Abstract
Heterochromatin Protein 1 (HP1a in Drosophila) is a conserved eukaryotic chromosomal protein that is prominently associated with pericentric heterochromatin and mediates the concomitant gene silencing. Mechanistic studies implicate HP1 family proteins as “hub proteins,” able to interact with a variety of chromosomal proteins through the chromo-shadow domain, as well as recognize key histone modification sites (primarily H3K9me2/3) through the chromo domain. Consequently, HP1 plays many important roles in chromatin architecture and impacts both gene expression and gene silencing, utilizing a variety of mechanisms. Clearly, HP1 function is altered by context, and potentially by post-translational modifications. Here, we report on recent ideas as to how this versatile protein accomplishes its diverse functions.
Keywords: HP1a, chromo domain, silencing, gene expression
The versatile HP1 family proteins
Heterochromatin Protein 1a (HP1a) was initially identified as a protein predominantly associated with heterochromatin by using immunofluorescent staining of the polytene chromosomes of Drosophila, screening monoclonal antibodies prepared against a collection of tight-binding nuclear proteins [1]. The gene encoding HP1a was first identified in mutagenesis screens for dominant suppressors of heterochromatic position effect variegation (reviewed in [2]). These and subsequent studies implicated HP1a in position-effect variegation (PEV), the heterochromatin-mediated silencing of euchromatic genes abnormally displaced to heterochromatin via rearrangement or transposition. Comparison with a second protein involved in stable silencing, Polycomb (Pc), identified a homologous domain, the “chromo” domain, subsequently shown to bind specific post-translational modifications of the histone H3 N-terminal tail: trimethylated lysine 27 (H3K27me3) in the case of Pc, and di- or trimethylated lysine 9 (H3K9me2/3) in the case of HP1a. Genetic and biochemical analyses indicate that direct binding of the modified histone tail by HP1a or Pc is utilized to promote gene silencing (Box 1). Analysis of the full HP1a sequence identified a second related motif, the chromo-shadow domain, which homodimerizes to form a platform capable of binding a variety of proteins. This two-domain structure allows HP1a to link a targeted histone post-translational modification (histone mark) physically with the non-histone proteins that it binds. We consider this to be the defining role of HP1 proteins, and hence for purposes of this review will define an HP1 protein as having an N-terminal chromo domain (CD), flexible hinge region (H), and C-terminal chromo-shadow domain (CSD) (see Fig. 1A). In addition to protein binding activities, the hinge region has been reported to bind nucleic acids, suggesting additional structural interactions. (See [2,3] for reviews with original citations.)
Box 1. Chromo domains interact directly with methylated histone H3 tails to cause silencing.
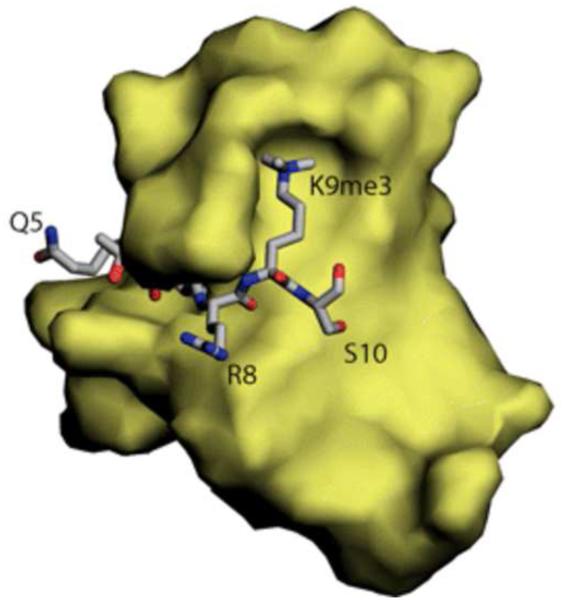
A combination of genetic manipulation and structural analysis has demonstrated that direct binding of the methylated H3 tail by the chromo domain protein is required for silencing by HP1a and by Pc. Structural studies show that the chromo domain folds with an alpha helix backed by a 3-strand beta-pleated sheet to form a crevice holding the histone tail with an aromatic cage capturing the methylated lysine (see Fig. 1B for the structure of HP1a). A V26M mutation that destabilizes the folded structure of HP1a results in a complete loss of H3K9me2 binding in vitro, and loss of PEV silencing in vivo ([76]; see Fig. 1C). Until recently, it has not been possible to do the reciprocal experiment, mutating the histone and assessing the in vivo consequences, because the core histone genes are present in multiple copies. However, a genetic system that deletes the wild type histone gene cluster and replaces it with a transgene cassette providing 12 copies has recently been developed in Drosophila melanogaster [77]. This has allowed a direct test showing that an H3 K27R mutation fails to repress transcription of genes that are normally silenced by the Pc complex 2; moreover, the mutant cells show homeotic changes that mimic those of Pc mutations [78]. Development of this system is a valuable contribution that will allow many other direct tests of the biological importance of histone modifications.
Figure 1.


(A) Schematic representation of HP1 family proteins; example shown is HP1a from D. melanogaster (see text for abbreviations). (B) A 3-D model of the chromo domain, showing binding of the H3 N-terminal tail and the aromatic cage for K9me3 (image from [74]). (C) Alignment of the chromo domain from Homo sapiens CBX1, CBX5, and CBX3 with S. pombe Swi6, D. melanogaster HP1a, C. elegans HPL-2, and Arabadopsis thaliana LPH1. Amino acids are color-coded for functional similarity. The conserved residues that make up the aromatic cage are marked with an asterisk, and the position of V26 in HP1a, site of a loss-of-function mutation, is marked with a dot.
As the genomes of Drosophila melanogaster and other eukaryotes were sequenced, it became evident that (a) the HP1 protein family is highly conserved, being found in organisms from the yeast Schizosaccharomyces pombe to humans, and that (b) most genomes have several homologues of this protein (reviewed in [4]). In D. melanogaster, five paralogues have been characterized: HP1a, the protein described above; HP1b, found in both heterochromatin and euchromatin; HP1c, primarily euchromatic; HP1d, the product of the rhino gene, expressed only in the female germline; and HP1e, expressed in the male germline. While orthologous genes for the first four are found in all Drosophila genomes sequenced through 2012, the HP1 family members expressed exclusively in the male germline differ in different species, suggesting a dynamic evolutionary process that could reflect karyotype evolution [5]. Genes encoding proteins containing only an HP1-related chromo domain (e.g., Oxpecker) or only an HP1-related chromo-shadow domain (e.g., Umbrea) have also been identified [5]. While these derived proteins (where tested, expressed in the germline) are certainly of interest, they do not meet the definition of HP1 family proteins, and should not be labeled as such.
