Abstract
The detailed investigation of variation in functionally important regions of the human genome is expected to promote understanding of genetically complex diseases. We resequenced 65 candidate genes for CNS disorders in an average of 85 European individuals. The minor allele frequency (MAF), an indicator of weak purifying selection, was lowest in radical amino acid alterations, whereas similar MAF was observed for synonymous variants and conservative amino acid alterations. In noncoding sequences, variants located in CpG islands tended to have a lower MAF than those outside CpG islands. The transition/transversion ratio was increased among both synonymous and conservative variants compared with noncoding variants. Conversely, the transition/transversion ratio was lowest among radical amino acid alterations. Furthermore, among nonsynonymous variants, transversions displayed lower MAF than did transitions. This suggests that transversions are associated with functionally important amino acid alterations. By comparing our data with public SNP databases, we found that variants with lower allele frequency are underrepresented in these databases. Therefore, radical variants obtain distinctively lower database coverage. However, those variants appear to be under weak purifying selection and thus could play a role in the etiology of genetically complex diseases.
Understanding the patterns and determinants of sequence variation that predispose to inherited disease has been an important goal of medical genetics for many years (Vogel 1972; Vogel and Kopun 1977). Recent technological progress and the availability of a human genome reference sequence have facilitated studies to systematically discover and analyze the genetic variation present in human populations. Most of the variation can be attributed to the presence of single nucleotide polymorphisms (SNPs), and >4 million SNPs have already been identified in the human genome (Sherry et al. 2001). It is generally agreed that SNPs form the basis for identifying susceptibility genes in genetically complex diseases. To enable an efficient and systematic search for disease-associated variants, a genome-wide haplotype map is considered to be a valuable tool (Patil et al. 2001; Gabriel et al. 2002). However, a recent study (Carlson et al. 2003) predicts that still -1 million SNPs will be needed for a positional genome-wide disease association scan. Alternatively, preferential assaying of functionally relevant variants is suggested to reduce genotyping load (Botstein and Risch 2003). Because particularly coding and regulatory SNPs are expected to cause a phenotypic effect, it will be important to understand variability in gene sequences. For this reason, several large-scale studies focused on gene-based analysis of sequence variability (Cambien et al. 1999; Cargill et al. 1999; Halushka et al. 1999; Stephens et al. 2001; Haga et al. 2002; Tiret et al. 2002; Carlson et al. 2003; Zhu et al. 2003).
To identify and analyze complete single nucleotide sequence variation of 65 genes in the European population, we resequenced a sample of healthy unrelated European individuals (on average 85). The selection of genes focused on candidates that have been hypothesized to play a role in neuropsychiatric diseases. In particular, genes coding for ion channels, neurotransmitter receptors, and proteins located downstream in the intracellular signaling pathways were investigated (Supplemental Table 1, available online at www.genome.org).
Minor allele frequency (MAF) and transition/transversion ratio were analyzed for different categories of SNPs to improve understanding of the characteristics and origin of genetic variation. In addition, we compared our SNP data with public databases to quantify database quality and completeness for coding regions.
RESULTS
Variation of Coding and Noncoding Regions
The coding regions of 65 genes, including flanking regions, were investigated. In total, we resequenced >150 kb of the human genome in a representative population-based sample from Europe (average number of individuals sequenced per gene was 85 [57 to 96]; for details, see Methods). Three hundred eighty-eight of 396 detected SNPs were located in sequences, which could be unambiguously aligned to the human genome assembly (build 30).
According to both Ensembl (Hubbard et al. 2002) and NCBI Mapview gene models (Wheeler et al. 2002), detected SNPs were categorized with regard to their genomic position. Among the 388 SNPs, 173 were located in regions being annotated as coding. When analyzing all coding SNPs, we found 79 to be nonsynonymous under the NCBI Mapview or the Ensembl gene models in at least one transcript, including a total of three premature stop codons. However, some differences existed between the NCBI Mapview (human genome build 30) and the Ensembl (version 9.30) gene models. In 22 cases, we found a SNP resulting in a nonsynonymous amino acid substitution that did occur exclusively under either Ensembl or NCBI Mapview gene models. In nine out of these 22 cases, differences resulted from additional alternative mRNA splicing products specified by only one of the two databases, and in the other 13 cases resulted from gene annotations specified by only one of the databases.
