Abstract
Recent research on long noncoding RNAs (lncRNAs) has expanded our understanding of gene transcription regulation and the generation of cellular complexity. Depending on their genomic origins, lncRNAs can be transcribed from intergenic or intragenic regions or from introns of protein-coding genes. We have recently reported more than 6000 intergenic lncRNAs in Arabidopsis. Here, we systematically identified long noncoding natural antisense transcripts (lncNATs), defined as lncRNAs transcribed from the opposite DNA strand of coding or noncoding genes. We found a total of 37,238 sense–antisense transcript pairs and 70% of annotated mRNAs to be associated with antisense transcripts in Arabidopsis. These lncNATs could be reproducibly detected by different technical platforms, including strand-specific tiling arrays, Agilent custom expression arrays, strand-specific RNA-seq, and qRT-PCR experiments. Moreover, we investigated the expression profiles of sense–antisense pairs in response to light and observed spatial and developmental-specific light effects on 626 concordant and 766 discordant NAT pairs. Genes for a large number of the light-responsive NAT pairs are associated with histone modification peaks, and histone acetylation is dynamically correlated with light-responsive expression changes of NATs.
Recent studies have uncovered, in both animals and plants, several thousand genomic loci that can be transcribed into long RNA molecules with little protein-coding potential (Guttman et al. 2009; Cabili et al. 2011; Liu et al. 2012; Li et al. 2013). Varying in length from 200 nt to >100 kb, these long noncoding RNAs (lncRNAs) are derived from intergenic regions, the opposite DNA strand of coding or noncoding genes or introns of protein-coding genes (Lavorgna et al. 2004). Genetic and molecular analyses of a selected number of lncRNAs have provided evidence for their functions in the regulation of metabolic processes, differentiation, and development (Cheetham et al. 2013; Leung et al. 2013; Ng et al. 2013a,b; Ulitsky and Bartel 2013).
One important class of noncoding RNAs is lncRNA generated from the opposite strand of coding or noncoding genes, the so-called natural antisense transcript (NAT). Sharing sequence complementarity with sense transcripts, NATs were first identified from viruses (Bovre and Szybalski 1969) and subsequently found in both prokaryotes and eukaryotes (Spiegelman et al. 1972; Wek and Hatfield 1986; Wong et al. 1987; Shah and Clancy 1992). Various techniques and genome-wide analyses of transcriptome data have been used to identify NATs (Faghihi and Wahlestedt 2009). These studies uncovered considerable antisense transcription for Saccharomyces cerevisiae (27% of genomic loci) (Faghihi and Wahlestedt 2009), Drosophila melanogaster (16.8%) (Zhang et al. 2006), mouse (72%) (Katayama et al. 2005), human (∼61%–72%) (Cheng et al. 2005; Katayama et al. 2005), and Arabidopsis (7.4%) (Wang et al. 2005; Henz et al. 2007), indicating NATs are a common feature of eukaryotic genomes. According to their coding potential, sense–antisense pairs can be categorized as coding–coding, coding–noncoding, or noncoding–noncoding pairs (Yin et al. 2007). In mammals, genome-wide analyses showed that most NATs do not have protein-coding potential (Chen et al. 2004; Katayama et al. 2005). Although previous studies in Arabidopsis mainly focused on mRNAs (Wang et al. 2005; Hazen et al. 2009), increasing lines of evidence indicate that the discovery of lncRNAs will significantly accelerate NAT identification. Matsui et al. (2008) and Okamoto et al. (2010) identified thousands of novel RNAs and 90% of them are potential NATs. Moreover, Wu et al. (2011) found many poly(A) addition sites corresponding to antisense regions that overlap with sense transcripts, suggesting pervasive transcription from both directions at a large number of genomic loci. We have recently characterized more than 6000 lncRNAs transcribed from intergenic regions of the Arabidopsis genome (Liu et al. 2012). Here, we focus on uncovering long noncoding NATs that are transcribed from the same locus of sense transcripts.
The accumulated results of specific genomic loci on sense–antisense pairs have established important physiological and pathological consequences of NATs regulation (Beiter et al. 2009; Ietswaart et al. 2012). NATs are involved in regulating responses to various abiotic and biotic stresses (Lavorgna et al. 2004; Werner 2005; Charon et al. 2010). Effects of antisense transcripts on the expression of sense transcripts can be classified into two groups: (1) concordant regulation in which sense and antisense transcripts are expressed coordinately; and (2) discordant regulation in which an antisense transcript is induced when the corresponding sense transcript is suppressed or vice versa.
NATs have been shown to deploy diverse mechanisms to regulate the expression of sense transcripts at the transcriptional or post-transcriptional level. With respect to transcriptional regulation, NAT-directed chromatin remodeling at target loci has emerged as an important mode of action (Camblong et al. 2007, 2009; Guell et al. 2009; Swiezewski et al. 2009). An antisense transcript might serve as a scaffold providing binding specificity for histone-modifying enzymes. In mammals, Xist is the first example of long noncoding antisense-mediated chromatin remodeling and gene silencing (Lee et al. 1999). Histone modifications have also been shown to be important for plant development (Loidl 2004; Chua et al. 2005; Zhang et al. 2007; Lu et al. 2011). For example, previous analyses revealed that light mediates a combinatorial interplay between histone modifications (Charron et al. 2009) and chromatin-based regulation of gene expression in light signaling pathways (Jang et al. 2011). However, there is as yet no systematic analysis on light-responsive NATs and their possible roles in histone modification.
In this study, we used a previously reported method (Liu et al. 2012) to identify a total of 37,238 NAT pairs in Arabidopsis. Using a custom-designed ATH NAT array, we detected and analyzed the expression of sense and antisense transcripts under dark and light conditions. We found spatial and temporal-specific light effects on 626 concordant and 766 discordant NAT pairs. Along with previously published genome-wide histone modification data, we observed a strong correlation between light-regulated NAT pairs and histone acetylation on their promoter and gene body regions.
Results
Identification of lncNATs in Arabidopsis
In the past few years, tiling arrays and RNA-seq experiments have emerged as the most popular platforms for transcriptomics analysis, including identification of novel transcripts and quantification of their expression levels. In the case of Arabidopsis, a large number of directional tiling array data sets are available from various experiments performed by different laboratories. These experiments generated transcriptome data of different organs/tissues and with plants were subjected to a variety of hormone treatments and biotic/abiotic stresses. However, these data sets have not yet been fully utilized to discover lncNATs. Therefore, we integrated 200 such array data sets to identify novel intragenic transcripts in Arabidopsis. Using the Reproducibility-based Tiling-array Analysis Strategy (RepTAS) (Liu et al. 2012), we uncovered more than 40,000 novel transcripts. A great majority of these transcripts (91.5%) do not encode long peptides (≥100 amino acids) and were identified as lncRNAs. We developed an identifier system to annotate all novel intragenic transcripts. Similar to the current TAIR identifier, we used the nomenclature “ATnTUxxxxxx,” in which “n” denotes the chromosome number and “xxxxxx” gives a unique number for each intragenic transcript.
Considering all novel transcripts and annotated mRNAs, we predicted a total of 37,238 NAT pairs in Arabidopsis, comprising 23,430 annotated mRNAs and 36,738 novel transcripts (Fig. 1A; Supplemental Table 1a; see Methods for details). Around 70% of Arabidopsis protein-coding genomic loci (23,430 of 33,602 representative genes; 69.7%) encode potential NAT pairs, suggesting that NATs are much more widespread than hitherto recognized. The length of lncNATs varies from 200 to 12,370 nt with an average length of 731 nt (Supplemental Fig. 1A). An overwhelming majority of NAT pairs (33,805 of 37,238 NAT pairs; 90.8%) consisted of an annotated mRNA and a newly identified lncNAT (Supplemental Table 1b). This observation suggests another layer of regulatory complexity on gene expression through lncNATs. We also found that 2186 NAT pairs were made up of two complementary lncNATs. The function of noncoding–noncoding NAT pairs remains unclear and requires further investigation. About 60% of NAT pairs were fully overlapping pairs (Supplemental Fig. 1B,C), whereas for the remaining 40% of NAT pairs, the sense and antisense transcripts may share a complementary sequence at either the 5′ end and/or the 3′ end.
