Abstract
The trials performed worldwide towards Non-Invasive Prenatal Diagnosis (NIPD) of Down syndrome (or Trisomy 21) have demonstrated the great commercial and medical potential of NIPD compared to the currently used invasive prenatal diagnostic procedures. Extensive investigation of methylation differences between the mother and the fetus has led to the identification of Differentially Methylated Regions (DMRs). In this study, we present a strategy using the Methylated DNA immunoprecipitation (MeDiP) methodology in combination with real-time qPCR to achieve fetal chromosome dosage assessment which can be performed non-invasively through the analysis of fetal-specific DMRs. We achieved non-invasive prenatal detection of trisomy 21 by determining the methylation ratio of normal and trisomy 21 cases for each tested fetal-specific DMR present in maternal peripheral blood, followed by further statistical analysis. The application of the above fetal-specific methylation ratio approach provided correct diagnosis of 14 trisomy 21 and 26 normal cases.
Down Syndrome (or Trisomy 21) (OMIM190685) is considered to be the most frequent etiology of mental retardation with an incidence of 1 in 700 child births in all populations worldwide 1. Prenatal genetic diagnosis of trisomy 21 is currently performed using conventional cytogenetic or DNA analyses, which require fetal genetic material to be obtained by amniocentesis, chorionic villus sampling or cordocentesis. However, the above procedures are invasive and are associated with a considerable risk of fetal loss 1. Therefore, there is a need for the development of Non-Invasive Prenatal Diagnostic (NIPD) strategies.
The discovery of free fetal DNA (ffDNA) in the maternal circulation during pregnancy 2 has become a focus for alternative approaches towards the development of NIPD. Recently, ffDNA has been successfully used for the determination of fetal sex 3 and fetal RhD status in maternal plasma 4,5. Nevertheless, direct analysis of the limited amount of ffDNA (3-6%) in the presence of excess of maternal DNA is a great challenge for the assessment of fetal chromosomal copy number 6. Recent advances in this field have demonstrated that ffDNA present in maternal circulation can be discriminated and/or enriched 7,8. One of the most interesting developments has been the investigation of Differentially Methylated Regions (DMRs) between fetal DNA and maternal peripheral blood8. Serpin peptidase inhibitor clade B member 5 (SERPINB5) has been the first gene identified to be fetal-specific hypomethylated in maternal plasma 9. A number of additional studies have focused on single gene promoter regions 9,10 or CpG islands on chromosome 21 11-13.
Current approaches developed using ffDNA for NIPD are subject to a number of limitations. The two main applications are the use of methylation-sensitive restriction enzymes and the use of sodium bisulfite. In the first application, the requirement for DMRs containing a restriction site 11 limits the number of loci suitable for testing whereas in the second application, the bisulfite conversion leads to DNA degradation14. To overcome the above limitations, we have recently used a newly developed technique called Methylated DNA immunoprecipitation (MeDiP) 15,16 to investigate the DNA methylation pattern of chromosomes 13, 18, 21, X and Y 16. The MeDiP methodology uses an antibody specific for 5-methylcytidine to capture methylated sites and therefore enriching for fetal-specific methylated DNA 17.
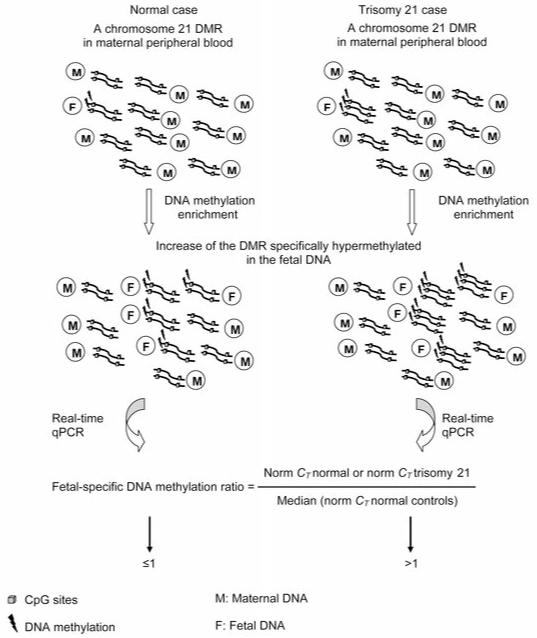
We have selected a subset of DMRs on chromosome 21 and we have applied the MeDiP methodology in combination with real-time qPCR in normal and trisomy 21 cases. To provide chromosome dosage information, the ffDNA has to be hypermethylated compared to the maternal DNA. This is essential to achieve fetal-specific methylation enrichment which is the key element in our study. We hypothesize that we would be able to discriminate normal from trisomy 21 cases by comparing the ratio values obtained from normal and trisomy 21 cases using fetal-specific methylated regions located on chromosome 21 (fetal-specific methylation ratio approach) (Fig. 1). Furthermore, we hypothesize that a combination of DMRs and not a single DMR may be able to give an accurate NIPD of normal and trisomy 21 cases.
