SUMMARY
Global downregulation of microRNAs (miRNAs) is commonly observed in human cancers and can have a causative role in tumorigenesis. The mechanisms responsible for this phenomenon remain poorly understood. Here we show that YAP, the downstream target of the tumor-suppressive Hippo signaling pathway regulates miRNA biogenesis in a cell density-dependent manner. At low cell density, nuclear YAP binds and sequesters p72 (DDX17), a regulatory component of the miRNA processing machinery. At high cell density, Hippo-mediated cytoplasmic retention of YAP facilitates p72 association with Microprocessor and binding to a specific sequence motif in pri-miRNAs. Inactivation of the Hippo pathway or expression of constitutively active YAP causes widespread miRNA suppression in cells and tumors and a corresponding post-transcriptional induction of MYC expression. Thus, the Hippo pathway links contact-inhibition regulation to miRNA biogenesis and may be responsible for the widespread miRNA repression observed in cancer.
Keywords: MicroRNA, Microprocessor, Hippo, Yap, p72, DDX17, Cancer
INTRODUCTION
MicroRNAs (miRNAs) comprise a large family of regulatory RNAs that repress expression of target messenger RNAs (mRNAs) and have important roles in development and disease. Processing to the mature ~22 nucleotide miRNA is executed by the stepwise cleavage of long primary miRNAs (pri-miRNAs) by the Microprocessor and Dicer complexes (Figure S1A). Microprocessor minimally comprises the ribonuclease DROSHA and its double-stranded RNA-binding partner DGCR8 (Denli et al., 2004; Gregory et al., 2004). Microprocessor recognizes pri-miRNA through the stem-loop (Zeng et al., 2005) and the stem-loop-ssRNA junction (Han et al., 2006), and cleaves both the 5′ and 3′ flanking segments to generate pre-miRNA. Various co-factors can associate with Microprocessor (Fukuda et al., 2007; Gregory et al., 2004; Siomi and Siomi, 2010). These regulatory proteins include hnRNP A1 (Guil and Cáceres, 2007), p68 and p72 (DDX5 and DDX17, respectively) (Fukuda et al., 2007), Smad (Davis et al., 2008), KHSRP (Trabucchi et al., 2009), BRCA1 (Kawai and Amano, 2012), and FUS/TLS (Morlando et al., 2013). Microprocessor can also be modulated by inhibitory factors including Lin28A/B (Piskounova et al., 2011), Musashi homolog 2 (MSI2) and Hu antigen R (HuR) (Choudhury et al., 2013), and NF90-NF45 (Sakamoto et al., 2009) binding to distinct subsets of pri-miRNAs. Pre-miRNAs are exported to the cell cytoplasm by Exportin-5 (XPO5) where they are further cleaved by a complex of the ribonuclease DICER and the double-stranded RNA-binding protein TRBP2, generating mature miRNA duplexes (Chendrimada et al., 2005). The 5′ or 3′ miRNA is selected and loaded into the RNA-induced silencing complex (RISC) that recognizes sites in the 3′ untranslated region (UTR) of target mRNAs to repress protein expression (Bartel, 2009)
Altered miRNA expression is a hallmark of cancer and individual miRNAs can have either tumor suppressive or oncogenic functions. Furthermore, a prevailing feature observed in human cancers is the global decrease in miRNA expression compared to the corresponding normal tissue (Lee et al., 2008; Lu et al., 2005; Maillot et al., 2009; Ozen et al., 2008; Thomson et al., 2006). This miRNA suppression has a causative role in tumorigenesis (Chang et al., 2008; Kumar et al., 2007, 2009), implying its potential as a therapeutic target but the underlying mechanism is unknown. Importantly, widespread miRNA repression in cancers is likely a result of defective miRNA processing as evidenced by the accumulation of pri-miRNAs and the corresponding depletion of mature miRNAs (Lee et al., 2008; Thomson et al., 2006). Although rare mutations in Dicer (Hill et al., 2009), TRBP2 (Melo et al., 2009) and XPO5 (Melo et al., 2010) have been reported, the pathways and mechanisms controlling miRNA expression remain poorly understood.
To investigate how miRNA expression might be dysregulated in tumors, we focused on the report that miRNA biogenesis is affected by cell density (Hwang et al., 2009). These observations are specially relevant considering that loss of cell-contact inhibition is a common feature of tumor cells. Connecting these previously reported phenomena we postulated that the observed global miRNA repression in tumors might be related to the cell density-dependent regulation of miRNA biogenesis. We focused on the Hippo signaling pathway as a potential regulator of cell density-dependent miRNA biogenesis, because (1) Hippo pathway activity is highly sensitive to cell density and cell-cell junctions (Kim et al., 2011; Schlegelmilch et al., 2011; Silvis et al., 2011; Zhao et al., 2007); (2) the Hippo pathway regulates the balance between differentiation and renewal of multiple stem and progenitor cell types (Camargo et al., 2007; Lian et al., 2010; Schlegelmilch et al., 2011); and (3) misregulation of Hippo signaling is a common feature of human solid tumors (Harvey et al., 2013; Zhao et al., 2010). The Hippo cascade is emerging as an essential pathway for the regulation of tissue homeostasis and organ size (Ramos and Camargo, 2012), and is characterized by responsiveness to physiological cues such as cellular crowding (Zhao et al., 2007), activation of G protein-coupled receptors (Yu et al., 2012), cell shape (Wada et al., 2011) and mechanical forces (Dupont et al., 2011; Halder et al., 2012). These cues culminate in differential subcellular localization of the transcriptional co-activator YAP. At low cell density, Hippo signaling is suppressed and YAP localizes in the nucleus where it promotes cellular proliferation through transcriptional mechanisms. As cellular crowding increases, cell-cell contacts form and YAP is phosphorylated and sequestered in the cytoplasm by adherens junction proteins E-cadherin (Kim et al., 2011) and α-catenin (Schlegelmilch et al., 2011; Silvis et al., 2011). Nuclear YAP induces the reversible overgrowth of multiple organs and tumorigenesis in mice (Camargo et al., 2007; Dong et al., 2007). Additionally, deregulation of the Hippo pathway has been reported at a high frequency in a broad range of different human carcinomas, and it often correlates with poor patient prognosis (Harvey et al., 2013).
Here we identify the Hippo signaling pathway as a regulator of Microprocessor activity. We show that YAP regulates miRNA biogenesis through sequestering the Microprocessor component p72 in a cell density-dependent manner. We furthermore find that perturbation of Hippo signaling causes widespread miRNA suppression in cells and tumors and may underlie the widespread miRNA repression in human tumors.
RESULTS
Hippo pathway component YAP regulates Microprocessor activity in a cell density-dependent manner
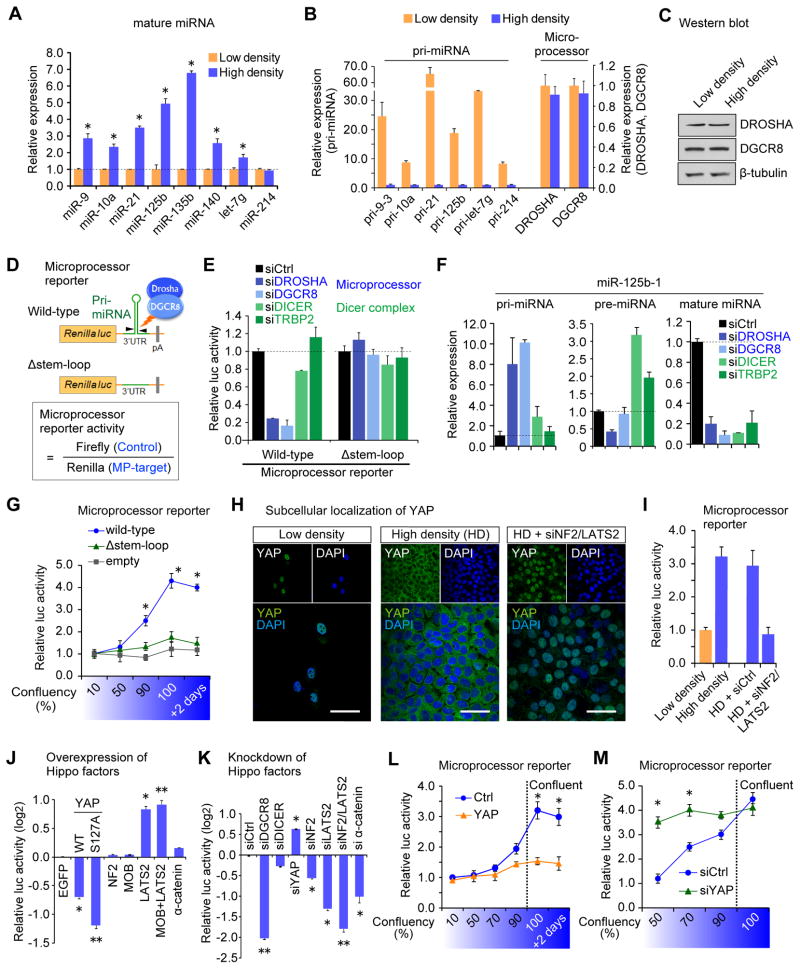
To investigate the potential mechanism of cell density-dependent miRNA biogenesis and gain insight into global miRNA suppression in tumors, we first characterized miRNA regulation in the non-transformed human keratinocyte HaCaT cell line. Consistent with published data, we observed elevated miRNA expression at high cell density (Figures 1A and S1B, Hwang et al., 2009). To avoid the selective loss of miRNAs with low GC content that reportedly occurs when extracting RNA from a small number of cells (Kim et al., 2012), we plated similar numbers of cells onto plates of different sizes. This then allowed us to culture cells at varying confluence without introducing the technical artifact caused by different RNA yields. The corresponding pri-miRNAs were upregulated at lower cell density (Figure 1B), implying a general blockade of miRNA processing at lower cell density. Expression of Microprocessor components DROSHA and DGCR8 was not altered by cell density (Figures 1B and 1C), suggesting the activity, not the quantity, of Microprocessor may underlie the altered miRNA biogenesis.
