SUMMARY
Broadly neutralizing antibodies to HIV are much sought-after (a) to guide vaccine design, both as templates and to inform on the authenticity of vaccine candidates, (b) to assist in structural studies and (c) as potential therapeutics. However, the number of targets on the viral envelope spike for such antibodies is limited. Here, we describe a set of human monoclonal antibodies that define a previously undefined target on HIV Env. The antibodies recognize a glycan-dependent epitope on the prefusion conformation of gp41 and unambiguously distinguish cleaved from uncleaved Env trimers, an important property given increasing evidence that cleavage is required for vaccine candidates that seek to mimic the functional HIV envelope spike. The availability of this set of antibodies expands the number of vaccine targets on HIV and provides reagents to characterize the native envelope spike.
INTRODUCTION
Broadly neutralizing antibodies (bnAbs) to highly antigenically variable viruses such as HIV, influenza virus and HCV have attracted much attention in recent years as they provide new opportunities to counter a particularly troublesome category of pathogens (Burton et al., 2012; Kwong and Mascola, 2012; Klein et al., 2013;). Such antibodies are being used to help guide vaccine design for these pathogens by defining conserved epitopes that can then be presented to the immune system under more favorable conditions than typically occur in natural infection or using most conventional vaccination approaches. For example, grafting of conserved epitopes into molecular scaffolds to serve as candidate immunogens is one approach under intense study (Jardine et al., 2013; Kulp and Schief, 2013). BnAbs have also been critical in native envelope protein structural studies, notably for HIV (Julien et al., 2013a; Lyumkis et al., 2013), HCV (L. Kong et al. Science in press), Ebola (Lee et al., 2008) and RSV (McLellan et al., 2013). Finally, there is increasing evidence of the potential of bnAbs as therapeutics (Klein et al., 2012b; Barouch et al., 2013; Shingai et al., 2013). However, the number of targets identified by bnAbs is limited and the discovery of more targets would be of considerable value for vaccine design, structural studies and for therapeutic applications.
For HIV, bnAbs have been identified that recognize a region of gp41 close to the virus membrane (the membrane proximal external region or MPER) (Muster et al., 1993; Zwick et al., 2001; Huang et al., 2012), the CD4 binding site (CD4bs) of gp120 (Burton et al., 1994; Zhou et al., 2010; Scheid et al., 2011; Wu et al., 2011) a region of the V2 loop of gp120 including the glycan at N160 (Walker et al., 2009; 2011) and a region of gp120 centered on the glycan at N332 (Buchacher et al., 1994; Trkola et al., 1996; Walker et al., 2011). Two other potential bnAb sites, the first, possibly involving the V3 loop and the coreceptor site of gp120 (Klein et al., 2012a), and the second, possibly involving the CD4bs of gp120 and the N-trimer structure on gp41 (Zhang et al., 2012) have been described but only partially characterized. We have previously isolated a number of potent broadly neutralizing monoclonal antibodies to the V2 and N332 regions from HIV-infected donors (Walker et al., 2009; 2011). These antibodies were isolated by direct functional screening rather than by antigen selection (Scheid et al., 2009). In this method, IgG+ memory B cells from the donor were plated at approximately a single B cell per well and, following activation for 7–8 days, supernates were screened for their ability to neutralize indicator viruses. Antibody variable region genes were rescued from positive wells by PCR and IgG molecules expressed for further studies.
Here, we applied this approach to an HIV-infected donor who had outstanding potency and breadth of serum that led to classification of the donor as an “elite neutralizer” (Simek et al., 2009; Walker et al., 2010). We describe a set of antibodies that specifically recognize cleaved HIV Env trimer via a glycan-dependent epitope expressed on the pre-fusion form of gp41. The antibodies bind a previously undefined region on Env, and thus provide an additional target when developing immunogens for HIV vaccine design. Further, the antibodies can be used to distinguish between native and non-native conformations of Env immunogens, facilitating the selection of appropriate immunogens.
RESULTS
Isolation of antibodies PGT151–158 that show broad neutralization of HIV
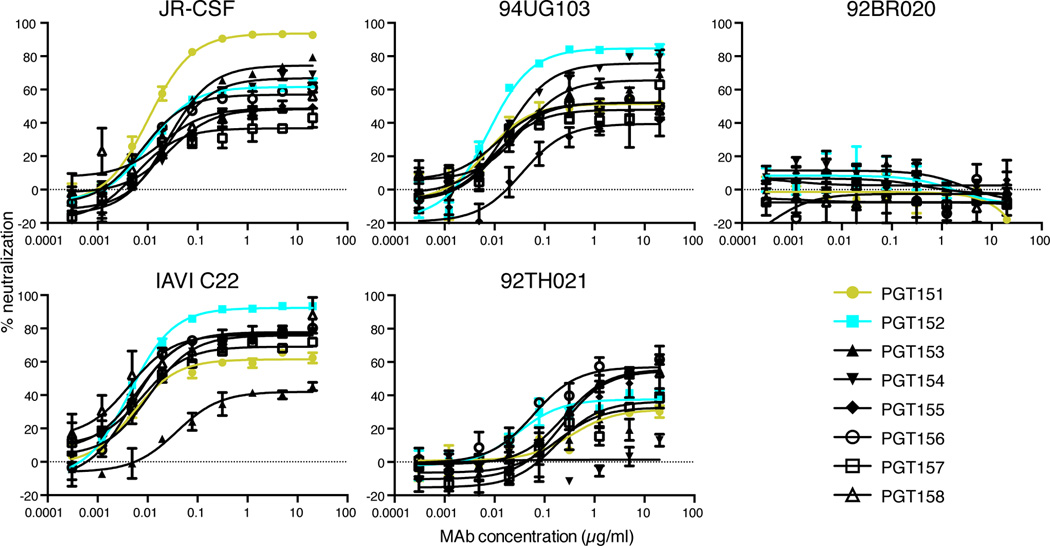
By direct functional screening of supernatants derived from activated memory B cells from an elite neutralizer infected with a clade C virus, we identified antibodies that showed notable neutralizing activity against a small set of viruses, JR-CSF (clade B), 93IN905 (clade C) and IAVI C22 (clade C), which were chosen because they were potently neutralized by the donor serum. Eight related antibody sequences, designated PGT151–158 (the PGT151 family, Table S1), were retrieved from positive wells, expressed and found to have cross-clade neutralization activity against a small panel of 5 indicator viruses in a TZM-bl pseudovirus neutralization assay (Simek et al., 2009; Walker et al., 2010). The heavy chain complementarity determining region 3s (HCDR3s) of the PGT151 family were found to be 28 amino acids in length, an unusual feature shared by several HIV-1 bnAbs targeting glycan-dependent epitopes and the MPER on gp41 (Mascola and Haynes, 2013). As shown in Figure 1, most of the antibodies neutralized 4 of the 5 viruses in the indicator panel, but none neutralized the clade B isolate 92BR020. A notable feature of the neutralization curves for some of the antibody-isolate combinations was that, although IC50 values indicated potent activity, 100% neutralization was not achieved. In a number of cases, neutralization saturation occurred in the range 50–80% with some cases of lower saturation, particularly against the clade AE isolate 92TH021.
Figure 1. PGT151–158 neutralization of an indicator panel of 5 pseudoviruses.
Serial dilutions of antibody were preincubated with pseudovirus for 1 hour and then added to TZM-bl cells. Three days post-infection luciferase values were measured and % neutralization was calculated. Data is one representative experiment of at least two replicate experiments and is presented as mean of two replicate wells +/− SEM.
Two of the antibodies with the most complete neutralization against the indicator panel of viruses, PGT151 and PGT152, were then evaluated for neutralization against a much larger panel of 117 cross-clade virus isolates in a TZM-bl pseudovirus neutralization assay. PGT151 neutralized approximately 66% of viruses with a median IC50 of 8 ng/ml and PGT152 neutralized 64% of viruses with a median IC50 of 12 ng/ml (Table 1). These bnAbs have IC50 values that are about 1-log lower than the V2 prototype bnAb PG9 (Table S2) and are comparable to the IC50 values that have been determined for the N332 glycan targeting bnAbs PGT121 and PGT128 on the same panel (Walker et al., 2011; Ferguson et al., 2013). However, it should be noted that, for a significant fraction of virus isolates, as above, complete neutralization was not achieved. PGT151 neutralized 26% of isolates and PGT152 neutralized 20% of isolates with a maximum neutralization of <80%. The percent of isolates that plateaued at <80% neutralization was much higher for PGT151 and PGT152 compared to PG9, which neutralized 1% of viruses with a plateau <80%. We have previously described such incomplete neutralization of HIV isolates by bnAbs PG9 and its somatic variant PG16, albeit at lower levels, and, in part, we ascribed this phenomenon to glycan heterogeneity (Doores and Burton, 2010). This also appears to be the case for PGT151 and PGT152 (see below).
Table 1.
PGT151 and PGT152 potency and breadth of neutralization against a 117-pseudovirus panel a
| MAb | PGT151 | PGT152 | ||
|---|---|---|---|---|
| median IC50/IC80 (µg/ml) | IC50 | IC80 | IC50 | IC80 |
| 0.008 | 0.024 | 0.012 | 0.034 | |
| breadth % | 66 | 44 | 64 | 47 |
See also Table S2.