Figure 1. Schematic illustration of the fetal-specific DNA methylation ratio approach.
The fetus with trisomy 21 has an extra copy of the fetal-specific methylated region compared to the normal fetus. DNA methylation enrichment followed by real-time qPCR of a fetal-specific methylated region, can lead to relative quantification of the amount of fetal DNA in normal and trisomy 21 cases.
Results
Fetal-specific DNA methylation ratio approach
To evaluate the efficiency of the MeDiP methodology, we tested a known hypermethylated (HYP113) and a hypomethylated (U122) control region. We observed a hypermethylated status for the HYP113 region with at least 315% higher values and a hypomethylated status for the U122 region with at least 49% lower values in both normal and trisomy 21 cases.
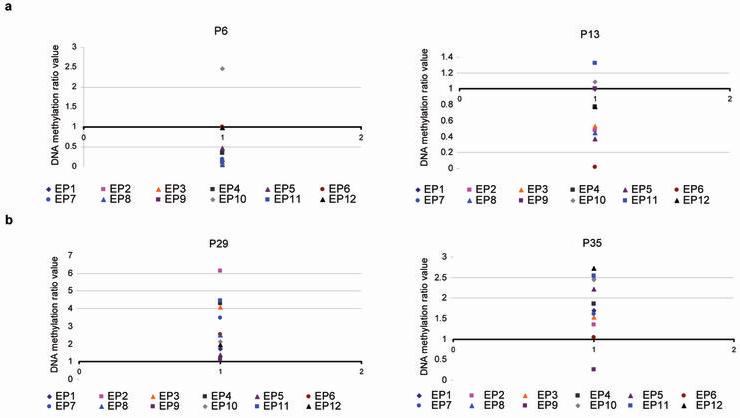
We developed an approach for determining the DNA methylation ratio of 12 selected DMRs (EP1 to EP12) in 20 normal and 20 trisomy 21 cases (Fig. 1), (Supplementary Table 1). We found that the majority of the ratio values in normal cases are at or below the value of 1 whereas in trisomy 21 cases are above the value of 1 (Supplementary Table 1). For example, in the normal cases P6 and P13, the ratio values are at or below the value of 1 with the exception of EP10 in both cases and EP11 in case P13 (Fig. 2a). The ratio values in the trisomy 21 cases P29 and P35, are above the value of 1 with the exception of EP9 in case P35 (Fig. 2b).
Figure 2. DNA methylation ratio values obtained from the 12 DMRs.
The y axis represents the methylation ratio value. Each color dot represents the ratio value obtained from one of the 12 DMRs tested (EP1 to EP12). (a) DNA methylation ratio values obtained from the normal cases P6 and P13. (b) DNA methylation ratio values obtained from the trisomy 21 cases P29 and P35.
Statistical analysis
Further to the evaluation of the fetal-specific methylation ratio values, we proceeded with a detailed univariate statistical analysis that revealed the statistical significance of each of the 12 DMRs. To achieve this we employed the Mann-Whitney U Test which estimates the significance of each of the DMRs in terms of P values (Supplementary Table 2).
