Abstract
The protein MeCP2 mediates epigenetic regulation by binding methyl-CpG (mCpG) sites on chromatin. MeCP2 consists of six domains of which one, the methyl binding domain (MBD), binds mCpG sites in duplex DNA. We show that solution conditions with physiological or greater salt concentrations or the presence of nonspecific competitor DNA is necessary for the MBD to discriminate mCpG from CpG with high specificity. The specificity for mCpG over CpG is >100-fold under these solution conditions. In contrast, the MBD does not discriminate hydroxymethyl-CpG from CpG. The MBD is unusual among site-specific DNA binding proteins in that (i) specificity is not conferred by the enhanced affinity for the specific site but rather by suppression of its affinity for generic DNA, (ii) its specific binding to mCpG is highly electrostatic, and (iii) it takes up as well as displaces monovalent cations upon DNA binding. The MBD displays an unusually high affinity for single-stranded DNA independent of modification or sequence. In addition, the MBD forms a discrete dimer on DNA via a noncooperative binding pathway. Because the affinity of the second monomer is 1 order of magnitude greater than that of nonspecific binding, the MBD dimer is a unique molecular complex. The significance of these results in the context of neuronal function and development and MeCP2-related developmental disorders such as Rett syndrome is discussed.
DNA methylation is a reversible epigenetic determinant regulating cellular differentiation.1 The effect of DNA methylation on gene expression is realized through specific regulators termed “methyl-CpG binding proteins” (MBPs). The three structural families that comprise the MBP are the MBDs, the zinc finger, and SRA.2 Disruption of the proteins that specifically recognize epigenetic methylation marks can cause disease. In Rett Syndrome, complex phenotypes, including language and motor skills, are caused by mutations in a MBD family member, methyl-CpG binding protein 2 (MeCP2).3 MeCP2 is a multifunctional protein localized to the cell nucleus that regulates gene expression, chromatin structure, and RNA splicing processes that together direct brain development.4−7 Common MeCP2 Rett Syndrome-associated mutations are missense localized in its DNA binding domain, the MBD.3 These Rett Syndrome mutations alter the ability of the MBD to bind DNA bearing the methyl-CpG (mCpG) modification.8−10
MeCP2 is highly expressed in neuronal tissues.4 In addition to its binding of mCpG islands, MeCP2 and linker histone H1 compete for DNA binding between nucleosomes.11 The mechanism of exchange between DNA-bound MeCP2 (whether mCpG, CpG, or random sequence) and histone H1 is undefined. The MBD is critical for the proper interaction of MeCP2 with chromatin,10,12 although other domains contribute to DNA binding and chromatin restructuring.13,14 The models of MeCP2 modulation of human and mouse BDNF gene expression considered here (activation, repression, or “dual” model) assume recognition of mCpG as the first step.15 Because discrimination of mCpG in genomic DNA is at the heart of the mechanism(s) by which MeCP2 exerts its epigenetic function, it is important to understand the physical basis of MBD binding and specificity to fully understand how the epigenetic signal propagates to biological regulation.
Because of its high level of expression, specific and nonspecific MeCP2 DNA binding must balance for the protein to exert its biological function. The classic conundrum of proteins that bind to DNA-specific sequences is that if the level of DNA sequence nonspecific binding is too high, the protein is sequestered “off target” and thus functionally impaired. This state of affairs may occur because of low specificity for the target site. Understanding specificity is particularly important for understanding the biology of MeCP2 because it is very highly expressed in neuronal tissues,4 bringing both specific and nonspecific interactions into play.16
“Binding specificity” is formally defined as Ks/Kn, the ratio of the affinity of a protein for a specific sequence (Ks) relative to its affinity for generic DNA (Kn) determined under common solution conditions.17 Variable binding affinities and stoichiometries have been reported for MeCP2 and its fragments in studies conducted under disparate solution conditions.11,18−22 Specificity values of 3 and 12–18 have been reported for human and Xenopus MeCP2, respectively.11,19 In contrast, protein–DNA regulators of cellular metabolism are typically specific for their target sites by multiple orders of magnitude.23 Explanations for the effect of the naturally occurring mutations responsible for Rett syndrome and other neurological disorders have been based upon binding studies conducted under a variety of solution conditions.5,6,11,19,24 In addition, MeCP2 domains C-terminal to the MBD are extensively disordered25,26 and restructure upon the protein binding mCpG sites.9,22,26 These conformational changes are postulated to allosterically influence interaction of MeCP2 with other regulatory proteins.25,26 Delineation of mCpG binding specificity will allow further insight into the molecular dysfunction caused by disease mutations and whether allosteric transitions are linked to the precise nature of the bound DNA.
In our studies, we explore discrimination of mCpG from CpG by MeCP2 by dissecting the balance of forces underlying these reversible association reactions. Our initial studies focus on the MBD because this domain mediates MeCP2 recognition of mCpG.8,26,27 A structure of the MBD bound to methylated DNA8,9 provides insight into methyl-specific binding; the degree to which hydration is reported to contribute to discrimination of the methylated residue in the structure is highly unusual. To our surprise, salt uptake rather than hydration appears to play a dominant role in the specificity of mCpG discrimination, electrostatics play an outsized role in binding to modified nucleotides, and the domain can assemble on DNA to form a discrete dimer. The implications of these findings for the function of full-length MeCP2 during neuronal activity and development are discussed.
Experimental Procedures
Protein, DNA, and Other Reagents
The DNA binding domain of MeCP2 (MBD, residues 76–167; Uniprot entry P51608) was inserted into a PET21 derivative by the Einstein Protein Production Facility to yield pMCSG7. Following transformation into Rosetta (DE3) cells, the protein was expressed following standard procedures in media containing 0.4% sorbitol. The MBD was purified as previously described28 except that HEPES buffer was used throughout and the His tag was cleaved by recombinant Tev protease as described by the manufacturer (Invitrogen). A molar extinction coefficient (εM) of 11460 M−1cm−1 was used to determine the MBD concentration from their 280 nm absorption spectra.
All buffer solution solutes were purchased from Sigma. Labeled and unlabeled DNA oligonucleotides were purchased from Integrated DNA Technologies, Inc. (Coralville, IA), and their concentration was optically determined using extinction coefficients calculated from sequence. The extinction was corrected for the fluorescein absorption at 260 nm when appropriate. The 20 bp oligonucleotide 5′-TCTGGAACGGAATTCTTCTA-3′, with C methylated or unmethylated, was used in our study. This sequence is taken from promoter III of the mouse BDNF gene and is the DNA present in the MBD cocrystal structure.8 A random sequence 20 bp oligonucleotide not bearing the CpG site (5′-TCTGGTATGAACTTCTA-3′) was also analyzed. The top strands of each duplex were 5′-labeled with fluorescein or unlabeled. The duplexes used in our studies have stabilities of −35.9 and −26.5 kcal/mol corresponding to melting temperatures of 40.7 and 32.5 °C, respectively, under our experimental conditions, resulting in an undetectable single-strand oligonucleotide at the lowest concentration used in binding assays (5 nM).
Fluorescence, Absorption, and Light Scattering Measurements
All experiments were performed in buffer containing 25 mM Tris-HCl, 6% glycerol, 0.1 mM EDTA, 0.1 mM TCEP, and either 25 or 150 mM KCI (pH 7.6) at 22 °C except for the salt-induced dissociation experiments for which the KCl concentration is specified. The presence or absence of 10 μg/mL nonspecific competitor poly(dA-dT) is noted in the figure and table legends. Absorption measurements were taken with a NanoDrop 2000 UV–vis spectrophotometer. Fluorescence measurements were taken with a Jobin Yvon (Edison, NJ) Fluoromax-3 spectrofluorometer. The intensity of the Raman scattering band of water was used as the internal standard of fluorometer sensitivity. Elastic light scattering (ELS) was also recorded using the Fluoromax-3 spectrofluorometer as a control for protein aggregation. The scattered 350 nm light was collected at an angle of 90° to the incident illumination.
Analytical Ultracentrifuge
Sedimentation equilibrium experiments were performed using the absorption optics of a Beckman XL-I analytical ultracentrifuge with six-channel centerpieces in the Ti-60 rotor. Three concentrations of the MBD in buffer were equilibrated at 20 °C for 24 h each at 10000, 20000 and 30000 rpm. The absorbance scans obtained at 280 nm were globally analyzed using HeteroAnalysis version 1.0.114 (J. L. Cole and J. W. Lary, Analytical Ultracentrifugation Facility, Biotechnology Services Center, University of Connecticut, Storrs, CT) for the weight-average molecular weight. The resolved molecular weight and the 95% joint confidence intervals are reported. The values of ν̅ (from the amino acid composition), density, and viscosity were calculated using Sednterp version 1.06 (B. Hayes, T. Laue, and J. Philo, Sedimentation Interpretation Program, 2003, University of New Hampshire, Durham, NH).
Equilibrium Binding
MBD binding isotherms were calculated and analyzed as described below. Fractional saturation (Y̅) is calculated for equilibrium titrations by
| 1 |
where Aobs, Amin, and Amax are the observed, minimum, and maximum values of the measured fluorescence anisotropy, respectively. For the equilibrium PfnH + Of ↔ POnH, where Pf and Of are the free concentrations of protein and DNA, respectively, PO is the protein–DNA complex, and nH is the Hill coefficient, fractional saturation is related to the binding polynomial by
| 2 |
where PO and Otot are the protein–DNA complex and total DNA concentrations, respectively. Substitution and transformation29 yield
| 3 |
where kd is the equilibrium dissociation constant and Ptot is the total protein concentration. Equation 3 can be transformed into a convenient form in which Ptot is substituted with the ratio PtotnH/Otot as the independent variable to yield
| 4 |
Equation 4 reduces to the single-site (Langmuir) binding model with modifications explicitly considering the total DNA concentration where nH = 1. Fitting eq 4 against Y̅ obtained from the experimental data accurately yields kd even when the concentration of the DNA is comparable to kd.
