Abstract
In a previous study, we established a one-step methylation-specific polymerase chain reaction (OS-MSP) assay for the detection of methylated DNA (met-DNA) and total DNA levels in serum. For the present study, this OS-MSP assay was used for patients with breast cancer treated with neoadjuvant chemotherapy (NAC) in order to investigate the prognostic significance of met-DNA and total DNA levels. Following treatment with NAC and prior to surgery, serum samples obtained from 120 patients with stage II/III breast cancer were subjected to the OS-MSP assay for analysis of the glutathione S-transferase pi 1, Ras association (RalGDS/AF-6) domain family member 1 and retinoic acid receptor β2 genes. The detection of methylation in a minimum of one of these genes indicated a positive outcome of the assay. The total DNA content of the serum was also determined. Of the 120 stage II/III patients, seven (6%) were positive for met-DNA in serum and showed a significantly worse overall survival (OS) time compared with patients negative for met-DNA (n=113) (5-year OS, 43 vs. 85%; P=0.002). The patients with high total DNA levels in serum (n=40) also showed a significantly worse OS compared with those with low total DNA levels (n=80) (65 vs. 91%; P<0.001). The presence of met-DNA and high total DNA levels in the serum were found to be significant prognostic factors that are independent of a pathological complete response by multivariate analysis. Following NAC, met-DNA and high total DNA levels in the serum detected with the OS-MSP assay constitute novel prognostic factors for patients with breast cancer; this may be clinically useful for the prognosis prediction for patients who do not achieve a pathological complete response following NAC.
Keywords: breast cancer, neoadjuvant chemotherapy, methylation, DNA, serum, prognosis
Introduction
In recent years, growing numbers of patients with breast cancer have been treated with neoadjuvant chemotherapy (NAC) to shrink the tumor size for an improved chance of breast conservation. In fact, it has been reported by specific studies that more patients treated with NAC undergo breast-conserving surgery than those not treated (1,2). Another advantage of NAC is that the patient prognosis can be estimated by means of a pathological evaluation of the surgical specimens, i.e., if a pathological complete response (pCR) is achieved, the patient prognosis can be expected to be excellent, while the prognosis of patients who do not achieve a pCR is reportedly worse, with a 5-year relapse-free survival rate of 50–70% (3,4). Since prognostic evaluation is extremely important in deciding whether or not further adjuvant therapy should be used, more effective prognostic factors are required for those patients who cannot achieve a pCR.
It has been hypothesized and is currently accepted that the detection of tumor-specific DNA methylation in serum is useful for prognosis prediction and for monitoring responses to systemic therapy in patients with breast cancer (5–13). Certain studies have assessed the prognostic value of the presence of methylated DNA (met-DNA) in the serum of patients with breast cancer (5–9). It has also been reported that breast cancer patients with met-DNA in the serum detected by one-step methylation-specific polymerase chain reaction (PCR) show a poorer prognosis compared with those without it (10,11). However, other studies have reported that met-DNA in the serum of patients with breast cancer treated with adjuvant therapy correlates with the pathological response (12,13). A study by Sharma et al (12) detected methylation of the glutathione S-transferase pi 1 (GSTP1) and breast cancer 1, early onset (BRCA1) genes in serum more frequently in non-responders to NAC than in responders, while Avraham et al (13) reported that none of the responders to NAC showed methylated Ras association (RalGDS/AF-6) domain family member 1 (RASSF1A) in the serum. However, no studies have been published on the correlation between met-DNA in serum and the prognosis for patients with breast cancer treated with NAC. Therefore, the present study investigated whether the presence of met-DNA in the serum, as detected by one-step methylation-specific PCR (OS-MSP), may be associated with a poor prognosis for patients with breast cancer treated with NAC. In addition, the prognostic significance of high total DNA levels in the serum was also investigated.
Materials and methods
Patients
Patients with invasive breast cancer (n=120) who underwent breast conserving surgery or mastectomy following NAC at the Osaka University Hospital (Suita-shi, Osaka, Japan) between March 2000 and May 2007 were retrospectively included in the present study. Informed consent was obtained from each patient. A total of 44 patients had been treated with paclitaxel (80 mg/m2) weekly for 12 cycles followed by 5-fluorouracil (500 mg/m2), epirubicin (75 mg/m2) and cyclophosphamide (500 mg/m2) every three weeks for 4 cycles (P-FEC). Another 37 patients had been treated with docetaxel (75 mg/m2) every three weeks for 4 cycles, while 29 had been treated with cyclophosphamide (600 mg/m2) and epirubicin (60 mg/m2) every three weeks for 4 cycles, followed by docetaxel (60 mg/m2) every three weeks for 4 cycles. Finally, 10 patients had been treated with other types of chemotherapy consisting of docetaxel (60 mg/m2) or cyclophosphamide (600 mg/m2) and epirubicin (60 mg/m2) every three weeks or paclitaxel (80 mg/m2) weekly.
