Abstract
Japanese encephalitis virus (JEV) contains a single positive-strand RNA genome nearly 11 kb in length and is not formally thought to generate subgenomic RNA molecules during replication. Here, we report the abundant accumulation of a 3′-terminal 521- to 523-nucleotide (nt) genome fragment, representing a major portion of the 585-nt 3′ untranslated region, in both mammalian (BHK-21) and mosquito (C6/36) cells infected with any of nine strains of JEV. In BHK-21 cells, the viral genome was detected as early as 24 h postinfection, the small RNA was detected as early as 28 h postinfection, and the small RNA was 0.25 to 1.5 times as abundant as the genome on a molar basis between 28 and 48 h postinfection. In C6/36 cells, the genome and small RNA were present 5 days postinfection and the small RNA was 1.25 to 5.14 times as abundant as the genome. The 3′-terminal 523-nt small RNA contains a 5′-proximal stable hairpin (nt 6 to 56) that may play a role in its formation and the conserved flavivirus 3′-cyclization motif (nt 413 to 420) and the 3′-terminal long stable hairpin structure (nt 440 to 523) that have postulated roles in genome replication. Abundant accumulation of the small RNA during viral replication in both mammalian and mosquito cells suggests that it may play a biological role, perhaps as a regulator of RNA synthesis.
Japanese encephalitis virus (JEV) is a mosquito-borne flavivirus found primarily in areas of Asia. The virus has a normal transmission cycle between birds and mosquitoes but also a zoonotic transmission cycle with swine serving as amplifier hosts from which infected mosquitoes transmit the virus to humans. In humans, JEV can cause acute meningioencephalomyelitis, resulting in fatality rates of 5 to 40% (6, 7). An effective formalin-inactivated vaccine is available, but multiple doses are required to confer protective immunity and some pathogenic side effects have been reported (21, 42). JEV is thus a continuing public health threat (28, 39).
The JEV genome is a positive, single-stranded RNA of 10,951 to 10,978 nucleotides (nt) that contains a type 1 5′ cap but no 3′ poly(A) tail and undergoes replication entirely in the cytoplasm. The genome contains a single large open reading frame flanked by a 95-nt 5′ untranslated region (UTR) and a 585-nt 3′ UTR in the 10,976-nt genome of a majority of JEV strains. The open reading frame is translated as a single polyprotein that undergoes co- and posttranslational processing to yield three structural proteins called capsid (C), premembrane (prM), and envelope (E) and seven nonstructural (NS) proteins called NS1, NS2A/B, NS3, NS4A/B, and NS5 (reviewed in reference 23). Although the details of JEV RNA replication have not yet been characterized, it is thought that most flaviviruses share a common replication strategy (23). Three viral RNA species found in flavivirus-infected cells have been characterized: a single-stranded 40 to 44S genomic RNA, a double-stranded genome-length replicative form, and a genome-length partially double-stranded replicative intermediate (5, 11, 12, 38, 40). The minus strand in the replicative form presumably functions as a recycling template for the generation of progeny positive-strand genomes.
Although subgenomic RNAs are not formally thought to be products of the flavivirus replication cycle, some studies have shown the existence of small (8 to 15S) viral RNA species in cell cultures infected with St. Louis encephalitis virus (29), West Nile virus (WNV) (43), and JEV (37, 38), but only those of WNV have been characterized in detail. Wengler and Gross (43) reported that WNV produces RNAs of 6.5 × 104 and 4.2 × 104 Da (∼197 and 127 nt in length, respectively) in BHK-21 cells, but only the larger species was detected in insect cells. The larger species was found to be a virus-specific positive-strand RNA with no cap or poly(A) tail that was inactive as a template in cell-free translation systems. It was postulated that the smaller RNA was probably nonfunctional subgenomic mRNA, but its origin, sequence, and function were not determined. Takegami and Hotta (38) reported a 10S plus-strand RNA species in both JEV-infected C6/36 and Vero cells by Northern analysis using an in vitro-transcribed RNA probe specific to genomic nt 9183 to 10883, but the RNA was not characterized further. Recently, an RNA of 600 nt which hybridizes to a 3′-terminal 1.7-kb fragment of Murray Valley encephalitis virus (MVEV) RNA was detected in the brains of infected mice (41); however, its structure, sequence, and function were not addressed.
In addition to the small RNAs, naturally occurring defective interfering RNA replicons (DI RNAs) have been reported for WNV (3, 4, 14), MVEV (22), and JEV (33), but only the molecular structure of MVEV DI RNA has been determined (22). The MVEV DI RNAs present in persistently infected Vero cells were all shown to have in-frame internal deletions of 2.3 to 2.6 kb from the prM, E, and NS1 genes of the virus genome. The DI RNAs had thus retained the presumed terminal replication promoter regions of the viral genome, the entire C gene, and genes for the nonstructural proteins.