This review focuses on HP1a of Drosophila melanogaster, the best-studied of these proteins, and its well-studied homologues in other species, primarily Swi6 from S. pombe and human HP1α/HP1β/HP1γ. No HP1 orthologs are found in the budding yeast Sacccharomyces cerevisiae, which does not utilize either H3K9 or H3K27 methylation, instead relying on the SIR protein complex for domain silencing [6]. HP1a orthologues are found in many fungi and most animals, and appear to have similar functions in heterochromatin structure in the organisms mentioned (S. pombe, flies, mammals). This role of HP1a is highly conserved, as shown by the observation that ectopic expression of two of the three human HP1-coding genes (Cbx3/HP1γ and Cbx5/HP1α) can significantly enhance heterochromatic silencing in flies [7]. The chromo domain of M31, a mouse homologue, can functionally replace that domain in Swi6, the S. pombe homologue [8]. However, in some species the homologous protein is best described as “HP1a like.” In Caenorhabditis elegans, the chromosomes are holocentric; consequently there is no equivalent of “pericentric heterochromatin,” but the distal portions of the chromosome arms show an enrichment for H3K9me1/2/3 [9]. The C. elegans HP1a homologue, HPL-2 (Heterochromatin Protein-Like 2), is required for developmental regulation of key genes, behaving more like Polycomb in this regard [10]. Nonetheless, HPL-2 plays a role in maintaining the long-term silencing triggered by a piRNA-dependent foreign RNA response, suggesting heterochromatin formation [11]. In vitro, HPL-2 binds both H3K27me2/3 and H3K9me2/3 [12], but the mechanism of HPL-2 targeting in vivo remains unclear. Rather than using the H3K9me3/HP1a and H3K27me3/Pc complexes as generally distinct and different silencing systems, as seen in flies [13], worms may have a more complex heterochromatin structure, as loss of any of several histone methyltransferase (HMT) activities (methylating K9, K27 or K36) causes partial release of silencing of a tandem repeat array [14]. Further, many of the mammalian Cbx proteins, as well as plant LHP1 (Like Heterochromatin Protein-1), fail to discriminate between H3K9me3 and H3K27me3 (reviewed in [15]). Arabidopsis LHP1 appears to be restricted to promoting Polycomb-group silencing [16]. While pericentric heterochromatin in Arabidopsis is enriched in H3K9me2 (and 5-methylcytosine; 5mC), no HP1a orthologue has been identified that localizes specifically to these domains in plants. Thus, the mechanism of HP1a/H3K9me3 domain silencing conserved among S. pombe, flies, and mammals, while widespread, appears not to apply in all eukaryotes.
HP1a is a versatile chromosomal protein. As mentioned above, it was first described in D. melanogaster as a protein localized to heterochromatin, and is required in a dosage-dependent manner for silencing of PEV reporters. Drosophila HP1a binds H3K9me2/3 through its chromo domain, and binds SU(VAR)3-9, one of the HMTs that methylates histone H3 on K9, through its chromo shadow domain. This combination suggests a mechanism for heterochromatin spreading and perhaps epigenetic inheritance, as first shown in S. pombe (reviewed in [3]). HP1a is essential in flies, necessary for genome integrity and the stability of repetitious sequences [17]. It plays multiple roles at telomeres, being involved in telomere capping, elongation, and silencing of telomere-associated repeats (reviewed in [18]). HP1 family proteins have been implicated in the mechanism of scheduled DNA replication, where they bind Chromatin Assembly Factor-1 [19,20], and in double strand break repair [21]. All of these roles can be interpreted as the result of HP1a contributing to a chromatin state that promotes a compact assembly and/or promotes transcriptional silencing, which we discuss next. However, HP1a can also contribute to a chromatin state that promotes gene expression, and a discussion of this will follow.
The Role of HP1a in Silencing
A major role of HP1a is in the assembly and maintenance of heterochromatin, a relatively condensed form of chromatin that promotes gene silencing. Indeed, formation of heterochromatin may be an ancient defense designed to silence transposable elements, which can otherwise wreak havoc in the genome (summarized in [22]). The most detailed evidence for the role of HP1 family proteins in controlling heterochromatin transcription comes from studies in fission yeast. S. pombe has two HP1 family proteins, Swi6 and Chp2. Swi6 is the more abundantly expressed of the two and contributes in greater degree to silencing at the heterochromatic mat locus in a dose-dependent fashion [23]. Interestingly, Chp2 overexpression displaces Swi6 and causes loss of silencing, suggesting that both proteins compete for the same binding sites. Both proteins participate in excluding RNA polymerase II (Pol II) from heterochromatic domains and in transcriptional silencing at centromeric repeats [24]. Similarly, HP1a has been shown to decrease gene expression in PEV in Drosophila, with a loss of associated Pol II (reviewed in [3]). How is this accomplished? A number of alternative mechanisms have recently been proposed.
Histone modification
Assembly of heterochromatin in most eukaryotes appears to depend on core histone deacetylation and histone H3K9 di- and tri-methylation. HP1a facilitates this process by reading the K9 methyl mark and recruiting relevant enzymes to the site. This can include 5mC DNA methylation, common in mammals. The process of heterochromatin formation can be considered a cascade of chromatin modifications; for example, H3K9 must be deacetylated before it can be methylated (the mark read by HP1a) (see [3] for an illustration). In Drosophila, the histone deacetylase and histone methyltransferase can be recovered as a complex [25]. However, there is growing evidence that these steps in histone modification can occur as distinct events. For example, the two S. pombe HP1a orthologues, Swi6 and Chp2, exist in distinct complexes, both of which promote silencing. Chp2 associates with SHREC histone deacetylase, required for H3K14 deacetylation, a shift that limits Pol II access to the template. Swi6 is required for the RNAi-dependent silencing mechanism, which utilizes local transcripts for targeting and recruits the H3K9 HMT Clr4 ([26]; see [27] for review). These dual pathways also occur in Neurospora crassa. Here, H3K9me3 binds HP1 in a complex that directs DNA methylation, resulting in silencing. A distinct HP1 complex works in parallel to cause silencing by maintaining histone hypoacetylation [28].
