Background: Prion strains exhibit distinct physical and biochemical repertoires, aggregation propensity, and biological properties.
Results: A biochemical approach is developed for defining the conformational features of prions with or without glycosylphosphatidylinositol (GPI)-anchor.
Conclusion: GPI anchorless prions are detected in human genetic prion diseases, but not in sporadic forms.
Significance: Unveiling the structure of GPI anchorless prions to predict pathological properties.
Keywords: Amyloid, Glycosyl Phosphatidylinositol Anchors, Neurodegenerative Diseases, Prions, Transgenic Mice
Abstract
The role of the GPI-anchor in prion disease pathogenesis is still a challenging issue. In vitro studies have shown that anchorless cellular prion protein (PrPC) undergoes aberrant post-translational processing and metabolism. Moreover, transgenic (Tg) mice overexpressing anchorless PrPC develop a spontaneous neurological disease accompanied with widespread brain PrP amyloid deposition, in the absence of spongiform changes. Generation of PrP forms lacking the GPI and PrP amyloidosis are striking features of human stop codon mutations in the PrP gene (PRNP), associated with PrP cerebral amyloid angiopathy (PrP-CAA) and Gerstmann-Sträussler-Scheinker (GSS) syndrome. More recently, the presence of anchorless PrP species has been also claimed in sporadic Creutzfeldt-Jakob disease (sCJD). Using a highly sensitive protein separation technique and taking advantage of reference maps of synthetic PrP peptides, we investigated brain tissues from scrapie-infected “anchorless PrP” Tg mice and wild type mice to determine the contribution of the GPI-anchor to the molecular mass and isoelectric point of PrP quasispecies under two-dimensional electrophoresis. We also assessed the conformational properties of anchorless and anchored prions under standard and inactivating conditions. These studies were extended to sCJD and GSS. At variance with GSS, characterization of PrP quasispecies in different sCJD subtypes ruled out the presence of anchorless prions. Moreover, under inactivating conditions, mice anchorless prions, but not sCJD prions, generated internal PrP fragments, cleaved at both N and C termini, similar to those found in PrP-CAA and GSS brain tissues. These findings show that anchorless PrPSc generates GSS-like PrP fragments, and suggest a major role for unanchored PrP in amyloidogenesis.
Introduction
Transmissible spongiform encephalopathies (TSEs),3 or prion diseases, include Creutzfeldt-Jakob disease (CJD), fatal familial insomnia (FFI), and GSS in humans, chronic wasting disease in cervids, scrapie in sheep, and bovine spongiform encephalopathy in cattle (1).
In TSEs, PrPC is converted to an abnormal isoform, or PrPSc, characterized by relative resistance to protease digestion and detergent insolubility (2). PrPC is a GPI-anchored glycoprotein, which in its mature form spans residues 23–231 after the removal of the N-terminal and C-terminal signal sequences, and attachment of the GPI-anchor at residue 231 (3, 4). Post-translational PrPC processing involves endosomal recycling of the cell surface full-length protein and proteolytic cleavages at residues 111/112 and ∼90, leading to the generation of N-terminally truncated C1 and C2 fragments (5); an additional proteolytic cleavage of PrPC is thought to occur at the C terminus, near the GPI-anchor, which results in detachment of the full-length protein from the cellular surface. Even though the bulk of brain PrPC is GPI-anchored, protease-mediated shedding of the PrPC ectodomain or enzymatic cleavage of the GPI phospholipid moiety have been reported in the cerebrospinal fluid (CSF), plasma, urine, and cell cultures (6, 7, 8). However, physiologically shed PrPC species display distinct features from PrPC experimentally exposed to the enzyme phosphatidylinositol-specific phospholipase C (PI-PLC). Indeed, upon delipidation of PrPC molecules with PI-PLC, the diacylglycerol hydrophobic tail is hydrolyzed, with exposure of negatively charged phosphate groups by the retained portion of the anchor. The detachment of the phospholipid moiety accounts for the paradoxical slower migration of PI-PLC-digested PrPC species under SDS-polyacrylamide gel electrophoresis (9). Expectedly, this effect is not observed upon PI-PLC treatment of native and recombinant anchorless PrPC molecules (10) and of post-translational modified PrPC species lacking the phospholipid component (8). On the other hand, in vitro studies have shown that anchorless PrPC is not tethered to the cell membrane, and is not recycled within the endosomal compartment, being instead secreted extracellularly (11).
Intriguingly, transgenic mice overexpressing anchorless PrPC develop a spontaneous GSS-like neurologic illness with widespread PrP amyloid deposition in brain tissues, as a result of aggregation and accumulation of an internal PrP fragment (12). Additionally, infection of “anchorless PrP” mice induces the formation of angiocentric amyloid plaques, as opposed to granular PrPSc deposition observed in wild-type mice (13). This suggests that GPI-anchored and anchorless PrP are converted at diverse subcellular and/or extracellular sites, and spread through different routes (14). Anchorless and anchored PrPSc molecules display marked variability and heterogeneity in their glycosylation profile, generation of protease-resistant quasispecies, and aggregation propensity, adding further complexity to their physicochemical features and strain properties. Although valuable information has been gained from experimental models, a clear definition of molecular properties of anchorless PrP in human prion disorders is still missing.
sCJD, the most common human prion disease, has an annual incidence of ∼1.5–2 per million worldwide, and a still unknown etiology. Prevailing hypotheses suggest that the disorder is triggered by spontaneous changes in PrPC conformation, although concern has been raised that some sCJD cases might occur as a consequence of environmental exposure, case-to-case transmission, or food contamination (15). On a molecular ground, sCJD PrPSc is characterized by two major types of PrP27–30 with unglycosylated peptides of 21 and 19 kDa, in addition to distinct C-terminal fragments, but not internal PrP truncated fragments.