In this study, we report a JEV-specific RNA molecule of 521 to 523 nt in both mammalian and mosquito cells infected with any of nine strains of JEV. The small RNA did not have properties of a DI RNA replicon but rather was found to be identical to the 3′-terminal 521 to 523 nt of the genome. This sequence represents the highly conserved 3′-terminal region in the 3′ UTRs of all characterized JEV isolates. We thus confirm and extend the observations of Takegami and Hotta (38) by establishing the structure of the small RNA and by showing that it is a consistent product of JEV replication in both mammalian and mosquito cells. We postulate, furthermore, that the small RNA plays a biological role, perhaps as a competitor of the virus genome 3′ terminus during genome replication.
MATERIALS AND METHODS
Cells and viruses.
BHK-21 mammalian cells were grown in RPMI 1640 medium supplemented with 2% fetal bovine serum (Gibco-BRL). C6/36 mosquito cells, derived from Aedes albopictus (18), were grown in Dulbecco's modified Eagle's medium supplemented with 2% fetal bovine serum (Gibco-BRL). The viruses used were JEV RP-2 and JEV RP-9, mutants selected from a γ-irradiated strain, JEV NT109, originally isolated from Culex tritaeniorhynchus (10). JEV TP-53 (Taipei County, Taiwan, 1994), JEV CH-1392 (Chang-Hwa County, Taiwan, 1990), and JEV CC-27 (Chao-Chow, Pingtung County, Taiwan, 1983) were isolates collected from mosquitoes in northern, central, and southern areas of Taiwan, respectively. JEV S-26 (Miao-Li County, Taiwan, 1994) and JEV PC-806 (Chao-Chow, Pingtung County, Taiwan, 1990) were isolated from infected pigs in the indicated counties. The vaccine strains Beijing-1 (China, 1949) and Nakayama (Japan, 1935) were originally isolated from infected human brain (16, 26). For a majority of the GenBank-catalogued JEV strains, including Beijing-1 and JEV RP9, the genome length is 10,976 nt.
Northern analysis.
For studies of BHK-21 cells, cells infected at the appropriate multiplicities of infection (MOI) were incubated at 37°C for 1 h and refed with RPMI 1640 medium supplemented with 2% fetal bovine serum, and total RNA was extracted with Trizol reagent (Invitrogen) at the appropriate times postinfection (at 4- or 8-h intervals from 24 to 48 h postinfection). The yield of cytoplasmic RNA from one 35-mm-diameter dish was ∼10 μg, and 2.5 μg was used per lane for formaldehyde-agarose gel electrophoresis as described previously (8, 34). For studies of C6/36 cells, the same procedure was used as for BHK-21 cells except that the cells were grown at 28°C and infected at an MOI of 0.1, and the cytoplasmic RNA was extracted 5 days postinfection. For probe labeling for Northern analysis, an ∼100-pmol oligonucleotide was tailed with digoxigenin (DIG)-ddUTP using a DIG Oligonucleotide 3′-End Labeling kit (Roche Molecular Biochemicals). Hybridization and chemoluminescence detection of the virus-specific bands with DIG-labeled oligonucleotides were done following the procedures of the manufacturer. Quantitation of the bands was done with the Phoretix (Newcastle upon Tyne, United Kingdom) 1D, version 3.0, imaging system.
Primer extension analysis.
The method used for primer extension analysis was that described by Sambrook et al. (32). Five pmol of 32P-labeled oligonucleotide 3JEV10543(−) was coprecipitated in ethanol with 5 μg of template RNA, and the dried RNA was dissolved in 10 μl of water. The RNA mixture was heated for 5 min at 90°C, incubated at 42°C for 30 min, and further incubated for 1 h at 42°C in a 30-μl reaction mixture containing 1× first-strand buffer (Invitrogen), 1 mM deoxynucleoside triphosphates, 30 U of RNasin (Promega), and 200 U of Superscript II (Invitrogen). The extended products were analyzed by electrophoresis on a 6% polyacrylamide gel and imaged using autoradiography.
RACE analysis.
5′ rapid amplification of cDNA ends (RACE) (Invitrogen) was carried out according to the manufacturer's protocol. Cytoplasmic RNA extracted using Trizol agent (Invitrogen) from JEV-infected BHK-21 cells at 48 h postinfection was used as a source of templates. For this, 5 μg of RNA was used for first-strand cDNA synthesis with oligonucleotide 3JEV10950(−), and the cDNA products were C tailed using dCTP and terminal deoxynucleotidyl transferase. Five microliters of C-tailed cDNA was used for PCR amplification (30 cycles of 94°C for 1 min, 55°C for 1 min, and 72°C for 1 min, followed by final extension at 72°C for 7 min) with primer 3JEV10929(−) and the 5′ RACE anchor primer. PCR products ∼520 nt in size were gel purified and subsequently cloned into a pGEM-T Easy vector (Promega) following the manufacturer's instructions. Plasmid DNA was sequenced commercially with an ABI PRISM 377-97 DNA sequencer (Protech Technology Enterprise Co., Ltd.).
Synthetic oligonucleotides.
