Abstract
The innate immune system has evolved under selective pressure since the radiation of multicellular life approximately six hundred million years ago. Because of this long history, innate immune mechanisms found in modern eukaryotic organisms today are highly complex, yet are built from common molecular strategies. It is now clear that evolution has selected a conserved set of anti-microbial peptides as well as Pattern Recognition Receptors (PRRs) that initiate cellular-based signals as a first line of defense against invading pathogens. Conversely, microbial pathogens employ their own strategies to evade, inhibit, or otherwise manipulate the innate immune response. Here, we discuss recent discoveries that have changed our view of immune modulatory mechanisms employed by bacterial pathogens, focusing specifically on the initial sites of microbial recognition and extending to host cellular signal transduction, pro-inflammatory cytokine production, and alteration of protein trafficking and secretion.
Introduction
Since the discovery of Toll-like Receptor 4 (TLR4) as the Lipopolysaccharide (LPS) receptor over 15 years ago (Poltorak et al, 1998a), we now recognize that invading bacteria, viruses, fungi, and eukaryotic parasites are sensed by a diverse set of cellular pattern recognition receptors (PRRs) that include Toll-like receptors (TLRs), C-type lectin-like receptors (CLRs), nucleotide binding and oligomerization domain-like receptors (NLRs), cytoplasmic double-stranded DNA (dsDNA) receptors, and RIG-I-like receptors (RLR) (Creagh & O'Neill, 2006; Fritz et al, 2006; Janeway, 1989; Poltorak et al, 1998b; Takaoka & Shinohara, 2008; Uematsu & Akira, 2007; van Vliet et al, 2007). This diverse set of receptors bridge a critical gap between the innate and adaptive immune response system as they function to detect pathogen-associated or danger-associated molecular patterns known as PAMPs and DAMPs, respectively. Upon recognition of these signals, PRRs trigger intracellular signal transduction cascades that connect information about pathogen invasion to expression and secretion of immune-modulatory chemokines and cytokines. Importantly, these molecules are critical for triggering both innate and adaptive immune responses in animals.
Despite the fast-acting intracellular signaling mechanisms induced by PRRs, microbial pathogens have evolved countermeasures to thwart innate immunity in order to promote their lifestyle in the infected host organism. Only recently, however, studies aimed at characterizing the bacterial systems that inhibit the host innate immune response have been defined in molecular detail. Although several important points pertaining to the properties and function of bacterial defense mechanisms remain to be clarified, recent discoveries represent a major breakthrough in the field of molecular microbiology. Here we highlight emerging themes from the recent literature, from evasion mechanisms by which pathogens avoid antimicrobial molecules, to the ability of pathogens to directly modulate innate immune signal transduction pathways, and immune receptor localization and cytokine secretion.
The First Line of Defense: Bacterial Subversion of Antimicrobial Peptides
While recent work on innate immunity has been primarily focused on elucidating signaling systems down stream of PRRs, antimicrobial peptides (AMPs) are one of the most ancient forms of protection against bacterial infection. For example, epithelial cells of the skin and lung, and Paneth cells in the gut, secrete defensins, gramicidins, and cathelicidins as a tissue protective mechanism against the continuous challenges of bacteria and fungi. In addition, circulating neutrophils also secrete AMPs at sites of acute microbial infection. Whether in the lung, intestine, or at the surface of the skin, most AMPs disrupt membrane integrity through pore formation, causing an efflux of essential molecules and ions from invading pathogens. Importantly however, we now recognize that bacterial pathogens have evolved specific strategies to avoid or abrogate the lytic activity of AMPs during infection.
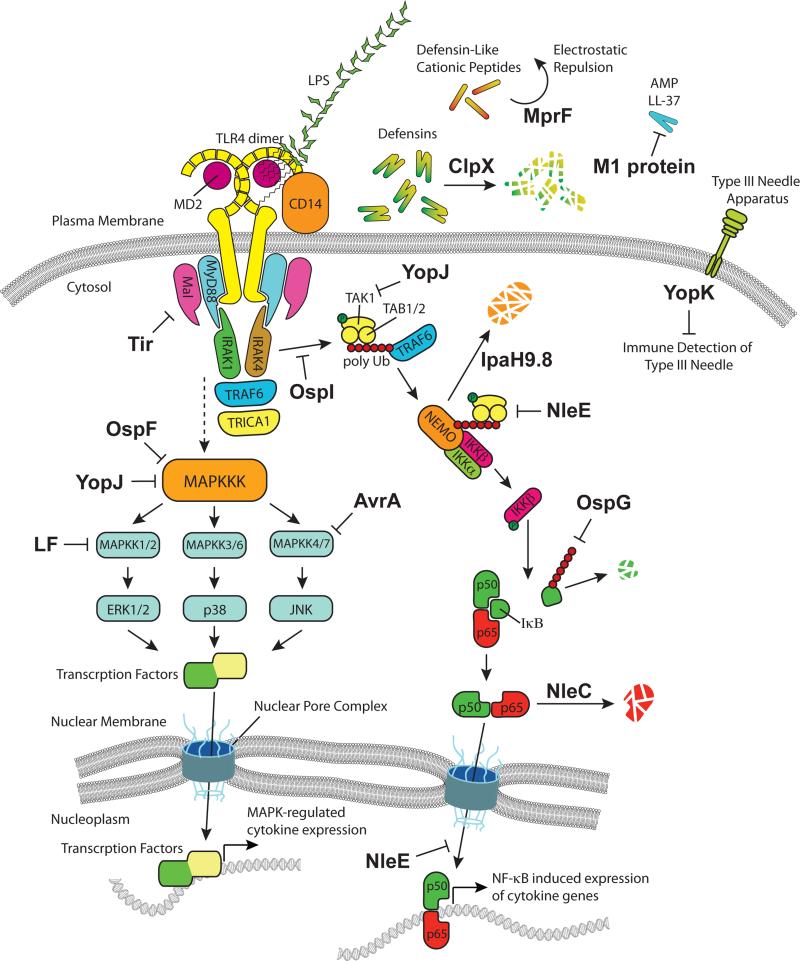
Perhaps the simplest mechanism bacteria use to combat the anti-microbial activity of AMPs is by physical avoidance through electrostatic repulsion. For example, recent studies suggest that the multiple peptide resistance factor protein, or MprF, provides Staphylococcus aureus with resistance to defensin-like cationic peptides (Figure 1) (Ernst et al, 2009). Here, the hydrophilic C-terminal domain of MprF is responsible for adding an L-Lysine to the phosphatidylglycerol lipid while in the inner leaflet of the bacterial membrane. Afterward, the membrane-bound N-terminal portion of MprF acts to flip the lipid to the outer leaflet so that the positively charged lysine residue can act as an electrostatic buffer that provides limited resistance to the cationic defensin molecules, primarily secreted by neutrophils, present in the host lumen.
Figure 1.