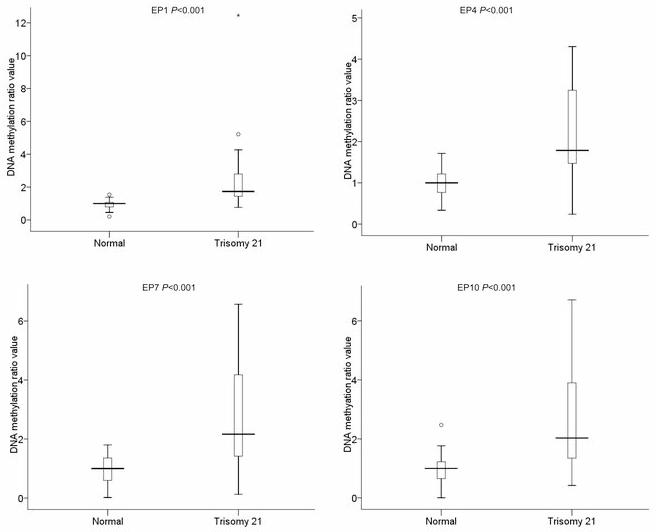
DMRs that can discriminate efficiently normal cases from trisomy 21 cases (P< 0,001) such as EP1, EP4, EP7 and EP10, showed a clear separation of the range of values obtained from normal cases compared to the ones obtained from trisomy 21 cases (Fig. 3). On the other hand, in DMRs which are not statistically significant (P> 0.05) such as EP8 and EP11, there was no clear separation between the normal and trisomy 21 cases as the ratio values obtained from both normal and trisomy 21 cases are close to the value of 1 (Supplementary Table 1).
Figure 3. BoxPlot representation of the results obtained from four DMRs.
EP1, EP4, EP7 and EP10 in 20 normal and 20 trisomy 21 cases. The boxplots depict the 5-number summaries, namely the minimum and maximum values, the upper (Q3) and lower (Q1) quartiles, and the median. The median is identified by a line inside the box. The length of the box represents the interquartile range (IQR = Q3 – Q1). Values more than three IQR’s from either end of the box are labeled as extremes and denoted by an asterisk (*).Values more than 1.5 IQR’s but less than 3 IQR’s from either end of the box are labeled as outliers (o).
Although a single DMR may not lead to the correct diagnosis of a normal or a trisomy 21 pregnancy with confidence, we hypothesize that a combination of DMRs may be able to achieve it. To test the above hypothesis we used statistical tools that would generate the ideal combination of DMRs, which can give the highest possible diagnostic sensitivity and specificity. We concluded that the application of the so called discriminant analysis provides a statistical approach, which determines accurately a normal or a trisomy 21 pregnancy.
The implementation of the discriminant analysis on 40 cases showed that eight out of 12 DMRs, entered the final model of the prediction equation and none was removed so that the stepwise analysis was consisted of eight steps in each of which a new predictor variable was chosen. The predictors were chosen in the following order according to the Wilks’ Lambda statistic: EP4 (0,646), EP12 (0,456), EP6 (0,340), EP5 (0,251), EP7 (0,210), EP11 (0,159), EP10 (0,133), and EP8 (0,122). The DMRs EP1, EP2, EP3 and EP9 were excluded from the model.
We then calculated the discriminant function coefficients for each of the DMRs. We used these values to construct the prediction equation which can be used to classify cases. The resulting prediction equation is shown below.
where XEPn = ratio valueSample; EPn, n = 1,…12
Classification of normal and trisomy 21 cases
Cases that give a D value above the cutting point are classified as “trisomy 21” while those with values below the cutting point are classified as “normal”. As the two groups of cases are of equal size, the cutting point is “0”. Hence, when D>0 the case is classified as “trisomy 21” otherwise is classified as “normal” (Table 1), (Supplementary Table 3). Statistical evaluation of the diagnostic efficiency of the discriminant analysis function using the original validatory method, showed a perfect classification for all normal and trisomy 21 cases which correspond to 100% specificity and 100% sensitivity of the methodology. In order to enhance the capability of the resulting discriminating function, we checked if all the assumptions were met (Supplementary Data).