We have previously shown JR-CSF gp120 released from virus grown in PBMCs contains a higher percentage of complex N-linked glycans (21%) than its pseudovirus counterpart (2%) (Bonomelli et al., 2011). Therefore, we wanted to determine whether the incomplete neutralization observed for the PGT151 family, using pseudovirus, translated into incomplete neutralization using the more relevant PBMC grown replication competent virus. PGT151 family neutralization of JR-CSF, JR-FL, 93IN905, and 92RW020 pseudovirus produced in 293T cells was compared to replication competent virus produced in PBMCs using TZM-bl as the target cells (Figure S1A–D, first and second columns). In parallel to the TZM-bl experiments, the replication competent virus was also assessed in neutralization of PBMCs (Figure S1A–D, third column). Neutralization of 92RW009 replication competent virus was assessed in TZM-bl and PBMC (Figure S1E). The luciferase values for the TZM-bl neutralization assays were read after a 3-day incubation, and the PBMC neutralization assays were assessed for p24 levels on days 4 and 7. In general, somewhat higher levels of saturation of neutralization were seen in assays when replication competent virus grown in PBMCs was used and when PBMCs were employed as target cells. As an example, PGT153, which did not neutralize 93IN905 pseudovirus entry into TZM-bl cells, was able to neutralize 93IN905 replication competent virus entry into these same cells, albeit modestly at 35% maximum neutralization (Figure S1C and Figure S1F). This plateau increased to 70% when replication competent viral entry into PBMCs was evaluated. These results suggest that the breadth and potency of PGT151 and PGT152 neutralization may be somewhat underestimated using a pseudovirus-TZM-bl assay relative to a PBMC grown replication competent virus-PBMC assay.
We next compared neutralization by the PGT151 family members with that of the donor serum on a smaller 25-virus panel (Table S3). The composition of this panel, derived from the 117-virus panel, was arrived at by selection of viruses showing a range of sensitivities to PGT151. As shown in Table S3, all but three of the viruses that were neutralized by the donor serum were also neutralized by individual members of the PGT151 family. The IC50 values of PGT151 (r = −0.54, P value = 0.0263), PGT152 (r = −.73, P value = 0.0013), PGT156 (r = −0.69, P value = 0.03), PGT157 (r = −0.73, P value = 0.03) and PGT158 (r= −0.69, P value = 0.288) had an inverse correlation to the donor serum 50% neutralization titer (1/dilution). Incomplete neutralization by the donor serum also correlated with incomplete neutralization by the PGT151 family members: PGT152, PGT153, PGT155, PGT156 and PGT158. The results indicate the PGT151 family can largely recapitulate the serum neutralization breadth and potency on this small virus panel.
PGT151 and PGT152 bnAbs bind specifically to cleaved and not to uncleaved Env trimers
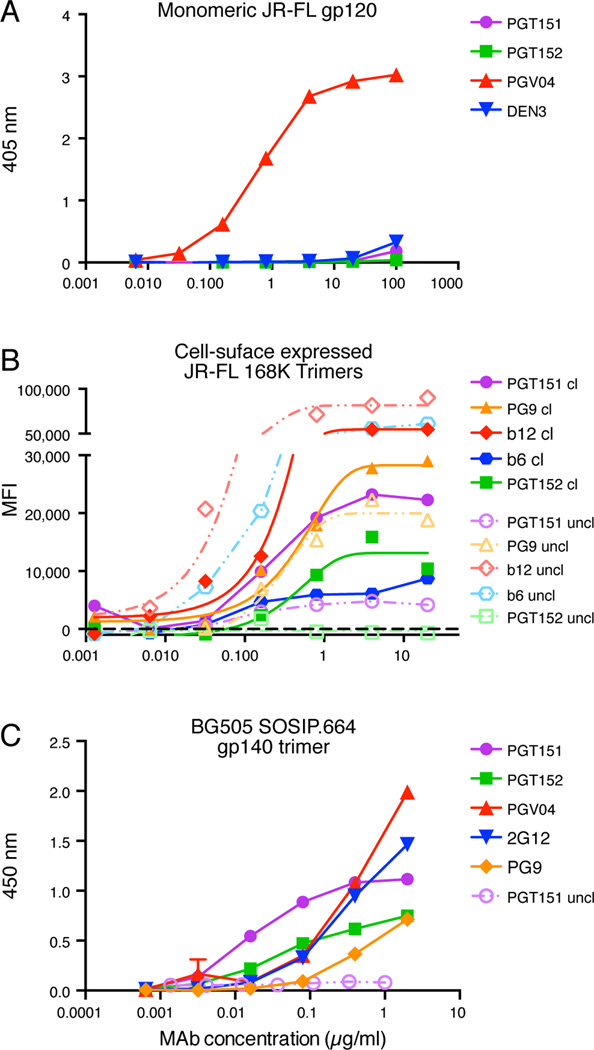
ELISA studies showed that PGT151 and PGT152 do not bind to monomeric gp120 from several viruses from multiple clades although they are able to potently neutralize the corresponding viruses (Figure 2A and Figure S2A). PGT151 and PGT152 also did not bind 293F cell–produced gp41 trimers (Figure S2B), which are most likely in a 6-helix bundle conformation. Surprisingly, PGT153 did show some binding to the gp41 trimeric protein and, of note, this antibody has the least sequence identity to PGT151 and other family members (Table S1). The results suggested that the antibodies may be largely specific for native Env trimer. In order to investigate this possibility, we measured the binding of PGT151 and PGT152 to native cleaved and uncleaved variant JRFL trimers on the surface of 293T cells by flow cytometry. This particular isolate was chosen as it has previously been shown that fully cleaved JR-FL Env is efficiently displayed on the surface of transfected 293T cells, whereas, for most other isolates (Pancera and Wyatt, 2005), relatively high proportions of uncleaved trimers are expressed. As shown in Fig 2B, PGT151 and PGT152 bind exclusively to variant JR-FL cleaved trimers and show no binding to the corresponding uncleaved timers on the surface of transfected cells. Cleavage was recently shown to be important for formation of native-like structure, and presentation of quaternary epitopes (Ringe et al., 2013). Together these data strongly argue that PGT151 and PGT152 are trimer specific and, furthermore, are able to distinguish cleaved from uncleaved trimers.
Figure 2. PGT151 and PGT152 binding to different forms of Env.
(A) PGT151 and PGT152 do not bind to JR-FL gp120 monomer as measured by ELISA. (B) PGT151 binds to native, cleaved (cl) but not to uncleaved (uncl) JR-FL E168K trimers expressed on the surface of 293T cells as measured by flow cytometry, and (C) PGT151 and PGT152 bind to cleaved soluble BG505 SOSIP.664 gp140 trimers but not to the corresponding uncleaved gp140 trimers as measured by ELISA. See also Figure S2. Data for (A) and (C) is presented as mean of two replicates +/− SEM. Data for (B) is a representative experiment of two replicate experiments.
PGT151 and PGT152 were shown not to bind to a number of recombinant, uncleaved, gp140 preparations (Figure S2B), which, although they contain three gp120 subunits attached to trimeric gp41 ectodomains, adopt conformations that do not resemble native Env trimers (Julien et al., 2013a; Lyumkis et al., 2013; Ringe et al., 2013). However, PGT151 and PGT152 do bind well to the stabilized recombinant trimeric molecule, BG505 SOSIP.664 gp140 trimer (Figure 2C), which is a close antigenic and structural mimic of the native Env trimer (Julien et al., 2013a; Lyumkis et al., 2013; Ringe et al., 2013; Sanders et al., 2013). PGT151 also distinguishes cleaved from uncleaved BG505 SOSIP.664 gp140 trimers (Figure 2C).
No antibody demonstrating such an unambiguous cleaved trimer reactivity profile has been described previously although trimer, e.g. (Walker et al., 2009; Sanders et al., 2013), and cleavage (Li et al., 2012) preferences have been reported.