Recently, the generation of anchorless PrP forms has been also claimed in sCJD, hence suggesting that in addition to anchored PrPSc conformers, anchorless molecules could contribute to the phenotypic heterogeneity of this disorder (16). This issue raises additional concerns regarding the neuroinvasive properties of these quasispecies and the potential infectivity of human body fluids and peripheral tissues of sCJD patients. In the present study, we used a highly sensitive protein separation technique to assess the electrophoretic coordinates of anchorless and anchored PrPSc isoforms, using a panel of different synthetic PrP peptides as a reference map. Further, we exploited the conformational properties of anchorless and anchored prions to investigate their expression in different sCJD subtypes.
EXPERIMENTAL PROCEDURES
Synthetic Peptides and Antibodies
Human synthetic PrP peptides spanning sequences 23–230, 90–230, 105–230, and 121–230 were purchased (Alicon AG, Zurich, CH); PrP peptide 82–146 was kindly donated by Dr. M. Salmona. The following mouse monoclonal antibodies recognizing different human PrP epitopes were used: 3F4, residues 108–111 (Signet Laboratories), 6D11, residues 93–109 (Signet Laboratories), ICSM-35, residues 93–102 (d-Gen UK), 12B2, residues 89–93 (kindly donated by Dr. J.P.M. Langeveld), SAF70, residues 142–160 (Cayman Chemicals), 6H4, residues 144–152 (Prionics, CH), 4G11, residues 199–216, and 3E2, residues 214–231 (kindly donated by Dr. L. Capucci).
Animal Inoculation
All mice were housed at the Rocky Mountain Laboratories (RML) in an AAALAC-accredited facility. Research protocols and experimentation were approved by the NIH RML Animal Care and Use Committee. Transgenic GPI anchorless PrP mice (tg44+/+) were generated as previously described (13). Four to six-week-old mice were inoculated intracerebrally with 50 μl of a 1% brain homogenate of RML scrapie containing 0.7–1.0 × 106 ID50. One ID50 is the dose causing infection in 50% of C57BL/10SnJ mice. Animals were observed daily for onset and progression of scrapie. Mice were euthanized when clinical signs were consistent and progressive.
sCJD Tissue Samples
Brain samples were obtained from 29 cases of definite sCJD, 11 methionine homozygous at codon 129 with type 1 PrPSc (MM1), 3 methionine/valine with type 1 PrPSc (MV1), 3 MM with type 2 PrPSc (MM2), 7 MV2, 5 VV2, and 1 case of variant CJD (vCJD), diagnosed according to current criteria (17). Post-mortem intervals ranged from 4 to 30 h. Genomic DNA, extracted from frozen brain tissues, was searched for PRNP mutations and M/V polymorphism at codon 129. Neuropathological and immunohistochemical studies were performed as described previously (18).
Immunoblot Analysis
Brain samples were homogenized in 9 volumes of lysis buffer (100 mm sodium chloride, 10 mm EDTA, 0.5% Nonidet P-40, 0.5% sodium deoxycholate, 10 mm Tris, pH 7.4). Aliquots were adjusted to a final concentration of 50 μg of proteinase K (Roche Applied Science, Germany) per milliliter and incubated at 37 °C for 60 min; protease digestion was quenched by adding PMSF to a final concentration of 2 mm (Boehringer, Manheim). In some experiments, PMFS was omitted and samples were dissolved in Laemmli buffer (3% SDS, 3% β-mercaptoethanol, 2 mm EDTA, 10% glycerol, 62.5 mm Tris, pH 6.8), before boiling at 100 °C for 5 min. The combined exposure of samples to the denaturing anionic detergent SDS and PK, was exploited to solubilize non-amyloid PrP complexes and assess the conformation of PrPSc species under conditions altering the quaternary and tertiary structure of PrPSc.
For N-deglycosylation, samples were treated with N-glycosidase F (PNGase-F) according to the manufacturer's instruction (Roche Applied Science) for 8 h at 37 °C. Samples were dissolved in Laemmli buffer and boiled for 5 min. An equivalent of 0.4 mg of wet tissue was loaded on 13% SDS-PAGE gels and proteins were transferred onto PVDF membrane (Immobilon P, Millipore) for 2 h at 60 V. Membranes were blocked with 1% nonfat dry milk in TBST (10 mm Tris, 150 mm NaCl, 0.1% Tween-20, pH 7.5) for 1 h at 37 °C and incubated overnight at 4 °C with anti-human PrP monoclonal antibodies (3F4, 1:5,000; 6D11, 1:3,000; ICSM-35, 1:1,000; 12B2, 1:8.000; SAF70, 1:1,000; 6H4, 1:5,000; 4G11, 1:1,000; 3E2, 1: 3,000). Blots were developed with an enhanced chemiluminescence system (ECL, Amersham Biosciences) and PrP visualized on autoradiographic films (Hyperfilm, Amersham Biosciences). Films were scanned by using a densitometer (GS-200, Bio-Rad).
Two-Dimensional Gel Electrophoresis (2D-PAGE)
For isoelectric focusing (IEF), using immobilized pH gradients (IPG) in the first dimension, pre-cast gels with a linear pH range of 3–10 were used (Bio-Rad). Before IEF, the dry gels were reswollen for 14–15 h in 125 μl of buffer (6 m urea, 2 m thiourea, 5% β-mercaptoethanol, 2% Nonidet P-40, and 2% Ampholytes) containing the equivalent of 2 mg of wet tissue. IEF was carried out at 20 °C for 4 h with raising voltage (500–8000 V), in a cooled horizontal electrophoresis unit (IPGphor, Pharmacia). For the second dimension, the IPG strips were equilibrated for 20 min in 50 mm Tris-HCl, 6 m urea, 10% glycerol, 2% SDS, and a trace of bromphenol blue and loaded on a 16% SDS-PAGE (19). Immunoblotting was performed as described above.