Two-Site Binding Model
If a molecule has two binding sites with different affinities for the same ligand, then eq 2 can be rewritten as
| 5 |
where Y̅ is the total saturation of the two sites (Y̅1 and Y̅2) and PO1, PO2, and Otot are the protein–DNA complex and total DNA concentrations, respectively. At low concentrations of the DNA target where Ptot ≈ Pfree, eq 5 can be transformed into the sum of two isotherms
| 5a |
where kd1 and kd2 are the dissociation constants for the first and second binding sites, respectively, and Ptot is the total protein concentration. Because the total anisotropy change is the sum of the change for the two binding events (Aobs = Aobs1 + Aobs2), eq 5a can be transformed into
| 6 |
where Aobs, Aobs1, Aobs2, Amin, and Amax are defined for two binding sites as in eq 1. A0 is the initial value of Aobs. If the measured parameter Aobs is a relative quantity, then A0 is equal to 0 and eq 6 is transformed into
| 7 |
We used the relative value of anisotropy Arel = (Aobs – ADNA)/ADNA as a binding parameter, where Aobs is as in eqs 6 and 7 and ADNA is the value of the anisotropy of the DNA in the absence of the MBD. Nonlinear least-squares fitting of eq 7 yields values of kd1, kd2, Amax1, and Amax2.
We have used oligonucleotides end labeled with fluorescein as in many other studies of protein–DNA complexes.11,30,31 In those studies and this study, we measure no significant change in the fluorescence intensity of the fluorescein probe upon protein binding under a given solution condition. In this study, we conducted control experiments in which labeled DNA was present in “tracer amounts” to which was added unlabeled DNA. Isotherms identical to those obtained using only the labeled DNA were obtained (data not shown).
Salt-Induced Dissociation Titrations
The electrostatic contribution to formation of the MBD–DNA complex was determined from the net number of ions (cations, m+, and anions, x-) released
| 8 |
where kd is the experimentally determined equilibrium dissociation constant, (kd)0 is the value of kd extrapolated to 1 M salt, and m+ is the cation concentration.32 Usually, m+ ≫ x– because of the polyanionic nature of DNA and the polyampholyte nature of proteins. Because the influence of water is negligible at high salt concentrations, SK reflects the net cation release or uptake upon formation of a protein–DNA complex.
SK can be conveniently obtained from salt displacement isotherms in which a protein–DNA complex is dissociated by titration with increasing salt concentrations.33 In these experiments
| 9 |
where
| 10 |
with (kd)0 transformed into mp, the salt concentration at which half of the initial protein–DNA complex is dissociated. The nonelectrostatic portion of ΔG is calculated from (kd)0 (eq 8) by ΔGnel = −RT ln[1/(Kd)0].33
Because the affinity of the protein for the different sequences analyzed differs, Y̅ for the complexes differs under the initial condition of the experiment. An impact of different Y̅ values on the resolved values of SK and mp is circumvented by setting the initial condition of the salt dissociation experiments at Pt = Ot. As was shown previously,33 there is no dependence on the concentrations of the reactants, Pt and Ot, under that condition. Thus, eq 10 can be used to measure relative rather than absolute values of Y̅.
Equimolar concentrations of the MBD and the unlabeled duplex (0.5–1.0 uM) along with the labeled oligonucleotide at 15 nM were mixed and incubated for 1 h. KCl was then added at the specified concentration and the solution incubated for 1 h prior to measurements. Because the fluorescence of fluorescein is sensitive to high salt concentrations, dissociation of the MBD from DNA was followed by measurement of the anisotropy of the intrinsic protein fluorescence at 330 nm and excitation of 280 nm. Anorml = (Aobs – A0)/(Amax – A0), where Aobs, A0, and Amax are the observed and fitted initial and final values of anisotropy, respectively, and Anorml is a measured parameter. The relative values of Y̅ analyzed range from unity to zero for each salt displacement isotherm (Figure 9B). Nonlinear least-squares fitting of salt displacement isotherms to eqs 8–10 yields values of (kd)0, mp, and SK at any macromolecule concentration.33
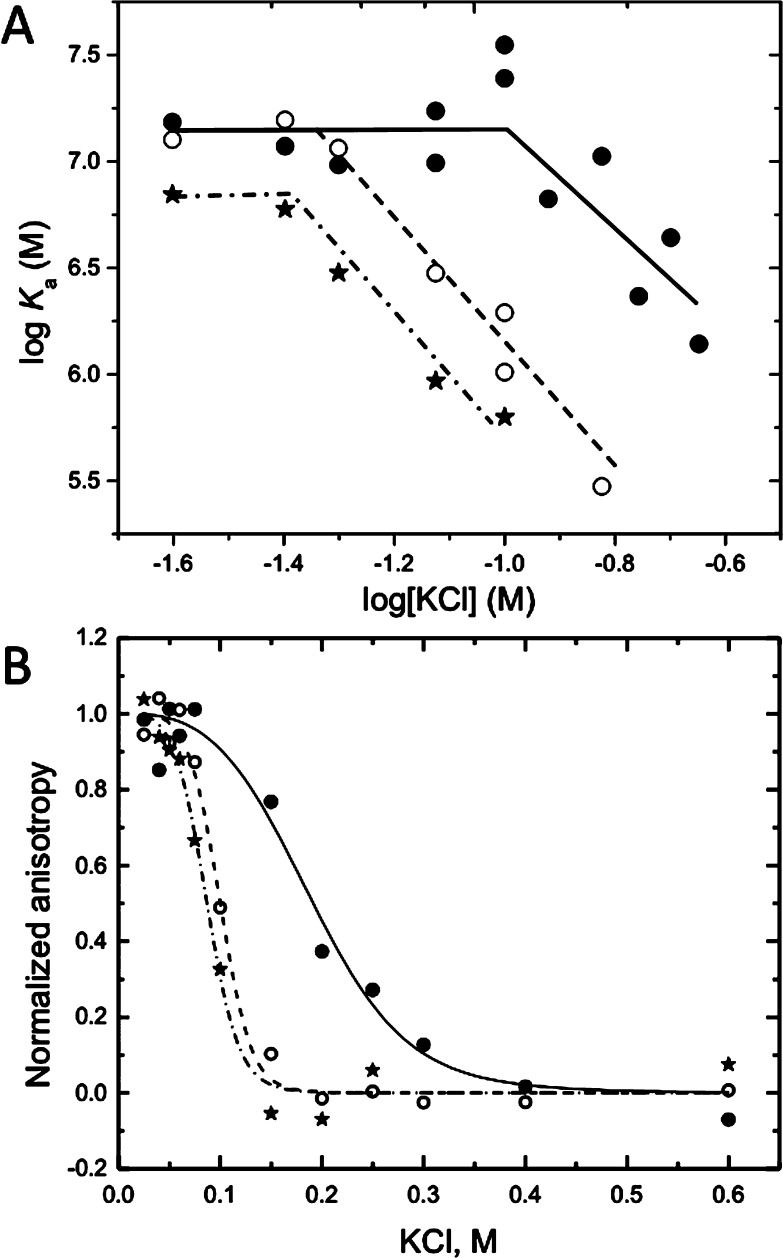
Figure 9.
(A) Wyman linkage analysis of the binding of the MBD to mCpG-bearing (●, —), CpG-bearing (○, ---), and random sequence (∗, −·−) oligonucleotides in standard buffer as a function of KCl concentration. Each value of the association constant (Ka) was determined from a binding isotherm such as shown in Figure 1 obtained at the indicated KCl concentration. (B) Salt-induced dissociation isotherms of complexes of the MBD with the mCpG, CpG, and random sequence oligonucleotides in standard buffer as a function of KCl concentration. The intrinsic tryptophan fluorescence of the MBD (ex280 and em330) was monitored as described in Experimental Procedures. The lines depict the fit to eqs 9 and 10 (Table 3). All designations are the same as in panel A.
Comparison of the Goodness of Fit of Alternative Binding Models
Comparison of the performance of models is typically characterized by the root-mean-square deviations (rmsds) between the theoretical and experimental estimates of the data
| 11 |
where Xi and Yi are the theoretical and experimental estimates, i, respectively, in N experimental points. The normalized rmsd (nrmsd) or error (nrmse) is the rmsd divided by the range of observed values of a variable being predicted
| 12 |
The quality of the fit of a model to the data is reflected in the rmsd and nrmsd values. Smaller values indicate a closer relationship between theory and experiment.
Results
MBD Binding Specificity
Key characteristics of a protein–DNA interaction that drive its biological function are its specificity for its target site and its stoichiometry. Our studies compare the binding of the MBD to the DNA duplex present in the MBD cocrystal structure8 bearing the CpG site with C either methylated or unmethylated and a “random” sequence oligonucleotide lacking any known determinant for MBD affinity. Figures 1 and 2 summarize the dramatic effect of salt concentration on the discrimination of mCpG from CpG. As noted previously,11 the MBD lacks specificity for mCpG at low salt concentrations. Consistent with this finding, we find that the MBD binds in a low-salt solution mCpG, CpG, and random sequence with comparable affinity within experimental error (Figure 1A and Table 1). Increasing the salt concentration to an approximately physiological concentration (150 mM KCl) slightly increases the affinity of the MBD for mCpG while dramatically weakening its binding to CpG and the random sequence (Figures 1B and Table 1). The observed increase in binding affinity for mCpG with an increasing salt concentration is very unusual for a protein–DNA interaction. In contrast, the suppression of CpG and random sequence binding by salt is canonical protein–DNA binding behavior.34
Figure 1.