Table 1. Prediction values obtained from 40 known samples.
| Status | Range of prediction values | Median prediction value |
|---|---|---|
| Normal | −4.29 to −1.05 | −2.63 Values > −1.00: None |
| Trisomy 21 | +0.58 to +4.35 | +2.54 Values <1.00: 0.58 & 0.96 |
We re-evaluated the specificity and sensitivity of the methodology by performing a blind study consisting of 40 samples. The ratio values obtained from the eight selected DMRs (Supplementary Table 4), were applied to the prediction equation for each sample separately in order to calculate the D value. A total of 26 cases were classified as normal whereas the remaining 14 were classified as trisomy 21 (Table 2). Cross referencing with the samples’ karyotype confirmed the above findings (Supplementary Table 5), indicating 100% specificity and 100% sensitivity of our approach.
Table 2. Prediction values obtained from 40 blind samples.
| Sample | Status | Prediction value |
|---|---|---|
| P41 | Normal | −2.34 |
| P42 | Normal | −1.16 |
| P43 | Normal | −3.54 |
| P44 | Normal | −2.79 |
| P45 | Normal | −3.19 |
| P46 | Normal | −2.88 |
| P47 | Normal | −3.66 |
| P48 | Normal | −3.24 |
| P49 | Normal | −4.66 |
| P50 | Normal | −3.86 |
| P51 | Normal | −4.30 |
| P52 | Normal | −4.35 |
| P53 | Normal | −2.81 |
| P54 | Normal | −2.37 |
| P55 | Normal | −2.41 |
| P56 | Normal | −2.47 |
| P57 | Normal | −2.10 |
| P58 | Normal | −3.36 |
| P59 | Normal | −2.26 |
| P60 | Normal | −2.99 |
| P61 | Normal | −2.80 |
Discussion
We hereby demonstrate that the application of a newly developed methodology known as MeDiP in combination with real-time qPCR using maternal peripheral blood, permits non-invasive prenatal detection of trisomy 21. The key enabling elements of this strategy are based on the direct assessment of fetal DNA within maternal circulation after methylation enrichment of fetal-specific methylated chromosome 21 regions. The employment of the above methodology provided efficient enrichment of ffDNA in maternal circulation for the 80 cases (40 known and 40 blind) included in this study. The fetal-specific DNA methylation ratio approach and further statistical analysis showed that a combination of eight specific DMRs out of 12 enabled the correct diagnosis of all the cases.
Interestingly, our results show a higher diagnostic sensitivity and specificity compared to a previous study which used the RNA-SNP strategy (90% and 96.5% respectively) 18. The RNA-SNP study included only ten trisomy 21 cases in the range of 12-20 weeks old with one trisomy 21 being incorrectly classified. In our study a total number of 40 known (20 normal and 20 trisomy 21) and 40 blind samples (26 normal and 14 trisomy 21) from pregnancies of 11.1–14.4 weeks old, were correctly classified providing 100% sensitivity and 100% specificity. Moreover, the diagnostic efficiency of our strategy is higher compared to currently applied first trimester screening protocols involving the use of nuchal translucency and biochemical markers 19,20.
An additional advantage of our methodology is observed when compared to the results obtained from next generation sequencing technologies which are of high cost and not easily accessible to diagnostic laboratories 21-23. Our approach involves the application of the MeDiP and real-time qPCR methodologies which are accessible in all basic diagnostic laboratories as they require no major and expensive infrastructure, are technically easier and of lower cost.
Moreover, our approach is an improvement in comparison to the currently applied methodologies such as the use of sodium bisulfite conversion of the DNA which can cause up to 96% DNA degradation 14 and can complicate even further the quantification of limited amounts of ffDNA. Additionally, our diagnostic approach is not affected by fetal gender or the presence of informative polymorphic sites in contrast to previous studies 18,24-27. This advantage is of high importance as the technology will be available population wide.
The approach described here has opened the way to NIPD of trisomy 21 to be potentially employed into the routine practice of all diagnostic laboratories and be applicable to all pregnancies. Such a non-invasive approach will avoid the risk of miscarriages of normal pregnancies due to current invasive procedures. Furthermore, it is speculated that this diagnostic strategy will probably replace in the future the current biochemical screening methods for trisomy 21. Nevertheless, a larger-scale study will need to be performed in order to assist the introduction of the diagnostic strategy into the clinical practice of prenatal diagnostic laboratories.