PGT151 and PGT152 bind to complex tri- and tetra-antennary glycans
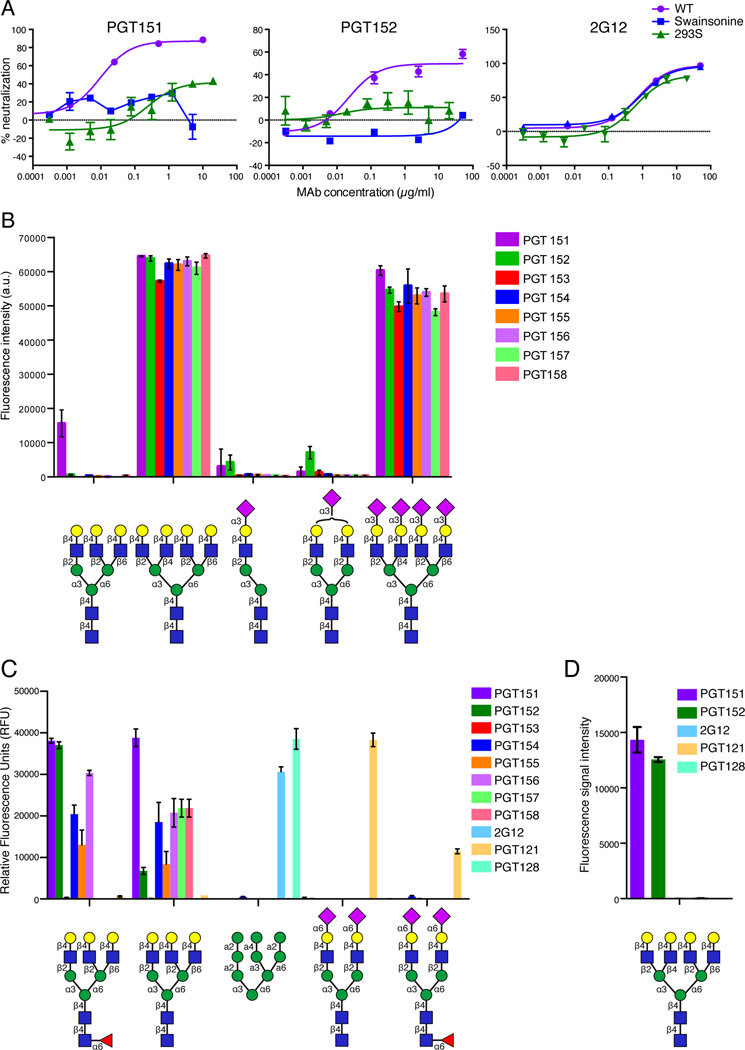
Given the less than 100% neutralization observed in a number of assays that is reminiscent of the previously described glycan dependency of binding for some anti-HIV antibodies, we investigated the ability of PGT151 and PGT152 to neutralize a number of glycan-modified viruses. The glycosidase inhibitor swainsonine prevents the formation of complex glycans, and pseudovirus made in the mutant GNT1-deficient HEK-293S cell line (GNT1−/−) also does not contain complex glycans. As shown in Fig. 3A, PGT151 or PGT152 neutralization of JR-CSF virus grown in the presence of swainsonine or in 293S cells was greatly reduced compared to neutralization of wild type virus. Moreover, although PGT151 reacted well with BG505 SOSIP.664 gp140 trimers expressed in 293T cells, it did not react with the same trimers derived from 293S cells (data not shown). These results suggest that PGT151 and PGT152 bind to complex glycans on Env.
Figure 3. PGT151-PGT158 bind complex carbohydrates.
(A) PGT151 and PGT152 neutralization of JR-CSF pseudovirus generated (i) in the presence of the glycosidase inhibitor swainsonine, which prevents the formation of complex glycans by inhibiting the trimming of mannose residues from the Man-α6 arm of the GlcNAcMan5GlcNAc2 structure or (ii) in 293S cells (GNT1−/− cells), a cell line that is deficient in N-acetylglucosaminyltransferase I and is unable to add a GlcNAc residue to the Man5GlcNAc2 structure to permit processing to complex glycans. Neutralization of JR-SCF pseudovirus by 2G12 is not affected by either (i) or (ii) since it binds exclusively to high-mannose glycans on the glycan shield of Env. Data is presented as mean +/− SEM. (B) Glycan microarray analysis reveals PGT151-PGT158 preferentially bind tetraantennary complex carbohydrates and PGT151 additionally binds a triantennary glycan as detected on the Wong glycan array. Data is presented as mean +/− SD. (C) Glycan microarray analysis using the CFG microarray additionally reveals that all PGT151 family MAbs except PGT153 preferentially bind triantennary complex carbohydrates; and (D) PGT151 and PGT152 bind a tetrantennary complex carbohydrate on the neoglycolipid microarray. The symbols for common monosaccharides are as follows: purple diamonds represent sialic acid, yellow circles represent galactose, red triangles represent fucose, blue squares represent N-acetyl glucosamine and green circles represent mannose. See also Tables S4–S5. Data for the CFG microarray is presented as mean +/− SEM.
To further investigate, we screened PGT151–158 for glycan specificity using several glycan microarrays. PGT151–158 bound tetrantennary complex type N-glycans with terminal galactose with and without terminal sialic acid residues (Figure 3B, Figure 3D and Table S5). While terminal sialic acid linked to the 6-position of galactose blocks binding of the antibodies, sialic acid linked to the 3-position appears to be accommodated. We note that in Figure 3B, the signal for tetrantennary N-glycan binding was saturated. In addition, all of the antibodies except PGT153 also bound triantennary complex type N-glycans with terminal galactose (Figure 3B–C), although, with lower avidity than the binding to tetra-antennary glycans. Blattner et al. further confirmed these results by showing that PGT151 and PGT152 bound to triantennary complex glycans using isothermal titration calorimetry (ITC) (Blattner et al., 2014). None of the antibodies were observed to interact with high-mannose glycans of the type demonstrated to interact with the antibodies 2G12 and PGT128 or the biantennary complex glycan shown to interact with PGT121 (Figure 3C–3D and Table S4–S5).
Overall, we concluded that the binding of the PGT151 family is glycan dependent but does not involve either of the high-mannose-containing regions that have been previously characterized to be important for broad neutralization. These regions are the high-mannose patch on gp120 around N332 recognized by antibodies such as 2G12, PGT121 and PGT128 and the V2 loop region around N160 recognized by antibodies such as PG9 and PG16 (Doores and Burton, 2010; McLellan et al., 2011; Walker et al., 2011; Julien et al., 2013b; Kong et al., 2013). Of note, PG16 has recently been shown to interact on glycan microarrays with a terminally sialylated tetrantennary glycan. However, this glycan has a different sialic acid linkage compared to the tetrantennary glycan that interacts with PGT151-PGT158 (Shivatare et al., 2013a). Taken together, these results indicate that the PGT151 family of antibodies bind a glycan-dependent epitope on Env that has not been previously characterized.
PGT151 does not compete with any known bnAbs for binding to Env spikes
We next epitope mapped PGT151, which is the somatic variant showing the most complete neutralization, by analyzing competition for binding to cleaved Env spikes with known bnAbs (Figure S3A). When PGT151 was prebound to JR-FL ΔCT Env trimers expressed on the surface of 293T cells, it did not compete with bnAbs targeting the V2 loop N160 glycan dependent epitope recognized by bnAb PGT145, or the region around N332 recognized by bnAbs PGT121, PGT128, PGT135, 2G12 and PGT130. This result was consistent with the glycan array data above. Additionally, when PGT151 was prebound to JR-FL E168K trimers, PGV04 (Figure S3A) and CD4-IgG (Figure S3B) were still able to bind the trimers suggesting its epitope did not significantly overlap with the CD4 binding site. PGT151 did show partial competition with CD4 prebound to cell surface expressed trimers (Figure S3C), which most likely is due to conformational changes that occur on the trimer after CD4 binding (Liu et al., 2008) leading to partial disruption of the PGT151 epitope.
It has previously been shown that MPER-targeting bnAbs, 2F5, 4E10, and 10E8 do not bind well to cleaved JR-FL trimers on the surface of 293T cells (Chakrabarti et al., 2011; Huang et al., 2012), and 2F5 and 4E10 have been shown in contrast to bind well to uncleaved JR-FL trimers (Chakrabarti et al., 2011). As PGT151 binds only to cleaved trimers, we were unable to perform satisfactory competition experiments using MPER-targeting bnAbs and PGT151. These results are further evidence to indicate that PGT151 binds to a previously unidentified epitope on Env.
PGT151 binds to a glycan-dependent epitope on gp41
Next, a large number of viruses containing alanine substitutions in gp120 and gp41 were evaluated in the context of JR-CSF neutralization by PGT151. As shown in Table S6A and Table 2, only substitutions in gp41 had a significant effect on PGT151 neutralization. The substitutions that had the greatest effect on IC50 and level of maximum neutralization of JR-CSF were N611D, N637K, E647A and E647G. The former two substitutions are expected to lead to the loss of the glycans at N611 and N637, occupied most likely by complex glycans (Go et al., 2008; 2009). Next, we investigated the ability of PGT151 to neutralize a panel of variants that had been generated in the LAI strain of HIV-1 (Table S6B). Again, the gp41 residues N611 and N637 were found to be important. In addition, an A501T substitution in gp120 showed a significant if moderate effect (Table 2 and Table S6B). Finally, in the context of the isolate JR-FL, the most marked effects occurred for substitutions leading to the loss of glycans at N611 and N637. Residues N611 and N637 are extremely highly conserved across the 30,324 viral sequences found in the Los Alamos database at 98.7% and 92.8%, respectively. E647 is 90.2% conserved across the viral sequences.
Table 2.
The effect of single amino acid substitutions on PGT151 neutralization for 3 HIV isolatesa
| PGT151 | ||||||
|---|---|---|---|---|---|---|
| Fold IC50 Increase relative to wild-type |
%Maximum neutralization |
Fold IC50 Increase relative to wild-type |
%Maximum neutralization |
Fold IC50 Increase relative to wild-type |
%Maximum neutralization |
|
| variant virus | JR-CSF | LAI | JR-FL | |||
| A501T | 6 | 75 | ||||
| N611A | 9 | 44 | 14 | 81 | >350 | 31 |
| N611D | >200 | 0 | 67 | 45 | >200 | 8 |
| S613A | 9 | 41 | ND | ND | ND | ND |
| N637A | 1 | 50 | 2 | 70 | 1 | 65 |
| N637K | >200 | 20 | >115 | 37 | >200 | 33 |
| T639A | 2 | 52 | ND | ND | ND | ND |
| E647A | >200 | 33 | 1 | 96 | 1 | 72 |
| E647G | >200 | 7 | 2 | 95 | 3 | 94 |
| WT | 1 | 93 | 1 | 100 | 1 | 99 |
Neutralization activity is reported as fold increase in IC50 value relative to wild-type and was calculated using the equation (IC50 mutant)/(IC50 wild-type). Percent maximum neutralization is the plateau of the neutralization curve reached by the MAb. Boxes are color coded as follows: blue, substitutions that had a negligible effect on IC50 or maximum neutralization; yellow, 4- to 9-fold increase in IC50; orange, 10- to 99-fold increase; red, >100-fold increase in IC50; lavender, maximum neutralization, ≥50%; brown, maximum neutralization, <50%; gray, virus not significantly infectious; and ND, not done. Data are presented as the mean of two independent experiments. See also Table S6.