The oligonucleotides used in this study are described in Table 1.
TABLE 1.
Mapping of small RNA by Northern blotting
| Oligonucleotidea | Oligonucleotide sequence (5′-3′)b | Genome map positionc | Oligonucleotide probe identifies small RNA by Northern blottingd |
|---|---|---|---|
| 5JEVT7RI(+) | ccggaattctaatacgactcactatagAGAAGTTTATCTGTGTGAACTTCTTG | 1-26 | No |
| JEV Core96(+) | ggaattcATGACTAAAAAACCAGGAGG | 96-115 | No |
| 5NS2a3681(+) | ACTTACACTGATTTGGCGAG | 3681-3700 | No |
| 5JEV9201(+) | GTGGAAGGCTCAGGCGTCCAAAAGCTG | 9201-9227 | No |
| 3JEV10950(+) | CTGGTGGTGAGGAAGAACACAGGATCT | 10950-10976 | No |
| 5JEV8(−) | AAGCCAAGAAGTTCACACAGATA | 8-30 | No |
| 3JEV5617(−) | CCCTGTCTGGTATCTCATCCT | 5617-5637 | No |
| 3JEV9431(−) | CATCACGGTCTTTCCTTCTGCTGCA | 9431-9455 | No |
| NS5/10371(−) | ttaaggatccCTAGATGACCCTGTCTTCCTGGAT | 10371-10394 | No |
| 3JEV10421(−) | GCATTTTCTCATTTACATTATTTACAT | 10421-10447 | No |
| 3JEV10453(−) | CAGCTTTTGCTGGCCTGACTCCATA | 10453-10477 | Yes |
| 3JEV10543(−) | AGGGCTTTCTCACTTTCTATTGTCA | 10543-10567 | Yes |
| JEV10731(−) | AGTCCTTACACCTCGTCTATT | 10731-10751 | Yes |
| JEV10809(−) | AGTCCTTCCACCTCCTCTACA | 10809-10829 | Yes |
| 3JEV10929(−) | GCTACATACTTCGGCGCTCTGT | 10929-10950 | Yes |
| 3JEV10950(−) | AGATCCTGTGTTCTTCCTCACCACCAG | 10950-10976 | Yes |
Polarity of oligonucleotide with plus or minus sense is indicated by + or −, respectively.
Uppercase letters represent JEV sequence. The underlined sequences represent restriction endonuclease sites, and italics represent the T7 promoter.
Sequence numbers correspond to bases in the positive-strand JEV RP9 genome.
Conditions of the Northern assay are described in the text.
RNA structure predictions.
Higher-order RNA structure predictions were made by the mfold program of M. Zuker (25, 45; http://bioinfo.rpi.edu/∼zukerm/).
Nucleotide sequence accession numbers.
JEV genome nucleotide positions correspond to those for JEV RP9 deposited in GenBank under accession number AF014161.
RESULTS
Detection of JEV small-RNA species whose kinetics of appearance parallels that of the genome.
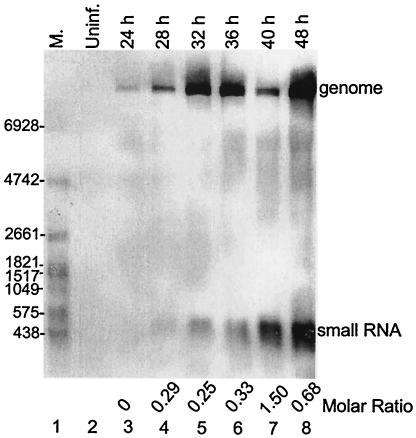
Initially, a small viral RNA species ∼500 nt in length was observed when RNA from JEV RP-9-infected BHK-21 cells was examined 48 h postinfection by Northern analysis with a probe detecting genomic 3′-terminal sequence (data not shown). To determine the kinetics of appearance and the relative abundance of the small RNA and the virus genome, cells were infected with JEV RP-9 at an MOI of 0.01 and the RNA was examined at 4- or 8-h intervals through 48 h postinfection, which is near the time of maximal cytopathic effect under these conditions. The probe used was 3JEV10950(−), which detects RNA containing the very 3′-terminal 27 nt of the JEV genome (Table 1 and Fig. 1). From Fig. 1, lane 3, it can be seen that genomic RNA appeared as early as 24 h postinfection, and its abundance continued to increase throughout the 48-h infection period, except for a diminution at 40 h postinfection. The small RNA species appeared as early as 28 h postinfection (Fig. 1, lane 4) and likewise continued to increase in abundance throughout the 48-h infection period. We concluded that the small RNA was of viral origin, since it was not present in mock-infected cells at 48 h (Fig. 1, lane 2). Furthermore, since the sequence of the small RNA is contained only once in the virus genome (described below), the molar ratios of the small RNAs to genomic RNA at the various times could be determined by a simple ratio analysis of image densities. Thus, the ratios of small RNA to genome ranged from 0.25 at 32 h postinfection to 1.5 at 40 h postinfection (Fig. 1).
