Abstract
The complete set of cDNAs encoding the enzymes of known metabolic pathways has not previously been available for any mammal. Here, transcripts encoding the metabolic pathways of the mouse (mouse metabolome) were reconstructed by making use of the KEGG metabolic pathway database and gene ontology (GO) assignment to the mouse representative transcript and protein set (RTPS), which contains all available mouse transcript sequences including the FANTOM set of RIKEN mouse cDNA clones. By assigning EC numbers extracted from the molecular function ontology in GO, the known mouse transcriptome was predicted to encode enzymes with 726 unique EC numbers. Of these, 648 EC numbers were newly assigned based on the FANTOM set. The mouse metabolome confirmed by cDNA analysis includes almost all of the enzymes of well known pathways such as the tricarboxylic acid cycle and urea cycle. On the other hand, analysis of enzymes required for the tryptophan metabolism pathway revealed a lack of connectivity, indicating that cDNAs/genes encoding several key enzymes remain to be identified. The information derived from coexpression from the cDNA microarray analysis of enzymes of known function may lead to identification of the missing components of the metabolome, and will add new insights into the connectivity of the mammalian metabolic pathways.
Many aspects of intermediary metabolism are shared among living organisms from Escherichia coli to human, whereas others are idiosyncratic to different kingdoms or classes. Although the enzymatic reactions of intermediary metabolism have been the basis of biochemical studies for many years, and in many cases the enzymes responsible have been purified and studied in detail, there are still many enzymes for which the reaction is known but neither the nucleotide sequence of the cDNA encoding the protein, nor the protein amino acid sequence has been determined. Many inborn errors of metabolism are known in humans, and there will undoubtedly be more identified when we know the identity, genomic location, and function of all of the mammalian metabolic enzymes. Although the term `metabolome' is used for all of the chemical compounds (and their concentrations) in a cell or a microbial organism, it is difficult to state all of the chemical compounds from the genomic view. In this study, we focused on the metabolic enzymes that are coded in the genome, and we used the term `metabolome' for our analysis. Our `metabolome' analysis presented here is an initial effort toward the complete understanding of all of the metabolic processes achieved by all of the chemical compounds including the metabolic enzymes. The major advance in mouse transcriptome sequencing resulting from the FANTOM clone set and its integration with public cDNA sequences to produce a representative transcript and protein set (RTPS; The FANTOM Consortium and the RIKEN Genome Exploration Research Group Phase II Team, 2002), which contains virtually all transcribed mRNA sequences in the world, permits a corresponding advance in our knowledge of the metabolome. Here, we report the reconstruction of all of the metabolic pathways in the mouse using the RTPS.
A similar effort to assemble and annotate genes encoding metabolic processes for microbial organisms has been ongoing in the KEGG project (Kanehisa et al. 2002). In the KEGG database, these protein sequences have been categorized, and the reconstruction of metabolic pathways for microbial genomes is provided on their Web site (http://www.genome.ad.jp/kegg/). Analysis for each new completed genome requires a complete set of proteomes generated from the extensive analysis of open reading frames (ORFs) and automatic translation of them into amino acid sequences. These analyses are commonly constrained by the accuracy of the DNA sequence and gene (ORF) predictions. Such constraints are magnified in the analysis of mammalian genomes because of the greater scope of sequence errors (by virtue of the sheer scale of the projects), the uncertainty of exon-intron predictions, and the presence of pseudogenes and duplicate genes. For example, there are 70 loci in the mouse genome that might be predicted to encode glyceraldehyde-3-phosphate dehydrogenase. For this reason, a completed transcriptome is an essential prerequisite for completion of genomic annotation of the metabolome.
The reconstruction of the mouse metabolome was attempted in the analysis of the first FANTOM set of 21,076 full-length cDNAs (The RIKEN Genome Exploration Research Group Phase II Team and the FANTOM Consortium 2001), but the coverage was insufficient. The FANTOM2 RTPS cDNA set includes 40,000 additional full-length cDNAs and integrates public transcriptome data. This metabolic enzyme set was compared with the predicted set of enzymes from the human proteome, and the orthologs were identified. In the present study we also performed a comprehensive cDNA microarray analysis of different mouse tissues. These analyses provide an insight into metabolic specialization in different tissues of a mammal, and may also permit the identification of missing components of the pathways among RIKEN transcripts for which the function is not yet known.