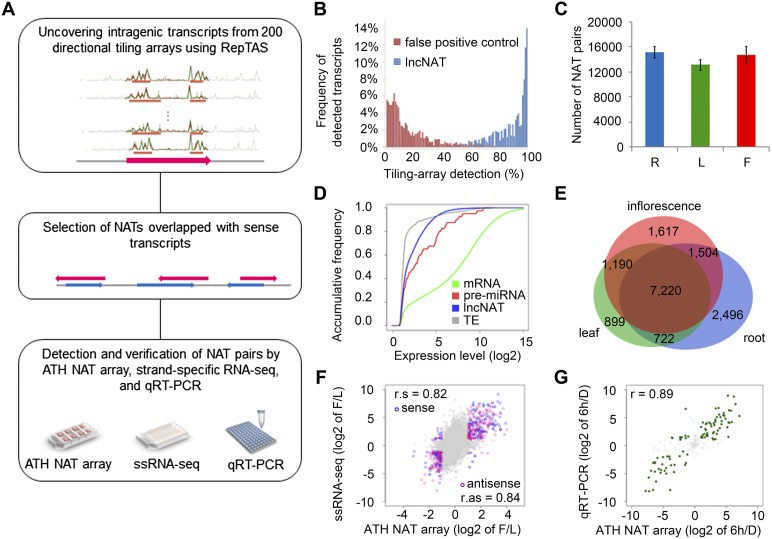
Figure 1.
Identification and profiling of lncNATs in Arabidopsis. (A) Pipeline for the identification of lncNATs in Arabidopsis. NAT pairs were identified from mRNAs and RepTAS-uncovered novel intragenic transcripts. NAT pairs were verified by different platforms, including ATH NAT custom array, strand-specific RNA-seq (ssRNA-seq), and quantitative RT-PCR (qRT-PCR). (B) Detection of lncNATs in 200 tiling arrays. The x-axis represents the percentage of tiling arrays in which the transcripts could be detected, and the y-axis represents the percentage of detectable transcripts. Blue bars show lncNATs and red bars show false positives. (C) Reproducible detection of NAT pairs by ATH NAT custom array. The y-axis represents the number of detectable NAT pairs in all three biological replicates of each sample. (R) Roots; (L) leaves; (F) inflorescences. (D) Accumulative frequency of mRNA expression levels (green line), pre-miRNAs (red line), lncNATs (blue line), and transposable element transcripts (gray line) in roots. The x-axis represents the log2 value of normalized signal intensity and the y-axis represents accumulative frequency. (E) Venn diagram showing the number of organ-specific NAT pairs in roots (blue), leaves (green), and inflorescences (red). (F) Correlation between ATH NAT array and ssRNA-seq in the detection of organ-specific NAT pairs. The fold-change between leaves and inflorescences of sense transcript levels is represented by blue circles, and antisense transcript levels by pink circles. Gray circles are transcripts that do not show expression differences (less than twofold) between the two organs. (r.s) Correlation for sense transcripts; (r.as) correlation for antisense transcripts. (G) Correlation between ATH NAT array and qRT-PCR in the detection of light-responsive NATs. The x-axis gives the log2 value of fold change detected by ATH NAT array and the y-axis gives the value detected by qRT-PCR. Gray circles represent NATs that do not change more than twofold in expression after light treatment. (6h) Samples treated with 6 h of continuous white light; (D) etiolated seedlings.
Among these sense/antisense transcripts, 95% of novel transcripts could be reproducibly detected by >60% of tiling arrays (the total number of tiling arrays was 200) (Fig. 1B). In contrast, the false positive controls, defined as 85–140-nt signal peaks or partial transcripts, were not reproducibly detected by tiling arrays. Compared with our previous results on Arabidopsis lincRNAs, we found that lncNATs are more reproducibly detected than lincRNAs and similar to annotated mRNAs (Liu et al. 2012). Thus, the vast majority of novel transcripts are reproducibly transcribed and not generated by spurious transcriptional noise.
To further verify our sense/antisense transcripts, we employed strand-specific RNA-seq (ssRNA-seq) technology to examine transcripts in leaf, inflorescence, and silique samples. Note that the three organ samples were derived from plants grown under normal conditions. We identified around 38–48 million 100-cycle single-end reads from each RNA library (Supplemental Table 2) and aligned them to the Arabidopsis genome sequence and annotation (TAIR10) with TopHat (Trapnell et al. 2009). Focusing on transcripts encoding NAT pairs, we found that 86.0% of mRNAs (20,158 of 23,430) and 42.9% of novel transcripts (15,778 of 36,738) could be detected by ssRNA-seq reads. This means that 45.1% of NAT pairs uncovered from our analysis of tiling array data could also be detected by another technical platform, RNA-seq technology, in selected samples.
Profiling of NAT pairs and their verification
To profile the expression of Arabidopsis NAT pairs, we designed a custom high-density, long-oligonucleotide expression array and used this platform to detect sense and antisense transcripts. We chose the Agilent SurePrint G3 format (8 × 60 K) to design our ATH NAT custom array (Supplemental Fig. 2; see Methods). This array contained 33,526 NAT pairs comprising 21,528 annotated mRNAs and 32,834 lncNATs, as well as singleton transcripts without antisense RNA, including 431 mRNAs and 2657 long intergenic noncoding RNAs (lincRNAs). Each transcript could be detected by one or two 60-mer probe(s). Ninety percent of our newly predicted NAT pairs were represented on this ATH NAT array. Using ATH NAT arrays, we analyzed RNA samples from Arabidopsis roots, leaves, and inflorescences, with three biological replicates each. Supplemental Figure 3 shows that the signal intensities of the spike-in probes were highly correlated with their relative RNA amounts (Pearson correlation coefficient >0.99). Moreover, after normalization for each organ, the three biological replicates showed an excellent correlation (Pearson product-moment correlation coefficient >0.9; P-value < 2.2 × 10−16) (Supplemental Fig. 4). These results confirmed the overall excellent quality of the ATH NAT arrays.
Approximately one-half of the NAT pairs were detectable in all three replicates in at least one organ, suggesting pervasive expression of NAT pairs in Arabidopsis. We first selected a detectable transcript, whose signal intensity was correlated with its relative RNA concentration and was significantly higher than those of the negative probes (for details, see Methods). We then defined the detectable NAT pair if (1) both the sense and antisense transcripts were detectable; and (2) expression of the NAT pair could be reproducibly detected in all three replicates in at least one organ. In total, 15,648 NAT pairs were detectable in at least one of the three organs: root, leaf, and inflorescence (Fig. 1C). Because our NATs were extracted from transcriptome data of different organs, plants at different stages, and plants treated with various conditions, NAT pairs not detected in any of the selected organs may be expressed in other organs or only under specific developmental stage or treatment conditions, e.g., abiotic stresses.
Expression levels of NATs were close to those of miRNA precursors (pre-miRNAs) and lower than those of mRNAs (Fig. 1D). The same result was seen in all three organs examined (Supplemental Fig. 5). The lower NAT expression level is similar to what we observed for lincRNAs (Liu et al. 2012). Although most NATs were not expressed at high levels, they exhibited a clear organ-specific profile (Fig. 1E). This result suggests specific regulation of NAT expression; and therefore, these transcripts are unlikely to be by-products of spurious transcription.
To confirm organ-specific expression of NATs, we selected NATs with different expression levels (more than twofold change) in leaf and inflorescence samples based on ATH NAT custom array results. We analyzed ssRNA-seq data of leaf and inflorescence samples and calculated reads per kilobase per million reads (RPKM) for each transcript. Our comparison showed that the detection of NATs by ATH NAT array was consistent with the ssRNA-seq result (r > 0.8; P-value < 2.2 × 10−16) (Fig. 1F), and the organ-specific expression of NAT pairs could be reproducibly detected by both platforms.
Moreover, we examined the abundance of NATs in etiolated seedlings and seedlings undergoing de-etiolation in continuous white light for 6 h. Among the transcripts detected by ATH NAT arrays, the expression of 50 lncNATs and 11 coding NATs were further verified by quantitative RT-PCR (qRT-PCR). Figure 1G shows a good correlation in transcript abundance detected by these two platforms (r = 0.89; P-value < 2.2 × 10−16). Taken together, our results show that the existence of NATs and their organ- and condition-specific expression could be reliably and reproducibly verified by different platforms. These results provide further evidence that many NATs are bona fide transcripts.