Finally, we speculate that evaluation and selection of DMRs from our previously generated microarray data 16 can be potentially used for NIPD of chromosomes 13, 18, X and Y associated aneuploidies.
Methods
Samples
For the purpose of the diagnostic strategy followed hereby, we initially used 40 maternal peripheral blood samples with known karyotype (20 from normal pregnancies and 20 from trisomy 21 pregnancies) (Supplementary Table 5). Furthermore, we used 40 additional samples in a blind fashion so that their identity and karyotype results were hidden from the investigator who carried out the test.
We collected 80 samples from 11.1-14.4 weeks of gestation in EDTA tubes and then stored them at −80 °C within 6 h of collection until further use. The experiments were approved by the Cyprus National Bioethics Committee. Informed consent was obtained from all participants.
Selection of DMRs
An in depth investigation of our previously identified DMRs 16, has led to the selection of 12 DMRs (EP1-EP12) located on chromosome 21 for further investigation. EP8 and EP12 can be found in Supplementary Table 3 of our previous study16. The 12 DMRs were selected based on three criteria. Firstly, they should demonstrate a hypermethylated status in placenta and hypomethylated status in peripheral blood in order to achieve fetal-specific DNA methylation enrichment and therefore increase the amount of ffDNA in maternal circulation. Secondly, they should have common methylation status between first and third trimester placentas in order to ensure the tissue specificity of the methylation status 16. Finally, the level of differential methylation observed for these DMRs should be above the value of 1 in a logarithmic scale in at least one of the oligonucleotide probes included in the region 16. Along with the 12 DMRs, we used two additional regions located on chromosomes 13 (HYP113) and 22 (U122) as hypermethylated and hypomethylated controls respectively 15,16.
Application of MeDiP and real-time qPCR
We extracted DNA from 1 ml of each sample using the QIAamp DNA blood Midi Kit (Qiagen). We then proceeded to immunoprecipitation of methylated sites as described previously 16. Briefly, we sheared by sonication 2.5 μg of DNA into fragments of approximately 300-1000 basepairs (bp) in size. The fragmented DNA was blunt-ended and then we added annealing linkers to both ends generated. We removed 50 ng of ligated DNA from each sample to be used as input genomic control DNA. We then applied the MeDiP methodology to the remaining ligated DNA samples (800 ng-1.2 μg) as described previously 16. Finally, we performed Ligation-Mediated PCR (LM-PCR) reactions using 10 ng of each input and immunoprecipitated DNA fraction with the aim to increase the amount of DNA.
We performed real-time qPCR reactions to all input and immunoprecipitated fragments for the selected DMRs on chromosome 21 and the two control regions. The procedure followed for primers’ design, optimization and the cycle conditions used have been described previously 16 (Supplementary Table 6).
Statistical analysis
We used the 40 samples with known karyotype to evaluate the degree of discrimination of the 12 selected DMRs. To achieve this, we compared the fetal-specific methylation ratio values of the 20 normal cases to the ratio values of the 20 trisomy 21 cases for each of the 12 DMRs (Supplementary Table 1), (Supplementary Methods). In theory, the ratio of normal cases should be 1 and 1.5 for trisomy 21 cases. However, the observed ratios are usually of a smaller value for normal cases and of a higher value for trisomy 21 cases as a result of inter-individual variability of the methylation levels16 as well as due to the presence of maternal DNA background.
Further statistical analysis was essential to determine with accuracy the statistical significance of each DMR. To achieve this, we used the Mann-Whitney U test. This test is a non-parametric test for assessing whether two independent sets of observations come from the same distribution.
The final aim of our analysis was to correctly interpret the results of a case and accurately classify it as normal or trisomy 21. To achieve this we applied the so called discriminant analysis. The discriminant analysis can generate functions from a set of cases for which group membership is known. These functions can then be applied to new cases with measurements for the predictor variables but unknown group membership. We constructed a linear discriminant equation such that the two groups differ as much as possible on the function (Supplementary Methods).
