SUMMARY
The GroEL/ES chaperonin system functions as a protein folding cage. Many obligate substrates of GroEL share the (βα)8 TIM-barrel fold, but how the chaperonin promotes folding of these proteins is not known. Here we analyzed the folding of DapA at peptide resolution using hydrogen/deuterium exchange and mass spectrometry. During spontaneous folding, all elements of the DapA TIM-barrel acquire structure simultaneously, in a process associated with a long search time. In contrast, GroEL/ES accelerates folding more than 30-fold by catalyzing segmental structure formation in the TIM-barrel. Segmental structure formation is also observed during the fast spontaneous folding of a structural homolog of DapA from a bacterium that lacks GroEL/ES. Thus, chaperonin-independence correlates with folding properties otherwise enforced by protein confinement in the GroEL/ES cage. We suggest that folding catalysis by GroEL/ES is required by a set of proteins to reach native state at a biologically relevant time-scale, avoiding aggregation or degradation.
INTRODUCTION
The chaperonins form nano-cages for single protein molecules to fold in isolation and are essential components of the protein folding machinery in bacteria, archaea and eukaryotic cells. The most widely studied chaperonin is GroEL and its co-factor GroES of E. coli (Kim et al., 2013; Saibil et al., 2013). GroEL receives its substrates from Trigger factor and the Hsp70 system (DnaK/DnaJ/GrpE), which chaperone a wide range of nascent polypeptides emerging from ribosomes (Calloni et al., 2012; Oh et al., 2011). The substrate interactome of GroEL comprises ~250 different proteins, including 50-80 proteins with an obligate GroEL requirement for folding (Fujiwara et al., 2010; Kerner et al., 2005). Approximately 30-50 % of these share the (βα)8 triose-phosphate isomerase (TIM)-barrel fold, a domain topology characterized by many long-range interactions (Fujiwara et al., 2010; Kerner et al., 2005). Why these proteins are GroEL-dependent and how exactly the chaperonin system promotes their folding is still unresolved (Azia et al., 2012; Gershenson and Gierasch, 2011; Jewett and Shea, 2010). One model suggests that the chaperonin cage acts solely by allowing folding to occur unimpaired by aggregation (passive-cage model) (Apetri and Horwich, 2008; Horwich et al., 2009). Another model posits that, in addition, encapsulation in the chaperonin cage may accelerate folding kinetics through confinement (active-cage model) (Brinker et al., 2001; Chakraborty et al., 2010; Tang et al., 2006) (Figure 1A). While multiple studies demonstrate the functional significance of the GroEL/ES-cage in vitro and in vivo (Brinker et al., 2001; Chen et al., 2013; Clare et al., 2012; Kerner et al., 2005; Martin and Hartl, 1997; Tang et al., 2008; Tang et al., 2006), a third model suggests that the function of GroEL/ES is to unfold misfolded states through iterative binding cycles, with subsequent folding occurring either inside the cage or in free solution (iterative-annealing model) (Thirumalai and Lorimer, 2001; Yang et al., 2013).
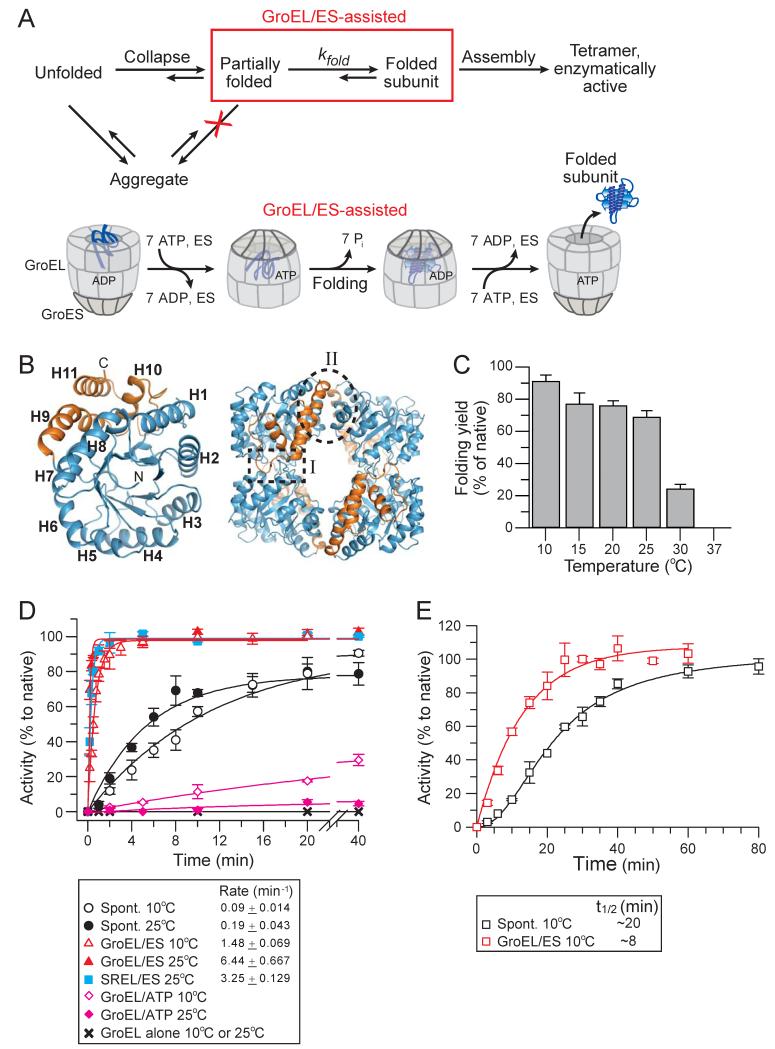
Figure 1. Spontaneous and Chaperonin-assisted Refolding of DapA.
(A) Schematic representation of DapA refolding/assembly. Top panel: Spontaneous refolding/assembly, highlighting the steps modulated by GroEL/ES in preventing off-pathway aggregation (red cross) and/or accelerating subunit folding (red box). Bottom panel: Model of the GroEL/ES mechanism of assisted refolding.
(B) Structure of E. coli DapA monomer (left) and tetramer (right) in ribbon representation (PDB 1YXC). The (βα)8 TIM-barrel domain is shown in blue and the C-terminal domain in gold. Helices H1 to H11 and locations of the strong and weak interfaces of the tetramer (interface I and II, respectively) are indicated.
(C) Yield of spontaneous DapA refolding at 10°C-37°C. Refolding was initiated by diluting GuHCl-denatured DapA into refolding buffer B to a final concentration of 200 nM monomer and yields analyzed by enzyme assay after 1.5 h (15°C-37°C) and after 16 h (10°C). Folding yields are plotted as DapA activities in % of native enzyme control incubated at the respective temperature.
(D) Rates of spontaneous and chaperonin-assisted DapA subunit folding at 10°C and 25°C. Refolding was measured at 200 nM DapA by diluting the denatured protein into refolding buffer as in (C) or into buffer containing 2 μM GroEL or SREL and 4 μM GroES. Assisted refolding was initiated by addition of ATP. In the case of SREL, urea-denatured DapA and low salt refolding buffer C was used (see Experimental Procedures). Spontaneous refolding was stopped by addition of 0.8 μM GroEL D87K and GroEL/ES-assisted refolding with 50 mM CDTA. In the case of SREL/ES, 50 mM CDTA and 60 mM GuHCl were added to stop folding. Reactions were incubated for 1 h at 25°C to allow for complete assembly prior to enzyme assay. Single exponential rates are indicated.
(E) Spontaneous and assisted assembly of DapA. Refolding reactions were performed as in (D) at 10°C with 2 μM DapA and 4 μM GroEL/8 μM GroES when indicated. The reactions were not stopped and enzyme activities were measured at the time points indicated. All standard deviations are from at least 3 independent experiments.
GroEL is an ATP-driven macromolecular machine of ~800 KDa consisting of two, heptameric rings of ~57 kDa subunits stacked back-to-back (Kim et al., 2013; Saibil et al., 2013). The apical domains of the GroEL subunits, forming the ring opening, expose hydrophobic amino acid residues for the binding of molten globule-like folding intermediates. Upon ATP binding to GroEL, GroES, a heptameric ring of ~10 kDa subunits, caps the GroEL ring that holds the substrate, resulting in its displacement into an enclosed cage large enough for proteins up to ~60 kDa (Figure 1A). This step is accompanied by a dramatic conformational change in GroEL that renders the inner lining of the cage hydrophilic and net-negatively charged. The enclosed protein is then free to fold during the time required for the hydrolysis of 7 ATP molecules in the GroEL ring. Subsequent ATP binding to the opposite ring causes the dissociation of GroES and substrate release. Not-yet folded protein is rapidly recaptured by GroEL for another folding attempt.