Figure 1. YAP regulates Microprocessor activity in a cell density-dependent manner.
(A) qRT-PCR analysis of miRNA expression in HaCaT cells. Data were normalized to U6. *p < 0.05, Student’s t test. (B) Relative expression of pri-miRNAs and DROSHA and DGCR8 at low and high densities. qRT-PCR data normalized to GAPDH. (C) Western blot analysis. (D) Schematic representation of the Microprocessor (MP) reporter. The Δstem-loop mutant lacks the pre-miRNA stem-loop crucial for the recognition by Microprocessor. (E) Microprocessor reporter assays. (F) Expression levels of pri-, pre- and mature miR-125b after indicated siRNA-mediated knockdown, normalized to GAPDH for the pri-miRNA and to U6 for the pre- and mature miRNA. (G) Microprocessor reporter activity at different cell densities. *p < 0.05 versus empty, Student’s t test. (H) Immunocytochemistry analysis of YAP localization. YAP nuclear translocation was induced by knockdown of NF2 and LATS2. Scale bar, 30μm. (I-M) Microprocessor reporter assays *p < 0.05, **p < 0.01, Student’s t test. Data are represented as mean +/− SEM. See also Figure S1.
To assess Microprocessor activity in cells, we engineered a luciferase reporter that utilizes portions of pri-miR-125b-1 or pri-miR-205 embedded in the 3′UTR of the Renilla luciferase gene (Figure 1D). A similar approach to monitor Microprocessor activity has been described (Tsutsui et al., 2008). Cleavage by Microprocessor is expected to destabilize the Renilla luciferase mRNA and lead to decreased Renilla luminescence. We measured Microprocessor activity by normalizing to the control Firefly luciferase value so that the calculated values positively correlated with the endogenous Microprocessor activity (Figure 1D). To validate these reporters we measured response to DROSHA or DGCR8 knockdown in HaCaT cells (Figures 1E and S1C) where pri-miR-125b, but not pre-miR-125b, accumulates and mature miR-125b is suppressed (Figure 1F). Validation was also performed using Dgcr8-knockout mouse embryonic stem cells (Figures S1D and S1E). The reporter was not affected by knockdown of DICER or TRBP2 (Figure 1E). To further confirm the specificity, we generated a control construct in which the pre-miRNA stem-loop was deleted (Figure 1D). Expression of this reporter was unresponsive to depletion of Microprocessor (Figure 1E). Altogether, these data verify that the reporter serves as a sensitive readout of Microprocessor activity in cells.
Using the reporter system we found Microprocessor activity was enhanced at higher cell densities compared to lower cell confluency (Figures 1G and S1F). To explore how this cell density-dependent Microprocessor activity could be regulated, we focused on the Hippo signaling pathway. YAP localizes in the nucleus at low confluency and translocates to the cytoplasm at high density (Figure 1H). This localization is dependent on the upstream kinase LATS2 and other upstream negative regulatory molecules such as the tumor suppressor NF2. Inactivation of NF2 and LATS2 abrogated the cytoplasmic sequestration of YAP at higher density (Figure 1H, “HD + siNF2/LATS2”). Using the Microprocessor reporter we found that knockdown of NF2 and LATS2 abrogated the enhanced Microprocessor activity observed at high density (Figure 1I), implying that the Hippo pathway regulates Microprocessor activity. Additionally, forced expression of either YAP or a nuclear-targeted phospho-mutant YAP S127A repressed Microprocessor activity, whereas overexpression of LATS2 resulted in enhanced reporter activity, presumably through YAP phosphorylation and cytoplasmic retention (Figure 1J). Individual knockdown of NF2, LATS2 or α-catenin had the reciprocal effect on Microprocessor activity (Figure 1K). We also tested Lats1- and Lats2-deficient mouse embryonic fibroblasts (MEFs) generated by transducing Lats1−/−;Lats2fl/fl MEFs with Cre-expressing adenovirus (Kim et al., 2013, Figure S1G) and observed suppressed Microprocessor reporter activity (Figure S1H) and lowered miRNA expression (Figures S1I and S1J). We further examined the consequences of YAP overexpression or knockdown at different cell densities. Both forced YAP expression (Figure 1L) and YAP knockdown (Figure 1M) abrogated the cell density-dependency of Microprocessor activity. Altogether, these results reveal that the Hippo signaling pathway and its downstream component YAP regulate Microprocessor activity.
YAP sequesters p72 (DDX17) from Microprocessor in a cell density-dependent manner
We next interrogated how YAP could control Microprocessor activity. Since YAP has an established role as a transcriptional co-activator, we first focused on the possible transcriptional role of YAP (Yagi et al., 1999). Microarray analyses showed none of the Microprocessor-related genes were significantly affected by YAP activation (Figure S2A). To further rule out a transcriptional role for YAP in Microprocessor regulation, we made use of a YAP S94A mutant unable to bind the TEAD-family of transcription factors (Schlegelmilch et al., 2011; Zhao et al., 2008). YAP S94A and its nuclear-targeted version YAP S94A/5SA suppressed Microprocessor activity to a similar extent as their wild-type counterparts (Figure S2B). These results imply that YAP regulates Microprocessor activity independent of its transcriptional activity. We therefore focused on the possibility that YAP might regulate Microprocessor posttranscriptionally.
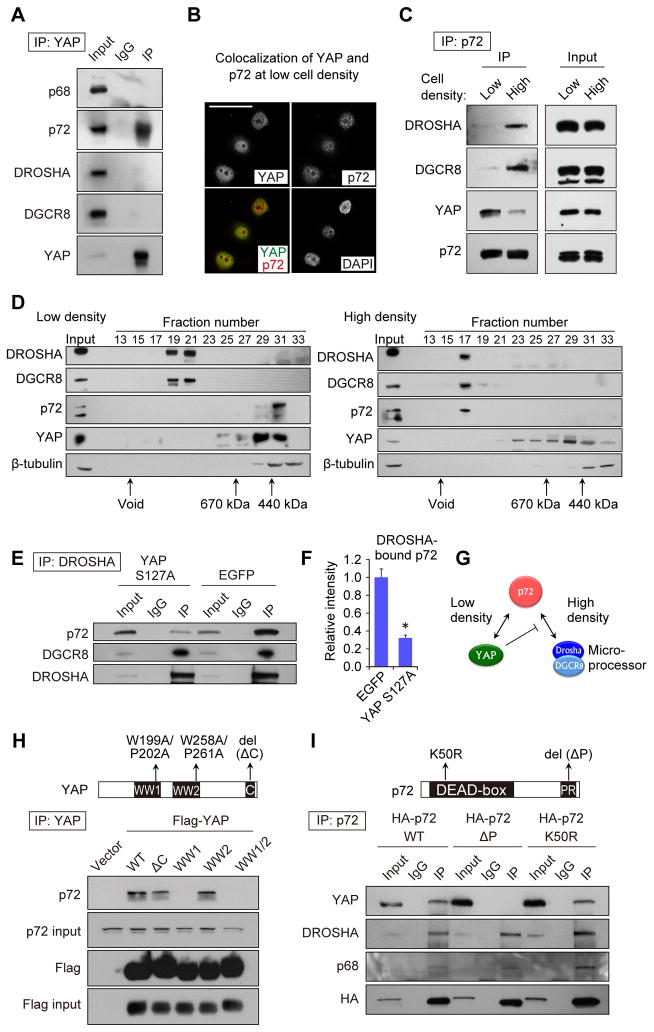
We tested whether YAP might physically interact with Microprocessor components. We did not detect an association between YAP and DROSHA or DGCR8 in co-immunoprecipitation (co-IP) assays (Figure 2A). We next considered that YAP might associate with the Microprocessor accessory proteins p68 (DDX5) and p72 (DDX17), DEAD (Asp-Glu-Ala-Asp)-box RNA helicases that are components of a large DROSHA-containing complex required for processing of a large subset of miRNAs (Fukuda et al., 2007; Gregory et al., 2004). It is emerging that several different cellular signaling pathways use the p68 and/or p72 association with Microprocessor to effect regulation of pri-miRNA processing (Newman and Hammond, 2010; Siomi and Siomi, 2010). Co-IPs indicated that p72, but not the structurally similar p68, specifically associates with endogenous YAP protein (Figure 2A).
Figure 2. YAP sequesters p72 from Microprocessor complex in a cell density-dependent manner.
(A) Co-immunoprecipitation assays (Co-IPs) with endogenous YAP in HaCaT cells. (B) Immunocytochemistry of YAP and p72 in HaCaT cells at low density. Nuclei were stained with DAPI. Scale bar, 30μm. (C) Co-IP with HA-p72 in HaCaT cells at low and high density. (D) Western blot analysis of Superose 6 gel-filtration fractions. Whole cell lysates from HaCaT cells cultured at low and high densities were fractionated. β-tubulin served as a control. (E) Co-IP with Flag-DROSHA in HaCaT cells transfected with YAP or control EGFP. (F) Densitometry measurement for the amount of p72 bound by DROSHA in the HaCaT cells transfected with YAP or control EGFP (n=3). *p < 0.05, Student’s t test. (G) The scheme of interactions among YAP, p72 and Microprocessor. (H) Co-IP with Flag-YAP and YAP mutants. Mutations in YAP are represented in the upper panel. WT, wild-type. (I) Co-IP with HA-p72 and p72 mutants. Mutations in p72 are represented in the upper panel. See also Figure S2.