RESULTS AND DISCUSSION
Overall Features of the Mouse Metabolome Reconstructed From the Mouse Transcriptome
The mouse metabolome was initially reconstructed from the FANTOM2 set by assigning EC numbers (NC-IUBMB 1992) to all of the cDNA sequences potentially coding enzymes and then matching these EC numbers to metabolic pathway maps. In the FANTOM2 set, there were 648 unique EC numbers predicted for 6888 clones (11.33% of the FANTOM2 set), which consist of 3182 gene clusters (9.56% of all 33,294 clusters in the FANTOM2 set). Although the FANTOM2 set is biased toward novel transcripts by avoiding the sequencing of the known genes (judged by the 3′ end sequence of cDNA clones), it seems to cover most of the metabolic pathways. For example, full-length cDNA sequences predicted to encode all of the enzymes in the tricarboxylic acid cycle were identified for the first time among the FANTOM2 set. The cDNAs encoding the majority of enzymes in other major metabolic pathways (e.g., the glycolysis and urea cycle) were also identified, but some transcripts encoding known enzymes in other metabolic pathways were still missing from the FANTOM2 set. For example, phenylalanine 4-monooxygenase (EC 1.14.16.1) in the degradation pathway of phenylalanine was not identified in the FANTOM2 set. Some missing transcripts such as this example arose from a deliberate strategy of the RIKEN Encyclopedia project, which aimed to identify novel cDNAs as the first priority. Known genes identified among first-pass 3′ ESTs were not carried through to full-length sequencing. For example, there is a RIKEN cDNA clone (cDNA cloneid: 0610009C13, not in the FANTOM set) that was judged as phenylalanine 4-monooxygenase (EC 1.14.16.1) by 3′ end sequence. This transcript was previously known because the gene is mutated in the common human inborn error of metabolism, phenylketonuria. For this reason, we assembled the RTPS, which brings together new FANTOM2 cDNAs and known full-length transcripts. There were 726 unique EC numbers predicted in 3583 gene clusters (9.66% of all 37,086 clusters in the FANTOM2 set). The increment in predicted metabolic enzymes of the RTPS compared to the FANTOM2 clone set alone is relatively small. Despite the subtraction strategy, around 90% of enzymatic genes are included in the FANTOM2 set, showing high coverage of the FANTOM2 set in the mouse transcriptome. These results are summarized in Table 1.
Table 1.
The Number of EC and Clusters in FANTOM2 and Representative Transcript Set
| EC numbers | Clusters | cDNA clones | |
|---|---|---|---|
| FANTOM2 set (33,409 clusters in 60,770 clones) | 648 | 3,182 | 6,888 |
| Representative transcript and protein set (RTPS, 37,086 clusters) | 726 | 3,583 | not available |
The number of cDNA clones is only available for FANTOM2 set as the public sequence information may not contain the information about cDNA clones
The complete lists of functional annotation of enzymatic genes with EC numbers for both the FANTOM set and the RTPS are accessible at http://fantom2.gsc.riken.go.jp/metabolome/, and the reconstruction of all metabolic pathways for both sets can be browsed from the Web links indicated in those tables.
Comparison With the Metabolome Sets in KEGG
The FANTOM2 set is the largest mammalian transcriptome that is supported by physical cDNA clones. Metabolome data sets for human, mouse, and other model organisms have been maintained in the KEGG project as a subset of computationally reconstructed metabolome by collecting evidence from existing databases in molecular biology and adding functional assignment to predicted ORFs. In the KEGG, the mouse set contained 480 EC numbers, whereas we have independently assigned 648 and 726 unique EC numbers in the FANTOM2 clone set and the RTPS, respectively. There are two major points in this article. One is that we have newly identified 275 unique EC numbers in the mouse, and the other is that 648 mouse EC numbers were supported by the physical clones.
It seemed that the human set contained more EC numbers because the public database contains well-annotated human sequences inferred from the human genome sequence. The main difference between our set and the KEGG human set is that we used only the information about confirmed transcripts with source tissue information, whereas the creators of the KEGG set included the whole set of genes annotated in the human genome sequence with protein coding potential.