RESULTS
Molecular Typing of Anchorless and Wild Type Prions
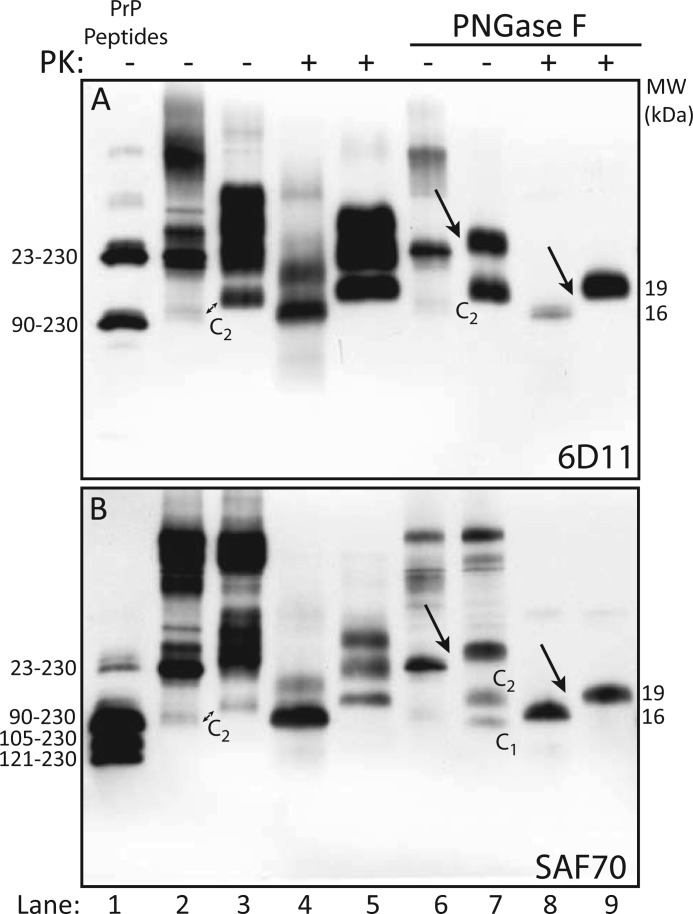
Immunoblots with 6D11 of non-PK-treated brain homogenates (BHs) from “anchorless PrP” Tg mice and “anchored PrP” C57BL mice, both infected with the RML scrapie strain, showed that anchorless PrP resolved in 2 bands (Fig. 1A, lane 2), representing mono- and unglycosylated forms, as opposed to the customary separation of anchored PrP under di-, mono- and unglycosylated species (Fig. 1A, lane 3). As expected, the unglycosylated anchorless PrP band co-migrated with the PrP23−230 synthetic peptide at 23 kDa (Fig. 1A, lanes 1 and 2). Higher molecular mass forms, consistent with PrP multimers, in addition to a barely detectable 16 kDa truncated fragment, were observed in “anchorless PrP” Tg mice; conversely, anchored PrP showed an additional C-terminal truncated fragment migrating in a 19-kDa zone, accounting for the C2 fragment, or the unglycosylated fragment generated by endogenous proteolysis (5). Following treatment with PK, anchorless PrPSc separated under mono- and unglycosylated isoforms, migrating at ∼ 21 and 16 kDa (Fig. 1A, lane 4), whereas anchored PrPSc glycoforms migrated at ∼28, 24, and 19 kDa (Fig. 1A, lane 5). Hence, the unglycosylated fragment of anchorless PrPSc migrated ∼3 kDa faster than the corresponding fragment of anchored PrPSc, in keeping with the estimated molecular mass of the GPI anchor. Enzymatic deglycosylation of anchorless PrP yielded a 23-kDa band, in addition to a barely visible 16 kDa band, and reduced anchored PrP to two distinct bands of 26 and 19 kDa, representing full-length PrP and the C2 fragment (Fig. 1A, lanes 6 and 7); further treatment with PK generated a core fragment of 16 kDa for anchorless PrP and 19 kDa for anchored PrP (Fig. 1A, lanes 8 and 9). Membranes probed with mAb SAF70 overlapped results observed with 6D11 (Fig. 1B, lanes 2–9), with the exception of an additional lower band detected in wild type mice, corresponding to the C1 fragment (Fig. 1B, lane 7). Taken together, the foregoing results were consistent with the GPI anchor contributing for ∼3 kDa to the orthogonal migration of PrP, either before or after PK proteolysis.
FIGURE 1.
Characterization of PrPSc in “anchorless PrP” transgenic mice and wild-type mice. A, immunoblot with 6D11 of synthetic PrP peptides (lane 1), and brain homogenates from “anchorless PrP” Tg mice (even lanes) and “anchored PrP” C57BL mice (odd lanes), untreated (−) or treated (+) with PK, before (lanes 2–5) and after deglycosylation with PNGase F (lanes 6–9). 6D11 recognizes synthetic PrP23−230 and PrP90−230 peptides migrating at 23 and 16kDa (lane 1). Anchorless PrP is resolved in two bands, including mono- and unglycosylated species, migrating at 26 and 23 kDa (lane 2) the latter co-migrating with PrP23−230 (lane 1). Anchored PrP separates as four 35 to 19-kDa bands, representing full length PrP isoforms and the N-terminally truncated fragment C2 (lane 3). After PK digestion, anchorless PrPSc migrates as two bands of 21- and16-kDa (lane 4), while anchored PrPSc shows the canonical triplet of differently glycosylated isoforms (lane 5). After deglycosylation, anchorless PrPSc is reduced to a single 23-kDa band (lane 6) and anchored PrPSc to two bands of 26- and 19-kDa representing full-length unglycosylated PrP and the C2 fragment, respectively (lane 7). PK treatment generates a core fragment of 16 kDa for anchorless PrPSc and 19 kDa for anchored PrPSs (lanes 8 and 9, arrow). B, immunoblot analysis with SAF 70 of the same samples as in A. As expected, SAF 70 also recognizes synthetic PrP105–230 and PrP121–230 peptides (lane 1). Patterns of PrP separation are comparable to those shown in panel A, except for the detection of C1 in wild type mice (lane 7).
