Significance
Specific gene-targeting nucleases are critical for genetic engineering and therapy. Meganucleases have been shown to harbor many properties, including small monomeric folds and high cleavage specificities, that are appropriate for this purpose. However, they are rarely used for genome modification because their DNA specificity requires an extensive network of up to 50 amino acids, making redesign difficult. Because extensively reprogramming molecular recognition across such a complex interface is a fundamental biotechnology challenge, meganucleases are used here as a model system for the development of an efficient approach to alter their specificity. The nucleases produced by this method, when tethered to transcription activator-like effector domains, resulted in highly active DNA targeting reagents with notable recognition properties.
Keywords: protein engineering, gene modification, gene therapy
Abstract
LAGLIDADG homing endonucleases (meganucleases) are sequence-specific DNA cleavage enzymes used for genome engineering. Recently, meganucleases fused to transcription activator-like effectors have been demonstrated to efficiently introduce targeted genome modifications. However, retargeting meganucleases to genomic sequences of interest remains challenging because it usually requires extensive alteration of a large number of amino acid residues that are situated in and near the DNA interface. Here we describe an effective strategy to extensively redesign such an extensive biomolecular interface. Well-characterized meganucleases are computationally screened to identify the best candidate enzyme to target a genomic region; that protein is then redesigned using iterative rounds of in vitro selections within compartmentalized aqueous droplets, which enable screening of extremely large numbers of protein variants at each step. The utility of this approach is illustrated by engineering three different meganucleases to cleave three human genomic sites (found in two exons and one flanking intron in two clinically relevant genes) and a fourth endonuclease that discriminates between single-nucleotide polymorphism variants of one of those targets. Fusion with transcription activator-like effector DNA binding domains significantly enhances targeted modification induced by meganucleases engineered in this study. Simultaneous expression of two such fusion endonucleases results in efficient excision of a defined genomic region.
Genome engineering is a discipline in which specific chromosomal loci are altered at precisely defined sites, producing cells and organisms with heritable DNA sequence modifications (1). Many tools have been developed for this purpose, including zinc finger nucleases (ZFNs) (2), transcription activator-like effector nucleases (TALENs) (3), CRISPR/Cas9 endonucleases (CRISPRs) (4, 5), and meganucleases (6). ZFNs can be purchased or (with effort) engineered using publicly available resources (7, 8) but often display measurable off-target activity (9, 10). In contrast, TALENs and CRISPRs can be more easily reprogrammed to a wider range of DNA sequences (11), but their lengthy reading frames may complicate packaging and delivery in certain contexts and applications (12). A recent study indicated that guide RNA–Cas9 complexes may induce significant off-target mutagenesis (13–16); however, the simultaneous generation of single-stranded breaks at closely spaced sites on opposing DNA strands, using a Cas9 nickase and multiple guide RNAs, may reduce off-target mutagenesis (17, 18).
Meganucleases are naturally occurring, compact DNA cleavage enzymes that recognize long (∼20 base pairs) DNA targets (19). Several studies have demonstrated that these nucleases can be retargeted to DNA sequences of interest, albeit with considerable effort (20). The challenge in altering their specificity lies in the same property that makes them excellent reagents for genome engineering: their DNA binding and cleavage functions are intimately coupled to one another, and a significant portion of their structure is engaged in a complex network of direct and indirect DNA contacts involving up to 50 separate amino acid residues. Although an industrial-scale engineering pipeline for altering meganuclease specificity has been developed (6), the specialized knowledge and technology required for that approach generally precludes their use by academic laboratories. In addition, a recent study has demonstrated that MegaTALs (fusions of TAL effector DNA binding regions to meganucleases) can induce a high level of targeted gene modification in human cells (21). However, the utility of this platform still depends upon the development of efficient strategies to engineer meganucleases with desired specificity.
Meganucleases and zinc finger arrays for ZFNs have been generally redesigned using cell-based selection systems, where cell culturing is time-consuming and the efficiency of transformation limits the number of mutants to be screened (6, 7, 22). In contrast, selection systems such as in vitro compartmentalization (IVC) and mRNA display (in which protein-coding libraries are readily generated by PCR-based approaches) allow for screening much more complex libraries (1010 to 1012 of variant genes) (23, 24). IVC has been exploited to isolate restriction enzymes from bacterial genome libraries (25) but not to extensively modify gene-targeting nucleases by multiple cycles of mutagenesis and selection. Here we describe a strategy for reprogramming meganuclease cleavage specificity, wherein (1) a genomic target region to be modified is computationally scanned to identify sites that are at least 50% identical to target sequences for well-characterized meganucleases and (2) variant meganucleases with desired specificity are then engineered using IVC (Fig. 1A and Fig. S1). This approach is facilitated by a growing collection of meganucleases that have been crystallized in complex with their DNA targets (26).
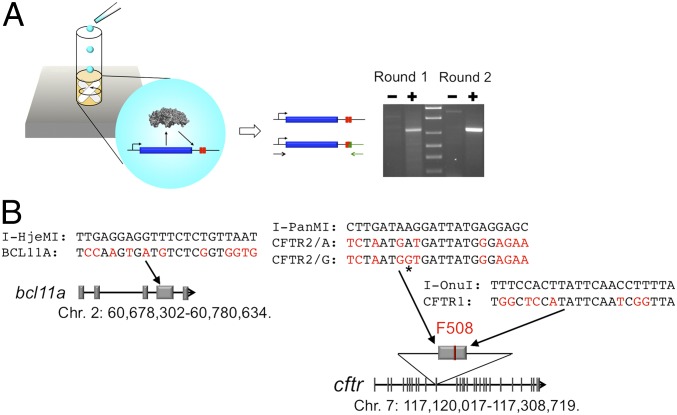
Fig. 1.