Why should these changes promote silencing? The chromatin fiber is continually in flux, with relatively unstable nucleosomes (generally marked by the core histone variants H2A.X and H3.3) around the transcription start sites (TSS), where accessibility is detectable as a nuclease hypersensitive site (HS site). Core histone acetylation (particularly in the N-terminal tails of histones H3 and H4) is prominently associated with the active state, presumably because this modification neutralizes electrostatic interaction between the acidic DNA and basic histones, facilitating unfolding of the chromatin fiber. Indeed, comparing the same transgene in heterochromatin to the gene in its normal home in euchromatin in Drosophila has shown a loss of HS site accessibility and a more regularly spaced nucleosome array at the heterochromatic copy. Accessibility is partially restored by HP1a depletion, confirming that the effect is due to heterochromatin packaging (see [3] for original citations). A model based on histone turnover makes sense here; the histone deacetylase Clr3 (required for silencing in S. pombe) can inhibit histone turnover and promote nucleosome occupancy across heterochromatin domains, while Epe1, which promotes the active state, promotes histone exchange [29]. Thus, histone hypoacetylation, by itself, can promote silencing. What about the role of H3K9me2/3? The presence of any methyl groups precludes acetylation at this residue, and binding of HP1a could interfere with demethylation and subsequent acetylation, thus contributing to a stable heterochromatin state. In addition, HP1 can provide a binding platform for proteins that further drive assembly and maintenance of the heterochromatin state, such as H3K9 methyltransferase. However, in some mammalian systems, the HP1 proteins are not required to maintain silencing of endogenous retroviruses (the test applied for heterochromatin formation in this study); the HMT is sufficient, suggesting that H3K9me3 might block transcription by other mechanisms [30].
Chromatin fiber condensation
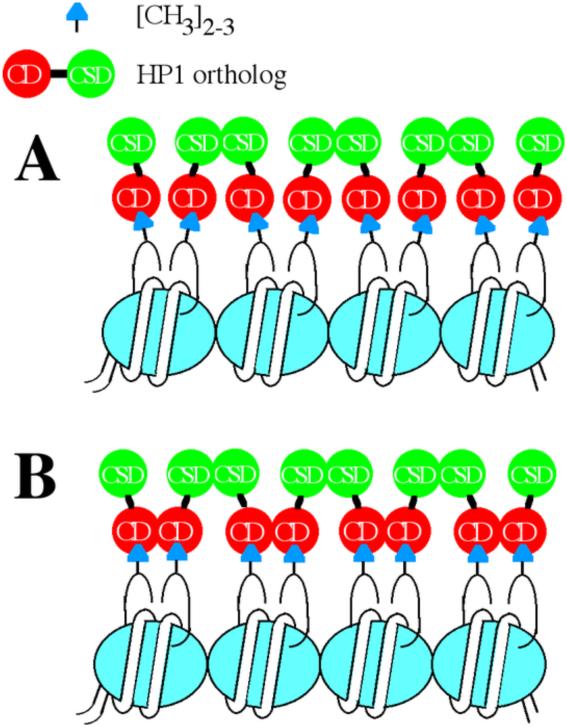
HP1a binding also suggests mechanisms for chromatin fiber condensation to promote silencing, presumably also by blocking pol II access. Because HP1a forms homodimers through the CSD, leaving the two CDs available to bind H3K9me3, HP1a dimers could crosslink the nucleosome array (Fig. 2A). Crosslinking adjacent nucleosomes could impose uniform nucleosome spacing, which is observed in heterochromatin; crosslinking two turns of a 30nm fiber could preclude the unwinding necessary for transcription. Indeed, sedimentation analysis comparing human HP1α binding to unmethylated or H3K9 trimethylated nucleosome arrays shows significantly higher array self-association with the methylated arrays, an effect requiring the CSD [31]. Recent work in S. pombe has suggested that Swi6 binds more effectively to H3K9me3 by dimerizing through CD-CD contacts [32]. This leaves the two CSDs available to dimerize with two additional Swi6 molecules, creating a tetramer that could bridge to nearby H3K9-methylated nucleosomes (see Fig. 2B). The importance of the CD-CD contacts in tetramer formation on the nucleosome was not anticipated based on earlier studies of metazoan HP1 CDs in solution; CD-CD dimerization was not observed in earlier solution studies or crystal structures [15]. In particular, a recent, detailed biophysical characterization of recombinant mammalian HP1β binding to recombinant mononucleosomes disclosed no other constraints on interaction, or multimerization interfaces, besides the H3K9me3 mark and the CD in creating flexible and dynamic nucleosome binding [33]. These results suggest that CD-CD interaction may be limited to Swi6. The CD-CD association postulated for Swi6 is supported by genetic analysis; missense mutations with increased CD interaction in vitro result in increased tetramerization of Swi6 (which is thought to occur on the nucleosome), and are correlated with increased silencing and heterochromatin spreading [32]. A conformational shift in Swi6 has been proposed to promote heterochromatin spreading [34].
Figure 2.
Cartoon models of a nucleosome array in which H3K9 is di- or trimethylated and bound by HP1. A. In this model, CSD-CSD homo-dimerization is indicated, which could facilitate chromatin condensation [31] and impose regular nucleosome positioning, as previously reported for HP1a-dependent transgene silencing in Drosophila [75]. B. In this model, based on studies on Swi6 in S. pombe, both CD-CD and CSD-CSD self-associations are indicated, which could facilitate further condensation of the chromatin fiber (model based on [32]).
Swi6/HP1 also contributes to higher order chromatin condensation by recruiting other factors, notably cohesin, which is required for mitotic sister chromatid cohesion [35]. While such a direct interaction has not been demonstrated in higher eukaryotes, recent studies in mammalian cells have shown that HP1 interaction with the HMT Suv4-20h2 leads to cohesin recruitment at pericentric heterochromatin, with associated compaction [36].
Transcript degradation
Heterochromatin domains generally show low levels of associated RNA Pol II, but many recent studies show that transcription can occur, and can be essential to targeting heterochromatin formation through the RNAi system (see below). Several reports have documented nucleic acid binding by HP1, most often involving the hinge region, but sometimes either the CD or CSD. A recent study in S. pombe, looking primarily at the MAT loci and telomeres, argues that Swi6 can capture transcripts from heterochromatic reporter genes and direct them to the nuclear exosome for degradation. Extensive genetic alteration of the hinge region that abolished RNA binding allowed persistence of the heterochromatic transcripts [37], but may have altered other Swi6 properties as well. An ability of Swi6 to facilitate silencing at several levels could potentially create a “fail safe” system where silencing is critical. It will be important to see if similar HP1 family protein-dependent transcript degradation is detected in metazoa as well. The HP1-family protein Rhino/HP1d is required for targeted post-transcriptional degradation of certain transposon RNAs in the Drosophila female germline, and is required for nuage organization, suggesting a role in RNA processing, albeit for piRNA production [38].