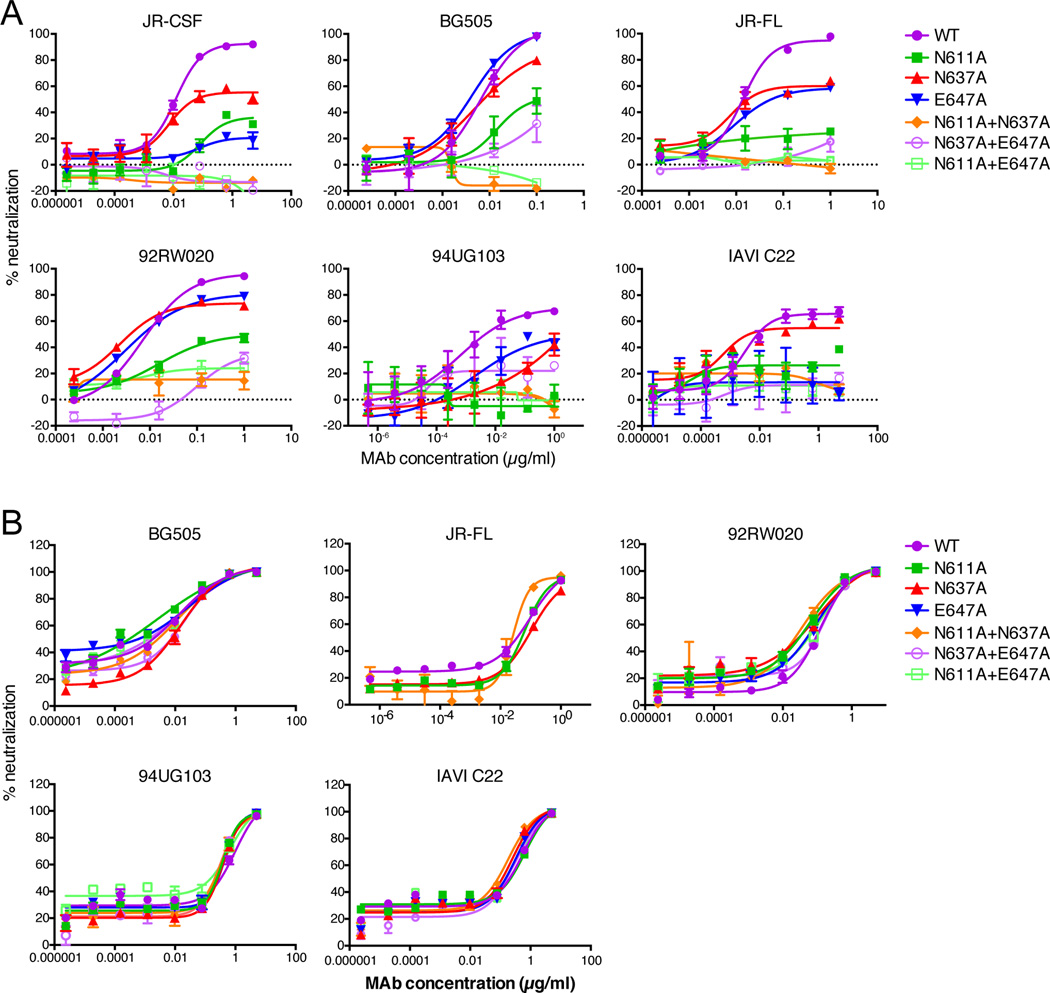
Given the evidence that some HIV glycan-dependent antibodies can interact with differing sets of glycans in an isolate-dependent context (McLellan et al., 2011) (D. Sok et al. manuscript in preparation), we tested combinations of double substitutions of N611A, N637A, and E647A in JR-CSF (clade B), BG505 (clade A), JR-FL (clade B), 92RW020 (clade A), 94UG103 (clade A) and IAVI C22 (clade C) in comparison with single substitutions in terms of their effects on PGT151 neutralization (Figure 4A). For all isolates, N611A consistently had the greatest effect on IC50 and maximum level of neutralization. The single N637A and E647A substitutions had little to no effect on the IC50 for most isolates and varying effects on the maximum level of neutralization by PGT151. However, the N611A+N637A and N611A+E647A double substitutions completely abrogated neutralization of all isolates by PGT151. PGV04 was used as a control to show that the substitutions did not affect the IC50 or maximum inhibition of this CD4bs bnAb (Figure 4B).
Figure 4. PGT151 neutralization of viruses containing N611A, N637A, E647A single and double alanine substitutions.
Serial dilutions of antibody (A) PGT151 and (B) PGV04, used as a control, were preincubated with pseudovirus for 1 hour and then added to TZM-bl cells. Three days post-infection luciferase values were measured and % neutralization was calculated. See also Figure S4. Data are from a representative experiment of at least two replicate experiments and are presented as mean values of two replicate wells +/− SEM.
To determine whether the glycans at N611 and N637 were the only combination of gp41 glycans that PGT151 uses for binding, we made double substitutions of all four (N611, N616, N625, N637) gp41 glycan combinations in JR-CSF (Figure S4A) and JR-FL (Figure S4C), and the four equivalent (N611, N618, N625, N637) gp41 glycan combinations in BG505 (Figure S4B). No other combination of glycan substitution other than N611A+N637A completely eliminated neutralization by PGT151. All other double substitution combinations that removed two glycans in gp41 (N616 or N618, N625) in combination with either N611 or N637 had no further effects on neutralization than the single N611 or N637 substitution alone. The double substitution N616A+625A had no effect at all on neutralization by PGT151. PGV04 was used as a control for these substitutions for BG505 (Figure S4C), and PGV04 and 10E8 were used as controls for the substitutions in JR-FL (Figure S4C). In addition, double substitutions leading to the removal of the 4 gp41 glycans in JR-CSF were made in combination with N88A, which would lead to the removal of a glycan that would not be expected to be close to the PGT151 epitope (Julien et al., 2013a; Lyumkis et al., 2013). These double substitutions had no effect on PGT151 neutralization in the same way as the double substitutions at N616 (or N618) and N625 had no effect on PGT151 neutralization.
Glycan sites in gp120 predicted to be at or close to the PGT151 epitope, based on structural work (Julien et al., 2013a; Lyumkis et al., 2013; Blattner et al., 2014), were also eliminated in combination with N611 or N637 for the JR-CSF isolate (N230, N241, N276 and N448) (Figure S4A) and the BG505 isolate (N234, N276, and N448) (Figure S4B). A number of combinations did indeed have effects on the maximum neutralization observed (Figure S4A, S4B). While the N234A+N637A substitution in BG505 lowered the PGT151 percent maximum neutralization, this result was isolate specific. However, the substitutions N448A+N611A and N448A+N637A in JR-CSF and N448A+N611A in BG505 increased the maximum neutralization of PGT151 relative to the single N611A and N637A substitutions alone. These results suggest that glycans in regions of gp120 in the PGT151 epitope or in close proximity to the PGT151 epitope can affect the ability of PGT151 to achieve 100% maximum neutralization.
Blattner et al. fitted the high resolution EM structure of BG505 SOSIP.664 into the reconstruction of the JR-FL:PGT151 Fab complex predicting the PGT151 LC to interact with N276 from one gp120 protomer, and N262 and N448 from a second gp120 protomer (Blattner et al., 2014). The removal of the glycan at N276 or N448 in BG505 or JR-CSF backgrounds had no effect on PGT151 neutralization. Furthermore, the removal of the combination of glycans at N276+N611 or N276+N611 had no further effects on PGT151 neutralization as compared to the effects of virus lacking only the glycan at N611 or N637 respectively. In contrast, the removal of the glycan at N448 in combination with N637 or N611 increased PGT151 maximum neutralization compared to the neutralization plateaus of virus lacking the single glycans at N611 or N637. This suggests that the glycan at N448 affects PGT151 neutralization only in the context of the loss of a glycan at N611 and/or N637 or of the presence of certain glycoforms at these positions. Our data paint a complex picture of Env recognition by PGT151. Many interactions appear to be isolate dependent and phenotypes may not manifest when single mutations are introduced (e.g. N234 and N637 in BG505, Fig. S4B). Thus, there may be redundant or compensatory interactions utilized by PGT151. Furthermore, at first glance our data may seem contradictory to that of Blattner et al., as the negative stain EM data fitted into the high resolution EM structure indicates that the glycans at N448 and N276 contact PGT151. However, we have previously shown that removal of the glycan at N276, which forms contacts to VRC01 as determined by crystal structure, actually increases VRC01 neutralization potency in functional assays (Falkowska et al., 2012). It appears that the VRC01 antibody has evolved to largely to avoid the obstruction of the N276 glycan rather than to use it for a positive binding interaction. Thus, functional and structural studies can be complementary in understanding the complex interaction of antibodies with epitopes involving glycans.