Immunocytochemistry showed that YAP and p72 colocalize in the nucleus of HaCaT cells at low density (Figure 2B) and not at high density (Figure S2C). We assessed the impact of cell density on this interaction by co-IPs with endogenous p72 protein. At higher density, p72 interacted with DROSHA and DGCR8, consistent with its role in pri-miRNA processing. Interestingly, the interaction between p72 and DROSHA/DGCR8 complex was significantly decreased at lower density and instead p72 was associated with YAP (Figure 2C). To gain deeper insight into these cell density-dependent interactions, we fractionated cell lysates collected at low and high densities using a gel-filtration column. At high density, p72 eluted in the same fractions as DROSHA and DGCR8, implying p72 association with the Microprocessor complex (Figure 2D, “High density”). Remarkably, at low density, p72 was not detected in the same fractions as DROSHA, but in the lower molecular weight fractions where YAP was also eluted, implying the interaction of p72 and YAP at low cell density (Figure 2D, “Low density”).
This dynamic cell density-dependent association of p72 with Microprocessor raised the possibility that nuclear YAP might inhibit Microprocessor activity at low cell density by binding and sequestering p72 from DROSHA and DGCR8. Overexpression of the constitutively active YAP S127A mutant led to a reduction in the relative amount of p72 associated with DROSHA in co-IPs, (Figures 2E–2G). Further analyses indicated that YAP WW domain 1 (W177–W199) and p72 C-terminal proline-rich sequence were essential for that interaction, while YAP WW domain 2 and p72 K50 residue, which is required for HDAC1 interaction (Mooney et al., 2010), were not (Figures 2H and 2I). We then tested YAP mutants with the Microprocessor reporter system. YAP WW domain mutant 1 (WW1) failed to inhibit Microprocessor activity (Figure S2D). We knocked down p72, and found that the density-dependent enhancement of Microprocessor activity was abrogated in the Microprocessor reporter system in a similar fashion as knockdown of NF2/LATS2 (Figure S2E). Combinatorial knockdown did not show any additive effect. qRT-PCR analyses of pri-, pre- and mature miR-125b corroborated these results (Figure S2F). Taken together, the association of p72 with Microprocessor is cell density-dependent, and nuclear YAP sequesters p72 through its WW1 domain at low cell density to suppress Microprocessor activity.
We further explored whether the TEAD proteins were also part of the protein complex containing YAP and p72. Co-IPs revealed that the YAP/p72 complex does not contain TEAD1 (Figure S2G). Additionally, we also observed interaction of TAZ, a YAP paralog, with p72 (Figure S2H) (Dupont et al., 2011; Halder et al., 2012). Forced expression of TAZ lowered mature miRNA expression (Figure S2I), which was accompanied with increased pri-miRNA expression (Figure S2J). Simultaneous knockdown of YAP and TAZ had an additive effect on Microprocessor reporter activity (Figures S2K and S2L). Thus, our results suggest a similar role of TAZ in miRNA biogenesis and further implicate another Hippo signaling molecule in the regulation of miRNA processing.
Inhibition of the Hippo signaling pathway suppresses Microprocessor activity
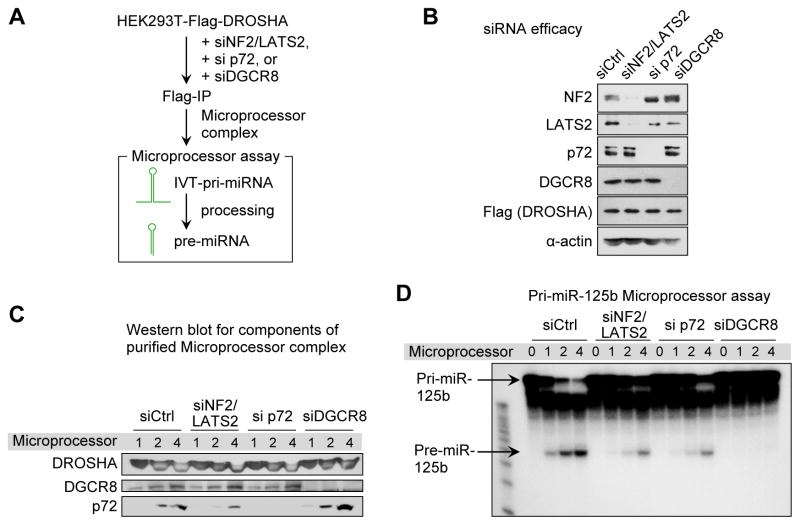
We next examined the significance of p72 and Hippo signaling for Microprocessor function using a Microprocessor biochemical assay (Figure 3A). We depleted DGCR8, p72 or NF2 and LATS2 in a stable HEK293T cell line expressing Flag-DROSHA (Figure 3B) and affinity-purified DROSHA-containing complexes (Figure 3C). When NF2 and LATS2 were depleted (Figure 3B), the amount of p72 associated with DROSHA was lowered (Figure 3C), consistent with our findings in HaCaT cells. We measured pri-miRNA processing activity of Microprocessor isolated from control and knockdown cells. The p72-depleted Microprocessor displayed compromised activity for pri-miR-125b-1 (Figure 3D). Upon depletion of NF2 and LATS2 Microprocessor activity was similarly impaired (Figure 3D). These findings imply that p72 depletion and YAP activation have a direct impact on pri-miRNA processing and strongly suggest that the Hippo pathway regulates miRNA expression through altering Microprocessor activity.
Figure 3. Hippo pathway and p72 regulate pri-miRNA processing efficiency of Microprocessor.
(A) Scheme of the Microprocessor assay. IVT, in vitro-transcribed. (B) Western blot analysis showing the siRNA efficacies and expression of Flag-DROSHA. (C) Western blot of purified protein complexes. (D) Microprocessor assays with in vitro transcribed pri-miR-125b. The numbers for Microprocessor indicate the relative amounts of Flag-IP products used for western blot (C) and Microprocessor assay (D).
Global impact of Hippo pathway on miRNA biogenesis
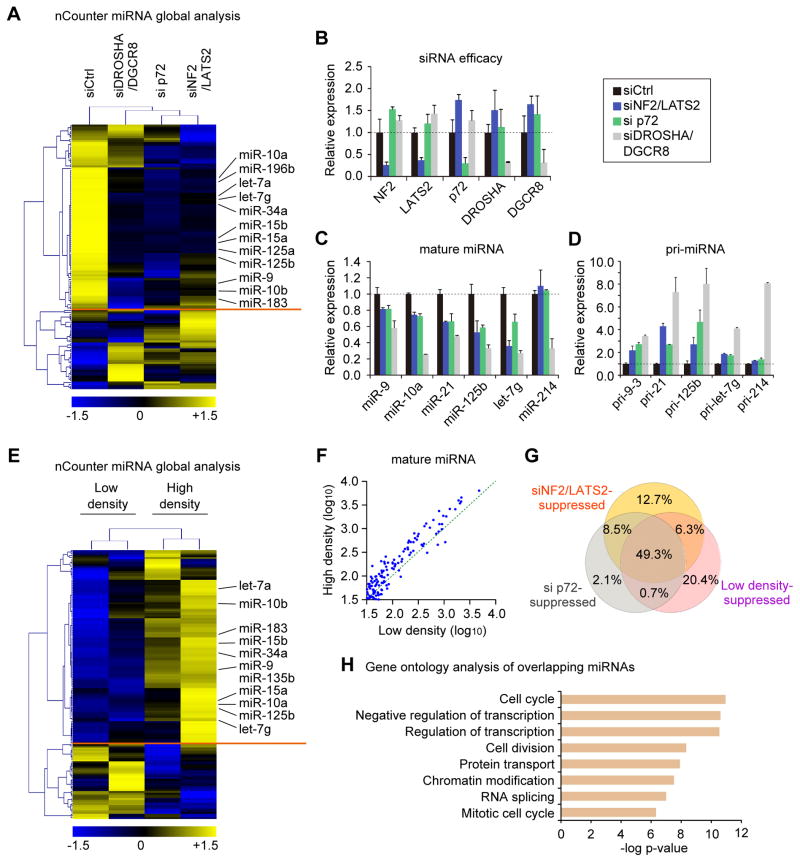
Our data above would predict that Hippo signaling inactivation, and consequent YAP nuclear translocation would result in general miRNA suppression. We sought to examine this by utilizing nCounter technology to profile more than 600 different miRNAs. Indeed, NF2/LATS2 knockdown at high cell density lowered (<0.8 fold compared to siCtrl) 61.0% of miRNAs in HaCaT cells (Figures 4A and 4B). We next addressed to what extent p72 explains global miRNA repression by YAP activation. As a result, 59.8% of miRNAs were suppressed by p72 knockdown, and 90.2% of p72-suppressed miRNAs overlapped with siNF2/LATS2-suppressed miRNAs. Quantification by qRT-PCR validated the repression of representative miRNAs (Figure 4C) and the corresponding accumulation of pri-miRNAs (Figure 4D). The expression of miR-214, which is independent of p72 in the mouse embryo (Fukuda et al., 2007), was not affected by either p72 or NF2/LATS2 knockdown (Figure 4C). We further examined the role of p72 as a mediator of miRNA repression via Hippo signaling by testing whether forced expression of p72 could rescue expression of the siNF2/LATS2-suppressed miRNAs. Indeed, p72 expression in addition to the siNF2/LATS2 transfection enhanced numerous miRNAs (Figure S3A), suggesting that p72 positively regulates global miRNA biogenesis downstream of NF2 and LATS2.
Figure 4. Global impact of Hippo pathway on miRNA biogenesis.
(A) Global miRNA expression analysis of HaCaT with indicated knockdown. The miRNAs <0.8 or 1.2< fold compared to the siCtrl were analyzed by hierarchical clustering. (B) The efficacy of siRNAs used in (A). The expression values were normalized to GAPDH. (C) qPCR-quantification of mature miRNAs normalized to U6. (D) Pri-miRNA expression levels measured by qRT-PCR. Data were normalized to GAPDH. (E) miRNA expression analysis in RNA samples from low- and high-density HaCaT cells. miRNAs changed by <0.8 or 1.2< fold were analyzed by hierarchical clustering. (F) Scatter plot of miRNA expression levels (log10) in the low and the high densities (G) Venn diagram showing the overlap of miRNAs repressed by siNF2/LATS2, si p72, or low density. (H) Gene ontology analysis of the overlapping miRNAs in (G). Bonferroni-corrected p-values were indicated. Data are represented as mean +/− SEM. See also Figure S3.