Here we present evidence that encapsulation in the GroEL/ES-cage modulates the mechanism of protein folding, as demonstrated with E. coli dihydrodipicolinate synthase (DapA), a GroEL-dependent TIM-barrel protein (Kerner et al., 2005; McLennan and Masters, 1998). Spontaneous and assisted folding was analyzed by hydrogen/deuterium exchange (H/DX) and mass spectrometry (MS) at peptide resolution. The slow spontaneous folding of DapA initiates from an ensemble of largely unstructured intermediates in a highly cooperative manner, with nearly all structural elements of the TIM-barrel acquiring H/DX protection simultaneously. Accordingly, this process is associated with a long search time and a significant entropic penalty. Strikingly, GroEL/ES accelerates DapA folding more than 30-fold, with the confining environment of the chaperonin cage lowering the entropic component of the activation barrier by promoting segmental structure formation in the TIM-barrel. Segmental structure formation is also observed during the fast spontaneous folding of MsNanA (N-acetylneuraminic acid aldolase), a close structural homolog of DapA from a bacterium that lacks GroEL/ES. Thus, chaperonin-independence of MsNanA correlates with folding properties that are enforced in the chaperonin-dependent homolog by confinement in the GroEL/ES cage.
RESULTS
Accelerated Folding of DapA by GroEL/ES
DapA is an essential tetrameric enzyme that catalyzes the condensation of L-aspartate-β-semialdehyde and pyruvate to dihydrodipicolinic acid, a metabolite required for lysine and peptidoglycan biosynthesis. The monomer (31.2 kDa) consists of a N-terminal (βα)8 TIM-barrel domain of 224 amino acids, containing the catalytic site, and a 68 amino acid C-terminal domain of three α-helices that contributes to the strong and weak interfaces of the tetramer (interface I and II, respectively) (Dobson et al., 2005) (Figure 1B). Formation of enzymatically active DapA involves subunit folding, followed by assembly to dimer and tetramer (Reboul et al., 2012). Upon dilution from guanidine-HCl (GuHCl), DapA refolded and assembled efficiently between 10°C and 25°C but aggregated at higher temperature (Figure 1C). To measure subunit folding independent of assembly, refolding reactions were stopped by addition of GroEL to trap not-yet folded subunits, followed by incubation for ~1 h prior to enzyme assay to allow for completion of assembly. The rate of spontaneous folding at 25°C was ~0.2 min−1 (t1/2 ~3.6 min) with a yield of ~75 % (Figures 1C and 1D). Spontaneous refolding was apparently not limited by prolyl-isomerization (DapA contains 2 cis-prolines in the C-terminal domain): rapid unfolding on ice to maintain the prolines in their native configuration (Schmid, 1986) did not accelerate refolding (data not shown). Strikingly, GroEL/ES accelerated the folding reaction ~30 fold to a rate of ~6.0 min−1 (t1/2 ~7 s) at 25°C and ~16-fold to a rate of ~1.5 min−1 (t1/2 ~28 s) at 10°C, with ~100 % yield (Figure 1D). Assembly of active tetramer, as measured by immediate enzyme assay during refolding, was ~2.5-fold accelerated by GroEL/ES (10°C; 2 μM DapA) (Figure 1E). DapA tetramer proved to be kinetically stable with a slow rate of spontaneous unfolding (t1/2 ~29 days at 10°C) (data not shown). Notably, the Hsp70 chaperone system (DnaK/DnaJ/GrpE) allowed only very slow refolding (t1/2 >1 h), even at 37°C, but efficiently maintained DapA in a non-aggregated state for transfer to GroEL/ES and completion of folding within seconds (Figure S1A). Thus, the ability to accelerate DapA folding appears to be unique to GroEL/ES.
To test whether accelerated DapA folding occurs inside the GroEL/ES cage, we used the single-ring mutant of GroEL (SREL) that allows only one round of ATP hydrolysis upon GroES binding, resulting in a SREL/ES complex with substrate encapsulated (Weissman et al., 1996). Refolding was performed with urea-denatured DapA, as the SREL/ES complex is sensitive to salt and GuHCl. We confirmed that DapA bound to SREL was stably encapsulated upon addition of ATP and GroES (Figures S1B and S1C). The folding reaction was stopped by adding magnesium chelator and 60 mM GuHCl which results in dissociation of GroES and capture of not-yet folded DapA by SREL. DapA folded at a rate of ~3.3 min−1 at 25°C (t1/2 ~13 s), i.e. ~17-fold faster than spontaneous folding (Figure 1D). Thus, a single-round of encapsulation in the GroEL/ES cage is sufficient to achieve accelerated DapA folding.
Catalysis of DapA Folding
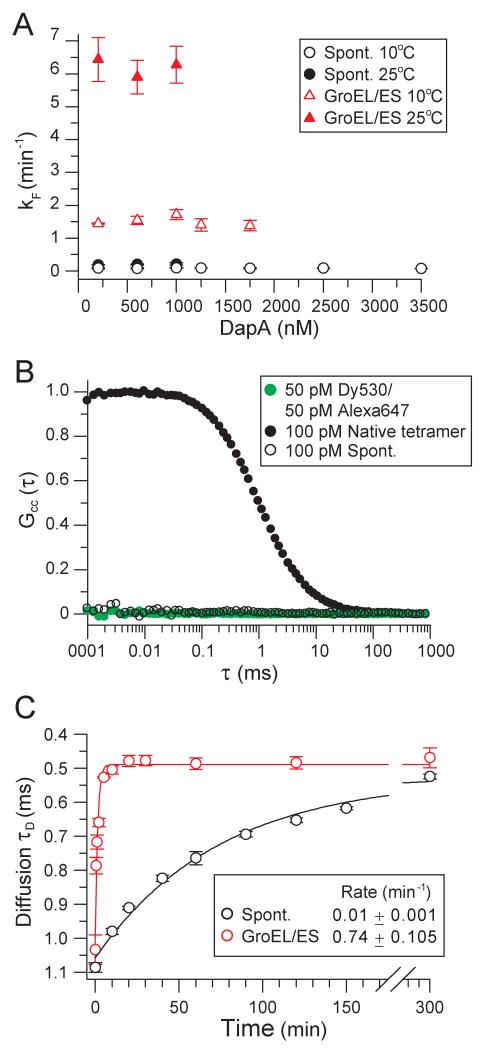
Does the acceleration of folding by GroEL/ES reflect a catalytic role of the chaperonin or merely a consequence of aggregation prevention (Figure 1A)? To test whether spontaneous DapA folding was limited by aggregation, we measured folding over a wide concentration range. The rate of spontaneous folding was concentration independent at 10°C and 25°C (Figures 2A and S2A). Partial aggregation at high concentrations resulted in a reduction in yield but did not slow the apparent folding rate (Figures S2A and S2B), indicating that aggregation was irreversible.
Figure 2. GroEL/ES Catalyzes DapA Refolding.
(A) Rates of spontaneous and GroEL/ES-assisted refolding are concentration independent. Spontaneous and GroEL/ES-assisted subunit refolding was performed at 10°C and 25°C as in Figure 1D over a range of DapA concentrations.
(B) Absence of inter-molecular association during refolding by FCCS. DapA-293C was labeled with either Dy530 or Alexa647 fluorophores. The labeled proteins were mixed, denatured and diluted into refolding buffer to a final concentration of 50 pM each. FCCS was recorded within the first 30 min of refolding. As a positive control, DapA-293C tetramer refolded and assembled from Dy530 and Alexa647 labeled subunits at 200 nM concentration was diluted to 100 pM. A 1:1 mixture of the free dyes at 50 pM each was used as a negative control.
(C) Spontaneous and assisted refolding of DapA-293C labeled with Alexa647 was measured at a final concentration of 100 pM at 20°C. GroEL and GroES, when present, were 2 μM and 4 μM, respectively. Refolding was stopped either by addition of 2 μM GroEL (spontaneous refolding) or by addition of Apyrase (assisted refolding). The difference in diffusion rate between not-yet folded DapA bound to GroEL and folded DapA monomer free in solution was monitored by FCS, resulting in rates of subunit refolding. All standard deviations are from at least 3 independent experiments.
To further rule out aggregation as the cause of slow spontaneous folding, we used fluorescence cross-correlation spectroscopy (FCCS) to analyze refolding reactions at very low DapA concentration (100 pM), where intermolecular association is excluded (Mukhopadhyay et al., 2007). A mutant of DapA was designed in which the surface exposed cysteines (C20, C141, C218) were changed to serine and a cysteine was added to the C-terminus (DapA-293C). DapA-293C was labeled with Alexa647 or Dy530 and the proteins mixed at equimolar amounts, denatured and allowed to refold at 100 pM total. No FCCS signal was observed during refolding, indicating the absence of intermolecular association (Figure 2B). In contrast, cross-correlation was observed for 100 pM native tetramer when the labeled proteins were first allowed to refold and assemble at 200 nM (Figure 2B). Using fluorescence correlation spectroscopy (FCS), we next measured the diffusion coefficient of spontaneously refolded DapA-293C-Alexa monomer (~102 ± 2 μm2s−1) and of GroEL-bound DapA-293C-Alexa (~49 ± 2 μm2s−1) (Figure S2C). Taking advantage of the slower diffusion speed of the GroEL-DapA complex and the ability of GroEL to trap not-yet folded DapA molecules, we monitored the time dependent decrease in the average diffusion time of DapA-293C-Alexa and extracted a refolding rate of 0.01 ± 0.001 min−1 (Figure 2C). Note that DapA-293C-Alexa is enzymatically active but its spontaneous refolding rate is ~13-times slower than that of wild-type DapA. Importantly, GroEL/ES accelerated the folding of DapA-293C-Alexa (100 pM) more than ~50-fold at 20°C (Figure 2C). These data demonstrate that GroEL/ES functions as a highly efficient catalyst of DapA folding under conditions where subunit association (due to aggregation or productive assembly) is excluded.
