Abstract
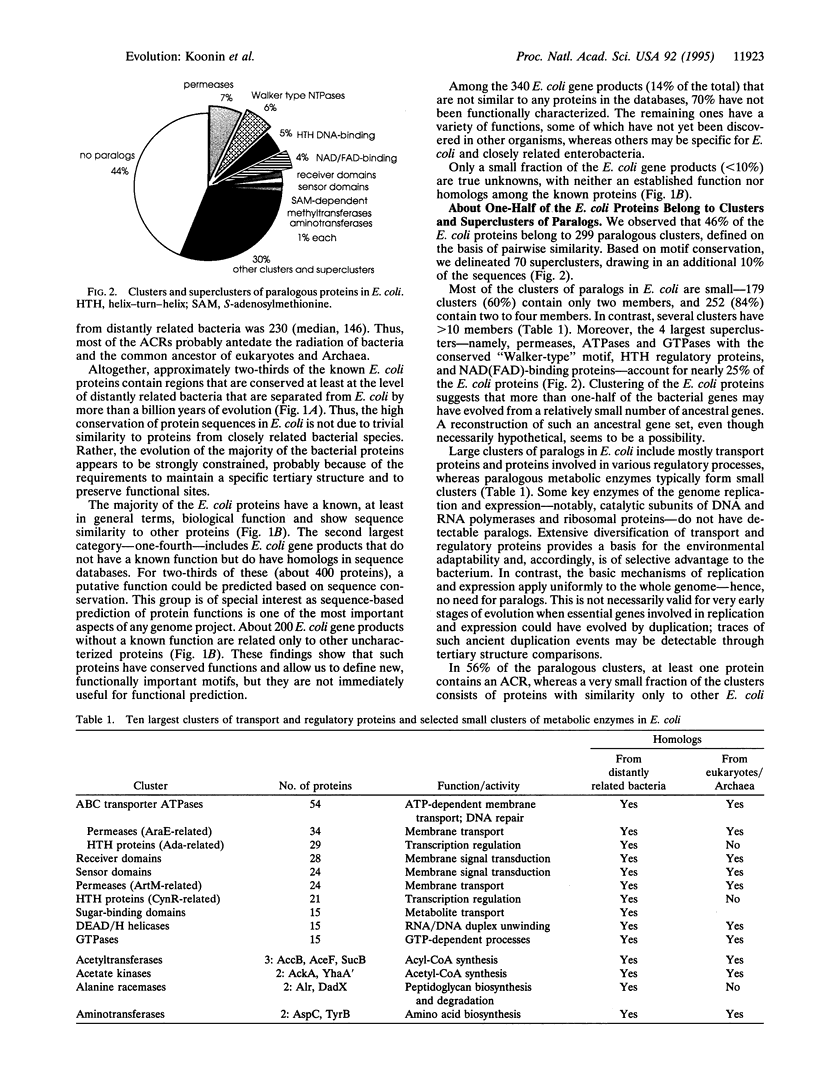
A computer analysis of 2328 protein sequences comprising about 60% of the Escherichia coli gene products was performed using methods for database screening with individual sequences and alignment blocks. A high fraction of E. coli proteins--86%--shows significant sequence similarity to other proteins in current databases; about 70% show conservation at least at the level of distantly related bacteria, and about 40% contain ancient conserved regions (ACRs) shared with eukaryotic or Archaeal proteins. For > 90% of the E. coli proteins, either functional information or sequence similarity, or both, are available. Forty-six percent of the E. coli proteins belong to 299 clusters of paralogs (intraspecies homologs) defined on the basis of pairwise similarity. Another 10% could be included in 70 superclusters using motif detection methods. The majority of the clusters contain only two to four members. In contrast, nearly 25% of all E. coli proteins belong to the four largest superclusters--namely, permeases, ATPases and GTPases with the conserved "Walker-type" motif, helix-turn-helix regulatory proteins, and NAD(FAD)-binding proteins. We conclude that bacterial protein sequences generally are highly conserved in evolution, with about 50% of all ACR-containing protein families represented among the E. coli gene products. With the current sequence databases and methods of their screening, computer analysis yields useful information on the functions and evolutionary relationships of the vast majority of genes in a bacterial genome. Sequence similarity with E. coli proteins allows the prediction of functions for a number of important eukaryotic genes, including several whose products are implicated in human diseases.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Altschul S. F., Boguski M. S., Gish W., Wootton J. C. Issues in searching molecular sequence databases. Nat Genet. 1994 Feb;6(2):119–129. doi: 10.1038/ng0294-119. [DOI] [PubMed] [Google Scholar]
- Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. Basic local alignment search tool. J Mol Biol. 1990 Oct 5;215(3):403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Bork P., Ouzounis C., Casari G., Schneider R., Sander C., Dolan M., Gilbert W., Gillevet P. M. Exploring the Mycoplasma capricolum genome: a minimal cell reveals its physiology. Mol Microbiol. 1995 Jun;16(5):955–967. doi: 10.1111/j.1365-2958.1995.tb02321.x. [DOI] [PubMed] [Google Scholar]
- Borodovsky M., Rudd K. E., Koonin E. V. Intrinsic and extrinsic approaches for detecting genes in a bacterial genome. Nucleic Acids Res. 1994 Nov 11;22(22):4756–4767. doi: 10.1093/nar/22.22.4756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chothia C. Proteins. One thousand families for the molecular biologist. Nature. 1992 Jun 18;357(6379):543–544. doi: 10.1038/357543a0. [DOI] [PubMed] [Google Scholar]
- Daniels D. L., Plunkett G., 3rd, Burland V., Blattner F. R. Analysis of the Escherichia coli genome: DNA sequence of the region from 84.5 to 86.5 minutes. Science. 1992 Aug 7;257(5071):771–778. doi: 10.1126/science.1379743. [DOI] [PubMed] [Google Scholar]
- Fleischmann R. D., Adams M. D., White O., Clayton R. A., Kirkness E. F., Kerlavage A. R., Bult C. J., Tomb J. F., Dougherty B. A., Merrick J. M. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science. 1995 Jul 28;269(5223):496–512. doi: 10.1126/science.7542800. [DOI] [PubMed] [Google Scholar]
- Gray M. W. The evolutionary origins of organelles. Trends Genet. 1989 Sep;5(9):294–299. doi: 10.1016/0168-9525(89)90111-x. [DOI] [PubMed] [Google Scholar]
- Green P., Lipman D., Hillier L., Waterston R., States D., Claverie J. M. Ancient conserved regions in new gene sequences and the protein databases. Science. 1993 Mar 19;259(5102):1711–1716. doi: 10.1126/science.8456298. [DOI] [PubMed] [Google Scholar]