Light regulates NAT pairs in a development-dependent and organ-specific manner
Light is one of the most important environmental factors regulating plant growth and development. Previous research mainly focused on protein factors regulating light signaling pathways and the identification of light-responsive transcription factors (Jiao et al. 2007). However, there is as yet no genome-wide analysis of light-regulated NAT pairs in Arabidopsis, especially for lncNATs.
In an attempt to identify NAT pairs associated with light responses, we sampled etiolated Columbia-0 (Col-0) WT seedlings and seedlings undergoing de-etiolation in continuous white light for 1 h and 6 h (Supplemental Fig. 6). To obtain information on organ-specific transcriptome profiles, we further dissected seedlings into cotyledons, hypocotyls, and roots. RNAs from three biological replicates of each of the three organs were separately hybridized to ATH NAT arrays to profile light-responsive NAT pairs.
We first identified light-responsive transcripts from NAT pairs and singletons by the following criteria: (1) a twofold change in expression levels; and (2) eBays ANOVA P-value <0.05. A total of 9235 mRNAs and 13,907 lncRNAs were identified as light-responsive transcripts, which together covered >42% of the genomic loci represented on the ATH NAT array (Fig. 2A). These light-regulated transcripts showed a clear organ preference; most light-responsive transcripts, including both mRNAs and lncRNAs, were preferentially expressed in cotyledons compared to the other two organs (Fig. 2B; Supplemental Fig. 7). The number of light-induced and light-repressed transcripts was comparable in each organ, suggesting no organ-specific bias toward positive and negative regulation. Moreover, there were more regulated mRNAs at the 6-h time point compared to 1 h.
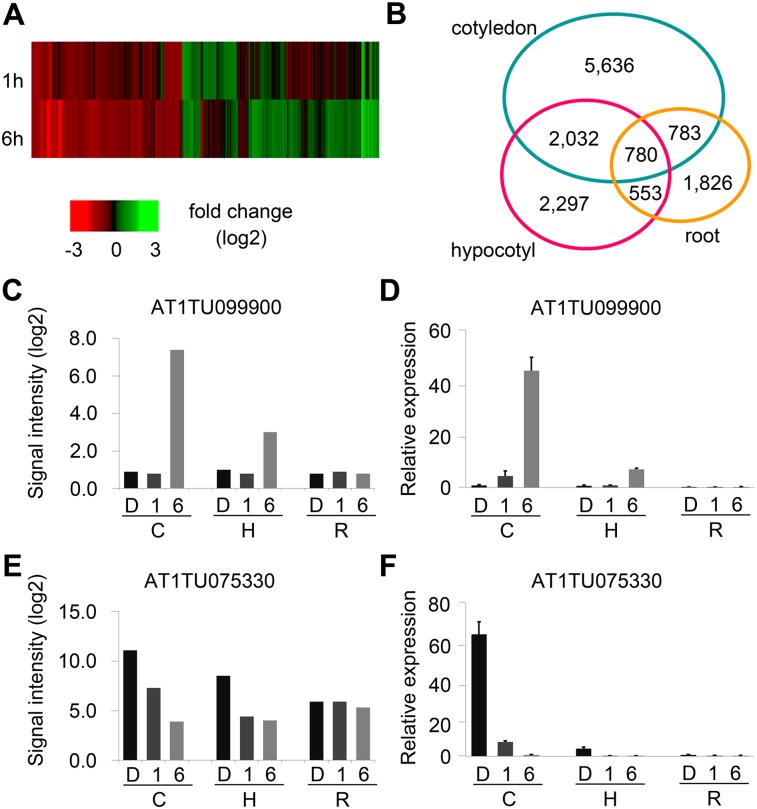
Figure 2.
Light-regulated coding and noncoding transcripts. (A) Heat map representing fold change (log2 value) of light-regulated transcript levels at 1 h and 6 h in cotyledons. (B) Organ-preferential expression of light-regulated lncRNAs. Venn diagrams show number of light-regulated transcripts in each organ. (C) Signal intensity and (D) validation by qRT-PCR of an organ-specific light-induced lncNAT. (E) Signal intensity and (F) validation by qRT-PCR of an organ-specific light-repressed lncNAT. Error bar gives standard error (SE) (n = 3). (C) cotyledon; (H) hypocotyl; (R) root; (D) dark; (1) 1-h light; (6) 6-h light.
We validated expression levels of a panel of representative light-induced/repressed lncNATs by qRT-PCR. Our qRT-PCR results confirmed that some lncNATs were significantly regulated by light, and their expression profiles were also consistent with the results from ATH NAT arrays (Fig. 2C–F; Supplemental Fig. 8; Supplemental Table 3). Together, these results provide evidence not only for light regulation of lncNATs, but also for the sensitivity and reliability of the ATH NAT arrays.
We further classified light-regulated NAT pairs into two groups: concordant regulation and discordant regulation. To be conservative, we used stringent criteria for the identification of light-regulated concordant NAT pairs. For concordant NAT pairs, one transcript should change in expression level more than eight times (P-value <0.05) after light treatment, and the other should change in expression level in the same direction as its partner no less than two times with a P-value <0.05. For discordant NAT pairs, expression levels of the sense and antisense transcripts should change in the opposite direction no less than two times with a P-value <0.05.
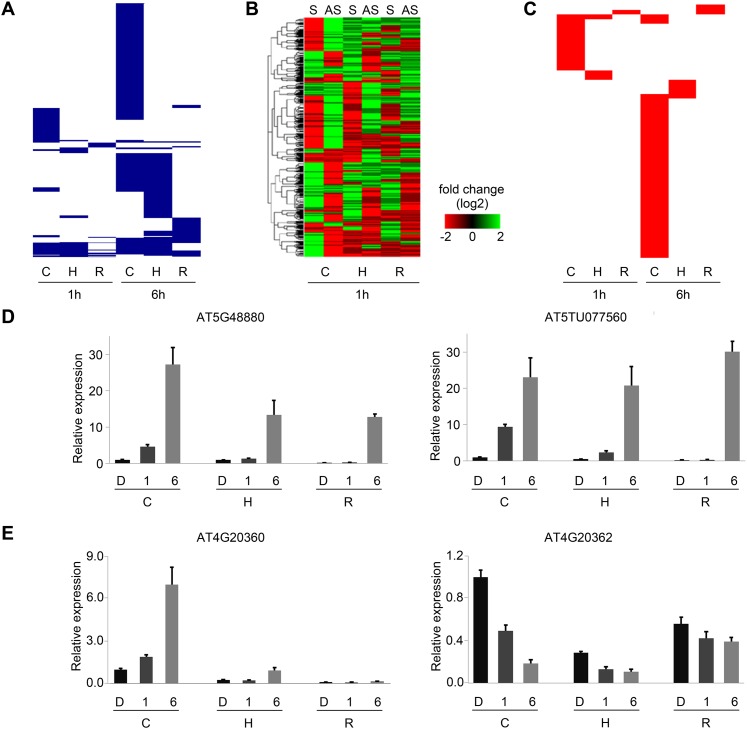
We identified a total of 626 concordant NAT pairs potentially involved in light signaling pathways (Fig. 3A,D). These NAT pairs were regulated by light in a spatial and developmental-specific pattern: (1) Expression of most of the light-responsive concordant NAT pairs was detected in cotyledons compared to hypocotyls and roots; (2) comparing transcriptome data of the three organs, we found light-responsive NAT pairs detected in one organ generally did not significantly change their expression levels in the other two organs, suggesting an organ-specific regulation of NAT pairs in light signaling pathways; and (3) the number of light-regulated NAT pairs at 1 h was much smaller than those at 6 h. The majority of light-regulated NAT pairs at the two time points were different, suggesting amplification of the light signal and utilization of specific NAT pairs in short- and long-term light response. For example, SPA1, which encodes a light signaling repressor, was up-regulated more than eight times after 1 h of light treatment in cotyledons. Its concordant antisense transcript, AT2TU076050, was also up-regulated more than four times at the same time point and in the same organ. Neither showed such strong induction at the other time point or organs. Another example is HYH, a homolog of HY5, and its concordant NAT, AT3TU028510. At 1 h, both HYH and AT3TU028510 were significantly induced in hypocotyls only; they were further induced at 6 h in roots as well as hypocotyls.