Chaperonin-Dependent and -Independent TIM-barrel Proteins
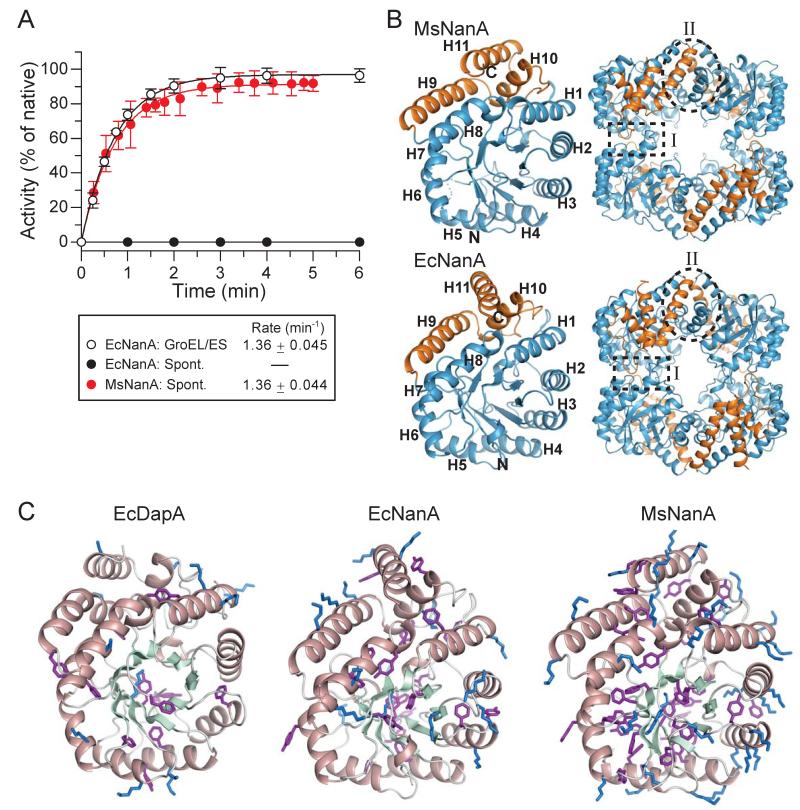
Why are only some TIM-barrel proteins chaperonin-dependent (Azia et al., 2012; Kerner et al., 2005)? To obtain insight into this question we compared the spontaneous folding of DapA and its close structural homolog N-acetylneuraminic acid aldolase (NanA) from M. synoviae (Ms), a bacterium lacking GroEL. Notably, the E. coli ortholog, EcNanA, is an obligate GroEL substrate (Fujiwara et al., 2010; Kerner et al., 2005) that refolds efficiently with GroEL/ES at a rate of ~1.36 min−1 (t1/2 ~30 s) at 25°C (Figure 3A). However, in contrast to DapA, no spontaneous refolding could be measured with EcNanA (at 100-400 nM) even at low temperature, due to pronounced aggregation (Figure 3A). Remarkably, MsNanA renatured spontaneously with a half-time similar to that measured for the assisted refolding of EcNanA (Figure 3A). GroEL was inefficient in binding MsNanA upon dilution from denaturant and no acceleration of folding was measured in the presence of GroEL/ES and ATP (Figure S3A). Bis-ANS binding experiments demonstrated that during renaturation at 10°C MsNanA buried ~80 % of its hydrophobic regions at a rate of ~2.2 min−1 (t1/2 ~20 s) (Figure S3B), i.e. more than an order of magnitude faster than refolding/assembly to active enzyme (t1/2 ~2.3 min at 10°C; 2 μM MsNanA) (data not shown). In contrast, DapA buried hydrophobicity at the slow rate of spontaneous subunit folding (t1/2 ~10 min at 10°C) (Figure S3B).
Figure 3. Chaperonin-Dependent and Independent TIM-Barrel Proteins.
(A) Assisted refolding of EcNanA and spontaneous refolding of MsNanA occur at similar rates. Spontaneous and GroEL/ES assisted refolding of EcNanA were analyzed at 25°C (400 nM EcNanA) in buffer D essentially as described in Figure 1D. Spontaneous renaturation of MsNanA (400 nM final concentration) in buffer D was analyzed by direct enzyme assay at the time points indicated. The observed kinetics reflects both subunit folding and assembly. Standard deviations are from at least 3 independent experiments.
B) Crystal structures of MsNanA (PDB 4N4P, this study) and EcNanA (PDB 2WO5) are shown as for DapA in Figure 1B. Left, monomer; right, tetramer. Helices H1 to H11 as well as the locations of the interfaces I and II of the tetramer are indicated.
C) Amino acid compositional bias in MsNanA. The monomer structures of EcDapA, EcNanA and MsNanA are shown in ribbon representations with α-helices and β-strands indicated in salmon and pale green, respectively. The side-chains of lysines and aromatic residues (Phe and Tyr) are highlighted in blue and purple, respectively.
Considering that EcNanA and MsNanA share only ~36 % sequence identity (~26 % pairwise identity with DapA), it was of interest to compare their structures. We solved the crystal structure of MsNanA at 1.8 Å resolution (Figure 3B and Table S1) and confirmed that the subunit structures of MsNanA, EcNanA and DapA are highly similar (pairwise r.m.s.d. of 1.4 to 1.8 Å) (Figures 3B and 1B). Interestingly, MsNanA contains more than twice the number of lysines (34) as the chaperonin-dependent EcNanA and DapA proteins (16 and 13, respectively) and less than half the number of arginines (5 compared to 12 and 13, respectively). Lysines are generally solvent exposed and confer helical propensity (Groebke et al., 1996). Indeed, the structure of MsNanA shows numerous lysines located on the solvent exposed surfaces of the amphiphilic TIM-barrel α-helices (such as H1, H2, H3) (Figure 3C). These lysine residues would be predicted to stabilize the α-helices and to facilitate the formation of the hydrophobic core (Figure 3C). Furthermore, the total number of aromatic residues Phe and Tyr in MsNanA is increased to 34 compared to 23 in EcNanA and 12 in DapA. The aromatic side chains contribute to the hydrophobic core of MsNanA and some engage in edge-face interactions (Figure 3C), stabilizing the fold (Burley and Petsko, 1985; Singh and Thornton, 1985). Thus, differences in amino acid composition may explain the efficient folding of MsNanA in the absence of chaperonin.
Comparing Spontaneous Folding of DapA and MsNanA
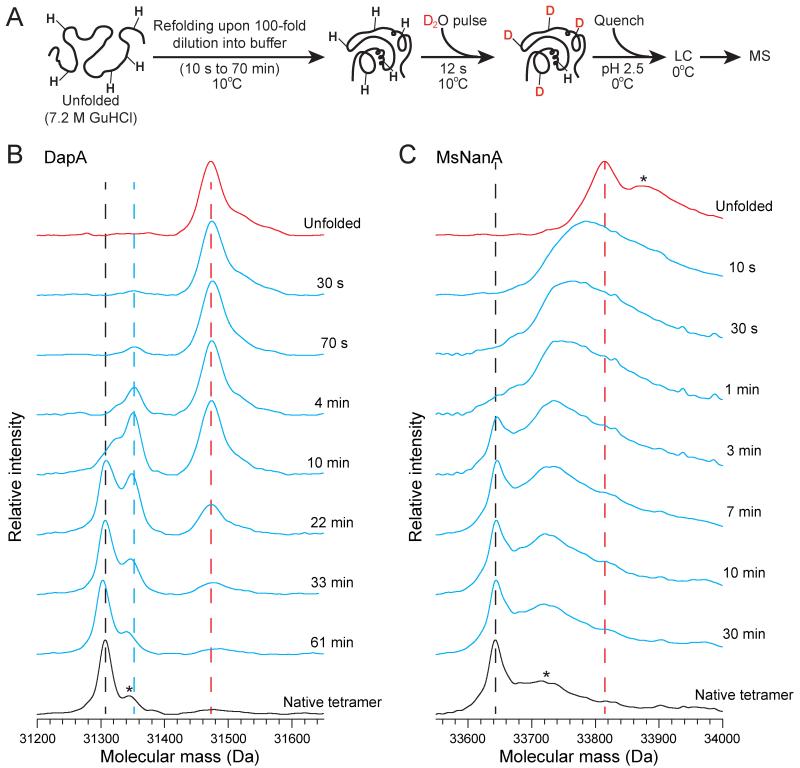
To characterize conformational states populated during spontaneous folding of DapA and MsNanA, we employed pulse-labeling H/DX coupled to liquid chromatography (LC) and MS (Miranker et al., 1993; Wales and Engen, 2006; Woodward and Hilton, 1979; Zhang and Smith, 1993). After various times of refolding, the protein was pulse-labeled for 12 s by 10-fold dilution into D2O buffer (Figure 4A). These experiments were performed at 10°C to improve time resolution of folding and exclude aggregation. Unfolded DapA incorporated 205 ± 2 deuterons (~74 % of total; 31475 Da, Figure 4B) and the native tetramer only 37 ± 2 deuterons (31307 Da, Figure 4B). During the first 70 s of refolding, DapA populated a distribution of molecules with an average mass essentially identical to that of the unfolded protein (Figure 4B), consistent with the presence of only ~20 % secondary structure by CD spectroscopy immediately upon dilution from denaturant (Figure S3C). The wide high-mass peak thus reflects a population of conformationally dynamic molecules lacking stable secondary and tertiary interactions. This population gradually decreased at a rate of 0.08 min−1 (Figures 4B and S3D), corresponding to the rate of folding (Figure 1D), and gave rise to a lower-mass peak at ~31350 Da. This peak most likely represents the folded monomer (with ~80 exchangeable hydrogens being deuterated); it disappeared with slower kinetics, reflecting assembly (Figure 1E) to give rise to native tetramer at ~31307 Da (Figure 4B). Thus, DapA folding follows two-state behavior with only unstructured intermediate and folded subunits being populated.