FIG. 1.
Detection of small RNA in JEV-infected mammalian cells. Cytoplasmic RNA was extracted from uninfected BHK-21 cells and from cells infected with JEV RP-9 at the indicated times postinfection, and Northern analysis was done with a probe detecting nt 10950 to 10976 in the 3′ UTR. M., DIG-labeled RNA molecular size marker with the number of bases indicated on the left; Uninf., uninfected cells. The positions of the viral genome and the small RNA are indicated on the right. The molar ratios of small RNA to genome RNA are indicated at the bottom.
The small RNA does not have properties of a DI RNA.
To test whether the small RNA was a DI RNA, two experimental approaches were taken. The first was biological and attempted to find a disappearance of the small RNA upon high dilution of the virus inoculum, and conversely, to find an increased inhibition of virus genome replication at lower dilutions of the inoculum. The rationale for this approach was based on the classic observations that DI RNAs lose their competitive advantage over the genome when the DI RNA-containing virus inoculum is diluted (17). The second approach was a structural analysis to determine whether the termini of the small RNA mimicked the terminal promoter regions of the viral genome.
To measure the potential effects of differing inoculum dilutions on small-RNA and genome abundances, BHK-21 cells were infected with JEV RP-9 at MOI of 1, 0.1, 0.01, and 0.001 and the molar ratios of the small and genomic RNAs were determined 24 and 48 h postinfection. In all, the abundance of the small RNAs relative to the genome stayed within the range of 0.11 to 0.35, indicating that the ratio did not change significantly as a function of the inoculum dilution (data not shown). The ratios also did not deviate from a range of 0.25 to 0.69 when supernatant from a dish inoculated at an MOI of 0.01 was diluted 1,000-fold at each of four 48-h serial passages (data not shown). These results together demonstrated that the small RNA does not behave biologically as a DI RNA.
The structure of the small-RNA species was determined by Northern, primer extension, and 5′ RACE analyses. For Northern analyses, 16 probes that would variously detect positive or negative strands in the regions of the 3′ and 5′ genomic termini and in various regions throughout the genome were used on RNA extracted 48 h postinfection from BHK-21 cells infected with JEV RP-9 at an MOI of 0.01 (the data are summarized in Table 1). None of the minus-strand-detecting probes identified a small RNA, whereas they did identify the minus strand of the genome, and only positive-strand-detecting probes mapping downstream of nt 10,453 identified the small RNA. These data together indicated that the small RNA was of plus-strand polarity and was probably located only within the 3′ UTR.
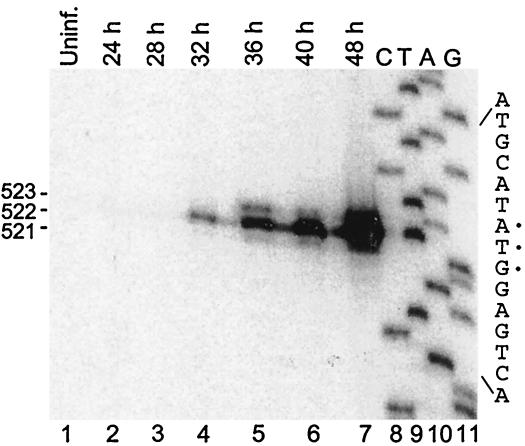
In primer extension analyses, the radiolabeled oligonucleotide 3JEV10543(−) was used to determine the map position of the 5′ termini of the small RNA. The results showed that the 5′ ends of most small RNA molecules started at nt 10,455 early in infection (28 to 32 h postinfection), and as the infection progressed, two other small RNAs appeared with 5′ nucleotides terminating at positions 10,454 and 10,456 (Fig. 2). The largest small-RNA species, therefore, was 523 nt in length.
FIG. 2.
Primer extension analysis of the small RNA. The cytoplasmic RNAs shown in Fig. 1 were used as templates for the extension of the 32P-labeled 3JEV10543(−) probe. The DNA used for preparing the sequencing ladder was pGEM-T vector containing a 1.7-kb 3′-proximal cDNA insert from the JEV genome. The sizes of the extended products are indicated as bases.
For complete nucleotide sequence determination of the small RNA, including the 5′-most base, the method of 5′ RACE was used. RACE PCR products ∼520 nt in size were gel purified and cloned into a pGEM-T Easy vector. Of 12 completely sequenced transformants, 5 contained the predicted insert of ∼520 nt, with 3 starting at nt 10,455 and 2 starting at nt 10,456. The seven other clones had 5′ ends starting at distant sites, indicating that they were probably derived from viral genomic RNA. Results from Northern analyses, primer extension, and 5′ RACE sequencing therefore determined that the small RNA was 521 to 523 nt in length and corresponded to the 3′ terminus of the genome.
The small RNA is generated in both mammalian and mosquito cells by all JEV isolates tested.