PGT151 and PGT152 mediate antibody-dependent cell-mediated cytotoxicity (ADCC) and are not polyreactive
Neutralizing antibodies of the appropriate isotype recognizing native Env spikes might be expected to mediate killing of HIV-infected cells by ADCC. PGT151 and PGT152 were investigated in an ADCC assay against target cells infected with NL4–3, YU2 or JR-CSF viruses and shown to mediate this effector activity (Figure 5). SIVmac239 was used as a negative control.
Figure 5. ADCC by PGT151 and PGT152.
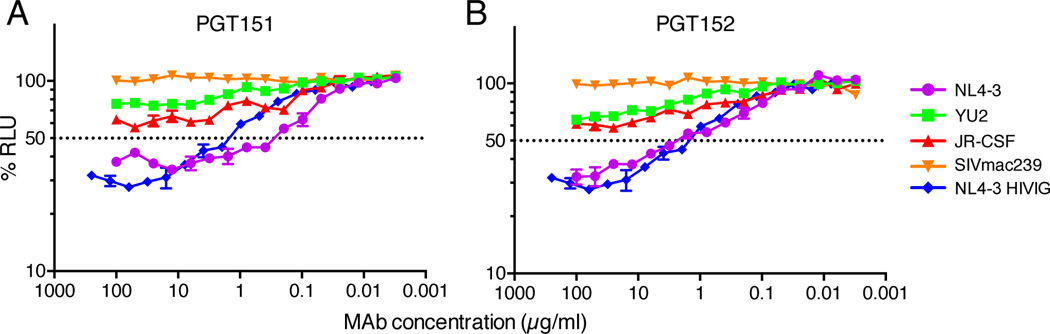
(A) PGT151 and (B) PGT152 were titered for ADCC activity against target cells infected with HIV-1 NL4–3, YU2, JR-CSF and SIVmac239, and using an NK cell line (KHYG-1) expressing CD16 as the effector cell. The killing of virus-infected cells by ADCC is indicated by a loss of relative light units (RLU). SIVmac239 is used as a negative control and HIVIG is used as a positive control. Data are presented as mean values of three replicates +/− SEM.
Finally, no evidence for polyreactivity for either PGT151 or PGT152 was found by screening against a panel of antigens (Figure S5A) and PGT151 did not bind to HEp-2 cells in an antinuclear antibody (ANA-HEp-2) indirect immunofluorescence assay indicating it does not bind to nuclear or cytoplasmic self-antigens (Figure S5B). These results indicate PGT151 and PGT152 have the capacity to mediate ADCC in addition to neutralizing free virus, which may be significant for the anti-viral activity of the antibodies in vivo, either vaccine-induced or administered passively.
DISCUSSION
The past few years have seen an explosion in the generation of bnAbs to HIV (Burton et al., 2012; Kwong and Mascola, 2012; Klein et al., 2013; Mascola and Haynes, 2013). Many of the new bnAbs are directed to epitopes defined earlier but have greater potency and/or breadth of neutralization against diverse isolates (Walker et al., 2009; 2011; Wu et al., 2010; Scheid et al., 2011; Huang et al., 2012). However, at least two epitope regions on gp120 involving protein and glycan recognition have been described (Walker et al., 2009; 2011). One of these regions is a V2 epitope dependent on a glycan at N160, whereas the other is a cluster of epitopes involving conserved parts of the V loops and centered on N332. Here, we describe a set of antibodies recognizing a previously undefined glycan-dependent epitope on gp41. The two prototype antibodies from this set, PGT151 and PGT152, are very potent and neutralize about 2/3 of a large panel of viral isolates with a median IC50 approximately 10 ng/ml, albeit showing incomplete neutralization with a significant fraction of viruses. The epitope recognized is centered on the highly conserved glycan site at N611 with the involvement of another highly conserved glycan site at N637 and the less conserved residue E647. The relative contributions of these individual residues appear to be isolate dependent. Substitution of N611 consistently had an adverse effect on PGT151 neutralization and, in combination with other substitutions at N637 or E647, invariably led to complete abrogation of antibody neutralization. Nevertheless, resistant viruses to PGT151 neutralization were identified in the large panel tested, which contained both N611 and N637 glycan sites. This is in line with previous studies on glycan-dependent gp120 antibodies for which crucial glycans have been identified but for which changes in nonglycan related amino acid residues lead to resistance even in the presence of these crucial glycans (Walker et al., 2009; 2011; McLellan et al., 2011; Doria-Rose et al., 2012). We also note that viruses lacking both the N611 and N637 glycan sites tend to show lower infectivity than wild type viruses suggesting they may be associated with a lower fitness in vivo that could be associated, at least in part, with their high levels of conservation.
For JR-CSF, the N611D substitution completely abolished PGT151 neutralization whereas removing the glycan at N611 by N611A or S613A substitutions had somewhat lesser effects on IC50 and percent maximum neutralization. Similarly, the N637K substitution abolished PGT151 neutralization whereas N637A or T639A substitutions only affected percent maximum neutralization. The stronger effect of N611D and N637K as compared to N611A and N637A were also seen in PGT151 neutralization of these variants in the LAI background. The effects of substitution thus appear to go beyond loss of glycans and suggest that PGT151 may bind to protein in addition to glycan components on gp41. For JR-FL, N611D and N611A substitutions produced comparable effects whereas the effects of N637K were more marked than those of N637A suggesting differences in the precise mode of recognition of PGT151 for different Envs.
Blattner et al. determined the ability of PGT151 to bind to variants of the BG505 SOSIP.664 trimer (Blattner et al., 2014). The N611Q+N637Q double substitution also eliminated binding of PGT151 in the context of the BG505 SOSIP.664 trimer. Some differences in the magnitudes of effects of single substitutions of glycan sites at N611 and N637 were observed that may reflect the presence of the MPER of gp41 and the proximity of the viral membrane in neutralization studies but not those using the recombinant SOSIP protein. Similarly, some differences were observed between neutralization studies and SOSIP binding studies in terms of the effects of a number of gp120 substitutions on PGT151 binding. Thus, a number of substitutions led to decreased binding of PGT151 to BG505 SOSIP (Blattner et al., 2014) but did not impact neutralization. The residues that lowered binding but did not impact neutralization might have had a general effect on the trimer structure of BG505 SOSIP.664 that did not affect trimer structure and/or accessibility of PGT151 to its epitope in the context of virus neutralization. Alternatively, it may be a strain specific effect as we made substitutions in JR-CSF and LAI.
The glycans involved in interaction with PGT151 and PGT152 are likely complex glycans, based on the fact that virus produced with high-mannose glycans only is not neutralized, and consistent with glycan array studies that show optimal binding to tri- and tetra-antennary N-linked glycans. The ability of PGT151 to recognize slightly different epitopes on different isolates may at first seem surprising but has in fact been noted for both V2 and N332-dependent HIV bnAbs (McLellan et al., 2011) (D. Sok et al. manuscript in preparation). It appears that bnAbs recognizing neighboring glycans can employ varying modes of recognition to the extent that alternate glycans can be used on different isolates in some cases. Detailed molecular understanding of how promiscuous binding is achieved for the PGT151 family of antibodies must await high-resolution structures of these antibodies in complex with Env.
Similar to the V2 glycan-dependent bnAbs such as PG9 and PG16, the PGT151 family of antibodies shows incomplete neutralization behavior in the TZM-bl and PBMC assays i.e. neutralization curves that plateau at less than 100% for a fraction of viral isolates. For PG9, incomplete neutralization has been ascribed, at least in part, to glycan heterogeneity (Doores and Burton, 2010). It seems to also be the case for the PGT151 antibodies as we have shown that glycans in gp120 that are in close proximity to the PGT151 epitope can affect PGT151 neutralization. Of note, the extent of incomplete neutralization appears to be generally less in an assay using replication competent viruses grown in PBMCs infecting PBMCs than in assays that use the corresponding pseudoviruses grown in 293T or replication competent viruses grown in PBMCs infecting TZM-bl cells. The impact of incomplete neutralization on the ability of antibodies to protect in vivo has not yet been clearly established.
One of the most striking features of PGT151 and PGT152 is that they are specific for cleaved Env trimer and do not recognize uncleaved Env trimer. This behavior is seen in the context of trimer expressed on the surface of infected cells and in the context of a recombinant trimer that mimics virion-expressed trimer. The gp41 components of the uncleaved soluble gp140 proteins have previously been shown to adopt a 6-helix bundle conformation, while, the gp41 components in the context of the cleaved, SOSIP trimers retain the prefusion form (Guttman and Lee, 2013; Julien et al., 2013a; Lyumkis et al., 2013; Ringe et al., 2013). Hence, PGT151 and PGT152 report on the conformation of gp41 and trimeric Env and should be valuable in distinguishing different forms of Env as found in a number of potential immunogens e.g. VLPs. The antibodies also appear to stabilize the cleaved Env trimer (C. Blattner et al, accompanying manuscript) and this property may be crucial in structural studies on functional spikes isolated from infected cell surfaces.
Finally, the PGT151 family of antibodies defines a previously unidentified broadly neutralizing target on the HIV envelope spike that, following molecular characterization, may be incorporated into vaccine designs. The prevalence of the PGT151 specificity in donors with broadly neutralizing sera is likely limited given earlier observations on the predominance of neutralizing specificities e.g. (Walker et al., 2010). However, more precise understanding of prevalence will require the use of the distinguishing features presented in this study to characterize sera from a number of cohorts of infected individuals.