Schematic of the IVC method used for selection of retargeted meganucleases (A) and human genome target sites for engineered meganucleases (B). (A) Individual PCR fragments containing variant meganuclease genes (blue box) and target sites (red boxes) are compartmentalized with in vitro transcription/translation components in oil–surfactant mixture. Successfully expressed endonuclease variants can only cleave target sites coupled to their own genes, allowing for maintenance of genotype–phenotype linkage during selection. Variant endonuclease genes coupled to cleaved target sites can be recovered by ligation to DNA adaptors containing a complementary sticky end to that generated by engineered meganucleases, followed by PCR amplification with a set of primers, one of which is specific to an adaptor sequence (green arrow). DNA fragments containing cleaved target sites are amplified from DNA libraries subjected to in vitro selection (+ lanes) but not from naive (unselected) ones (− lanes) (see Fig. S1 for more detail). (B) Nucleotide sequences that are substituted from the original target sites recognized by wild-type meganucleases are highlighted in red. The asterisk indicates a position of the SNP observed in the cftr exon 11 of the human population, where an A:T or G:C base pair has been observed at an ∼50:50 ratio.
Results
Identification of Genomic Sequences to Be Targeted by Engineered Meganucleases.
To develop a strategy to efficiently retarget multiple types of meganucleases to genomic targets of interest, we selected two genome loci from the human genome for targeted modification on the basis of our research interest in potential therapeutic applications for hemoglobinopathies and cystic fibrosis (Fig. 1B): the exon 4 of bcl11a [which encodes a transcription regulator involved in B lymphopoiesis, erythropoiesis, and regulation of globin expression (27)] and the exon 11 in cftr [which encodes a transmembrane conductance regulator which is dysfunctional in individuals with cystic fibrosis, most often as a result of deletion of codon 508 within that exon (28)].
Using a publically available bioinformatic meganuclease Web server (LAHEDES; www.homingendonuclease.net) (26), we searched each of these regions for 22 base pair sequences that displayed significant identity to DNA targets for wild-type, monomeric meganucleases that have been crystallized in complex with their DNA targets (Fig. S2). An increasing number of such structures are now available, including I-OnuI (PDB ID 3QQY), I-LtrI (3R7P), I-PanMI (4JEE), I-GzeMII (4EFJ), I-HjeI (3UVF) I-LtrWI (4LQ0), and I-SmaMI (4LOX). We excluded candidate target sites that contained multiple nucleotide mismatches within their central four base pairs compared with the wild-type endonuclease targets because base substitutions at those base positions may lead to DNA backbone distortion that significantly compromises meganuclease specificity and activity (29).
We ultimately chose three target sites within the two target loci that exhibited 50–70% identity against the target sequences recognized by different wild-type meganucleases (I-OnuI, I-PanMI, and I-HjeMI) (Fig. 1B). Two of the three sites are situated in exons, and the other is in a flanking intron region. A site for a redesigned I-PanMI meganuclease (CFTR2) spans a well-known human single-nucleotide polymorphism (SNP) (A/G in cftr exon 11, rs213950; www.ensembl.org/). We therefore also sought to create two variant endonucleases that each act respectively at only one of the SNP variants (to assess whether this approach can produce reagents to discriminate between such closely related DNA sequences).
Reprogramming of Meganuclease Specificity.
Structural studies of meganucleases have demonstrated that two to four consecutive nucleotide base pairs within a DNA target site are generally recognized by a cluster of six to nine amino acid residues (26). These protein regions are termed “contact modules” and are cataloged for each meganuclease in the LAHEDES Web server. We randomized amino acid residues within those modules to alter the specificity of each meganuclease at altered base positions and constructed variant endonuclease libraries by overlap extension PCR using degenerate primers. Oligonucleotides used for redesign of meganucleases and amino acid positions subjected to randomization are shown in Dataset S1.
Shuffling protein residues at up to nine positions gives rise to a large theoretical number of unique variant endonucleases (as large as 209, or ∼5 × 1011). To screen as many unique protein variants as possible from randomized libraries, we modified a previously described in vitro compartmentalization system (23, 25), where DNA fragments encoding variant endonucleases are individually expressed in compartmentalized, aqueous droplets that contain an in vitro transcription/translation reaction mixture (Fig. 1A and Fig. S1). In each droplet, a variant endonuclease has the opportunity to bind and cleave a target site that is placed in the same DNA construct as for its own gene, allowing us to couple the cleavage activity against the target site to the corresponding meganuclease gene.
To identify variant enzymes that are capable of cleaving a target site containing a cluster of altered base pairs, we carried out two to three rounds of selection under increasing levels of stringency (by reducing time periods for in vitro protein synthesis and DNA strand cleavage and/or by elevating temperatures during each round; see Materials and Methods for more detail). We continued with an iterative approach, in which variant genes enriched after the first rounds of site-directed mutagenesis were used as templates for further randomization of additional amino acids and subsequent selections against a target site containing more base pair substitution(s) (as illustrated in Fig. S3). Using this strategy, we redesigned the N- and C-terminal half domains of meganucleases to engineer variant enzymes that cleaved chimeric target sites composed of one half of human genomic target sites and one half of the original meganuclease target sites. We then shuffled together the engineered N- and C-terminal half domains, and identified a collection of variant endonucleases that recognized human genomic target sequences in the IVC selection system.
We then filtered these populations of active meganucleases through cleavage assays in bacteria (30) to select individual clones that displayed substantial activity in living cells. In this assay, meganucleases that are expressed in individual bacterial cells can rescue the cell growth only when they recognize and cleave their target sequences on reporter plasmids that bacterial cells harbor, leading to elimination of the reporter plasmid that can induce expression of a toxic gene. If necessary, random mutagenesis was introduced to incorporate additional point mutations in the entire ORFs of variant endonuclease genes that might increase overall activity and/or stability.