Bacterial effectors that target and manipulate the innate immune and inflammatory pathways. Upon LPS stimulation, the TLR4 dimer associates with MD2 and forms a complex with CD14 to signal through the cytoplasmic tails of TLR4. Stimulated TLR4 binds MyD88 and Mal (also TIRAP), which recruits IRAK1/4, TRAF6, and TRICA1 to stimulate inflammatory cell survival pathways (MAPK and NF-κB). Bacterial effector proteins inhibit this process in many different ways. MprF, from Staphylococcus aureus, repels cationic defensin peptides electrostatically. ClpX proteolytically degrades defensin molecules. The M1 protein binds and sequesters the AMP LL-37. YopK shields the Type III translocon from being recognized by host intracellular sensors. Tir, while functioning to maintain intimate contact between A/E pathogens and host, also utilizes ITIM-like motifs to downregulate TLR signaling. OspF possesses phosphothreonine lyase activity, which potently downregulates the MAPK cascade through covalent modification. OspI prevents TRAF6 auto-polyubiquitinylation to reduce NF-κB signaling. YopJ is an acetyl transferase that targets the MAPK pathway and also has activity against TAK1. LF cleaves the amino terminus of MAPKK1/2. AvrA inhibits the JNK pathway through acetyltransferase activity toward MAPKKs. IpaH9.8 targets the NEMO complex for proteasomal degradation. OspG binds to ubiquitinylated E2 enzymes to prevent the eventual ubiquitinylation of IκBα. NleC is a protease that degrades the p65 subunit of NF-κB. NleE modifies TAB2 and TAB3 to regulate NF-κB signaling.
In addition to this cloaking mechanism, many bacteria secrete proteases such as the caseinolytic enzymes that can eliminate soluble AMPs. One such protease, ClpXP, and more specifically, the ATPase subunit ClpX from Bacillus anthracis, was shown to have extracellular protease activity toward cathelicidin and alpha-defensin antimicrobial peptides (Figure 1) (McGillivray et al, 2009). Functional ClpX was essential for B. anthracis to disrupt host innate immune clearance mechanisms. In a similar manner, the M1 protein from S. pyogenes was found to contribute to overall resistance of Group A Streptococcus to the cathelicidin AMP LL-37 (Lauth et al, 2009). Interestingly, instead of repelling LL-37, the M1 protein was shown to bind and sequester the AMP, thus rendering it harmless against the invading pathogen (Figure 1). Importantly, these recent examples indicate that AMP inhibition is a critical anti-innate immune mechanism employed by several pathogens, and that this area of investigation is ripe for new discovery.
Stealth and Evasion: Bacterial Pathogens Disguise Their PAMPs
If a bacterial pathogen is able to successfully evade destruction by antimicrobial peptides, most host organisms have evolved a second line of defense centered on microbial recognition of PAMPs by PRRs and the subsequent production of cell-intrinsic immune mechanisms and/or recruitment of immune cells. In response to these challenges, many bacterial pathogens have modified the molecular structure of their PAMPs, thereby avoiding immune detection through stealth and evasion. For example, lipopolysaccharide (LPS) is a ubiquitous component of Gram-negative bacterial cell wall, and is composed of diverse O-antigen side chains that are anchored to the outer leaflet of the bacterial envelope by Lipid A. Importantly, Lipid A is directly recognized by the mammalian TLR4-MD2-CD14 PRR complex to activate innate immune signaling pathways (Figure 1). Some bacterial species have evolved another form of LPS that has undergone modification of the fatty acid side chain of Lipid A from 12-16 to over 20 carbons in length, thereby augmenting its detectability by TLRs (Matsuura, 2013). Yersinia pestis, the causative agent of the Bubonic plague of the 14th century, possess the ability to reduce the acylation status of Lipid A from hexa-acylated to tetra-acylated in response to an increase in temperature from 21°C (flea temperature) to 37°C (human temperature) (Montminy et al, 2006; Rebeil et al, 2004). This temperature-dependent hypo-acylation activity affords Yersinia the ability to evade host defenses by stealth, as tetra-acylated Lipid A is significantly less recognizable than hexa-acylated LPS by TLR4. Similarly, Shigella that have been internalized and multiply within the host cytoplasm hypo-acylate Lipid A such that upon escape from infected cells, these pathogens possessing the poorly immunogenic form do not alert the innate immune system, and thus stealthily spread and disseminate within the gut microenvironment (Paciello et al, 2013).
As with the example of Lipid A, we now recognize that innate immune evasion strategies may depend on molecular adaptation of highly conserved bacterial cellular components. However, it is also clear that virulence factors, and particularly molecules that directly target host cell physiological systems, are detected by the innate immune system. As discussed in more detail below, many Gram-negative bacterial pathogens utilize protein secretory systems (e.g. Type III/IV/VI secretion systems) to deliver virulence factors, termed ‘effector’ proteins, directly into the host cell. Several groups have now demonstrated that the Type III translocon is a pro-inflammatory PAMP, capable of being recognized by both the NLRP3 and NLRC4 inflammasome (Brodsky et al, 2010; Miao et al, 2010; Zhao et al, 2011). Because bacterial secretory systems are among the most important virulence determinants, it is also reasonable to suspect that pathogens have evolved mechanisms to block immune recognition of these essential systems. Indeed, it was recently discovered that Yersinia spp. masks pore forming attachment of the Type III translocon to the host cell, thereby cloaking the major virulence system of Yersinia from immune recognition (Brodsky et al, 2010). After being secreted by the Type III needle apparatus, Brodsky et al. demonstrated that YopK interacts with the distal end of the Type III translocon to prevent host cellular recognition and inflammasome activation (Figure 1). This is a unique strategy, as it is normally thought that Type III effector proteins have specific eukaryotic target(s). It will be interesting to determine if YopK alters the structure of the needle apparatus directly, or alternatively, competitively inhibits inflammasome activation. In either case however, YopK targets and cloaks a newly recognized PAMP, highlighting the evolutionary arms race that takes place between host and pathogen. Surely as new host immune strategies are uncovered, we expect to find additional mechanisms of masking bacterial PAMPs as immune evasion strategies.
The Inside Story: Bacterial Pathogens Target Intracellular Signal Transduction Cascades
While our discussion has so far focused on initial points of interaction between bacterial pathogens and host cells, its is clear that the power of the immune systems is based on the complex and highly dynamic cellular signal transduction systems the relay information from PRRs to nuclear transcription of pro-inflammatory modulators. Thus, it stands to reason that signal transduction is a high value target of bacterial pathogens. Indeed, as the field of innate immunity has exploded over the past 15 years, the molecular details by which pathogens modulate host immune response systems have also come to light. A majority of our understanding has come from the various ways in which pathogens target the Mitogen Activated Protein Kinase (MAPK) signaling axis and the NEMO-IKK-NF-κB pathway, which we will explore in more detail here.
Effector Molecules Target the MAPK Cascade
As outlined in Figure 1, the MAPK signaling cascade involves sequential activation and amplification of downstream kinases that govern a bevy of transcriptional responses required for proper cellular metabolism, growth, and division. While MAPK signaling is essential for normal operation of the eukaryotic cell, some branches are critical for decoding information obtained from PRRs, and transmitting the appropriate response signal to induce inflammation as an antimicrobial defense mechanism. Because the MAPK pathway response to PAMPs or DAMPs is carefully tuned at multiple sites, this cascade represents a major pressure point that has been successfully exploited by bacterial toxins and effector proteins.