EXPERIMENTAL PROCEDURES
Isolation of monoclonal antibodies
The method for isolating human monoclonal antibodies from memory B cells in circulation has previously been described (Walker et al., 2009).
Pseudovirus generation
The generation of pseudoviruses incorporating Envs from different HIV-1 strains and/or containing single alanine substitutions is fully described elsewhere (Pantophlet et al., 2003). Swainsonine was added at the time of transfection for the glycosidase inhibitor experiments, and was used at 20 µM.
Neutralization assay
The neutralization activity of antibodies against pseudovirus was measured using a TZM-bl assay as previously described (Li et al., 2005). Neutralization of PBMC grown replication competent virus was evaluated by using the TZM-bl assay as above or human PBMCs. For neutralization using PBMCs, antibody and virus were pre-incubated for 1 hr at 37°C before being added to the stimulated PBMCs in a 96-well plate. The cells were incubated for 24 hrs, washed 3 times, and incubated for 4–7 days. p24 was read on day 4 and day 7 post-infection.
Enzyme-linked immunosorbent assay (ELISA)
96-well plates were coated overnight at 4°C with anti-gp120 antibody D7324 (International Enzymes, Inc.) at 5 µg/ml in PBS. Plates were washed 4 times with PBS, 0.05% Tween, and either gp120 protein or lysed virus (931N905 or 94UG103) was added and incubated for 2 hrs at 37°C. The remainder of the experiment was conducted as previously described (Falkowska et al., 2012). The gp140 and gp41 proteins were directly coated onto the ELISA plates. The polyreactivity ELISA was previously described (Walker et al., 2009).
BG505 SOSIP.664 trimer ELISA
The procedure for the Ni-NTA ELISA has been previously described (Bontjer et al., 2010). Briefly, 0.1 µg/ml of His-tagged BG505 SOSIP gp140 was added in PBS to Ni-NTA His Sorb 96-well plates (Qiagen) for 1 hr at 37°C. Wells were washed 4 times with PBS 0.05% Tween 20. MAbs were added at a serial dilution in 1% BSA in PBS for 2 hrs at 37°C. Wells were washed and anti-human IgG F(ab’)2 -HRP was added at 1:5,000 dilution for 45 min at room temperature. Wells were washed and 1:1 mixed TMB substrate solution (Pierce) was added for colorimetric detection. The colorimetric reaction was stopped using 2N H2SO4 and absorption was measured at 450 nm. Uncleaved tagged BG505 SOSIP was generated by replacing the optimized cleavage site (RRRRRR) by an uncleavable motif (SEKS) (Ringe et al., 2013). The ELISA for uncleaved D7324-tagged BG505 SOSIP trimers and corresponding cleaved D7324-tagged BG505 SOSIP was previously described (Ringe et al., 2013). In Figure 2C, MAb binding to uncleaved D7324-tagged BG505 SOSIP or cleaved His-tagged BG505 SOSIP is shown. The data for cleaved D7324-tagged BG505 SOSIP is not shown but is comparable to Histagged BG505 SOSIP.
Flow cytometry
Serial dilutions (1:5) of PGT151, PGT152, PG9, b12, and b6 were added starting at 20 µg/ml to JR-FL E168KΔCT WT or uncleaved (REKR replaced with SEKS)-transfected 293T cells and incubated for 1 hr at 37°C on a plate rocker. The plate was washed 2 times in FACS buffer (PBS, 10% FBS, 0.01% sodium azide) and stained with a 1:200 dilution of R-phycoerythrin (PE) conjugated AffiniPure F(ab’)2 fragment goat anti-human IgG1 F(ab’)2 (Jackson ImmunoResearch). Binding was analyzed using flow cytometry as previously described (Walker et al., 2009).
The Consortium for Functional Glycomics (CFG) glycan microarray analysis
PGT151 and PGT152 were screened on a printed glycan microarray version 5.0 from the CFG as described previously (Blixt et al., 2004). Antibodies were used at 30 µg/ml and were precomplexed with 15 µg/ml secondary antibody (goat anti-human-Fc-rPE, Jackson Immunoresearch) before addition to the slide. Complete glycan array data sets for all antibodies can be found at http://www.functionalglycomics.org through the public database of the Consortium for Functional Glycomics under resource request number 2944.
Neoglycolipid (NGL) microarray analysis
Analysis with neoglycolipid arrays (Feizi and Chai, 2004) was carried out as described previously (Pejchal et al., 2011). In brief, PGT151, and PGT121 were analyzed at 50 µg/ml followed by biotinylated anti-human IgG (Vector) at 5 µg/ml. The results of PGT128 and 2G12 were taken from earlier experiments performed using different versions of microarrays as described (Pejchal et al., 2011). The analyses were performed at 20°C. The full array data with 38 oligosaccharide probes are in Table S4.
Wong glycan microarray analysis
Amine-functional glycans were printed in replicates of three onto NHS-activated glass slides at a 100 mM concentration as previously described (Liang et al., 2011; Shivatare et al., 2013b). Printed slides were incubated for 1hr at 80% humidity followed by o/n desiccation. Slides were blocked with ethanolamine (50 mM ethanolamine in borate buffer, pH 9.2). Antibodies, 10 µg, were pre-complexed with DyLight 649-AffiniPure mouse antihuman IgG-Fc (5 µg, Jackson) for 1 hr at 4°C. The sample was added to glycan array and incubated at 1 hr at 4°C. The slides were washed sequentially in PBS 0.05% Tween-20, PBS and then water. Arrays were scanned on a ProScanArray HT (PerkinElmer) confocal slide scanner. Image analysis was carried out with Genepix Pro 6.0 analysis software (Molecular Devices Corporation, Union City, CA). Error bars represent the average percentage error for all data points reported. The oligosaccharide probes and array data are shown in Table S5.
ADCC
ADCC activity was measured as previously described (Alpert et al., 2012). CEM.NKR-CCR5-sLTR-Luc cells (1 × 106 cells), which contain a Tat-inducible luciferase reporter gene, were infected with HIV-1NL4–3 (200 ng p24), HIV-1YU2 (1 µg p24), HIV-1JR-CSF (1 µg p24) or SIVmac239 (500 ng p27) by spinoculation in the presence of polybrene (40 µg/ml) (EMD Millipore). Four days post-infection, CEM.NKR-CCR5-sLTR-Luc cells were washed three times and incubated in 96-well plates with an NK cell line (KHYG-1) expressing CD16 at a 10:1 E:T ratio in the presence of serial dilutions of monoclonal antibodies or purified immunoglobulin from HIV-1+ donors (HIVIG; NIH AIDS Reagent Program, Division of AIDS, NIAID, NIH: Catalog #3957, HIV-IG from NABI and NHLBI). After an 8-hour incubation, cells were mixed with luciferase substrate (BriteLite Plus; Perkin Elmer), and luciferase activity in relative light units (RLU) was measured using a Victor X4 plate reader (Perkin Elmer). ADCC activity was calculated from the mean RLU for triplicate wells at each antibody concentration relative to the means for background and maximal RLU from replicate wells containing uninfected and infected cells, respectively, incubated with NK cells, but without antibody.
Statistics
Statistical analyses were done with Prism 6.0 for Mac (GraphPad, La Jolla, CA).
Supplementary Material
HIGHLIGHTS.
PGT151–158 constitute a family of broad and potent HIV-1 neutralizing antibodies.
PGT151–152 bind specifically to cleaved and not to uncleaved HIV Env trimers.
PGT151–152 bind to complex tri- and tetra-antennary glycans.
PGT151–152 bind a conserved glycan-dependent epitope on gp41.