Targeting of heterochromatin formation
Clearly, heterochromatin formation is a potent silencing mechanism; thus the cell must target domains that should be silenced, while maintaining appropriate access for transcription. Hypotheses as to targeting mechanisms generally turn on either protein recognition of specific DNA sequences or RNA-based recognition (often invoking RNAi-based recognition of transient transcripts) to localize either HP1 or appropriate histone modification enzymes to initiate the heterochromatin formation cascade. An RNAi-based mechanism for heterochromatin targeting is well-established in some fungi (e.g., S. pombe) and plants based on the recognition of transcribed repetitious sequences. In general, the data indicate that Argonaute family proteins carrying small RNAs can target heterochromatin proteins, including H3K9 HMTs and HP1, by sequence recognition of transient transcripts (reviewed in [39]). Similarly, growing evidence implicates a piRNA system in silencing of transposable elements (TEs) and their remnants in most metazoa, particularly in the female germline [40]. For example, depletion of either HP1a or PIWI (an Argonaute protein that can bind piRNAs and enter the nucleus) in the Drosophila female germline leads to increased expression of the same set of TEs [41]. De-repression of TEs upon PIWI depletion correlates with increased Pol II occupancy, whereas expression of piRNAs that target a reporter construct results in increased association of H3K9me3 and HP1a [42]. Analysis of Drosophila ovarian somatic cells has also demonstrated Piwi-dependent transcriptional silencing of transposons, through a mechanism dependent on Maelstrom (an HMG protein) to block RNA Pol II recruitment [43]. In vitro, PIWI binds directly to HP1a through a PxVxL domain [44], but whether or not this occurs in vivo remains an open question, as a PIWI missense mutation that disrupts the in vitro interaction fails to compromise function in vivo [41].
An RNAi-based system seems well-adapted to recognizing TEs, which can change rapidly, are quite variable, and must be silenced to maintain genome integrity. What about regions with satellite DNA? Many mammalian transcription factors (e.g. Pax3 and Pax9) have binding sites in pericentric heterochromatin; indeed, a high concordance has been found between Suv39h-dependent heterochromatic repeat regions and transcription factor binding sites in mouse. Pax3 and Pax9 depletion results in de-repression of major satellite transcripts, impairment of heterochromatic marks, and defects in chromosome segregation [45]. While in this case targeting may be determined by specific protein binding, loss of Pax3/Pax9 results in bidirectional transcription up-regulation, resulting in dsRNA [45], which could once again feed into the piRNA system.
In some cases, binding of specific transcription factors to euchromatic sites appears to trigger formation of small blocks of heterochromatin to achieve silencing. For example, phosphorylated HERS (histone gene-specific epigenetic repressor in late S phase) binds to the histone gene regulatory regions in Drosophila, and anchors HP1a and Su(var)3-9 to induce silencing of this repeated gene cluster [46]. Similarly, the TIF1β/KAP1 co-repressor targets HP1 to specific loci in the euchromatic arms to silence those genes [47,48]. In contrast to the PEV associated with pericentric heterochromatin, there is very little spreading of the silencing marks in these cases, suggesting a silencing state that differs in some key characteristics from heterochromatin formation. However, analysis of the modENCODE data makes clear that as Drosophila cells mature, large “facultative heterochromatin” domains enriched for H3K9me2/3 and HP1a accumulate in the euchromatic arms [49,50]; unfortunately, there has been little study of this process to date.
In summary, the evidence points to an HP1-based silencing mechanism in which HP1 binds histone tails in a way that constrains nucleosome arrays to occlude the chromatin template. The mechanism for HP1 targeting depends on the H3K9 methyl mark and, at least in some cases, on the specificity of the RNAi pathway. The mechanism by which these interactions contribute to the defining cytological appearance of heterochromatin is unknown.
The Role of HP1a in Gene Activation
While HP1a is prominently associated with silencing, there is growing evidence that it can also promote gene expression. HP1a dosage can impact the expression of subsets of genes residing in euchromatin, as well as affecting heterochromatic gene expression (reviewed in [51]). How can a protein that plays a major role in silencing also promote gene activation? No doubt the versatility of HP1a in binding to a variety of effector proteins is critical (reviewed in [52]). In addition, HP1 family proteins may not always be interacting with H3 silencing marks; the chromodomain of Cbx3/HP1γ can also bind to H1K26me2, trimethylated lysine 185 of G9a, and no doubt other modified polypeptides [53]. Whether the observed HP1a dosage effects are direct or indirect is difficult to establish (Box 2), although if HP1a is found at significant levels at the impacted gene, the effect is inferred to be direct. What sort of direct effects should be considered? Two mechanisms have been proposed: an impact on overall heterochromatin structure, and a role in transcript elongation.
BOX 2. Dynamic chromatin.
Chromatin is a multi-faceted assembly, and many of the proteins involved form multiple complexes. Even HP1 family proteins, which are associated with stable heterochromatin, are dynamic. Using GFP-tagged proteins and fluorescence recovery after photobleaching (FRAP), it was found that 50% of mammalian HP1 was exchanged within 2.5 sec. in heterochromatin and 0.6 sec. in euchromatin [79]. This implies that the distribution of a given chromosomal protein can shift from one chromatin compartment to another, depending on its own concentration and the local concentrations of available partners. Hence it can be difficult to ascertain whether the effect on a particular locus of depletion of a given chromosomal protein is direct or indirect. Effects that suggest shifts in HP1a can be seen using variegating reporters present in different heterochromatic domains. For example, while Su(var)3-9 mutations lead to a loss of silencing at reporters in the pericentric heterochromatin, they can lead to enhanced silencing on the fourth chromosome of D. melanogaster [80]. A plausible explanation is that this mutation causes a greater loss of H3K9me2/3 in the pericentric heterochromatin than it does on the fourth chromosome, where the HMT dSETDB1 plays a major role. The loss of H3K9me2/3 in pericentric heterochromatin will cause a loss of HP1a from that domain; more can now accumulate on the fourth chromosome, where H3K9me2/3 levels remain high. Thus every event that perturbs chromatin structure in one compartment should be assumed to have perturbed the structure in other compartments, resulting in indirect effects.