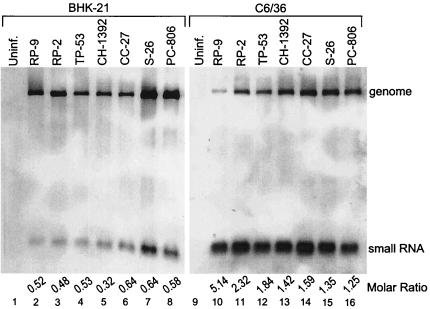
The small RNA was initially detected in BHK-21 cells with isolates JEV RP-2 and JEV RP-9 (Fig. 1 and 3, and data not shown), both laboratory-derived strains. To determine whether generation of the small RNA in mammalian cells is a general property of JEV and not just a property of laboratory-derived strains, wild-type JEV isolates from various regions of Taiwan, namely, JEV TP-53, JEV CH-1392, JEV CC-27, JEV S-26, and JEV PC-806, were tested in BHK-21 cells. From RNA extracted at 48 h postinfection, it can be seen that the small RNA was generated by each of the five JEV field isolates (Fig. 3, lanes 4 to 8). In addition, the small RNA was detected in BHK-21 cells infected with the JEV vaccine strains Beijing-1 and Nakayama isolated from China and Japan, respectively (data not shown). These results, therefore, demonstrated that generation of the small RNA is a general property of JEV and is not an idiosyncrasy of laboratory strains of JEV.
FIG. 3.
Detection of the small RNA in both mammalian (BHK-21) and mosquito (C6/36) cells infected with different isolates of JEV. Cytoplasmic RNA was extracted from JEV-infected BHK-21 cells 48 h postinfection (lanes 2 to 8) and from JEV-infected C6/36 cells 5 days postinfection (lanes 10 to 16). Lanes 1 and 9 contain cytoplasmic RNA extracted from uninfected (Uninf.) cells. Northern analysis was done with a probe detecting nt 10950 to 10976 in the 3′ UTR.
To determine whether generation of the small RNA is a special property of mammalian cells, its existence in mosquito cells was sought. Since growth of JEV in C6/36 cells is slower than in BHK-21 cells, RNA for Northern analysis was extracted 5 days postinfection from cells infected with an MOI of 0.1. This is near the time of maximal cytopathic effect in mosquito cells under these conditions. Analysis of both the laboratory strain and five field strains of JEV in mosquito cells demonstrated the generation of the small RNA (Fig. 3, lanes 10 to 16). Interestingly, the small-RNA/genomic-RNA molar ratios were greater in mosquito cells (ranging from 1.25 to 5.14) than in mammalian cells (ranging from 0.25 to 1.50) when measured at a time just prior to cell death.
DISCUSSION
In this paper, we identify and determine the structure of a small-RNA species generated in JEV-infected cells of mammalian or mosquito origin. The small RNA represents the very 3′-terminal 521 to 523 nt of the JEV 585-nt 3′ UTR and is present in the range of 0.25 to 5.14 molar ratio relative to the virus genome. Because of the cytolytic nature of JEV infection in mammalian cells and the possibility of isopycnic copurification of cellular replication complexes with virions, we were unable to conclusively determine whether the small RNA observed by Northern analysis in purified virion preparations is packaged. Certainly the small RNA was present in material purified by this method (data not shown), but further experimentation will be required to rigorously answer the question of packaging.
What is the origin of the small RNA? Inasmuch as the abundance of the small RNA was not influenced by inoculum dilution, we conclude that it does not represent a defective or DI RNA replicon. This conclusion was supported by its structure, which shows it to represent only the 3′-terminal 521 to 523 nt of the virus genome. On the basis of the experiments presented here, we conclude that the small RNA is synthesized de novo in each infection and that the mechanism of its origin is not an idiosyncrasy of any particular JEV strain or host cell type.
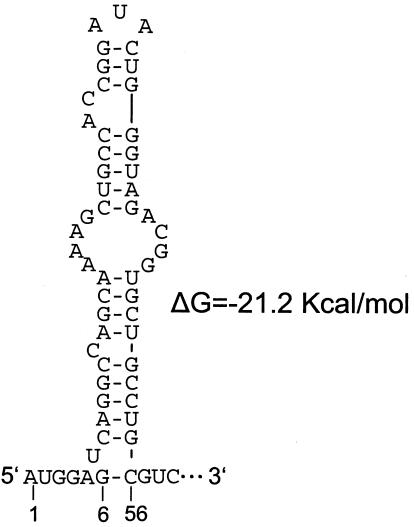
We speculate that the small RNA is made either by transcription from a full-length minus strand through an internal promoter or by replication of the viral genome followed by RNase-specific cleavage. The first scenario seems unlikely, since transcripts of this size would require an internal promoter located upstream of nt 10454, and no sequence has been found by inspection that would appear to mimic the 5′ end of the genome or otherwise suggest a promoter. The second scenario seems more likely, since nt 6 through 56 in the 523-nt RNA are predicted to fold into a stable stem-loop structure (free energy, −21.2 kcal/mol) (Fig. 4) that could in principle serve as a region of RNase resistance or could associate with viral or cellular protein(s) to form an RNase-resistant complex.