Figure 3.
Light-regulated NAT pairs. (A) Clustering of light-regulated concordant (positively correlated) NAT pairs. Each blue line represents a light-regulated concordant NAT pair in cotyledon, hypocotyl, and root at 1 h and 6 h. (B) Heat map of discordant (negatively correlated) NAT pairs at 1 h. Expression levels of discordant sense (S) and antisense (AS) transcripts in cotyledon, hypocotyl, and root. (C) Clustering of light-regulated discordant (negatively correlated) NAT pairs. Each red line represents a light-regulated discordant NAT pair in cotyledon, hypocotyl, and root at 1 h and 6 h. (D,E) Validation of light-regulated concordant and discordant NAT pairs by qRT-PCR. Bar plot represents relative expression level. Error bar gives standard error (SE) (n = 3). (C) cotyledon; (H) hypocotyl; (R) root; (D) dark; (1) 1-h light; (6) 6-h light.
We also found 766 light-responsive discordant NAT pairs (Fig. 3B,E; Supplemental Fig. 9). Here, the large number of light-responsive, discordant NAT pairs that were identified suggested a potentially widespread occurrence of negative regulation by NATs. The regulation of these discordant NAT pairs displayed a similar pattern as those of concordant pairs in terms of both organ- and developmental-stage specificity (Fig. 3C). As we were interested in the function of discordant pairs, we performed GO term enrichment analysis for genes encoding these NAT pairs using a hypergeometric test. Genes coding for transcription factors were significantly overrepresented after 1 h of white light, whereas metabolism-related genes were overrepresented after 6 h of white light (P-value <0.05) (Supplemental Table 4a,b). For example, an mRNA coding the phototropic-responsive NPH3 family protein, AT3G49970, was specifically down-regulated in hypocotyls after 1 h of light accompanied by its up-regulated lncNAT, AT3TU075200. In other organs, this discordant pair did not show more than twofold expression differences. It will be interesting to check the function of this lncNAT in phototropic responses. However, we did not detect such functional enrichment for genes encoding light-responsive concordant NAT pairs (Supplemental Table 4c).
Dynamic association of light responsive histone modifications on NAT pairs
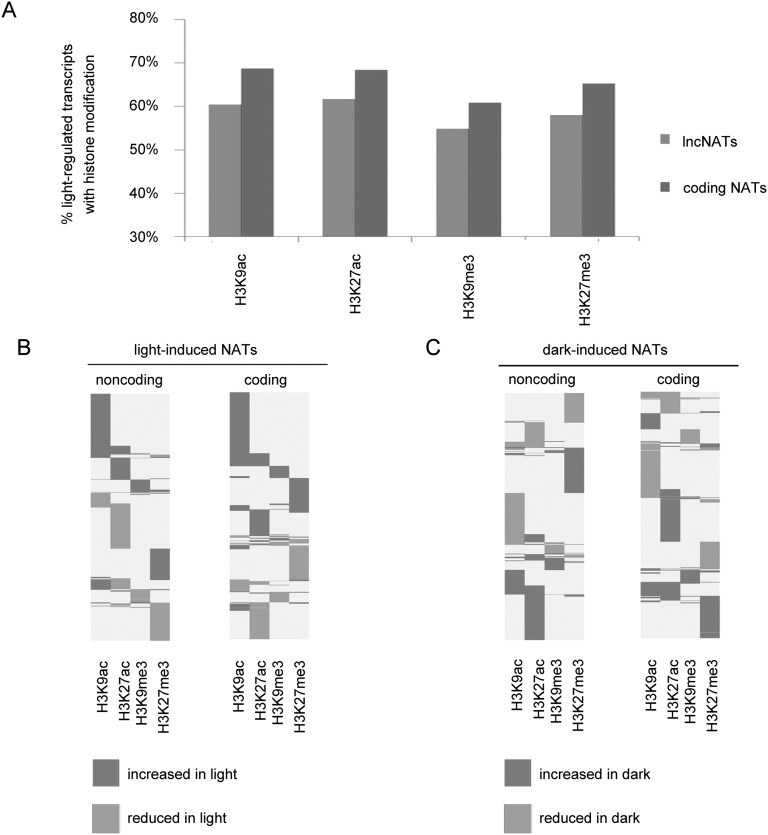
Previous genome-wide profiling of histone modifications revealed that H3K9ac and H3K27ac are specific to genes not related to transposable elements, whereas H3K9me3 and H3K27me3 are associated with transposable elements and also intergenic regions (Charron et al. 2009). However, histone modifications of genes for noncoding transcripts derived from the antisense strand of coding genes have not yet been investigated. To investigate light effects on histone modification of NAT pairs, we used public ChIP-chip data of four kinds of histone modifications, including H3K9ac, H3K9me3, H3K27ac, and H3K27me3 (Charron et al. 2009) and scanned for enrichment of histone marks using TileMap (Charron et al. 2009; Ji et al. 2011). This ChIP-chip data set was generated from 7-d-old seedlings exposed to either 0 or 6 h of white light, using experimental conditions similar to those used for our ATH NAT arrays. Therefore, we aligned histone mark enrichments at the promoter (defined as upstream 500-nt regions) and gene body regions of coding and noncoding NAT pairs responsive to 6 h of light. Our results showed that genes for a large number of light-regulated lncNATs (∼60%) were associated with histone modifications in darkness and/or light conditions; meanwhile, 66% of genes for protein-coding NATs were associated with modified regions (Supplemental Table 5). Genes for both coding and noncoding NATs showed a slight preference toward histone H3 acetylation (Fig. 4A). Focusing on light-regulated NATs, we concluded that genes for both coding and lncNATs were associated with histone modifications in darkness and/or light; therefore, light may regulate the expression of NATs through histone modifications.
Figure 4.
Light effects on histone modifications associated with genes encoding NATs. (A) Association of four kinds of histone modifications and light-regulated genes for NATs. Percentage of histone modification associated with genes for noncoding and coding NATs. (B) Histone modifications of genes for light-induced NATs. Each line represents a gene for the corresponding light-induced transcript. (C) Histone modifications of genes for dark-induced NATs. Each line represents a gene for the corresponding dark-induced transcript.
Next, we attempted to correlate modifications of histone marks to changes of NAT expression levels in response to light and examined whether these light responsive histone modifications were associated with transcription changes of light-regulated NATs. For each kind of histone modification, we defined (1) “increased in light,” if the gene was methylated or acetylated under light condition and was demethylated or deacetylated in darkness; and (2) “reduced in light,” if the gene was methylated or acetylated in darkness but not under light treatment. Compared to H3K9me3 and H3K27me3, acetylation of histone H3 tails was more significantly correlated with light regulation of genes for both coding and noncoding NATs. More than 40% of genes for light-induced lncNATs (genes for 295 of 732 transcripts) were associated with an enrichment of either H3K9ac and/or H3K27ac in the light, and 77% (228 of 295 genes) were associated with H3K9ac enrichment (Fig. 4B). For chromatin regions containing genes for light-induced coding NATs, 49% (560 of 1138) were acetylated at H3K9 and/or H3K27 in the light but not in darkness. Acetylation of H3K9 was also a major contributor. Similar observations could be made for genes encoding dark-induced NATs (Fig. 4C). To strengthen our observation, we performed a hypergeometric test for possible correlation between H3K9ac/H3K27me3 and 6-h light-induced NATs in cotyledons, using as a control total transcripts covered by our ATH NAT array. Total transcripts included light-responsive transcripts, non-light-responsive transcripts, NATs, and non-NAT transcripts in this control data set. We found that H3K9ac modification was significantly correlated with the expression induction of NATs (P-value = 4.689986 × 10−17). However, the correlation between a decrease of H3K27me3 and an increase of the expression of NATs was not significant (P-value = 0.78). Moreover, we also examined this correlation using light-induced total transcripts as a control in order to see if H3 acetylation would preferentially correlate with transcriptional activation of light-responsive NATs. Based on the result from a hypergeometric test, the enrichment of H3K9ac was preferentially correlated with the induction of NAT abundance (P-value = 4.23786 × 10−6). On the other hand, we did not find H3K27me3 depletion to be preferentially correlated with the induction of NAT abundance (P-value = 0.9917614). Thus, histone H3 acetylation was correlated with transcriptional regulation of light-responsive NATs, with H3K9ac being the major contributor for this.