Serum samples were obtained following NAC and prior to surgery, and stored at −80°C until use. The median follow-up period was 73 months (range, 3–134 months) and the median age of the patients at the time of surgery was 51 years (range, 26–75 years). The clinicopathological characteristics of the patients are summarized in Table I. Adjuvant hormonal therapy was administered to 79 patients: Tamoxifen for 27, goserelin plus tamoxifen for 13 and anastrozole for 39 patients, all essentially in accordance with the St. Gallen recommendations (14–16). Subsequent to the surgery, the patients were followed up every 3 months for 1–2 years, every 6 months for 3–5 years and once every year thereafter. This study was approved by the Ethics Committee of Osaka University Graudate School of Medicine (Suita, Osaka, Japan).
Table I.
Correlations between the presence of methylated DNA or levels of total DNA in serum and the clinicopathological parameters prior to NACa.
| Met-DNAb in serum | Total DNA levels in serum | ||||||
|---|---|---|---|---|---|---|---|
|
|
|
||||||
| n | Met (−)c, n (%) | Met (+)d, n (%) | P-valuee | Low, n (%) | High, n (%) | P-valuef | |
| Total patients | 120 | 113 (94) | 7 (6) | 80 (67) | 40 (33) | ||
| Menopausal status | |||||||
| Premenopausal | 62 | 60 (97) | 2 (3) | 0.261 | 44 (71) | 18 (29) | 0.301 |
| Postmenopausal | 58 | 53 (91) | 5 (9) | 36 (62) | 22 (38) | ||
| Tumor size | |||||||
| T1 | 3 | 3 (100) | 0 (0) | 0.424g | 2 (67) | 1 (33) | 0.763g |
| T2 | 71 | 68 (96) | 3 (4) | 47 (66) | 24 (34) | ||
| T3 | 22 | 21 (95) | 1 (5) | 15 (68) | 7 (32) | ||
| T4 | 23 | 20 (87) | 3 (13) | 16 (70) | 7 (30) | ||
| Unknown | 1 | 1 (100) | 0 (0) | 0 (0) | 1 (100) | ||
| Lymph node metastasis | |||||||
| N0 | 35 | 35 (100) | 0 (0) | 0.105 | 24 (69) | 11 (31) | 0.776 |
| N1 | 85 | 78 (92) | 7 (8) | 56 (66) | 29 (34) | ||
| Histological grade | |||||||
| G1 | 26 | 26 (100) | 0 (0) | 0.116h | 20 (77) | 6 (23) | 0.031h |
| G2 | 64 | 61 (95) | 3 (5) | 44 (69) | 20 (31) | ||
| G3 | 25 | 22 (88) | 3 (12) | 12 (48) | 13 (52) | ||
| Unknown | 5 | 4 (80) | 1 (20) | 4 (80) | 1 (20) | ||
| Histological type | |||||||
| Invasive ductal carcinoma | 111 | 106 (95) | 5 (5) | 0.125 | 73 (66) | 38 (34) | 0.811e |
| Invasive lobular carcinoma | 8 | 6 (75) | 2 (25) | 6 (75) | 2 (25) | ||
| Others | 1 | 1 (100) | 0 (0) | 1 (100) | 0 (0) | ||
| ER | |||||||
| Negative | 53 | 48 (91) | 5 (9) | 0.239 | 35 (66) | 18 (34) | 0.897 |
| Positive | 67 | 65 (97) | 2 (3) | 45 (67) | 22 (33) | ||
| PR | |||||||
| Negative | 72 | 66 (92) | 6 (8) | 0.240 | 46 (64) | 26 (36) | 0.429 |
| Positive | 48 | 47 (98) | 1 (2) | 34 (71) | 14 (29) | ||
| HER2 | |||||||
| Negative | 88 | 83 (94) | 5 (6) | 1.000 | 61 (69) | 27 (31) | 0.398 |
| Positive | 28 | 27 (96) | 1 (4) | 17 (61) | 11 (39) | ||
| Unknown | 4 | 3 (75) | 1 (25) | 2 (50) | 2 (50) | ||
| CEA | |||||||
| Negative | 103 | 97 (94) | 6 (6) | 1.000 | 70 (68) | 33 (32) | 0.263d |
| Positive | 15 | 14 (93) | 1 (7) | 8 (53) | 7 (47) | ||
| Unknown | 2 | 2 (100) | 0 (0) | 2 (100) | 0 (0) | ||
| CA15-3 | |||||||
| Negative | 107 | 101 (94) | 6 (6) | 0.505 | 71 (66) | 36 (34) | 1.000d |
| Positive | 11 | 10 (91) | 1 (9) | 7 (64) | 4 (36) | ||
| Unknown | 2 | 2 (100) | 0 (0) | 2 (100) | 0 (0) | ||
| Pathological response | |||||||
| Non-pCR | 101 | 94 (93) | 7 (7) | 0.595 | 68 (67) | 33 (33) | 0.724 |
| pCR | 19 | 19 (100) | 0 (0) | 12 (63) | 7 (37) | ||
Neoadjuvant chemotherapy;
methylated DNA (met-DNA);
negative for met-DNA in serum;
positive for met-DNA in serum;
Fisher’s exact test;
χ2 test;
T1 + T2 vs. T3 + T4;
G1 + G2 vs. G3.
