shedding and higher HIV DNA levels. Also, it is not clear if CMV replication in the male genital tract is a surrogate for lower-level replication systemically or if localized genital CMV shedding is driving inflammation systemically through increased traffic of activated T cells from the genital compartment to the blood. Despite these limitations, this study provides some insights regarding a possible connection between asymptomatic CMV replication and the latent HIV reservoir. Specifically, it demonstrates that even during suppressive ART, CMV reactivation in the male genital tract is associated with increased levels of T cell activation and proliferation and higher HIV DNA levels. This mechanism might be important, given that most HIV-infected individuals and almost all MSM are also infected with CMV (35). Future studies are needed to determine if persistent CMV replication could be targeted as a strategy to reduce the size of the latent HIV reservoir and reduce persistent immune activation.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to all the participants in the California Collaborative Treatment Group (CCTG) and to the CFAR Genomic, Translational Virology, and Flow Cytometry Cores. We acknowledge all the nurses at all the enrollment sites and Matt Strain and Julian Blanco for very helpful discussions. The HIV RNA quantification standard was obtained through the NIH AIDS Research and Reference Reagent Program, DAIDS, NIAID, and the HIV VQA RNA Quantification Standard from the DAIDS Virology Quality Assurance Program. The primer and probe for quantification of herpesviruses, as well as the plasmids and quantification standards, were kindly provided by Fred Lakeman and Rich Whitley.
S.G. participated in the study design, performed the laboratory experiments, participated in the data analyses for the study, and wrote the primary version of the manuscript; M.M. participated in the study design, designed laboratory experiments, performed all the analysis of the immunological data, and revised the manuscript; D.M.S. participated in the study design, participated in the data analyses, and wrote the revised manuscript; M.V.V. performed the laboratory experiments; S.R.M. participated in study design, performed statistical analysis, and wrote the primary version of the manuscript; C.A.S. participated in the study design and data analysis and revised the manuscript; D.D.R., R.H.H., E.S.D., and M.P.D. participated in study design and revised the manuscript, S.R.M., E.S.D., M.P.D., and R.H.H. designed the clinical trial, enrolled participants, and collected and archived the samples. We all read and approved the final manuscript.
S.G., M.M., S.J.L., and M.V.V. do not have any commercial or other associations that might pose a conflict of interest. D.D.R. has served as a consultant for Chimerix, BMS, Gilead, Gen-Probe, Monogram, Sirenas, and Prism. D.M.S. has received grant support from ViiV Pharmaceuticals and consultant fees from Gen-Probe and Testing Talent Services. M.P.D. has received grant support from Merck, Gilead, and ViiV and has served as a consultant to Astra Zeneca. E.S.D. has received grant support from Abbott, Gilead, Merck, Pfizer. and ViiV and has acted as a consultant for Bristol Myers Squibb, Gilead, Merck. and ViiV. R.H.H. reports having received honoraria or consultant fees from BMS, Gilead Sciences. and Janssen and research support (to UCSD) from Abbott, GlaxoSmithKline, Pfizer, and Merck.
This work was supported by the Department of Veterans Affairs; the James Pendleton Charitable Trust; amfAR grant 108537 with support from FAIR; the U.S. National Institutes of Health (NIH) awards 7UM1AI068636-07, AI090970 (IAVI), AI69432, AI096113 (CARE), AI043638, MH62512, MH083552, AI100665, AI077304, AI36214, AI047745, AI74621, GM093939AI080353, AI306214 (CFAR), AI27670 (ACTU), and AI064086 (K24 to R.H.H.), and AI43638; a grant from the National Institutes of Health, University of California, San Francisco-Gladstone Institute of Virology and Immunology Center for AIDS Research (P30-AI027763); the California HIV/AIDS Research Program (RN07-SD-702, MC08-SD-700, and EI-11-SD-005); the National Center for Advancing Translational Sciences through UCLA CTSI grant UL1TR000124 and the National Institute of General Medical Sciences grant GM093939; and an Interdisciplicinary Research Fellowship in NeuroAIDS (R25-MH081482). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Published ahead of print 30 April 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.00831-14.