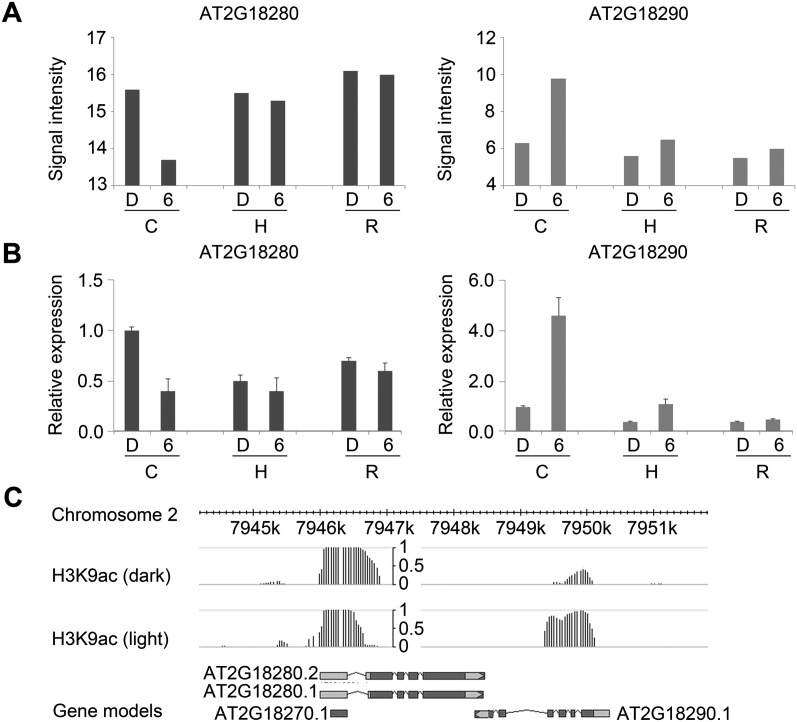
Considering the possible role of H3K9ac in regulating light-responsive NATs, we further linked H3K9ac to genes for light-regulated NAT pairs. We investigated concordant and discordant NAT pairs that were expressed in cotyledons after 6 h of continuous white light treatment. In total, genes for 23% concordant pairs and 28% discordant NAT pairs were associated with H3K9ac. Among the H3K9ac-associated NAT pair loci, ∼60% contained genes for both sense and antisense transcripts overlapping with H3K9ac modified regions, and 40% contained genes for either sense or antisense transcripts associated with H3K9ac. Moreover, we found H3K9 acetylation was positively correlated with transcript expression changes in 23% concordant NAT pairs and 38% discordant pairs. For example, we found a light-responsive discordant NAT pair from the ATH NAT array (Fig. 5A), which is composed of an mRNA coding TUBBY LIKE PROTEIN 2, ATTPL2 (AT2G18280) and an mRNA coding ANAPHASE PROMOTING COMPLEX 10, APC10 (AT2G18290). Expression changes of this NAT pair were validated by qRT-PCR (Fig. 5B). According to TAIR10 annotation, ATTPL2 may function in transcriptional regulation, whereas APC10 plays an essential role in cell proliferation during leaf development. In response to light, induction of APC10 was positively correlated with an increase of H3K9 acetylation in its transcription region (Fig. 5C). On the other hand, expression of ATTPL2 was repressed, accompanied by a decrease of H3K9ac at its 5′ end. These results strengthen our assumption that H3K9ac may be involved in the regulation of light-responsive NAT pairs.
Figure 5.
H3K9ac is associated with the regulation of a light-responsive discordant NAT pair. (A,B) Detection of a discordant NAT pair by ATH NAT array and qRT-PCR. Error bar gives standard error (SE) (n = 3). (C) cotyledon; (H) hypocotyl; (R) root; (D) dark; (6) 6-h light. (C) Gene structures of AT2G18280 and AT2G18290 and H3K9ac peaks at this locus in darkness and light (6 h).
Effects of light on NAT pairs are largely independent of siRNA pathways
siRNA-mediated NAT regulation has been documented in both animals and plants (Borsani et al. 2005; Chellappan et al. 2010; Smalheiser et al. 2011). Since NAT pairs may generate a double-stranded RNA duplex to produce siRNAs, the question arises whether the latter are indeed produced in response to light.
To investigate small RNAs (smRNAs) associated with NAT pairs, we used our previously published smRNA deep sequencing data derived from flowers, leaves, roots, and seedlings (Wang et al. 2011). All 20- to 25-nt smRNAs mapped to the complementary regions of NAT pairs were taken as potential nat-siRNAs. In total, only 230 NAT pairs may serve as precursors of nat-siRNAs (Supplemental Table 6). Since HYL1 is involved in nat-siRNAs biogenesis in plants (Borsani et al. 2005; Chellappan et al. 2010), we expected levels of nat-siRNA-associated sense and antisense transcripts to be changed in the hyl1 mutant, and this correlation was more significant than transcripts without nat-siRNA. Transcript expression levels in WT and hyl1 were processed from strand-specific tiling array data (Kurihara et al. 2009). Compared to transcripts without related nat-siRNAs, expression of nat-siRNA- associated NAT pairs was significantly regulated by HYL1 (P-value of Mann-Whitney U-test <0.05). Thus, expression of these 230 NAT pairs may be regulated by a nat-siRNA-mediated pathway involving HYL1. However, none of the 230 nat-siRNA-associated NAT pairs were regulated by light. These results suggest that nat-siRNA-mediated regulation of NAT pairs does not contribute to light signaling pathways in general.
Discussion
Long noncoding NATs are a pervasive feature of the Arabidopsis genome
The proportion of antisense transcripts in the Arabidopsis genome was previously reported to be much lower than that in mammalian genomes (Wang et al. 2005; Matsui et al. 2008). Two possible reasons may have contributed to this observed discrepancy. First, work on Arabidopsis NATs has so far focused only on mRNAs without consideration of ncRNAs (Matsui et al. 2008). Second, recent identification of long noncoding RNAs in Arabidopsis was limited to those arising from the intergenic regions (Liu et al. 2012). Here, using a robust method, RepTAS, to identify long noncoding antisense transcripts, we have uncovered more than 37,200 NAT pairs in Arabidopsis; and notably, ∼70% of Arabidopsis genes produce one or more antisense transcripts. The total number of sense/antisense transcripts is not equal to two times the predicted NAT pairs, because some transcripts may be involved in two or more pairs, e.g., AT1G05291 shares complementary sequences with two lncNATs, AT1TU005650 and AT1TU005660, at the 5′ and the 3′ end, respectively.
Compared with published work in Arabidopsis (Wang et al. 2005; Matsui et al. 2008; Okamoto et al. 2010), ∼70%–80% of previously identified NAT pairs were included in our results (Supplemental Table 7). We updated the published results to the current version of the Arabidopsis genome annotation (TAIR 10) and compared them with our NAT pairs. Around 80% of those NAT pairs (6585 of 8336 NAT pairs; 79.0%) were included in our data set. Similarly, ∼78% circadian NAT pairs (1355 of 1742 NAT pairs; 77.8%) reported by Hazen et al. (2009) and 67% root NAT pairs (1159 of 1733 NAT pairs; 66.9%) reported by Li et al. (2013) were covered by our results. Therefore, based on current transcriptome data, our results represent the most comprehensive findings to date, and extend our knowledge of antisense transcripts in plants. Moreover, our results suggest that a large group of lncRNAs transcribed from the opposite strand of genes may regulate expression of the sense transcripts.
Meanwhile, we noted that ∼20%–30% of the previously identified NAT pairs were not represented in our data set. This may be due to (1) different criteria used for the identification of novel transcripts; (2) different definitions of NAT pair; and (3) the specific transcriptome data used for analysis. Taking these into consideration, it is likely our result may not cover all the NAT pairs in Arabidopsis, and novel NATs from specific organs or from plants subject to specific treatments remain to be identified by future work.