We next interrogated the cell density-dependent global alteration in miRNA expression. As reported in other types of cells (Hwang et al., 2009), HaCaT cells also showed widespread variation of miRNA expression in a cell density-dependent manner. At lower cell density, 57.3% of miRNAs were suppressed (<0.8 fold) relative to higher density (Figures 4E and 4F). This density-dependent miRNA suppression could be rescued by YAP knockdown (Figure S3B). To examine to what extent this density-dependent miRNA modulation is regulated by YAP and p72, we compared the miRNAs repressed in lower density to the miRNAs repressed by p72 knockdown and NF2/LATS2 knockdown. In this analysis 49.3% of miRNAs overlapped among the three conditions of NF2/LATS2 knockdown, p72 knockdown, and low density (Figure 4G). Gene ontology analysis revealed that the predicted mRNA targets for the overlapping miRNAs were highly enriched in cell cycle control (Figure 4H). Together our data support that the Hippo signaling pathway, through the YAP-mediated control of p72 availability, is responsible for widespread cell density-dependent miRNA regulation.
p72 recognizes a sequence motif in pri-miRNAs
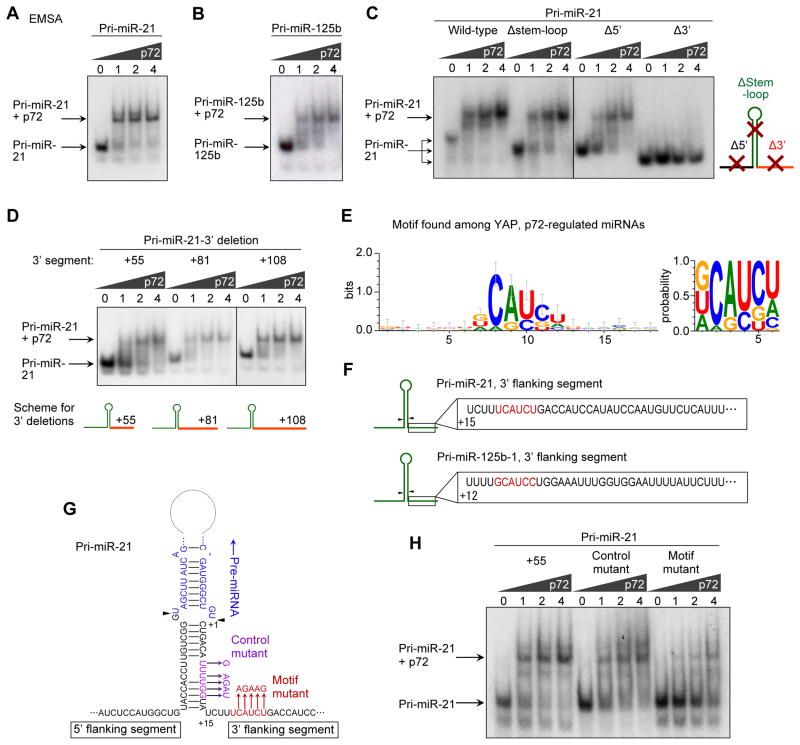
p72 harbors a DEAD box domain and is regarded as an RNA helicase. Although p72 is essential for normal miRNA expression in the developing mouse embryo (Fukuda et al., 2007), the precise role of p72 in pri-miRNA processing is unknown. We sought to dissect how p72 contributes to pri-miRNA processing. We hypothesized p72 might recognize a specific secondary structure or a sequence of pri-miRNAs to enhance the processing by Microprocessor. Electrophoretic mobility shift assays (EMSA) performed with recombinant p72 protein (Figure S4A) and in vitro transcribed pri-miRNAs showed a stable interaction between p72 and pri-miR-21 (Figure 5A) and pri-miR-125b-1 (Figure 5B). To examine the relevance of the secondary structure of pri-miRNAs, we deleted the stem-loop (Δstem-loop), 5′ flanking segment (FS, Δ5′) or 3′ FS (Δ3′) of pri-miR-21. The deletion of the stem-loop did not significantly affect the interaction, implying that the recognition of pri-miRNA by p72 is independent of the stem-loop structure (Figure 5C). The Δ5′ mutant showed slightly impaired interaction, but deletion of the 3′ flanking segment almost totally abolished the interaction, suggesting p72 interacts through the 3′ FS of pri-miR-21 (Figure 5C). Sequential shortening of the 3′FS suggested the distal part of the 3′ FS was dispensable (+81, +108, Figure 5D), though the interaction was minimally impaired with the shortest mutant (+55, Figure 5D). To test whether p72 recognizes a specific sequence in the 3′ FS of pri-miRNA, we searched for an overrepresented sequence motif in the 3′FS. As the input sequences, we utilized the sequences of pre-miRNA with of approximately 55 nt of 5′ and 3′ FS for the subset of the miRNAs repressed by both p72 and NF2/LATS2 knockdown. As the background data, those of non-suppressed pre-miRNAs were used. A VCAUCH sequence was identified in the 3′ FS of the subgroup of pri-miRNAs that are repressed by both p72 and NF2/LATS2 knockdown (Figure 5E). The motif was present at +19 and +16 of 3′ FS of pri-miR-21 and pri-miR-125b-1, respectively (Figure 5F). We examined the functional relevance of the motif by introducing mutations in the motif (Figure 5G, “motif mutant”) or in the adjacent sequence (“control mutant”). EMSA revealed an impaired interaction of p72 with the motif mutant, demonstrating the functional relevance of the motif in the 3′ FS of pri-miR-21 (Figure 5H). To further test the relevance of the motif sequence in a cellular context, we induced deletion mutations to the Microprocessor reporter plasmid. Deletions of the motif sequence significantly impaired the density sensitivity of the Microprocessor reporter (Figure S4B) and reduced the responsiveness to YAP activation through NF2/LATS knockdown (Figure S4C). These findings suggest p72 selectively binds the defined sequence in the 3′ FS of pri-miRNAs to enhance processing by the Microprocessor.
Figure 5. p72 DEAD box RNA helicase binds to a sequence motif in the 3′ flanking segment of pri-miRNA.
(A–D) EMSA with recombinant p72 protein and in vitro transcribed pri-miR-21, pri-miR-125b-1 or deletion mutants of pri-miR-21. Stem loop (Δstem-loop), 5′ flanking segment (Δ5′) or 3′ flanking segment (Δ3′) were deleted. (E) Identification of a sequence motif in the miRNAs repressed by both knockdown of NF2/LATS2 and p72. (F) Schematics showing the motif in the 3′ flanking segments of pri-miR-21 and pri-miR-125b-1. (G) Pri-miR-21 schematic indicating the motif mutations introduced and the control mutant. Arrowheads indicate cleavage sites by the Microprocessor. (H) EMSA with recombinant p72 protein and the +55 mutant, the control mutant and the motif mutant of the pri-miR-21. See also Figure S4.
YAP-regulated miRNAs target MYC
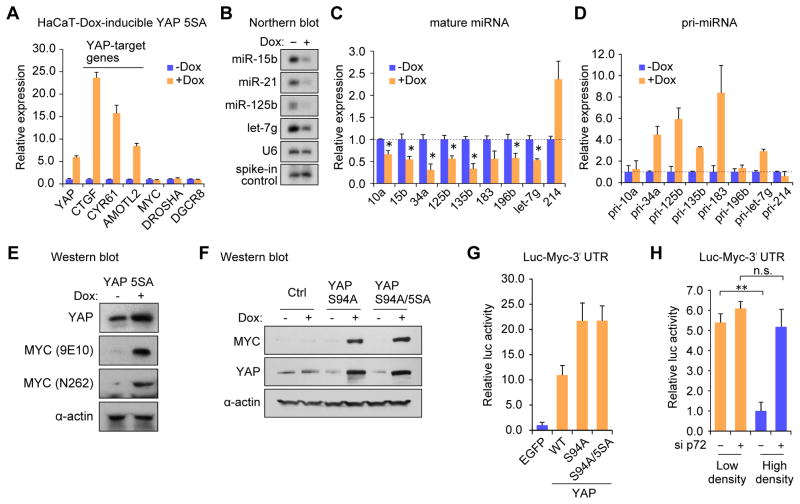
Although global miRNA suppression is suggested to have a causal role in tumorigenesis, the specific mechanisms underlying this are not fully understood. We considered that repression of a certain subset of miRNAs by YAP might lead to post-transcriptional enhancement of target gene(s) crucial for tumorigenesis and growth. To examine this, we established a HaCaT stable cell line expressing a YAP 5SA mutant in a doxycycline (Dox)-inducible manner. YAP 5SA has four serine residues substituted to alanine in addition to S127 and it displays enhanced nuclear localization (Zhao et al., 2007). After Dox treatment for 4 days, YAP and YAP-target genes CTGF, CYR61 and AMOTL2 were induced without affecting the mRNA levels of MYC (Figure 6A). Numerous mature miRNAs were repressed (Figures 6B and 6C) with a corresponding accumulation or sustained pri-miRNA expression (Figures 6D and S5A). Among the growth-related proteins tested, we found enhanced expression of MYC protein (Figure 6E) after YAP induction. MYC induction was also observed in HaCaT cell lines inducibly expressing YAP S94A and YAP S94A/5SA, suggesting MYC was post-transcriptionally induced (Figures 6F and S5B).
Figure 6. YAP-regulated miRNAs repress MYC expression.
(A) qRT-PCR analysis with data normalized to GAPDH. (B) miRNA Northern blot performed with spike-in of luciferase siRNA for normalization. (C) qRT-PCR analysis of mature miRNA levels normalized to U6. *p < 0.05, Student’s t test. (D) Relative pri-miRNA expression measured by qRT-PCR normalized to GAPDH. (E–F) Western blot analysis using indicated antibodies. (G–H) Luciferase assays with a MYC 3′UTR reporter. HaCaT cells were co-transfected with the luciferase and the expression plasmids for YAP or control EGFP. Luciferase activity was normalized to that of pRL-Tk. **p < 0.01, Student’s t test. n.s., not significant. Data are represented as mean +/− SEM. See also Figure S5.