REFERENCES
- 1.Opravil M, Cone RW, Fischer M, Vernazza PL, Bassetti S, Lorenzi P, Bisset LR, Ott P, Huber W, Knuchel MC, Roos M, Luthy R, Weber R. 2000. Effects of early antiretroviral treatment on HIV-1 RNA in blood and lymphoid tissue: a randomized trial of double versus triple therapy. Swiss HIV Cohort Study. J. Acquir. Immune Defic. Syndr. 23:17–25. 10.1097/00126334-200001010-00003 [DOI] [PubMed] [Google Scholar]
- 2.Gulick RM, Mellors JW, Havlir D, Eron JJ, Gonzalez C, McMahon D, Richman DD, Valentine FT, Jonas L, Meibohm A, Emini EA, Chodakewitz JA. 1997. Treatment with indinavir, zidovudine, and lamivudine in adults with human immunodeficiency virus infection and prior antiretroviral therapy. N. Engl. J. Med. 337:734–739. 10.1056/NEJM199709113371102 [DOI] [PubMed] [Google Scholar]
- 3.Wong JK, Hezareh M, Gunthard HF, Havlir DV, Ignacio CC, Spina CA, Richman DD. 1997. Recovery of replication-competent HIV despite prolonged suppression of plasma viremia. Science 278:1291–1295. 10.1126/science.278.5341.1291 [DOI] [PubMed] [Google Scholar]
- 4.Strain MC, Little SJ, Daar ES, Havlir DV, Gunthard HF, Lam RY, Daly OA, Nguyen J, Ignacio CC, Spina CA, Richman DD, Wong JK. 2005. Effect of treatment, during primary infection, on establishment and clearance of cellular reservoirs of HIV-1. J. Infect. Dis. 191:1410–1418. 10.1086/428777 [DOI] [PubMed] [Google Scholar]
- 5.Chun TW, Engel D, Berrey MM, Shea T, Corey L, Fauci AS. 1998. Early establishment of a pool of latently infected, resting CD4(+) T cells during primary HIV-1 infection. Proc. Natl. Acad. Sci. U. S. A. 95:8869–8873. 10.1073/pnas.95.15.8869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Finzi D, Blankson J, Siliciano JD, Margolick JB, Chadwick K, Pierson T, Smith K, Lisziewicz J, Lori F, Flexner C, Quinn TC, Chaisson RE, Rosenberg E, Walker B, Gange S, Gallant J, Siliciano RF. 1999. Latent infection of CD4+ T cells provides a mechanism for lifelong persistence of HIV-1, even in patients on effective combination therapy. Nat. Med. 5:512–517. 10.1038/8394 [DOI] [PubMed] [Google Scholar]
- 7.Ramratnam B, Mittler JE, Zhang L, Boden D, Hurley A, Fang F, Macken CA, Perelson AS, Markowitz M, Ho DD. 2000. The decay of the latent reservoir of replication-competent HIV-1 is inversely correlated with the extent of residual viral replication during prolonged anti-retroviral therapy. Nat. Med. 6:82–85. 10.1038/71577 [DOI] [PubMed] [Google Scholar]
- 8.Strain MC, Gunthard HF, Havlir DV, Ignacio CC, Smith DM, Leigh-Brown AJ, Macaranas TR, Lam RY, Daly OA, Fischer M, Opravil M, Levine H, Bacheler L, Spina CA, Richman DD, Wong JK. 2003. Heterogeneous clearance rates of long-lived lymphocytes infected with HIV: intrinsic stability predicts lifelong persistence. Proc. Natl. Acad. Sci. U. S. A. 100:4819–4824. 10.1073/pnas.0736332100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Richman DD, Margolis DM, Delaney M, Greene WC, Hazuda D, Pomerantz RJ. 2009. The challenge of finding a cure for HIV infection. Science 323:1304–1307. 10.1126/science.1165706 [DOI] [PubMed] [Google Scholar]
- 10.Buzon MJ, Massanella M, Llibre JM, Esteve A, Dahl V, Puertas MC, Gatell JM, Domingo P, Paredes R, Sharkey M, Palmer S, Stevenson M, Clotet B, Blanco J, Martinez-Picado J. 2010. HIV-1 replication and immune dynamics are affected by raltegravir intensification of HAART-suppressed subjects. Nat. Med. 16:460–465. 10.1038/nm.2111 [DOI] [PubMed] [Google Scholar]