Figure 4. Different Properties of DapA and MsNanA Refolding by H/DX.
(A) Schematic representation of the H/DX pulse experiment. After different times of spontaneous refolding, proteins are pulse-labeled with D2O buffer E for 12 s, followed by acid quenching of the H/DX reaction and LC-MS analysis. See Extended Experimental Procedures for details.
(B and C) Mass spectra during spontaneous refolding of DapA (B) and MsNanA (C). The positions of the unfolded proteins and the folded tetramers in the mass spectra are indicated by red and black dotted lines, respectively. In the case of DapA, the blue dotted line marks the position of folded monomer. Asterisk in the native tetramer in B indicates a potassium adduct. Asterisks on the broad peaks of unfolded MsNanA and MsNanA tetramer in C represent the presence of potassium and sodium adducts (1 sodium, 1 potassium, 2 sodium and 2 potassium).
In contrast, MsNanA (208 ± 3 deuterons incorporated in the unfolded state) immediately upon dilution from denaturant populated a broader range of folding intermediates with varying numbers of deuterons incorporated (Figure 4C). CD spectroscopy showed that these early intermediates contain ~60 % of the secondary structure of the native protein (Figure S3E), in contrast to DapA where secondary structure forms in parallel with acquisition of the native state (data not shown). Furthermore, unlike DapA, this heterogeneous population eventually converged into a peak around 33640 Da (38 ± 3 exchangeable deuterons), corresponding to assembled tetramer, without formation of discernible folded monomers (Figure 4C). Thus, in the case of MsNanA, subunit folding and assembly appear to be coupled.
The population of highly dynamic folding intermediates by DapA suggested the presence of a significant entropic component of the kinetic barrier to folding. Consistent with this notion, the spontaneous folding rate of DapA proved temperature independent between 15°C and 25°C (Figure S3F and Table S2) (Bicout and Szabo, 2000; Dobson et al., 1998; Matagne et al., 2000). Below 15°C the Arrhenius plot displayed a constant slope, reflecting a transition state with both enthalpic and entropic components (Dobson et al., 1998) (Table S2).The Arrhenius plot of GroEL/ES-assisted folding displayed a linear slope over the entire temperature range from 7.5-25°C (Figure S3F), indicating that the activation barrier has gained a significant enthalpic component and the entropic contribution is reduced (Table S2). To extend this analysis to 37°C, we performed refolding experiments with single-molecule detection (100 pM DapA; see Figure 2C). Identical rates of spontaneous refolding of DapA-293C-Alexa were measured at 20°C and 37°C (Figures 2C and S3G), suggesting that the non-Arrhenius behavior extends to 37°C. In contrast, the rate of assisted folding increased ~2-fold from 20°C to 37°C, resulting in a ~130-fold acceleration of folding by GroEL/ES over the spontaneous rate at the physiological temperature (Figures 2C and S3G).
The spontaneous renaturation of MsNanA displayed a similar temperature dependence as the assisted refolding of DapA (Figure S3F and Table S2), suggesting that GroEL/ES shifts the folding properties of DapA towards those of the chaperonin-independent MsNanA.
Analysis of DapA Folding at Peptide Resolution
H/DX analysis of full-length DapA upon refolding in the presence of GroEL/ES was not feasible because the signals for GroEL/ES and DapA extensively overlapped. However, detailed structural information on the spontaneous and assisted folding of DapA could be obtained by monitoring H/DX protection at peptide resolution. Again, we compared spontaneous and GroEL/ES-mediated refolding at 10°C to obtain improved time resolution and exclude aggregation. In addition, refolding was analyzed upon stable encapsulation of DapA in SREL/ES and compared to the cycling GroEL/ES reaction. These experiments were performed at 25°C because the SREL/ES complex is unstable at low temperature.
GroEL-DapA complexes were first isolated by gel-filtration prior to initiating folding by addition of GroES/ATP. After different times of refolding, pulse-labeling with D2O for 12 s was performed as above (Figure 4A), the labeling reaction quenched, subjected to pepsin digestion and the deuterium incorporation into individual peptides measured by LC-MS (Hu et al., 2013; Wales and Engen, 2006; Zhang and Smith, 1993). The deuterium level found in the peptides serves as a signature of DapA conformation during refolding, providing a snapshot of the co-existing molecule populations (Hu et al., 2013; Miranker et al., 1993; Zhang and Smith, 1993). Pepsin digestion of DapA produced 81 unique and overlapping peptides; we analyzed 28 peptides, covering 91 % of the sequence (Figures S4A and S4B), for which data quality was high in both spontaneous and assisted folding experiments (Figure S5). Note that peptide P176-180 forms a loop that did not change in protection (Figure S5) and therefore was not included in subsequent analyses.
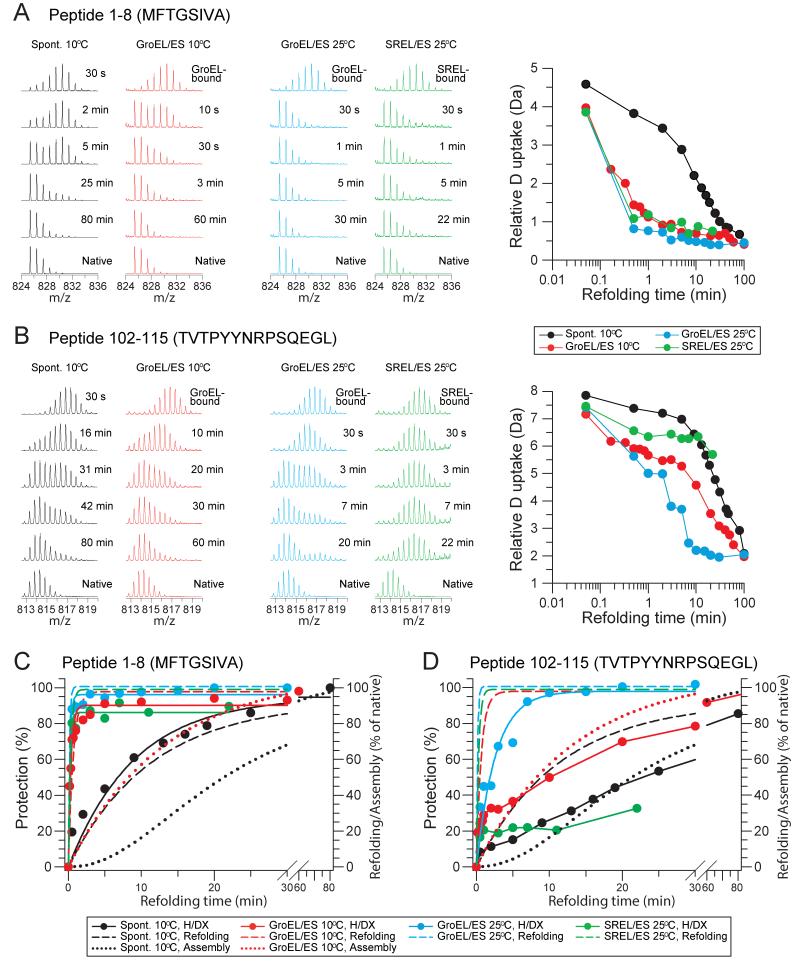
The isotope distributions for nearly all peptides during refolding (either spontaneous or assisted) were bimodal, indicating that peptides were either unfolded or folded, but not partially folded. This is illustrated for peptides P1-8 (MFTGSIVA) and P102-115 (TVTPYYNRPSQEGL): only two states are apparent, displaying either the same amount of deuterium incorporation as in the unfolded state (represented by the 30 s time point of spontaneous refolding) or limited deuterium incorporation as in the native protein (Figures 5A, 5B and S5). The mass spectra showed time-dependent transitions from an all-exchangeable to an all-protected population (Figures 5C and 5D). P1-8, belonging to the first β-strand of the TIM-barrel domain, acquired ~50 % protection after ~9 min of spontaneous folding at 10°C, at a rate similar to subunit folding (Figures 5C and 1D). In contrast, P102-115, forming part of interface I of the tetramer (Figure 1B), required more than 20 min to reach 50 % protection, which is similar to the rate of tetramer assembly (Figures 5D and 1E). DapA bound to GroEL showed only minor protection in a few peptides (Figure S5), consistent with previous observations that GroEL-bound proteins lack stable structure (Chen et al., 2001; Horst et al., 2005; Robinson et al., 1994). Strikingly, GroEL/ES accelerated the rate at which P1-8 acquired protection by at least 50-fold (t1/2 ~12 s, the duration of the D2O pulse) compared to spontaneous folding (t1/2 ~9 min) (Figures 5A and 5C). P102-115 acquired protection much more slowly (t1/2 ~9 min) with GroEL/ES, but still ~2-fold faster than in spontaneous folding (t1/2 ~20 min) (Figures 5B and 5D), consistent with assembly being enhanced due to accelerated subunit folding (Figures 5D and 1E). Acquisition of protection in distinct peptides generally correlated either with the rate of subunit folding or assembly measured by enzymatic assay, arguing against non-specific effects due to interaction of DapA with the wall of the GroEL/ES cage.