Isotherms determined by fluorescence anisotropy (ex490 and em520) for the binding of the MBD to the fluorescein-labeled duplex 5′-TCTGGAACGGAATTCTTCTA-3′ with C symmetrically methylated (●) or unmethylated (○) and fluorescein-labeled duplex 5′-TCTGGTATGAACTTCTTCTA-3′ lacking any MBD binding determinants (“random”, ∗) determined in the standard buffer solution containing 25 mM Tris, 6% glycerol, 0.1 mM EDTA, 100 μg/mL BSA, 0.1 mM TCEP (pH 7.6), and either 25 (A) or 150 mM KCl (B). The DNA concentration is 5 nM. The solid lines depict global fits to the Langmuir model (Table 1).
Figure 2.
Isotherms determined by fluorescence anisotropy (ex490 and em520) for the binding of the MBD to different DNAs at 150 mM KCl over a range of MBD concentrations broader than that shown in Figure 1. The reaction conditions and symbol designations are the same as in Figure 1.
Table 1. MBD Binding to mCpG, CpG, and Random Sequence DNA Oligonucleotides at Low and High Salt Concentrationsa.
| [KCl] (mM) | sequence | one-site analysis KD (μM) | two-site analysis KD1 and KD2 (μM) | nrmsd, one- vs two-site analysis |
|---|---|---|---|---|
| 25 | mCpG-mCpG | 0.15 ± 0.08 | 0.007 ± 0.003 | 0.085 vs 0.022 |
| 1.46 ± 0.57 | ||||
| CpG-CpG | 0.19 ± 0.09 | 0.011 ± 0.004 | 0.088 vs 0.022 | |
| 1.56 ± 0.64 | ||||
| random | 0.32 ± 0.08 | 0.07 ± 0.02 | 0.054 vs 0.019 | |
| 3.7 ± 2.5 | ||||
| 150 | mCpG-mCpG | 0.12 ± 0.01 | 0.075 ± 0.01 | 0.054 vs 0.019 |
| 3.7 ± 2.6 | ||||
| hmCpG-hmCpG | 2.1 ± 1.5 | na | na | |
| CpG-CpG | ≥10.3 | na | na | |
| random | ≥17.7 | na | na | |
| 25 (+) | mCpG-mCpG | 0.028 ± 0.015 | na | na |
| CpG-CpG | na | – | – | |
| random | na | – | – | |
| 150 (+) | mCpG-mCpG | 0.19 ± 0.01 | 0.14 ± 0.04 | 0.014 vs 0.016 |
| 1.7 ± 2.8 | ||||
| CpG-CpG | ND | – | – | |
| random | ND | – | – |
Binding isotherms were determined and analyzed as described in Experimental Procedures in solutions containing either 25 or 150 mM KCl. KD, KD1, and KD2 denote the equilibrium binding constants determined from the indicated model. The modified oligonucleotide was symmetrically methylated. (+) denotes the presence of poly(dA-dT). na denotes not applicable. ND denotes not determined.
The 150 mM KCl binding isotherms shown in Figure 1B were extended to higher MBD concentrations in an attempt to define their upper plateaus (Figure 2). Even so, an upper plateau could not be determined, allowing calculation of only lower limits for these binding affinities, ≥10.3 and 17.7 μM, respectively (Table 1). The mCpG isotherm encompasses both specific and nonspecific binding, an observation consistent with the literature.35 Under these conditions, the MBD displays ≥86-fold specificity for mCpG over CpG and ≥147-fold specificity over random sequence DNA (Table 1).
MBD Binding Stoichiometry
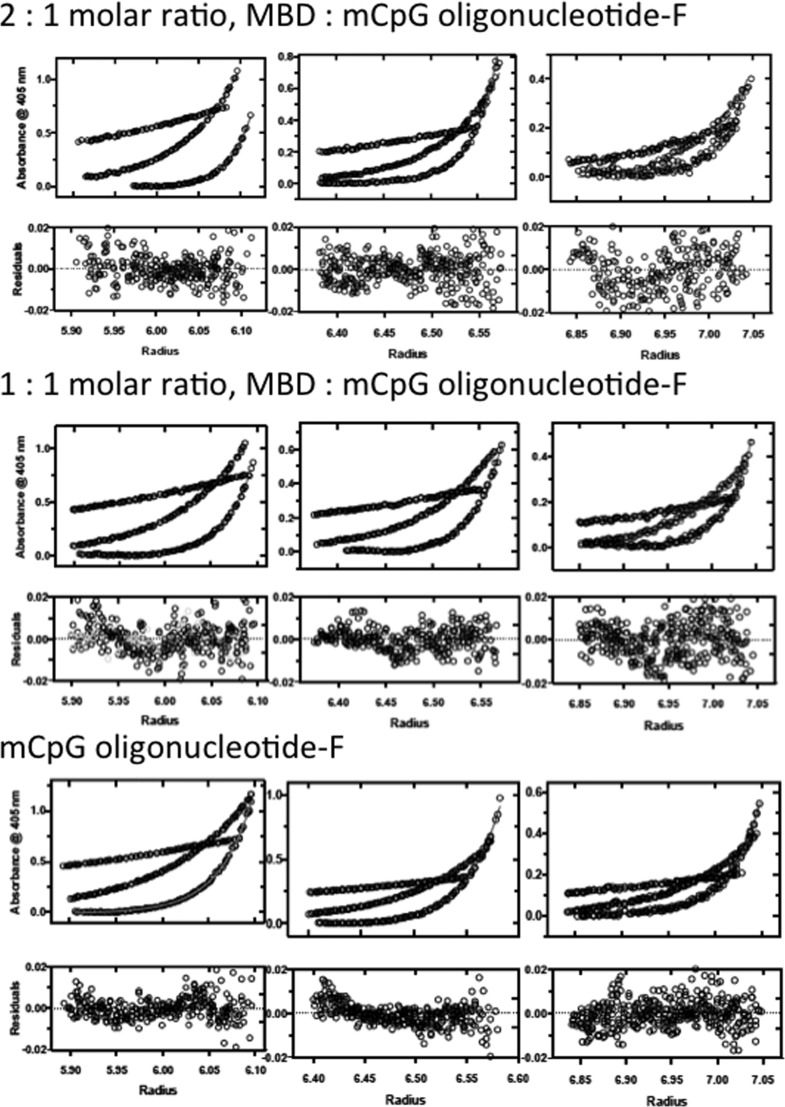
We routinely conduct titrations at DNA concentrations greater than the KD of the reaction to determine the stoichiometry of a binding reaction.28,33 In light of the structures of the 1:1 MBD–mCpG complexes determined by NMR and crystallography from equimolar mixtures of protein and DNA,8,9 we were quite surprised to measure a 2:1 stoichiometry for the MBD–mCpG complex (Figure 3). This result was confirmed using analytical ultracentrifugation by monitoring the dye absorption of the fluorescein-labeled mCpG oligonucleotide (Figure 4). The sedimentation equilibrium analysis yielded Mw values for the free DNA, and complexes assembled from 1:1 and 2:1 molar ratios of the MBD and DNA are within error of the values calculated from sequence (square brackets): 12.6 (12.1, 13.0), [12.8]; 24.3 (23.1, 25.4), [23.2]; and 33.1 (31.1, 35.3), [33.6] kDa, respectively. Each of the data sets is described as a single species with no evidence of heterogeneity or active self-association. The homogeneity of the 1:1 and 2:1 complexes was further confirmed by sedimentation velocity analysis (data not shown). Together, stoichiometric titration and analytical ultracentrifugation results make an unassailable case that the MBD binds as a dimer to mCpG-bearing DNA.
Figure 3.
Stoichiometric titrations of the binding of the MBD to the mCpG oligonucleotide duplex followed by anisotropy of the DNA-coupled probe in standard buffer and 150 mM KCl. The total DNA concentration is 0.5 μM using 5 nM labeled DNA as a tracer.
Figure 4.
Sedimentation equilibrium analysis of fluorescein end-labeled mCpG oligonucleotide (lower) and 1:1 (middle) and 2:1 (top) molar ratios of MBD:mCpG oligonucleotide-F by the 490 nm absorbance of the fluorescein label. The solid lines are the global fits to the model of a single monodisperse particle. The residuals for the fits to each channel are shown below the concentration distributions.
A Two-Site “Sequential” Binding Mechanism
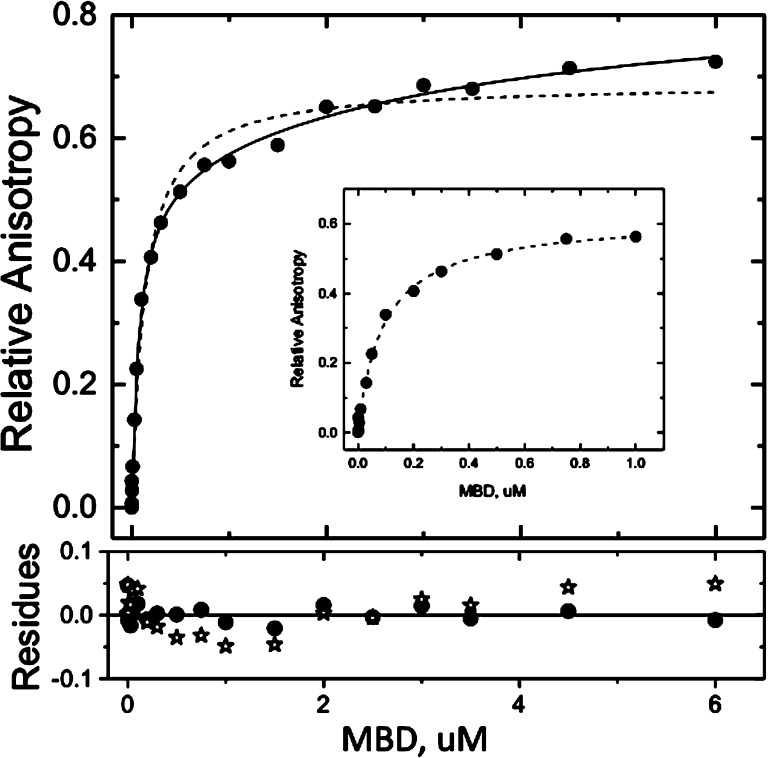
How can the 2:1 stoichiometry be reconciled with the published monomeric structures and our binding analyses? To answer this question, we conducted data-dense titrations at 150 mM KCl so that the shape of the isotherms could be precisely determined. It is readily apparent in these isotherms that the Langmuir binding model inadequately describes the binding of the MBD to mCpG (Figure 5, dotted line). Without sufficient data density and careful analysis, this poor fit is easy to either miss or ascribe to experimental error (e.g., Figure 1B; analysis not shown). Indeed, the initial portion of the isotherm (<1 μM) is perfectly described by the Langmuir binding model (Figure 5, inset). [In retrospect, a clue that the binding was more complex than Langmuir was that fits of the Hill equation (eq 4) to isotherms such as that shown in Figure 1 were typically described with coefficients below unity. Such behavior can indicate anticooperativity or heterogeneity.] Thus, the complexity of MBD–mCpG binding is not evident if either the number of data points is too small or the MBD concentration range is too narrow.