We aimed to further validate our results by performing a blind study. We used 40 blind samples and tested the methylation enrichment of the eight selected DMRs. The ratio values obtained from each sample were applied to the prediction equation and determined their status (normal or trisomy 21).
Supplementary Material
Acknowledgements
E. Papageorgiou. and P. Patsalis were supported by the Special Non-Invasive Advances in Fetal and Neonatal Evaluation (SAFE) Network of Excellence European Commission Funded 6th Framework Package Project Number: LSHBCT-2004-503243 and The Cyprus Institute of Neurology and Genetics. A. Karagrigoriou was supported by the University of Cyprus. E. Tsaliki was supported by the State Scholarships Foundation of Greece. V. Velissariou was supported by the Mitera Hospital in Athens, Greece. N. Carter. was supported by the Wellcome Trust. We thank C. Sismani, L. Kousoulidou and G. Koumbaris for extensive discussions, as well as M. Hulten for her encouragement to initiate this project.
Footnotes
Competing financial interest: P.C.P. and E.A.P. declare conflict of interest as they have filed a U.S. provisional patent for the approach (Application No. 61/405,421).
References
- 1.Hulten MA, Dhanjal S, Pertl B. Rapid and simple prenatal diagnosis of common chromosome disorders: advantages and disadvantages of the molecular methods FISH and QF-PCR. Reproduction. 2003;126:279–297. doi: 10.1530/rep.0.1260279. [DOI] [PubMed] [Google Scholar]
- 2.Lo YM, et al. Presence of fetal DNA in maternal plasma and serum. Lancet. 1997;350:485–487. doi: 10.1016/S0140-6736(97)02174-0. [DOI] [PubMed] [Google Scholar]
- 3.Honda H, et al. Fetal gender determination in early pregnancy through qualitative and quantitative analysis of fetal DNA in maternal serum. Hum. Genet. 2002;110:75–79. doi: 10.1007/s00439-001-0649-3. [DOI] [PubMed] [Google Scholar]
- 4.Bianchi DW, Avent ND, Costa JM, van der Schoot CE. Noninvasive prenatal diagnosis of fetal Rhesus D: ready for Prime(r) Time. Obstetrics and gynecology. 2005;106:841–844. doi: 10.1097/01.AOG.0000179477.59385.93. [DOI] [PubMed] [Google Scholar]
- 5.Lo YM, et al. Prenatal diagnosis of fetal RhD status by molecular analysis of maternal plasma. The New England journal of medicine. 1998;339:1734–1738. doi: 10.1056/NEJM199812103392402. [DOI] [PubMed] [Google Scholar]
- 6.Lo YM, et al. Quantitative analysis of fetal DNA in maternal plasma and serum: implications for noninvasive prenatal diagnosis. American journal of human genetics. 1998;62:768–775. doi: 10.1086/301800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chan KC, et al. Size distributions of maternal and fetal DNA in maternal plasma. Clinical chemistry. 2004;50:88–92. doi: 10.1373/clinchem.2003.024893. [DOI] [PubMed] [Google Scholar]
- 8.Poon LL, Leung TN, Lau TK, Chow KC, Lo YM. Differential DNA methylation between fetus and mother as a strategy for detecting fetal DNA in maternal plasma. Clinical chemistry. 2002;48:35–41. [PubMed] [Google Scholar]
- 9.Chim SS, et al. Detection of the placental epigenetic signature of the maspin gene in maternal plasma. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:14753–14758. doi: 10.1073/pnas.0503335102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chiu RW, et al. Hypermethylation of RASSF1A in human and rhesus placentas. The American journal of pathology. 2007;170:941–950. doi: 10.2353/ajpath.2007.060641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Old RW, Crea F, Puszyk W, Hulten MA. Candidate epigenetic biomarkers for non-invasive prenatal diagnosis of Down syndrome. Reproductive biomedicine online. 2007;15:227–235. doi: 10.1016/s1472-6483(10)60713-4. [DOI] [PubMed] [Google Scholar]