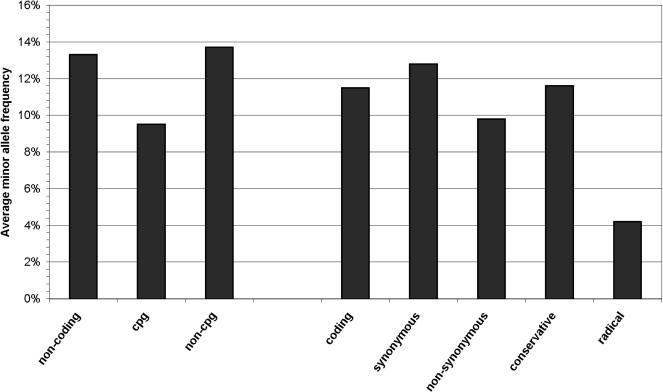
To compare our results with data from previous studies, we quantified variation of coding and noncoding regions by estimating both the nucleotide heterozygosity π and the level of nucleotide polymorphism θ (Supplemental Table 2). The nucleotide heterozygosity π is the average proportion of nucleotide differences between all pairs of sequences in a population. The level of nucleotide polymorphism θ is the proportion of nucleotide sites that are expected to be polymorphic in a population sample, corrected for the sample size (Hartl and Clark 1997). For coding regions, we estimated a nucleotide heterozygosity π of 4.2 × 10-4 and a level of nucleotide polymorphism θ of 4.5 × 10-4; for noncoding regions, a π of 5.0 × 10-4 and a θ of 4.8 × 10-4. These estimates are consistent with those reported earlier (Cargill et al. 1999; Halushka et al. 1999; Stephens et al. 2001). In a next step, we compared MAF between different SNP categories (Fig. 1) as an indicator of weak purifying selection on a certain category of variant. Consistent with biological intuition and earlier results (Tiret et al. 2002), nonsynonymous SNPs showed a significantly lower MAF (9.8%) compared with that of synonymous SNPs (12.8%; P = 0.021, one-sided Mann-Whitney U test). MAF of noncoding SNPs located within CpG islands (n = 31) was lower (9.5%) than was MAF of SNPs located in the remaining noncoding regions (n = 184; 13.8%), possibly reflecting that mutations in CpG islands are subject to a selective pressure. The statistical comparison, however, showed only a nonsignificant trend (P = 0.08, one-sided Mann-Whitney U test).
Figure 1.
Average minor allele frequency of SNPs according to their respective functional category. Noncoding variants are categorized according to their location in CpG islands; coding variants are categorized according to their impact on the encoded amino acid.
To discriminate between conservative and radical amino acid alteration, a measure of pairwise physicochemical amino acid dissimilarity is needed, as given by Grantham's distance (Grantham 1974). Based on Grantham's distance and data about occurrences of polymorphisms in pseudogenes, a higher risk of functional impact had been estimated for radical compared with conservative variants (Stephens et al. 2001). We compared MAF between synonymous variants (n = 94), conservative amino acid alterations (Grantham's distance ≤ 100; n = 58), and radical alterations (n = 18; Grantham's distance > 100). Interestingly, MAF of conservative alterations (11.6%) was not found to be significantly different from that of synonymous variants (12.4%). In contrast, a significant lower MAF was observed for radical alterations (4%) than for conservative alterations (P = 0.037, one-sided Mann-Whitney U test). Furthermore, all observed premature stop codons (n = 3) were singletons (observed in only one individual).
Transition/Transversion Ratio
About 71% of all polymorphisms were transitions, which is known to be typical for human genes (Stephens et al. 2001). When analyzing the ratio of transitions to transversions (Table 1), the ratio was found to be greater in coding (3.02) than in noncoding variants (1.99; χ2 = 3.43, P = 0.074, 1 df). Among coding SNPs, the transition/transversion ratio was found to be greater in synonymous variants (4.1) than in nonsynonymous variants (2.17; χ2 = 3.28, P = 0.078, 1df). In accordance with previous studies (Cargill et al. 1999; Halushka et al. 1999), we observed a markedly increased diversity at degenerate sites (π = 10.3 × 10-4, θ = 9.9 × 10-4) when the sequence length was corrected for the fraction of those sites. This observation can be explained by the selective analysis of transitions at degenerate sites, where transitions occur with a higher a priori likelihood.
Table 1.