To further examine the YAP-mediated post-transcriptional induction of MYC, we investigated the relevance of the MYC 3′UTR which contains potential targeting sites for several miRNAs that were repressed by YAP overexpression and p72 depletion (Figure S5C). Among them, let-7 (Kumar et al., 2007) and miR-34a (Christoffersen et al., 2010) were reported to target MYC 3′UTR. We utilized a luciferase gene harboring the MYC 3′UTR (Kumar et al., 2007) and compared luciferase activity to a control plasmid. YAP 5SA overexpression induced luciferase activity more than 10-fold compared to a control EGFP (Figure 6G). TEAD-binding deficient YAP mutants, YAP S94A and YAP S94A/5SA, also strongly activated the luciferase reporter, reinforcing the role of YAP in the posttranscriptional regulation of MYC expression (Figure 6G). The activity of Luc-MYC 3′UTR was also suppressed at higher cell density and correlated with the accumulation of miRNAs targeting MYC (Figure 6H). Knockdown of p72 at higher cell density rescued the repression of luciferase activity, suggesting that cell density-dependent regulation of MYC 3′UTR was mediated by p72 (Figure 6H). Collectively, the post-transcriptional induction of MYC protein is a functional outcome of YAP-mediated cell density-dependent global miRNA repression.
YAP mediates global miRNA suppression in tumors
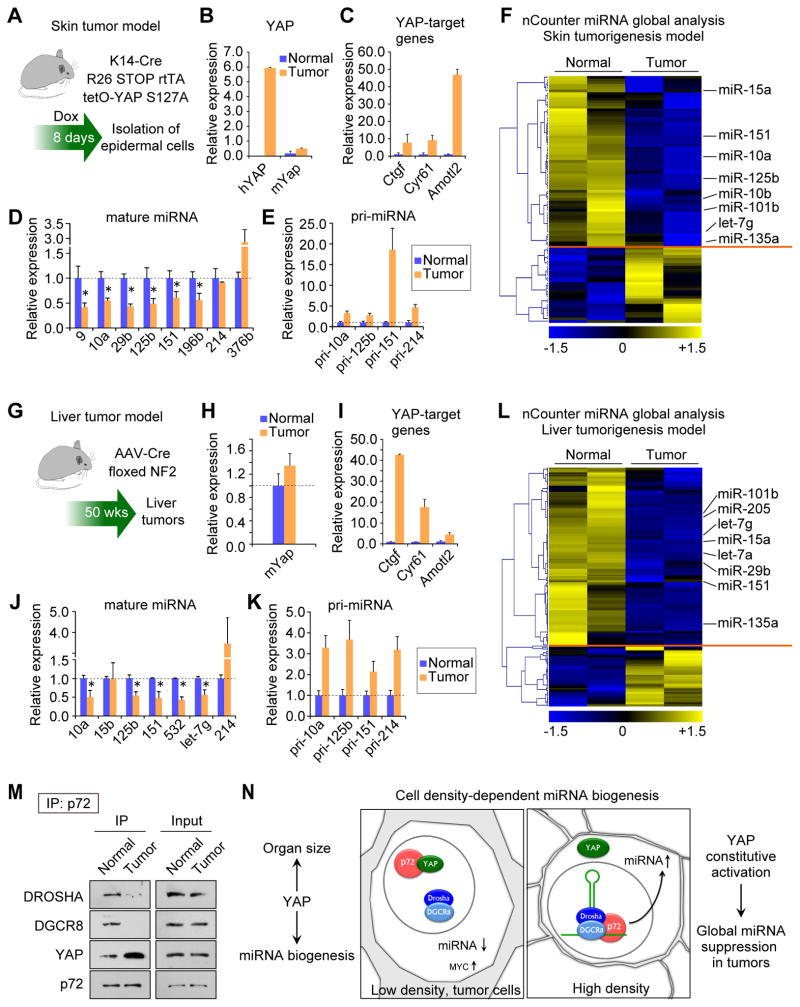
A large number of solid human cancers demonstrate impaired Hippo signaling and exhibit constitutive nuclear YAP localization (Harvey et al., 2013). Additionally, YAP activation leads to rapid tumor development in mice (Benhamouche et al., 2010; Camargo et al., 2007; Dong et al., 2007; Schlegelmilch et al., 2011; Zhang et al., 2010). Our findings in vitro imply that YAP-driven tumors might exhibit global miRNA repression. We tested this prediction in mouse models of YAP-induced tumorigenesis. We evaluated this in two distinct contexts of YAP activation: acute (8 days) YAP induction in the epidermis (Figure 7A) and chronic (50 weeks) YAP activation in the liver (Figure 7G). Short-term expression of a transgenic YAP S127A in the Keratin 14 positive (K14+) epidermal progenitor cells induces in situ squamous cell carcinoma-like tumors in mice which can produce invasive growth upon transplantation into nude mice (Schlegelmilch et al., 2011). Gene expression analyses of the oncogenic epidermal cells revealed potent induction of transgenic YAP S127A (hYAP, Figure 7B) and YAP target genes (Figure 7C). As predicted, YAP induction led to the repression of numerous mature miRNAs (Figure 7D) and the accumulation of pri-miRNAs (Figure 7E), recapitulating our in vitro findings. Global analysis revealed that 52.5% of miRNAs were suppressed at least 0.8 fold in the tumorigenic cells as compared to the normal epidermal cells (Figure 7F).
Figure 7. YAP mediates the global repression of miRNA biogenesis in tumors.
(A) Mouse model of YAP-induced skin tumorigenesis. (B) Expression of exogenous human YAP S127A and endogenous mouse Yap normalized to Hprt1 in isolated epidermal cells. (C) Expression levels of YAP-target genes normalized to Hprt1. (D) Mature miRNA expression levels normalized to sno142. *p < 0.05, Student’s t test. (E) Expression levels of the pri-miRNAs normalized to Hprt1. (F) Global miRNA analysis. (G) Mouse model of YAP-induced tumorigenesis in the liver. (H) Expression levels of mouse Yap normalized to Hprt1. (I) The expression levels of YAP-target genes normalized to Hprt1. (J) Mature miRNA expression levels normalized to sno142. *p < 0.05, Student’s t test. (K) Relative expression levels of the pri-miRNAs normalized to Hprt1. (L) Global miRNA analysis in the liver tissues. (M) Co-IP with p72 in the normal tissues and tumors from the mouse livers. (N) Proposed model. Data are represented as mean +/− SEM. See also Figure S6.
The liver tumor model relied on hepatocyte-specific deletion of Nf2 in adult mice (Figures 7G and 7H). This resulted in hepatomegaly, cholangiocarcinoma-like tumor formation (Figure S6A–C), and induction of YAP target genes (Figure 7I). The expression of mature miRNAs was repressed in the liver tumors compared to control tissue (Figure 7J), which was accompanied by the accumulation of pri-miRNAs (Figure 7K). The miRNA global analysis revealed 61.0% of miRNAs were repressed in the liver tumors as compared to normal tissue (Figure 7L). To explore the relevance of p72 in the context of YAP-induced tumorigenesis, we examined the association of p72 with Microprocessor and YAP. Co-IP revealed that the interaction between p72 and Microprocessor observed in normal livers was significantly decreased in tumor tissues, whereas the association between p72 and YAP was increased in Nf2-deficient tumors (Figure 7M). The interaction between p72 and YAP was also observed in the skin tumors (Figure S6D). Overall these results demonstrate that YAP-driven tumorigenesis is associated with widespread miRNA suppression and that YAP activation promotes the dissociation of p72 from Microprocessor complex in tumor cells.
Finally, we examined whether this transcription-independent function of YAP plays a causative role in cellular growth. We chose to study the consequences of expressing the TEAD-binding defective mutant YAP S94A/5SA, given that current dogma suggests that most of YAP’s effects are mediated by transcription through the TEAD proteins. YAP S94A/5SA repressed miRNAs with the p72-binding motifs (Figure S6E), and also showed significant acceleration of cell growth in HaCaT cells, although this effect was less potent than that of YAP 5SA. Co-expression of p72 in this cellular context fully counteracted the effect of YAP S94A/5SA (Figure S6F). A similar effect was observed in HepG2 human hepatocellular carcinoma cells, where YAP S94A/5SA expression significantly promoted their anchorage-independent growth in a p72-dependant manner (Figures S6G and S6H). Our results here support a functional role for YAP in mediating cellular proliferation independent of its canonical transcriptional partners and dependent on the Microprocessor component p72.