Figure 5. Deuterium Incorporation into Peptides of DapA During Spontaneous and Assisted Refolding.
(A and B) Deuterium incorporation into P1-8 of the TIM-barrel domain (A) and P102-115 of interface I (B) of DapA. Left panels: Examples of mass spectra for DapA peptides P1-8 and P102-115 at different times during spontaneous, GroEL/ES-assisted and SREL/ES-assisted refolding at 10°C and 25°C, as indicated. Amino acid sequences of the peptides are indicated in single letter code. Right panels: Deuterium uptake in Da is plotted versus refolding time (see Figure S5 for full dataset).
(C and D) Time courses of H/DX protection during refolding for P1-8 (C) and P102-115 (D). For comparison, subunit refolding and assembly based on enzymatic assay (Figures 1D and 1E) are also shown (dashed and dotted lines, respectively).
To distinguish between structure formation upon subunit folding versus assembly, we performed H/DX measurements during stable encapsulation of DapA in SREL/ES at 25°C. For comparison, folding with GroEL/ES was also analyzed at 25°C (Figures 5 and S5). In both systems, P1-8 acquired protection at essentially the same rate, demonstrating that a single round of protein encapsulation is sufficient to catalyze folding (Figures 5A, 5C and 1D). In contrast, P102-115 of interface I acquired only ~20 % protection in SREL/ES (Figures 5B, 5D and S5), demonstrating that full protection of this peptide results from subunit assembly after protein release from the chaperonin cage. Similar behavior was observed for other peptides involved in assembly (see below), further excluding H/DX protection as a result of protein binding to the cage wall.
Effect of Chaperonin on the Folding Pathway of DapA
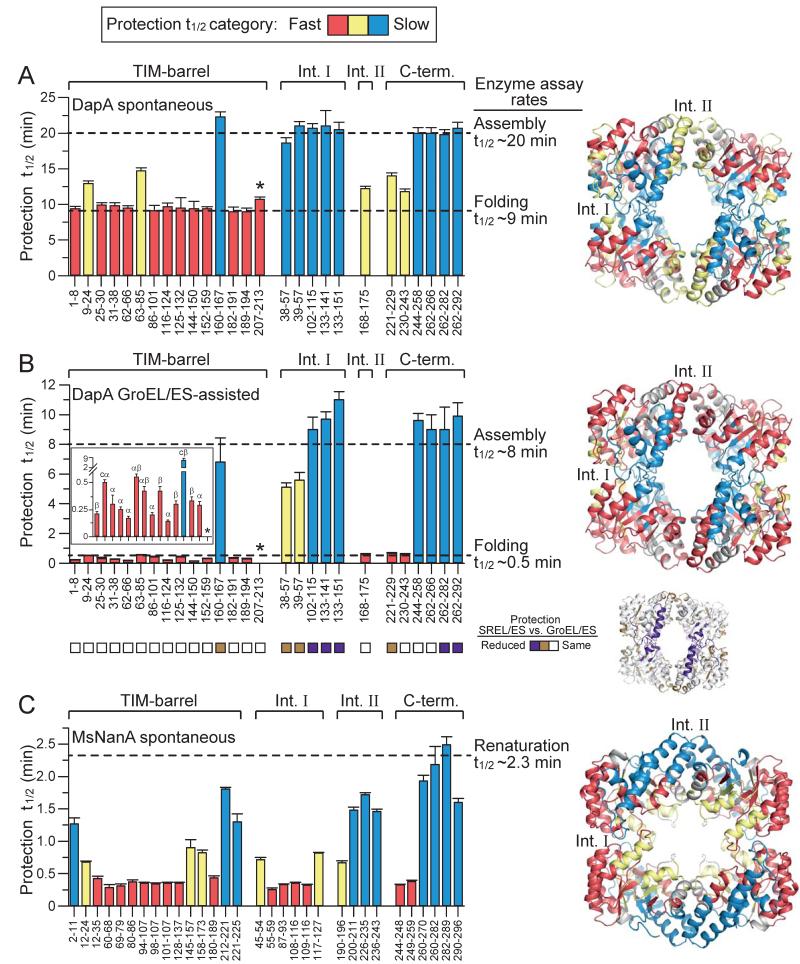
Our analysis of time-dependent H/DX protection of DapA peptides revealed different categories of protection (Figures 6 and S5). During spontaneous refolding at 10°C, the peptides in the fastest category were all located in the TIM-barrel domain and acquired protection with a t1/2 of ~9.5 min, consistent with the rate of subunit folding (Figures 6A, 1D and S6A). Three TIM-barrel peptides (P9-24; P63-85; P160-167) acquired protection more slowly. P160-167, containing the conserved K161 involved in binding the substrate pyruvate, has the slowest protection (t1/2 ~22 min). This is similar to the rate at which peptides of the C-terminal domain and regions at the tight dimer interface (interface I) acquire protection (Figures 6A and S6A), and corresponds to the rate of tetramer assembly and acquisition of enzymatic activity (Figure 1E). P63-85 has 3 of its residues located in interface I (Figure S5), possibly contributing to its slower protection. During GroEL/ES-assisted folding, all TIM-barrel peptides, except P160-167, acquired protection with a t1/2 of 30 s or less, while several of the C-terminal domain and subunit interface peptides reached protection with a t1/2 of 6-11 min (Figures 6B and S6B), the rate of assembly in the presence of GroEL/ES (Figure 1E). Upon encapsulation in the non-cycling SREL/ES complex, all TIM-barrel peptides (except P160-167) reached protection at similar rates as with the cycling GroEL/ES, while regions involved in assembly showed markedly reduced protection (Figure 6B).
Figure 6. Comparison of DapA and MsNanA Refolding at Peptide Resolution.
Apparent half-times of H/DX protection for peptides along the amino acid sequence during spontaneous (A) and GroEL/ES-assisted refolding/assembly (B) of DapA, and spontaneous renaturation of MsNanA (C) at 10°C are presented in the bar graphs and are mapped on the tetramer structures of DapA and MsNanA (right panels). Peptides are assigned either to TIM-barrel domain, C-terminal domain or interfaces I and II (when at least 25 % of the sequence is in the interface; see Figure S5). Half-times of protection are color-coded: Red bars indicate peptides with half-times as fast or faster as subunit refolding in enzymatic refolding assays (Figure 1D); blue bars denote peptides with half-times of protection as slow as assembly (Figure 1E), and yellow bars denote peptides with intermediate half-times of protection. Note that P207-213 (asterisk) in GroEL/ES-assisted folding is already fully protected in the GroEL-bound state. The insert in (B) highlights the differences in protection of TIM-barrel domain peptides (x-axis labeling as in main figure of panel B) and the secondary structure of the peptides is indicated: α, α-helix; β, β-strand; cα, coil and α-helix; cβ, coil and β-strand. Squares below the bar graph in (B) indicate protection properties of peptides upon refolding with SREL/ES in comparison to GroEL/ES at 25°C. Peptides with reduced protection with SREL/ES are highlighted in the tetramer structure. In the case of MsNanA, refolding and assembly are coupled and half-times of protection are colored from fast (red <0.5 min) to slow (blue >1.0 min).
Our analysis suggests that during spontaneous folding the structure of the TIM-barrel evolves in a highly concerted process, with almost all its segments not involved in assembly acquiring H/DX protection simultaneously (t1/2 ~9.5 min) (Figures 6A, S5 and S6A). During folding with GroEL/ES, these peptides acquire protection 20- to 50-fold faster (Figures 6B insert). Moreover, protection no longer develops simultaneously for all TIM-barrel peptides. Specifically, P62-66, P116-124 and P144-150 mapping to α-helices H2, H4 and H6, respectively, acquire protection 2- to 3-fold faster than peptides with mixed secondary elements: P9-24 (coil and α-helix), P63-85 (αβ), P86-101 (αβ) and P125-132 (coil and β-strand) (Figures 6B insert and S6B). These differences in protection were highly reproducible (Figure S5) and suggest that confinement by chaperonin catalyzes folding by promoting local structure formation in amphiphilic α-helices onto which β-strands can dock (Figures 6B, insert and S6B). This is consistent with GroEL/ES reducing the entropic component of the folding energy barrier (Table S2).