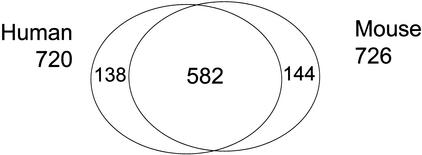
These two sets overlap considerably in EC numbers, but despite the requirement for experimental confirmation, the mouse RTPS-derived set is marginally larger than the KEGG human set (Fig. 1). In comparison with the human metabolome predicted in the KEGG (720 unique EC numbers), 582 unique EC numbers were included in both human and mouse. The difference between these two sets derived from the differences in the fourth digit in EC numbers, indicating that the ligand specificity of each enzyme is still difficult to predict by only computational assignment of gene function.
Figure 1.
Comparison of EC numbers in human and mouse. Enzymatic genes coded in human and transcribed in mouse are compared in EC number. The difference comes mainly from the variation in ligand specificity in enzymes that are difficult to predict only from computational sequence analyses.
Because these functional annotations were inferred mainly from sequence similarity analyses and those enzymes with similar function have the same structural motives, it is not straightforward to determine substrate specificity. For example, it was difficult to distinguish aminotransferases (EC2.6.1.x, where “x” is an arbitrary number) from each other when the bacterial metabolomes were reconstructed (Bono et al. 1998). Thus, we further analyzed the human and mouse metabolomes by ignoring the fourth part of the EC number. For example, EC numbers 1.1.1.1 and 1.1.1.2 are degenerated to 1.1.1.x. As a result, enzymes in EC 3.6.4.x (hydrolases, acting on acid anhydrides; involved in cellular and subcellular movement), such as myosin ATPase (EC 3.6.4.1), were distinctly found in the mouse metabolome, while no such enzymes were currently contained in the human set. On the other hand, no distinct enzymatic groups were found in the mouse metabolome that are only contained in the human metabolome data set. This observation indicates that almost all transcripts predicted in human have functional counterparts in the mouse RTPS.
Overall, the comparative analysis of human and mouse metabolomes is helpful in checking false-positive and false-negative assignment of EC numbers. Some GO annotations in enzymatic assignment were improved using the missing enzyme information in certain metabolic pathways.
Reconstruction of Metabolic Pathways: Amino Acid Metabolism Pathways
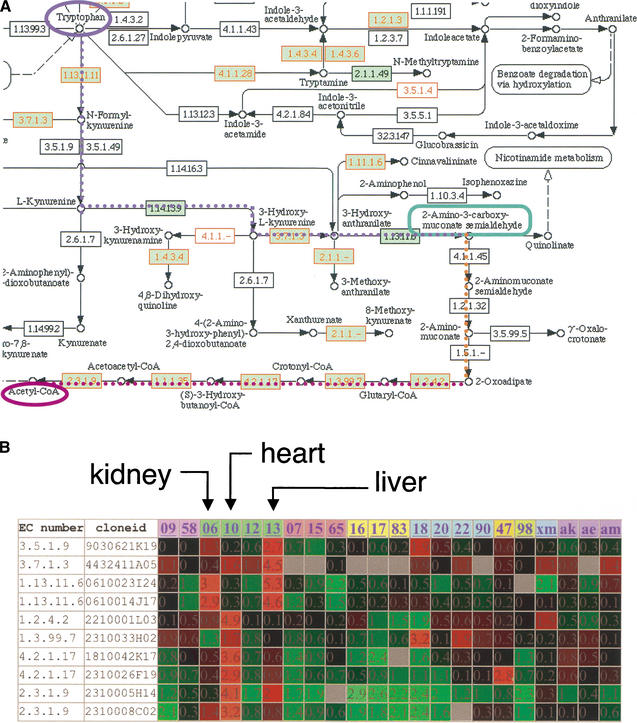
Metabolic pathways can be reconstructed by matching EC numbers in the metabolic pathway map of possible metabolic reactions. All of the reconstruction in silico can be seen at the Metabolomapper Web interface (http://fantom2.gsc.riken.go.jp/metabolome/). As a practical application of such a resource, the mammalian amino acid metabolism pathways are outlined below. In mammals, 20 amino acids are classified as essential or nonessential amino acids. The former cannot be synthesized from other amino acids and must be obtained from the diet. For nonessential amino acids, the molecules are produced by transfer of other amino acids. Based solely upon the automatic EC number assignment, several enzymes appeared to be missing in amino acid degradative pathways, but most of them could be found after careful investigation of orthologous genes in other organisms. As an interesting example, the tryptophan metabolism pathway is shown in Figure 2A. There, the dotted line (shown in blue, red, and purple) depicts the degradative pathway. Although the enzyme EC 3.5.1.9 was found in neither human nor mouse by the computational search, the corresponding protein was recently cloned (Pabarcus and Casida 2002), and the clone was identified in the FANTOM2 set by human curation. The time lag of inclusion of gene associations in GO was the reason for the failure in the computational assignment of this clone (Table 2). Two enzymes, EC 1.14.13.9 and EC 1.10.11.6, were also found in the RTPS after an intensive human curation (Table 2). Three other enzymes (ECs 4.1.1.45, 1.2.1.32, 1.5.1.