DISCUSSION
Here we uncover a novel role for the Hippo signaling pathway in the regulation of miRNA biogenesis. Our results provide mechanistic understanding for two unexplained phenomena - cell density-dependent activation of miRNA biogenesis, and widespread decrease in miRNA expression in tumors (Hwang et al., 2009; Lu et al., 2005). We found that YAP, the downstream Hippo signaling transducer, induces widespread miRNA repression by sequestering p72 from the Microprocessor in a cell density-dependent manner (Figure 7N). At low cell density YAP is nuclear, promotes cell proliferation, and represses miRNA biogenesis. At higher cell density, YAP is inactivated by exclusion from the cell nucleus, thereby allowing p72 to associate with Microprocessor and pri-miRNAs resulting in enhanced miRNA biogenesis. The association of the related protein, p68 (DDX5), with the Microprocessor is also dynamically regulated. p68 is directly phosphorylated by MAPK-activated protein kinase 2 (MK2), and p68 phosphorylation is necessary for its nuclear localization (Hong et al., 2013). Therefore an emerging theme for controlling Microprocessor activity is through the accessibility of these related co-factors, either by phosphorylation-dependent control of nuclear localization (for p68) or through the sequestration of p72 in the nucleus by YAP. Cell signaling pathways can impact other components of the miRNA biogenesis machinery including the mitogen-activated protein kinase (MAPK) Erk–mediated phosphorylation of TRBP (Paroo et al., 2009), and the epidermal growth factor receptor (EGFR)–mediated phosphorylation of Ago2 (Shen et al., 2013).
p72 enhances pri-miRNA processing by the Microprocessor and recognizes a VCAUCH sequence motif in the pri-miRNA 3′ flanking region (3′FS). A recent in vitro selection and high-throughput sequencing approach for functional pri-miRNA variants or for Microprocessor-binding variants of human pri-miRNAs identified a CNNC motif in the 3′FS conserved amongst vertebrates for a large subset of human pri-miRNAs. This functional motif, located ~16–20 nt downstream of the Drosha-cleavage site is required for efficient pri-miRNA processing and overlaps with the p72-binding motif that we identified. In that study the authors identified SRp20/SRSF3 as a factor that binds the CNNC motif. Although the relevance of this splicing regulator in miRNA biogenesis was not tested it is possible that multiple different factors may converge at this 3′ FS site to mediate pri-miRNA processing. It will be interesting to examine the interplay between p72 and other possible regulators in miRNA biogenesis (Auyeung et al., 2013).
The mechanism we characterized represents a novel transcription-independent function of the YAP protein. YAP has another transcription-independent role in the growth control of intestinal stem cells, where YAP sequesters Dishevelled protein in the cytoplasm thereby repressing Wnt signaling (Barry et al., 2012). The major transcriptional role of YAP is mediated through TEAD DNA-binding proteins (Zhao et al., 2008) and therefore YAP S94A, which is deficient in TEAD binding, has a deficit in the transcription of crucial target genes. The finding that YAP S94A and its nuclear-targeted version YAP S94A/5SA repressed Microprocessor activity provides evidence that YAP represses miRNAs independent of its transcriptional activity. Furthermore, YAP can induce cellular proliferation independent of TEAD and can be rescued by p72. MYC globally suppresses miRNA through transcription (Chang et al., 2008). Our findings cannot be explained by the transcriptional repression of miRNA by MYC since we observe accumulation of pri-miRNAs and corresponding decrease in mature miRNAs upon manipulation of the YAP/p72/Microprocessor pathway. Also, this post-transcriptional control of miRNA biogenesis corresponds well with the reported widespread blockade of pri-miRNA processing observed in various human cancers.
Cell proliferation and differentiation need to be coordinated for the dynamic control of organ growth and repair. The molecular and cellular mechanisms responsible for integrating these processes remain poorly understood. Since the Hippo signaling pathway plays an important role in organ size control, it will be of interest to examine the relevance of miRNA expression changes in that context. Elevated miRNA expression likely serves to repress cell proliferation and promote cell differentiation (Kanellopoulou et al., 2005; Yi et al., 2009). Failure of this switching may lead to uncontrolled cell expansion and widespread repression of miRNAs, which are hallmarks of tumors. Our findings that the Hippo pathway synchronizes cellular expansion and miRNA biogenesis illuminate the potential for new therapeutics that target miRNA biogenesis for the treatment of human cancers.
EXPERIMENTAL PROCEDURES
Cell lines
HaCaT cells were cultured in DMEM + 10% FBS. At low-density, cells existed as single cells or small colonies. For high-density conditions similar numbers of cells were seeded in a smaller culture dish than the low-density condition, and cultured to reach confluency. Percent confluence was estimated by microscopic observation. pInducer20 (Meerbrey et al., 2011) -YAP 5SA, -YAP S94A or -YAP S94A/5SA was transduced to generate doxycycline (Dox)–inducible HaCaT and HepG2 cell lines. The transduced cells were selected with G418 (400 ng/ml) for 2 weeks. For YAP induction, Dox was added at 1000 ng/ml for 4 days. HEK293T-Flag-DROSHA cells (Gregory et al., 2004) were cultured in DMEM + 10% FBS with puromycin (2 μg/ml). For proliferation assays, cells were plated at 1.5 × 105 cells/ml in triplicate in 6 well-plates and were counted at the indicated time points. SV40 LT-immortalized Lats1−/−;Lats2fl/fl MEFs were described previously (Kim et al., 2013). For Lats2 deletion, Ad5-CMV-Cre (Gene Transfer Vector Core, University of Iowa) was infected.
Plasmids
Plasmids for YAP, YAP S127A, WW1-, WW2-, WW1/WW2-mutants, ΔC and α-catenin were described previously (Schlegelmilch et al., 2011). Plasmids for YAP S94A, S94A/5SA (Zhao et al., 2008), HA-p72-WT and K50R (Mooney et al., 2010) were kindly provided. pRL-MYC-3′UTR (Kumar et al., 2007) was from Addgene (Plasmid 14806). Luciferase assays were performed using Dual-luciferase reporter system (Promega). Lipofectamine 2000 (Invitrogen) was used for transfections.
Microprocessor reporter
For the Microprocessor reporter, the human pre-miR-125b stem-loop with the flanking upstream and downstream sequences were inserted to the 3′ UTR of Renilla luciferase gene in psiCHECK2 plasmid (Table S1). For the mutated control (Δstem-loop), the stem-loop was deleted (Table S1). For the motif deletions, GCATCC (+16 to +21 in the 3′FS, “Δmotif”) or the proximal sequence of 3′FS (+1 to +65 in the 3′FS, “Δproximal”) was deleted.
Gene expression analysis
For miRNA RNA extraction was with TRIzol (Invitrogen). TaqMan miRNA assays (Applied Biosystems) were used to quantify mature miRNA expression. Pri-miRNA levels were analyzed by qPCR using Fast SYBR Green Master Mix (Applied Biosystems). For pre-miRNA quantification, small RNAs were enriched using mirVana (Ambion). Primers used for qPCR are listed in Table S2. TaqMan probes (Applied Biosystems) were used for mRNA quantification For knockdown experiments, Lipofectamine RNAiMAX (Invitrogen) was used to transfect siRNAs (sequences in Table S3) at 10 nM.
Northern blot
Total RNA was isolated from 5 × 105 cell HaCaT cells cultured at either low or high densities. 100 fmoles of control RNA (GL2 siRNA, 5′-CGUACGCGGAAUACUUCG-3′) was spiked into cell lysates. Northern blots were performed as described (Gregory et al., 2004).
Immunoprecipitation and western blot
Cells were lysed with NETN buffer (100 mM NaCl, 20 mM Tris-Cl pH 8.0, 0.5 mM EDTA, 0.5% Nonidet P-40). After centrifugation at 20,000g at 4°C for 5 min, lysates were pretreated with Protein A/G Sepharose beads (Sigma) and incubated with antibodies at 4°C overnight. The protein-antibody complexes were incubated with protein A/G sepharose at 4°C for 1 hr. For the IP with Flag or HA tag, the pretreated lysates were incubated with anti-Flag M2 Affinity Gel (A2220, Sigma-Aldrich), EZview Red Anti-HA Affinity Gel (Sigma) or control IgG AC (Santa Cruz) at 4°C for 1hr. The beads were washed 3 times with NETN 200 buffer (200 mM NaCl, 20 mM Tris-Cl pH 8.0, 0.5 mM EDTA and 0.5% Nonidet P-40). The sample buffer was added and incubated at 95°C for 5 min. After centrifugation at 20,000g for 1 min, the supernatants were collected for western blot analysis using antibodies in Table S4.
Fractionation of protein complexes
Whole cell lysates of HaCaT cells cultured at the low and high densities were fractionated with Superose 6 gel filtration column as described previously (Gregory et al., 2004). Fractions from the gel-filtration chromatography were concentrated with Amicon Ultra centrifugal filters (Millipore) and analyzed by SDS–PAGE and Western blot.
Immunocytochemistry
HaCaT cells were fixed with 4% paraformaldehyde for 10 min at RT, permeabilized with 0.1% TritonX-100 for 1 min at RT, blocked with 2% FBS, and incubated with antibody against YAP (1:200, Cell Signaling, #4912) and p72 (1:200, Bethyl Laboratories, A300-509A) at 4°C overnight. After washing PBS, cells were incubated with anti-mouse-Alexa Fluor 488 and anti-rabbit-Alexa Fluor 546 (1:1,000, Invitrogen). Nuclei were stained with DAP (Invitrogen).
Microprocessor assay
Microprocessor was purified from HEK293T-Flag-DROSHA stable cell line 96 hrs after transfection of siRNA for NF2, LATS2, p72 or DGCR8, or negative control. In vitro transcription of pri-miR-21 and miR-125b-1 and Microprocessor assays using affinity purified Flag-Drosha complexes was performed as described previously (Gregory et al., 2004).
miRNA global expression analysis
nCounter miRNA assay (nanoString, Geiss et al., 2008) was used for global miRNA analysis. miRNAs with normalized expression levels more than those of negative control probes were analyzed. The miRNA expression was normalized to all miRNAs except for liver analysis which was normalized with the top 100 genes. For hierarchical clustering analysis the normalized values for each miRNA were z-transformed and Multiple Experiment Viewer (Saeed et al., 2006) was used for computing the complete linkage hierarchical clustering algorithm with the Pearson correlation metric. The gene ontology enrichment analysis for miRNAs were performed with starBase (Yang et al., 2011). Bonferroni-corrected p-values were presented.
Motif analysis
For discovering potential p72 recognition sites in the pri-miRNA, the pre-miRNA sequences with flanking regions approximately 55 nt upstream and 55 nt downstream were obtained from the Ensemble database. The sequences were analyzed with Improbizer (http://users.soe.ucsc.edu/~kent/improbizer/improbizer.html) sequence logos were generated using WebLogo (Crooks et al., 2004).