NAC, neoadjuvant chemotherapy; ER, estrogen receptor; PR, progesterone receptor; HER2, human epidermal growth factor receptor 2; CEA, carcinoembryonic antigen; CA, cancer antigen; pCR, pathological complete response.
OS-MSP assay for GSTP1, RASSF1A and retinoic acid receptor β2 (RARβ2) promoter hypermethylation in serum
The OS-MSP assay was conducted as previously described (10). In brief, a total of 1,000 μl of serum from each patient was solubilized by incubation with lysis buffer (5 M guanidine-HCl and 0.65 mg/ml proteinase K) at 50°C for 1 h and then incubated with a 5 M bisulfite solution at 80°C for 40 min. Bisulfite-modified DNA was purified with a DNA purification kit (Qiagen, Valencia, CA, USA) and eluted in 50 μl dH2O. Modification by bisulfite was completed by treatment with 0.3 M NaOH for 5 min at room temperature, subsequent to which bisulfite-modified DNA was extracted by gel filtration (GE Healthcare, Princeton, NJ, USA) for use as the PCR template. Promoter hypermethylation of GSTP1, RASSF1A and RARβ2 was then evaluated by OS-MSP. The primer and probe sequences were as follows: GSTP1 forward primer, 5′-CGTCGTGATTTAGTATTGGGGC-3′ and reverse primer, 5′-CTAATAACGAAAACTACGACGACGAAA-3′; GSTP1 probe, 5′-FAM-ATAAGGTTCGGAGGTCGCGAGGTTTTC GT-DDQ1-3′; RASSF1A forward primer, 5′-ATAGTTTTTGTA TTTAGGTTTTTATTGCGC-3′ and reverse primer, 5′-GCT AACAAACGCGAACCG-3′; RASSF1A probe, 5′-FAM-TTG AAGTCGGGGTTCGTTTTGTGGTTTCGT-DDQ1-3′; RARβ2 forward primer, 5′-GAATATCGTTTTTTAAGTTAAGTC GTC-3′ and reverse primer, 5′-GAAACGCTACTCCTAACT CACG-3′; and RARβ2 probe, 5′-FAM-AGGCGTAAAGGG AGAGAAGTTGGTGTTTA-DDQ1-3′. For the PCR amplifications, the Light Cycler 480 Real-Time PCR System (Roche Applied Science, Madison, WI, USA) was used under the following conditions: 1 cycle at 95°C for 10 min, followed by 50 cycles of 95°C for 30 sec, 60°C for 30 sec and 72°C for 30 sec. Every sample was analyzed in a single assay for each gene. Finally, the PCR products were also analyzed by means of 3% agarose gel electrophoresis, staining with ethidium bromide and visualization under UV illumination.
Assay for total DNA levels in serum
Total DNA levels were quantified as previously described (10). In brief, in order to quantify total DNA levels in serum following bisulfite treatment, a genomic locus lacking a cytosine base was selected, since such a locus is not affected by bisulfite treatment. This locus was amplified by PCR using the primers, probe and standard oligoDNA. The primer and probe sequences were as follows: Forward primer, 5′-AGGGAGTAGAGAAAAAGT AGGAAGATGAGT-3′ and reverse primer, 5′-TCCAACATC ACATCCAATCCA-3′; probe, 5′-FAM-AGGGTGATAATG AGTGTGTTGGGAAATAGA-DDQ1-3′; and standard oligo DNA, 5′-AGGGAGTAGAGAAAAAGTAGGAAGATGAGT CCAGGGTGATAATGAGTGTGTTGGGAAATAGACCTG GATTGGATGTGATGTTGGA-3′. The PCR conditions were as aforementioned. The patients were divided into three tertiles (low, middle and high) according to the level of total DNA in serum. Those in the low and middle tertiles were then combined and treated as the low total DNA group and those in the high tertile were treated as the high total DNA group.
DNA extraction from tumor tissues
DNA was also extracted as previously described (17) from breast tumor tissues obtained prior to (core-needle biopsy specimens) or following (surgical specimens) NAC from the patients who showed positive results for the OS-MSP assay in at least one of the three genes. The specimens were then examined as to whether the corresponding genes were methylated in the tumor tissues. For this examination, 1 μg of DNA was subjected to sodium bisulfite treatment using the EpiTect Bisulfite kit (48; Qiagen) according to the manufacturer’s instructions, and analyzed by the OS-MSP assay for promoter hypermethylation of GSTP1, RASSF1A and RARβ2, as previously described (10).