-) were not found in the RTPS. The sequence corresponding to the EC 4.1.1.45 (2-amino-3-carboxymuconate-6-semialdehyde decarboxylase) was just recently cloned in rat (Tanabe et al. 2002) and then, in human and mouse (Fukuoka et al. 2002). Although we also have the corresponding clone in the RIKEN mouse encyclopedia clone set (cloneid; F530007C24, DDBJ/EMBL/GenBank accession nos.: BB745194 [3′ EST sequence] BB847230 [5′EST sequence]), this clone was not included in the FANTOM2 set and thus was not represented in the current RTPS. This clone, obtained from kidney, was cloned only after the extensive subtraction strategy used for collecting mouse cDNAs, suggesting that the expression level was low. The remaining two enzymes seem to be completely absent in the currently available transcriptome set. The low abundance suggests that this metabolic pathway toward acetyl-CoA is not commonly used in mouse, or that there is a separate enzyme yet to be identified. A line of evidence that supports the former hypothesis was obtained from the microarray data. We examined the expression profile of the genes on this pathway using the data of the RIKEN 20k mouse cDNA microarray (Nos. 1–3, a total of ∼ 60K clones are represented; Miki et al. 2001; Bono et al. 2002). The expression profile for genes coding enzymes in the tryptophan metabolism pathway revealed that the enzymes required for the upper part of this pathway (blue dotted line in Fig. 2A), leading to the production of 2-amino-3-carboxy-muconate semialdehyde, showed high expression in liver and kidney, whereas the downstream region of this pathway (red dotted line) showed highest expression in the heart (Fig. 2B). This could mean that the two organ systems cooperate metabolically, or that this pathway to acetyl-CoA is not used in the mouse. The pathway to nicotinate (map00760 in KEGG/PATHWAY) could be an alternative pathway for tryptophan metabolism in mouse.
Figure 2.
Reconstruction of mouse tryptophan metabolism pathway in silico. (A) An example of an automatically reconstructed pathway using the KEGG Web interface by specifying multiple EC numbers in the query. The EC number in the green box was found in the human metabolome set, and the EC number drawn in red was covered by the RTPS. There were missing enzymes on the pathway that were assigned computationally. These enzymes are described in Table 2. Four of six missing clones were finally identified after intensive human curation. (B) Gene expression of mRNAs coding enzymes in the tryptophan metabolism pathway by microarray. Genes are sorted by order of the cascade of tryptophan degradation to Acetyl-CoA, not by clustering by similar expression profiles. It is apparent that the genes in the upper stream of this cascade are highly expressed in liver and kidney, and those downstream of the cascade are highly expressed in heart. 09, spleen; 58, thymus; 06, kidney; 10, heart; 12, lung; 13, liver; 07, brain; 15, cerebellum; 65, cerebellum neonate 10 day; 16, placenta; 17, testis; 83, uterus; 18, pancreas; 20, small intestine; 22, stomach; 90, colon; 47, skin neonate 10 day; 98, bone; xm, muscle; ak, adipose dorsal kidney; ae, adipose epididymal; am, adipose mesenteric.
Table 2.