Figure 5.
Isotherms of the binding of the MBD to methylated DNA in standard buffer at 150 mM KCl as determined by fluorescence anisotropy (ex490 and em520) fit to the two-site (—) or single-site (---) models as described in Experimental Procedures. The bottom panel represents the residuals with respect to the fitting models [two-site (●) and single-site (∗)]. The DNA duplex analyzed is the same as that in Figure 1. The inset shows the same data truncated at 1 μM that were independently fit to the single-site model.
The simplest binding model that describes these isotherms is a noninteracting two-site model (Figure 5, solid line). The MBD binds tightly to the first site (KD1 = 75 ± 10 nM) and 50-fold less so (KD2 = 3.7 ± 2.6 μM) to the second site (Table 1). Random residuals and lower nrmsd values demonstrate the superiority of the two-site model for formation of the MBD–mCpG complex (Figure 5 and Table 1). Because KD2 is 1 order of magnitude tighter than nonspecific binding, the second binding event is a distinct and unique molecular interaction.
Because of the wide separation of the resolved affinities, the two binding events occur in sequence. Only after the first site is almost fully occupied does the second site become bound. This conclusion is supported by three independent observations. First, we observe that the MBD is monomeric in solution [Mw = 10.6 (9.8, 11.2), [10.4] kDa] as assayed by sedimentation equilibrium and velocity analysis consistent with literature observations.36 Second, each of the two binding events is separately well described by the Langmuir polynomial. Third, the 1:1 complex is unequivocally monodisperse as assayed by sedimentation equilibrium and velocity analysis. If the binding were at all cooperative or the two binding events were closer in affinity, a mixture of 1:1 and 2:1 complexes would be observed at substoichiometric protein concentrations. It is the last observation that rationalizes the existence of the published 1:1 MBD–mCpG structures.8,9 A 1:1 protein:DNA ratio was used to form the complexes that were structurally analyzed. Because binding of the second monomer follows complete occupancy of the first, the 1:1 complex is observed to be homogeneous (Figure 4, middle panel).
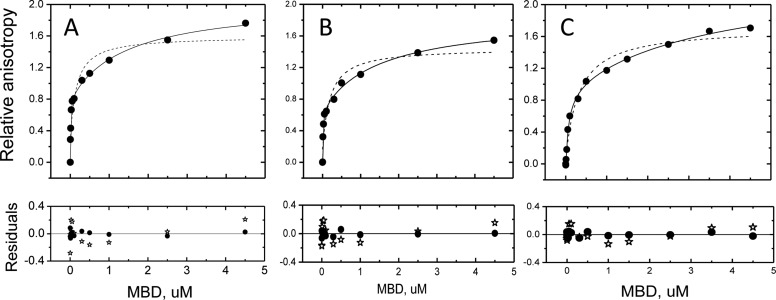
To test whether dimeric binding of the MBD was unique to modified DNA, we determined “data-dense” isotherms at 25 mM KCl, conditions under which the binding affinity of the MBD for mCpG, CpG, and random sequence DNA is comparably high. The two-site binding model better fits all three isotherms (Figure 6). Again, the values of KD1 and KD2 determined are readily distinguishable (Table 1). From these results, we conclude that the assembly of MBD monomers to form a discrete DNA-bound dimer is not unique to its interaction with modified DNA.
Figure 6.
Isotherms of the binding of the MBD to methylated (A), unmethylated (B), and random (C) DNA obtained in standard buffer at 25 mM KCl as determined by fluorescence anisotropy (ex490 and em520). The isotherms were fit to the two-site (—) or single-site (---) models as described in Experimental Procedures. The bottom panels show the residuals with respect to the fitting models [two-site (●) and single-site (∗)].
A Complementary Probe of mCpG Binding Specificity
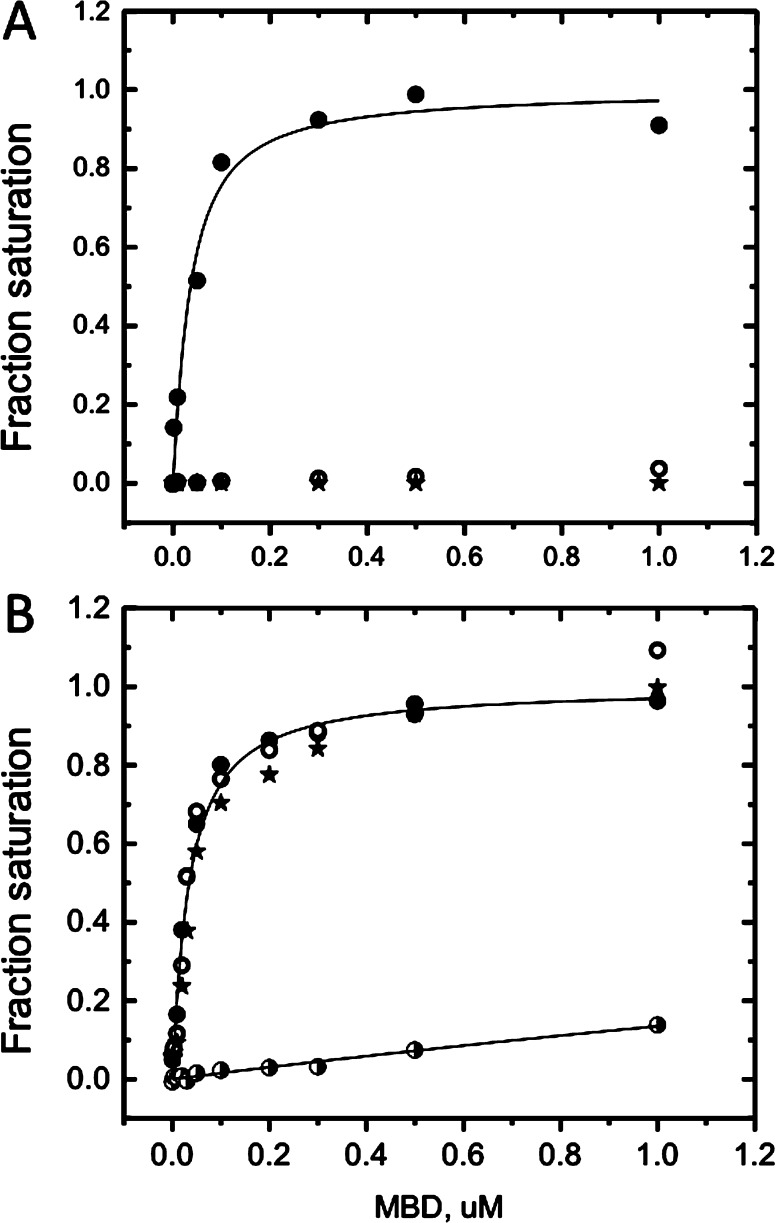
The addition of competitor DNA to “soak up” nonspecifically bound protein is also used to distinguish specific binding.37 When we added the poly(dA-dT) oligonucleotide to 25 mM KCl MBD binding titrations, the affinity of the MBD for mCpG increases several-fold and dramatically decreases for CpG and random sequence oligonucleotides to undetectable (Figure 7A and Table 1). This behavior is comparable to that observed at 150 mM KCl (Figure 1B and Table 1). The presence of poly(dA-dT) has little effect at 150 mM KCl on either binding affinity or dimer formation (Figure 7 and Table 1).
Figure 7.
(A) Isotherms determined by fluorescence anisotropy (ex490 and em520) of the binding of the MBD to the mCpG (●), CpG (○), and random sequence (∗) oligonucleotides (Figure 1 legend) in standard buffer at 150 mM KCl and 10 μg/mL poly(dA-dT) competitor. (B) Isotherms of the binding of the MBD to single-stranded oligonucleotides in standard buffer at 25 mM KCl (designated as in panel A) or to mCpG at 150 mM KCl (◑). No competitor DNA is present. The solid lines depict the best fit to the Langmuir binding model in both panels.
The dependence of mCpG binding specificity on either salt or the presence of competitor shows that suppression of nonspecific binding underlies the ability of the MBD to discriminate sites of modified nucleotides at physiological salt concentrations. The fact that the poly(dA-dT) competitor obliterates binding of the MBD to CpG and random DNA at 25 mM KCl shows that these reactions are entirely electrostatic in nature. However, one aspect of the binding of the MBD to mCpG suggests additional complexity to its binding mechanism. That aspect is the increase in binding affinity when the KCl concentration is increased from 25 to 150 mM. The canonical behavior of DNA binding proteins is for the affinity to decrease with an increasing counterion concentration; this is true for both specific and nonspecific interactions, albeit with different magnitudes for the dependencies.32 We will return to this issue following the presentation of two other unexpected binding behaviors.