Maintaining heterochromatin structure
As mentioned above, genes normally packaged in heterochromatin appear to require that HP1-rich environment for optimal expression. Drosophila genes normally found in pericentric heterochromatin can be silenced by juxtaposition (through rearrangement) with a euchromatic domain. Protein factors found in heterochromatin are required for optimal expression of these genes in their native state (reviewed in [51]). Similarly, most of the genes on the heterochromatic fourth chromosome lose expression when HP1a is depleted [54]. It has been argued [55] that heterochromatin formation is required for piRNA production, based on the loss of expression upon depletion of dSETDB1 from a piRNA cluster that maps to a heterochromatic domain. These results suggest that HP1a is required to maintain a necessary feature of the chromatin structure at and around active genes that are surrounded by heterochromatin. One clue comes from direct studies of two fourth chromosome genes, Dyrk3 and Caps, which show a loss of HS sites (indicating loss of promoter accessibility) on HP1a depletion [56]. But how this might be accomplished remains unknown.
Promoting transcript elongation and processing
In addition to its association with heterochromatin, Drosophila HP1a is also associated with a small set of sites in the euchromatic arms of the major autosomes, including conspicuous chromosome puffs, sites with high transcription levels [57]. Chromosome-wide binding studies using cultured cells find HP1a enriched across the entire transcription units of certain exon-rich active genes in euchromatin [58]. HP1a stimulates transcript elongation at a small set of euchromatic sites, and is associated with heterogeneous nuclear ribonuclear proteins (hnRNPs) [59], suggesting a role for HP1a in transcript processing in flies. The mammalian HP1-family protein CBX3/HP1γ is required for the full activity of some euchromatic genes, as evidenced by reduced expression upon CBX3/HP1γ knockdown. The knockdown leads to defective recruitment of splicing factors, with accumulation of unspliced transcripts for these genes [60]. These results argue for a positive role for HP1a in transcript elongation/processing for some euchromatic genes, the opposite of the result reported above from S. pombe looking at heterochromatic transcripts [61].
Taken together, the context-dependent effects of HP1 family proteins on gene expression may be understood mechanistically by thinking of these proteins and their associated complexes as chromatin organizers. In some cases, the resulting chromatin occludes promoters and/or regulatory DNA, while in other cases, the resulting chromatin facilitates access to promoters and/or regulatory DNA for activators. By extension, Pol II elongation could be either impeded or enhanced by HP1-dependent architecture, and RNA processing efficiency directly or indirectly affected [62].
HP1 post-translational modifications and chromatin assembly
The stable epigenetic silencing associated with HP1-enriched heterochromatin contrasts with the remarkably dynamic chromatin-binding behavior of mammalian HP1 family proteins (see Box 2). Such dynamic behavior may be regulated by post-translational modifications (PTMs), which could tip the balance towards or away from heterochromatin assembly. Recent systematic mass spectrometry analysis has disclosed evidence for a variety of posttranslational modifications of mammalian HP1 family proteins, reminiscent of the histone code [63]. These include phosphorylation, acetylation, formylation, monomethylation, sumoylation and ubiqutinylation. The enzymes controlling most of these modifications, and the functional significance of most of these modifications, are unknown. Table 1 summarizes several modifications of HP1 and the observed properties conferred by the modifications.
Table I.
A sample of HP1 family protein modifications and associated properties
| Modification | Property | Refs |
|---|---|---|
| Phosphorylation of CBX5/HP1α and CBX3/HP1γ |
Increased phosphorylation correlates with mitosis |
[59] |
| Pim-1 phosphorylation in the hinge region of CBX3/HP1γ |
Inhibits transcriptional repression | [60] |
| Casein kinase II phosphorylation N-terminal to the CBX1/HP1β chromodomain |
Loss of HeK9me2 binding | [61] |
| Casein kinase II phosphorylation N-terminal to the HP1a chromo domain and C-terminal to the chromo shadow domain |
Promotes heterochromatin binding and PEV silencing |
[62,63] |
| Protein kinase A sites in the HP la hinge domain | Enhanced dimerization and binding to H3K9me2 |
[64] |
| Phosphorylation of peptide binding surface of the HP1a chromo shadow domain |
Destabilizes dimerization; increases binding to HP2 and PIWI |
[40] |
| Ubiquitinylation of CBX5/HP1α and CBX1/HP1β |
Faster turnover than CBX3/HPlγ | [65,66] |
| SUMOylation of the CBX5/HP1α hinge domain |
Targeting to satellite repeats via RNA binding |
[67] |
| SUMO deconjugation of CBX5/HP1α | Retention in heterochromatin | [68] |
Testing the functional significance of PTMs involves expressing mutant forms of the HP1 family protein in which the modified residue is replaced with alanine or some other amino acid that cannot be modified (or, in the case of phosphoserine, a phosphomimetic amino acid like glutamate) and testing the ability of the mutant protein to replace the wild-type form in a silencing assay. S. pombe and D. melanogaster, with their facile genetics, would be the best organisms in which to test hypotheses about HP1 family protein PTMs. As with the histones, changes in PTMs could be key to the alternative functions discussed above.
Concluding remarks
HP1, a small protein with three major domains (CD, H, and CSD), is remarkably influential in promoting distinct forms of packaging within eukaryotic genomes, impacting expression of the genes within those chromatin regions. While not universal among eukaryotes, the HP1a/H3K9me2/3 silencing mechanism is very common and highly conserved in metazoa. Despite exciting progress during that last decade, much remains to be learned about how HP1 is selectively targeted for heterochromatin formation, how the resulting heterochromatin assembly actually achieves silencing, and how HP1 carries out other “off label” functions, including promoting transcript elongation in some cases (see Box 3). A common thread may be found in its ability to generate compact structures. However, HP1a interacts with a wide range of partner proteins, and the findings above suggest that every HP1 family protein will have a variety of isoforms due to PTMs. Such modifications may create or destroy interaction sites with other proteins and/or may alter conformation or turnover rates at specific genomic sites. Understanding the PTMs of HP1 proteins may well help resolve many of the mysteries described above.
Box 3. Outstanding questions.
What is the mechanism of HP1-mediated silencing?
Do other HP1 family proteins form tetramers, as was reported for Swi6 in S. pombe??
Can HP1 family proteins be shown to cross-link nucleosome arrays? How general is the HP1-dependent RNA degradation model?