Histological grade and estrogen receptor (ER), progesterone receptor (PR) and human epidermal growth factor receptor 2 (HER2) expression
Histological grade was determined using the Scarff-Bloom-Richardson grading system (18). ER and PR were classified as positive when ≥10% of the tumor cells showed immunohistochemically-positive staining (ER, clone 6F11; and PR, clone 16; Ventana Japan K.K., Yokohama and SRL, Inc., Tokyo, Japan, respectively). HER-2 expression was determined immunohistochemically using anti-human c-erbB-2 polyclonal antibody (Nichirei Biosciences, Inc., Tokyo, Japan) or by means of fluorescence in situ hybridization (FISH) using PathVysion Her2 DNA Probe kits (SRL, Inc.). When a tumor showed +3 immunostaining (positive tumor cells >30%) or a FISH ratio (HER2 gene signals to chromosome 17 centromere signals) of ≥2.0, it was considered HER2-positive.
Evaluation of the response to chemotherapy
The pathological response to chemotherapy was evaluated by examining the surgical specimens obtained during the surgery. The specimens were cut at 5-mm intervals for the preparation of hematoxylin and eosin sections. A complete loss of invasive tumor cells in the primary tumor site without any lymph node metastasis was classified as a pCR.
Statistical analysis
Associations of met-DNA or total DNA levels in serum with various clinicopathological parameters were assessed by means of Pearson’s χ2 test. Disease-free survival (DFS) was calculated as the time from surgery until the date of any recurrence of breast cancer (local or distant) or mortality from any cause. Overall survival (OS) was calculated as the time from surgery to the date of mortality from any cause. DFS and OS were assessed with the Kaplan-Meier method and log-rank tests, while univariate and multivariate analyses (Cox regression models) were used to evaluate the prognostic significance of various parameters. Multivariate analyses included the parameters P<0.1 or known prognostic factor (pCR). All the statistical analyses were two-sided and P<0.05 was considered to indicate a statistically significant difference. SPSS software (SPSS, Inc., Chicago, IL, USA) was used for all statistical analyses.
Results
Associations between clinicopathological parameters and met-DNA or total DNA levels in serum
Promoter hypermethylation of GSTP1, RASSF1A and RARβ2 in serum was detected with the OS-MSP assay in 4, 1 and 2% of the 120 patients, respectively. When promoter hypermethylation was found in at least one of these three genes, the result of the OS-MSP assay for met-DNA in serum was considered to be positive for the subsequent analysis. A total of seven patients (6%) were found to be positive for met-DNA in serum. No significant association was observed between met-DNA in serum and any clinicopathological characteristics (Table I). The promoter methylation status of the genes methylated in the serum of the seven patients who were positive for met-DNA was also examined in the tumor specimens obtained from the same patients. The same genes that were methylated in the serum were also found to be methylated in the tumor tissue of each of the seven patients.
The median total DNA level in the serum was 1.9 ng/ml (range, 0–63.3 ng/ml). The patients were divided into tertiles (high, middle and low) according to the level of total DNA in the serum. The middle and low tertiles were combined and treated as the low total DNA group (n=80), and the high tertile was treated as the high total DNA group (n=40). The cut-off value for the high and low total DNA groups was 3.3 ng/ml. Correlations between the total DNA levels in the serum and the clinicopathological characteristics are shown in Table I. The patients in the high total DNA group were significantly more likely to have tumors of high histological grade (52 vs. 29%, P=0.031) and to be positive for met-DNA in the serum (71 vs. 31%, P=0.028). By contrast, pCR was not significantly associated with either met-DNA or total DNA levels in the serum.
Association of prognosis with met-DNA or total DNA levels in serum
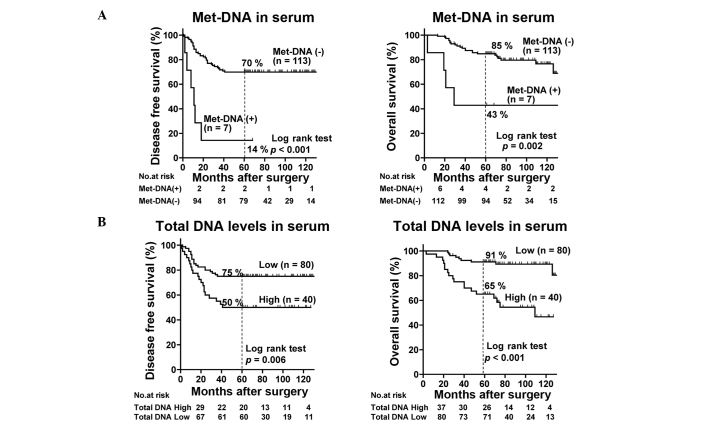
The seven patients with serum positive for met-DNA exhibited significantly worse DFS and OS rates compared with those negative for met-DNA (n=113) (P<0.001 and P=0.002, respectively; Fig. 1A), while the patients with high total DNA levels in the serum (n=40) also showed significantly worse DFS and OS rates compared with those with low levels of total DNA (n=80) (P=0.006 and P<0.001, respectively; Fig. 1B). Subsequently, univariate and multivariate analyses were conducted to determine whether met-DNA and total DNA levels in the serum are independent prognostic factors for the other parameters (Tables II and III). The multivariate analysis showed that met-DNA in the serum was significantly associated with DFS (P=0.003) and OS (P=0.009), as was total DNA levels in the serum (P=0.045 and P=0.001, respectively).