Occurrence of SNPs According to Functional Category and Type of Nucleotide Replacement
| Transitions | Transversions | Ratio | |
|---|---|---|---|
| Total | 273 | 115 | 2.37 |
| Noncoding | 143 | 72 | 1.99 |
| Noncoding non-CpG | 123 | 61 | 2.02 |
| Noncoding CpG | 20 | 11 | 1.82 |
| Coding | 130 | 43 | 3.02 |
| Synonymous | 78 | 19 | 4.1 |
| Nonsynonymous | 52 | 24 | 2.17 |
| Conservative | 43 | 15 | 2.9 |
| Radical | 9 | 9 | 1 |
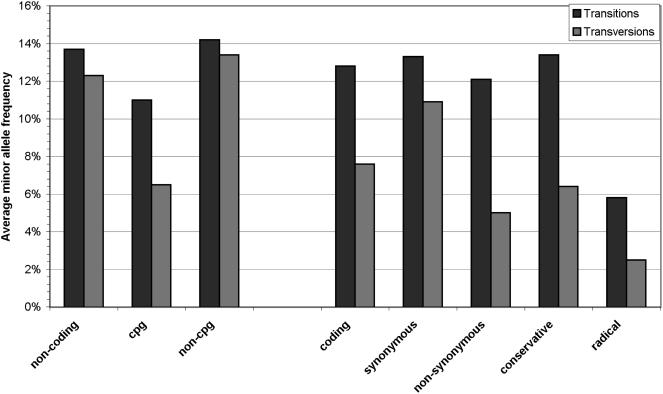
The comparison of transition/transversion ratios observed in conservative substitutions (2.9) and radical substitutions (1.0; χ2 = 3.7, P = 0.081, 1 df) suggests that radical alterations are more likely to result from transversions, whereas conservative alterations are preferentially induced by transitions. We tested this hypothesis by comparing MAF between transitions and transversions in different categories of polymorphisms (Fig. 2). Most notably, among nonsynonymous SNPs, transversions showed a significantly lower MAF (5.0%) than did transitions (12.1%; P = 0.023, one-sided Mann-Whitney U test). These results show that transversions are more likely to induce radical alterations, whereas transitions preferentially account for synonymous and conservative substitutions.
Figure 2.
Minor allele frequency of SNPs according to their functional category and type of nucleotide replacement. Noncoding variants are categorized according to their location in CpG islands; coding variants are categorized according to their impact on the encoded amino acid.
Database Representation
In a final step, we compared the 388 SNPs identified in the present study to polymorphisms annotated to the human genome reference sequence (Supplemental Table 3). Of all 388 SNPs, 41% were known as polymorphic sites to dbSNP (build 112; Sherry et al. 2001), 32% were known to HGVBAS (Fredman et al. 2002), and 12% were known to the TSC database (Sachidanandam et al. 2001). Vice versa, in our sample of Europeans, representing only part of the world population, the validation rate of SNPs was 38% for dbSNP, 38% for HGVBAS, 49% for the TSC data set, and 60% for the subset of dbSNP submitted more than once. A recent analysis of variation in coding regions (Carlson et al. 2003) found higher confirmation rates for both dbSNP (59%) and TSC SNPs (65%). This difference in confirmation rate of TSC SNPs is likely to result from their different population sample, which includes both European and African Americans. The more substantial difference for dbSNPs might result both from the higher variety of methods underlying data in dbSNP or the more recent dbSNP version used in our study (build 112 versus build 104). Another recent study (Reich et al. 2003) reported a validation rate of 89% for TSC SNPs. This high validation rate observed by Reich et al. (2003) could be further explained by the fact that mainly noncoding genomic regions were investigated, whereas our study and the study of Carlson et al. (2003) focused on coding regions.