How does an HP1a-driven heterochromatin structure contribute to maintenance of gene expression for heterochromatic genes?
How does HP1 contribute to HS site formation in these domains?
How is HP1a targeted to a small set of euchromatic loci? In many cases this is a silencing mechanism, but in other cases it provides positive effects; at these latter sites, can it enhance transcript elongation, or transcript processing?
Do post-translational modifications determine HP1a partners, and hence HP1a activities?
Highlights.
We propose a structure-based definition of HP1 family proteins
We review evidence that HP1 is a context-dependent modifier of gene expression
Post-translational modifications of HP1 family proteins alter functionality
Acknowledgements
We apologize to the many workers in the field whose papers could not be cited directly due to space limitations. Work in the Elgin lab is supported by NSF grant MCB-1243724. We thank our modENCODE colleagues and members of the Elgin lab for thoughtful comments on the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.James TC, Elgin SCR. Identification of a nonhistone chromosomal protein associated with heterochromatin in Drosophila and its gene. Mol. Cell. Biol. 1986;6:3862–3872. doi: 10.1128/mcb.6.11.3862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Eissenberg JC, Reuter G. Cellular mechanism for targeting heterochromatin formation in Drosophila. Int. Rev. Cell. Mol. Biol. 2009;273:1–47. doi: 10.1016/S1937-6448(08)01801-7. [DOI] [PubMed] [Google Scholar]
- 3.Elgin SCR, Reuter G. Position-effect variegation, heterochromatin formation, and gene silencing in Drosophila. Cold Spring Harbor Perspect. Biol. 2013;5:a017780. doi: 10.1101/cshperspect.a017780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lomberk G, et al. Evidence for the existence of an HP1-mediated subcode within the histone code. Nat. Cell Biol. 2006;8:407–415. doi: 10.1038/ncb1383. [DOI] [PubMed] [Google Scholar]
- 5.Levine MT, et al. Phylogenomic analysis reveals dynamic evolutionary history of the Drosophila Heterochromatin Protein 1 (HP1) gene family. PLoS Genet. 2012;8(6):e1002729. doi: 10.1371/journal.pgen.1002729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Millar CB, Grunstein M. Genome-wide patterns of histone modifications in yeast. Nat. Rev. Mol. Cell. Biol. 2006;7:657–666. doi: 10.1038/nrm1986. [DOI] [PubMed] [Google Scholar]
- 7.Ma J, et al. Expression and functional analysis of three isoforms of human heterochromatin-associated protein HP1 in Drosophila. Chromosoma. 2001;109:36–44. doi: 10.1007/s004120000113. [DOI] [PubMed] [Google Scholar]
- 8.Wang G, et al. Conservation of heterochromatin protein 1 function. Mol. Cell. Biol. 2000;20:6970–6983. doi: 10.1128/mcb.20.18.6970-6983.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu T, et al. Broad chromosomal domains of histone modification patterns in C. elegans. Genome Res. 2011;21:227–236. doi: 10.1101/gr.115519.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Meister P, et al. Caenorhabditis elegans Heterochromatin protein 1 (HPL-2) links developmental plasticity, longevity and lipid metabolism. Genome Biol. 2011;12:R123. doi: 10.1186/gb-2011-12-12-r123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ashe A, et al. piRNAs can trigger a multigenerational epigenetic memory in the germline of C. elegans. Cell. 2012;150:88–99. doi: 10.1016/j.cell.2012.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Studencka M, et al. Transcriptional repression of Hox genes by C. elegans HP1/HPL and H1/HIS-24. PLoS Genet. 2012;8:e1002940. doi: 10.1371/journal.pgen.1002940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Riddle NC, et al. Enrichment of HP1a on Drosophila chromosome 4 genes creates an alternate chromatin structure critical for regulation in this heterochromatic domain. PLoS Genet. 2012;8(9):e1002954. doi: 10.1371/journal.pgen.1002954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Towbin BD, et al. Step-wise methylation of histone H3K9 positions heterochromatin at the nuclear periphery. Cell. 2012;150:934–947. doi: 10.1016/j.cell.2012.06.051. [DOI] [PubMed] [Google Scholar]
- 15.Blus BJ, et al. Epigenetic virtues of chromodomains. Crit. Rev. Biochem. Mol. Biol. 2011;46:507–526. doi: 10.3109/10409238.2011.619164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Derkacheva M, et al. Arabidopsis MSI1 connects LHP1 toPRC2 complexes. EMBO J. 2012;32:2073–2085. doi: 10.1038/emboj.2013.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Peng JC, Karpen GH. Epigenetic regulation of heterochromatic DNA stability. Curr. Opin. Genet. Dev. 2008;18:204–211. doi: 10.1016/j.gde.2008.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fanti L, et al. HP1: a functionally multifaceted protein. Curr. Opin. Genet. Dev. 2008;18:169–174. doi: 10.1016/j.gde.2008.01.009. [DOI] [PubMed] [Google Scholar]
- 19.Murzina N, et al. Heterochromatin dynamics in mouse cells: interaction between chromatin assembly factor 1 and HP1 proteins. Mol. Cell. 1999;4:529–540. doi: 10.1016/s1097-2765(00)80204-x. [DOI] [PubMed] [Google Scholar]
- 20.Huang H, et al. Drosophila CAF-1 regulates HP1-mediated epigenetic silencing and pericentric heterochromatin stability. J. Cell Sci. 2010;123:2853–2861. doi: 10.1242/jcs.063610. [DOI] [PubMed] [Google Scholar]
- 21.Chiolo I, et al. Double-strand breaks in heterochromatin move outside of a dynamic HP1a domain to complete recombinational repair. Cell. 2011;144:732–744. doi: 10.1016/j.cell.2011.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Federoff NV. Transposable elements, epigenetics, and genome evolution. Science. 2012;338:758–767. doi: 10.1126/science.338.6108.758. [DOI] [PubMed] [Google Scholar]