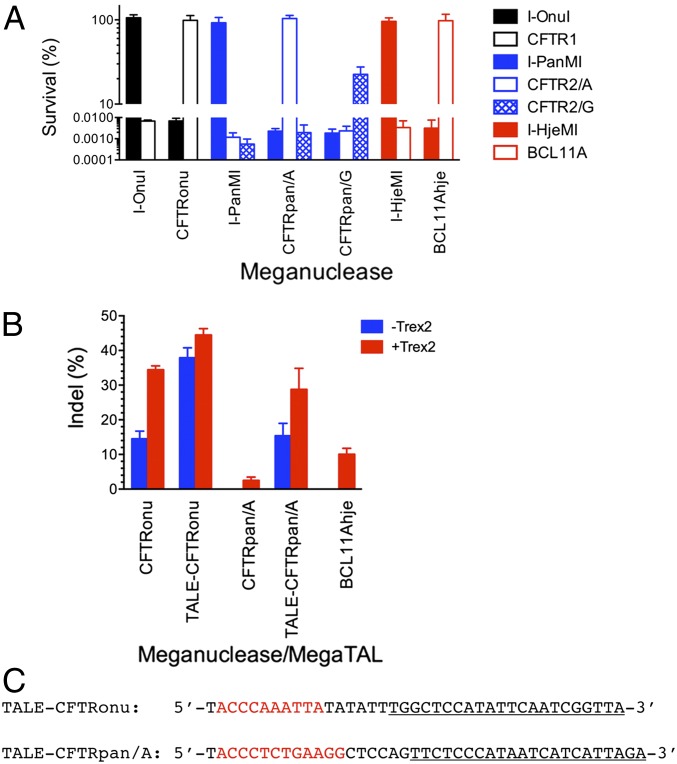
The final engineered endonucleases all efficiently increased the survival rate of bacterial cells that harbored reporter plasmids containing their target sites but not those containing the original target sites recognized by their parental meganucleases (Fig. 2A). Sequencing of the retargeted meganucleases indicated that 21–28 amino acid substitutions were introduced into each wild-type endonuclease scaffold, with a small number of substitutions (0–5) found outside of the protein–DNA interface (Fig. S4). The two I-PanMI–derived meganucleases (termed “CFTRpan/A” and “CFTRpan/G”) that targeted two SNP variants within the CFTR2 site displayed excellent discrimination between those targets. Out of total of nine amino acid residues that differ between those redesigned meganucleases, six were situated on the DNA interface adjacent to the SNP base position.
Fig. 2.
Evaluation of retargeted meganucleases and MegaTALs in bacterial and human cells. (A) Activity of wild-type and engineered meganucleases was assessed using the two-plasmid cleavage assay in bacterial cells (30), where hydrolysis of target sites on a reporter plasmid encoding a toxic protein rescues the cell growth on the selective plates. Error bars represent SDs of two to four independent experiments. Target sites correspond to those shown in Fig. 1B. I-OnuI, the wild-type I-OnuI meganuclease; CFTRonu, the I-OnuI variant endonuclease retargeted to the CFTR1 site; I-PanMI, the wild-type I-PanMI meganuclease; CFTRpan/A, the I-PanMI variant endonuclease retargeted to the CFTR2/A site; CFTRpan/G, the I-PanMI variant endonuclease retargeted to the CFTR2/G site; I-HjeMI, the wild-type I-HjeMI meganuclease; BCL11Ahje, the I-HjeMI variant endonuclease retargeted to the BCL11A site. (B) HEK 293T cells were transfected with an expression plasmid for engineered meganucleases or MegaTALs only or together with the Trex2-expressing plasmid. Genomic regions spanning endogenous genome target sites for engineered meganucleases were PCR-amplified, and the percentage of indels (used as indicators of targeted modification) was estimated using T7 endonuclease I (31). No band that was specifically cleaved by this endonuclease was detected in PCR fragments amplified from cells transfected with the nonexpression plasmid (pUC19) only and those cotransfected with pUC19 and the Trex2-expressing plasmid. Blue bars, expression of meganucleases only; red bars, coexpression of meganucleases and Trex2. Error bars represent SDs of three independent experiments. TALE-CFTRonu, the fusion of CFTRonu to the N∆150/C+63 TAL effector (TALE); TALE-CFTRpan/A, the fusion of CFTRpan/A to the TALE. (C) Sequences of MegaTAL target sites. Binding sites for TALEs are in red, and target sites recognized by engineered meganucleases are underlined. Note that the CFTR2/A site is reverse complement of that shown in Fig. 1B.
Targeted Mutagenesis by Redesigned Meganucleases at Their Endogenous Target Sites in Human Cells.
We next examined if the retargeted meganucleases could induce targeted modification at their endogenous sites in human embryonic kidney (HEK) 293T cells. Sequencing of the cftr exon 11 indicated that this cell line harbored only the SNP variant A; we therefore excluded the CFTRpan/G endonuclease from this analysis. All three engineered endonucleases were expressed at similar levels (Fig. S5A). We carried out T7 endonuclease I assays (31) to estimate the accumulation of small nucleotide insertions and deletions (indels) induced by the respective meganucleases. These assays revealed that expression of each endonuclease [either alone or in concert with the Trex2 exonuclease (32)] resulted in indels at the endogenous target sites in unsorted cell populations, with frequencies ranging from 2.5 to 34% (Fig. 2B).
The range of indel frequencies induced by the three redesigned enzymes at their corresponding target sites displayed considerably greater variation than their corresponding activities in the bacterial gene elimination assay (Fig. 2A). Most notably, the two cftr-targeting meganucleases (CFTRonu and CFTRpan/A) induce indels at their respective human genomic target sites (that are only 250 base pairs apart) with at least 10-fold different efficiencies. Additional assays in the same cell line, using an Episomal Direct Repeat (DR)-GFP reporter (33), also indicated that CFTRonu was more active (Fig. S5B), suggesting that the wide range of activities displayed by these redesigned meganucleases in human cells is not caused solely by differences in chromatin structures and/or nucleosome positioning at their target sites (34). Bioinformatic search for sites that were similar to the on-targets for the engineered meganucleases indicated that the human genome contains far more closely related sites (which contain no more than three base pair differences) for CFTRpan/A (a total of 32 sequences) than for CFTRonu (only eight such sites) (Table S1). It is therefore possible that a subset of closely related off-target sites may trap meganucleases and thereby act as competitive inhibitors because meganucleases have been shown to display more promiscuous binding specificity than DNA strand cleavage specificity (22, 35). However, we cannot rule out the possibility that these two meganucleases may have significant differences in activity that are not fully recapitulated in bacterial nuclease assays.
The same assays conducted using episomal DR-GFP reporter plasmids verified that the two I-PanMI–derived endonucleases discriminated between the SNP variants of the CFTR2 site (Fig. S5B), although the signal to noise ratio in these assays is relatively low because of spontaneous homologous recombination occurring within the episomal reporters.
Impact of Tethered TAL Effectors on Targeted Mutagenesis by Retargeted Meganucleases.