Two-Dimensional Mapping of Anchorless and Wild Type Prions
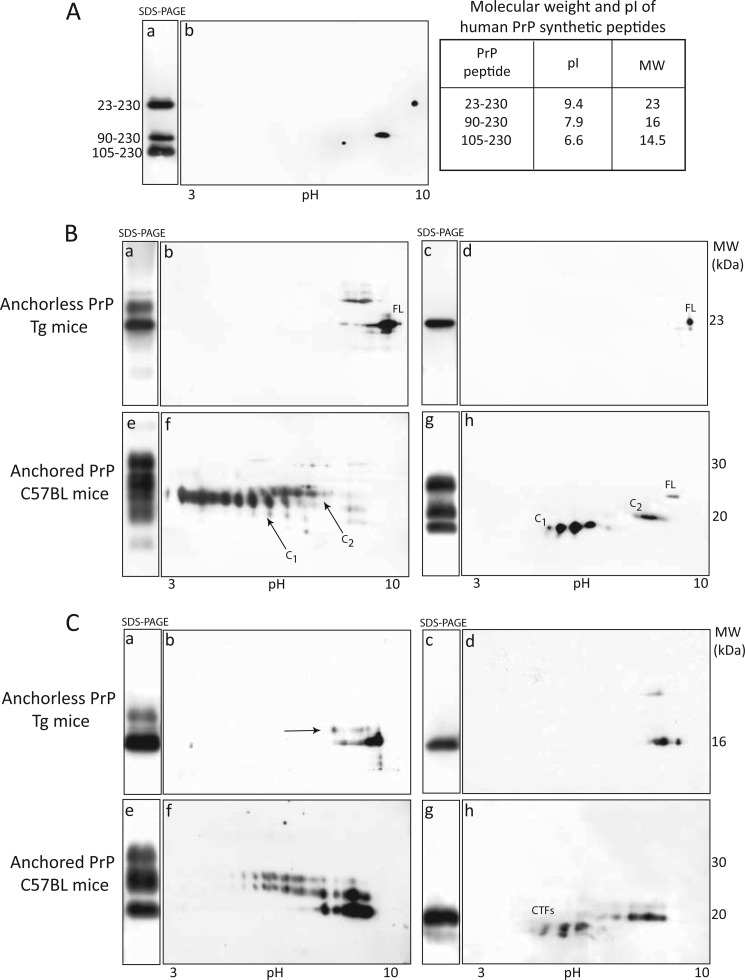
By 2D-PAGE, we next compared the isoelectric point (reflecting the net charge) and the mobility (determined by the molecular mass) of anchorless PrP and anchored PrP glycoforms. Reference SDS-PAGE and 2D-PAGE maps of human synthetic PrP peptides were as follows: the 23–230 peptide migrated at 23 kDa, pI 9.4, the 90–230 at 16 kDa, pI 7.9, and the 105–230 at 14.5 kDa, pI 6.6 (Fig. 2A, panels a, b, and table).
FIGURE 2.
SDS-PAGE and two-dimensional analyses of PrP in RML-infected “anchorless PrP” transgenic mice and “anchored PrP” C57BL mice. A, SDS-PAGE (a) and 2D-PAGE (b) separation of human synthetic PrP23–230, PrP90–230, and PrP105–230 peptides (table). B, immunoblots with SAF 70 of brain homogenates from “anchorless PrP” Tg mice (panels a–d) and “anchored PrP” mice (panels e–h). Monoglycosylated and unglycosylated anchorless PrP species migrate at 26 kDa, pI 8–9.2, and 23 kDa, pI 9.4, (panels a and b), both reduced to a single deglycosylated 23-kDa isoform (panels c and d). Anchored PrP separates in a 35-to-18 kDa zone (panel e), within a wide pI range (panel f). After glycans removal, full-length PrP migrates at ∼26 kDa, pI 8.4, C2 at 20 kDa, pI 7.5, and C1 separates as four 18-kDa spots, pI range 5.5–4.0 (panels g and h). C, PK-treated anchorless PrPSc separates in two zones at 19- and 16-kDa, corresponding to monoglycosylated and unglycosylated PrP isoforms (panels a and b); monoglycosylated PrP species are poorly resolved due to immature glycosylation (panel b, arrow); after glycans removal, anchorless PrPSc core fragment migrates at 16kDa (panel c), as multiple basic spots, pI 7.5–8.0, indicative of N-terminal ragged ends (panel d). PK-treated anchored PrPSc migrates in a 30-to-19-kDa zone, 3.0 to 7.5 pH range (panels e and f), reduced to 19- and 16-kDa isoforms after PNGase-F treatment (panel g); the pI range of the spots of the PrPSc core fragment is a pI range of 6.2–7.6, whereas 16-kDa C-terminal truncated fragments separates in a 4.5-to-5.5 pI zone (panel h, CTFs).
Immunoblots with 6D11 (data not shown) and SAF70 of BHs from anchorless PrP Tg mice showed that the unglycosylated PrP isoform migrated at 23 kDa, pI 9.4, matching the orthogonal and horizontal mobilities of the synthetic PrP peptide 23–230, whereas monoglycosylated isoforms showed a string pattern in a 28-kDa zone, pI 8–9.2 (Fig. 2B, panels a and b), all reduced to a single 23-kDa spot upon enzymatic deglycosylation (Fig. 2B, panels c and d). In overexposed films a 16-kDa spot, matching the migration of the 90–230 peptide, was seen.