FIG. 4.
mfold-predicted 5′-terminal hairpin on the small RNA. The stable bulged 5′-terminal hairpin (nt 6 to 56 of the small RNA, corresponding to nt 10459 to 10509 in the JEV RP9 genome), as it is predicted by the m-fold algorithm of Zuker, is depicted.
What is the function of the small RNA? Previous studies of JEV infection in mosquito C6/36 cells have shown an early peaking of negative-strand synthesis, i.e., at 12 h postinfection, with subsequent shut-off of minus-strand synthesis and a continued synthesis of positive-strand RNA (38). A similar temporal regulation of RNA synthesis in porcine kidney cells was not observed, apparently due to extensive rapid cell death (40). Assuming that early inhibition of minus-strand synthesis is a universal pattern in JEV replication, we suggest that the small RNA described here may play a role in its inhibition.
The 3′ UTRs of mosquito-borne flaviviruses vary from ∼400 to 600 nt in length, and within each subgroup, the 3′ UTR can be divided into a highly variable 5′-terminal subdomain and a highly conserved 3′-terminal subdomain that in turn harbors a number of conserved higher-order structural elements (15, 24, 27, 30, 31). In JEV, on the basis of alignment of all 28 GenBank-deposited JEV genome sequences, these subdomains are formed by sequences that are 64 and 521 nt in length, respectively (19, 24, 30). It is therefore striking that the small RNA precisely represents the highly conserved region of the 3′ UTR. Based on documented functions of the conserved 3′-terminal subdomain in flaviviruses, two functions might be expected for which the small RNA could mechanistically compete. (i) In JEV, an 8-nt cyclization motif is found, mapping at genome nt 10,866 to 10,873, that potentially interacts with genome nt 136 to 143 (15). A similar interaction in yellow fever virus between genome nt 10,751 to 10,758 and 156 to 163 has been shown to be required for genome replication (13), and genome cyclization has been shown to be required for replication in Kunjin (20) and dengue (44) viruses. (ii) A 3′-long stable hairpin structure located within the 3′-terminal 83 nt interacts with both viral and cellular proteins and is required for RNA synthesis (1, 2, 9, 36). In JEV, viral NS3 and NS5 proteins and the cellular protein Mov 34 bind to the 3′ long stable hairpin (9, 36). Thus, in principle, inhibition of either of these functions by the small RNA could lead to a down regulation of minus-strand RNA synthesis and continued synthesis of positive-strand RNA.
In recent years, many small noncoding RNA molecules that play novel unpredicted roles in virus replication processes have been found. These include gene silencing, regulation of RNA processing, and regulation of transcription (reviewed in reference 35). It is therefore possible that the JEV small RNA could have an effect on virus production, persistence, or pathogenesis in ways not directly connected to genome replication, as speculated above. Further research is required to determine the biological significance of the JEV small RNA. Certainly its abundance alone implies importance as a regulatory molecule.
Acknowledgments
We thank Li-Kuang Chen and Ching-Len Liao for kindly providing the JEV RP-2 and RP-9 mutants and Chwan-Chuen King for providing the TP-53, S-26, PC-806, CH-1392, CC-27, Beijing-1, and Nakayama viral isolates. We thank David Brian, Gwong-Jen J. Chang, and Chwan-Chuen King for many helpful discussions and critical reading of the manuscript.
This work was supported by grant NHRI-EX91-8936SC from the National Health Research Institute and grant NSC 91-2311-B-259-002 from the National Science Council, Taipei, Taiwan, Republic of China.
REFERENCES
- 1.Blackwell, J. L., and M. A. Brinton. 1995. BHK cell proteins that bind to the 3′ stem-loop structure of the West Nile virus genome RNA. J. Virol. 69:5650-5658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blackwell, J. L., and M. A. Brinton. 1997. Translation elongation factor-1 alpha interacts with the 3′ stem-loop region of West Nile virus genomic RNA. J. Virol. 71:6433-6444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brinton, M. A. 1983. Analysis of extracellular West Nile virus particles produced by cell cultures from genetically resistant and susceptible mice indicates enhanced amplification of defective interfering particles by resistant cultures. J. Virol. 46:860-870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brinton, M. A. 1982. Characterization of West Nile virus persistent infections in genetically resistant and susceptible mouse cells. I. Generation of defective nonplaquing virus particles. Virology 116:84-98. [DOI] [PubMed] [Google Scholar]
- 5.Brinton, M. A. 1986. Replication of flaviviruses, p. 327-374. In S. Schlesinger and M. J. Schlesinger (ed.), The Togaviridae and Flaviviridae. Plenum Press, Inc., New York, N.Y.