We next examined whether tethering engineered meganucleases to TAL effectors (as additional DNA binding domains) would improve the efficiency of targeted genome modification in human cells as recently described (21). The two redesigned meganucleases that targeted the cftr locus were each tethered to a TAL effector DNA binding domain (termed “N∆148/C+63,” a construct that spans from residue 149 of the canonical TAL effector N-terminal sequence to residue +63 beyond the end of the TAL effector repeat sequences). The TAL effectors were individually designed to target a 10- or 12-nucleotide sequence that was six base pairs upstream of each meganuclease target (Fig. 2C). The frequencies of indels generated by both engineered meganucleases at the same endogenous target sites were increased after incorporation of the nuclease into the MegaTAL architecture, and at least a 10-fold improved efficiency was observed in the case of CFTRpan/A (Fig. 2B).
Targeted Deletion by Multiplexed Coexpression of Two MegaTALs.
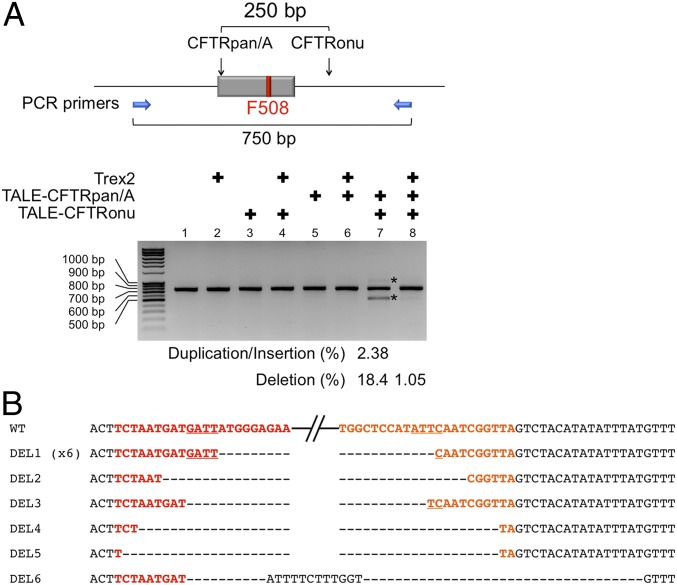
Previous studies have demonstrated that introduction of site-specific double strand breaks at two genomic loci promotes targeted deletion and insertion (36–38), which expand the range of genome engineering applications. We therefore examined whether a pair of MegaTALs were capable of inducing targeted chromosomal deletions. Expression of two MegaTALs that targeted sites ∼250 base pairs apart gave rise to three DNA bands when a genomic region spanning their target sites was PCR-amplified using a single set of primers (Fig. 3A). Sequencing of the smallest DNA band revealed that approximately a half of the targeted deletions (DEL1: 6 clones out of 12) were generated through three-base microhomology-mediated end joining between 3′, four-base overhangs generated by the two engineered meganucleases without any end processing (Fig. 3B). Sequencing of the larger DNA band identified two independent events: one was a chromosomal duplication, whereas the other was a targeted inversion and insertion of a small fragment derived from an expression plasmid for the two MegaTALs (Fig. S6A). Sequencing of clones harboring the middle-sized DNA band identified one inversion event (out of 30 clones sequenced: Fig. S6B). Deletion, duplication, and insertion promoted by the MegaTALs were substantially suppressed by coexpression of Trex2 (Fig. 3A), in a good agreement with a previous study (39).
Fig. 3.
Targeted genomic deletion by coexpression of the two MegaTALs. (A) A schematic of the cftr gene exon 11, its flanking introns, target sites for TALE-CFTRpan/A and TALE-CFTRonu, and binding sites for primers used is shown in Upper. Targeted deletion and duplication/insertion were detected only in cells expressing two MegaTALs (lane 7 in Lower) and suppressed by overexpression of the Trex2 exonuclease (lane 8). Asterisks indicate PCR fragments containing duplication/insertion or deletion. (B) Sequencing analysis of deletion junctions generated by coexpression of the two MegaTALs. Target sequences for CFTRpan/A and CFTRonu are in red and orange, respectively, and their central four base positions are underlined. The DEL1 sequence was obtained six times, and the rest of the DEL sequences were found once.
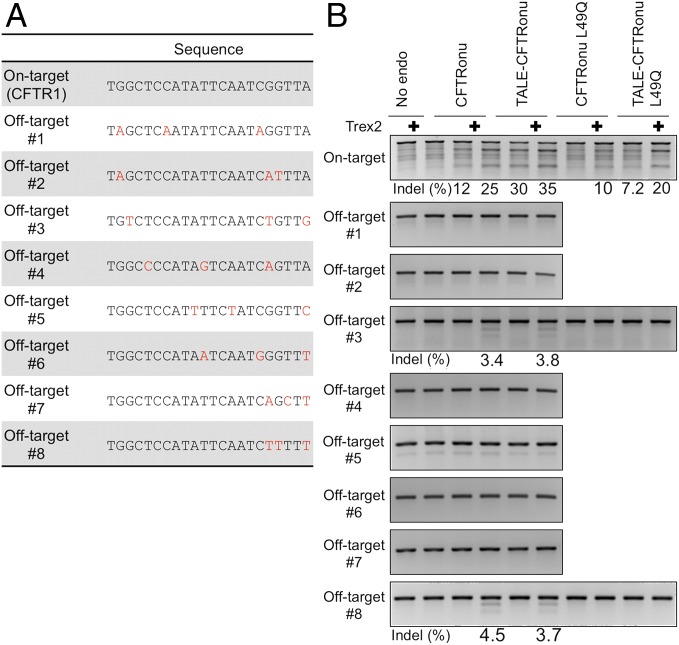
Off-Target Cleavage Activity by an Engineered Meganuclease.
To assess off-target mutagenesis by engineered meganucleases, we assayed indels in cells expressing the CFTRonu meganuclease at 8 genomic sites that were the closest to its on-target sequence (all containing 3 base substitutions from the CFTR1 target site: Fig. 4A). When Trex2 was coexpressed, both the stand-alone CFTRonu and its corresponding MegaTAL promoted low but detectable levels of indels at two of the off-target sites (Fig. 4B). Comparable frequencies of off-target mutagenesis were induced by the two types of meganuclease constructs, suggesting that the fused TAL effector had little impact on binding and cleavage activity of an engineered meganuclease at off-target sites, presumably because of high binding affinity of the stand-alone nuclease toward the noncognate DNA target sequences. These results are consistent with our earlier study of MegaTALs (21).