The Characteristics of the Missing Enzymes in Tryptophan Metabolism
| EC number | Enzyme name | Inferred cDNA clone | Why the enzyme seemed to be missing |
|---|---|---|---|
| 3.5.1.9 | arylformamidase | 9030621K19 | Recently cloned, so not in GO yet |
| 1.14.13.9 | kynurenine 3-monooxygenase | 4930567E01 | Possible error in the assignment of GO terms |
| 1.13.11.6 | 3-hydroxyanthranilate 3,4-dioxygenase | 0610012J07 | Enzyme name was registered, but EC number was not in GO |
| *4.1.1.45 | aminocarboxymuconate-semialdehyde decarboxylase | F530007C24 | Recently cloned, so not in GO yet |
| *1.2.1.32 | aminomuconate-semialdehyde dehydrogenase | Not found | |
| *1.5.1.- | 2-aminomuconate reductase | Not found |
Enzymes with asterisk show no reference sequences in the public database. See details in the text
Conclusions
EC number mapping to metabolic pathway has clearly provided the first comprehensive overview of a mammalian metabolome based upon the existence of actual transcripts. The example of the tryptophan catabolic pathway illustrates the fact that the existence of a pathway cannot be inferred solely from genome annotation. In particular, gene expression array information can be used to add significant additional insight into likely connectivity. It will also provide indications as to which members of functionally related enzyme classes are likely to catalyze specific reactions in a particular pathway. Ongoing studies in the RIKEN Encyclopedia project are generating such information for all of the FANTOM2 cDNAs, and information derived from such experiments will be incorporated into the Metabolomapper site.
METHODS
Data Set
Mouse cDNA sequences from the FANTOM2 set were used for the analysis. Some well known genes are missing in this set because they avoided selecting cDNA clones for full-length sequencing that have 3′ and/or 5′ end sequences matched to known mouse genes at that time. Known enzymatic genes were thus added to the list of enzymes when checking the connectivity of metabolic pathways. Details are described in the text above in the section entitled “Overall Features of the Mouse Metabolome Reconstructed From the Mouse Transcriptome.” The human set of predicted EC numbers was obtained on August 13, 2002 from the KEGG/GENES database ftp site (ftp://ftp.genome.ad.jp/pub/kegg/) and then processed with appropriate UNIX commands.
Reconstruction of Metabolic Pathways
We use EC numbers to match enzymes to pathways (Bono et al. 1998). These EC numbers were obtained from GO (Gene Ontology; The Gene Ontology Consortium 2001) assignment to FANTOM cDNA clones, systematically assigned in the FANTOM analysis pipeline (The FANTOM Consortium and the RIKEN Genome Exploration Research Group Phase II Team 2002). This assignment comes from GO annotations in the MGD (Blake et al. 2002), SWISSPROT (Bairoch and Apweiler 2000), InterPro (Apweiler et al. 2001), and SCOP superfamily (Murzin 1996) databases (computational assignment of GO terms, http://fantom2.gsc.riken.go.jp/fantom2/SI/sup16_go.pdf). GO assignments from SCOP superfamilies with evidence code IEA (inferred from electronic annotation) using data resources from the Superfamily: HMM library and genome assignments server (Gough et al. 2001) contained too many GO assignments, which could be the false-positive annotations; thus GO annotations from there were not used to extract EC numbers. These EC numbers from GO were primarily used as a reference to compare the human set of EC numbers described above, and to map them to specific metabolic pathways of interest. The connectivity of these pathways was then checked manually, because the references for metabolic pathway were still incomplete for mammalian cells, and nonenzymatic reactions can play an important role in the pathway.
Acknowledgments
We especially thank D. Hume of the Univ. of Queensland for valuable advice and English editing. This study was supported by a Research Grant for the RIKEN Genome Exploration Research Project, and by a Grant-in-Aid for Scientific Research on Priority Areas (C) “Genome Information Science,” both from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.974603.
Footnotes
[Supplemental material is available online at www.genome.org.]