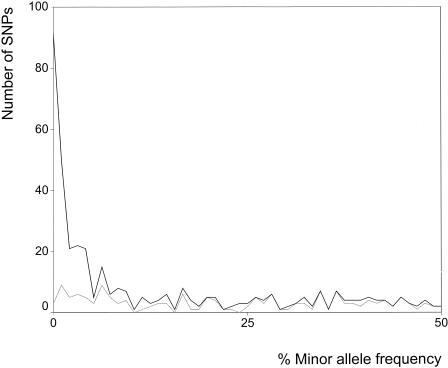
As expected, diversity in genomic regions investigated by Reich et al. (2003) was substantially higher (7.1 × 10-4) than those reported for coding regions here and elsewhere (Cargill et al. 1999; Halushka et al. 1999). Consistently, our observed MAF distribution (Fig. 3) for coding regions appears to be skewed toward low-frequency SNPs compared with the genome-wide distribution (Reich et al. 2003). On the other hand, because of the lower MAF of SNPs being functionally more relevant, the rate of SNPs missing in databases is increased for coding regions: We found 41.8% of noncoding, 44.0% noncoding non-CpG, 29.0% of noncoding CpG, 38.7% of coding, 42.3% of synonymous, 43.1% of conservative, and only 5.6% of radical variants represented in dbSNP.
Figure 3.
Distributions of observed SNPs (black line) and SNPs described in dbSNP (build 112; gray line) over their minor allele frequency.
DISCUSSION
We performed an extensive experimental and computational analysis of single nucleotide sequence variation in a set of candidate genes for neuropsychiatric diseases. Overall diversity for coding and noncoding regions was found in agreement with earlier studies of candidate genes for blood pressure homeostasis (Halushka et al. 1999) and a diverse set of genes (Cargill et al. 1999).
Under the assumption that a lower MAF indicates the presence of weak purifying selection, we found radical variants to evolve clearly differently from conservative variants. On the other hand, we found conservative variants close to synonymous variants. Consequently, inferences about natural selection based solely on nonsynonymous to synonymous substitution ratios should be treated with caution. In contrast to our results, a previous study (Tiret et al. 2002) had reported a clear difference in MAF between synonymous and conservative SNPs but, on the other hand, did not find a different MAF of conservative and nonconservative SNPs. In seeming contradiction, another study (Cargill et al. 1999) reported a difference of nucleotide diversity between conservative and nonconservative alterations. Both these earlier studies (Cargill et al. 1999; Tiret et al. 2002) had used BLOSUM scores (Henikoff and Henikoff 1992) to discriminate between conservative and nonconservative variants. When we applied BLOSUM scores to our data, we found similar MAF of conservative and nonconservative SNPs consistent to that of Tiret et al. (2002) whereas comparison of nucleotide diversity was consistent to that of Cargill et al. (1999). This can be explained by the fact that BLOSUM scores are empirically derived mutability scores reflecting a combination of amino acid similarity, design of the genetic code, and codon usage bias. Therefore, the equal MAF of conservative and nonconservative alterations observed earlier (Tiret et al. 2002) is likely to result from chemically conservative alterations, classified as nonconservative by BLOSUM scores (BLOSUM < 0). In contrast, the different nucleotide diversity of conservative and nonconservative alterations observed earlier (Cargill et al. 1999) results from more diversity produced by common alterations (BLOSUM ≥ 0) and less diversity produced by seldom alterations (BLOSUM < 0). Unlike BLOSUM scores, Grantham's distance (Grantham 1974) is a measure of pairwise amino acid dissimilarity solely based on physicochemical properties, which we believe is more appropriate to discriminate between conservative and radical alterations for the purpose of disease association studies.
For outside coding regions, we observed a trend toward a lower MAF for SNPs located within CpG islands compared with those located outside CpG islands. This result is not unexpected, because CpG islands are associated with regulatory functions, and consequently, mutations in CpG islands may be subject to weak purifying selection. However, this result was statistically not significant, presumably due to poor statistical power resulting from the relatively low number of such variants in our data set. Future and more detailed studies of larger data sets will shed more light on this issue.
The investigation of transition/transversion ratios has a long history of scientific interest in interspecies comparisons (Collins and Jukes 1994; Graur and Li 2000). Our data support the previously formulated hypothesis, that among nonsynonymous polymorphisms radical amino acid alterations preferentially result from transversions (Zhang 2000). This is sensible under the assumption that conservative alterations are functionally more similar to synonymous substitutions than to radical alterations, as concluded above.
About 80% of the highly frequent variants (MAF > 20%) that we identified in the course of our study were already deposited in public databases. On the other hand, infrequent variants were underrepresented in public SNP databases: Only 20% of rare variants (MAF < 5% except singletons) and three out of 90 singleton SNPs were described before. Because variants of functional impact are likely to belong to these low-frequency variants, they might nevertheless be relevant for the development of common complex diseases, as has been predicted from theoretical considerations (Pritchard and Cox 2002). This emphasizes the importance of resequencing efforts in the search for disease-associated genes.