Fig. 4.
Analysis of off-target mutagenesis induced by the CFTRonu meganuclease. (A) Sequences of human genomic sites that are similar to the CFTR1 site. Base substitutions from the CFTR1 site are shown in red. (B) The T7EI assays (31) were carried out, and indels were detected at two closely related sites (off-targets #3 and #8) in cells coexpressing Trex2. The higher background of nonspecific cleavage products produced by T7 endonuclease I for the on-target site is due to a larger PCR fragment spanning the genomic target (∼850 base pairs) than those that were generated for the off-target sites (∼250 base pairs).
To reduce the level of undesired off-target mutagenesis, further introduction of mutagenesis into the engineered meganuclease and counterselection against the off-target sites might be useful. However, an alternative (and perhaps more straightforward) approach that could also improve specificity might be the simple incorporation of a point mutation in the nuclease domain that attenuates its overall cleavage activity, to increase the dependence of cleavage activity on simultaneous, cooperative recognition of the full-length target by the two DNA binding domains (i.e., the TAL effector and the meganuclease). Such point mutations can be easily identified based on crystal structures. This concept is illustrated by the results of experiments using the CFTRonu variant meganuclease that contains a single substitution (L49Q) in the vicinity of the active site: this point mutation suppressed the formation of indels at both the on- and off-target sites (Fig. 4B). Targeted modification was specifically enhanced at the on-target site by incorporating the CFTRonu L49Q meganuclease into the MegaTAL platform, although the efficiency of on-target mutagenesis was significantly compromised relative to that induced by constructs containing the original CFTRonu meganuclease.
Discussion
The ability to reliably and efficiently redesign biomolecular interfaces and thereby drive predictable and controllable genotypic and/or phenotypic changes is a challenge of great importance in many areas of synthetic biology and bioengineering. The type of recognition mechanism used by meganucleases, involving a highly networked interface of up to 20% of the protein (i.e., as many as 50 residues) and a DNA target of 20 base pairs, is an excellent representative system for the type of problem in molecular engineering that is difficult to address experimentally.
In this study, we retargeted three wild-type meganucleases to human genome sequences that differed from their original targets at up to 50% of the base positions (a process that eventually required the identification of 25–30 simultaneous point mutations in each protein) and verified that the engineered meganucleases all induced targeted genome modification. Given that this approach allows for targeting any 22-bp sequence where the central 4-bp sequence is conserved and where up to 11 nucleotides are mismatched at any of the remainder of the base positions, the theoretical probability of finding sites that can be targeted using this strategy is 1 in 1,842 base pairs when using a single meganuclease. Using as few as six monomeric meganucleases that cleave target sites with entirely unique central 4-bp sequences (I-HjeMI, I-OnuI, I-GzeMII, I-PanMI I-SmaMI, and I-LtrI/I-LtrWI), this probability is increased to 1 in 307 base pairs. This may be a conservative approximation because nearly the entire DNA interface of wild-type endonucleases (except regions adjacent to the central four base positions) appear to be amenable to randomization and selection (Fig. S4B). The growing availability of crystal structures of wild-type meganucleases, combined with continued refinement of the IVC selection method, should further expand the sequence space that can be addressed using meganucleases and MegaTALs.
We showed that combining TAL effector DNA binding regions with engineered meganucleases into a single polypeptide led to an increase in the frequency of targeted modification. Not only could this MegaTAL platform significantly reduce the burden of optimizing meganuclease activity during engineering process, but it also might improve the off-target to on-target ratio, particularly when an engineered meganuclease that hardly induces targeted modification alone is fused instead of a highly active meganuclease that may raise an off-targeting issue.
Although the development of easily programmable genome engineering tools enables their routine use in a wide range of cell types and model organisms, concerns about off-target mutagenesis, particularly in therapeutic applications, must be resolved. In addition to DSBs at single sites that primarily lead to indels, those generated simultaneously at two chromosomal loci, albeit rare events, can promote even more significant changes of chromosomes including translocation (40). Although further experiments need to be conducted, this study suggests that an engineered meganuclease tethered to a sequence-specific DNA binding domain such as a TAL effector and zinc finger array (which recognizes extremely long target sequences) may be one of platforms that can be exploited for therapeutic applications requiring exceptionally high demands for sequence specificity.
Materials and Methods
IVC.
The ORFs of meganucleases, I-OnuI E178D, I-HjeMI, and I-PanMI, were cloned between NcoI and NotI sites of pET21d(+) (EMD Millipore). The sequences are shown in Dataset S1. To introduce site-directed saturation mutagenesis into the ORFs, sequences containing their partial ORFs with regions (∼20 base pairs) that overlapped adjacent PCR fragments at both ends were amplified in separate tubes, using primers that contained degenerate codon NNK (coding all 20 amino acids) (Fig. S1A). The positions of amino acid residues shuffled and PCR primers used are also shown in Dataset S1. PCR products were purified by extraction from agarose gels and assembled in the subsequent round of PCR with a sequence containing two copies of target sites for variant endonucleases to construct DNA libraries (Fig. S1B). A successfully assembled DNA fragment was again purified by gel extraction (Fig. S1C).