- Henikoff S., Henikoff J. G. Amino acid substitution matrices from protein blocks. Proc Natl Acad Sci U S A. 1992 Nov 15;89(22):10915–10919. doi: 10.1073/pnas.89.22.10915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingrosso D., Fowler A. V., Bleibaum J., Clarke S. Sequence of the D-aspartyl/L-isoaspartyl protein methyltransferase from human erythrocytes. Common sequence motifs for protein, DNA, RNA, and small molecule S-adenosylmethionine-dependent methyltransferases. J Biol Chem. 1989 Nov 25;264(33):20131–20139. [PubMed] [Google Scholar]
- Koonin E. V., Bork P., Sander C. Yeast chromosome III: new gene functions. EMBO J. 1994 Feb 1;13(3):493–503. doi: 10.1002/j.1460-2075.1994.tb06287.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin E. V. Computer-assisted identification of a putative methyltransferase domain in NS5 protein of flaviviruses and lambda 2 protein of reovirus. J Gen Virol. 1993 Apr;74(Pt 4):733–740. doi: 10.1099/0022-1317-74-4-733. [DOI] [PubMed] [Google Scholar]
- Koonin E. V. Prediction of an rRNA methyltransferase domain in human tumor-specific nucleolar protein P120. Nucleic Acids Res. 1994 Jul 11;22(13):2476–2478. doi: 10.1093/nar/22.13.2476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogh A., Mian I. S., Haussler D. A hidden Markov model that finds genes in E. coli DNA. Nucleic Acids Res. 1994 Nov 11;22(22):4768–4778. doi: 10.1093/nar/22.22.4768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labedan B., Riley M. Widespread protein sequence similarities: origins of Escherichia coli genes. J Bacteriol. 1995 Mar;177(6):1585–1588. doi: 10.1128/jb.177.6.1585-1588.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen G. J., Woese C. R., Overbeek R. The winds of (evolutionary) change: breathing new life into microbiology. J Bacteriol. 1994 Jan;176(1):1–6. doi: 10.1128/jb.176.1.1-6.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer J. D. Comparative organization of chloroplast genomes. Annu Rev Genet. 1985;19:325–354. doi: 10.1146/annurev.ge.19.120185.001545. [DOI] [PubMed] [Google Scholar]
- Razin S. Peculiar properties of mycoplasmas: the smallest self-replicating prokaryotes. FEMS Microbiol Lett. 1992 Dec 15;100(1-3):423–431. doi: 10.1111/j.1574-6968.1992.tb14072.x. [DOI] [PubMed] [Google Scholar]
- Riley M. Functions of the gene products of Escherichia coli. Microbiol Rev. 1993 Dec;57(4):862–952. doi: 10.1128/mr.57.4.862-952.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robison K., Gilbert W., Church G. M. Large scale bacterial gene discovery by similarity search. Nat Genet. 1994 Jun;7(2):205–214. doi: 10.1038/ng0694-205. [DOI] [PubMed] [Google Scholar]
- Schimmang T., Tollervey D., Kern H., Frank R., Hurt E. C. A yeast nucleolar protein related to mammalian fibrillarin is associated with small nucleolar RNA and is essential for viability. EMBO J. 1989 Dec 20;8(13):4015–4024. doi: 10.1002/j.1460-2075.1989.tb08584.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuler G. D., Altschul S. F., Lipman D. J. A workbench for multiple alignment construction and analysis. Proteins. 1991;9(3):180–190. doi: 10.1002/prot.340090304. [DOI] [PubMed] [Google Scholar]
- Tatusov R. L., Altschul S. F., Koonin E. V. Detection of conserved segments in proteins: iterative scanning of sequence databases with alignment blocks. Proc Natl Acad Sci U S A. 1994 Dec 6;91(25):12091–12095. doi: 10.1073/pnas.91.25.12091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tollervey D., Lehtonen H., Jansen R., Kern H., Hurt E. C. Temperature-sensitive mutations demonstrate roles for yeast fibrillarin in pre-rRNA processing, pre-rRNA methylation, and ribosome assembly. Cell. 1993 Feb 12;72(3):443–457. doi: 10.1016/0092-8674(93)90120-f. [DOI] [PubMed] [Google Scholar]
- Wahl R., Rice P., Rice C. M., Kröger M. ECD--a totally integrated database of Escherichia coli K12. Nucleic Acids Res. 1994 Sep;22(17):3450–3455. doi: 10.1093/nar/22.17.3450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipkas D., Riley M. Proposal concerning mechanism of evolution of the genome of Escherichia coli. Proc Natl Acad Sci U S A. 1975 Apr;72(4):1354–1358. doi: 10.1073/pnas.72.4.1354. [DOI] [PMC free article] [PubMed] [Google Scholar]