- 11.Chun TW, Nickle DC, Justement JS, Large D, Semerjian A, Curlin ME, O'Shea MA, Hallahan CW, Daucher M, Ward DJ, Moir S, Mullins JI, Kovacs C, Fauci AS. 2005. HIV-infected individuals receiving effective antiviral therapy for extended periods of time continually replenish their viral reservoir. J. Clin. Invest. 115:3250–3255. 10.1172/JCI26197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mahnke YD, Greenwald JH, DerSimonian R, Roby G, Antonelli LR, Sher A, Roederer M, Sereti I. 2012. Selective expansion of polyfunctional pathogen-specific CD4(+) T cells in HIV-1-infected patients with immune reconstitution inflammatory syndrome. Blood 119:3105–3112. 10.1182/blood-2011-09-380840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chomont N, El-Far M, Ancuta P, Trautmann L, Procopio FA, Yassine-Diab B, Boucher G, Boulassel MR, Ghattas G, Brenchley JM, Schacker TW, Hill BJ, Douek DC, Routy JP, Haddad EK, Sekaly RP. 2009. HIV reservoir size and persistence are driven by T cell survival and homeostatic proliferation. Nat. Med. 15:893–900. 10.1038/nm.1972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Trono D, Van Lint C, Rouzioux C, Verdin E, Barre-Sinoussi F, Chun TW, Chomont N. 2010. HIV persistence and the prospect of long-term drug-free remissions for HIV-infected individuals. Science 329:174–180. 10.1126/science.1191047 [DOI] [PubMed] [Google Scholar]
- 15.Bosque A, Famiglietti M, Weyrich AS, Goulston C, Planelles V. 2011. Homeostatic proliferation fails to efficiently reactivate HIV-1 latently infected central memory CD4+ T cells. PLoS Pathog. 7:e1002288. 10.1371/journal.ppat.1002288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hatano H. 2013. Immune activation and HIV persistence: considerations for novel therapeutic interventions. Curr. Opin. HIV AIDS 8:211–216. 10.1097/COH.0b013e32835f9788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chun TW, Murray D, Justement JS, Hallahan CW, Moir S, Kovacs C, Fauci AS. 2011. Relationship between residual plasma viremia and the size of HIV proviral DNA reservoirs in infected individuals receiving effective antiretroviral therapy. J. Infect. Dis. 204:135–138. 10.1093/infdis/jir208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chun TW, Nickle DC, Justement JS, Meyers JH, Roby G, Hallahan CW, Kottilil S, Moir S, Mican JM, Mullins JI, Ward DJ, Kovacs JA, Mannon PJ, Fauci AS. 2008. Persistence of HIV in gut-associated lymphoid tissue despite long-term antiretroviral therapy. J. Infect. Dis. 197:714–720. 10.1086/527324 [DOI] [PubMed] [Google Scholar]
- 19.Day CL, Kaufmann DE, Kiepiela P, Brown JA, Moodley ES, Reddy S, Mackey EW, Miller JD, Leslie AJ, DePierres C, Mncube Z, Duraiswamy J, Zhu B, Eichbaum Q, Altfeld M, Wherry EJ, Coovadia HM, Goulder PJ, Klenerman P, Ahmed R, Freeman GJ, Walker BD. 2006. PD-1 expression on HIV-specific T cells is associated with T-cell exhaustion and disease progression. Nature 443:350–354. 10.1038/nature05115 [DOI] [PubMed] [Google Scholar]
- 20.Trautmann L, Janbazian L, Chomont N, Said EA, Gimmig S, Bessette B, Boulassel MR, Delwart E, Sepulveda H, Balderas RS, Routy JP, Haddad EK, Sekaly RP. 2006. Upregulation of PD-1 expression on HIV-specific CD8+ T cells leads to reversible immune dysfunction. Nat. Med. 12:1198–1202. 10.1038/nm1482 [DOI] [PubMed] [Google Scholar]
- 21.Gianella S, Strain MC, Rought SE, Vargas MV, Little SJ, Richman DD, Spina CA, Smith DM. 2012. Associations between virologic and immunologic dynamics in blood and in the male genital tract. J. Virol. 86:1307–1315. 10.1128/JVI.06077-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hunt PW. 2012. HIV and inflammation: mechanisms and consequences. Curr. HIV/AIDS Rep. 9:139–147. 10.1007/s11904-012-0118-8 [DOI] [PubMed] [Google Scholar]
- 23.Gianella S, Anderson CM, Vargas MV, Richman DD, Little SJ, Morris SR, Smith DM. 2013. CMV DNA in semen and blood is associated with higher levels of proviral HIV DNA. J. Infect. Dis. 207:898–902. 10.1093/infdis/jis777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Koelsch KK, Liu L, Haubrich R, May S, Havlir D, Gunthard HF, Ignacio CC, Campos-Soto P, Little SJ, Shafer R. 2008. Dynamics of total, linear nonintegrated, and integrated HIV-1 DNA in vivo and in vitro. J. Infect. Dis. 197:411. 10.1086/525283 [DOI] [PubMed] [Google Scholar]