Two to three rounds of IVC were conducted following each round of site-directed saturation mutagenesis to enrich active variant genes. The oil–surfactant mixture [2% (vol/vol) ABIL EM 90 (gift from Evonik Industries AG Personal Care) and 0.05% Triton X-100 in light mineral oil] was thoroughly mixed with the saturation buffer [100 mM potassium glutamate (pH 7.5), 10 mM magnesium acetate (pH 7.5), 1 mM DTT, and 5 mg/mL BSA], incubated at 37 °C for 20 min, and centrifuged at 16,000 × g for 15 min at 4 °C. Five hundred microliters of the upper phase was used to emulsify 30 µL of the in vitro protein synthesis mixture [25 μL of PURExpress (New England Biolabs), 20 units of RNase inhibitor, 1 mg/mL BSA, and a DNA library] by constant stirring at 1,400 r.p.m. for 3.5 min on ice (Fig. 1A and Fig. S1D). Eight nanograms of a DNA library were added in the aqueous phase in the first round of IVC and reduced to 1 ng or 0.5 ng in the second or third round, respectively. The emulsion was incubated at 30 °C for 4 h in the first round of IVC and then heated at 75 °C for 15 min. After an addition of 170 µL of 10 mM Tris⋅HCl (pH 8.0), emulsified droplets were collected by centrifugation at 16,000 × g for 15 min at 4 °C and broken by an addition of phenol/chloroform/isoamyl alcohol. Nucleic acids were recovered by isopropanol precipitation and treated with 5 µL of RNase Mixture Enzyme Mix (Life Technologies) at 37 °C for 30 min. After purification using QIAquick PCR purification kit (Qiagen), a gene library was ligated to a more than 100-fold excess of a (unphosphorylated) DNA adaptor with a 4-base 3′ overhang sequence complementary to a sticky end of target sites generated by variant endonucleases during IVC (Fig. S1E) and added to PCR mixtures containing a pair of primers, one of which was specific to a DNA adaptor to enrich ORFs of variant genes coupled to cleaved target sites (Fig. S1F). Sequences of adaptors and adaptor-specific primers used in this study are shown in Dataset S1. PCR products were gel-purified, and sequences encoding variant endonucleases were further PCR-amplified (Fig. S1G) and assembled with a sequence containing the T7 promoter and one containing two copies of a target site (Fig. S1H).
In the second round of selection, compartmentalized reaction mixtures were incubated at 42 °C for 75 min, and the third round of IVC was carried out at the same temperature for 30 min.
Two-Plasmid Cleavage Assay in Bacteria.
The assays were carried out based on previous studies (30, 41). Briefly, the NovaXGF′ competent cells harboring a pCcdB reporter plasmid (that contained four copies of a target site) were transformed with the Endo plasmid encoding a meganuclease by electroporation. The transformants were grown in the 2 × YT medium (16 g/L tryptone, 10 g/L yeast extract, 5.0 g/L NaCl) supplemented with 0.02% L-arabinose at 37 °C for an hour to induce expression of meganuclease genes and spread on both the nonselective plates (1 × M9 salt, 1% glycerol, 0.8% tryptone, 1 mM MgSO4, 1 mM CaCl2, 2 µL/mL thiamine, and 100 µg/mL carbenicillin) and the selective plates (the nonselective plates supplemented with 0.02% L-arabinose and 0.4 mM isopropyl β-D-1-thiogalactopyranoside to induce expression of the toxic CcdB protein). After incubation at 37 °C for 18–24 h, colonies were counted to calculate the survival rates on the selective plates.
I-OnuI variant endonucleases that were engineered through IVC were cloned between NcoI and NotI sites of pEndo and subjected to two rounds of selection under the conditions described above. The pEndo plasmids were extracted from individual colonies and sequenced.
When engineered meganucleases derived from I-HjeMI and I-PanMI were screened, a pool of variant genes were subjected to selection under the conditions where meganuclease genes were expressed in the 2 × YT medium containing 0.02% L-arabinose and 100 µg/mL carbenicillin at 30 °C for 15 h before plating as described in our previous study (41). Surviving colonies were scraped off from the selective plates, and random mutagenesis was introduced into the variant genes using Gene Morph II Random Mutagenesis Kit (Agilent Technologies). The resultant PCR fragments were cloned into the same vector and screened with an increasing stringency through four rounds of selection. Expression of endonuclease genes was induced at 30 °C for 4 h before plating in the first two rounds (plates were incubated at 30 °C) and at 37 °C for an hour in the subsequent two rounds (plates were incubated at 37 °C). ORFs of variant endonucleases were PCR-amplified from survival clones and cloned into the pEndo vector containing the C-terminal tag (AANDENYAAAV) that facilitates proteolytic degradation in Escherichia coli as described previously (42). The sequence of the plasmid (termed “pEndo/Degron”) is shown in Dataset S1. Two more rounds of selection were carried out under the same conditions as for the last two rounds, and ORFs of retargeted meganuclease genes recovered from survival colonies were sequenced. We expected that incorporation of the C-terminal degradation tag would accelerate turnover of a meganuclease, facilitating identification of engineered enzymes with higher activity; however, relatively little improvement was observed.
Plasmids Used for Transfection of HEK 293T Cells.
Meganuclease and MegaTAL genes with the N-terminal HA epitope tag and nuclear localization signal and the mCherry fluorescent gene were individually cloned into pExodus (32) (Addgene plasmid 39991) using Gibson Assembly Master Mix (New England Biolabs) or standard molecular cloning techniques. Double-stranded oligonucleotides containing each of the target sites for meganucleases and a stop codon were inserted into the DR-GFP reporter plasmid (33). Sequences of individual plasmids used in this study are shown in Dataset S1.
T7EI Assay for Estimating Indels.
HEK 293T cells were seeded in 0.9 mL of Opti-MEM (Life Technology) supplemented with 5% (vol/vol) FBS at 1.6–1.8 × 105 cells per well in 12-well plates 1 d before transfection and transfected with 0.4 μg each of the pExodus plasmid for expression of a retargeted meganuclease or MegaTAL and pExodus CMV.Trex2, using X-tremeGene 9 transfection reagent (Roche). The pUC19 plasmid was used as a control. Three days after transfection, genomic DNA was extracted using Quick-gDNA miniprep kit (Zymo Research), and genomic regions spanning a target site were PCR-amplified using Phusion DNA polymerase (New England Biolabs). Primers used are shown in Table S2. After purification using DNA clean and concentrator-5 kit (Zymo Research), 200–400 ng of double-stranded fragments were heated in 10 μL of 1 × NEBuffer 2 at 95 °C for 5 min and slowly cooled down (95 °C to 85 °C at −2 °C/s and 85 °C to 25 °C at −0.1 °C/s). Reannealed DNA was incubated with 5 units of T7EI (New England Biolabs) at 37 °C for 60 min, and the reaction was terminated by an addition of 4 × loading dye [40 mM Tris⋅HCl (pH8.0), 4 mM EDTA, 0.4% SDS, 20% (vol/vol) glycerol, 0.1% orange G, and 0.4 mg/mL proteinase K]. DNA fragments were separated on a 2% (wt/vol) agarose-Tris-borate-EDTA (TBE) gel. The frequency of indel was estimated using the following equation as described previously (43): (% indel) = 100 × (1 − (1 − fraction cleaved)1/2).