ACKNOWLEDGEMENTS
This work was supported by IAVI through the NAC (DRB, PP, IAW, ABW, JPM), NIH grant AI33232 (DRB), the CHAVI-ID UM1AI100663 (DRB, IAW, ABW), and The Ragon Institute (DRB and EF); the Aids fonds Netherlands, grants #2011032 (RD) and #2012041 (MJvG); HIVRAD, grant # P01 AI82362 (JPM); NIH R01 #s AI098484 and AI095098 (DE); a DAAD Research fellowship (C.B.); a Dissertation Fellowship from the CA HIV/AIDS Research Program (J.H.L); a Vidi grant NWO and ERC-StG-2011-280829-SHEV (RWS); grant #’s 101-CDA-L07 (CYW), NSC 101-2321-B-001-024 and NSC 102-2321-B-001-017 (CHW and CYW); AI69993 and AI98602 (MBZ) and T32 AI007244 (A Ramirez); GRS/79268, EP/G037604/1, WT093378MA, and U01 CA128416 (TF); and GM62116 and GM098791 support the CFG glycan microarrays. We thank A Cuevas, D Parton, J Russell, A Cupo, J Korzun and PJ Klasse for technical assistance and/or advice, and C Corbaci for her help in figure preparation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Alpert MD, Heyer LN, Williams DEJ, Harvey JD, Greenough T, Allhorn M, Evans DT. Articles of Significant Interest Selected from This Issue by the Editors. J. Virol. 2012;86:11955–11955. [Google Scholar]
- Barouch DH, Whitney JB, Moldt B, Klein F, Oliveira TY, Liu J, Stephenson KE, Chang H-W, Shekhar K, Gupta S, et al. Therapeutic efficacy of potent neutralizing HIV-1-specific monoclonal antibodies in SHIV-infected rhesus monkeys. Nature. 2013;503:224–228. doi: 10.1038/nature12744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blattner C, Lee JH, Sliepen K, Derking R, Falkowska E, Torrents de la Peña A, Cupo A, Julien JP, van Gils M, et al. Structural delineation of a quaternary, cleavage-dependent epitope at the gp41-gp120 interface on intact HIV-1 Env trimers. Immunity. 2014 doi: 10.1016/j.immuni.2014.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blixt O, Head S, Mondala T, Scanlan C, Huflejt ME, Alvarez R, Bryan MC, Fazio F, Calarese D, Stevens J, et al. Printed covalent glycan array for ligand profiling of diverse glycan binding proteins. Proc. Natl. Acad. Sci. U.S.a. 2004;101:17033–17038. doi: 10.1073/pnas.0407902101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonomelli C, Doores KJ, Dunlop DC, Thaney V, Dwek RA, Burton DR, Crispin M, Scanlan CN. The glycan shield of HIV is predominantly oligomannose independently of production system or viral clade. PLoS ONE. 2011;6:e23521. doi: 10.1371/journal.pone.0023521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bontjer I, Melchers M, Eggink D, David K, Moore JP, Berkhout B, Sanders RW. Stabilized HIV-1 envelope glycoprotein trimers lacking the V1V2 domain, obtained by virus evolution. J. Biol. Chem. 2010;285:36456–36470. doi: 10.1074/jbc.M110.156588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchacher A, Predl R, Strutzenberger K, Steinfellner W, Trkola A, Purtscher M, Gruber G, Tauer C, Steindl F, Jungbauer A. Generation of human monoclonal antibodies against HIV-1 proteins; electrofusion and Epstein-Barr virus transformation for peripheral blood lymphocyte immortalization. AIDS Res. Hum. Retroviruses. 1994;10:359–369. doi: 10.1089/aid.1994.10.359. [DOI] [PubMed] [Google Scholar]
- Burton DR, Pyati J, Koduri R, Sharp SJ, Thornton GB, Parren PW, Sawyer LS, Hendry RM, Dunlop N, Nara PL. Efficient neutralization of primary isolates of HIV-1 by a recombinant human monoclonal antibody. Science. 1994;266:1024–1027. doi: 10.1126/science.7973652. [DOI] [PubMed] [Google Scholar]
- Burton DR, Poignard P, Stanfield RL, Wilson IA. Broadly neutralizing antibodies present new prospects to counter highly antigenically diverse viruses. Science. 2012;337:183–186. doi: 10.1126/science.1225416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarti BK, Pancera M, Phogat S, O'Dell S, McKee K, Guenaga J, Robinson J, Mascola J, Wyatt RT. HIV type 1 Env precursor cleavage state affects recognition by both neutralizing and nonneutralizing gp41 antibodies. AIDS Res. Hum. Retroviruses. 2011;27:877–887. doi: 10.1089/aid.2010.0281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doores KJ, Burton DR. Variable loop glycan dependency of the broad and potent HIV-1-neutralizing antibodies PG9 and PG16. J. Virol. 2010;84:10510–10521. doi: 10.1128/JVI.00552-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doria-Rose NA, Georgiev I, O'Dell S, Chuang G-Y, Staupe RP, McLellan JS, Gorman J, Pancera M, Bonsignori M, Haynes BF, et al. A short segment of the HIV-1 gp120 V1/V2 region is a major determinant of resistance to V1/V2 neutralizing antibodies. J. Virol. 2012;86:8319–8323. doi: 10.1128/JVI.00696-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falkowska E, Ramos A, Feng Y, Zhou T, Moquin S, Walker LM, Wu X, Seaman MS, Wrin T, Kwong PD, et al. PGV04, an HIV-1 gp120 CD4 binding site antibody, is broad and potent in neutralization but does not induce conformational changes characteristic of CD4. J. Virol. 2012;86:4394–4403. doi: 10.1128/JVI.06973-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feizi T, Chai W. Innovation: Oligosaccharide microarrays to decipher the glyco code. Nat Rev Mol Cell Biol. 2004;5:582–588. doi: 10.1038/nrm1428. [DOI] [PubMed] [Google Scholar]
- Ferguson AL, Falkowska E, Walker LM, Seaman MS, Burton DR, Chakraborty AK. Computational Prediction of Broadly Neutralizing HIV-1 Antibody Epitopes from Neutralization Activity Data. PLoS ONE. 2013;8:e80562. doi: 10.1371/journal.pone.0080562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Go EP, Chang Q, Liao H-X, Sutherland LL, Alam SM, Haynes BF, Desaire H. Glycosylation Site-Specific Analysis of Clade C HIV-1 Envelope Proteins. J. Proteome Res. 2009;8:4231–4242. doi: 10.1021/pr9002728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Go EP, Irungu J, Zhang Y, Dalpathado DS, Liao H-X, Sutherland LL, Alam SM, Haynes BF, Desaire H. Glycosylation site-specific analysis of HIV envelope proteins (JR-FL and CON-S) reveals major differences in glycosylation site occupancy, glycoform profiles, and antigenic epitopes' accessibility. J. Proteome Res. 2008;7:1660–1674. doi: 10.1021/pr7006957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Lee KK. A functional gp41-gp120 interaction is observed in monomeric but not oligomeric, uncleaved HIV-1 Env gp140. J. Virol. 2013;86:8750–8764. doi: 10.1128/JVI.01681-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Ofek G, Laub L, Louder MK, Doria-Rose NA, Longo NS, Imamichi H, Bailer RT, Chakrabarti B, Sharma SK, et al. Broad and potent neutralization of HIV-1 by a gp41-specific human antibody. Nature. 2012;491:406–412. doi: 10.1038/nature11544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jardine J, Julien J-P, Menis S, Ota T, Kalyuzhniy O, McGuire A, Sok D, Huang P-S, MacPherson S, Jones M, et al. Rational HIV immunogen design to target specific germline B cell receptors. Science. 2013;340:711–716. doi: 10.1126/science.1234150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julien J-P, Cupo A, Sok D, Stanfield RL, Lyumkis D, Deller MC, Klasse PJ, Burton DR, Sanders RW, Moore JP, et al. Crystal Structure of a Soluble Cleaved HIV-1 Envelope Trimer. Science. 2013a doi: 10.1126/science.1245625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julien J-P, Sok D, Khayat R, Lee JH, Doores KJ, Walker LM, Ramos A, Diwanji DC, Pejchal R, Cupo A, et al. Broadly Neutralizing Antibody PGT121 Allosterically Modulates CD4 Binding via Recognition of the HIV-1 gp120 V3 Base and Multiple Surrounding Glycans. PLoS Pathog. 2013b;9:e1003342. doi: 10.1371/journal.ppat.1003342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein F, Gaebler C, Mouquet H, Sather DN, Lehmann C, Scheid JF, Kraft Z, Liu Y, Pietzsch J, Hurley A, et al. Broad neutralization by a combination of antibodies recognizing the CD4 binding site and a new conformational epitope on the HIV-1 envelope protein. J. Exp. Med. 2012a;209:1469–1479. doi: 10.1084/jem.20120423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein F, Halper-Stromberg A, Horwitz JA, Gruell H, Scheid JF, Bournazos S, Mouquet H, Spatz LA, Diskin R, Abadir A, et al. HIV therapy by a combination of broadly neutralizing antibodies in humanized mice. Nature. 2012b;492:118–122. doi: 10.1038/nature11604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein F, Mouquet H, Dosenovic P, Scheid JF, Scharf L, Nussenzweig MC. Antibodies in HIV-1 vaccine development and therapy. Science. 2013;341:1199–1204. doi: 10.1126/science.1241144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong L, Lee JH, Doores KJ, Murin CD, Julien J-P, McBride R, Liu Y, Marozsan A, Cupo A, Klasse PJ, et al. Supersite of immune vulnerability on the glycosylated face of HIV-1 envelope glycoprotein gp120. Nat. Struct. Mol. Biol. 2013;20(7):796–803. doi: 10.1038/nsmb.2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulp DW, Schief WR. Advances in structure-based vaccine design. Curr Opin Virol. 2013;3:322–331. doi: 10.1016/j.coviro.2013.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwong PD, Mascola JR. Human Antibodies that Neutralize HIV-1: Identification, Structures, and B Cell Ontogenies. Immunity. 2012;37:412–425. doi: 10.1016/j.immuni.2012.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JE, Fusco ML, Hessell AJ, Oswald WB, Burton DR, Saphire EO. Structure of the Ebola virus glycoprotein bound to an antibody from a human survivor. Nature. 2008;454:177–182. doi: 10.1038/nature07082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Gao F, Mascola JR, Stamatatos L, Polonis VR, Koutsoukos M, Voss G, Goepfert P, Gilbert P, Greene KM, et al. Human immunodeficiency virus type 1 env clones from acute and early subtype B infections for standardized assessments of vaccine-elicited neutralizing antibodies. J. Virol. 2005;79:10108–10125. doi: 10.1128/JVI.79.16.10108-10125.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, O'Dell S, Wilson R, Wu X, Schmidt SD, Hogerkorp C-M, Louder MK, Longo NS, Poulsen C, Guenaga J, et al. HIV-1 neutralizing antibodies display dual recognition of the primary and coreceptor binding sites and preferential binding to fully cleaved envelope glycoproteins. J. Virol. 2012;86:11231–11241. doi: 10.1128/JVI.01543-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang C-H, Wang S-K, Lin C-W, Wang C-C, Wong C-H, Wu C-Y. Effects of neighboring glycans on antibody-carbohydrate interaction. Angew. Chem. Int. Ed. Engl. 2011;50:1608–1612. doi: 10.1002/anie.201003482. [DOI] [PubMed] [Google Scholar]