In conclusion, we showed that in coding regions conservative alterations are more similar to synonymous variants than to radical variants. In noncoding sequences, variants located in CpG islands appear to be of higher functional relevance than are variants outside CpG islands. Our study further shows that nonsynonymous transversions are more likely to cause functional consequences than are transitions, as is reflected by weak purifying selection against these variants. As a result of purifying selection, variants of phenotypic relevance display lower MAF, which leads to their underrepresentation in databases. However, the existence of many rare variants under weak selective pressure indicates that those variants indeed could play a role in the etiology of genetically complex diseases.
METHODS
Establishment of the DNA Bank
A DNA bank representative of the current European population was established. After informed consent had been obtained, 150 blood samples were collected from healthy unrelated individuals, originating from 30 European countries (Supplemental Table 4). Gender and country of origin of participants were recorded. Only those individuals were included whose grandparents were born in the respective participant's country of origin. Blood samples were used for immediate isolation of DNA as well as for transformation of B lymphocytes to provide a permanent source of DNA for future studies. For each country represented in the DNA bank, at least 30% more samples than needed were gathered. For the final establishment of the DNA bank, 96 samples were randomly selected, with the number of individuals included from each country corresponding to its population size.
The probability P to detect a certain variant, v, having a minimal population frequency, f, follows as P(v) = 1 -(1 - f)2N, where N is the number of individuals. Thus, a variant having a population frequency of 1% in the European population is detected with a probability of -85% in a sample of 96 individuals.
Selection of Candidate Genes
Candidate genes were chosen according to both positional and functional criteria. Genes coding for ion channels, receptors of neurotransmitters, proteins playing a possible role in neurodevelopment, as well as proteins located downstream in the intracellular signaling pathways are considered as functional candidates. Positional candidates are those genes that are located in genomic regions considered relevant for neuropsychiatric diseases by means of linkage analysis studies.
Genomic sequences were obtained from the clone sequence entries of the NCBI GenBank database (Wheeler et al. 2002). Expression information was obtained from UniGene Clusters. For genes without contig annotations, gene models were identified by aligning the respective mRNA or cDNA sequences to genomic clones using BLAST (Altshul et al. 1997). Only candidate genes were selected for which genomic sequence data upstream and downstream of all exons were available.
PCR Amplification of Coding Regions and DNA Sequencing
According to the respective annotation of coding sequence parts, primers were designed manually or by using the Primer3 Program (Rozen and Skaletsky 2000). PCR fragments contained at least 50 bases of the 5′ and 3′ flanking regions for each exon. To obtain specific products, PCR conditions were optimized by using gradient PCR Programs, DMSO, or special Taq-polymerase. The optimized PCRs were then performed to amplify DNA fragments on 96 samples in microtiterplates. Double-strand sequencing was performed by multiple commercial institutions (AGOWA [Berlin], BGI LifeScience [Beijing], Medigenomix [Munich], and Seqlab [Göttingen]) using ABI3100, ABI377, and ABI3730 sequencers. Internal sequencing primers were used to increase reaction specificity.
Data Analysis and Software
Primary evaluation of sequencing traces was performed by using the software package consisting of the programs Phred (Ewing et al. 1998), Phrap, Polyphred (Nickerson et al. 1997), and Consed (Gordon et al. 1998) from Washington University. All SNPs detected by Polyphred were checked by a human expert. Local storage of results was implemented by using the relational database management system MySQL. SNP information and genotype of samples generated by Polyphred was parsed into the database table format automatically. Inconsistencies between forward and reverse reads in genotype outputs, indicating Polyphred errors, were edited before genotypes were loaded into the database. Consensus sequences of resequenced fragments were aligned to the human genome by using Megablast (Zhang et al. 2000). Analysis of experimentally obtained variation data with respect to human genome annotations were performed by using a custom-made software package.
Acknowledgments
We thank the anonymous DNA sample donors and, in alphabetical order, Dr. D. Di Bella, Dr. F. Bellivier, and Dr. V.M. Steen, who helped us with collecting the sample. We further would like to thank the anonymous reviewers for their helpful comments on the manuscript. This work was supported by grants from the German Human Genome Project (DHGP) and the German National Genome Research Network (NGFN).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
[Supplemental material is available online at www.genome.org. The sequence data from this study have been submitted to dbSNP under accession nos. ss12586678-ss12587076.]