Chromosomal Modifications Generated by Coexpression of the Two MegaTALs.
HEK 293T cells were transfected with 0.2 μg each of the two pExodus plasmids expressing MegaTALs and 0.4 μg of pExodus CMV.Trex2, as described above. Three days after transfection, genomic DNA was extracted, and the genomic target region was PCR-amplified using a pair of primers (5′-TCT GAA TCA TGT GCC CCT TCT C-3′ and 5′-AAC CAT TGA GGA CGT TTG TCT C-3′). PCR fragments were separated on a 1.5% (wt/vol) agarose-TBE gel. Gel-extracted PCR fragments were cloned using TOPO TA cloning kit (Life Technology) and sequenced.
Episomal GFP Reporter Gene Recombination Assay.
HEK 293T cells were transfected with 0.36 μg each of the pExodus plasmid for meganucleases and a DR-GFP reporter plasmid containing a target site and 0.08 μg of pExodus-mCherry, as described above. Two days after transfection, cells were analyzed by flow cytometry, and the transfection efficiency was normalized based on a fraction of mCherry-positive cells.
Western Blotting.
Transfected cells were lysed in 20 mM Tris⋅HCl (pH 6.8), 500 mM NaCl, 1 mM EDTA, and 1% Triton X-100 and centrifuged at 16,000 × g for 30 min at 4 °C following incubation on ice for 20 min. Three micrograms of each supernatant were separated on a 15% (wt/vol) polyacrylamide-SDS gel and transferred to a PVDF membrane. HA-tagged proteins and β-tubulin were detected using anti-HA polyclonal antibody (1:200; Y-11; Santa Cruz) and anti-β-tubulin monoclonal antibody (1:500; clone TUB 2.1; Sigma Aldrich) as primary antibodies, and ECL anti-rabbit IgG HRP-linked whole antibody (1:25,000) and ECL anti-mouse IgG HRP-linked whole antibody (1:4,000; GE Healthcare) as secondary antibodies. Protein–antibody complexes were detected using ECL Western Blotting Detection reagent (GE Healthcare Life Sciences) and X-ray films (Thermo Scientific).
Supplementary Material
Acknowledgments
The I-PanMI homing endonuclease and target site were provided for this study by Dr. Abigail Lambert. Ms. Laura Taylor contributed to IVC selection experiments against the cftr target locus. The expression plasmid for the mouse Trex2 exonuclease, pExodus CMV.Trex2 (Addgene plasmid 40210), was a gift from Kamila Gwiazda and Dr. Andrew Scharenberg (Seattle Children’s Research Institute). This work was funded in part by a fellowship from the Japan Science Foundation (to R.T.) and by a grant from the National Institutes of Health (R01 GM49857 to B.L.S.).
Footnotes
Conflict of interest statement: B.L.S. is a founder of a biotechnology startup company (Pregenen, Inc.) that manufactures and applies engineered meganucleases for research and medicine. None of the reagents or materials in this study are related to the work at that company.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1321030111/-/DCSupplemental.
References
- 1.Segal DJ, Meckler JF. Genome engineering at the dawn of the golden age. Annu Rev Genomics Hum Genet. 2013;14:135–158. doi: 10.1146/annurev-genom-091212-153435. [DOI] [PubMed] [Google Scholar]
- 2.Urnov FD, Rebar EJ, Holmes MC, Zhang HS, Gregory PD. Genome editing with engineered zinc finger nucleases. Nat Rev Genet. 2010;11(9):636–646. doi: 10.1038/nrg2842. [DOI] [PubMed] [Google Scholar]
- 3.Christian M, et al. Targeting DNA double-strand breaks with TAL effector nucleases. Genetics. 2010;186(2):757–761. doi: 10.1534/genetics.110.120717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cong L, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339(6121):819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mali P, et al. RNA-guided human genome engineering via Cas9. Science. 2013;339(6121):823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arnould S, et al. Engineered I-CreI derivatives cleaving sequences from the human XPC gene can induce highly efficient gene correction in mammalian cells. J Mol Biol. 2007;371(1):49–65. doi: 10.1016/j.jmb.2007.04.079. [DOI] [PubMed] [Google Scholar]
- 7.Maeder ML, Thibodeau-Beganny S, Sander JD, Voytas DF, Joung JK. Oligomerized pool engineering (OPEN): An ‘open-source’ protocol for making customized zinc-finger arrays. Nat Protoc. 2009;4(10):1471–1501. doi: 10.1038/nprot.2009.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wright DA, et al. Standardized reagents and protocols for engineering zinc finger nucleases by modular assembly. Nat Protoc. 2006;1(3):1637–1652. doi: 10.1038/nprot.2006.259. [DOI] [PubMed] [Google Scholar]
- 9.Gabriel R, et al. An unbiased genome-wide analysis of zinc-finger nuclease specificity. Nat Biotechnol. 2011;29(9):816–823. doi: 10.1038/nbt.1948. [DOI] [PubMed] [Google Scholar]
- 10.Pattanayak V, Ramirez CL, Joung JK, Liu DR. Revealing off-target cleavage specificities of zinc-finger nucleases by in vitro selection. Nat Methods. 2011;8(9):765–770. doi: 10.1038/nmeth.1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gaj T, Gersbach CA, Barbas CF., 3rd ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol. 2013;31(7):397–405. doi: 10.1016/j.tibtech.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Holkers M, et al. Differential integrity of TALE nuclease genes following adenoviral and lentiviral vector gene transfer into human cells. Nucleic Acids Res. 2013;41(5):e63. doi: 10.1093/nar/gks1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cradick TJ, Fine EJ, Antico CJ, Bao G. CRISPR/Cas9 systems targeting β-globin and CCR5 genes have substantial off-target activity. Nucleic Acids Res. 2013;41(20):9584–9592. doi: 10.1093/nar/gkt714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fu Y, et al. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat Biotechnol. 2013;31(9):822–826. doi: 10.1038/nbt.2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hsu PD, et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat Biotechnol. 2013;31(9):827–832. doi: 10.1038/nbt.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pattanayak V, et al. High-throughput profiling of off-target DNA cleavage reveals RNA-programmed Cas9 nuclease specificity. Nat Biotechnol. 