Figure 1.
Prognosis of NAC-treated breast cancer patients in association with (A) met-DNA or (B) total DNA levels in the serum. Disease-free survival (DFS) and overall survival (OS) rates were evaluated in all the NAC-treated patients in association with the presence of met-DNA and total DNA levels in the serum. NAC, neoadjuvant chemotherapy; Met-DNA (−), patients negative for methylated DNA in the serum; Met-DNA (+), patients positive for methylated DNA in the serum; high, patients in the high tertile of total DNA; low, combination of patients in the low and middle tertiles of total DNA.
Table II.
Univariate and multivariate analysis of various prognostic factors for DFS of 120 patients.
| Univariate analysis | Multivariate analysis | |||||
|---|---|---|---|---|---|---|
|
|
|
|||||
| Parameters | HR | 95% CI | P-value | HR | 95% CI | P-value |
| Met-DNA in serum (positive vs. negative) | 6.650 | 2.74–16.17 | <0.001 | 4.226 | 1.63–10.95 | 0.003 |
| Total DNA levels in serum (high vs. low) | 2.336 | 1.26–4.34 | 0.007 | 1.926 | 1.02–3.654 | 0.045 |
| Tumor size (3,4 vs. 1,2) prior to NAC | 1.422 | 0.76–2.68 | 0.276 | |||
| Histological grade (3 vs. 1,2) prior to NAC | 1.560 | 0.76–3.21 | 0.227 | |||
| LN (positive vs. negative) prior to NAC | 2.856 | 1.20–6.81 | 0.018 | 2.864 | 1.17–7.01 | 0.021 |
| CEA (positive vs. negative) prior to NAC | 2.050 | 0.94–4.45 | 0.070 | 2.074 | 0.94–4.59 | 0.072 |
| CEA (positive vs. negative) following NAC | 1.815 | 0.56–5.89 | 0.321 | |||
| CA15-3 (positive vs. negative) prior to NAC | 1.099 | 0.39–3.09 | 0.858 | |||
| CA15-3 (positive vs. negative) following NAC | 0.433 | 0.06–3.15 | 0.408 | |||
| NAC (P-FEC vs. others) | 0.528 | 0.26–1.08 | 0.080 | 0.589 | 0.28–1.24 | 0.163 |
| Pathological response (pCR vs. non-pCR) | 0.234 | 0.06–0.97 | 0.045 | 0.260 | 0.06–1.12 | 0.071 |
DFS, disease-free survival; HR, hazard ratio; CI, confidence interval; Met-DNA, methylated DNA; NAC, neoadjuvant chemotherapy; LN, lymph node metastasis; CEA, carcinoembryonic antigen; CA, cancer antigen; P-FEC, paclitaxel 80 mg/m2 for 12 cycles followed by 5-fluorouracil (500 mg/m2), epirubicin (75 mg/m2) and cyclophosphamide (500 mg/m2) every three weeks for 4 cycles; pCR, pathological complete response.
Table III.
Univariate and multivariate analysis of various prognostic factors for OS of 120 patients.
| Univariate analysis | Multivariate analysis | |||||
|---|---|---|---|---|---|---|
|
|
|
|||||
| Parameters | HR | 95% CI | P-value | HR | 95% CI | P-value |
| Met-DNA in serum (positive vs. negative) | 4.834 | 1.65–14.15 | 0.004 | 4.805 | 1.49–15.52 | 0.009 |
| Total DNA levels in serum (high vs. low) | 5.119 | 2.28–11.49 | <0.001 | 4.112 | 1.77–9.57 | 0.001 |
| Tumor size (3,4 vs. 1,2) | 1.366 | 0.63–2.97 | 0.430 | |||
| Histological grade (3 vs. 1,2) prior to NAC | 2.530 | 1.16–5.53 | 0.020 | 2.302 | 1.02–5.21 | 0.046 |
| LN (positive vs. negative) prior to NAC | 2.179 | 0.82–5.77 | 0.117 | |||
| CEA (positive vs. negative) prior to NAC | 2.298 | 0.93–5.70 | 0.073 | 3.497 | 1.34–9.11 | 0.010 |
| CEA (positive vs. negative) following NAC | 0.643 | 0.09–4.75 | 0.666 | |||
| CA15-3 (positive vs. negative) prior to NAC | 2.087 | 0.72–6.04 | 0.175 | |||
| CA15-3 (positive vs. negative) following NAC | 0.046 | 0.00–65.53 | 0.405 | |||
| NAC (P-FEC vs. others) | 0.744 | 0.31–1.80 | 0.510 | |||
| Pathological response (pCR vs. non-pCR) | 0.418 | 0.10–1.77 | 0.235 | 0.258 | 0.06–1.18 | 0.081 |
OS, overall survival; HR, hazard ratio; CI, confidence interval; Met-DNA, methylated DNA; NAC, neoadjuvant chemotherapy; LN, lymph node metastasis; CEA, carcinoembryonic antigen; CA, cancer antigen; P-FEC, paclitaxel 80 mg/m2 for 12 cycles followed by 5-fluorouracil (500 mg/m2), epirubicin (75 mg/m2) and cyclophosphamide (500 mg/m2) every three weeks for 4 cycles; pCR, pathological complete response.