Natural antisense transcripts are likely to be bona fide transcripts
The majority of NAT pairs were reproducibly detected by tiling arrays and ATH NAT custom arrays. In addition, we collected ssRNA-seq data from only three plant organs grown under normal light/dark conditions. Nevertheless, using this limited ssRNA-seq data set, we detected >40% of the RepTAS-identified NAT pairs. Moreover, we confirmed a selected set of lncNATs (∼50) by qRT-PCR experiments. Thus, a large number of NATs could be detected by several different technical platforms, including tiling array, custom ATH NAT array, ssRNA-seq, and qRT-PCR, providing expression evidence that antisense transcripts are bone fide transcripts. Clear organ-specific expression of NAT pairs and their dynamic changes during de-etiolation also argue that the expression of antisense transcripts is under elaborate regulation.
Indeed, it is not surprising that some NATs could not be detected in our experiments. Most mammalian lncRNAs are expressed in a tissue-specific manner (Cabili et al. 2011), and many of the NATs identified here are also expressed in a tissue-specific manner. Thus, we believe that the undetected antisense transcripts may be expressed only under specific conditions (e.g., abiotic stress) or at certain developmental stages of Arabidopsis.
NATs are regulated by light in a spatial and development-specific manner
Previous reports using Affymetrix microarrays estimated that light could affect the expression of 20%–30% of the Arabidopsis genomic loci (Jiao et al. 2005). Here, using a custom-designed array to capture both coding and noncoding RNAs, we identified a total of 9236 light-responsive mRNAs and 13,907 light-responsive ncRNAs. Together, these transcripts are derived from >42% of the genomic loci represented on the ATH NAT array, suggesting close to one-half of the Arabidopsis genomic loci are responsive to light. This discrepancy between our results and previous data may be explained by the identification of a large number of light-regulated lncNATs that have hitherto escaped detection.
We found more than 1000 NAT pairs are regulated by light in a spatial and developmental-specific manner, suggesting their involvement in seedling light responses.
Organ-specific analysis showed more active transcriptome regulation in cotyledons than in the other two organs. This is not surprising as greening occurs primarily in the cotyledons in which the major event is the conversion of etioplasts to chloroplasts. In time course experiments during the de-etiolation process, we found that 1-h-responsive NAT pairs were different from 6-h-responsive pairs both in number and in species. The biological function of mRNAs for 1-h-reponsive NAT pairs was enriched in transcription factors and signaling components, whereas mRNAs for 6-h-responsive NAT pairs were mostly involved in metabolic processes. One possibility is that upon initial exposure to light, a relatively smaller group of mRNA genes, like those coding transcription factors and signaling components, is immediately either up- or down-regulated. The effects of these immediate-early genes that trigger primary light responses are subsequently amplified, leading to changes of a greater number of downstream genes that encode metabolism-related functions responsible for light adaptation. According to the function of mRNAs, it is possible that the corresponding lncNATs may play roles in similar biological processes with the immediate-early lncNATs being involved in transducing early light signals.
Mechanism under the regulation of NAT pairs in response to light
There are a number of proposed mechanisms for antisense-mediated regulation of sense transcripts. Research in the last several years has shown that certain antisense transcripts may direct histone modification at specific genomic loci. Here, we found 40%–50% light-responsive sense/antisense transcripts have their expression changes correlated with H3K9ac and/or H3K27ac. Moreover, we collected another set of ChIP-seq data of nine histone modifications in 2-wk-old Arabidopsis aerial tissues (Luo et al. 2013) and scanned histone mark enrichment peaks with MACS at P < 10−5 (Zhang et al. 2008). Similar to their observation, we also found sense and antisense transcripts to be associated with positive histone marks (Supplemental Fig. 10). These results provide insight into possible antisense-mediated histone modification of sense transcripts in response to light. However, we could not distinguish whether the change in abundance of NAT is the result of light-responsive histone modification or if light regulates NAT expression first followed by NAT-guided histone modification on the locus of sense transcripts. We hypothesized that a group of Arabidopsis antisense transcripts may direct histone modification and regulate the expression of sense transcripts. However, further experiments are needed to provide evidence for this hypothesis.
Methods
Identification of natural antisense transcript pairs
Novel transcripts were identified from 200 tiling arrays using RepTAS (Liu et al. 2012). Potential open reading frames (ORFs) of novel transcripts were predicted by GENSCAN using Arabidopsis parameters (Burge and Karlin 1997). By comparing novel transcripts to Arabidopsis gene structures, potential NAT pairs were defined as two transcripts that (1) are derived from opposite DNA strands; and (2) with a complementary sequence of >50 nt. Potential NAT pairs include pairs composed of two mRNAs, or one mRNA and one lncRNA, or two lncRNAs.
Design of ATH NAT array
ATH NAT array with a layout of 8 × 60 K was designed using Agilent eArray software with standard procedure (https://earray.chem.agilent.com/earray/). The array contained probes for 57,450 targets, which covered 33,526 predicted NAT pairs and 3088 singletons. For each target, one or two 60-mer probe(s) were assigned. Array design information was uploaded to the Gene Expression Omnibus (GEO) database with accession number GPL17515.
Plant materials and growth conditions
Arabidopsis thaliana ecotype Col-0 plants were used in this study. For sampling etiolated and de-etiolated seedlings, seeds were surface-sterilized in 70% ethanol briefly and placed directly into 30% Clorox with 0.1% Triton X-100 for 10 min and rinsed five times with sterilized water. Treated seeds were resuspended in sterilized 0.15% (w/v) agarose and sown in rows onto Murashige and Skoog (MS; MB biomedical, LLC, 263324) medium with 1% sucrose, Myo-inositol (Sigma, I3011), MES (Sigma, M8250), and 0.8% Bactoagar. Seeds on plates were stratified for 3 d at 4°C and exposed to white light (100 μmol m−2 s−1) for 1 h at 22°C to initiate germination before being transferred to continuous darkness for 4 d at 22°C. After a sampling of etiolated seedlings in the dark, seedlings on plates were exposed to continuous white light (100 μmol m−2 s−1) for 1 h and 6 h. Prior to sampling, de-etiolated seedlings were divided into cotyledons, hypocotyls, and roots, and the three organ samples were harvested and frozen immediately in liquid nitrogen and stored at −80°C.
Transcriptome detection by custom array
We performed RNA labeling, hybridization, and scanning according to the manufacturer's instructions (Agilent Technologies). Cyanine-3 (Cy3)–labeled cRNA was prepared from 100 ng total RNA using the One-Color Low RNA Input Linear Amplification Kit PLUS (Agilent Biotechnologies). Cy3-labeled cRNA was fragmented for 30 min at 60°C in 25 µL containing 2× Blocking Agent and 1× Fragmentation Buffer, and 25 µL of 2× Agilent Hybridization Buffer was added to the mixture and hybridized to ATH NAT array for 17 h at 65°C in a rotating Agilent hybridization oven at 10 rpm. After hybridization, the microarray slides were scanned (Agilent Technologies; model G2505C). For each time point (0, 1, and 6 h light), three biological replicates for cotyledons, hypocotyls, and roots in darkness (0 h) and light (1 h, 6 h) were analyzed resulting in a total of 27 hybridizations.
cDNA synthesis and qRT-PCR
Total RNA was isolated from cotyledons, hypocotyls, and roots using an RNeasy Mini Kit (Qiagen, 74904) following the manufacturer's instructions. For cDNA synthesis, 2.5 µg total RNA was incubated with SuperScript III Reverse Transcriptase (Invitrogen, 18080-051) according to the manufacturer's instructions. For quantitative RT-PCR analysis, cDNA was amplified using SYBR Premix ExTaq (Takara). Data were collected and analyzed by Bio-Rad CFX96 real-time system. ACTIN 2 levels were used for normalization.
ATH NAT array data analysis: expression of transcripts and NAT pairs
We used ATH NAT arrays to detect NAT-pair expression in specific organs or in plants after certain treatments. First, signal intensities were normalized by the Quantile method using GeneSpring software. We used two criteria to define probes with positive detection: (1) We used a number of spike-in probes that hybridized to a concentration series of RNAs. The lowest signal intensity of these probes was selected as a cutoff to define probes with positive detection; (2) the signal intensity of a probe should also be significantly higher than that of negative probes (P-value of Mann-Whitney U-test <0.05). Next, if two probes were assigned to a target, the signal intensities of both probes were compared to those of negative controls, and the one with the smaller P-value was selected as the representative probe. All representative probes were used in further analysis, e.g., identification of differentially expressed transcripts. Differentially expressed transcripts were selected if there was a twofold change in expression levels between control and treatment with a P-value <0.05 by eBays ANOVA using R with limma package.