The MBD Binds Tightly to Single-Stranded DNA (ssDNA)
The MBD binds with high affinity to single strands of the duplexes discussed above, methylated, unmethylated, and random (Figure 7B and Table 2). The binding of the MBD to ssDNA is mostly electrostatic and can be detected only at low salt concentrations and independent of CpG methylation. Remarkably, the MBD binds ssDNA more tightly than the corresponding duplex DNA sequence (Tables 1 and 2). The binding isotherms are perfectly fit to the Langmuir equation, suggesting that these reactions also lack cooperativity.
Table 2. MBD Binding to mCpG, CpG, and Random Sequence Single-Stranded DNA Oligonucleotides at Low and High Salt Concentrationsa.
| [KCl] (mM) | sequence | KD (μM) | nrmsd |
|---|---|---|---|
| 25 | mCpG | 0.03 ± 0.004 | 0.033 |
| CpG | 0.038 ± 0.009 | 0.06 | |
| random | 0.066 ± 0.016 | 0.053 | |
| 150 | mCpG | 6.4 ± 0.3 | 0.006 |
| CpG | nd | – | |
| random | nd | – |
Binding isotherms were determined and analyzed as described in Experimental Procedures in solutions containing either 25 or 150 mM KCl. KD denotes the equilibrium binding constants determined for the single-site Langmuir model. nd denotes not detectable.
The MBD Binds Poorly to Hydroxymethylated CpG
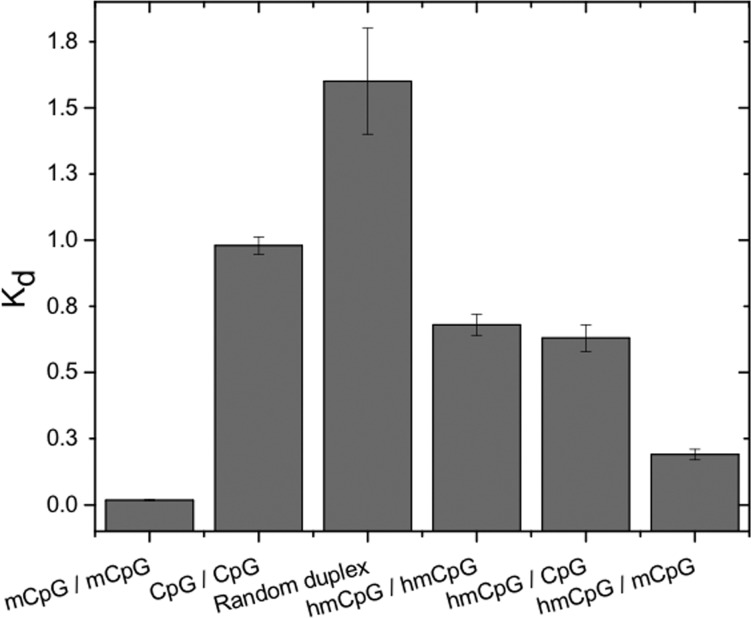
The discovery of high levels of 5-hydroxymethylcytosine (hmC) in neuronal genomes suggests that this epigenetic mark plays a cell-type-specific role in gene regulation.38 Published in vitro studies of MeCP2 and the binding of the MBD to hmCpG provide contradictory results; both high-affinity binding39 and low-affinity binding40,41 to hmCpG have been reported. In light of the salt dependence of the specificity of the MBD for mCpG discussed above, we determined isotherms for the binding of the MBD to both symmetrically and hemimodified hydroxymethylated oligonucleotides under both conditions.
At 150 mM KCl, the MBD binds 5hmCpG-containing DNA 20-fold less tightly than mCpG and only several-fold tighter than CpG (Table 1). Because the affinity of the MBD for hmCpG is so low at high salt concentrations, it was convenient to explore the binding of the MBD to symmetric and hemimodified targets at 100 mM KCl. The relative affinity of the MBD for mCpG and hmCpG is comparable to that at 150 mM KCl (Figure 8). The MBD binds hmCpG much less tightly than mCpG; there is a slight preference for the binding of symmetric hmCpG over CpG (0.5-fold) and random sequence (2-fold). The binding of the MBD to symmetric hmCpG and hemimodified (hmCpG/CpG) is the same within experimental error. Interestingly, the MBD binds the asymmetrically modified hmCpG/mCpG with greater affinity, although still weaker than symmetric mCpG. These data suggest that the asymmetrically modified hmCpG/mCpG may represent a distinct epigenetic state with regard to MeCP2 binding.
Figure 8.
Equilibrium dissociation constants (Kd, micromolar) determined in standard buffer and 100 mM KCl to the indicated symmetrically or asymmetrically modified DNA duplexes designated mCpG (methylated), CpG (unmethylated), and hmCpG (5-hydroxymethylated).
Quantitation of MBD Binding Electrostatics
Although nonelectrostatic interactions typically dominate DNA sequence-specific binding, electrostatics also plays an essential role in specific protein–DNA interactions.34,42,43 Thus, it is unsurprising that cations are required to neutralize some of this charge during the formation of a specific complex. A standard tool for partitioning a reaction into its electrostatic and nonelectrostatic components is the measurement of binding affinity as a function of salt concentration (linkage analysis). A typical protein–DNA interaction displays linear log–log dependence with the steeper slope of nonspecific binding resulting in intersection with the specific binding curve at low salt concentrations.42
The MBD is atypical of protein–DNA interactions in that it displays nonlinear linkage (Figure 9A). Formation of the MBD–mCpG complex is minimally salt-dependent from 25 to 150 mM. We interpret this observation to reflect a net cation uptake that offsets the displacement of condensed counterions typically observed when proteins bind to DNA. At ∼150 mM, the uptake reaction saturates in mCpG binding and the canonical cation displacement is observed. The binding of the MBD to CpG- and random sequence-containing DNA shows a similar salt dependence except that the inflection points are shifted to lower KCl concentrations (Figure 9A). Thus, cation uptake is not restricted to mCpG binding.
A complementary approach to partitioning the electrostatic and nonelectrostatic contributions of protein–DNA interactions is the salt displacement isotherm.33 Salt displacement isotherms for MBD complexes with mCpG-, CpG-, and random sequence-containing DNA are shown in Figure 9B. Because this assay is conducted at stoichiometric concentrations of the protein and DNA, it is minimally sensitive to the enhanced protein binding at low cation concentrations where the rate of dissociation of the complex is low. A slight plateau in the isotherm with an increased salt concentration is observed at low KCl concentrations for mCpG. Plateaus for CpG and random DNA could not be resolved in these salt displacement isotherms.
The energetic portioning calculated from these data is summarized in Table 3. The binding of the MBD to mCpG DNA displaces an average of six cations with an unusually large electrostatic component of binding of 73%. As expected, the binding of the MBD to random sequence DNA is completely electrostatic and accompanied by displacement of a slightly larger number of cations. Although the binding of the MBD to CpG is slightly favored over that to random sequence DNA, a nonelectrostatic component to its binding was not resolved. The energetics of the binding of the MBD to mCpG DNA is minimally affected by the presence of poly(dA-dT).
Table 3. Thermodynamic Parameters Derived from the Salt Displacement Isotherms for Association of the MBD with Duplex DNAa.
| mCpG | CpG | random | mCpG (+) | |
|---|---|---|---|---|
| mpb | 0.19 ± 0.008 | 0.1 ± 0.003 | 0.09 ± 0.004 | 0.2 ± 0.005 |
| nc | 6.1 ± 1.1 | 10.0 ± 2.0 | 8.2 ± 2.0 | 5.4 ± 0.5 |
| –ΔGtotd (kcal mol–1 K–1) | 9.3 ± 0.05 | 6.7 ± 0.08 | 6.4 ± 0.09 | 9.1 ± 0.03 |
| –ΔGnele (kcal mol–1 K–1) | 2.6 ± 0.6 | nd | nd | 3.4 ± 0.3 |
Salt displacement isotherms (Figure 9B) were determined and analyzed as described in Experimental Procedures for the MBD complexed to oligonucleotides bearing mCpG, CpG, or random sequence. The plus sign indicates the presence of 10 μg/mL poly(dA-dT). nd denotes not detectable.
The midpoint of a salt displacement isotherm.
The thermodynamic average number of ions released upon protein binding the DNA.
The Gibbs free energy of binding calculated from the KD values calculated from the binding isotherms determined at 150 mM KCl (Figure 1B and Table 1).
The nonelectrostatic component of the Gibbs free energy of binding.
Discussion
MeCP2 is a member of the MBD family of proteins whose biological function is to bind to sites of methylation in the genome.2,44 MeCP2-mediated epigenetic regulation starts with recognition of mCpG by its DNA binding domain, the MBD,44 and continues with multiple processes involving the other domains of the protein.16 However, many questions remain concerning the mechanism of mCpG binding specificity.
We explore in this paper a series of related questions concerning DNA binding by the MBD. (i) How well can the MBD discriminate mCpG from CpG or random sequence DNA? (ii) What is the thermodynamic driving force behind mCpG-specific binding? (iii) Does the published MBD–DNA crystal structure provide a complete and correct correlate for structure–function comparisons? (iv) Does the MBD specifically bind hmCpG? The answers to these questions are related and shed light on the mechanism by which MeCP2 exerts its biological function.
(i) Proteins that regulate cellular metabolism by recognizing and binding specific sequences of DNA typically display specificity of multiple orders of magnitude for their targets. Well-studied examples include gene regulatory proteins such as lac repressor,45 RNA polymerase,34 restriction endonucleases such as EcoRI,46 and general transcription factors such as the TATA binding protein.47 Our demonstration that at physiological salt concentrations the MBD binds mCpG with 100-fold specificity places MeCP2 within the established paradigm for regulatory proteins that bind or process genomic DNA (Figures 1 and 2 and Table 1). Clearly, the full-length protein is not required for mCpG recognition and specific binding, although the other domains of MeCP2 may attenuate and/or regulate the protein’s DNA binding.