- Liu J, Bartesaghi A, Borgnia MJ, Sapiro G, Subramaniam S. Molecular architecture of native HIV-1 gp120 trimers. Nature. 2008;455:109–113. doi: 10.1038/nature07159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyumkis D, Julien J-P, de Val N, Cupo A, Potter CS, Klasse PJ, Burton DR, Sanders RW, Moore JP, Carragher B, et al. Cryo-EM Structure of a Fully Glycosylated Soluble Cleaved HIV-1 Envelope Trimer. Science. 2013 doi: 10.1126/science.1245627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mascola JR, Haynes BF. HIV-1 neutralizing antibodies: understanding nature's pathways. Immunol. Rev. 2013;254:225–244. doi: 10.1111/imr.12075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLellan JS, Chen M, Joyce MG, Sastry M, Stewart-Jones GBE, Yang Y, Zhang B, Chen L, Srivatsan S, Zheng A, et al. Structure-based design of a fusion glycoprotein vaccine for respiratory syncytial virus. Science. 2013;342:592–598. doi: 10.1126/science.1243283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLellan JS, Pancera M, Carrico C, Gorman J, Julien J-P, Khayat R, Louder R, Pejchal R, Sastry M, Dai K, et al. Structure of HIV-1 gp120 V1/V2 domain with broadly neutralizing antibody PG9. Nature. 2011;480:336–343. doi: 10.1038/nature10696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muster T, Steindl F, Purtscher M, Trkola A, Klima A, Himmler G, Rüker F, Katinger H. A conserved neutralizing epitope on gp41 of human immunodeficiency virus type 1. J. Virol. 1993;67:6642–6647. doi: 10.1128/jvi.67.11.6642-6647.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pancera M, Wyatt R. Selective recognition of oligomeric HIV-1 primary isolate envelope glycoproteins by potently neutralizing ligands requires efficient precursor cleavage. Virology. 2005;332:145–156. doi: 10.1016/j.virol.2004.10.042. [DOI] [PubMed] [Google Scholar]
- Pantophlet R, Ollmann Saphire E, Poignard P, Parren PWHI, Wilson IA, Burton DR. Fine mapping of the interaction of neutralizing and nonneutralizing monoclonal antibodies with the CD4 binding site of human immunodeficiency virus type 1 gp120. J. Virol. 2003;77:642–658. doi: 10.1128/JVI.77.1.642-658.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pejchal R, Doores KJ, Walker LM, Khayat R, Huang P-S, Wang S-K, Stanfield RL, Julien J-P, Ramos A, Crispin M, et al. A potent and broad neutralizing antibody recognizes and penetrates the HIV glycan shield. Science. 2011;334:1097–1103. doi: 10.1126/science.1213256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ringe RP, Sanders RW, Yasmeen A, Kim HJ, Lee JH, Cupo A, Korzun J, Derking R, van Montfort T, Julien J-P. Cleavage strongly influences whether soluble HIV-1 envelope glycoprotein trimers adopt a nativelike conformation. Proc. Natl. Acad. Sci. U.S.A. 2013;110(45):18256–18261. doi: 10.1073/pnas.1314351110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders RW, Derking R, Cupo A, Julien J-P, Yasmeen A, de Val N, Kim HJ, Blattner C, la Peña, de AT, Korzun J, et al. A Next-Generation Cleaved, Soluble HIV-1 Env Trimer, BG505 SOSIP.664 gp140, Expresses Multiple Epitopes for Broadly Neutralizing but Not Non-Neutralizing Antibodies. PLoS Pathog. 2013;9:e1003618. doi: 10.1371/journal.ppat.1003618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheid JF, Mouquet H, Feldhahn N, Seaman MS, Velinzon K, Pietzsch J, Ott RG, Anthony RM, Zebroski H, Hurley A, et al. Broad diversity of neutralizing antibodies isolated from memory B cells in HIV-infected individuals. Nature. 2009;458:636–640. doi: 10.1038/nature07930. [DOI] [PubMed] [Google Scholar]
- Scheid JF, Mouquet H, Ueberheide B, Diskin R, Klein F, Oliveira TYK, Pietzsch J, Fenyo D, Abadir A, Velinzon K, et al. Sequence and structural convergence of broad and potent HIV antibodies that mimic CD4 binding. Science. 2011;333:1633–1637. doi: 10.1126/science.1207227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shingai M, Nishimura Y, Klein F, Mouquet H, Donau OK, Plishka R, Buckler-White A, Seaman M, Piatak M, Lifson JD, et al. Antibody-mediated immunotherapy of macaques chronically infected with SHIV suppresses viraemia. Nature. 2013;503:277–280. doi: 10.1038/nature12746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shivatare SS, Chang S-H, Tsai T-I, Ren C-T, Chuang H-Y, Hsu L, Lin C-W, Li S-T, Wu C-Y, Wong C-H. Efficient Convergent Synthesis of Bi-, Tri-, and Tetra-antennary Complex Type N-Glycans and Their HIV-1 Antigenicity. J. Am. Chem. Soc. 2013a;135:15382–15391. doi: 10.1021/ja409097c. [DOI] [PubMed] [Google Scholar]
- Shivatare SS, Chang S-H, Tsai T-I, Ren C-T, Chuang H-Y, Hsu L, Lin C-W, Li S-T, Wu C-Y, Wong C-H. Efficient Convergent Synthesis of Bi-, Tri-, and Tetraantennary Complex Type N-Glycans and Their HIV-1 Antigenecity. J. Am. Chem. Soc. 2013b;135:15382–15391. doi: 10.1021/ja409097c. [DOI] [PubMed] [Google Scholar]
- Simek MD, Rida W, Priddy FH, Pung P, Carrow E, Laufer DS, Lehrman JK, Boaz M, Tarragona-Fiol T, Miiro G, et al. Human immunodeficiency virus type 1 elite neutralizers: individuals with broad and potent neutralizing activity identified by using a high-throughput neutralization assay together with an analytical selection algorithm. J. Virol. 2009;83:7337–7348. doi: 10.1128/JVI.00110-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trkola A, Purtscher M, Muster T, Ballaun C, Buchacher A, Sullivan N, Srinivasan K, Sodroski J, Moore JP, Katinger H. Human monoclonal antibody 2G12 defines a distinctive neutralization epitope on the gp120 glycoprotein of human immunodeficiency virus type 1. J. Virol. 1996;70:1100–1108. doi: 10.1128/jvi.70.2.1100-1108.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker LM, Huber M, Doores KJ, Falkowska E, Pejchal R, Julien J-P, Wang S-K, Ramos A, Chan-Hui P-Y, Moyle M, et al. Broad neutralization coverage of HIV by multiple highly potent antibodies. Nature. 2011;477:466–470. doi: 10.1038/nature10373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker LM, Phogat SK, Chan-Hui P-Y, Wagner D, Phung P, Goss JL, Wrin T, Simek MD, Fling S, Mitcham JL, et al. Broad and potent neutralizing antibodies from an African donor reveal a new HIV-1 vaccine target. Science. 2009;326:285–289. doi: 10.1126/science.1178746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker LM, Simek MD, Priddy F, Gach JS, Wagner D, Zwick MB, Phogat SK, Poignard P, Burton DR. A limited number of antibody specificities mediate broad and potent serum neutralization in selected HIV-1 infected individuals. PLoS Pathog. 2010;6:e1001028. doi: 10.1371/journal.ppat.1001028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Yang Z-Y, Li Y, Hogerkorp C-M, Schief WR, Seaman MS, Zhou T, Schmidt SD, Wu L, Xu L, et al. Rational design of envelope identifies broadly neutralizing human monoclonal antibodies to HIV-1. Science. 2010;329:856–861. doi: 10.1126/science.1187659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Zhou T, Zhu J, Zhang B, Georgiev I, Wang C, Chen X, Longo NS, Louder M, McKee K, et al. Focused evolution of HIV-1 neutralizing antibodies revealed by structures and deep sequencing. Science. 2011;333:1593–1602. doi: 10.1126/science.1207532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M-Y, Yuan T, Li J, Rosa Borges A, Watkins JD, Guenaga J, Yang Z, Wang Y, Wilson R, Li Y, et al. Identification and characterization of a broadly cross-reactive HIV-1 human monoclonal antibody that binds to both gp120 and gp41. PLoS ONE. 2012;7:e44241. doi: 10.1371/journal.pone.0044241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou T, Georgiev I, Wu X, Yang Z-Y, Dai K, Finzi A, Kwon YD, Scheid JF, Shi W, Xu L, et al. Structural basis for broad and potent neutralization of HIV-1 by antibody VRC01. Science. 2010;329:811–817. doi: 10.1126/science.1192819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwick MB, Labrijn AF, Wang M, Spenlehauer C, Saphire EO, Binley JM, Moore JP, Stiegler G, Katinger H, Burton DR, et al. Broadly neutralizing antibodies targeted to the membrane-proximal external region of human immunodeficiency virus type 1 glycoprotein gp41. J. Virol. 2001;75:10892–10905. doi: 10.1128/JVI.75.22.10892-10905.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.