- 6.Burke, D. S., and C. J. Leake. 1988. Japanese encephalitis, p. 63-92. In T. P. Monath (ed.), The arboviruses: epidemiology and ecology, vol. III. CRC Press, Inc., Boca Raton, Fla. [Google Scholar]
- 7.Burke, D. S., and T. P. Monath. 2001. Flaviviruses, p. 1043-1126. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed., vol. I. Lippincott William & Wilkins, Philadephia, Pa. [Google Scholar]
- 8.Chang, R. Y., M. A. Hofmann, P. B. Sethna, and D. A. Brian. 1994. A cis-acting function for the coronavirus leader in defective interfering RNA replication. J. Virol. 68:8223-8231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen, C. J., M. D. Kuo, L. J. Chien, S. L. Hsu, Y. M. Wang, and J. H. Lin. 1997. RNA-protein interactions: involvement of NS3, NS5, and 3′ noncoding regions of Japanese encephalitis virus genomic RNA. J. Virol. 71:3466-3473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen, L. K., Y. L. Lin, C. L. Liao, C. G. Lin, Y. L. Huang, C. T. Yeh, S. C. Lai, J. T. Jan, and C. Chin. 1996. Generation and characterization of organ-tropism mutants of Japanese encephalitis virus in vivo and in vitro. Virology 223:79-88. [DOI] [PubMed] [Google Scholar]
- 11.Chu, P. W., and E. G. Westaway. 1985. Replication strategy of Kunjin virus: evidence for recycling role of replicative form RNA as template in semiconservative and asymmetric replication. Virology 140:68-79. [DOI] [PubMed] [Google Scholar]
- 12.Cleaves, G. R., T. E. Ryan, and R. W. Schlesinger. 1981. Identification and characterization of type 2 dengue virus replicative intermediate and replicative form RNAs. Virology 111:73-83. [DOI] [PubMed] [Google Scholar]
- 13.Corver, J., E. Lenches, K. Smith, R. A. Robison, T. Sando, E. G. Strauss, and J. H. Strauss. 2003. Fine mapping of a cis-acting sequence element in yellow fever virus RNA that is required for RNA replication and cyclization. J. Virol. 77:2265-2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Debnath, N. C., R. Tiernery, B. K. Sil, M. R. Wills, and A. D. Barrett. 1991. In vitro homotypic and heterotypic interference by defective interfering particles of West Nile virus. J. Gen. Virol. 72:2705-2711. [DOI] [PubMed] [Google Scholar]
- 15.Hahn, C. S., Y. S. Hahn, C. M. Rice, E. Lee, L. Dalgarno, E. G. Strauss, and J. H. Strauss. 1987. Conserved elements in the 3′ untranslated region of flavivirus RNAs and potential cyclization sequences. J. Mol. Biol. 198:33-41. [DOI] [PubMed] [Google Scholar]
- 16.Hashimoto, H., A. Nomoto, K. Watanabe, T. Mori, T. Takezawa, C. Aizawa, T. Takegami, and K. Hiramatsu. 1988. Molecular cloning and complete nucleotide sequence of the genome of Japanese encephalitis virus Beijing-1 strain. Virus Genes 1:305-317. [DOI] [PubMed] [Google Scholar]
- 17.Holland, J. J. 1990. Defective viral genomes, p. 156-165. In B. N. Fields and D. M. Knipe (ed.), Fields virology. Raven Press Ltd., New York, N.Y.
- 18.Igarashi, A. 1978. Isolation of a Singh's Aedes albopictus cell clone sensitive to Dengue and Chikungunya viruses. J. Gen. Virol. 40:531-544. [DOI] [PubMed] [Google Scholar]
- 19.Jan, L. R., K. L. Chen, C. F. Lu, Y. C. Wu, and C. B. Horng. 1996. Complete nucleotide sequence of the genome of Japanese encephalitis virus ling strain: the presence of a 25-nucleotide deletion in the 3′-nontranslated region. Am. J. Trop. Med. Hyg. 55:603-609. [DOI] [PubMed] [Google Scholar]
- 20.Khromykh, A. A., H. Meka, K. J. Guyatt, and E. G. Westaway. 2001. Essential role of cyclization sequences in flavivirus RNA replication. J. Virol. 75:6719-6728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ku, C. C., C. C. King, C. Y. Lin, H. C. Hsu, L. Y. Chen, Y. Y. Yueh, and G. J. Chang. 1994. Homologous and heterologous neutralization antibody responses after immunization with Japanese encephalitis vaccine among Taiwan children. J. Med. Virol. 44:122-131. [DOI] [PubMed] [Google Scholar]
- 22.Lancaster, M. U., S. I. Hodgetts, J. S. Mackenzie, and N. Urosevic. 1998. Characterization of defective viral RNA produced during persistent infection of Vero cells with Murray Valley encephalitis virus. J. Virol. 72:2474-2482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lindenbach, B. D., and C. M. Rice. 2001. Flaviviridae: the viruses and their replication, p. 991-1042. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed., vol. I. Lippincott Williams & Wilkins, Philadelphia, Pa. [Google Scholar]
- 24.Liu, J.-L. 2001. Phylogenetic analysis of E gene and 3′ untranslated region of Japanese encephalitis virus isolated in Taiwan. M.S. thesis. National Taiwan University, Taipei, Taiwan.