Folding Mechanism of GroEL/ES-independent MsNanA
Next we analyzed the folding/assembly of MsNanA by H/DX at peptide resolution (Figures S4C S4D and S7) to obtain insight into the mechanism underlying its chaperonin-independence. We detected non-uniform rates of protection for elements within the TIM-barrel domain, with up to 4-fold rate differences (t1/2 ~15 s to ~1.8 min at 10°C) (Figures 6C and S7), consistent with the population of a broad range of folding intermediates as detected by H/DX of the full-length protein (Figure 4C). Folding appears to initiate in a nucleus formed by the amphiphilic α-helices H1 to H3 with their corresponding β-strands and H10 in the C-terminal domain, which is adjacent to H1. Structure formation then proceeds in a wave around the TIM-barrel and reaches completion with formation of H8 and the C-terminal domain helices H9 and H11 (Figures 6C and S6C). Helices H1 to H3 are enriched in solvent exposed lysines and their early formation may be coupled to organization of the hydrophobic TIM-barrel core which is stabilized by numerous aromatic (Tyr and Phe) residues (Figure 3C). Although both the assisted folding of DapA and the spontaneous renaturation of MsNanA involve local structure formation in the TIM-barrel, the folding regimes differ. In the former we observed multiple foci of initial structure formation, as might be expected for a protein confined in the chaperonin-cage, whereas structure initiates asymmetrically in the latter. Another notable feature of MsNanA is that peptides located in interface I acquire protection at the fast rate of TIM-barrel folding (Figure 6C), which may facilitate subunit assembly to occur coupled with folding. In contrast, interface I of DapA acquires protection from exchange slower than the TIM-barrel, indicating that subunit folding and assembly are sequential steps (Figures 6A and 6B).
DISCUSSION
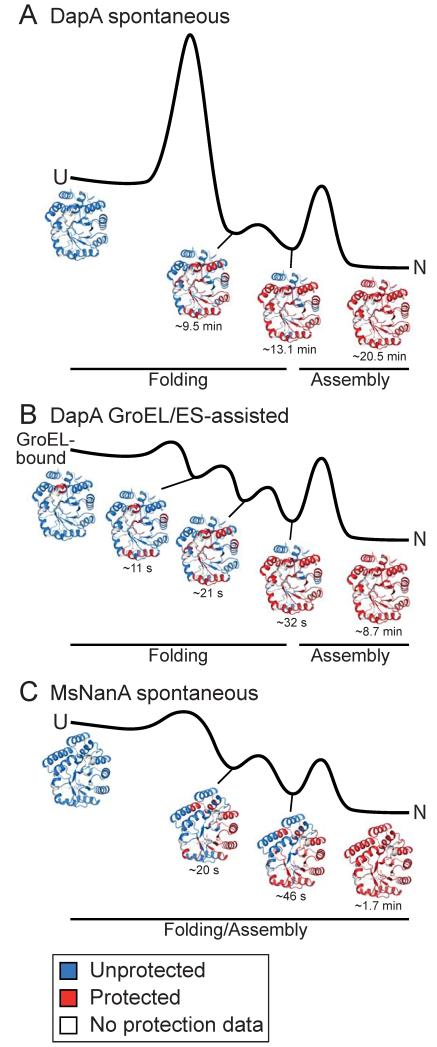
Whether GroEL/ES actively promotes folding beyond preventing aggregation has remained controversial. In this study, we analyzed the spontaneous and assisted refolding of DapA, an obligate in vivo substrate of GroEL. We find that the chaperonin accelerates DapA folding more than 30-fold over its spontaneous folding rate. Analysis by H/DX-MS at peptide resolution demonstrates that GroEL/ES catalyzes folding of the DapA TIM-barrel domain. The slow spontaneous folding of the TIM-barrel in the absence of GroEL/ES involves a concerted transition from an ensemble of dynamic folding intermediates to the native state that is associated with a high free energy barrier (Figure 7A). In contrast, folding in the confining environment of the chaperonin cage is characterized by rapid stepwise, i.e. less cooperative, structure formation, which effectively lowers the entropic component of the energy barrier (Figure 7B). The spontaneous folding of MsNanA, a GroEL-independent protein virtually identical in structure, also employs a segmental folding regime (Figure 7C). We propose that the chaperonin cage acts as a powerful folding catalyst for a set of proteins which otherwise fail to reach native state at a biologically relevant time scale.
Figure 7. Mechanisms of Spontaneous and GroEL/ES-Assisted TIM-Barrel Folding.
Free energy diagrams, schematically summarizing the salient features of spontaneous (A) folding and GroEL/ES-assisted folding (B) of DapA and spontaneous folding of MsNanA (C). Intermediate states populated during folding with the approximate half-times indicated, as determined by H/DX-MS at peptide resolution, are tentatively assigned to different phases of the energy diagrams. Ribbon diagrams show acquisition of H/DX protection during folding in red. U, unfolded state; N, native tetramer. See Discussion for detail.
Catalysis of Folding is Biologically Relevant
The observed rate enhancement of folding by chaperonin cannot be explained by prevention of aggregation that might otherwise slow the spontaneous folding reaction. Importantly, 50- to 130-fold accelerated folding (at 25-37°C) was measured by FCS at a very low concentration of DapA (100 pM), conditions excluding intermolecular association (aggregation or assembly) as determined by dual color FCCS. This dramatic catalysis of folding is biologically highly relevant. About 30-50 % of the obligate GroEL substrates, including DapA, share the TIM-barrel domain fold (Fujiwara et al., 2010; Kerner et al., 2005) and many of these proteins aggregate or are degraded in E. coli cells when GroEL/ES is depleted (Calloni et al., 2012; Kerner et al., 2005; Powers et al., 2012). At physiological temperature (37°C), GroEL/ES allows folding of proteins like DapA to be completed faster than the time of synthesis (~14 s, assuming translation at 20 amino acids/s), thus avoiding the build-up of unfolded protein and making efficient use of available chaperonin capacity.
GroEL/ES Modifies Folding of Encapsulated Protein
Our analysis by H/DX coupled to MS showed that the spontaneous folding of DapA proceeds in an apparent two-state manner with only kinetically trapped folding intermediate and largely folded subunits being significantly populated (Figure 7A). The folding intermediate represents an ensemble of collapsed states lacking stable structure. These species bind ANS, indicative of a fluctuating hydrophobic core (Dobson et al., 1998), contain only ~20 % secondary structure by CD and have virtually no H/DX protection compared to the unfolded state in denaturant. H/DX-MS at peptide resolution showed that nearly all peptides within the (βα)8 TIM-barrel acquire protection with an identical half-time of ~9.5 min at 10°C, equivalent to the rate of subunit folding (Figures 7A and 6A). This indicates that the TIM-barrel domain folds in a highly concerted manner. This process is associated with a long search time as the (βα)8 barrel contains many long-range interactions and at a length of 224 amino acids exceeds the theoretical size limit for productive hydrophobic collapse (Lin and Zewail, 2012).
GroEL/ES affects predominantly the folding of the TIM-barrel domain, accelerating structure formation at peptide level 20- to 50-fold compared to spontaneous folding (at 10°C) (Figure 6). Different secondary structure elements of the TIM-barrel acquire H/DX protection at up to 3-fold different rates, with the fastest speed being measured for structure formation in α-helices. Thus, in the GroEL/ES catalyzed reaction folding nucleates locally, building up the structure in a segmental manner and thereby reducing the entropic penalty (Figure 7B). Moreover, the difference in protection rate between TIM-barrel segments and peptides located in the C-terminal domain is magnified to more than 15-fold in assisted folding (only ~2-fold in spontaneous folding), reducing possible effects of inter-domain interference that may retard spontaneous folding.
How does GroEL/ES catalyze the folding of the DapA TIM-barrel? Theory predicts that steric confinement of unfolded protein in a repulsive (net-negatively charged) cage can accelerate folding by 1-2 orders of magnitude by restricting the conformational freedom of folding intermediates and making the formation of local and long-range contacts, including those present in the transition state, more favorable (Baumketner et al., 2003; Hayer-Hartl and Minton, 2006; Sirur and Best, 2013). Our results with the single ring variant of GroEL, SREL, show that folding catalysis is achieved upon a single round of protein encapsulation within the SREL/ES cage. This excludes repetitive binding and unfolding of misfolded states by GroEL (Lin et al., 2008; Sharma et al., 2008; Thirumalai and Lorimer, 2001) as a requirement of accelerated folding, at least for DapA. The negative net charge of the GroEL/ES cavity wall likely plays an additional role through an ordering effect on water structure that may enhance hydrophobic core packing of encapsulated protein (England and Pande, 2008; Tang et al., 2006). However, accelerated folding is not generally observed for all substrates. For example, in the case of the model substrate rhodanese, spontaneous and GroEL/ES-assisted refolding occur at similar rates (Brinker et al., 2001; Hofmann et al., 2010). We suggest that, as a result of co-evolution, the physical properties of the GroEL/ES-cage are particularly suited to achieve folding catalysis for a subset of TIM-barrel domain proteins, which occupy ~45 % of GroEL capacity in vivo (Kerner et al., 2005).
Escape from Chaperonin Dependence
A small group of bacteria, including M. synoviae, lack GroEL/ES but contain a number of orthologs of GroEL-dependent E. coli proteins. Our analysis of MsNanA, the ortholog of EcNanA and a close structural homolog of DapA, provided insight into the strategies that have been employed in evolution to render a protein GroEL-independent. Unlike DapA, MsNanA does not form a largely unstructured intermediate during renaturation. Instead, hydrophobic collapse is closely coupled with the gain of secondary structure during refolding, as measured by CD and H/DX. The secondary structural elements of the TIM-barrel form sequentially, propagating “as a wave” from a nucleus initiating at α-helices H1-H3 (Figure 7C). The crystal structure of MsNanA showed that these amphiphilic helices are enriched in solvent exposed lysines which confer strong α-helical propensity. Burial of hydrophobic residues coupled with the formation of native structure is apparently facilitated by a hydrophobic core that is enriched in aromatic residues. These structural properties may explain the ability of MsNanA to nucleate TIM-barrel folding in distinct segments, a feature otherwise induced by the confining environment of the GroEL/ES cage. Moreover, folding and assembly of MsNanA appears to be coupled, suggesting a mechanism of ‘self-chaperoning’. MsNanA’s independence of the chaperonin cage for folding would have facilitated the evolution of such a mechanism.