Recombinant p72 protein purification and EMSA
His-tagged p72 was expressed and purified from BL21-CodonPlus Competent bacteria (Stratagene). EMSA with internally labeled pri-miR-125b, pri-miR-21 or mutated pri-miR-21 (sequences in Table S5) was performed in binding buffer (50 mM Tris pH 7.5, 100 mM NaCl, 10 mM 2-mercaptoethanol, 20 U RNasin [Promega], 1 mM ATP) with 1 nM pri-miRNA and incubating for 45 min at RT. Bound complexes were resolved on native 3.5% polyacrylamide gels and visualized by radiography.
Soft agar assay
HepG2 cell lines were suspended in DMEM with 10% FBS, 0.3% SeaPlaque agarose (Lonza, #50101) and 1000 ng/ml Dox, and were plated at 3000 cells/well in a six-well culture dish on a layer of 0.6% agar containing the same medium. DMEM with 10% FBS and 1000 ng/ml Dox was added on the gels. Cell colonies were stained with crystal violet after 14 days in culture and quantified with Image J.
Mouse models
Mouse experiments were approved by the BCH Animal Care and Use Committee and were performed in accordance with all relevant guidelines and regulations.
Skin tumorigenesis model
Adult R26stoprtTA/+ Col-tetO-YAPS127A/+ K14-Cre (“+Cre” group) and R26stoprtTA/+ Col-tetO-YAPS127A/+ (“-Cre” control group, n=6) were treated for 8 days with Dox (1 mg/ml) administered in drinking water. The epidermal cells, which are enriched for the skin progenitor cells, were collected as described.(Schlegelmilch et al., 2011).
Liver tumorigenesis model
Nf2fl/fl (Benhamouche et al., 2010) female mice (n=3) were administered PBS or AAV2/8-Cre (AV-8-PV1091, University of Pennsylvania Vector Core, MOI = 1011) to induce hepatocyte-specific deletion of Nf2 gene. Livers were inspected after 50 weeks tumors were collected for analysis.
Statistical Analysis
For all quantified data, mean ± standard error of the mean (SEM) is presented. Statistical significance between two experimental groups is indicated by an asterisk and comparisons were made using the Student’s t-test. P-values less than 0.05 were considered significant.
Supplementary Material
HIGHLIGHTS.
Hippo pathway regulates microRNA biogenesis.
YAP sequesters p72 (DDX17) from the Microprocessor in a cell density-dependent manner.
p72 (DDX17) recognizes a sequence motif in pri-miRNAs.
YAP mediates widespread microRNA suppression in tumors.
Acknowledgments
Thanks to Ralf Janknecht for HA-p72 plasmid, Tyler Jacks for pRL MYC 3′UTR plasmid, Kun-Liang Guan for YAP S94A and S94A/5SA plasmids, and Dae-Sik Lim for Lats1/2 knockout MEFs. M.M is supported by Japan Heart Foundation / Bayer Yakuhin Research Grant Abroad. R.T. was supported by a Simeon Burt Wolbach Fellowship from Boston Children’s Hospital. R.I.G was supported by a grant from the US National Institute of General Medical Sciences (NIGMS) (R01GM086386). F.D.C is supported by awards from Stand Up to Cancer-AACR initiative, NIH grant R01 CA131426, and DOD. FDC is a Pew Scholar in the Biomedical Sciences.
Footnotes
Accession Numbers
The GEO accession numbers for nCounter and microarray analyses are GSE52276 and GSE49384.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errorsmaybe discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Auyeung VC, Ulitsky I, McGeary SE, Bartel DP. Beyond secondary structure: primary-sequence determinants license pri-miRNA hairpins for processing. Cell. 2013;152:844–858. doi: 10.1016/j.cell.2013.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barry ER, Morikawa T, Butler BL, Shrestha K, De la Rosa R, Yan KS, Fuchs CS, Magness ST, Smits R, Ogino S, et al. Restriction of intestinal stem cell expansion and the regenerative response by YAP. Nature. 2012;493:106–110. doi: 10.1038/nature11693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benhamouche S, Curto M, Saotome I, Gladden AB, Liu CH, Giovannini M, McClatchey AI. Nf2/Merlin controls progenitor homeostasis and tumorigenesis in the liver. Genes Dev. 2010;24:1718–1730. doi: 10.1101/gad.1938710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camargo FD, Gokhale S, Johnnidis JB, Fu D, Bell GW, Jaenisch R, Brummelkamp TR. YAP1 increases organ size and expands undifferentiated progenitor cells. Curr Biol. 2007;17:2054–2060. doi: 10.1016/j.cub.2007.10.039. [DOI] [PubMed] [Google Scholar]
- Chang TC, Yu D, Lee YS, Wentzel EA, Arking DE, West KM, Dang CV, Thomas-Tikhonenko A, Mendell JT. Widespread microRNA repression by Myc contributes to tumorigenesis. Nat Genet. 2008;40:43–50. doi: 10.1038/ng.2007.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chendrimada TP, Gregory RI, Kumaraswamy E, Norman J, Cooch N, Nishikura K, Shiekhattar R. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature. 2005;436:740–744. doi: 10.1038/nature03868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhury NR, Alves FDL, Andes-Aguayo L, de Graf T, Caseres JF, Rappsilber J, Michlewski G. Tissue-specific control of brain-enriched miR-7 biogenesis. Genes Dev. 2013;27:24–38. doi: 10.1101/gad.199190.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christoffersen NR, Shalgi R, Frankel LB, Leucci E, Lees M, Klausen M, Pilpel Y, Nielsen FC, Oren M, Lund aH. p53-independent upregulation of miR-34a during oncogene-induced senescence represses MYC. Cell Death Differ. 2010;17:236–245. doi: 10.1038/cdd.2009.109. [DOI] [PubMed] [Google Scholar]
- Davis BN, Hilyard AC, Lagna G, Hata A. SMAD proteins control DROSHA-mediated microRNA maturation. Nature. 2008;454:56–61. doi: 10.1038/nature07086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denli AM, Tops BBJ, Plasterk RHA, Ketting RF, Hannon GJ. Processing of primary microRNAs by the Microprocessor complex. Nature. 2004;432:231–235. doi: 10.1038/nature03049. [DOI] [PubMed] [Google Scholar]
- Dong J, Feldmann G, Huang J, Wu S, Zhang N, Comerford SA, Gayyed MF, Anders RA, Maitra A, Pan D. Elucidation of a universal size-control mechanism in Drosophila and mammals. Cell. 2007;130:1120–1133. doi: 10.1016/j.cell.2007.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupont S, Morsut L, Aragona M, Enzo E, Giulitti S, Cordenonsi M, Zanconato F, Le Digabel J, Forcato M, Bicciato S, et al. Role of YAP/TAZ in mechanotransduction. Nature. 2011;474:179–183. doi: 10.1038/nature10137. [DOI] [PubMed] [Google Scholar]
- Fukuda T, Yamagata K, Fujiyama S, Matsumoto T, Koshida I, Yoshimura K, Mihara M, Naitou M, Endoh H, Nakamura T, et al. DEAD-box RNA helicase subunits of the Drosha complex are required for processing of rRNA and a subset of microRNAs. Nat Cell Biol. 2007;9:604–611. doi: 10.1038/ncb1577. [DOI] [PubMed] [Google Scholar]
- Gregory RI, Yan KP, Amuthan G, Chendrimada T, Doratotaj B, Cooch N, Shiekhattar R. The Microprocessor complex mediates the genesis of microRNAs. Nature. 2004;432:235–240. doi: 10.1038/nature03120. [DOI] [PubMed] [Google Scholar]
- Guil S, Cáceres JF. The multifunctional RNA-binding protein hnRNP A1 is required for processing of miR-18a. Nat Struct Mol Biol. 2007;14:591–596. doi: 10.1038/nsmb1250. [DOI] [PubMed] [Google Scholar]
- Halder G, Dupont S, Piccolo S. Transduction of mechanical and cytoskeletal cues by YAP and TAZ. Nat Rev Mol Cell Biol. 2012;13:591–600. doi: 10.1038/nrm3416. [DOI] [PubMed] [Google Scholar]
- Han J, Lee Y, Yeom KH, Nam JW, Heo I, Rhee JK, Sohn SY, Cho Y, Zhang BT, Kim VN. Molecular basis for the recognition of primary microRNAs by the Drosha-DGCR8 complex. Cell. 2006;125:887–901. doi: 10.1016/j.cell.2006.03.043. [DOI] [PubMed] [Google Scholar]
- Harvey KF, Zhang X, Thomas DM. The Hippo pathway and human cancer. Nat Rev Cancer. 2013;13:246–257. doi: 10.1038/nrc3458. [DOI] [PubMed] [Google Scholar]
- Hill DA, Ivanovich J, Priest JR, Gurnett CA, Dehner LP, Desruisseau D, Jarzembowski JA, Wikenheiser-Brokamp KA, Suarez BK, Whelan AJ, et al. DICER1 mutations in familial pleuropulmonary blastoma. Science. 2009;325:965. doi: 10.1126/science.1174334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong S, Noh H, Chen H, Padia R, Pan ZK, Su SB, Jing Q, Ding HF, Huang S. Signaling by p38 MAPK stimulates nuclear localization of the microprocessor component p68 for processing of selected primary microRNAs. Sci Signal. 2013;6:ra16. doi: 10.1126/scisignal.2003706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang HW, Wentzel Ea, Mendell JT. Cell-cell contact globally activates microRNA biogenesis. Proc Natl Acad Sci U S A. 2009;106:7016–7021. doi: 10.1073/pnas.0811523106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanellopoulou C, Muljo SA, Kung AL, Ganesan S, Drapkin R, Jenuwein T, Livingston DM, Rajewsky K. Dicer-deficient mouse embryonic stem cells are defective in differentiation and centromeric silencing. Genes Dev. 2005;19:489–501. doi: 10.1101/gad.1248505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai S, Amano A. BRCA1 regulates microRNA biogenesis via the DROSHA microprocessor complex. J Cell Biol. 2012;197:201–208. doi: 10.1083/jcb.201110008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M, Kim M, Lee S, Kuninaka S, Saya H, Lee H, Lee S, Lim DS. cAMP/PKA signalling reinforces the LATS-YAP pathway to fully suppress YAP in response to actin cytoskeletal changes. EMBO J. 2013;32:1543–1555. doi: 10.1038/emboj.2013.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim N, Koh E, Chen X, Gumbiner BM. E-cadherin mediates contact inhibition of proliferation through Hippo signaling-pathway components. Proc Natl Acad Sci U S A. 2011;108:11930–11935. doi: 10.1073/pnas.1103345108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YK, Yeo J, Kim B, Ha M, Kim VN. Short structured RNAs with low GC content are selectively lost during extraction from a small number of cells. Mol Cell. 2012;46:893–895. doi: 10.1016/j.molcel.2012.05.036. [DOI] [PubMed] [Google Scholar]