- 25.Gianella S, Smith DM, Vargas MV, Little SJ, Richman DD, Daar ES, Dube MP, Zhang F, Ginocchio CC, Haubrich RH, Morris SR, CCTG 592 Team. 2013. Shedding of HIV and human herpesviruses in the semen of effectively treated HIV-1 infected men who have sex with men. Clin. Infect. Dis. 57:441–447. 10.1093/cid/cit252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gianella S, Morris SR, Tatro E, Vargas MV, Haubrich RH, Daar ES, Dube MP, Richman DD, Little SJ, Smith DM. 2014. Virologic correlates of anti-CMV IgG levels in HIV-1 infected men. J. Infect. Dis. 209:452–456. 10.1093/infdis/jit434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Butler DM, Delport W, Kosakovsky Pond SL, Lakdawala MK, Cheng PM, Little SJ, Richman DD, Smith DM. 2010. The origins of sexually transmitted HIV among men who have sex with men. Sci. Transl. Med. 2:18re11. 10.1126/scitranslmed.3000447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gianella S, Strain MC, Rought SE, Vargas M, Little SJ, Richman DD, Spina CA, Smith DM. 2012. Associations between the virologic and immunologic dynamics in blood and in the male genital tract. J. Virol. 86:1307–1315. 10.1128/JVI.06077-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Strain MC, Lada SM, Luong T, Rought SE, Gianella S, Terry VH, Spina CA, Woelk CH, Richman DD. 2013. Highly precise measurement of HIV DNA by droplet digital PCR. PLoS One 8:e55943. 10.1371/journal.pone.0055943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mulder J, McKinney N, Christopherson C, Sninsky J, Greenfield L, Kwok S. 1994. Rapid and simple PCR assay for quantitation of human immunodeficiency virus type 1 RNA in plasma: application to acute retroviral infection. J. Clin. Microbiol. 32:292–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schmid A, Gianella S, von Wyl V, Metzner KJ, Scherrer AU, Niederöst B, Althaus CF, Rieder P, Grube C, Joos B, Weber R, Fischer M, Günthard HF. 2010. Profound depletion of HIV-1 transcriptionally active PBMC by early cART during primary HIV-1 infection but not by treatment during chronic infection: results of the Zurich Primary HIV Infection Study. PLoS One 5:e13310. 10.1371/journal.pone.0013310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Parrinello CM, Sinclair E, Landay AL, Lurain N, Sharrett AR, Gange SJ, Xue X, Hunt PW, Deeks SG, Hodis HN, Kaplan RC. 2012. Cytomegalovirus immunoglobulin G antibody is associated with subclinical carotid artery disease among HIV-infected women. J. Infect. Dis. 205:1788–1796. 10.1093/infdis/jis276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kaiser P, Joos B, Niederöst B, Weber R, Günthard HF, Fischer M. 2007. Productive human immunodeficiency virus type 1 infection in peripheral blood predominantly takes place in CD4/CD8 double-negative T lymphocytes? J. Virol. 81:9693–9706. 10.1128/JVI.00492-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Boulassel MR, Chomont N, Pai NP, Gilmore N, Sekaly RP, Routy JP. 2012. CD4 T cell nadir independently predicts the magnitude of the HIV reservoir after prolonged suppressive antiretroviral therapy. J. Clin. Virol. 53:29–32. 10.1016/j.jcv.2011.09.018 [DOI] [PubMed] [Google Scholar]