One of the earliest reported direct inhibition of MAPKs by a bacterial pathogen was the discovery that Anthrax lethal toxin (lethal factor; LF) of Bacillus anthracis cleaves the amino terminus of MAPKK1 and MAPKK2 (Figure 1) (Duesbery et al, 1998). This seminal finding was shortly followed by the discovery that a Type III secreted effector protein, YopJ of Yersinia pestis (YopP in Y. enterocolotica and YopY in Y. pseudotuberculosis, hereafter referred to as YopJ), also inhibits the activation of MAPK signaling pathways (Orth et al, 1999). Although some discrepancies exist as to the exact enzymatic activity of YopJ and its homologs, it is clear that several isoforms, including YopJ itself, functions as an acetyl transferase that covalently modifies key serine and threonine residues of MAP kinases (Mukherjee et al, 2006). This acetylation reaction results in the inability of upstream kinases to trans-phosphorylate the down stream kinase, thus preventing the MAPK-mediated cytokine transcriptional response to Yersinia infection (Mukherjee et al, 2006; Mukherjee et al, 2008). Importantly, there is now good evidence that YopJ targets several MAP kinases, including MAPK 1, 3, 4, and 5 and TGF-β-activated kinase 1 (TAK1) (Figure 1) (Mukherjee et al, 2006; Paquette et al, 2012). It is also important to note that this general catalytic mechanism has been evolved for specific pathogen purposes. Recently, the YopJ homologue from Salmonella, AvrA, was found to harbor acetyl transferase activity and to inhibit MAPK4 and MAPK7 of the c-Jun NH2-terminal kinase (JNK) signaling pathway (Figure 1), thereby altering the transcription of pro survival genes during infection (Jones et al, 2008; Wu et al, 2012).
Historically, the identification and molecular characterization of LF and YopJ ushered in a new era of anti-innate immune signaling research. From numerous studies over the past 5-7 years, it is now clear that the irreversible modification of host signaling enzymes represents an effective and widely used strategy for manipulating the immune response system. Because the post-translational modifications of host enzymes are unique to a given pathogen, and often represent completely new catalytic mechanisms, they have been difficult to discover. However, the widespread development of mass spectrometry as tool for biological discovery has aided in this effort. For example, the Shigella flexneri Type III effector protein OspF (which is homologous to the Salmonella SpvC and Pseudomonas syringae HopAI1) was initially described to possess phosphatase-like activity toward activated MAPK. Subsequent mass spectrometry analysis shed light on this reaction, as it was discovered that OspF catalyzes a β-elimination reaction of phosphothreonine to a β-methyldehydroalanine, thus rendering the MAP kinase cascade permanently inactive (Figure 1) (Li et al, 2007; Zhu et al, 2007). Phosphorylation and dephosphorylation are pervasive signaling mechanisms in biology, and the phosphothreonine lyase activity of the OspF family represents the only known enzyme class capable of carrying out this permanent and irreversible modification. Therefore, OspF defines a novel phosphothreonine lyase capable of permanently dephosphorylating host MAPK enzymes. Perhaps equally striking is that OspF, and its homologues in Salmonella and Pseudomonas, take advantage of the a conserved MAPK docking motif (D motif) in mammals in order to specifically recognize and target host substrates for covalent modification (Wei et al, 2012). This level of target specificity by a class of bacterial effector proteins demonstrates the elegance by which pathogens inhibit inflammatory activation.
Effector Molecules Target the NF-κB Pathway
The canonical NF-κB pathway has been the focus of intense investigation for nearly three decades. Significant progress has been made, and although full understanding of particular nuances remains, the function of NF-κB and its transcriptional regulation of immunomodulatory genes have been determined with great detail. In general, the NF-κB pathway involves recognition of PAMPs via TLRs (and other PRRs) and subsequent signal transduction through TIRAP and MyD88 that activates the kinase activity of the NEMO/IKKα/IKKβ complex. This, in turn, leads to the phosphorylation and subsequent ubiquitinylation of IκBα, freeing the NF-κB components (represented in Figure 1 as p65 and p50) to migrate to the nucleus and begin transcription of the various NF-κB-regulated genes. Because this summary does not justify the amazing complexity of this system, we recommend several excellent reviews that provide in-depth perspective on this critical pathway (Dev et al, 2011; Hayden & Ghosh, 2008; Kovalenko & Wallach, 2006). Importantly, the NF-κB signal transduction pathway is a target-rich environment for many pathogens that successfully invade, colonize and disseminate within the host organism.
In addition to manipulating the innate immune responses of epithelial cells, bacterial pathogen inhibition of NF-κB signaling pathways greatly reduces macrophages’ ability to induce inflammation, undergo apoptosis, and communicate with other immune cells. One of the best characterized pathogens, Shigella flexneri, harbors several unique mechanism for NF-κB signaling inhibition which, together, allow them to propagate within macrophages and to reduce acute inflammation during invasion. This is accomplished in part by the Type III effector OspI, which suppresses the tumor-necrosis factor receptor-associated factor 6 (TRAF6)-mediated signaling pathway (Sanada et al, 2012). TRAF6 auto-polyubiquitinylation, subsequent to the activity of an E2 ligase, is responsible for initiating a cascade that culminates in the NF-κB dependent transcription of pro-inflammatory genes. OspI was found to deamidate glutamine-100 of the E2 ubiquitin ligase UBC13 to glutamic acid thus rendering it inactive, which prohibits TRAF6 auto-polyubiquitinylation. This ingeniously simple modification leads to a reduction of the NF-κB proinflammatory pathway to prolonged infection by Shigella (Figure 1).
Manipulation of ubiquitinylation pathways within the host is not an isolated mechanism of NF-κB subversion. Shigella also secretes IpaH9.8, which functions as an E3 ubiquitin ligase to dampen the host immune response by targeting the NEMO complex for proteasomal degradation (Ashida et al, 2010). As mentioned previously, NEMO is critical for downstream activation of NF-κB. IpaH9.8 activity induces Lys27-mediated polyubiquitylation of NEMO, which in turn promotes its proteasome-dependent degradation, and results in reduced NF-κB activation during infection (Figure 1). The IpaH family is of particular interest since many bacterial species that target vertebrate and plant hosts contain proteins that share sequence similarity within this family (Rohde et al, 2007).
Clearly, ubiquitinylation represents an important signaling and regulatory mechanism of NF-κB signaling, and it is therefore no surprise that pathogens have developed multiple mechanisms to exploit this post-translation modification. The Shigella effector OspG was shown to bind ubiquitin and ubiquitinylated E2s to reduce E3-mediated ubiquitinylation of IκBα, thereby preventing the dissociation of IκBα/p50/RelA and inhibiting NF-κB-mediated transcription (Figure 1) (Kim et al, 2005; Zhou et al, 2013). Interestingly, OspG possess kinase activity in addition to that of ubiquitin binding activity (Kim et al, 2005). Recent studies indicate that the binding to ubiquitin activates kinase activity through an unusual allosteric mechanism involving and E2~Ub conjugate (Kim et al, 2005; Pruneda et al, 2014; Zhou et al, 2013). In fact, the crystal structure of OspG bound to UbcH5c~Ub revealed that OspG binds to the ubiquitinylated form of UbcH5c and that this binding was sufficient to reorganize the kinase active site into a catalytically competent state. Because these studies provide significant insight into the kinase regulatory mechanism at the molecular level, we are now poised to discover the host substrate of the kinase-active form of OspG that regulates NF-κB signal transduction.