EXPERIMENTAL PROCEDURES
Proteins
Chaperone and substrate proteins were purified as previously described (Brinker et al., 2001; Hayer-Hartl et al., 1996; Kerner et al., 2005) (see Extended Experimental Procedures).
Refolding, Assembly and Enzymatic Assays
Spontaneous refolding of DapA was initiated by 100- to 200-fold dilution from denaturant into refolding buffer B (20 mM Tris-HCl pH 7.5, 100 mM KCl, 10 mM MgCl2, 10 mM pyruvate) at the temperatures and final monomer concentrations indicated in figure legends. Refolding was stopped by addition of excess GroEL D87K (GroEL Trap). For GroEL/ES-assisted refolding of DapA, unfolded substrate was diluted into buffer B containing chaperones as specified in figure legends and was stopped by CDTA or apyrase as indicated. For SREL/ES-assisted refolding, DapA was unfolded in urea and refolding performed in low salt buffer C (20 mM Tris-HCl pH 7.5, 10 mM KCl, 5 mM MgCl2, 10 mM pyruvate). In the case of MsNanA, spontaneous renaturation was performed in refolding buffer D (20 mM Tris-HCl pH 7.5, 100 mM KCl, 10 mM MgCl2) and enzyme activity measured immediately. Enzyme activities were measured as previously described ((Kerner et al., 2005) and Extended Experimental Procedures).
FCS and dcFCCS Experiments
DapA-293C was labeled with either Alexa647 (Invitrogen) or Dy530 (Dyomics) using maleimide chemistry. Fluorescence correlation spectroscopy (FCS) and dual color fluorescence cross-correlation spectroscopy (dcFCCS) (Muller et al., 2005) were performed to investigate the oligomeric state of DapA during refolding and to measure the rates of refolding at 100 pM DapA concentration. See Extended Experimental Procedures for further details.
Hydrogen/Deuterium Exchange (H/DX)
Refolding reactions were essentially performed as above. Aliquots were withdrawn at different times and pulse-labeled for 12 s by 10-fold dilution with buffer E (20 mM Tris-HCl, 20 mM KCl, pD 7.5, 99.9% D2O) to a final concentration of 90 % D2O, followed by acid quenching (Figure 4A). Intact proteins were immediately analyzed by LC-MS on a Waters Synapt G1 mass spectrometer. To obtain H/DX data at peptide resolution, acid quenched samples were injected into an H/DX Waters nanoACQUITY UPLC (Wales et al., 2008) and passed through a Poroszyme immobilized pepsin cartridge (Applied Biosystems). Peptic peptides eluting from the pepsin column were trapped, desalted and then separated in 6 min with a 8-40 % acetonitrile gradient in 0.1 % formic acid pH 2.5. All chromatographic elements were held at 2.5°C. The average amount of back-exchange was 20-25 % (Wales et al., 2008). All experiments were performed between 2-4 times. Mass spectra were processed with DynamX software (Waters Corp., Milford, MA, USA). See Extended Experimental Procedures for detailed description.
Miscellaneous
Bis-ANS fluorescence, CD spectroscopy and X-ray crystallography were performed using standard procedures as described in Extended Experimental Procedures.
Supplementary Material
ACKNOWLEDGMENTS
We thank L. Moroder and H.J. Musiol for the synthesis of β-semialdehyde. Expert technical assistance by N. Wischnewski and A.R. Lange is acknowledged, as is assistance with the H/DX experiments from T.E. Wales, R.E. Iacob and R. Körner. We thank the MPIB crystallization facility and the personnel at beamline X10SA of SLS Villigen, Switzerland for their assistance. We gratefully acknowledge the financial support from the Munich Center for Integrated Protein Science (CiPSM). J.R.E. was on sabbatical at the Department of Cellular Biochemistry, Max Planck Institute of Biochemistry from 06/2012-06/2013 and was partially supported during this time by NIH R01-GM101135 and a research collaboration with the Waters Corporation. F.G. was supported by a postdoctoral fellowship from La Fondation pour la Recherche Medicale (code FRM SPE20071211473).
Footnotes
Supplemental Information includes Extended Experimental Procedures, 2 Tables and 7 Figures.
REFERENCES
- Apetri AC, Horwich AL. Chaperonin chamber accelerates protein folding through passive action of preventing aggregation. Proc. Natl. Acad. Sci. U.S.A. 2008;105:17351–17355. doi: 10.1073/pnas.0809794105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azia A, Unger R, Horovitz A. What distinguishes GroEL substrates from other Escherichia coli proteins? FEBS J. 2012;279:543–550. doi: 10.1111/j.1742-4658.2011.08458.x. [DOI] [PubMed] [Google Scholar]
- Baumketner A, Jewett A, Shea JE. Effects of confinement in chaperonin assisted protein folding: Rate enhancement by decreasing the roughness of the folding energy landscape. J. Mol. Biol. 2003;332:701–713. doi: 10.1016/s0022-2836(03)00929-x. [DOI] [PubMed] [Google Scholar]
- Bicout DJ, Szabo A. Entropic barriers, transition states, funnels, and exponential protein folding kinetics: A simple model. Protein Sci. 2000;9:452–465. doi: 10.1110/ps.9.3.452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinker A, Pfeifer G, Kerner MJ, Naylor DJ, Hartl FU, Hayer-Hartl M. Dual function of protein confinement in chaperonin-assisted protein folding. Cell. 2001;107:223–233. doi: 10.1016/s0092-8674(01)00517-7. [DOI] [PubMed] [Google Scholar]
- Burley SK, Petsko GA. Aromatic-aromatic interaction: a mechanism of protein structure stabilization. Science. 1985;229:23–28. doi: 10.1126/science.3892686. [DOI] [PubMed] [Google Scholar]
- Calloni G, Chen T, Schermann SM, Chang H-C, Genevaux P, Agostini F, Tartaglia GG, Hayer-Hartl M, Hartl FU. DnaK functions as a central hub in the E. coli chaperone network. Cell Rep. 2012;1:251–264. doi: 10.1016/j.celrep.2011.12.007. [DOI] [PubMed] [Google Scholar]
- Chakraborty K, Chatila M, Sinha J, Shi Q, Poschner BC, Sikor M, Jiang G, Lamb DC, Hartl FU, Hayer-Hartl M. Chaperonin-catalyzed rescue of kinetically trapped states in protein folding. Cell. 2010;142:112–122. doi: 10.1016/j.cell.2010.05.027. [DOI] [PubMed] [Google Scholar]
- Chen D-H, Madan D, Weaver J, Lin Z, Schroder GF, Chiu W, Rye HS. Visualizing GroEL/ES in the act of encapsulating a folding protein. Cell. 2013;153:1354–1365. doi: 10.1016/j.cell.2013.04.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JW, Walter S, Horwich AL, Smith DL. Folding of malate dehydrogenase inside the GroEL-GroES cavity. Nat. Struct. Biol. 2001;8:721–728. doi: 10.1038/90443. [DOI] [PubMed] [Google Scholar]
- Clare DK, Vasishtan D, Stagg S, Quispe J, Farr GW, Topf M, Horwich AL, Saibil HR. ATP-triggered conformational changes delineate substrate-binding and - folding mechanics of the GroEL chaperonin. Cell. 2012;149:113–123. doi: 10.1016/j.cell.2012.02.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobson CM, Sali A, Karplus M. Protein Folding - a Perspective From Theory and Experiment. Angew. Chem. Int. Edit. 1998;37:868–893. doi: 10.1002/(SICI)1521-3773(19980420)37:7<868::AID-ANIE868>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- Dobson RC, Griffin MD, Jameson GB, Gerrard JA. The crystal structures of native and (S)-lysine-bound dihydrodipicolinate synthase from Escherichia coli with improved resolution show new features of biological significance. Acta Crystallogr. Sect. D Biol. Crystallogr. 2005;61:1116–1124. doi: 10.1107/S0907444905016318. [DOI] [PubMed] [Google Scholar]
- England JL, Pande VS. Potential for modulation of the hydrophobic effect inside chaperonins. Biophys. J. 2008;95:3391–3399. doi: 10.1529/biophysj.108.131037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara K, Ishihama Y, Nakahigashi K, Soga T, Taguchi H. A systematic survey of in vivo obligate chaperonin-dependent substrates. EMBO J. 2010;29:1552–1564. doi: 10.1038/emboj.2010.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gershenson A, Gierasch LM. Protein folding in the cell: challenges and progress. Curr. Opin. Struct. Biol. 2011;21:32–41. doi: 10.1016/j.sbi.2010.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groebke K, Renold P, Tsang KY, Allen TJ, McClure KF, Kemp DS. Template-nucleated alanine-lysine helices are stabilized by position-dependent interactions between the lysine side chain and the helix barrel. Proc. Natl. Acad. Sci. U.S.A. 1996;93:4025–4029. doi: 10.1073/pnas.93.9.4025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayer-Hartl M, Minton AP. A simple semiempirical model for the effect of molecular confinement upon the rate of protein folding. Biochemistry. 2006;45:13356–13360. doi: 10.1021/bi061597j. [DOI] [PubMed] [Google Scholar]