2013;31(9):839–843. doi: 10.1038/nbt.2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mali P, et al. CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nat Biotechnol. 2013;31(9):833–838. doi: 10.1038/nbt.2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ran FA, et al. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell. 2013;154(6):1380–1389. doi: 10.1016/j.cell.2013.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stoddard BL. Homing endonucleases: from microbial genetic invaders to reagents for targeted DNA modification. Structure. 2011;19(1):7–15. doi: 10.1016/j.str.2010.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Silva G, et al. Meganucleases and other tools for targeted genome engineering: perspectives and challenges for gene therapy. Curr Gene Ther. 2011;11(1):11–27. doi: 10.2174/156652311794520111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Boissel S, et al. megaTALs: A rare-cleaving nuclease architecture for therapeutic genome engineering. Nucleic Acids Res. 2014;42(4):2591–2601. doi: 10.1093/nar/gkt1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jarjour J, et al. High-resolution profiling of homing endonuclease binding and catalytic specificity using yeast surface display. Nucleic Acids Res. 2009;37(20):6871–6880. doi: 10.1093/nar/gkp726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miller OJ, et al. Directed evolution by in vitro compartmentalization. Nat Methods. 2006;3(7):561–570. doi: 10.1038/nmeth897. [DOI] [PubMed] [Google Scholar]
- 24.Seelig B. mRNA display for the selection and evolution of enzymes from in vitro-translated protein libraries. Nat Protoc. 2011;6(4):540–552. doi: 10.1038/nprot.2011.312. [DOI] [PubMed] [Google Scholar]
- 25.Zheng Y, Roberts RJ. Selection of restriction endonucleases using artificial cells. Nucleic Acids Res. 2007;35(11):e83. doi: 10.1093/nar/gkm410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Taylor GK, et al. LAHEDES: The LAGLIDADG homing endonuclease database and engineering server. Nucleic Acids Res. 2012;40(Web Server issue):W110–W116. doi: 10.1093/nar/gks365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sankaran VG. Targeted therapeutic strategies for fetal hemoglobin induction. Hematology Am Soc Hematol Educ Program. 2011;2011:459–465. doi: 10.1182/asheducation-2011.1.459. [DOI] [PubMed] [Google Scholar]
- 28.Griesenbach U, Geddes DM, Alton EW. Gene therapy progress and prospects: cystic fibrosis. Gene Ther. 2006;13(14):1061–1067. doi: 10.1038/sj.gt.3302809. [DOI] [PubMed] [Google Scholar]
- 29.Molina R, et al. Non-specific protein-DNA interactions control I-CreI target binding and cleavage. Nucleic Acids Res. 2012;40(14):6936–6945. doi: 10.1093/nar/gks320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Doyon JB, Pattanayak V, Meyer CB, Liu DR. Directed evolution and substrate specificity profile of homing endonuclease I-SceI. J Am Chem Soc. 2006;128(7):2477–2484. doi: 10.1021/ja057519l. [DOI] [PubMed] [Google Scholar]
- 31.Kim HJ, Lee HJ, Kim H, Cho SW, Kim JS. Targeted genome editing in human cells with zinc finger nucleases constructed via modular assembly. Genome Res. 2009;19(7):1279–1288. doi: 10.1101/gr.089417.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Certo MT, et al. Coupling endonucleases with DNA end-processing enzymes to drive gene disruption. Nat Methods. 2012;9(10):973–975. doi: 10.1038/nmeth.2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pierce AJ, Johnson RD, Thompson LH, Jasin M. XRCC3 promotes homology-directed repair of DNA damage in mammalian cells. Genes Dev. 1999;13(20):2633–2638. doi: 10.1101/gad.13.20.2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Daboussi F, et al. Chromosomal context and epigenetic mechanisms control the efficacy of genome editing by rare-cutting designer endonucleases. Nucleic Acids Res. 2012;40(13):6367–6379. doi: 10.1093/nar/gks268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thyme SB, et al. Exploitation of binding energy for catalysis and design. Nature. 2009;461(7268):1300–1304. doi: 10.1038/nature08508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bauer DE, et al. An erythroid enhancer of BCL11A subject to genetic variation determines fetal hemoglobin level. Science. 2013;342(6155):253–257. doi: 10.1126/science.1242088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee HJ, Kim E, Kim JS. Targeted chromosomal deletions in human cells using zinc finger nucleases. Genome Res. 2010;20(1):81–89. doi: 10.1101/gr.099747.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee HJ, Kweon J, Kim E, Kim S, Kim JS. Targeted chromosomal duplications and inversions in the human genome using zinc finger nucleases. Genome Res. 2012;22(3):539–548. doi: 10.1101/gr.129635.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bennardo N, Gunn A, Cheng A, Hasty P, Stark JM. Limiting the persistence of a chromosome break diminishes its mutagenic potential. PLoS Genet. 2009;5(10):e1000683. doi: 10.1371/journal.pgen.1000683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brunet E, et al. Chromosomal translocations induced at specified loci in human stem cells. Proc Natl Acad Sci USA. 2009;106(26):10620–10625. doi: 10.1073/pnas.0902076106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Takeuchi R, et al. Tapping natural reservoirs of homing endonucleases for targeted gene modification. Proc Natl Acad Sci USA. 2011;108(32):13077–13082. doi: 10.1073/pnas.1107719108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Andersen JB, et al. New unstable variants of green fluorescent protein for studies of transient gene expression in bacteria. Appl Environ Microbiol. 1998;64(6):2240–2246. doi: 10.1128/aem.64.6.2240-2246.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Guschin DY, et al. A rapid and general assay for monitoring endogenous gene modification. Methods Mol Biol. 2010;649:247–256. doi: 10.1007/978-1-60761-753-2_15. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.