While Shigella has evolved several unique mechanisms to dampen the host innate immune response through NF-κB signaling inhibition, extracellular pathogens such as members of the Attaching and Effacing (A/E) pathogen group that include enteropathogenic E. coli (EPEC), enterohemorrhagic E. coli O157:H7 (EHEC), and Citrobacter rodentium, inhibit innate immune responses while maintaining intimate contact with the host plasma membrane. One of the first Type III effector proteins discovered in A/E pathogens was the Translocated Intimin Receptor (Tir), which integrates into the host plasma membrane and binds directly to the bacterial cell surface protein intimin (Kenny et al, 1997). Although Tir is essential for the strong bacterial attachment to host cells, and is a key molecule involved in the attaching and effacing lesion observed during infection, it has since been discovered to suppress innate immune signaling pathways. Yan and colleagues demonstrated that Tir shares sequence similarities with the immunoreceptor tyrosine-based inhibition motifs (ITIMs), conserved sequences often found in the cytoplasmic tails of inhibitory receptors of the immune system (Yan et al, 2012). After secretion into the host, ITIM-like motifs present in the Tir effector promote recruitment of Src homology region 2 domain-containing phosphatase 1 (SHP-1), which is known to have a suppressive role in cellular immune responses. SHP-1 has been demonstrated to inhibit the activation of NF-κB and MAPK by directly dephosphorylating them (Nandan et al, 1999), and when in complex with TRAF6, prevents its polyubiquitinylation and subsequent activation of TAK1, thereby further inhibiting the MAPK and NF-κB pathways (Zhang et al, 2003). Here, Tir directly associates with SHP-1 and nucleates a Tir-SHP-1-TRAF6 complex to suppress host cell innate immune responses to A/E lesions, thus providing a striking example of a bacterial effector protein that mimics an endogenous innate immune regulatory mechanism (Figure 1).
Beyond Tir, A/E pathogens have evolved an arsenal of effector proteins that target the NF-κB pathway. NleB, which was first reported to suppress p65 nuclear localization (Newton et al, 2010), was subsequently shown to reduce TNFα-dependent polyubiquitinylation of TRAF2 and stabilize IκB in response to TNFα stimulation. Gao and colleagues first identified Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) as an interacting partner of NleB, although the affinity was somewhat modest (Kd of 36 μM). Nonetheless, this interaction disrupted the GAPDH-TRAF2 interaction, and thus suppressed TRAF2-mediated polyubiquitinylation and subsequent NF-κB activation (Gao et al, 2013). More recently, two back-to-back reports indicated that NleB also targets the death domain (DD) containing adaptor protein TRADD in order to inhibit NF-κB activation (Li et al, 2013; Pearson et al, 2013). TRADD was identified as a host interacting partner of NleB, and further analysis showed that NleB was capable of disrupting TRADD death domain oligomerization. Mass spectrometry analysis revealed that NleB catalyzes the GlcNacylation of the TRADD DD at Arginine 235, and this post-translational modification was responsible for inhibiting functional oligomerization of the DDs necessary for the host cell to mount an apoptotic response. This was surprising, since most known GlcNacylation occurs on serine or threonine residues (Hurtado-Guerrero et al, 2008). In addition, TRADD was not the only NleB target identified in these studies, as this bacterial effector is capable of targeting FAS-associated protein with death domain (FADD) and the receptor-interacting serine/threonine-protein kinase 1 (RIPK1) proteins. GlcNacylation of TRADD and FADD arginines prevented the maturation of Caspase-8, a necessary component of the host apoptotic pathway, thus affording A/E pathogens the ability to thwart innate immune defense mechanisms of host cells.
While NleB functions upstream of NF-κB and modulates death receptor signaling pathways, EPEC secretes another effector protein, NleC, which possesses zinc protease activity, to delay host immune detection (Figure 1) (Yen et al, 2010). NleC catalyzed the cleavage of p65 between the 10th and 11th amino acids and therefore rendered it unable to partner with p50, and was subsequently degraded through normal host clearance mechanisms. As a result, NleC efficiently suppresses the host pro-inflammatory response to infection and provides A/E bacterial pathogens additional time and space to multiply. In addition to proteolytic inhibition, A/E pathogens also block nuclear trafficking of NF-κB as innate immunity inhibitory mechanism. The EPEC effector NleE, which shares significant sequence and functional homology to Shigella OspZ, plays a key role in this reduction of host immune response. Upon transfection of cells with NleE and subsequent stimulation by IL-1β or TNF-α, the variable NF-κB components p65 and c-Rel were prevented from translocating into the nucleus, and release of IL-8 was greatly reduced as a result (Figure 1) (Newton et al, 2010). Therefore, it seems that NleE is specific for arresting the traffic of p65 and c-Rel, as the nuclear translocation of p50 was still permitted (Newton et al, 2010). Interestingly, it was recently shown that NleE possesses S-adenosyl-L-methionine-dependent methyltransferase activity against TAB2 and TAB3, ubiquitin-chain sensory proteins involved in the NF-κB pathway. NleE specifically modified the zinc-coordination site in the zinc-finger domains of TAB2 and TAB3, causing these proteins to lose the zinc ion as well as ubiquitin-chain binding ability (Zhang et al, 2012). Disruption of the ubiquitin-chain binding by TAB2 and TAB3 results in their inability to activate TAK1 and IKK, thereby preventing the requisite NF-κB components from being released to translocate to the nucleus and mount an inflammatory response (Figure 1).
Because direct molecular mechanisms have been elucidated in detail, our analysis of the NF-κB pathway has largely focused on the Shigella and A/E pathogen group. Importantly however, NF-κB is certainly targeted by many pathogens, alternative mechanisms of signaling inhibition are likely to have evolved, and we suspect that many new and exciting details will emerge in the coming years.
Organelle Dysfunction: The Host Secretory Pathway as an Immune Modulatory Target of Bacterial Pathogens
As we have described so far, bacterial pathogens harbor a variety of elegant mechanisms to inhibit cellular signal transduction pathways that dampen the host inflammatory response. However, recent evidence reveals that pathogens have evolved alternative strategies to inhibit innate immunity by blocking the secretion of cytokines and trafficking of cell surface receptors. Here we will discuss the recent advancements in understanding how bacterial effectors target, engage, and inhibit molecules that regulate the host general secretory pathway (GSP).