- 25.Mathews, D. H., J. Sabina, M. Zuker, and D. H. Turner. 1999. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J. Mol. Biol. 288:911-940. [DOI] [PubMed] [Google Scholar]
- 26.McAda, P. C., P. W. Mason, C. S. Schmaljohn, J. M. Dalrymple, T. L. Mason, and M. J. Fournier. 1987. Partial nucleotide sequence of the Japanese encephalitis virus genome. Virology 158:348-360. [DOI] [PubMed] [Google Scholar]
- 27.Men, R., M. Bray, D. Clark, R. M. Chanock, and C. J. Lai. 1996. Dengue type 4 virus mutants containing deletions in the 3′ noncoding region of the RNA genome: analysis of growth restriction in cell culture and altered viremia pattern and immunogenicity in rhesus monkeys. J. Virol. 70:3930-3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Monath, T. P. 1988. Japanese encephalitis—a plague of the Orient. N. Engl. J. Med. 319:641-643. [DOI] [PubMed] [Google Scholar]
- 29.Naeve, C. W., and D. W. Trent. 1978. Identification of Saint Louis encephalitis virus mRNA. J. Virol. 25:535-545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Poidinger, M., R. A. Hall, and J. S. Mackenzie. 1996. Molecular characterization of the Japanese encephalitis serocomplex of the flavivirus genus. Virology 218:417-421. [DOI] [PubMed] [Google Scholar]
- 31.Proutski, V., E. A. Gould, and E. C. Holmes. 1997. Secondary structure of the 3′ untranslated region of flaviviruses: similarities and differences. Nucleic Acids Res. 25:1194-1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 33.Schmaljohn, C., and C. D. Blair. 1977. Persistent infection of cultured mammalian cells by Japanese encephalitis virus. J. Virol. 24:580-589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sethna, P. B., S. L. Hung, and D. A. Brian. 1989. Coronavirus subgenomic minus-strand RNAs and the potential for mRNA replicons. Proc. Natl. Acad. Sci. USA 86:5626-5630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Storz, G. 2002. An expanding universe of noncoding RNAs. Science 296:1260-1263. [DOI] [PubMed] [Google Scholar]
- 36.Ta, M., and S. Vrati. 2000. Mov34 protein from mouse brain interacts with the 3′ noncoding region of Japanese encephalitis virus. J. Virol. 74:5108-5115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Takeda, H., A. Oya, K. Hashimoto, T. Yasuda, and M. A. Yamada. 1978. Association of virus specific replicative ribonucleic acid with nuclear membrane in chick embryo cells infected with Japanese encephalitis virus. J. Gen. Virol. 38:281-291. [DOI] [PubMed] [Google Scholar]
- 38.Takegami, T., and S. Hotta. 1990. Synthesis and localization of Japanese encephalitis virus RNAs in the infected cells. Microbiol. Immunol. 34:849-857. [DOI] [PubMed] [Google Scholar]
- 39.Tsai, T. F., G.-J. Chang, and Y. X. Yu. 1999. Japanese encephalitis vaccines, p. 672-710. In A. P. Stanley and W. A. Orenstein (ed.), Vaccines, 3rd ed. W. B. Saunders, Philadelphia, Pa.
- 40.Uchil, P. D., and V. Satchidanandam. 2003. Characterization of RNA synthesis, replication mechanism, and in vitro RNA-dependent RNA polymerase activity of Japanese encephalitis virus. Virology 307:358-371. [DOI] [PubMed] [Google Scholar]
- 41.Urosevic, N., M. van Maanen, J. P. Mansfield, J. S. Mackenzie, and G. R. Shellam. 1997. Molecular characterization of virus-specific RNA produced in the brains of flavivirus-susceptible and -resistant mice after challenge with Murray Valley encephalitis virus. J. Gen. Virol. 78:23-29. [DOI] [PubMed] [Google Scholar]
- 42.Vaughn, D. W., and C. H. Hoke, Jr. 1992. The epidemiology of Japanese encephalitis: prospects for prevention. Epidemiol. Rev. 14:197-221. [DOI] [PubMed] [Google Scholar]
- 43.Wengler, G., and H. J. Gross. 1978. Studies on virus-specific nucleic acids synthesized in vertebrate and mosquito cells infected with flaviviruses. Virology 89:423-437. [DOI] [PubMed] [Google Scholar]
- 44.You, S., B. Falgout, L. Markoff, and R. Padmanabhan. 2001. In vitro RNA synthesis from exogenous dengue viral RNA templates requires long range interactions between 5′- and 3′-terminal regions that influence RNA structure. J. Biol. Chem. 276:15581-15591. [DOI] [PubMed] [Google Scholar]
- 45.Zuker, M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31:3406-3415. [DOI] [PMC free article] [PubMed] [Google Scholar]