“Specificity” as opposed to affinity is the determinant of the biological function of a DNA binding protein as it reflects the protein’s ability to discriminate its target within the genomic context. The observation that the MBD displays no specificity for mCpG over CpG at low salt concentrations in the absence of a competitor is not surprising. DNA site-specific protein binding and nonspecific protein binding typically have comparable sign dependence on salt concentration, albeit with different magnitudes; nonelectrostatic contributions weaken the salt dependence of site-specific binding. Thus, the specific binding affinity and nonspecific binding affinity by a protein typically converge at very low salt concentrations and diverge at high salt concentrations as we observe for the MBD (Table 1).
(ii) However, the convergence of the MBD’s specific and nonspecific binding affinity occurs by an atypical mechanism. The direct titration and salt displacement approaches that we have used share the theoretical foundation that the favorable entropy from counterion release drives the formation of complexes between proteins and DNA.32,48 Electrostatics contributes the lion’s share of the Gibbs free energy of formation of the MBD–mCpG complex (Figure 9B and Table 3). Specificity for mCpG emerges only when salt dampens the electrostatic contribution to DNA binding by the MBD.
The importance of electrostatics in MBD binding is confirmed by the ability of the poly(dA-dT) competitor to ameliorate the influence of salt.23 The nonspecific DNA competitor is an alternate probe of electrostatic interactions. The finding that DNA competitor influences the binding of the MBD to all three sequences at low salt concentrations is unsurprising (Table 1). However, the magnitude of the effect is unusual. Nonspecific binding is completely suppressed, highlighting the dominant electrostatic contribution to MBD binding revealed by the salt displacement analysis (Table 3). Nonspecific electrostatic interaction of the MBD with the DNA backbone phosphates constitutes a significant portion of the total MBD interactions even for DNA containing mCpG.8,9
Thus, the relative affinity of the MBD for methyl, sequence, and nonspecific DNA is critically dependent on the electrostatic binding contribution. This property of the MBD makes binding of the protein potentially very sensitive to subtle local changes in the cellular milieu that in turn may modulate the binding of MeCP2 to sites within chromatin.
The salt dependence of MBD binding is atypical. The MBD binding affinity for mCpG increases slightly as the salt concentration increases from 25 to 150 mM, in contrast to the expected decrease reflecting a net uptake of cations for DNA binding by the MBD below physiological salt concentrations (Figure 9 and Table 1). Cation uptake has been documented for only a few protein–DNA interactions, the TATA binding protein (TBP) from the hyperthermophilic archaeal organism Pyrococcus woesei(49) and for the papillomavirus E2 protein.33 In the first case, cation uptake is linked to the protein itself. In the second case, cation uptake is linked to a specific sequence of “linker DNA” that is not contacted by the protein. Charge neutralization is clearly one contribution of cations to the mCpG binding specificity of the MBD. We discuss a second alternative in the next section within the context of the determined MBD–mCpG crystal structure.
The MBD displays a small but detectable preference for CpG-containing DNA over random sequence DNA at 150 mM KCl. In contrast to the dramatic salt sensitivity of the specificity between mCpG and CpG binding, the salt sensitivity of CpG binding relative to random sequence is minimal, indicative of only a small nonelectrostatic contribution to CpG recognition (Table 1). Thus, CpG binding may result from the subtle sequence-dependent differences in the conformation of the phosphodiester backbone rather than direct interactions between the protein and the bases. Base sequence recognition via local changes in backbone conformation is not unprecedented but is unusual as the sole mechanism of specificity. Minimal CpG affinity is important in epigenetic regulation. If the MBD bound CpG tightly, it would compete with binding to mCpG marks, thus diminishing the amplitude of the epigenetic signal.
While it is unsurprising that the MBD binds ssDNA at low salt concentrations, the MBD’s higher affinity for ssDNA than for duplex DNA is surprising (Tables 1 and 2). This behavior likely reflects the large electrostatic contribution to binding as well as the fact that the binding of symmetrically methylated DNA is not highly cooperative, i.e., not “all or none”. Because MeCP2 is abundantly expressed,4 the protein may bind to ssDNA or RNA in neuronal cells, thereby stabilizing alternative chromatin structures, and perhaps be involved (as other SSB proteins) in telomere end maintenance, DNA replication, recombination, and repair.50 Such novel roles for MeCP2 function deserve further exploration.
(iii) Monovalent cations condensed to DNA are not immobile; they readily exchange with solution, and thus, any particular DNA site is only partially occupied at a given time. A coalition of partially occupied sites is thermodynamically indistinguishable from a single fully occupied site.51,52 Linkage and salt displacement analysis report the thermodynamic average of the number of ions released or taken up upon formation of protein–DNA complexes. Monovalent ions in general cannot be readily identified or distinguished from water molecules in crystal structures, a generalization that can be applied to the MBD–DNA complex.8
While the mechanism of cation uptake for the binding of mCpG by the MBD remains obscure, inferences can be made on the basis of the complex structures8,9 and the thermodynamic principles of cation binding to DNA. In the determined crystal structure, the interface between the MBD and mCpG DNA is reported to be highly hydrophilic. Its few classically hydrophobic contacts are offset by a larger than average number of hydrogen bonds. Twenty-three of the 25 “specific” MBD–DNA interactions noted in the determined structure are hydrogen bonds between the protein and DNA. The sole determinant of specific recognition of the methyl group observed in the crystal structure is hydrogen bonds mediated by five water molecules.8
It is difficult to distinguish monovalent cations from water molecules in protein and nucleic acid X-ray crystal structures.51 This is especially true for Na+ as it has the same number of electrons as water and, therefore, an equivalent scattering cross section.8 Thus, it is possible that one or more sites of the five methyl group-coordinating water molecules observed in the crystal structure are in fact partly or fully occupied by monovalent cations that directly mediate the MBD–mCpG-specific interaction. Another possible mechanism of mCpG-specific recognition is cation−π interaction. Cation−π interactions between the aromatic ring and a positive charge are commonly observed in protein–DNA complexes and strongly depend on the type of base and the position of the monovalent cations.53 The mCpG dinucleotide is recognized at the MBD–DNA interface by two arginine residues through hydrogen bonding and cation−π interactions.54
Alternative indirect mechanisms would affect the MBD–mCpG interface but not direct contacts with the methyl group. Cations rather than water within the interface would suppress the strong nonspecific electrostatic potential and thereby nonspecific binding. Another possibility is that although hydrogen bonds themselves are not salt-dependent, they are highly sensitive to the orientation of the constituent atoms. For instance, symmetrical hydrogen bonds are formed between the arginine “fingers” (R111 and R133) and the guanine bases of the mCpG duplex; the arginine fingers lie in a plane with the guanine bases and are locked in position by salt bridges with the carboxylates of D121 and E137.8 E137 is the only one of the four residues locked by salt bridges that is not perturbed by the binding of DNA55 and is the only glutamic acid residue among the 22 documented Rett syndrome-linked mutations located within the MBD.3
The aforementioned P. woesei TATA binding protein (TBP) shows that glutamic acid side chains can mediate cation uptake; mutation of glutamic acid residues converted ion uptake to ion release upon formation of this protein–DNA complex.49 Because the stability of glutamic acid-mediated electrostatic bridges will be sensitive to competition by cations, recognition of mCpG by the MBD might be indirectly salt-dependent, even though binding the neutral methyl group is itself not electrostatic in nature. Further study is required to distinguish the mechanism that accounts for the cation uptake that profoundly affects formation of the MBD–mCpG and thereby recognition of epigenetic marks.
All of the MBD–mCpG complexes that have been determined have been assembled from 1:1 ratios of protein to DNA.8,9,55 Despite the MBD being steadfastly monomeric in solution, we had decided to take a fresh look at this stoichiometry in light of a report that MeCP2 binds DNA as a cooperative dimer.11 Three independent analyses unequivocally confirm that the MBD binds to DNA as a dimer; stoichiometric titration (Figure 3), analytical sedimentation equilibrium (Figure 4), and binding isotherm analysis (Figure 5). The MBD is unusual among DNA binding proteins in that most others that bind their target sites as dimers detectably form dimers in solution. Proteins that we have studied with this characteristic include the cI, Gal, and Lac repressors and papillomavirus E2 protein.33,56−59 If a protein dimerizes in solution and the dimer binds to DNA, a Langmuir isotherm is observed if self-association is tight or a sigmoidal isotherm is observed if dimerization is weak.58 This binding mechanism clearly does not apply to the MBD. Rather, MBD binding is reminiscent of that of the steroid receptors whose solution dimerization is inversely coupled to the cooperativity of their binding on the DNA.61
The homogeneity of the 1:1 complex in the sedimentation analysis (Figure 4) was an important clue in understanding the MBD binding mechanism; cooperative binding of MBD monomers would yield a mixture of free DNA and 1:1 and 2:1 complexes, not the observed monodisperse 1:1 complex. This observation reconciles the measured 2:1 stoichiometry with the 1:1 determined structure because the complex was formed from equimolar protein and DNA.8 Analysis of the MBD–DNA binding isotherms clearly shows two binding events in sequence. Dimer formation is not linked to mCpG recognition (Figures 5 and 6). Although energetically coupling between the two binding events could not be resolved (analysis not shown), it is likely that it does exist because the second binding event is 1 order of magnitude tighter than nonspecific binding.
The structure of the MBD–mCpG complex clearly rationalizes some MBD point mutations linked to the dysfunctions that characterize Rett syndrome.8,9,55 These rationalized mutations diminish the stability of the domain or interfere with its DNA binding. However, the mechanisms by which other mutations cause pathology are unexplained by the complex structure. It is possible that some mutations within the bound dimer disrupt biologically important protein–protein interactions. Structural and mutagenesis studies are underway to test this hypothesis.