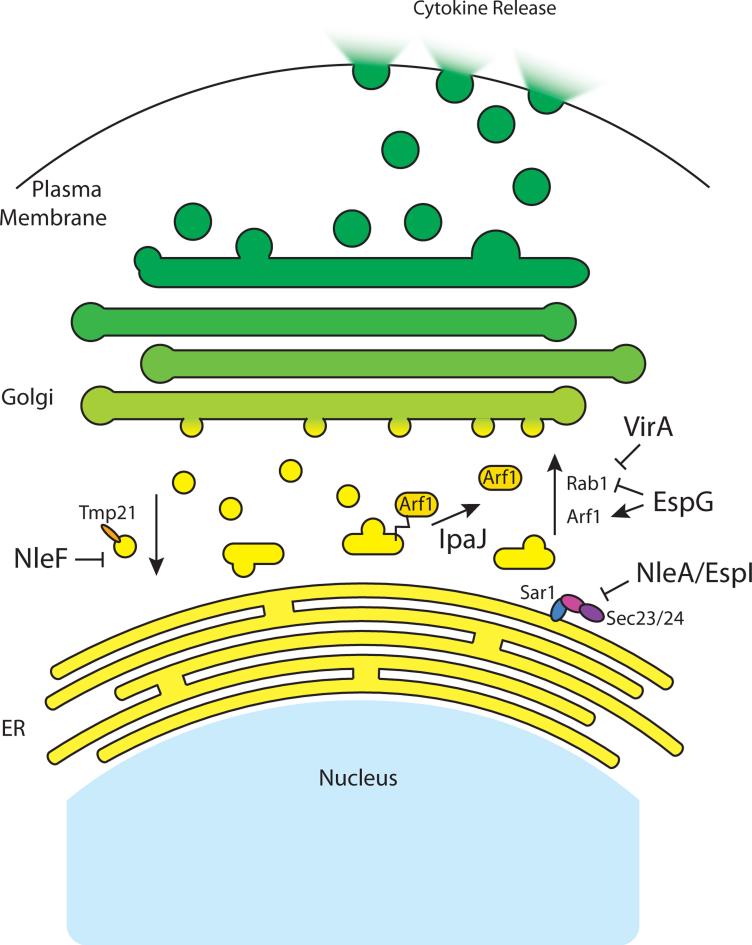
Initiating at the Endoplasmic reticulum (ER), the GSP traffics molecular cargo (e.g. cell surface receptors and cytokines) through a series of membrane-bound compartments, including ER-Golgi intermediate compartment (ERGIC) and the Golgi proper, and releasing this cargo through vesicle fusion with the plasma membrane (Figure 2). Vesicular trafficking between these compartments is supervised in large part by small GTPases including Arf1 and Sar1 that nucleate coat protein complex I and II (COPI and COPII) on vesicles involved in retrograde or anterograde transport, respectively. One of the most prolific GSP inhibitors are the A/E pathogens (EHEC, EPEC, and Citrobacter) that secrete the Type III effector proteins NleA/EspI, NleF, and EspG. These effector proteins specifically target the early steps of ER to Golgi trafficking through distinct molecular mechanisms. The first step of cargo packaging and vesicle budding from the ER is regulated by NleA/EspI, which acts to prevent normal COPII mediated vesicle trafficking, thereby inhibiting ER to Golgi transport (Figure 2) (Kim et al, 2007). Although this mechanism of regulation is not fully understood, it is clear that NleA/EspI binds to Sec23/24, a heterodimeric subunit of the CopII coat, in a Sar1 and lipid-dependent manner and decreases the rate of GTP hydrolysis by Sar1 (Thanabalasuriar et al, 2012). This interaction has been shown to stabilize COPII, which presumably leads to a defect in recycling and thus an abnormal buildup of COPII coated vesicles that slows proper sorting of anterograde trafficked vesicular cargo. In contrast to this mechanism, the GSP is also inhibited by the effector protein NleF which impedes the activity of COPI by binding to the transmembrane protein Tmp21, a COPI docking partner (Figure 2) (Olsen et al, 2013). Blocking COPI type vesicles would inhibit retrograde transport, which is essential for Golgi function. In both cases however, additional biochemical and structural analyses will be needed to fully understand these important host-pathogen interactions at the molecular level.
Figure 2.
Bacterial effectors that inhibit the GSP to suppress innate immune secretory mechanisms. NleF binds to the transmembrane protein Tmp21 to prevent COPI docking and thereby disrupt retrograde traffic. IpaJ cleaves the N-terminal myristoyl tail from Arf1, thus removing it from the membrane and promoting Golgi destruction. NleA/EspI prevent the uncoating of CopII vesicles, thus inhibiting their fusion during anterograde traffic. EspG disrupts the GSP through its Arf binding and Rab1GAP activities. VirA possesses Rab1GAP activity that acts to disrupt Golgi architecture.
While originally thought to alter host microtubule dynamics, it was later revealed that EspG acts as a scaffold to bring together GTP-bound Arf GTPases and active p21-associated kinase (PAK) resulting in the subsequent fragmentation of the Golgi apparatus (Figure 2) (Selyunin et al, 2011). Indeed, crystal structures show that EspG binds to Arf6 in the GTP-bound state and protects the bound GTP-binding protein from inactivation by ArfGAP (ASAP1). In addition, EspG directly activates PAK by disrupting the autoinhibitory interactions in this multi-domain kinase. While these studied indicated that EspG is a multifaceted effector molecule capable of bridging critical signaling pathways within cells, more recent studies have provided additional levels of complexity. For example, Dong et al. found that EspG (and its Shigella homolog VirA) function as Rab1-specific GTPase activating protein (GAP) (Dong et al, 2012). Further crystallographic evidence revealed that EspG harbors a catalytic TBC (Tre-2/Bub2/Cdc16) GAP catalytic core, yet the sequence and structural fold of this bacterial GAP domain has almost no similarity to host GAPs that target Rab family GTPases. Perhaps most remarkably, the binding site of Rab1 on EspG perfectly overlaps that of PAK, suggesting that this is a site of molecular evolution toward new effector functions (Selyunin et al, 2014, Cell Reports, in press). In addition, EspG binds and protects activated Arf1 leading to Golgi disruption while at the same time acting as a Rab1GAP thereby greatly reducing ER to Golgi vesicular trafficking (Figure 2) (Selyunin et al, 2014, Cell Reports, in press). Together, these multivalent interactions inhibit cytokine secretion during EPEC infection (Dong et al, 2012)
EHEC/EPEC is not the only pathogen that has evolved elegant mechanisms to inhibit the host secretory system. Shigella flexneri was recently found to use two different strategies for Golgi destruction and GSP inhibition. First, the effector VirA, which is an EspG homolog, was shown to also have TBC-mediated Rab1 GAP activity (Dong et al, 2012). This activity was sufficient to disrupt intra-Golgi vesicular movements and also confer protection against the host autophagic pathway, thus enabling Shigella to promote its intracellular lifestyle (Figure 2). Interestingly however, VirA is sufficient, but not necessary, to disrupt Golgi architecture during bacterial infection. Experiments carried out using a virA strain of Shigella were shown to cause Golgi disruption similar to wild type bacteria (Mounier et al, 2012). Recently, the missing component was identified through a gain-of-function screen for bacterial inhibitors of the GSP. Burnaevskiy et al. found that the Shigella Type III effector IpaJ was both necessary and sufficient to inhibit cytokine secretion and disrupt Golgi morphology during infection (Burnaevskiy et al, 2013). Bioinformatics analysis revealed that IpaJ shared secondary structural similarity with members of the C39-peptidase-like family, and contained the invariant catalytic triad consisting of Cys64, His206, and Asp218. Using a combination of yeast genetics and mass spectrometry, IpaJ was shown to specifically cleave the myristoylated glycine from Arf1, as well as numerous lipid-modified substrates, thus preventing host signal transduction from specific membrane microdomains (Figure 2). Together, these studies reveal an alternative mechanism of innate immune pathway inhibition based on global cellular disruption of host organelle structure and function. It will be important to determine if these bacterial effector proteins also block non-canonical secretion systems (such as the release of IL-1) that are not dependent on the GSP. In addition, further studies will be needed to elucidate the similarities and differences in VirA and IpaJ cellular function, as well as their role in immune function during infection.
