Schizophrenia, a severe brain disease with a prominent genetic basis, has been suggested to have a neurodevelopmental origin.1 Although many candidate susceptibility genes for schizophrenia have been identified,2 access to human neural progenitors and neurons to study the biology of disease genes is very limited. Induced pluripotent stem cells (iPSCs), reprogrammed from somatic cells of healthy subjects and patients,3 offer an unprecedented opportunity to recapitulate both normal and pathologic human development, thereby enabling a new approach to understanding human disease mechanisms.
Rare, large pedigrees in which a single genetic locus is likely to confer disease susceptibility have proven to be invaluable for the study of complex disorders.4 Disrupted-in-Schizophrenia 1 (DISC1) was initially identified at the breakpoint of a balanced t(1;11)(q42;q14) chromosomal translocation that segregates with schizophrenia, bipolar disorder and recurrent major depression in a large Scottish family.5,6 Linkage and association studies of additional populations have provided independent evidence to support the DISC1 locus as a risk factor for major mental disorders.2 How DISC1 contributes to a spectrum of mental disorders is unknown. Studies from rodents have implicated DISC1 in many neurodevelopmental processes.7 The splicing and expression of DISC1 in postmortem brains from individuals with schizophrenia is abnormal.8 Little is known about the function of human DISC1, which is significantly different from the rodent gene.8 We have previously identified an American family with schizophrenia and a 4 bp deletion in DISC1 that appears to result in diminished DISC1 expression.9 Here, we report the derivation of multiple iPSC lines from two affected siblings of this family with the DISC1 mutation, and from a healthy individual.
We collected skin biopsies from a male patient (III:5; named D1) and his sister (III:7, named D2) who were diagnosed with chronic undifferentiated schizophrenia and chronic paranoid schizophrenia, respectively.9 Both patients experienced auditory and visual hallucinations, multiple delusions and had formal thought disorder.9 Male neonatal foreskin fibroblasts from American Type Culture Collection were used in parallel (CRL-2097; named C1). We confirmed normal karyotypes of all fibroblasts and the presence of the 4 bp deletion at the exon-intron 12 region in D1 and D2, but not C1 fibroblasts (Supplementary Figure 1; Supplementary Table 1).
To avoid integration of foreign DNA into the host genome associated with classic virus-based methods of iPSC derivation,3 we explored the episomal vector approach, which was previously used to generate integration-free iPSCs from fetal human skin fibro-blasts.10 Because of its low efficacy, no iPSC lines have yet been reported to be derived from adult patients using this approach. We transfected a mixture of reprogramming plasmids into fibroblasts by electroporation and plated them onto freshly prepared feeder layers (see Supplementary Information). Colonies of iPSCs were manually picked after 3–6 weeks for expansion and subjected to extensive characterization. Multiple lines were examined and a minimum of two lines from each fibroblast that exhibited characteristic properties of iPSCs were obtained (Figure 1).
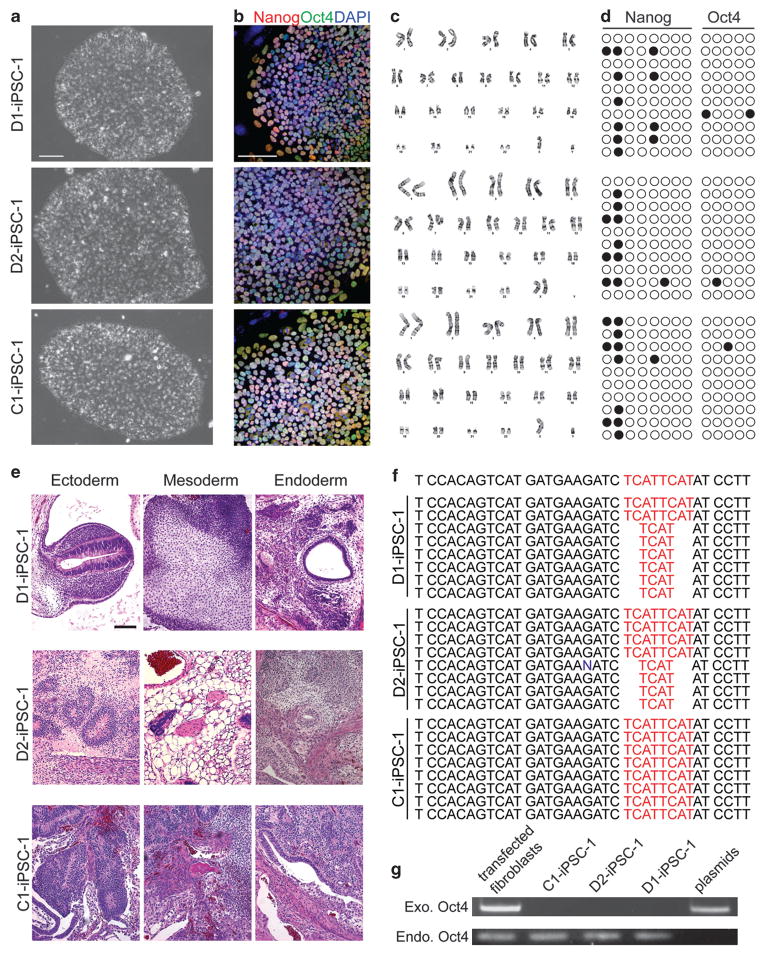
Figure 1.