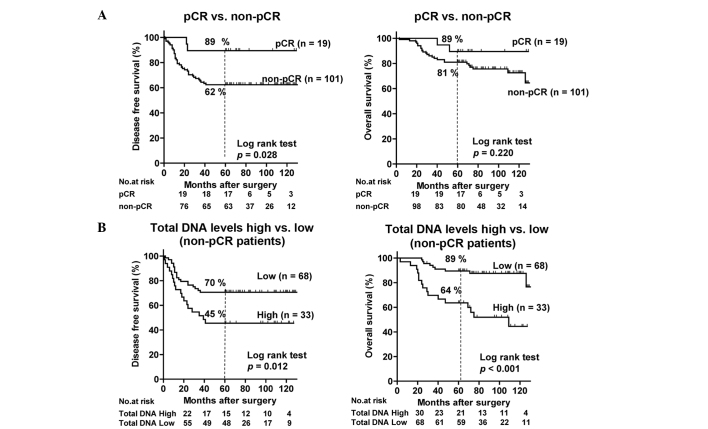
The patients who achieved pCR (n=19) showed significantly improved DFS rates compared with those who did not (n=101) (P=0.028; Fig. 2A), and the prognostic significance for the latter group of total DNA levels in the serum was evaluated. The patients with high total DNA levels in the serum (n=33) again showed significantly worse DFS and OS rates compared with those with low total DNA levels (n=68) (P=0.012 and P<0.001, respectively; Fig. 2B).
Figure 2.
Prognosis of NAC-treated breast cancer patients in association with the pathological response. Disease-free survival (DFS) and overall survival (OS) rates were assessed for (A) NAC-treated patients (n=120) according to pathological response and (B) NAC-treated patients (n=101) without pCR according to total DNA levels in the serum. NAC, neoadjuvant chemotherapy; pCR, pathological complete response; high, patients in the high tertile of total DNA in the serum; low, combination of patients in the low and middle tertiles of total DNA in the serum.
Discussion
The present study investigated whether detection of met-DNA and high total DNA levels in the serum by means of the OS-MSP assay could serve as novel prognostic factors for patients with breast cancer treated with NAC. By using multivariate analysis it was shown that met-DNA and total DNA levels in the serum following NAC are independent prognostic factors for DFS and OS, independent of pCR, which is a well-established prognostic factor for NAC-treated patients. The results indicate that met-DNA and total DNA levels in the serum may be clinically useful prognostic factors.
Met-DNA is believed to represent circulating tumor genomes, but not necessarily total DNA levels, as it can not only originate from tumor cells, but also from inflammatory cells, endothelial cells and fibroblasts in tumor tissues (19–21). Although the reason why total DNA levels in the serum is associated with prognosis remains unclear, it is possible that high total DNA levels in the serum may reflect specific tumor biology that is associated with tumor metastasis and tumor-induced inflammation. It is also possible that circulating tumor cells (22,23) and micrometastatic deposits of distant organs, including the bone marrow and liver, may contribute to total DNA levels (19,24).
The OS-MSP assay showed that the serum was positive for met-DNA in 6% of patients in the present study. This positivity appears to be slightly lower compared with our previous study, in which it was found that met-DNA was positively detected in 10% of stage I and II patients without NAC treatment (11). These results indicate that positivity of met-DNA in serum decreases following NAC. In fact, studies by Sharma et al (12) and Avraham et al (13) reported a significant decline in met-DNA levels following NAC. However, in the present study, the serum samples were not obtained prior to NAC and thus the change in the positivity of met-DNA prior to and following NAC could not be investigated.
While a significant correlation between met-DNA and pCR was not detected, none of the patients that were positive for met-DNA achieved pCR, which is consistent with the findings of other studies (12,13). Sharma et al (12) reported that all non-responders were positive for met-DNA (GSTP1 and BRCA1) following NAC, and Avraham et al (13) reported that all patients that were positive for met-DNA (RASSF1A) following NAC could not achieve pCR. These results appear to indicate that positivity for met-DNA following NAC is associated with non-pCR.