Immunoblots with 6D11 of wt mice BHs showed that PrP separated as trains of spots migrating between 35 and 19kDa, pIs ∼4–8.5, including glycosylated and unglycosylated isoforms of the full-length PrP and glycoforms of the C2 fragment (data not shown); upon deglycosylation, PrP isoforms resumed to two main 26-kDa and 19-kDa spots, pIs 8.4 and 7.2–8.2, accounting for the unglycosylated full-length PrP and the C2 fragment. Membranes probed with SAF70, which preferentially binds glycosylated forms of C1, disclosed a PrP pattern dominated by a set of spots with an acidic migration (Fig. 2B, panels e and f), all reduced to three set of spots after deglycosylation, including full-length PrP, C2, and C1 fragments (Fig. 2B, panels g and h). Notably, intra-sample differences in PrP decoration, observed between SDS-PAGE and 2D-PAGE, reflect the facilitation or restriction of mAbs binding to PrP molecules denatured by alternate protocols.
Taken together, using a high sensitive proteomic approach we confirmed that lack of the GPI anchor induces an alkaline shift of ∼1 pH unit in the horizontal migration of PrP, and also that anchorless PrP is characterized by immature glycosylation and altered endogenous proteolytic processing (13).
After PK digestion, anchorless PrPSc separated into minor glycosylated spots in a 19-kDa zone, in addition to well-represented unglycosylated 16 kDa isoforms, consistent with multiple ragged N-terminal ends of the unglycosylated PrP27–30 fragment (Fig. 2C, panels a and b), all reduced to a single train of 16-kDa spots, pIs 7.5–8, after PNGase treatment (Fig. 2C, panels c and d).
As expected, immunoblots with SAF70 of PK-digested BHs from C57BL mice showed the typical separation of PrPSc in three series of variably glycosylated isoforms (panels e and f), all resumed to three contiguous 19-kDa spots upon deglycosylation (Fig. 2C, panels e and f). After deglycosylation C-terminal truncated fragments (CTFs), migrating at 14–16 kDa, pI range 4–6, were observed (Fig. 2C, panels g and h). Taken together, besides differences in migration, anchorless PrPSc does not generate CTFs under protease treatment.
Molecular Characteristics of PrPSc in sCJD Subtypes and vCJD
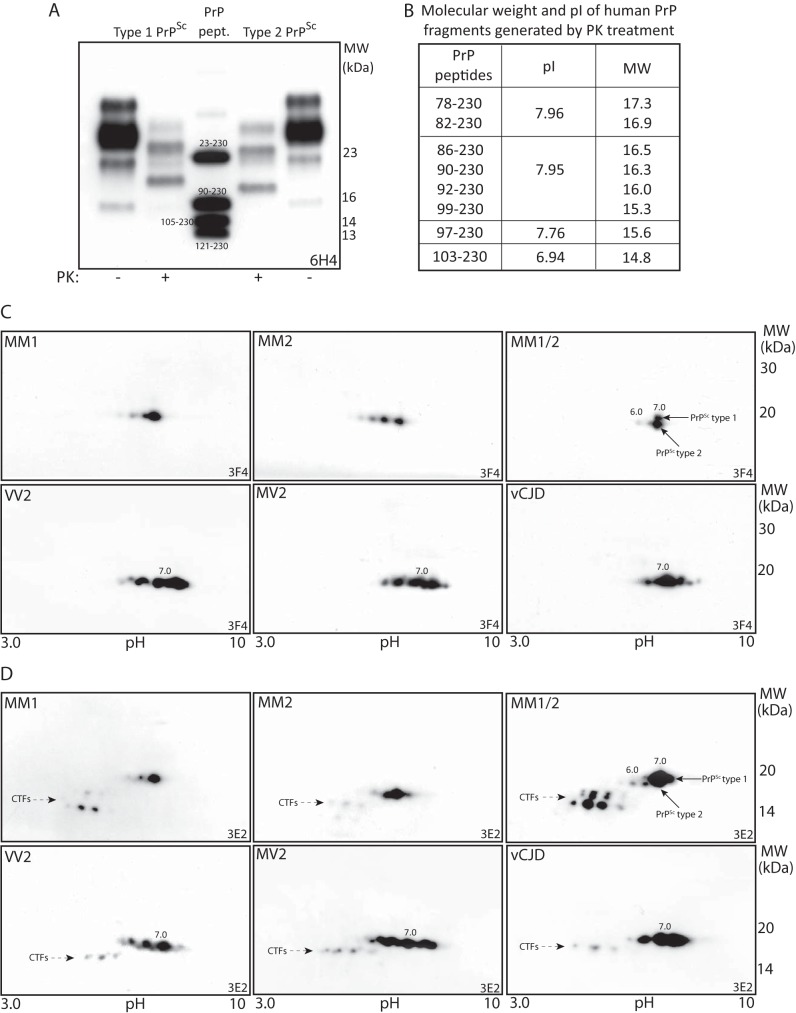
In previous 2D-PAGE studies of PrPSc in different sCJD subtypes and vCJD, we were unable to detect molecular forms consistent with anchorless PrP species. Here, we first assessed the SDS-PAGE migration of type-1 and type-2 PrPSc, as compared with the map of human synthetic PrP peptides. Accordingly, the unglycosylated PrP27–30 fragment migrates at 19.5-kDa in type-1 PrPSc, and 17-kDa in type-2 PrPSc (Fig. 3, A and B), which is at variance with their conventionally reported migration at 21 and 19 kDa (20). Next, we reassessed the 2D-PAGE migration of the core fragment in different sCJD subtypes using anti-PrP mAbs 3F4 and 3E2. In sCJD MM1, the PrP27–30 core fragment migrates as a single tailed 19.5-kDa spot centered at pI 7.0, whereas in sCJD MM2 the core fragment separated as three major 17.5-kDa spots, pI range 6.0–7.0, as an effect of multiple cleavage sites (Fig. 3C). Differences in migration between PrPSc of MM1 and MM2 subtypes were better visualized in a sCJD case with co-occurrence of both PrP types (MM1 + 2), where the core fragment of type-1 PrPSc and the most basic spot of the type-2 PrPSc train shared a spot at pI 7.0, with diverging molecular masses. In contrast to sCJD MM1, in MV2 and VV2 subtypes PrPSc separated as multiple spots in a wider pI range, spanning from 6.6 to 8.0, indicating a larger heterogeneity of PrP N-terminal variants, whereas in vCJD, PrPSc resolved in three major spots (18, 21).