- Kumar MS, Lu J, Mercer KL, Golub TR, Jacks T. Impaired microRNA processing enhances cellular transformation and tumorigenesis. Nat Genet. 2007;39:673–677. doi: 10.1038/ng2003. [DOI] [PubMed] [Google Scholar]
- Kumar MS, Pester RE, Chen CY, Lane K, Chin C, Lu J, Kirsch DG, Golub TR, Jacks T. Dicer1 functions as a haploinsufficient tumor suppressor. Genes Dev. 2009;23:2700–2704. doi: 10.1101/gad.1848209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee EJ, Baek M, Gusev Y, Brackett DJ, Nuovo GJ, Schmittgen TD. Systematic evaluation of microRNA processing patterns in tissues, cell lines, and tumors. RNA. 2008;14:35–42. doi: 10.1261/rna.804508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian I, Kim J, Okazawa H, Zhao J, Zhao B, Yu J, Chinnaiyan A, Israel MA, Goldstein LSB, Abujarour R, et al. The role of YAP transcription coactivator in regulating stem cell self-renewal and differentiation. Genes Dev. 2010;24:1106–1118. doi: 10.1101/gad.1903310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Getz G, Miska Ea, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando Aa, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- Maillot G, Lacroix-Triki M, Pierredon S, Gratadou L, Schmidt S, Bénès V, Roché H, Dalenc F, Auboeuf D, Millevoi S, et al. Widespread estrogen-dependent repression of microRNAs involved in breast tumor cell growth. Cancer Res. 2009;69:8332–8340. doi: 10.1158/0008-5472.CAN-09-2206. [DOI] [PubMed] [Google Scholar]
- Melo SA, Ropero S, Moutinho C, Aaltonen LA, Yamamoto H, Calin GA, Rossi S, Fernandez AF, Carneiro F, Oliveira C, et al. A TARBP2 mutation in human cancer impairs microRNA processing and DICER1 function. Nat Genet. 2009;41:365–370. doi: 10.1038/ng.317. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Melo SA, Moutinho C, Ropero S, Calin Ga, Rossi S, Spizzo R, Fernandez AF, Davalos V, Villanueva A, Montoya G, et al. A genetic defect in exportin-5 traps precursor microRNAs in the nucleus of cancer cells. Cancer Cell. 2010;18:303–315. doi: 10.1016/j.ccr.2010.09.007. [DOI] [PubMed] [Google Scholar]
- Mooney SM, Grande JP, Salisbury JL, Janknecht R. Sumoylation of p68 and p72 RNA helicases affects protein stability and transactivation potential. Biochemistry. 2010;49:1–10. doi: 10.1021/bi901263m. [DOI] [PubMed] [Google Scholar]
- Morlando M, Dini Modigliani S, Torrelli G, Rosa A, Di Carlo V, Caffarelli E, BI FUS stimulates microRNA biogenesis by facilitating co-transcriptional Drosha recruitment. EMBO J. 2013;31:4502–4510. doi: 10.1038/emboj.2012.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman MA, Hammond SM. Emerging paradigms of regulated microRNA processing. Genes Dev. 2010;24:1086–1092. doi: 10.1101/gad.1919710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozen M, Creighton CJ, Ozdemir M, Ittmann M. Widespread deregulation of microRNA expression in human prostate cancer. Oncogene. 2008;27:1788–1793. doi: 10.1038/sj.onc.1210809. [DOI] [PubMed] [Google Scholar]
- Paroo Z, Ye X, Chen S, Liu Q. Phosphorylation of the human microRNA-generating complex mediates MAPK/Erk signaling. Cell. 2009;139:112–122. doi: 10.1016/j.cell.2009.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piskounova E, Polytarchou C, Thornton JE, LaPierre RJ, Pothoulakis C, Hagan JP, Iliopoulos D, Gregory RI. Lin28A and Lin28B inhibit let-7 microRNA biogenesis by distinct mechanisms. Cell. 2011;147:1066–1079. doi: 10.1016/j.cell.2011.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos A, Camargo FD. The Hippo signaling pathway and stem cell biology. Trends Cell Biol. 2012;22:339–346. doi: 10.1016/j.tcb.2012.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto S, Aoki K, Higuchi T, Todaka H, Morisawa K, Tamaki N, Hatano E, Fukushima A, Taniguchi T, Agata Y. The NF90–NF45 complex functions as a negative regulator in the microRNA processing pathway. Mol Cell Biol. 2009;29:3754–3769. doi: 10.1128/MCB.01836-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlegelmilch K, Mohseni M, Kirak O, Pruszak J, Rodriguez JR, Zhou D, Kreger BT, Vasioukhin V, Avruch J, Brummelkamp TR, et al. Yap1 acts downstream of α-catenin to control epidermal proliferation. Cell. 2011;144:782–795. doi: 10.1016/j.cell.2011.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen J, Xia W, Khotskaya YB, Huo L, Nakanishi K, Lim SO, Du Y, Wang Y, Chang WC, Chen CH, et al. EGFR modulates microRNA maturation in response to hypoxia through phosphorylation of AGO2. Nature. 2013;497:383–387. doi: 10.1038/nature12080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silvis MR, Kreger BT, Lien WH, Klezovitch O, Rudakova GM, Camargo FD, Lantz DM, Seykora JT, Vasioukhin V. α-catenin is a tumor suppressor that controls cell accumulation by regulating the localization and activity of the transcriptional coactivator Yap1. Sci Signal. 2011;4:ra33. doi: 10.1126/scisignal.2001823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siomi H, Siomi MC. Posttranscriptional regulation of microRNA biogenesis in animals. Mol Cell. 2010;38:323–332. doi: 10.1016/j.molcel.2010.03.013. [DOI] [PubMed] [Google Scholar]
- Thomson JM, Newman M, Parker JS, Morin-Kensicki EM, Wright T, Hammond SM. Extensive post-transcriptional regulation of microRNAs and its implications for cancer. Genes Dev. 2006;20:2202–2207. doi: 10.1101/gad.1444406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trabucchi M, Briata P, Garcia-Mayoral M, Haase AD, Filipowicz W, Ramos A, Gherzi R, Rosenfeld MG. The RNA-binding protein KSRP promotes the biogenesis of a subset of microRNAs. Nature. 2009;459:1010–1014. doi: 10.1038/nature08025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsutsui M, Hasegawa H, Adachi K, Miyata M, Huang P, Ishiguro N, Hamaguchi M, Iwamoto T. Establishment of cells to monitor Microprocessor through fusion genes of microRNA and GFP. Biochem Biophys Res Commun. 2008;372:856–861. doi: 10.1016/j.bbrc.2008.05.141. [DOI] [PubMed] [Google Scholar]
- Wada K, Itoga K, Okano T, Yonemura S, Sasaki H. Hippo pathway regulation by cell morphology and stress fibers. Development. 2011;138:3907–3914. doi: 10.1242/dev.070987. [DOI] [PubMed] [Google Scholar]
- Yagi R, Chen LF, Shigesada K, Murakami Y, Ito Y. A WW domain-containing yes-associated protein (YAP) is a novel transcriptional co-activator. EMBO J. 1999;18:2551–2562. doi: 10.1093/emboj/18.9.2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi R, Pasolli HA, Landthaler M, Hafner M, Ojo T, Sheridan R, Sander C, O’Carroll D, Stoffel M, Tuschl T, et al. DGCR8-dependent microRNA biogenesis is essential for skin development. Proc Natl Acad Sci U S A. 2009;106:498–502. doi: 10.1073/pnas.0810766105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu FX, Zhao B, Panupinthu N, Jewell JL, Lian I, Wang LH, Zhao J, Yuan H, Tumaneng K, Li H, et al. Regulation of the Hippo-YAP pathway by G-protein-coupled receptor signaling. Cell. 2012;150:780–791. doi: 10.1016/j.cell.2012.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng Y, Yi R, Cullen BR. Recognition and cleavage of primary microRNA precursors by the nuclear processing enzyme Drosha. EMBO J. 2005;24:138–148. doi: 10.1038/sj.emboj.7600491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang N, Bai H, David KK, Dong J, Zheng Y, Cai J, Giovannini M, Liu P, Anders RA, Pan D. The Merlin/NF2 tumor suppressor functions through the YAP oncoprotein to regulate tissue homeostasis in mammals. Dev Cell. 2010;19:27–38. doi: 10.1016/j.devcel.2010.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Wei X, Li W, Udan RS, Yang Q, Kim J, Xie J, Ikenoue T, Yu J, Li L, et al. Inactivation of YAP oncoprotein by the Hippo pathway is involved in cell contact inhibition and tissue growth control. Genes Dev. 2007;21:2747–2761. doi: 10.1101/gad.1602907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Ye X, Yu J, Li L, Li W, Li S, Yu J, Lin JD, Wang CY, Chinnaiyan AM, et al. TEAD mediates YAP-dependent gene induction and growth control. Genes Dev. 2008;22:1962–1971. doi: 10.1101/gad.1664408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Li L, Lei Q, Guan KL. The Hippo-YAP pathway in organ size control and tumorigenesis: an updated version. Genes Dev. 2010;24:862–874. doi: 10.1101/gad.1909210. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.