A limitation of the present study is that the serum samples could be obtained following, but not prior to, NAC, so that the prognostic significance of met-DNA and total DNA levels in serum prior to NAC could not be addressed. However, the available data appears to indicate that met-DNA following NAC is a highly significant prognostic indicator for poor prognosis, as none of the patients that were positive for met-DNA attained pCR, and as many as 86% developed recurrence. High total DNA levels in the serum following NAC were also significantly associated with a poor prognosis. It has been reported that the quantity of disseminated or circulating tumor cells detected following, but not prior to, chemotherapy can predict prognosis, as the numbers detected following chemotherapy may reflect the residual presence of micro-metastatic tumors (25,26). In analogy to this finding, the presence of met-DNA and high total DNA levels may represent the residual presence of micro-metastatic tumors. Patients with met-DNA and/or high total DNA levels in the serum can thus be considered to be essentially resistant to NAC and to have a poor prognosis, necessitating the use of non-cross resistant, post-operative adjuvant chemotherapy.
In conclusion, the present study was able to demonstrate that the presence of met-DNA and high levels of total DNA detected with the OS-MSP assay is a significant and independent prognostic factor for patients with breast cancer treated with NAC. The OS-MSP assay may thus be clinically useful for the selection of NAC-treated patients who are at a high risk of relapse and thus require treatment with additional post-operative adjuvant chemotherapy. To the best of our knowledge, this is the first study to assess the association between met-DNA or total DNA levels in the serum and prognosis following NAC. However, future studies including larger numbers of patients with sufficient follow-up times are required to further validate the clinical utility of the OS-MSP assay for patients treated with NAC.
Acknowledgements
This study was supported in part by a Grants-in-Aid from the Comprehensive 10-Year Strategy for Cancer Control Program of the Ministry of Health, Labor and Welfare, Japan and a research fund from Sysmex Corporation. The funders had no role in the study design, data analysis, decision to publish or preparation of the manuscript. Shinzaburo Noguchi received research funding from Pfizer, Bristol-Myers Squibb and Sysmex Corporation, and Honoraria from Pfizer. Seung Jin Kim received honoraria from Sysmex Corporation, Pfizer and Bristol-Myers Squibb.
References
- 1.van der Hage JA, van de Velde CJ, Julien JP, Tubiana-Hulin M, Vandervelden C, Duchateau L. Preoperative chemotherapy in primary operable breast cancer: results from the European Organization for Research and Treatment of Cancer trial 10902. J Clin Oncol. 2001;19:4224–4237. doi: 10.1200/JCO.2001.19.22.4224. [DOI] [PubMed] [Google Scholar]
- 2.Singletary SE. Neoadjuvant chemotherapy in the treatment of stage II and III breast cancer. Am J Surg. 2001;182:341–346. doi: 10.1016/s0002-9610(01)00724-3. [DOI] [PubMed] [Google Scholar]
- 3.Wolmark N, Wang J, Mamounas E, Bryant J, Fisher B. Preoperative chemotherapy in patients with operable breast cancer: nine-year results from National Surgical Adjuvant Breast and Bowel Project B-18. J Natl Cancer Inst Monogr. 2001:96–102. doi: 10.1093/oxfordjournals.jncimonographs.a003469. [DOI] [PubMed] [Google Scholar]
- 4.Bear HD, Anderson S, Smith RE, et al. Sequential preoperative or postoperative docetaxel added to preoperative doxorubicin plus cyclophosphamide for operable breast cancer: National Surgical Adjuvant Breast and Bowel Project Protocol B-27. J Clin Oncol. 2006;24:2019–2027. doi: 10.1200/JCO.2005.04.1665. [DOI] [PubMed] [Google Scholar]
- 5.Müller HM, Widschwendter A, Fiegl H, et al. DNA methylation in serum of breast cancer patients: an independent prognostic marker. Cancer Res. 2003;63:7641–7645. [PubMed] [Google Scholar]
- 6.Göbel G, Auer D, Gaugg I, et al. Prognostic significance of methylated RASSF1A and PITX2 genes in blood- and bone marrow plasma of breast cancer patients. Breast Cancer Res Treat. 2011;130:109–117. doi: 10.1007/s10549-010-1335-8. [DOI] [PubMed] [Google Scholar]
- 7.Sharma G, Mirza S, Parshad R, et al. Clinical significance of promoter hypermethylation of DNA repair genes in tumor and serum DNA in invasive ductal breast carcinoma patients. Life Sci. 2010;87:83–91. doi: 10.1016/j.lfs.2010.05.001. [DOI] [PubMed] [Google Scholar]