- 35.Gianella S, Morris SR, Anderson C, Spina CA, Vargas MV, Young JA, Richman DD, Little SJ, Smith DM. 2013. Herpesviruses and HIV-1 drug resistance mutations influence the virologic and immunologic milieu of the male genital tract. AIDS 27:39–47. 10.1097/QAD.0b013e3283573305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Seshamma T, Bagasra O, Trono D, Baltimore D, Pomerantz RJ. 1992. Blocked early-stage latency in the peripheral blood cells of certain individuals infected with human immunodeficiency virus type 1. Proc. Natl. Acad. Sci. U. S. A. 89:10663–10667. 10.1073/pnas.89.22.10663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pasternak AO, Lukashov VV, Berkhout B. 2013. Cell-associated HIV RNA: a dynamic biomarker of viral persistence. Retrovirology 10:41. 10.1186/1742-4690-10-41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hsue PY, Deeks SG, Hunt PW. 2012. Immunologic basis of cardiovascular disease in HIV-infected adults. J. Infect. Dis. 205(Suppl. 3):S375–S382. 10.1093/infdis/jis200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gartner S, Liu Y. 2002. Insights into the role of immune activation in HIV neuropathogenesis. J. Neurovirol. 8:69–75. 10.1080/13550280290049525 [DOI] [PubMed] [Google Scholar]
- 40.Waldrop SL, Pitcher CJ, Peterson DM, Maino VC, Picker LJ. 1997. Determination of antigen-specific memory/effector CD4+ T cell frequencies by flow cytometry: evidence for a novel, antigen-specific homeostatic mechanism in HIV-associated immunodeficiency. J. Clin. Invest. 99:1739–1750. 10.1172/JCI119338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sylwester AW, Mitchell BL, Edgar JB, Taormina C, Pelte C, Ruchti F, Sleath PR, Grabstein KH, Hosken NA, Kern F, Nelson JA, Picker LJ. 2005. Broadly targeted human cytomegalovirus-specific CD4+ and CD8+ T cells dominate the memory compartments of exposed subjects. J. Exp. Med. 202:673–685. 10.1084/jem.20050882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gianella S, Smith DM, Vargas MV, Little SJ, Richman DD, Daar ES, Dube MP, Zhang F, Ginocchio CC, Haubrich RH, Morris SR, CCTG 592 Team. 2013. Shedding of HIV and human herpesviruses in the semen of effectively treated HIV-1-infected men who have sex with men. Clin. Infect. Dis. 57:441–447. 10.1093/cid/cit252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Casazza JP, Brenchley JM, Hill BJ, Ayana R, Ambrozak D, Roederer M, Douek DC, Betts MR, Koup RA. 2009. Autocrine production of beta-chemokines protects CMV-Specific CD4 T cells from HIV infection. PLoS Pathog. 5:e1000646. 10.1371/journal.ppat.1000646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Douek DC, Brenchley JM, Betts MR, Ambrozak DR, Hill BJ, Okamoto Y, Casazza JP, Kuruppu J, Kunstman K, Wolinsky S, Grossman Z, Dybul M, Oxenius A, Price DA, Connors M, Koup RA. 2002. HIV preferentially infects HIV-specific CD4+ T cells. Nature 417:95–98. 10.1038/417095a [DOI] [PubMed] [Google Scholar]
- 45.Hatano H, Strain MC, Scherzer R, Bacchetti P, Wentworth D, Hoh R, Martin JN, McCune JM, Neaton JD, Tracy RP, Hsue PY, Richman DD, Deeks SG. 2013. Increase in 2-long terminal repeat circles and decrease in D-dimer after raltegravir intensification in patients with treated HIV infection: a randomized, placebo-controlled trial. J. Infect. Dis. 208:1436–1442. 10.1093/infdis/jit453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hunt PW, Martin JN, Sinclair E, Epling L, Teague J, Jacobson MA, Tracy RP, Corey L, Deeks SG. 2011. Valganciclovir reduces T cell activation in HIV-infected individuals with incomplete CD4+ T cell recovery on antiretroviral therapy. J. Infect. Dis. 203:1474–1483. 10.1093/infdis/jir060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Catalfamo M, Di Mascio M, Hu Z, Srinivasula S, Thaker V, Adelsberger J, Rupert A, Baseler M, Tagaya Y, Roby G, Rehm C, Follmann D, Lane HC. 2008. HIV infection-associated immune activation occurs by two distinct pathways that differentially affect CD4 and CD8 T cells. Proc. Natl. Acad. Sci. U. S. A. 105:19851–19856. 10.1073/pnas.0810032105 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.