Positively correlated or concordant NAT pairs were defined as those in which the expression level of both transcripts changed in the same direction with the expression level of one transcript changing no less than eightfold and that of the other no less than twofold. For negatively correlated or discordant NAT pairs, the expression levels of the two transcripts should change in the opposite directions with both transcripts changing more than twofold in their expression level.
Plant material, RNA extraction, library construction, and sequencing for ssRNA-seq
Col-0 seedlings were grown on MS plates for 2 wk and transferred to soil under long day conditions (16 h light). Three-week-old seedlings were bombarded with uncoated 0.6 µm gold particles using a Biolistic PDS-1000/He Particle Delivery System (Bio-Rad), to simulate plasmid transformation, although no plasmid was used for these samples. Plants were grown for 18 d after bombarding. Leaves, flowers (inflorescences), and siliques were collected from these 5.5-wk-old plants and immediately frozen in liquid nitrogen. RNA was extracted using the Qiagen RNeasy Plant Mini kit. Extracted RNA was treated with Turbo DNase I (Life Technologies AM2238) according to product specification. DNase I treated RNA was then applied again to an RNeasy spin column with 0.5 volumes of ethanol, washed, and eluted according to kit instructions.
The quality of the purified RNA was assessed using the Agilent 2100 Bioanalyzer. cDNA libraries for ssRNA-seq were prepared using the Illumina TruSeq Stranded mRNA Sample Preparation Kit, according to the low sample protocol guidelines. The quality and size of the libraries from each sample was tested using an Agilent High Sensitivity D1K ScreenTape System. The average size of the cDNA fragments was between 296 and 306 bp, and the most abundant fragments were between 243 and 256 bp. The samples were pooled into one well (six samples total per well were used) and sequenced on an Illumina HiSeq 2500 with 100-cycle single reads per multiplexed sample.
ssRNA-seq data analysis
Strand-specific RNA-seq reads were mapped to the Arabidopsis genome (TAIR10) using TopHat (version 2.0.8). The mapped reads were then compared to NATs using homemade Perl scripts. The abundance of NATs was calculated by the number of mapped reads per kilobase per million reads (RPKM).
Tiling array data analysis
We obtained strand-specific tiling array data sets of hyl1-2 mutant from the GEO database under accession number GSE15699 (Kurihara et al. 2009). Briefly, signal intensities were normalized by the Quantile method and expression levels were calculated by a summation of signal intensities of positive probes using Tukey's Median Polish (Liu et al. 2012).
smRNA sequencing data analysis
WT smRNAs were downloaded from the GEO database under accession numbers GSM707678-GSM707681 (Wang et al. 2011). smRNAs were mapped to the complementary region of all predicted NAT pairs using Perl script.
Data access
All related ATH NAT array data, including raw data and normalized signal intensity, have been submitted to the NCBI Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/) under accession numbers GSE49381 and GSE49382. Strand-specific RNA-seq data have also been submitted to the GEO under accession number GSE49950. The predicted NATs can be found in the Supplemental Material and at http://chualab.rockefeller.edu/cgi-bin/gb2/gbrowse/arabidopsis/ by selecting the “NAT” track.
Supplementary Material
Acknowledgments
We thank Connie Zhao and Bin Zhang (Genomics Resource Center, Rockefeller University) for their technical support and members of the Chua laboratory for discussion. This work was supported by the Cooperative Research Program for Agriculture Science and Technology Development (Project No. PJ906910), Rural Development Administration, Republic of Korea, by a National Research Foundation of Korea Grant funded by the Korean Government (NRF-2011-357-F00015); and by a grant from Klein Wanzlebener Saatzucht (KWS) SAAT AG, Germany. M.J.K. is supported in part by a Natural Sciences and Engineering Research Council of Canada (NSERC) Postdoctoral Fellowship.
Author contributions: H.W., P.J.C., and N.-H.C. designed experiments. H.W. and J.L. collected data and performed bioinformatics and statistical analysis. H.W. and J.L. designed ATH NAT arrays. P.J.C., I.-C.J., and J.X. performed array hybridizations. P.J.C. and I.-C.J. conducted qRT-PCR verifications. M.J.K. performed the strand-specific RNA-seq experiment, and H.W. analyzed the data. H.W., P.J.C., and N.-H.C. wrote the article, which was reviewed and approved by all authors.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.165555.113.
References
- Beiter T, Reich E, Williams RW, Simon P 2009. Antisense transcription: A critical look in both directions. Cell Mol Life Sci 66: 94–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borsani O, Zhu J, Verslues PE, Sunkar R, Zhu JK 2005. Endogenous siRNAs derived from a pair of natural cis-antisense transcripts regulate salt tolerance in Arabidopsis. Cell 123: 1279–1291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bovre K, Szybalski W 1969. Patterns of convergent and overlapping transcription within the b2 region of coliphage λ. Virology 38: 614–626 [DOI] [PubMed] [Google Scholar]
- Burge C, Karlin S 1997. Prediction of complete gene structures in human genomic DNA. J Mol Biol 268: 78–94 [DOI] [PubMed] [Google Scholar]
- Cabili MN, Trapnell C, Goff L, Koziol M, Tazon-Vega B, Regev A, Rinn JL 2011. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev 25: 1915–1927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camblong J, Iglesias N, Fickentscher C, Dieppois G, Stutz F 2007. Antisense RNA stabilization induces transcriptional gene silencing via histone deacetylation in S. cerevisiae. Cell 131: 706–717 [DOI] [PubMed] [Google Scholar]
- Camblong J, Beyrouthy N, Guffanti E, Schlaepfer G, Steinmetz LM, Stutz F 2009. Trans-acting antisense RNAs mediate transcriptional gene cosuppression in S. cerevisiae. Genes Dev 23: 1534–1545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charon C, Moreno AB, Bardou F, Crespi M 2010. Non-protein-coding RNAs and their interacting RNA-binding proteins in the plant cell nucleus. Mol Plant 3: 729–739 [DOI] [PubMed] [Google Scholar]
- Charron JB, He H, Elling AA, Deng XW 2009. Dynamic landscapes of four histone modifications during deetiolation in Arabidopsis. Plant Cell 21: 3732–3748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheetham SW, Gruhl F, Mattick JS, Dinger ME 2013. Long noncoding RNAs and the genetics of cancer. Br J Cancer 108: 2419–2425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chellappan P, Xia J, Zhou X, Gao S, Zhang X, Coutino G, Vazquez F, Zhang W, Jin H 2010. siRNAs from miRNA sites mediate DNA methylation of target genes. Nucleic Acids Res 38: 6883–6894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Sun M, Kent WJ, Huang X, Xie H, Wang W, Zhou G, Shi RZ, Rowley JD 2004. Over 20% of human transcripts might form sense-antisense pairs. Nucleic Acids Res 32: 4812–4820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng J, Kapranov P, Drenkow J, Dike S, Brubaker S, Patel S, Long J, Stern D, Tammana H, Helt G, et al. 2005. Transcriptional maps of 10 human chromosomes at 5-nucleotide resolution. Science 308: 1149–1154 [DOI] [PubMed] [Google Scholar]
- Chua YL, Channelière S, Mott E, Gray JC 2005. The bromodomain protein GTE6 controls leaf development in Arabidopsis by histone acetylation at ASYMMETRIC LEAVES1. Genes Dev 19: 2245–2254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faghihi MA, Wahlestedt C 2009. Regulatory roles of natural antisense transcripts. Nat Rev Mol Cell Biol 10: 637–643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guell M, van Noort V, Yus E, Chen WH, Leigh-Bell J, Michalodimitrakis K, Yamada T, Arumugam M, Doerks T, Kuhner S, et al. 2009. Transcriptome complexity in a genome-reduced bacterium. Science 326: 1268–1271 [DOI] [PubMed] [Google Scholar]
- Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, et al. 2009. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature 458: 223–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazen SP, Naef F, Quisel T, Gendron JM, Chen H, Ecker JR, Borevitz JO, Kay SA 2009. Exploring the transcriptional landscape of plant circadian rhythms using genome tiling arrays. Genome Biol 10: R17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henz SR, Cumbie JS, Kasschau KD, Lohmann JU, Carrington JC, Weigel D, Schmid M 2007. Distinct expression patterns of natural antisense transcripts in Arabidopsis. Plant Physiol 144: 1247–1255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ietswaart R, Wu Z, Dean C 2012. Flowering time control: Another window to the connection between antisense RNA and chromatin. Trends Genet 28: 445–453 [DOI] [PubMed] [Google Scholar]
- Jang IC, Chung PJ, Hemmes H, Jung C, Chua NH 2011. Rapid and reversible light-mediated chromatin modifications of Arabidopsis phytochrome A locus. Plant Cell 23: 459–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji H, Jiang H, Ma W, Wong WH 2011. Using CisGenome to analyze ChIP-chip and ChIP-seq data. Curr Protoc Bioinformatics 33: 2.13.1–2.13.45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Y, Ma L, Strickland E, Deng XW 2005. Conservation and divergence of light-regulated genome expression patterns during seedling development in rice and Arabidopsis. Plant Cell 17: 3239–3256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Y, Lau OS, Deng XW 2007. Light-regulated transcriptional networks in higher plants. Nat Rev Genet 8: 217–230 [DOI] [PubMed] [Google Scholar]
- Katayama S, Tomaru Y, Kasukawa T, Waki K, Nakanishi M, Nakamura M, Nishida H, Yap CC, Suzuki M, Kawai J, et al. 2005. Antisense transcription in the mammalian transcriptome. Science 309: 1564–1566 [DOI] [PubMed] [Google Scholar]
- Kurihara Y, Kaminuma E, Matsui A, Kawashima M, Tanaka M, Morosawa T, Ishida J, Mochizuki Y, Shinozaki K, Toyoda T, et al. 2009. Transcriptome analyses revealed diverse expression changes in ago1 and hyl1 Arabidopsis mutants. Plant Cell Physiol 50: 1715–1720 [DOI] [PubMed] [Google Scholar]
- Lavorgna G, Dahary D, Lehner B, Sorek R, Sanderson CM, Casari G 2004. In search of antisense. Trends Biochem Sci 29: 88–94 [DOI] [PubMed] [Google Scholar]
- Lee JT, Davidow LS, Warshawsky D 1999. Tsix, a gene antisense to Xist at the X-inactivation centre. Nat Genet 21: 400–404 [DOI] [PubMed] [Google Scholar]
- Leung A, Trac C, Jin W, Lanting L, Akbany A, Saetrom P, Schones DE, Natarajan R 2013. Novel long noncoding RNAs are regulated by angiotensin II in vascular smooth muscle cells. Circ Res 113: 266–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Liberman L, Mukherjee N, Benfey P, Ohler U 2013. Integrated detection of natural antisense transcripts using strand-specific RNA sequencing data. Genome Res 23: 1730–1739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Jung C, Xu J, Wang H, Deng S, Bernad L, Arenas-Huertero C, Chua NH 2012. Genome-wide analysis uncovers regulation of long intergenic noncoding RNAs in Arabidopsis. Plant Cell 24: 4333–4345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loidl P 2004. A plant dialect of the histone language. Trends Plant Sci 9: 84–90 [DOI] [PubMed] [Google Scholar]
- Lu F, Cui X, Zhang S, Jenuwein T, Cao X 2011. Arabidopsis REF6 is a histone H3 lysine 27 demethylase. Nat Genet 43: 715–719 [DOI] [PubMed] [Google Scholar]
- Luo C, Sidote DJ, Zhang Y, Kerstetter RA, Michael TP, Lam E 2013. Integrative analysis of chromatin states in Arabidopsis identified potential regulatory mechanisms for natural antisense transcript production. Plant J 73: 77–90 [DOI] [PubMed] [Google Scholar]
- Matsui A, Ishida J, Morosawa T, Mochizuki Y, Kaminuma E, Endo TA, Okamoto M, Nambara E, Nakajima M, Kawashima M, et al. 2008. Arabidopsis transcriptome analysis under drought, cold, high-salinity and ABA treatment conditions using a tiling array. Plant Cell Physiol 49: 1135–1149 [DOI] [PubMed] [Google Scholar]
- Ng SY, Bogu GK, Soh BS, Stanton LW 2013a. The long noncoding RNA RMST interacts with SOX2 to regulate neurogenesis. Mol Cell 51: 349–359 [DOI] [PubMed] [Google Scholar]
- Ng SY, Lin L, Soh BS, Stanton LW 2013b. Long noncoding RNAs in development and disease of the central nervous system. Trends Genet 29: 461–468 [DOI] [PubMed] [Google Scholar]
- Okamoto M, Tatematsu K, Matsui A, Morosawa T, Ishida J, Tanaka M, Endo TA, Mochizuki Y, Toyoda T, Kamiya Y, et al. 2010. Genome-wide analysis of endogenous abscisic acid-mediated transcription in dry and imbibed seeds of Arabidopsis using tiling arrays. Plant J 62: 39–51 [DOI] [PubMed] [Google Scholar]
- Shah JC, Clancy MJ 1992. IME4, a gene that mediates MAT and nutritional control of meiosis in Saccharomyces cerevisiae. Mol Cell Biol 12: 1078–1086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smalheiser NR, Lugli G, Thimmapuram J, Cook EH, Larson J 2011. Endogenous siRNAs and noncoding RNA-derived small RNAs are expressed in adult mouse hippocampus and are up-regulated in olfactory discrimination training. RNA 17: 166–181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiegelman WG, Reichardt LF, Yaniv M, Heinemann SF, Kaiser AD, Eisen H 1972. Bidirectional transcription and the regulation of phage λ repressor synthesis. Proc Natl Acad Sci 69: 3156–3160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swiezewski S, Liu F, Magusin A, Dean C 2009. Cold-induced silencing by long antisense transcripts of an Arabidopsis Polycomb target. Nature 462: 799–802 [DOI] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL 2009. TopHat: Discovering splice junctions with RNA-Seq. Bioinformatics 25: 1105–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulitsky I, Bartel DP 2013. lincRNAs: Genomics, evolution, and mechanisms. Cell 154: 26–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang XJ, Gaasterland T, Chua NH 2005. Genome-wide prediction and identification of cis-natural antisense transcripts in Arabidopsis thaliana. Genome Biol 6: R30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Zhang X, Liu J, Kiba T, Woo J, Ojo T, Hafner M, Tuschl T, Chua NH, Wang XJ 2011. Deep sequencing of small RNAs specifically associated with Arabidopsis AGO1 and AGO4 uncovers new AGO functions. Plant J 67: 292–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wek RC, Hatfield GW 1986. Nucleotide sequence and in vivo expression of the ilvY and ilvC genes in Escherichia coli K12. Transcription from divergent overlapping promoters. J Biol Chem 261: 2441–2450 [PubMed] [Google Scholar]
- Werner A 2005. Natural antisense transcripts. RNA Biol 2: 53–62 [DOI] [PubMed] [Google Scholar]
- Wong F, Yuh ZT, Schaefer EL, Roop BC, Ally AH 1987. Overlapping transcription units in the transient receptor potential locus of Drosophila melanogaster. Somat Cell Mol Genet 13: 661–669 [DOI] [PubMed] [Google Scholar]
- Wu X, Liu M, Downie B, Liang C, Ji G, Li QQ, Hunt AG 2011. Genome-wide landscape of polyadenylation in Arabidopsis provides evidence for extensive alternative polyadenylation. Proc Natl Acad Sci 108: 12533–12538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Y, Zhao Y, Wang J, Liu C, Chen S, Chen R, Zhao H 2007. antiCODE: A natural sense-antisense transcripts database. BMC Bioinformatics 8: 319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu XS, Liu QR, Wei L 2006. Genome-wide in silico identification and analysis of cis natural antisense transcripts (cis-NATs) in ten species. Nucleic Acids Res 34: 3465–3475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Clarenz O, Cokus S, Bernatavichute YV, Pellegrini M, Goodrich J, Jacobsen SE 2007. Whole-genome analysis of histone H3 lysine 27 trimethylation in Arabidopsis. PLoS Biol 5: e129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, et al. 2008. Model-based analysis of ChIP-Seq (MACS). Genome Biol 9: R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.