FIGURE 3.
SDS-PAGE and 2D-PAGE analyses of PrPSc in sCJD and vCJD. A, immunoblot with 6H4 of BH from frontal cortexes of sCJD subtypes MM1, MV2, and synthetic PrP peptides, show that the PK-resistant unglycosylated PrPSc fragment is 19.5 kDa for type 1 and 17 kDa for type 2 PrPSc. B, molecular masses and pIs of synthetic PrP peptides matching major N-terminal variants of type 1 and type 2 PrPSc detected in human prion diseases. C, 2D-PAGE immunoblots with 3F4 of PrPSc core fragments in different molecular sCJD subtypes. In MM1 sCJD subtype, PrPSc core fragment consists of a single 19.5 kDa spot, pI 7.0, whereas in MM2 it is composed by three 17-kDa spots, pI range 6.0–7.0; distinct migration of PrPSc core fragment is shown in a MM1/2 sCJD case (arrows indicate different PrPSc types. In VV2, MV2, and vCJD, PrPSc separates as multiple 17-kDa spots, pIs range 6.6–8.0. D, immunoblots with 3E2 of BHs reported in panel C, show an overlapping pattern of PrPSc core fragments, in addition to additional sets of acidic spots representing CTFs (arrows).
By probing membranes with mAb 3E2 (epitope 214–231), the 2D-patterns of PrPSc core fragments in different sCJD subtypes and in vCJD overlapped those observed with 3F4; additionally, acidic C-terminal truncated fragments were seen in distinct sCJD subtypes, as already reported (Fig. 3D). Taken together, in MM1, MM2, and MM1 + 2 sCJD subtypes, each single spot of the core fragment is 1.0 pH unit more acidic than the corresponding synthetic peptide and ∼2.5 kDa slower, as expected for anchored PrP species. At variance, MV2, VV2, and vCJD show the presence of additional basic spots with migration between 7.0 and 8.0. Although caution is needed in assigning these species to anchorless PrP forms, the lack of the expected 2.5 kDa shift in their orthogonal migration and the recognition by mAb3E2 rule out the hypothesis that they could represent anchorless PrP species, rather suggesting the presence of molecular forms with modified GPI composition.
Antibody Mapping of PrPSc in sCJD and vCJD: No Evidence for Anchorless Species
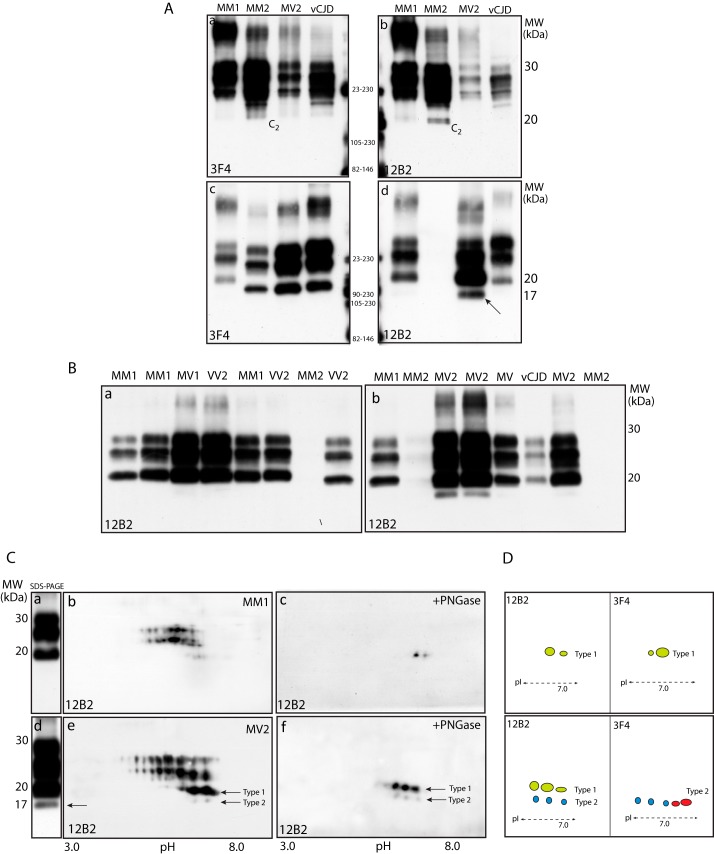
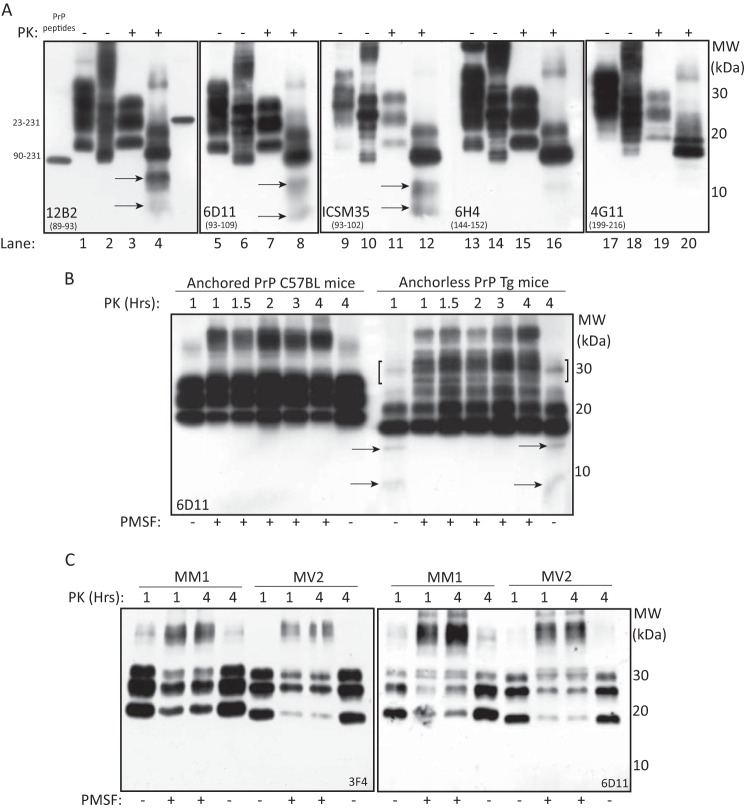
Previously, Notari et al. reported on the constitutive expression of anchorless PrPSc species in all sCJD subtypes, and their enrichment after PK digestion (16). Anchorless PrP quasispecies were tentatively identified based on their migration, ∼2 kDa faster than the core fragment, and their detection with mAbs directed to epitopes along PrP residues 89–221, including 12B2 and 3F4, but not with a rabbit antiserum directed to residues 220–231.