References
- Apweiler, R., Attwood, T.K., Bairoch, A., Bateman, A., Birney, E., Biswas, M., Bucher, P., Cerutti, L., Corpet, F., Croning, M.D., et al. 2001. The InterPro database, an integrated documentation resource for protein families, domains and functional sites. Nucleic Acids Res. 29: 37-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bairoch, A. and Apweiler, R. 2000. The SWISS-PROT protein sequence database and its supplement TrEMBL in 2000. Nucleic Acids Res. 28: 45-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blake, J.A., Richardson, J.E., Bult, C.J., Kadin, J.A., and Eppig, J.T. 2002. The Mouse Genome Database (MGD): The model organism database for the laboratory mouse. Nucleic Acids Res. 30: 113-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bono, H., Ogata, H., Goto, S., and Kanehisa, M. 1998. Reconstruction of amino acid biosynthesis pathways from the complete genome sequence. Genome Res. 8: 203-210. [DOI] [PubMed] [Google Scholar]
- Bono, H., Kasukawa, T., Hayashizaki, Y., and Okazaki, Y. 2002. READ: RIKEN Expression Array Database. Nucleic Acids Res. 30: 211-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The FANTOM Consortium and the RIKEN Genome Exploration Research Group Phase I and II Team. 2002. Analysis of the mouse transcriptome based on functional annotation of 60,770 full-length cDNAs. Nature 420: 563-573. [DOI] [PubMed] [Google Scholar]
- Fukuoka, S.I., Ishiguro, K., Yanagihara, K., Tanabe, A., Egashira, Y., Sanada, H., and Shibata, K. 2002. Identification and expression of a cDNA encoding human a-amino-b-carboxymuconate-e -semialdehyde decarboxylase (ACMSD): A key enzyme for the tryptophan-niacine pathway and quinolinate hypothesis. J. Biol. Chem. 24: 24. [DOI] [PubMed] [Google Scholar]
- The Gene Ontology Consortium. 2001. Creating the gene ontology resource: Design and implementation. Genome Res. 11: 1425-1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gough, J., Karplus, K., Hughey, R., and Chothia, C. 2001. Assignment of homology to genome sequences using a library of hidden Markov models that represent all proteins of known structure. J. Mol. Biol. 313: 903-919. [DOI] [PubMed] [Google Scholar]
- Kanehisa, M., Goto, S., Kawashima, S., and Nakaya, A. 2002. The KEGG databases at GenomeNet. Nucleic Acids Res. 30: 42-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miki, R., Kadota, K., Bono, H., Mizuno, Y., Tomaru, Y., Carninci, P., Itoh, M., Shibata, K., Kawai, J., Konno, H., et al. 2001. Delineating developmental and metabolic pathways in vivo by expression profiling using the RIKEN set of 18,816 full-length enriched mouse cDNA arrays. Proc. Natl. Acad. Sci. 98: 2199-2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murzin, A.G. 1996. Structural classification of proteins: New superfamilies. Curr. Opin. Struct. Biol. 6: 386-394. [DOI] [PubMed] [Google Scholar]
- NC-IUBMB. 1992. Enzyme nomenclature recommendations of the Nomenclature Committee of the International Union of Biochemistry and Molecular Biology on the Nomenclature and Classification of Enzymes Academic Press, New York, NY.
- Pabarcus, M.K. and Casida, J.E. 2002. Kynurenine formamidase: Determination of primary structure and modeling-based prediction of tertiary structure and catalytic triad. Biochim. Biophys. Acta 1596: 201-211. [DOI] [PubMed] [Google Scholar]
- The RIKEN Genome Exploration Research Group Phase II Team and the FANTOM Consortium. 2001. Functional annotation of a full-length mouse cDNA collection. Nature 409: 685-690. [DOI] [PubMed] [Google Scholar]
- Tanabe, A., Egashira, Y., Fukuoka, S., Shibata, K., and Sanada, H. 2002. Purification and molecular cloning of rat 2-amino-3-carboxymuconate-6–semialdehyde decarboxylase.Biochem. J. 361: 567-575. [DOI] [PMC free article] [PubMed] [Google Scholar]
WEB SITE REFERENCES
- http://fantom2.gsc.riken.go.jp/metabolome/; Metabolomapper.
- http://www.genome.ad.jp/kegg/; KEGG project.
- http://fantom2.gsc.riken.go.jp/fantom2/SI/sup16_go.pdf; Computational Assignment of GO terms.