(iv) The MBD binds hmCpG with minimal specificity relative to CpG at salt concentrations where the protein differentiates mCpG by 100-fold. The effect of hemi (hmCpG-mCpG) versus symmetrically (hmCpG-hmCpG) modified DNA on the MBD is roughly additive.41 Similar discrimination of hemimodified DNA has been shown for DNA restriction endonuclease.62 This discrimination of hemimodification has likely regulatory consequences. Binding of MeCP2 to hemimodified hmCpG/mCpG compared to either symmetrically modified DNA is a distinct state of MeCP2 binding that in turn may differentially modulate the establishment and/or maintenance of repressive chromatin structures.40 Given the weak binding of the protein to symmetrically modified hmCpG, it likely that much of the reported in vivo MeCP2 binding is to hemimodified DNA.
What Are the Biological Implications of These Results?
The studies presented here demonstrate that the MBD alone is necessary and sufficient for MeCP2 to discriminate mCpG sites in genomic DNA. Because MeCP2 is strongly expressed in neuronal tissues, its binding to modified, unmodified, and random sequence DNA may all contribute to biological function, perhaps in different ways. Salt plays a critical and unprecedented role in mediating binding. A striking aspect of the salt dependence of MBD specificity for binding to mCpG is its invariance up to ion concentrations that are considered physiological. Thus, the salt independence of binding of MeCP2 to DNA at low salt concentrations may reflect a buffer against these fluctuations or provide a specific response to them. In addition, the MBD binds to DNA as a dimer. Thus, the published structural studies do not provide a complete description of this protein–DNA interaction. Because MeCP2 is present at high concentrations in neuronal tissues4 and the full-length protein is reported to cooperatively bind DNA in chromatin as a dimer,11 it is possible that some of the structurally or thermodynamically unannotated Rett syndrome-linked mutations impact the dimer interface.
Glossary
Abbreviations
- mCpG
5-methylcytosine-guanine dinucleotide
- MeCP2
methyl-CpG binding protein
- MBD
methyl binding domain.
The authors declare no competing financial interest.
This work was supported by Grant 1R01-GM079618 from the National Institutes of Health and a pilot project award from The Rose F. Kennedy Intellectual and Developmental Disabilities Research Center (RFK IDDRC), Albert Einstein College of Medicine (P30 HD071593).
This article was published ASAP on May 23, 2014. Figure 6 and the funding statement have been updated. The corrected version was reposted on June 3, 2014.
Funding Statement
National Institutes of Health, United States
References
- Lister R.; Pelizzola M.; Kida Y. S.; Hawkins R. D.; Nery J. R.; Hon G.; Antosiewicz-Bourget J.; O’Malley R.; Castanon R.; Klugman S.; Downes M.; Yu R.; Stewart R.; Ren B.; Thomson J. A.; Evans R. M.; Ecker J. R. (2011) Hotspots of aberrant epigenomic reprogramming in human induced pluripotent stem cells. Nature 471, 68–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck-Koehntop B. A.; Defossez P. A. (2013) On how mammalian transcription factors recognize methylated DNA. Epigenetics 8, 131–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agarwal N.; Becker A.; Jost K. L.; Haase S.; Thakur B. K.; Brero A.; Hardt T.; Kudo S.; Leonhardt H.; Cardoso M. C. (2011) MeCP2 Rett mutations affect large scale chromatin organization. Hum. Mol. Genet. 20, 4187–4195. [DOI] [PubMed] [Google Scholar]
- Skene P. J.; Illingworth R. S.; Webb S.; Kerr A. R. W.; James K. D.; Turner D. J.; Andrews R.; Bird A. P. (2010) Neuronal MeCP2 Is Expressed at Near Histone-Octamer Levels and Globally Alters the Chromatin State. Mol. Cell 37, 457–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hite K. C.; Adams V. H.; Hansen J. C. (2009) Recent advances in MeCP2 structure and function. Biochem. Cell Biol. 87, 219–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guy J.; Cheval H.; Selfridge J.; Bird A. (2011) The Role of MeCP2 in the Brain. Annu. Rev. Cell Dev. Biol. 27, 11.11–11.22. [DOI] [PubMed] [Google Scholar]
- Bassani S.; Zapata J.; Gerosa L.; Moretto E.; Murru L.; Passafaro M. (2013) The neurobiology of X-linked intellectual disability. Neuroscientist 19, 541–552. [DOI] [PubMed] [Google Scholar]
- Ho K. L.; McNae I. W.; Schmiedeberg L.; Klose R. J.; Bird A. P.; Walkinshaw M. D. (2008) MeCP2 Binding to DNA Depends upon Hydration at Methyl-CpG. Mol. Cell 29, 525–531. [DOI] [PubMed] [Google Scholar]
- Ohki I.; Shimotake N.; Fujita N.; Jee J.; Ikegami T.; Nakao M.; Shirakawa M. (2001) Solution Structure of the Methyl-CpG Binding Domain of Human MBD1 in Complex with Methylated DNA. Cell 105, 487–497. [DOI] [PubMed] [Google Scholar]
- Free A.; Wakefield R. I.; Smith B. O.; Dryden D. T.; Barlow P. N.; Bird A. P. (2001) DNA recognition by the methyl-CpG binding domain of MeCP2. J. Biol. Chem. 276, 3353–3360. [DOI] [PubMed] [Google Scholar]
- Ghosh R. P.; Horowitz-Scherer R. A.; Nikitina T.; Shlyakhtenko L. S.; Woodcock C. L. (2010) MeCP2 Binds Cooperatively to Its Substrate and Competes with Histone H1 for Chromatin Binding Sites. Mol. Cell. Biol. 30, 4656–4670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuss D. P.; Cheema M.; Ng M. K.; Martinez de Paz A.; Williamson B.; Missiaen K.; Cosman J. D.; McPhee D.; Esteller M.; Hendzel M.; Delaney K.; Ausio J. (2013) Impaired in vivo binding of MeCP2 to chromatin in the absence of its DNA methyl-binding domain. Nucleic Acids Res. 41, 4888–4900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brink M. C.; Piebes D. G.; de Groote M. L.; Luijsterburg M. S.; Casas-Delucchi C. S.; van Driel R.; Rots M. G.; Cardoso M. C.; Verschure P. J. (2013) A role for MeCP2 in switching gene activity via chromatin unfolding and HP1γ displacement. PLoS One 8, e69347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker S. A.; Chen L.; Wilkins A. D.; Yu P.; Lichtarge O.; Zoghbi H. Y. (2013) An AT-hook domain in MeCP2 determines the clinical course of Rett syndrome and related disorders. Cell 152, 984–996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W.; Pozzo-Miller L. (2014) BDNF deregulation in Rett syndrome. Neuropharmacology 76(Part C), 737–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen J. C.; Ghosh R. P.; Woodcock C. L. (2010) Binding of the Rett Syndrome Protein, MeCP2, to Methylated and Unmethylated DNA and Chromatin. IUBMB Life 62, 732–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalodimos C. G.; Biris N.; Bonvin A. M.; Levandoski M. M.; Guennuegues M.; Boelens R.; Kaptein R. (2004) Structure and flexibility adaptation in nonspecific and specific protein-DNA complexes. Science 305, 386–389. [DOI] [PubMed] [Google Scholar]
- Ballestar E.; Yusufzai T. M.; Wolffe A. P. (2000) Effects of Rett syndrome mutations of the methyl-CpG binding domain of the transcriptional repressor MeCP2 on selectivity for association with methylated DNA. Biochemistry 39, 7100–7106. [DOI] [PubMed] [Google Scholar]
- Fraga M. F.; Ballestar E.; Montoya G.; Taysavang P.; Wade P. A.; Esteller M. (2003) The affinity of different MBD proteins for a specific methylated locus depends on their intrinsic binding properties. Nucleic Acids Res. 31, 1765–1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inomata K.; Ohki I.; Tochio H.; Fujiwara K.; Hiroaki H.; Shirakawa M. (2008) Kinetic and thermodynamic evidence for flipping of a methyl-CpG binding domain on methylated DNA. Biochemistry 47, 3266–3271. [DOI] [PubMed] [Google Scholar]
- Lao V. V.; Darwanto A.; Sowers L. C. (2010) Impact of base analogues within a CpG dinucleotide on the binding of DNA by the methyl-binding domain of MeCP2 and methylation by DNMT1. Biochemistry 49, 10228–10236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarsdale J. N.; Webb H. D.; Ginder G. D.; Williams D. C. Jr. (2011) Solution structure and dynamic analysis of chicken MBD2 methyl binding domain bound to a target-methylated DNA sequence. Nucleic Acids Res. 39, 6741–6752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Hippel P. H. (2007) From “simple” DNA-protein interactions to the macromolecular machines of gene expression. Annu. Rev. Biophys. Biomol. Struct. 36, 79–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer J. M.; Van Bokhoven H. (2009) Genetic and epigenetic defects in mental retardation. Int. J. Biochem. Cell Biol. 41, 96–107. [DOI] [PubMed] [Google Scholar]
- Adkins N. L.; Georgel P. T. (2011) MeCP2: Structure and function. Biochem. Cell Biol. 89, 1–11. [DOI] [PubMed] [Google Scholar]