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.1299703.
References
- Altshul, S.F., Madden, T.L., Schaffer, A.A., Zhang, J., Miller, W., and Lipman, D. 1997. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25: 3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botstein, D. and Risch, N. 2003. Discovering genotypes underlying human phenotypes: Past successes for mendelian disease, future approaches for complex disease. Nat. Genet. 33: 228-237. [DOI] [PubMed] [Google Scholar]
- Cambien, F., Poirier, O., Nicaud, V., Herrmann, S.M., Mallet, C., Ricard, S., Behague, I., Hallet, V., Blanc, H., Loukaci, V., et al. 1999. Sequence diversity in 36 candidate genes for cardiovascular disorders. Am. J. Hum. Gene 65: 183-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cargill, M., Altshuler, D., Ireland, J., Sklar, P., Ardlie, K., Patil, N., Shaw, N., Lane, C.R., Lim, E.P., Kalyanaraman, N., et al. 1999. Characterization of single-nucleotide polymorphisms in coding regions of human genes. Nat. Genet. 22: 231-238. [DOI] [PubMed] [Google Scholar]
- Carlson, C.S., Eberle, M.L., Rieder, M.J., Smith, J.D., Kruglyak, L., and Nickerson, D.A. 2003. Additional SNPs and linkage-disequilibrium analyses are necessary for whole-genome association studies in humans. Nat. Genet. 33: 518-521. [DOI] [PubMed] [Google Scholar]
- Collins, D.W. and Jukes, T.H. 1994. Rates of transitions and transversions in coding sequences since the human-rodent divergence. Genomics 20: 386-396. [DOI] [PubMed] [Google Scholar]
- Ewing, B., Hillier, L., Wendl, M., and Green, P. 1998. Basecalling of automated sequences traces using phred: Accuracy assessment. Genome Res. 8: 175-185. [DOI] [PubMed] [Google Scholar]
- Fredman, D., Siegfried, M., Yuan, Y.P., Sarkar, C.M., Bork, P., Lehväslaiho, H., and Brookes, A.J. 2002. HGVBASE: A human sequence variation database emphasizing data quality and a broad spectrum of data sources: Nucleic Acids Res. 30: 387-391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabriel, S.B., Schaffner, S.F., Nguyen, H., Moore, J.M., Roy, J., Blumenstiel, B., Higgins, J., DeFelice, M., Lochner, A., Faggart, M., et al. 2002. The structure of haplotype blocks in the human genome. Science 296: 2225-2229. [DOI] [PubMed] [Google Scholar]
- Gordon, D.C., Abajian, C., and Green, P. 1998. Consed: A graphical tool for sequence finishing. Genome Res. 8: 195-202. [DOI] [PubMed] [Google Scholar]
- Grantham, R. 1974. Amino acid difference formula to help explain protein evolution. Science 185: 862-864. [DOI] [PubMed] [Google Scholar]
- Graur, D. and Li, W.H. 2000. Fundamentals of molecular evolution. Sinauer Associates, Inc., Sunderland, MA.
- Haga, H., Yamada, R., Ohrishi, Y., Nakamura, Y., and Tanaka, T. 2002. Gene-based SNP discovery as part of the Japanese Millennium Genome Project: Identification of 190,562 genetic variations in the human genome. Single-nucleotide polymorphism. J. Hum. Genet. 47: 605-610. [DOI] [PubMed] [Google Scholar]
- Halushka, M.K., Fan, J.B., Kimberly, B., Hsie, L., Naiping, S., Alan, W., Cooper, R., Lipshutz, R., and Chakravarti, A. 1999. Patterns of single nucleotide polymorphsims in candidate genes for blood pressure homeostasis. Nat. Genet. 22: 239-247. [DOI] [PubMed] [Google Scholar]
- Hartl, D.L. and Clark, A.G. 1997. Principles of population genetics. Sinauer Associates, Inc., Sunderland, MA.