- 8.Lo PK, Sukumar S. Epigenomics and breast cancer. Pharmacogenomics. 2008;9:1879–1902. doi: 10.2217/14622416.9.12.1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Martens JW, Margossian AL, Schmitt M, Foekens J, Harbeck N. DNA methylation as a biomarker in breast cancer. Future Oncol. 2009;5:1245–1256. doi: 10.2217/fon.09.89. [DOI] [PubMed] [Google Scholar]
- 10.Yamamoto N, Nakayama T, Kajita M, et al. Detection of aberrant promoter methylation of GSTP1, RASSF1A, and RARβ2 in serum DNA of patients with breast cancer by a newly established one-step methylation-specific PCR assay. Breast Cancer Res Treat. 2012;132:165–173. doi: 10.1007/s10549-011-1575-2. [DOI] [PubMed] [Google Scholar]
- 11.Fujita N, Nakayama T, Yamamoto N, et al. Methylated DNA and total DNA in serum detected by one-step methylation-specific PCR is predictive of poor prognosis for breast cancer patients. Oncology. 2012;83:273–282. doi: 10.1159/000342083. [DOI] [PubMed] [Google Scholar]
- 12.Sharma G, Mirza S, Parshad R, Gupta SD, Ralhan R. DNA methylation of circulating DNA: a marker for monitoring efficacy of neoadjuvant chemotherapy in breast cancer patients. Tumour Biol. 33:1837–1843. doi: 10.1007/s13277-012-0443-y. [DOI] [PubMed] [Google Scholar]
- 13.Avraham A, Uhlmann R, Shperber A, et al. Serum DNA methylation for monitoring response to neoadjuvant chemotherapy in breast cancer patients. Int J Cancer. 131:E1166–E1172. doi: 10.1002/ijc.27526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goldhirsch A, Glick JH, Gelber RD, Coates AS, Senn HJ. Meeting highlights: International Consensus Panel on the treatment of primary breast cancer. Seventh International Conference on Adjuvant Therapy of Primary Breast Cancer. J Clin Oncol. 2001;19:3817–3827. doi: 10.1200/JCO.2001.19.18.3817. [DOI] [PubMed] [Google Scholar]
- 15.Goldhirsch A, Wood WC, Gelber RD, Coates AS, Thürlimann B, Senn HJ. Meeting highlights: updated international expert consensus on the primary therapy of early breast cancer. J Clin Oncol. 2003;21:3357–3365. doi: 10.1200/JCO.2003.04.576. [DOI] [PubMed] [Google Scholar]
- 16.Goldhirsch A, Glick JH, Gelber RD, Coates AS, Thürlimann B, et al. Panel members. Meeting highlights: international expert consensus on the primary therapy of early breast cancer 2005. Ann Oncol. 2005;16:1569–1583. doi: 10.1093/annonc/mdi326. [DOI] [PubMed] [Google Scholar]
- 17.Arai T, Miyoshi Y, Kim SJ, et al. Association of GSTP1 expression with resistance to docetaxel and paclitaxel in human breast cancers. Eur J Surg Oncol. 2008;34:734–738. doi: 10.1016/j.ejso.2007.07.008. [DOI] [PubMed] [Google Scholar]
- 18.Robbins P, Pinder S, de Klerk N, et al. Histological grading of breast carcinomas: a study of interobserver agreement. Hum Pathol. 1995;26:873–879. doi: 10.1016/0046-8177(95)90010-1. [DOI] [PubMed] [Google Scholar]
- 19.Schwarzenbach H, Hoon DS, Pantel K. Cell-free nucleic acids as biomarkers in cancer patients. Nat Rev Cancer. 2011;11:426–437. doi: 10.1038/nrc3066. [DOI] [PubMed] [Google Scholar]
- 20.Fleischhacker M, Schmidt B. Circulating nucleic acids (CNAs) and cancer - a survey. Biochim Biophys Acta. 2007;1775:181–232. doi: 10.1016/j.bbcan.2006.10.001. [DOI] [PubMed] [Google Scholar]
- 21.Kowalewska M, Nowak R, Chechlinska M. Implications of cancer-associated systemic inflammation for biomarker studies. Biochim Biophys Acta. 2010;1806:163–171. doi: 10.1016/j.bbcan.2010.06.002. [DOI] [PubMed] [Google Scholar]
- 22.Matuschek C, Bölke E, Lammering G, et al. Methylated APC and GSTP1 genes in serum DNA correlate with the presence of circulating blood tumor cells and are associated with a more aggressive and advanced breast cancer disease. Eur J Med Res. 2010;15:277–286. doi: 10.1186/2047-783X-15-7-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Van der Auwera I, Elst HJ, Van Laere SJ, et al. The presence of circulating total DNA and methylated genes is associated with circulating tumour cells in blood from breast cancer patients. Br J Cancer. 2009;100:1277–1286. doi: 10.1038/sj.bjc.6605013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schwarzenbach H, Alix-Panabières C, Müller I, et al. Cell-free tumor DNA in blood plasma as a marker for circulating tumor cells in prostate cancer. Clin Cancer Res. 2009;15:1032–1038. doi: 10.1158/1078-0432.CCR-08-1910. [DOI] [PubMed] [Google Scholar]
- 25.Mathiesen RR, Borgen E, Renolen A, et al. Persistence of disseminated tumor cells after neoadjuvant treatment for locally advanced breast cancer predicts poor survival. Breast Cancer Res. 14:R117. doi: 10.1186/bcr3242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xenidis N, Ignatiadis M, Apostolaki S, et al. Cytokeratin-19 mRNA-positive circulating tumor cells after adjuvant chemotherapy in patients with early breast cancer. J Clin Oncol. 2009;27:2177–2184. doi: 10.1200/JCO.2008.18.0497. [DOI] [PubMed] [Google Scholar]