- Hayer-Hartl MK, Weber F, Hartl FU. Mechanism of chaperonin action: GroES binding and release can drive GroEL-mediated protein folding in the absence of ATP hydrolysis. EMBO J. 1996;15:6111–6121. [PMC free article] [PubMed] [Google Scholar]
- Hofmann H, Hillger F, Pfeil SH, Hoffmann A, Streich D, Haenni D, Nettels D, Lipman EA, Schuler B. Single-molecule spectroscopy of protein folding in a chaperonin cage. Proc. Natl. Acad. Sci. U.S.A. 2010;107:11793–11798. doi: 10.1073/pnas.1002356107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horst R, Bertelsen EB, Fiaux J, Wider G, Horwich AL, Wuthrich K. Direct NMR observation of a substrate protein bound to the chaperonin GroEL. Proc. Natl. Acad. Sci. U.S.A. 2005;102:12748–12753. doi: 10.1073/pnas.0505642102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horwich AL, Apetri AC, Fenton WA. The GroEL/GroES cis cavity as a passive anti-aggregation device. FEBS Lett. 2009;583:2654–2662. doi: 10.1016/j.febslet.2009.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu W, Walters BT, Kan ZY, Mayne L, Rosen LE, Marqusee S, Englander SW. Stepwise protein folding at near amino acid resolution by hydrogen exchange and mass spectrometry. Proc. Natl. Acad. Sci. U.S.A. 2013;110:7684–7689. doi: 10.1073/pnas.1305887110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jewett AI, Shea J-E. Reconciling theories of chaperonin accelerated folding with experimental evidence. Cell Mol. Life Sci. 2010;67:255–276. doi: 10.1007/s00018-009-0164-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerner MJ, Naylor DJ, Ishihama Y, Maier T, Chang HC, Stines AP, Georgopoulos C, Frishman D, Hayer-Hartl M, Mann M, et al. Proteome-wide analysis of chaperonin-dependent protein folding in Escherichia coli. Cell. 2005;122:209–220. doi: 10.1016/j.cell.2005.05.028. [DOI] [PubMed] [Google Scholar]
- Kim YE, Hipp MS, Bracher A, Hayer-Hartl M, Hartl FU. Molecular chaperone functions in protein folding and proteostasis. Annu. Rev. Biochem. 2013;82:323–355. doi: 10.1146/annurev-biochem-060208-092442. [DOI] [PubMed] [Google Scholar]
- Lin MM, Zewail AH. Hydrophobic forces and the length limit of foldable protein domains. Proc. Natl. Acad. Sci. U.S.A. 2012;109:9851–9856. doi: 10.1073/pnas.1207382109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Z, Madan D, Rye HS. GroEL stimulates protein folding through forced unfolding. Nat. Struct. Mol. Biol. 2008;15:303–311. doi: 10.1038/nsmb.1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin J, Hartl FU. The effect of macromolecular crowding on chaperonin-mediated protein folding. Proc. Natl. Acad. Sci. U.S.A. 1997;94:1107–1112. doi: 10.1073/pnas.94.4.1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matagne A, Jamin M, Chung EW, Robinson CV, Radford SE, Dobson CM. Thermal unfolding of an intermediate is associated with non-Arrhenius kinetics in the folding of hen lysozyme. J. Mol. Biol. 2000;297:193–210. doi: 10.1006/jmbi.2000.3540. [DOI] [PubMed] [Google Scholar]
- McLennan N, Masters M. GroE Is Vital For Cell-Wall Synthesis. Nature. 1998;392:139. doi: 10.1038/32317. [DOI] [PubMed] [Google Scholar]
- Miranker A, Robinson CV, Radford SE, Aplin RT, Dobson CM. Detection of transient protein folding populations by mass spectrometry. Science. 1993;262:896–900. doi: 10.1126/science.8235611. [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay S, Krishnan R, Lemke EA, Lindquist S, Deniz AA. A natively unfolded yeast prion monomer adopts an ensemble of collapsed and rapidly fluctuating structures. Proc. Natl. Acad. Sci. U.S.A. 2007;104:2649–2654. doi: 10.1073/pnas.0611503104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller BK, Zaychikov E, Bräuchle C, Lamb DC. Pulsed interleaved excitation. Biophys. J. 2005;89:3508–3522. doi: 10.1529/biophysj.105.064766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh E, Becker AH, Sandikci A, Huber D, Chaba R, Gloge F, Nichols RJ, Typas A, Gross CA, Kramer G, et al. Selective ribosome profiling reveals the cotranslational chaperone action of trigger factor in vivo. Cell. 2011;147:1295–1308. doi: 10.1016/j.cell.2011.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers ET, Powers DL, Gierasch LM. FoldEco: A Model for Proteostasis in E. coli. Cell Rep. 2012;1:265–276. doi: 10.1016/j.celrep.2012.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reboul CF, Porebski BT, Griffin MD, Dobson RC, Perugini MA, Gerrard JA, Buckle AM. Structural and dynamic requirements for optimal activity of the essential bacterial enzyme dihydrodipicolinate synthase. PLOS Comput. Biol. 2012;8:e1002537. doi: 10.1371/journal.pcbi.1002537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson CV, Gross M, Eyles SJ, Ewbank JJ, Mayhew M, Hartl FU, Dobson CM, Radford SE. Conformation of GroEL-bound alpha-lactalbumin probed by mass spectrometry. Nature. 1994;372:646–651. doi: 10.1038/372646a0. [DOI] [PubMed] [Google Scholar]
- Saibil HR, Fenton WA, Clare DK, Horwich AL. Structure and Allostery of the Chaperonin GroEL. J. Mol. Biol. 2012;425:1476–1486. doi: 10.1016/j.jmb.2012.11.028. [DOI] [PubMed] [Google Scholar]
- Schmid FX. Fast-folding and slow-folding forms of unfolded proteins. Methods Enzymol. 1986;131:70–82. doi: 10.1016/0076-6879(86)31035-8. [DOI] [PubMed] [Google Scholar]
- Sharma S, Chakraborty K, Mueller BK, Astola N, Tang YC, Lamb DC, Hayer-Hartl M, Hartl FU. Monitoring protein conformation along the pathway of chaperonin-assisted folding. Cell. 2008;133:142–153. doi: 10.1016/j.cell.2008.01.048. [DOI] [PubMed] [Google Scholar]
- Singh J, Thornton JM. The interaction between phenylalanine rings in proteins. FEBS Lett. 1985;191:1–6. [Google Scholar]
- Sirur A, Best RB. Effects of interactions with the GroEL cavity on protein folding rates. Biophys. J. 2013;104:1098–1106. doi: 10.1016/j.bpj.2013.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang YC, Chang HC, Chakraborty K, Hartl FU, Hayer-Hartl M. Essential role of the chaperonin folding compartment in vivo. EMBO J. 2008;27:1458–1468. doi: 10.1038/emboj.2008.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang YC, Chang HC, Roeben A, Wischnewski D, Wischnewski N, Kerner MJ, Hartl FU, Hayer-Hartl M. Structural features of the GroEL-GroES nano-cage required for rapid folding of encapsulated protein. Cell. 2006;125:903–914. doi: 10.1016/j.cell.2006.04.027. [DOI] [PubMed] [Google Scholar]
- Thirumalai D, Lorimer GH. Chaperonin-mediated protein folding. Annu. Rev. Biophys. Biomolec. Struct. 2001;30:245–269. doi: 10.1146/annurev.biophys.30.1.245. [DOI] [PubMed] [Google Scholar]
- Wales TE, Engen JR. Hydrogen exchange mass spectrometry for the analysis of protein dynamics. Mass Spectrom. Rev. 2006;25:158–170. doi: 10.1002/mas.20064. [DOI] [PubMed] [Google Scholar]
- Wales TE, Fadgen KE, Gerhardt GC, Engen JR. High-speed and high-resolution UPLC separation at zero degrees Celsius. Anal. Chem. 2008;80:6815–6820. doi: 10.1021/ac8008862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissman JS, Rye HS, Fenton WA, Beechem JM, Horwich AL. Characterization of the Active Intermediate of a GroEL-GroES-Mediated Protein Folding Reaction. Cell. 1996;84:481–490. doi: 10.1016/s0092-8674(00)81293-3. [DOI] [PubMed] [Google Scholar]
- Woodward CK, Hilton BD. Hydrogen exchange kinetics and internal motions in proteins and nucleic acids. Annu. Rev. Biophys. Bioeng. 1979;8:99–127. doi: 10.1146/annurev.bb.08.060179.000531. [DOI] [PubMed] [Google Scholar]
- Yang D, Ye X, Lorimer GH. Symmetric GroEL:GroES2 complexes are the protein-folding functional form of the chaperonin nanomachine. Proc Natl Acad Sci U S A. 2013 doi: 10.1073/pnas.1318862110. DOI:10.1073/pnas.1318862110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Smith DL. Determination of amide hydrogen exchange by mass spectrometry: a new tool for protein structure elucidation. Protein Sci. 1993;2:522–531. doi: 10.1002/pro.5560020404. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.