- 12.Chim SS, et al. Systematic search for placental DNA-methylation markers on chromosome 21: toward a maternal plasma-based epigenetic test for fetal trisomy 21. Clinical chemistry. 2008;54:500–511. doi: 10.1373/clinchem.2007.098731. [DOI] [PubMed] [Google Scholar]
- 13.Fazzari MJ, Greally JM. Epigenomics: beyond CpG islands. Nature reviews. 2004;5:446–455. doi: 10.1038/nrg1349. [DOI] [PubMed] [Google Scholar]
- 14.Grunau C, Clark SJ, Rosenthal A. Bisulfite genomic sequencing: systematic investigation of critical experimental parameters. Nucleic acids research. 2001;29:E65–65. doi: 10.1093/nar/29.13.e65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rakyan VK, et al. An integrated resource for genome-wide identification and analysis of human tissue-specific differentially methylated regions (tDMRs) Genome research. 2008;18:1518–1529. doi: 10.1101/gr.077479.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Papageorgiou EA, et al. Sites of differential DNA methylation between placenta and peripheral blood: molecular markers for noninvasive prenatal diagnosis of aneuploidies. The American journal of pathology. 2009;174:1609–1618. doi: 10.2353/ajpath.2009.081038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Weber M, et al. Chromosome-wide and promoter-specific analyses identify sites of differential DNA methylation in normal and transformed human cells. Nature genetics. 2005;37:853–862. doi: 10.1038/ng1598. [DOI] [PubMed] [Google Scholar]
- 18.Lo YM, et al. Plasma placental RNA allelic ratio permits noninvasive prenatal chromosomal aneuploidy detection. Nat. Med. 2007;13:218–223. doi: 10.1038/nm1530. [DOI] [PubMed] [Google Scholar]
- 19.Wald NJ, Watt HC, Hackshaw AK. Integrated screening for Down’s syndrome on the basis of tests performed during the first and second trimesters. The New England journal of medicine. 1999;341:461–467. doi: 10.1056/NEJM199908123410701. [DOI] [PubMed] [Google Scholar]
- 20.Weisz B, Rodeck CH. An update on antenatal screening for Down’s syndrome and specific implications for assisted reproduction pregnancies. Hum. Reprod. Update. 2006;12:513–518. doi: 10.1093/humupd/dml021. [DOI] [PubMed] [Google Scholar]
- 21.Chiu RW, et al. Noninvasive prenatal diagnosis of fetal chromosomal aneuploidy by massively parallel genomic sequencing of DNA in maternal plasma. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:20458–20463. doi: 10.1073/pnas.0810641105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fan HC, Blumenfeld YJ, Chitkara U, Hudgins L, Quake SR. Noninvasive diagnosis of fetal aneuploidy by shotgun sequencing DNA from maternal blood. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:16266–16271. doi: 10.1073/pnas.0808319105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chiu RW, et al. Maternal plasma DNA analysis with massively parallel sequencing by ligation for noninvasive prenatal diagnosis of trisomy 21. Clinical chemistry. 2010;56:459–463. doi: 10.1373/clinchem.2009.136507. [DOI] [PubMed] [Google Scholar]
- 24.Tsui NB, et al. Detection of trisomy 21 by quantitative mass spectrometric analysis of single-nucleotide polymorphisms. Clinical chemistry. 2005;51:2358–2362. doi: 10.1373/clinchem.2005.056978. [DOI] [PubMed] [Google Scholar]
- 25.Tsui NB, Lo YM. A microarray approach for systematic identification of placental-derived RNA markers in maternal plasma. Methods Mol. Biol. 2008;444:275–289. doi: 10.1007/978-1-59745-066-9_22. [DOI] [PubMed] [Google Scholar]
- 26.Tong YK, et al. Noninvasive Prenatal Detection of Trisomy 21 by an Epigenetic-Genetic Chromosome-Dosage Approach. Clinical chemistry. 2009;56:90–98. doi: 10.1373/clinchem.2009.134114. [DOI] [PubMed] [Google Scholar]
- 27.Tsui NB, et al. Non-invasive prenatal detection of fetal trisomy 18 by RNA-SNP allelic ratio analysis using maternal plasma SERPINB2 mRNA: a feasibility study. Prenat. Diagn. 2009;29:1031–1037. doi: 10.1002/pd.2340. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.