- 23.Sadaie M, et al. Balance between distinct HP1 family proteins controls heterochromatin assembly in fission yeast. Mol. Cell. Biol. 2008;28:6973–6988. doi: 10.1128/MCB.00791-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fischer T, et al. Diverse roles of HP1 proteins in heterochromatin assembly and functions in fission yeast. Proc. Natl. Acad. Sci. USA. 2009;106:8998–9003. doi: 10.1073/pnas.0813063106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Czermin B, et al. Physical and functional association of SU(VAR)3-9 and HDAC1 in Drosophila. EMBO Rep. 2001;2:915–919. doi: 10.1093/embo-reports/kve210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Motamedi MR, et al. HP1 proteins form distinct complexes and mediate heterochromatic gene silencing by non-overlapping mechanisms. Mol. Cell. 2008;32:778–790. doi: 10.1016/j.molcel.2008.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Alper BJ, et al. Centromeric heterochromatin assembly in fission yeast-balancing transcription, RNA interference, and chromatin modification. Chromosome Res. 2012;20:521–534. doi: 10.1007/s10577-012-9288-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Honda S, et al. Heterochromatin protein 1 forms distinct complexes to direct histone deacetylation and DNA methylation. Nat. Struct. Mol. Biol. 2012;19:471–477. doi: 10.1038/nsmb.2274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Aygun O, et al. HDAC-mediated suppression of histone turnover promotes epigenetic stability of heterochromatin. Nat. Struct. Mol. Biol. 2013;20:547–554. doi: 10.1038/nsmb.2565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maksakova IA, et al. H3K9me3-binding proteins are dispesable for SETDB1/H3K9me3-dependent retroviral silencing. Epigenet. Chromatin. 2011;4:12. doi: 10.1186/1756-8935-4-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Azzaz AM, et al. HP1Hsα promotes nucleosome associations that drive chromatin condensation. J. Biol. Chem. 2014 doi: 10.1074/jbc.M113.512137. jbc.M113.512137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Canzio D, et al. Chromodomain-mediated oligomerization of HP1 suggests a nucleosome-bridging mechanism for heterochromatin assembly. Mol. Cell. 2011;41:67–81. doi: 10.1016/j.molcel.2010.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Munari F, et al. Methylation of lysine 9 in histone H3 directs alternative modes of highly dynamic interaction of heterochromatin protein hHP1β with the nucleosome. J. Biol. Chem. 2012;287:33756–33765. doi: 10.1074/jbc.M112.390849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Canzio D, et al. A conformational switch in HP1 releases auto-inhibition to drive heterochromatin assembly. Nature. 2013;496:377–381. doi: 10.1038/nature12032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nonaka N, et al. Recruitment of cohesin to heterochromatic regions by Swi6/HP1 in fission yeast. Nat. Cell Biol. 2002;4:89–93. doi: 10.1038/ncb739. [DOI] [PubMed] [Google Scholar]
- 36.Hahn M, et al. Suv4-20h2 mediates chromatin compaction and is important for cohesin recruitment to heterochromatin. Genes Dev. 2013;27:859–872. doi: 10.1101/gad.210377.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Keller C, et al. HP1Swi6 mediates the recognition and destruction of heterochromatic RNA transcripts. Mol. Cell. 2012;47:215–227. doi: 10.1016/j.molcel.2012.05.009. [DOI] [PubMed] [Google Scholar]
- 38.Klattenhoff C, et al. The Drosophila HP1 homologue Rhinois required for transposon silencing and piRNA production by dual strand clusters. Cell. 2009;138:1137–1149. doi: 10.1016/j.cell.2009.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Castel SE, Martiensson RA. RNA interference in the nucleus: roles for small RNAs in transcription, epigenetics and beyond. Nat. Rev. Genet. 2013;14:100–112. doi: 10.1038/nrg3355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Casteñeda J, et al. piRNAs, transposon silencing, and germline genome integrity. Mutat. Res. 2011;714:95–104. doi: 10.1016/j.mrfmmm.2011.05.002. [DOI] [PubMed] [Google Scholar]
- 41.Wang S, Elgin SCR. Drosophila Piwi functions downstream of piRNA production mediating a chromatin-based transposon silencing mechanism in female germline. Proc. Natl. Acad. Sci. USA. 2011;108:21164–21169. doi: 10.1073/pnas.1107892109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Le Thomas A, et al. Piwi induces piRNA-guided transcriptional silencing and establishment of a repressive chromatin state. Genes Dev. 2013;27:390–399. doi: 10.1101/gad.209841.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sienski G, et al. Transcriptional silencing of transposons by Piwi and Maelstrom and its impact on chromatin state and gene expression. Cell. 2012;151:964–980. doi: 10.1016/j.cell.2012.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mendez DL, et al. The HP1a disordered C terminus and chromo shadow domain cooperate to select target peptide partners. ChemBiochem. 2011;12:1084–1096. doi: 10.1002/cbic.201000598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bulut-Karslioglu A, et al. A transcription factor-based mechanism for mouse heterochromatin formation. Nat. Struct. Mol. Biol. 2012;19:1023–1030. doi: 10.1038/nsmb.2382. [DOI] [PubMed] [Google Scholar]
- 46.Ito S, et al. Epigenetic silencing of core histone genes by HERS in Drosophila. Mol. Cell. 2012;45:494–504. doi: 10.1016/j.molcel.2011.12.029. [DOI] [PubMed] [Google Scholar]
- 47.Cammas F, et al. Association of the transcriptional corepressor TIF1beta with heterochromatin protein 1 (HP1): an essential role for progression throughdifferentiation. Genes Dev. 2004;18:2147–2160. doi: 10.1101/gad.302904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schultz DC, et al. SETDB1: a novel KAP-1-associated histone H3, lysine 9-specific methyltransferase that contributes to HP1-mediated silencing of euchromatic genes by KRAB zinc-finger proteins. Genes Dev. 2002;16:919–932. doi: 10.1101/gad.973302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kharchenko PV, et al. Comprehensive analysis of the chromatin landscape in Drosophila melanogaster. Nature. 2011;471:480–485. doi: 10.1038/nature09725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Riddle NC, et al. Plasticity in patterns of histone modifications and chromosomal proteins in Drosophila heterochromatin. Genome Res. 2011;21:147–163. doi: 10.1101/gr.110098.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yasuhara JC, Wakimoto BT. Oxymoron no more: the expanding worldof heterochromatic genes. Trends Genet. 2006;22:330–338. doi: 10.1016/j.tig.2006.04.008. [DOI] [PubMed] [Google Scholar]
- 52.Grewal SI, Jia S. Heterochromatin revisited. Nat. Rev. Genet. 2007;8:35–46. doi: 10.1038/nrg2008. [DOI] [PubMed] [Google Scholar]