In the present study, PrPSc characterization in different sCJD subtypes and vCJD, did not show additional bands, consistent with anchorless PrP species, either before (Fig. 4A, panels a and b) or after PK treatment (Fig. 4A, panels c and d). In particular, immunoblots with anti-PrP mAbs 6D11 (epitope 93–109) and ICSM-35 (epitope 93–102) showed only a different affinity and variability of the PrPSc glycosylation profile, but no additional faster migrating bands (data not shown). At variance, immunoblots with 12B2, recognizing an epitope at position 89–93 of human PrP, showed that the unglycosylated PrP27–30 fragment co-migrated in a 19.5 kDa zone in all sCJD subtypes, either with type-1 or type-2 PrPSc and vCJD. An exception to this pattern was the lack of recognition of PrP27–30 in CJD MM2 subtype, and the detection of a PrP band migrating in a 17 kDa zone in four out seven sCJD MV2 cases (Fig. 4B, panels a and b). These findings further support previous demonstration that type-1 PrPSc is variably expressed in vCJD and sCJD cases with type-2 PrPSc, either homozygous or heterozygous at codon 129. The lack of PrP27–30 staining in MM2 cases is consistent with this molecular subtype being mostly composed by the N-terminal variant of PrP27–30 starting at S97, hence lacking the 12B2 epitope (21).
FIGURE 4.
Antibody mapping of PrPSc in sCJD and vCJD: absence of unanchored PrPSc forms. A, immunoblots with mAbs 3F4 and 12B2 of native (panels a and b) and PK-treated (panels c and d) BHs from different sCJD subtypes and vCJD. In PK-untreated samples, PrP species separate in a 35-to-19.5-kDa zone in MM1, and 35-to-17-kDa in MM2, the faster band accounting for the C2 fragment. After PK digestion, the conventional pattern of PrP27–30 is shown with 3F4; conversely, in sCJD MV2 subtype, immunoblot with 12B2 shows the customary di- and monoglycosylated bands, in addition to two unglycosylated bands, the slower co-migrating with the unglycosylated fragment of MM1 subtype, and the faster at 17-kDa (d, arrow). B, immunoblot with 12B2 of PK-treated BH from MM1, MV1, and VV2 sCJD subtypes shows co-migration of PrP27–30 glycoforms (panel a); conversely, the PrP27–30 pattern in MV2 subtype includes 4 bands, the faster migrating in 17 kDa zone (panel b). C, SDS-PAGE and 2D-PAGE immunoblots with 12B2 of PK-treated MM1 BHs (panels a–c) and MV2 sCJD subtypes (panels d–f) shows that in MM1, PrP27–30 separates as three 30-to-19.5-kDa bands (panel a) that resolve into three sets of spots (panel b) accounting for differently glycosylated species, all reduced to a main 19.5-kDa spot, pI 7.0, after deglycosylation (panel c). In MV2 subtype, 12B2 decorates three major 30-to-19.5-kDa bands, in addition to a 17-kDa band (panel d, arrow), that separates into four sets of spots; following PNGase F treatment, two train of spots at 19.5 and 17-kDa are seen, accounting for type-1 and type-2 core fragments (panel f, arrows). D, schematic representation of the core fragment patterns obtained with 3F4 and 12B2 in MM1 and MV2 sCJD subtypes, combining results shown in Figs. 3C and 4C.
After 2D-PAGE separation, immunoblots with 12B2 confirmed the co-expression of type-1 and type-2 unglycosylated PrP27–30 spots in four MV2 cases. As expected, these spots shared a similar horizontal migration, within a 6.0–7.0 pI range, but differed in their orthogonal mobility at 19.5 and 17 kDa (Fig. 4C). As compared with the pattern obtained with mAbs 3E2 and 3F4 (see Fig. 3, C and D), in MV2 cases the reduced number of spots, and in particular the absence of basic 17.5-kDa isoforms, is consistent with mAb 12B2 (epitope 89–93) missing the capture of G92, S97, and W99 N-terminal species that are highly expressed in this sCJD subtype (21). Therefore, using an extensive panel of anti-PrP mAbs and a high sensitive separation technique, we were unable to confirm the presence of anchorless PrP species, as suggested by Notari et al. using a conventional SDS-PAGE technique and an anti-PrP rabbit antiserum (16).