Conclusions
Microbial pathogens have evolved elegant yet often complex mechanisms to target, subvert, and/or thwart the innate immune response of the host. Although the Gram-negative bacterial pathogens that have been discussed have distinct life styles and exploit various niches within the host, they share in common a repertoire of effector proteins that function to inhibit a myriad of host innate immune defenses. During this discussion, we have mainly focused on the action of individual effectors and their enzyme activities, however pathogens secrete numerous effectors concomitantly during an infection. Thus, invading pathogens may commandeer various control points of the host immune response with both spatial and temporal precision. In addition, our discussion has focused on specific subsets of innate immune mechanisms, and no doubt these examples only scratch the surface of this fascinating field. We predict that many new discoveries are forthcoming, and that these insights will heighten our understanding of the complex molecular battle between host and pathogens.
Acknowledgements
We would like to thank members of the Alto Lab for suggestions and insights, especially Maarten F. de Jong, Nikolay Burnaevskiy, and Patrick Woida. We also thank Leonardo D. Estrada for critical reading of the manuscript. This work was supported by grants from the National Institutes of Health (NIAID; 1F32AI098384) to L.E.R. and (NIAID; R01AI083359 and NIGMS; R01GM100486), the Welch Foundation (I-1704), the Burroughs Wellcome Fund to N.M.A.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ashida H, Kim M, Schmidt-Supprian M, Ma A, Ogawa M, Sasakawa C. A bacterial E3 ubiquitin ligase IpaH9.8 targets NEMO/IKKgamma to dampen the host NF-kappaB-mediated inflammatory response. Nat Cell Biol. 2010;12(sup):66–73. 61–69. doi: 10.1038/ncb2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodsky IE, Palm NW, Sadanand S, Ryndak MB, Sutterwala FS, Flavell RA, Bliska JB, Medzhitov R. A Yersinia effector protein promotes virulence by preventing inflammasome recognition of the type III secretion system. Cell Host Microbe. 2010;7:376–387. doi: 10.1016/j.chom.2010.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnaevskiy N, Fox TG, Plymire DA, Ertelt JM, Weigele BA, Selyunin AS, Way SS, Patrie SM, Alto NM. Proteolytic elimination of N-myristoyl modifications by the Shigella virulence factor IpaJ. Nature. 2013;496:106–109. doi: 10.1038/nature12004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creagh EM, O'Neill LA. TLRs, NLRs and RLRs: a trinity of pathogen sensors that cooperate in innate immunity. Trends Immunol. 2006;27:352–357. doi: 10.1016/j.it.2006.06.003. [DOI] [PubMed] [Google Scholar]
- Dev A, Iyer S, Razani B, Cheng G. NF-kappaB and innate immunity. Curr Top Microbiol Immunol. 2011;349:115–143. doi: 10.1007/82_2010_102. [DOI] [PubMed] [Google Scholar]
- Dong N, Zhu Y, Lu Q, Hu L, Zheng Y, Shao F. Structurally distinct bacterial TBC-like GAPs link Arf GTPase to Rab1 inactivation to counteract host defenses. Cell. 2012;150:1029–1041. doi: 10.1016/j.cell.2012.06.050. [DOI] [PubMed] [Google Scholar]
- Duesbery NS, Webb CP, Leppla SH, Gordon VM, Klimpel KR, Copeland TD, Ahn NG, Oskarsson MK, Fukasawa K, Paull KD, Vande Woude GF. Proteolytic inactivation of MAP-kinase-kinase by anthrax lethal factor. Science. 1998;280:734–737. doi: 10.1126/science.280.5364.734. [DOI] [PubMed] [Google Scholar]
- Ernst CM, Staubitz P, Mishra NN, Yang SJ, Hornig G, Kalbacher H, Bayer AS, Kraus D, Peschel A. The bacterial defensin resistance protein MprF consists of separable domains for lipid lysinylation and antimicrobial peptide repulsion. PLoS Pathog. 2009;5:e1000660. doi: 10.1371/journal.ppat.1000660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritz JH, Ferrero RL, Philpott DJ, Girardin SE. Nod-like proteins in immunity, inflammation and disease. Nat Immunol. 2006;7:1250–1257. doi: 10.1038/ni1412. [DOI] [PubMed] [Google Scholar]
- Gao X, Wang X, Pham TH, Feuerbacher LA, Lubos ML, Huang M, Olsen R, Mushegian A, Slawson C, Hardwidge PR. NleB, a bacterial effector with glycosyltransferase activity, targets GAPDH function to inhibit NF-kappaB activation. Cell Host Microbe. 2013;13:87–99. doi: 10.1016/j.chom.2012.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayden MS, Ghosh S. Shared principles in NF-kappaB signaling. Cell. 2008;132:344–362. doi: 10.1016/j.cell.2008.01.020. [DOI] [PubMed] [Google Scholar]
- Hurtado-Guerrero R, Dorfmueller HC, van Aalten DM. Molecular mechanisms of OGlcNAcylation. Curr Opin Struct Biol. 2008;18:551–557. doi: 10.1016/j.sbi.2008.09.005. [DOI] [PubMed] [Google Scholar]
- Janeway CA., Jr. Approaching the asymptote? Evolution and revolution in immunology. Cold Spring Harb Symp Quant Biol 54 Pt. 1989;1:1–13. doi: 10.1101/sqb.1989.054.01.003. [DOI] [PubMed] [Google Scholar]
- Jones RM, Wu H, Wentworth C, Luo L, Collier-Hyams L, Neish AS. Salmonella AvrA Coordinates Suppression of Host Immune and Apoptotic Defenses via JNK Pathway Blockade. Cell Host Microbe. 2008;3:233–244. doi: 10.1016/j.chom.2008.02.016. [DOI] [PubMed] [Google Scholar]
- Kenny B, DeVinney R, Stein M, Reinscheid DJ, Frey EA, Finlay BB. Enteropathogenic E. coli (EPEC) transfers its receptor for intimate adherence into mammalian cells. Cell. 1997;91:511–520. doi: 10.1016/s0092-8674(00)80437-7. [DOI] [PubMed] [Google Scholar]