- 53.Ruan J, et al. Structural basis of the chromodomain of Cbx3 bound to methylated peptides from histone H1 and G9a. PLoS ONE. 2012;7(4):e35376. doi: 10.1371/journal.pone.0035376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Riddle NC, et al. Enrichment of HP1a on Drosophila chromosome 4 genes creates an alternate chromatin structure critical for regulation in this heterochromatic domain. PLoS Genet. 2012;8(9):e1002954. doi: 10.1371/journal.pgen.1002954. 2012 Sep. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rangan P, et al. piRNA production requires heterochromatin formation in Drosophila. Curr. Biol. 2011;21:1373–1379. doi: 10.1016/j.cub.2011.06.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cryderman DE, et al. Heterochromatin protein 1a is required for an open chromatin structure. Transcription. 2011;2:95–99. doi: 10.4161/trns.2.2.14687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.James TC, et al. Distribution patterns of HPI, a heterochromatin-associated chromosomal protein of Drosophila. Eur. J. Cell Biol. 1989;50:170–180. [PubMed] [Google Scholar]
- 58.de Wit E, et al. High-resolution mapping reveals links of HP1with active and inactive chromatin components. PLoS Genet. 2007;3(3):e38. doi: 10.1371/journal.pgen.0030038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Piacentini L, et al. Heterochromatin Protein 1 (HP1a) positively regulates euchromatic gene expression through RNA transcript association and interaction with hnRNPs in Drosophila. PLoS Genet. 2009;5(10):e1000670. doi: 10.1371/journal.pgen.1000670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Smallwood A, et al. CBX3 regulates efficient RNA processing genome-wide. Genome Res. 2012;22:1426–1436. doi: 10.1101/gr.124818.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bühler M, Hiller S. Dynamic nature of heterochromatin highlighted by a HP1Swi6-dependent gene silencing mechanism. Cell Cycle. 2012;11:3907–3908. doi: 10.4161/cc.22234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ip JY, et al. Global impact of RNA polymerase II elongation inhibition on alternative splicing regulation. Genome Res. 2011;21:390–401. doi: 10.1101/gr.111070.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.LeRoy G, et al. Heterochromatin protein 1 is extensively decorated with histone code-like post-translational modifications. Mol Cell Proteomics. 2009;8:2432–2442. doi: 10.1074/mcp.M900160-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Minc E, et al. Localization and phosphorylation of HP1 proteinsduring the cell cycle in mammalian cells. Chromosoma. 1999;108:220–234. doi: 10.1007/s004120050372. [DOI] [PubMed] [Google Scholar]
- 65.Koike N, et al. Identification of heterochromatin protein 1 (HP1) as a phosphorylation target by Pim-1 kinase and the effect of phosphorylation on the transcriptional repression function of HP1. FEBS Lett. 2000;467:17–21. doi: 10.1016/s0014-5793(00)01105-4. [DOI] [PubMed] [Google Scholar]
- 66.Ayoub N, et al. HP1-beta mobilization promotes chromatin changes that initiate the DNA damage response. Nature. 2008;453:682–686. doi: 10.1038/nature06875. [DOI] [PubMed] [Google Scholar]
- 67.Zhao T, Eissenberg JC. Phosphorylation of Heterochromatin Protein 1 by casein kinase II is required for efficient heterochromatin binding in Drosophila. J. Biol. Chem. 1999;274:15095–15100. doi: 10.1074/jbc.274.21.15095. [DOI] [PubMed] [Google Scholar]
- 68.Zhao T, et al. Phosphorylation site mutations in Heterochromatin Protein 1 (HP1) reduce or eliminate silencing activity. J. Biol. Chem. 2001;276:9512–9518. doi: 10.1074/jbc.M010098200. [DOI] [PubMed] [Google Scholar]
- 69.Badugu R, et al. Mutations in the heterochromatin protein 1 (HP1) hinge domain affect HP1 protein interactions and chromosomal distribution. Chromosoma. 2005;113:370–384. doi: 10.1007/s00412-004-0324-2. [DOI] [PubMed] [Google Scholar]
- 70.Chaturvedi P, Parniak VK. Lamin A rod domain mutants target Heterochromatin Protein 1α and β for proteasomal degradation by activation of F-box protein, FBXW10. PLoS ONE. 2010;13(5):e10620. doi: 10.1371/journal.pone.0010620. 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chaturvedi P, et al. Ubiquitin ligase RNF123 mediates degradation of Heterochromatin Protein 1a and b in lamin A/C knock-down cells. PLoS ONE. 2012;7(10):e47558. doi: 10.1371/journal.pone.0047558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Maison C, et al. SUMOylation promotes de novo targeting of HP1alpha to pericentric heterochromatin. Nat. Genet. 2011;43:220–227. doi: 10.1038/ng.765. [DOI] [PubMed] [Google Scholar]
- 73.Maison C, et al. The SUMO protease SENP7 is a critical component to ensure HP1 enrichment at pericentric heterochromatin. Nat. Struct. Mol. Biol. 2012;19:458–460. doi: 10.1038/nsmb.2244. [DOI] [PubMed] [Google Scholar]
- 74.Hughes RM, et al. Recognition of trimethyllysine by a chromodomain is not driven by the hydrophobic effect. Proc. Natl. Acad. Sci. USA. 2007;104:11184–11188. doi: 10.1073/pnas.0610850104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sun FL, et al. Long-range nucleosome ordering is associated with gene silencing in Drosophila melanogaster pericentric heterochromatin. Mol. Cell. Biol. 2001;21:2867–2879. doi: 10.1128/MCB.21.8.2867-2879.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Jacobs SA, et al. Specificity of HP1 chromo domain for methylated N-terminus of histone H3. EMBO J. 2001;20:5232–5241. doi: 10.1093/emboj/20.18.5232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gunesdogan U, et al. A genetic system to assess in vivo the functions of histones and histone modifications in higher eukaryotes. EMBO Rep. 2010;11:772–776. doi: 10.1038/embor.2010.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Pengelly AR, et al. A histone mutant reproduces the phenotype caused by loss of histone-modifying factor Polycomb. Science. 2013;339:698–699. doi: 10.1126/science.1231382. [DOI] [PubMed] [Google Scholar]
- 79.Cheutin T, et al. Maintenance of stable heterochromatin domainsby dynamic HP1 binding. Science. 2003;299:721–725. doi: 10.1126/science.1078572. [DOI] [PubMed] [Google Scholar]
- 80.Brower-Toland B, et al. Multiple SET methyltransferases are required to maintain normal heterochromatin domains in the genome of Drosophila melanogaster. Genetics. 2009;181:1303–1319. doi: 10.1534/genetics.108.100271. [DOI] [PMC free article] [PubMed] [Google Scholar]