- Ghosh R. P.; Nikitina T.; Horowitz-Scherer R. A.; Gierasch L. M.; Uversky V. N.; Hite K.; Hansen J. C.; Woodcock C. L. (2010) Unique physical properties and interactions of the domains of methylated DNA binding protein 2 (MeCP2). Biochemistry 49, 4395–4410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klose R. J.; Sarraf S. A.; Schmiedeberg L. S.; McDermott S. M.; Stancheva I.; Bird A. P. (2005) DNA Binding Selectivity of MeCP2 Due to a Requirement for A/T Sequences Adjacent to Methyl-CpG. Mol. Cell 19, 667–678. [DOI] [PubMed] [Google Scholar]
- Gupta S.; Cheng H.; Mollah A. K.; Jamison E.; Morris S.; Chance M. R.; Khrapunov S.; Brenowitz M. (2007) DNA and protein footprinting analysis of the modulation of DNA binding by the N-terminal domain of the Saccharomyces cerevisiae TATA binding protein. Biochemistry 46, 9886–9898. [DOI] [PubMed] [Google Scholar]
- Khrapunov S.; Brenowitz M. (2004) Comparison of the effect of water release on the interaction of the Saccharomyces cerevisiae TATA binding protein (TBP) with “TATA Box” sequences composed of adenosine or inosine. Biophys. J. 86, 371–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baud S.; Margeat E.; Lumbroso S.; Paris F.; Sultan C.; Royer C.; Poujol N. (2002) Equilibrium binding assays reveal the elevated stoichiometry and salt dependence of the interaction between full-length human sex-determining region on the Y chromosome (SRY) and DNA. J. Biol. Chem. 277, 18404–18410. [DOI] [PubMed] [Google Scholar]
- Dragan A. I.; Frank L.; Liu Y.; Makeyeva E. N.; Crane-Robinson C.; Privalov P. L. (2004) Thermodynamic signature of GCN4-bZIP binding to DNA indicates the role of water in discriminating between the AP-1 and ATF/CREB sites. J. Mol. Biol. 343, 865–878. [DOI] [PubMed] [Google Scholar]
- Record M. T. Jr.; Ha J. H.; Fisher M. A. (1991) Analysis of equilibrium and kinetic measurements to determine thermodynamic origins of stability and specificity and mechanism of formation of site-specific complexes between proteins and helical DNA. Methods Enzymol. 208, 291–343. [DOI] [PubMed] [Google Scholar]
- Blakaj D. M.; Kattamuri C.; Khrapunov S.; Hegde R. S.; Brenowitz M. (2006) Indirect Readout of DNA Sequence by Papillomavirus E2 Proteins Depends Upon Net Cation Uptake. J. Mol. Biol. 358, 224–240. [DOI] [PubMed] [Google Scholar]
- deHaseth P. L.; Lohman T. M.; Burgess R. R.; Record M. T. Jr. (1978) Nonspecific interactions of Escherichia coli RNA polymerase with native and denatured DNA: Differences in the binding behavior of core and holoenzyme. Biochemistry 17, 1612–1622. [DOI] [PubMed] [Google Scholar]
- Hashimoto H.; Liu Y.; Upadhyay A. K.; Chang Y.; Howerton S. B.; Vertino P. M.; Zhang X.; Cheng X. (2012) Recognition and potential mechanisms for replication and erasure of cytosine hydroxymethylation. Nucleic Acids Res. 40, 4841–4849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams V. H.; McBryant S. J.; Wade P. A.; Woodcock C. L.; Hansen J. C. (2007) Intrinsic disorder and autonomous domain function in the multifunctional nuclear protein, MeCP2. J. Biol. Chem. 282, 15057–15064. [DOI] [PubMed] [Google Scholar]
- Smith A. J.; Humphries S. E. (2009) Characterization of DNA-binding proteins using multiplexed competitor EMSA. J. Mol. Biol. 385, 714–717. [DOI] [PubMed] [Google Scholar]
- Branco M. R.; Ficz G.; Reik W. (2012) Uncovering the role of 5-hydroxymethylcytosine in the epigenome. Nat. Rev. Genet. 13, 7–13. [DOI] [PubMed] [Google Scholar]
- Mellen M.; Ayata P.; Dewell S.; Kriaucionis S.; Heintz N. (2012) MeCP2 binds to 5hmC enriched within active genes and accessible chromatin in the nervous system. Cell 151, 1417–1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frauer C.; Hoffmann T.; Bultmann S.; Casa V.; Cardoso M. C.; Antes I.; Leonhardt H. (2011) Recognition of 5-Hydroxymethylcytosine by the Uhrf1SRA Domain. PLoS One 6, e21306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valinluck V.; Tsai H. H.; Rogstad D. K.; Burdzy A.; Bird A.; Sowers L. C. (2004) Oxidative damage to methyl-CpG sequences inhibits the binding of the methyl-CpG binding domain (MBD) of methyl-CpG binding protein 2 (MeCP2). Nucleic Acids Res. 32, 4100–4108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- deHaseth P. L.; Lohman T. M.; Record M. T. Jr. (1977) Nonspecific interaction of lac repressor with DNA: An association reaction driven by counterion release. Biochemistry 16, 4783–4790. [DOI] [PubMed] [Google Scholar]
- Record M. T. Jr.; deHaseth P. L.; Lohman T. M. (1977) Interpretation of monovalent and divalent cation effects on the lac repressor-operator interaction. Biochemistry 16, 4791–4796. [DOI] [PubMed] [Google Scholar]
- Baubec T.; Ivanek R.; Lienert F.; Schubeler D. (2013) Methylation-dependent and -independent genomic targeting principles of the MBD protein family. Cell 153, 480–492. [DOI] [PubMed] [Google Scholar]
- Frank D. E.; Saecker R. M.; Bond J. P.; Capp M. W.; Tsodikov O. V.; Melcher S. E.; Levandoski M. M.; Record M. T. Jr. (1997) Thermodynamics of the interactions of lac repressor with variants of the symmetric lac operator: Effects of converting a consensus site to a non-specific site. J. Mol. Biol. 267, 1186–1206. [DOI] [PubMed] [Google Scholar]
- Sapienza P. J.; Dela Torre C. A.; McCoy W. H. t.; Jana S. V.; Jen-Jacobson L. (2005) Thermodynamic and kinetic basis for the relaxed DNA sequence specificity of “promiscuous” mutant EcoRI endonucleases. J. Mol. Biol. 348, 307–324. [DOI] [PubMed] [Google Scholar]
- Librizzi M. D.; Brenowitz M.; Willis I. M. (1998) The TATA element and its context affect the cooperative interaction of TATA-binding protein with the TFIIB-related factor, TFIIIB70. J. Biol. Chem. 273, 4563–4568. [DOI] [PubMed] [Google Scholar]
- Privalov P. L.; Dragan A. I.; Crane-Robinson C. (2011) Interpreting protein/DNA interactions: Distinguishing specific from non-specific and electrostatic from non-electrostatic components. Nucleic Acids Res. 39, 2483–2491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien R.; DeDecker B.; Fleming K. G.; Sigler P. B.; Ladbury J. E. (1998) The effects of salt on the TATA binding protein-DNA interaction from a hyperthermophilic archaeon. J. Mol. Biol. 279, 117–125. [DOI] [PubMed] [Google Scholar]
- Shereda R. D.; Kozlov A. G.; Lohman T. M.; Cox M. M.; Keck J. L. (2008) SSB as an organizer/mobilizer of genome maintenance complexes. Crit. Rev. Biochem. Mol. Biol. 43, 289–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hud N. V.; Polak M. (2001) DNA-cation interactions: The major and minor grooves are flexible ionophores. Curr. Opin. Struct. Biol. 11, 293–301. [DOI] [PubMed] [Google Scholar]
- Varnai P.; Zakrzewska K. (2004) DNA and its counterions: A molecular dynamics study. Nucleic Acids Res. 32, 4269–4280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wintjens R.; Lievin J.; Rooman M.; Buisine E. (2000) Contribution of cation-π interactions to the stability of protein-DNA complexes. J. Mol. Biol. 302, 395–410. [DOI] [PubMed] [Google Scholar]
- Zou X.; Ma W.; Solov’yov I. A.; Chipot C.; Schulten K. (2012) Recognition of methylated DNA through methyl-CpG binding domain proteins. Nucleic Acids Res. 40, 2747–2758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakefield R. I.; Smith B. O.; Nan X.; Free A.; Soteriou A.; Uhrin D.; Bird A. P.; Barlow P. N. (1999) The solution structure of the domain from MeCP2 that binds to methylated DNA. J. Mol. Biol. 291, 1055–1065. [DOI] [PubMed] [Google Scholar]
- Strahs D.; Brenowitz M. (1994) DNA conformational changes associated with the cooperative binding of cI-repressor of bacteriophage lambda to OR. J. Mol. Biol. 244, 494–510. [DOI] [PubMed] [Google Scholar]
- Brenowitz M.; Jamison E.; Majumdar A.; Adhya S. (1990) Interaction of the Escherichia coli Gal repressor protein with its DNA operators in vitro. Biochemistry 29, 3374–3383. [DOI] [PubMed] [Google Scholar]
- Brenowitz M.; Mandal N.; Pickar A.; Jamison E.; Adhya S. (1991) DNA-binding properties of a lac repressor mutant incapable of forming tetramers. J. Biol. Chem. 266, 1281–1288. [PubMed] [Google Scholar]
- Blakaj D. M.; Fernandez-Fuentes N.; Chen Z.; Hegde R.; Fiser A.; Burk R. D.; Brenowitz M. (2009) Evolutionary and biophysical relationships among the papillomavirus E2 proteins. Front. Biosci. 14, 900–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bain D. L.; Yang Q.; Connaghan K. D.; Robblee J. P.; Miura M. T.; Degala G. D.; Lambert J. R.; Maluf N. K. (2012) Glucocorticoid receptor-DNA interactions: Binding energetics are the primary determinant of sequence-specific transcriptional activity. J. Mol. Biol. 422, 18–32. [DOI] [PubMed] [Google Scholar]
- Rice M. R.; Blumenthal R. M. (2000) Recognition of native DNA methylation by the PvuII restriction endonuclease. Nucleic Acids Res. 28, 3143–3150. [DOI] [PMC free article] [PubMed] [Google Scholar]