- Kim DW, Lenzen G, Page AL, Legrain P, Sansonetti PJ, Parsot C. The Shigella flexneri effector OspG interferes with innate immune responses by targeting ubiquitin-conjugating enzymes. Proc Natl Acad Sci U S A. 2005;102:14046–14051. doi: 10.1073/pnas.0504466102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Thanabalasuriar A, Chaworth-Musters T, Fromme JC, Frey EA, Lario PI, Metalnikov P, Rizg K, Thomas NA, Lee SF, Hartland EL, Hardwidge PR, Pawson T, Strynadka NC, Finlay BB, Schekman R, Gruenheid S. The bacterial virulence factor NleA inhibits cellular protein secretion by disrupting mammalian COPII function. Cell Host Microbe. 2007;2:160–171. doi: 10.1016/j.chom.2007.07.010. [DOI] [PubMed] [Google Scholar]
- Kovalenko A, Wallach D. If the prophet does not come to the mountain: dynamics of signaling complexes in NF-kappaB activation. Mol Cell. 2006;22:433–436. doi: 10.1016/j.molcel.2006.05.002. [DOI] [PubMed] [Google Scholar]
- Lauth X, von Kockritz-Blickwede M, McNamara CW, Myskowski S, Zinkernagel AS, Beall B, Ghosh P, Gallo RL, Nizet V. M1 protein allows Group A streptococcal survival in phagocyte extracellular traps through cathelicidin inhibition. J Innate Immun. 2009;1:202–214. doi: 10.1159/000203645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Xu H, Zhou Y, Zhang J, Long C, Li S, Chen S, Zhou JM, Shao F. The phosphothreonine lyase activity of a bacterial type III effector family. Science. 2007;315:1000–1003. doi: 10.1126/science.1138960. [DOI] [PubMed] [Google Scholar]
- Li S, Zhang L, Yao Q, Li L, Dong N, Rong J, Gao W, Ding X, Sun L, Chen X, Chen S, Shao F. Pathogen blocks host death receptor signalling by arginine GlcNAcylation of death domains. Nature. 2013;501:242–246. doi: 10.1038/nature12436. [DOI] [PubMed] [Google Scholar]
- Matsuura M. Structural Modifications of Bacterial Lipopolysaccharide that Facilitate Gram-Negative Bacteria Evasion of Host Innate Immunity. Front Immunol. 2013;4:109. doi: 10.3389/fimmu.2013.00109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGillivray SM, Ebrahimi CM, Fisher N, Sabet M, Zhang DX, Chen Y, Haste NM, Aroian RV, Gallo RL, Guiney DG, Friedlander AM, Koehler TM, Nizet V. ClpX contributes to innate defense peptide resistance and virulence phenotypes of Bacillus anthracis. J Innate Immun. 2009;1:494–506. doi: 10.1159/000225955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao EA, Mao DP, Yudkovsky N, Bonneau R, Lorang CG, Warren SE, Leaf IA, Aderem A. Innate immune detection of the type III secretion apparatus through the NLRC4 inflammasome. Proc Natl Acad Sci U S A. 2010;107:3076–3080. doi: 10.1073/pnas.0913087107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montminy SW, Khan N, McGrath S, Walkowicz MJ, Sharp F, Conlon JE, Fukase K, Kusumoto S, Sweet C, Miyake K, Akira S, Cotter RJ, Goguen JD, Lien E. Virulence factors of Yersinia pestis are overcome by a strong lipopolysaccharide response. Nat Immunol. 2006;7:1066–1073. doi: 10.1038/ni1386. [DOI] [PubMed] [Google Scholar]
- Mounier J, Boncompain G, Senerovic L, Lagache T, Chretien F, Perez F, Kolbe M, Olivo-Marin JC, Sansonetti PJ, Sauvonnet N. Shigella effector IpaB-induced cholesterol relocation disrupts the Golgi complex and recycling network to inhibit host cell secretion. Cell Host Microbe. 2012;12:381–389. doi: 10.1016/j.chom.2012.07.010. [DOI] [PubMed] [Google Scholar]
- Mukherjee S, Keitany G, Li Y, Wang Y, Ball HL, Goldsmith EJ, Orth K. Yersinia YopJ acetylates and inhibits kinase activation by blocking phosphorylation. Science. 2006;312:1211–1214. doi: 10.1126/science.1126867. [DOI] [PubMed] [Google Scholar]
- Mukherjee S, Negi VS, Keitany G, Tanaka Y, Orth K. In vitro activation of the IkappaB kinase complex by human T-cell leukemia virus type-1 Tax. J Biol Chem. 2008;283:15127–15133. doi: 10.1074/jbc.M704831200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nandan D, Lo R, Reiner NE. Activation of phosphotyrosine phosphatase activity attenuates mitogen-activated protein kinase signaling and inhibits c-FOS and nitric oxide synthase expression in macrophages infected with Leishmania donovani. Infect Immun. 1999;67:4055–4063. doi: 10.1128/iai.67.8.4055-4063.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton HJ, Pearson JS, Badea L, Kelly M, Lucas M, Holloway G, Wagstaff KM, Dunstone MA, Sloan J, Whisstock JC, Kaper JB, Robins-Browne RM, Jans DA, Frankel G, Phillips AD, Coulson BS, Hartland EL. The type III effectors NleE and NleB from enteropathogenic E. coli and OspZ from Shigella block nuclear translocation of NF-kappaB p65. PLoS Pathog. 2010;6:e1000898. doi: 10.1371/journal.ppat.1000898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen RL, Echtenkamp F, Cheranova D, Deng W, Finlay BB, Hardwidge PR. The enterohemorrhagic Escherichia coli effector protein NleF binds mammalian Tmp21. Vet Microbiol. 2013;164:164–170. doi: 10.1016/j.vetmic.2013.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orth K, Palmer LE, Bao ZQ, Stewart S, Rudolph AE, Bliska JB, Dixon JE. Inhibition of the mitogen-activated protein kinase kinase superfamily by a Yersinia effector. Science. 1999;285:1920–1923. doi: 10.1126/science.285.5435.1920. [DOI] [PubMed] [Google Scholar]
- Paciello I, Silipo A, Lembo-Fazio L, Curcuru L, Zumsteg A, Noel G, Ciancarella V, Sturiale L, Molinaro A, Bernardini ML. Intracellular Shigella remodels its LPS to dampen the innate immune recognition and evade inflammasome activation. Proc Natl Acad Sci U S A. 2013 doi: 10.1073/pnas.1303641110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paquette N, Conlon J, Sweet C, Rus F, Wilson L, Pereira A, Rosadini CV, Goutagny N, Weber AN, Lane WS, Shaffer SA, Maniatis S, Fitzgerald KA, Stuart L, Silverman N. Serine/threonine acetylation of TGFbeta-activated kinase (TAK1) by Yersinia pestis YopJ inhibits innate immune signaling. Proc Natl Acad Sci U S A. 2012;109:12710–12715. doi: 10.1073/pnas.1008203109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson JS, Giogha C, Ong SY, Kennedy CL, Kelly M, Robinson KS, Lung TW, Mansell A, Riedmaier P, Oates CV, Zaid A, Muhlen S, Crepin VF, Marches O, Ang CS, Williamson NA, O'Reilly LA, Bankovacki A, Nachbur U, Infusini G, Webb AI, Silke J, Strasser A, Frankel G, Hartland EL. A type III effector antagonizes death receptor signalling during bacterial gut infection. Nature. 2013;501:247–251. doi: 10.1038/nature12524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poltorak A, He X, Smirnova I, Liu MY, Van Huffel C, Du X, Birdwell D, Alejos E, Silva M, Galanos C, Freudenberg M, Ricciardi-Castagnoli P, Layton B, Beutler B. Defective LPS signaling in C3H/HeJ and C57BL/10ScCr mice: mutations in Tlr4 gene. Science. 1998a;282:2085–2088. doi: 10.1126/science.282.5396.2085. [DOI] [PubMed] [Google Scholar]