Characterization of iPSC lines from two schizophrenia patients with a DISC1 mutation and a healthy subject. (a) Sample phase images of iPSCs derived from two schizophrenia patients with a DISC1 mutation (D1 and D2) and from a control subject (C1). Scale bar: 100 μm. (b) Sample confocal images of immunostaining of pluripotency markers, Nanog and Oct4, and DAPI. Scale bar: 100 μm. (c) Normal karyotypes of derived iPSCs. (d) Methylation status of CpGs in the promoter regions of Nanog and Oct4. Open and close circles represent unmethylated and methylated CpGs, respectively. (e) Sample H&E staining images of teratomas formed by iPSCs injected into adult SCID/Beige mice. Scale bar: 100 μm. (f) Genomic sequencing analysis of the 4 bp deletion in iPSCs. (g) PCR-based demonstration of the lack of plasmid sequences in the genomic DNA of iPSCs.
All iPSCs exhibited typical morphology of human embryonic stem cells (hESCs), normal karyotypes and expression of a panel of markers for human pluripo-tent stem cells, including Nanog, Oct4, Sox2, SSEA3, SSEA4, TRA-1-60, TRA-1-81 and TRA-2-49 (Figures 1a–c; Supplementary Figure 2). Successful reprogramming requires demethylation of CpGs in promoter regions of pluripotent genes, which are normally heavily methylated in fibroblasts.3 Bisulfite sequencing analysis of genomic DNA from iPSCs demonstrated the largely unmethylated status of CpGs in Nanog and Oct4 promoters (Figure 1d). The teratoma assay was used to determine the pluripotency of iPSCs after injection into adult SCID/Beige mice. Tumors were harvested 1–3 months later and histo-logical analysis showed the differentiation of iPSCs into ectodermal, mesodermal and endodermal tissues (Figure 1e; Supplementary Figure 3). Sequencing analysis confirmed the 4 bp deletion in D1 and D2, but not C1 iPSCs (Figure 1f). Using an established PCR-based detection method,10 no signal for reprogramming plasmids was found in genomic DNA of iPSCs (Figure 1g). Together, our analyses demonstrated that these cell lines are bona fide iPSCs that maintain genetic characteristics of original subjects.
To our knowledge, this study presents the first iPSC lines derived from schizophrenia patients. In addition, it presents the first cases of generation of integration-free iPSCs from adult patients using the episomal vector approach.10 Although significant effort has been made towards the derivation of iPSCs free of foreign DNA integration to avoid the disruption of host genome and potential reactivation of oncogenes used for reprogramming, these methods are generally much less efficient.10 Our study demonstrates the feasibility of generating high quality integration-free iPSCs from adult patients. These established iPSC lines from schizophrenia patients with a defined DISC1 mutation will provide a useful resource for investigating the function of DISC1 in human neurodevelopment. Future studies using these iPSCs may serve as an entry point to clarify the molecular and cellular pathogenesis of schizophrenia.
Supplementary Material
Footnotes
Conflict of interest
The authors declare no conflict of interest.
Supplementary Information accompanies the paper on the Molecular Psychiatry website (http://www.nature.com/mp)
Contributor Information
RL Margolis, Email: rmargolis@jhmi.edu.
H Song, Email: shongju1@jhmi.edu.
G-l Ming, Email: gming1@jhmi.edu.
References
- 1.Weinberger DR. Arch Gen Psychiatry. 1987;44:660–669. doi: 10.1001/archpsyc.1987.01800190080012. [DOI] [PubMed] [Google Scholar]
- 2.Ross CA, Margolis RL, Reading SA, Pletnikov M, Coyle JT. Neuron. 2006;52:139–153. doi: 10.1016/j.neuron.2006.09.015. [DOI] [PubMed] [Google Scholar]
- 3.Park IH, Arora N, Huo H, Maherali N, Ahfeldt T, Shimamura A, et al. Cell. 2008;134:877–886. doi: 10.1016/j.cell.2008.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McClellan JM, Susser E, King MC. Br J Psychiatry. 2007;190:194–199. doi: 10.1192/bjp.bp.106.025585. [DOI] [PubMed] [Google Scholar]
- 5.St Clair D, Blackwood D, Muir W, Carothers A, Walker M, Spowart G, et al. Lancet. 1990;336:13–16. doi: 10.1016/0140-6736(90)91520-k. [DOI] [PubMed] [Google Scholar]
- 6.Millar JK, Wilson-Annan JC, Anderson S, Christie S, Taylor MS, Semple CA, et al. Hum Mol Genet. 2000;9:1415–1423. doi: 10.1093/hmg/9.9.1415. [DOI] [PubMed] [Google Scholar]
- 7.Chubb JE, Bradshaw NJ, Soares DC, Porteous DJ, Millar JK. Mol Psychiatry. 2008;13:36–64. doi: 10.1038/sj.mp.4002106. [DOI] [PubMed] [Google Scholar]
- 8.Nakata K, Lipska BK, Hyde TM, Ye T, Newburn EN, Morita Y, et al. Proc Natl Acad Sci USA. 2009;106:15873–15878. doi: 10.1073/pnas.0903413106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sachs NA, Sawa A, Holmes SE, Ross CA, DeLisi LE, Margolis RL. Mol Psychiatry. 2005;10:758–764. doi: 10.1038/sj.mp.4001667. [DOI] [PubMed] [Google Scholar]
- 10.Yu J, Hu K, Smuga-Otto K, Tian S, Stewart R, Slukvin II, et al. Science. 2009;324:797–801. doi: 10.1126/science.1172482. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.