- Henikoff, S. and Henikoff, J.G. 1992. Amino acid substitution matrices from protein blocks. Proc. Natl. Acad. Sci. 89: 10915-10919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard, T., Barker, D., Birney, E., Cameron, G., Chen, Y., Clark, L., Cox, T., Cuff, J., Curwen, V., Down, T., et al. 2002. The Ensembl genome database project. Nucleic Acids Res. 30: 38-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickerson, D.A., Tobe, V.O., and Taylor, S.L. 1997. Polyphred: automating the detection and genotyping of single nucleotide substitutions using fluorescence based resequencing. Nucleic Acids Res. 25: 2745-2751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patil, N., Berno, A.J., Hinds, D.A., Wade, A.B., Jigna, M.D., Hacker, C.R., Kautzer, C.R., Lee, D.H., Marjoribanks, C., McDonough, D.P., et al. 2001. Blocks of limited haplotype diversity revealed by high resolution scanning of human chromosome 21. Science 294: 1719-1723. [DOI] [PubMed] [Google Scholar]
- Pritchard, J.K. and Cox, N.J. 2002. The allelic architecture of human disease genes: Common disease-common variant... or not? Hum. Mol. Genet. 11: 2417-2423. [DOI] [PubMed] [Google Scholar]
- Reich, D.E., Gabriel, S.B., and Altshuler, D. 2003. Quality and completeness of SNP databases. Nat. Genet. 33: 457-458. [DOI] [PubMed] [Google Scholar]
- Rozen, S. and Skaletsky, H.J. 2000. Primer3 on WWW for general users and for biologist programmers. In Bioinformatics methods and protocols: Methods in molecular biology (eds. S. Krawetz and S. Misener), pp. 365-386. Humana Press, Totowa, NJ. [DOI] [PubMed]
- Sachidanandam, R., Weissman, D., Schmidt, S.C., Kakol, J.M., Stein, L.D., Marth, G., Sherry, S., Mullikin, J.C., Mortimore, B.J., Willey, D.L., et al. 2001. A map of human genome sequence variation containing 1.42 million single nucleotide polymorphisms. Nature 409: 928-933. [DOI] [PubMed] [Google Scholar]
- Sherry, S.T., Ward, M.H., Kholodov, M., Baker, J., Phan, L., Smigielski, E.M., and Sirotkin, K. 2001. dbSNP: The NCBI database of genetic variation. Nucleic Acids Res. 29: 308-311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens, J.C., Schneider, J.A., Tanguay, D.A., Choi, J., Acharya, T., Stanley, S.E., Jiang, R., Messer, C.J., Chew, A., Han, J.H., et al. 2001. Haplotype variation and linkage disequilibrium in 313 human genes. Science 293: 489-493. [DOI] [PubMed] [Google Scholar]
- Tiret, L., Poirier, O., Nicaud, V., Barbaux, S., Herrmann, S.M., Perret, C., Raoux, S., Francomme, C., Lebard, G., Tregouet, D., et al. 2002. Heterogeneity of linkage disequilibrium in human genes has implications for association studies of common diseases. Hum. Mol. Genet. 11: 419-429. [DOI] [PubMed] [Google Scholar]
- Vogel, F. 1972. Non-randomness of base replacement in point mutation. J. Mol. Evol. 1: 334-367. [DOI] [PubMed] [Google Scholar]
- Vogel, F. and Kopun, M. 1977. Higher frequencies of transitions among point mutations. J. Mol. Evol. 9: 159-180. [DOI] [PubMed] [Google Scholar]
- Wheeler, D.L., Church, D.M., Lash, A.E., Leipe, D.D., Madden, T.L., Pontius, J.U., Schuler, G.D., Schriml, L.M., Tatusova, T.A., Wagner, L., et al. 2002. Database resources of the National Center for Biotechnology Information: 2002 update. Nucleic Acids Res. 30: 13-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, J. 2000. Rates of conservative and radical non-synonymous nucleotide substitutions in mammalian nuclear genes. J. Mol. Evol. 50: 56-68. [DOI] [PubMed] [Google Scholar]
- Zhang, Z., Schwartz, S., Wagner, L., and Miller, W. 2000. A greedy algorithm for aligning DNA sequences. J. Comput. Biol. 7: 203-214. [DOI] [PubMed] [Google Scholar]
- Zhu, X., Yan, D., Cooper, R.S., Luke, A., Ikeda, M.A., Chang, Y.C., Weder, A., and Chakravarti, A. 2003. Linkage disequilibrium and haplotype diversity in the genes of the renin-angiotensin system: Findings from the family blood pressure program. Genome Res. 13: 173-181. [DOI] [PMC free article] [PubMed] [Google Scholar]