- Poltorak A, Smirnova I, He X, Liu MY, Van Huffel C, McNally O, Birdwell D, Alejos E, Silva M, Du X, Thompson P, Chan EK, Ledesma J, Roe B, Clifton S, Vogel SN, Beutler B. Genetic and physical mapping of the Lps locus: identification of the toll-4 receptor as a candidate gene in the critical region. Blood Cells Mol Dis. 1998b;24:340–355. doi: 10.1006/bcmd.1998.0201. [DOI] [PubMed] [Google Scholar]
- Pruneda JN, Smith FD, Daurie A, Swaney DL, Villen J, Scott JD, Stadnyk AW, Le Trong I, Stenkamp RE, Klevit RE, Rohde JR, Brzovic PS. E2~Ub conjugates regulate the kinase activity of Shigella effector OspG during pathogenesis. EMBO J. 2014 doi: 10.1002/embj.201386386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebeil R, Ernst RK, Gowen BB, Miller SI, Hinnebusch BJ. Variation in lipid A structure in the pathogenic yersiniae. Mol Microbiol. 2004;52:1363–1373. doi: 10.1111/j.1365-2958.2004.04059.x. [DOI] [PubMed] [Google Scholar]
- Rohde JR, Breitkreutz A, Chenal A, Sansonetti PJ, Parsot C. Type III secretion effectors of the IpaH family are E3 ubiquitin ligases. Cell Host Microbe. 2007;1:77–83. doi: 10.1016/j.chom.2007.02.002. [DOI] [PubMed] [Google Scholar]
- Sanada T, Kim M, Mimuro H, Suzuki M, Ogawa M, Oyama A, Ashida H, Kobayashi T, Koyama T, Nagai S, Shibata Y, Gohda J, Inoue J, Mizushima T, Sasakawa C. The Shigella flexneri effector OspI deamidates UBC13 to dampen the inflammatory response. Nature. 2012;483:623–626. doi: 10.1038/nature10894. [DOI] [PubMed] [Google Scholar]
- Selyunin AS, Sutton SE, Weigele BA, Reddick LE, Orchard RC, Bresson SM, Tomchick DR, Alto NM. The assembly of a GTPase-kinase signalling complex by a bacterial catalytic scaffold. Nature. 2011;469:107–111. doi: 10.1038/nature09593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takaoka A, Shinohara S. DNA sensors in innate immune system. Uirusu. 2008;58:37–46. doi: 10.2222/jsv.58.37. [DOI] [PubMed] [Google Scholar]
- Thanabalasuriar A, Bergeron J, Gillingham A, Mimee M, Thomassin JL, Strynadka N, Kim J, Gruenheid S. Sec24 interaction is essential for localization and virulence-associated function of the bacterial effector protein NleA. Cell Microbiol. 2012;14:1206–1218. doi: 10.1111/j.1462-5822.2012.01789.x. [DOI] [PubMed] [Google Scholar]
- Uematsu S, Akira S. Toll-like receptors and Type I interferons. J Biol Chem. 2007;282:15319–15323. doi: 10.1074/jbc.R700009200. [DOI] [PubMed] [Google Scholar]
- van Vliet SJ, den Dunnen J, Gringhuis SI, Geijtenbeek TB, van Kooyk Y. Innate signaling and regulation of Dendritic cell immunity. Curr Opin Immunol. 2007;19:435–440. doi: 10.1016/j.coi.2007.05.006. [DOI] [PubMed] [Google Scholar]
- Wei P, Wong WW, Park JS, Corcoran EE, Peisajovich SG, Onuffer JJ, Weiss A, Lim WA. Bacterial virulence proteins as tools to rewire kinase pathways in yeast and immune cells. Nature. 2012;488:384–388. doi: 10.1038/nature11259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Jones RM, Neish AS. The Salmonella effector AvrA mediates bacterial intracellular survival during infection in vivo. Cell Microbiol. 2012;14:28–39. doi: 10.1111/j.1462-5822.2011.01694.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan D, Wang X, Luo L, Cao X, Ge B. Inhibition of TLR signaling by a bacterial protein containing immunoreceptor tyrosine-based inhibitory motifs. Nat Immunol. 2012;13:1063–1071. doi: 10.1038/ni.2417. [DOI] [PubMed] [Google Scholar]
- Yen H, Ooka T, Iguchi A, Hayashi T, Sugimoto N, Tobe T. NleC, a type III secretion protease, compromises NF-kappaB activation by targeting p65/RelA. PLoS Pathog. 2010;6:e1001231. doi: 10.1371/journal.ppat.1001231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Ding X, Cui J, Xu H, Chen J, Gong YN, Hu L, Zhou Y, Ge J, Lu Q, Liu L, Chen S, Shao F. Cysteine methylation disrupts ubiquitin-chain sensing in NF-kappaB activation. Nature. 2012;481:204–208. doi: 10.1038/nature10690. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Jimi E, Bothwell AL. Receptor activator of NF-kappa B ligand stimulates recruitment of SHP-1 to the complex containing TNFR-associated factor 6 that regulates osteoclastogenesis. J Immunol. 2003;171:3620–3626. doi: 10.4049/jimmunol.171.7.3620. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Yang J, Shi J, Gong YN, Lu Q, Xu H, Liu L, Shao F. The NLRC4 inflammasome receptors for bacterial flagellin and type III secretion apparatus. Nature. 2011;477:596–600. doi: 10.1038/nature10510. [DOI] [PubMed] [Google Scholar]
- Zhou Y, Dong N, Hu L, Shao F. The Shigella type three secretion system effector OspG directly and specifically binds to host ubiquitin for activation. PLoS One. 2013;8:e57558. doi: 10.1371/journal.pone.0057558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Li H, Long C, Hu L, Xu H, Liu L, Chen S, Wang DC, Shao F. Structural insights into the enzymatic mechanism of the pathogenic MAPK phosphothreonine lyase. Mol Cell. 2007;28:899–913. doi: 10.1016/j.molcel.2007.11.011. [DOI] [PubMed] [Google Scholar]