GSS-like Internal PrPSc Fragments Are Generated in “Anchorless PrP” Tg Mice, but Not in sCJD and vCJD, following PK Digestion under Inactivating Conditions
We next investigated the influence of GPI anchor in PrPSc conformation under conditions where PMSF was omitted. Using this protocol, PK-digested BHs from anchorless PrP Tg mice, showed, in addition to PrP27–30, bands migrating at 14 and 8 kDa, both recognized by mAbs binding to PrP residues 89–109, including 12B2, 6D11, and ICSM-35 (Fig. 5A). Conversely, mAb 6H4 faintly stained the 14-kDa band, and 4G11 detected barely visible bands in a higher molecular zone, corresponding to previously identified C-terminal fragments (Fig. 5A). At variance, the 14 and 8-kDa bands were not observed in anchored PrP C57BL mice. Interestingly, omission of PMSF induced a reduction of large PrP aggregates, migrating in a high zone, suggesting that these aggregates originate the PrP27–30 in wild type mice and internal fragments in “anchorless PrP” mice, either at 1 or 4 h of PK digestion (Fig. 5B). Under the same experimental conditions, no truncated fragments migrating in the 14–8 kDa range were observed in brain homogenates from different sCJD subtypes (Fig. 5C).
FIGURE 5.
PK digestion under inactivating conditions generates GSS-like internal PrPSc fragments in “anchorless PrP” Tg mice, but not in sCJD and vCJD. A, immunoblot analysis of wild type (odd numbers) and “anchorless PrP” Tg mice (even numbers) BHs, before (−) and after (+) PK treatment. In “anchorless PrP mice” PK treatment under inactivating conditions, generates two bands migrating at 14- and 8-kDa, recognized by 12B2, 6D11, and ICSM-35 (lanes 4, 8, 12, arrows), but not 4G11; on the contrary, 6H4 decorates a faint 14-kDa band. B, immunoblots with 6D11 of BHs from “anchored PrP” mice and “anchorless PrP” Tg mice digested with 50 μg/ml PK at different time intervals. At 1 and 4 h, PK digestion, under inactivating conditions, generates PrP fragments of 14- and 8-kDa in “anchorless PrP” Tg mice but not in “anchored PrP” mice (arrows), paralleled by a decrease of high molecular mass PrP aggregates (brackets). C, immunoblots with 3F4 and 6D11 of sCJD MM1 and MV2 BHs digested with 50 μg/ml PK for 1 and 4 h under standard and inactivating conditions, show the absence of internal PrP fragments.
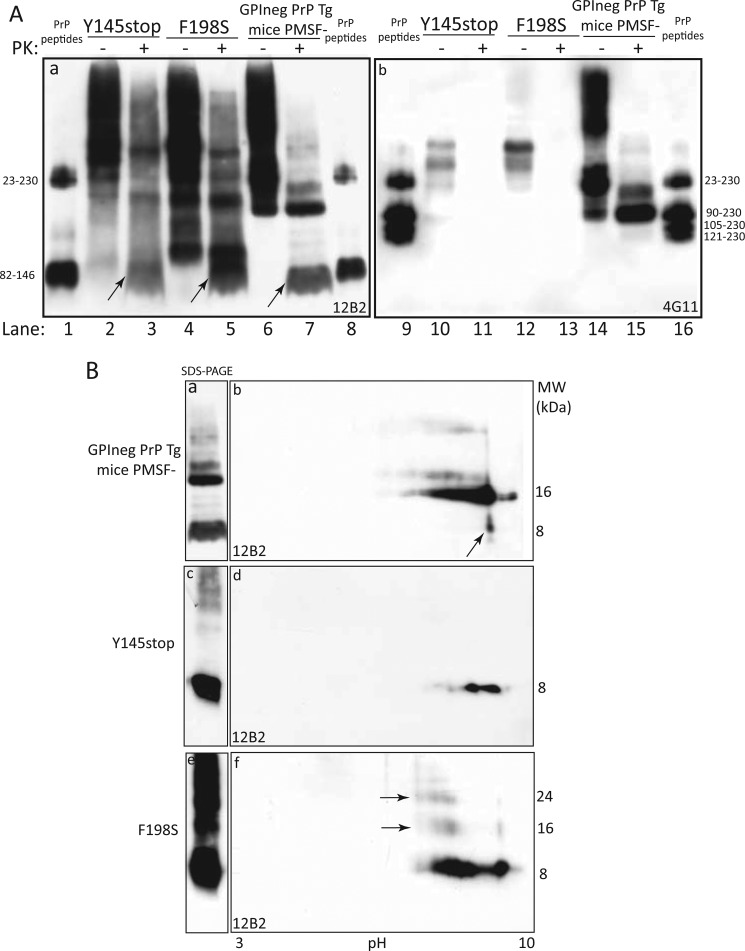
In human prion disorders, the detection of N- and C-terminally PrPSc truncated fragments is a peculiarity of GSS, and, therefore, we compared the electrophoretic migration of PrP fragments detected in “anchorless PrP” Tg mice with brain samples from PrP-CAA Y145Stop and GSS F198S mutations. Under inactivating conditions, immunoblots with 12B2 of PK-digested BHs from Y145Stop, showed the presence of major bands, accounting for the monomeric 8-kDa fragment and multimeric forms thereof, whereas in F198S an additional band of 11 kDa was observed (Fig. 6A, panel a). Intriguingly, the 8-kDa band co-migrated with the faster anchorless PrP internal fragment detected in “anchorless PrP” Tg mice as well as with the synthetic peptide spanning PrP sequence 82–146. As expected, 4G11 failed to recognize internal PrP fragments (Fig. 6A, panel b). Taken together, generation of the internal PrP fragment is a feature of anchorless prions, but not of anchored prions.
FIGURE 6.
The 8-kDa internal fragment detected in “anchorless PrP” transgenic mice shares its electrophoretic migration with the internal fragments generated by PK digestion in Y145Stop and F198S mutations. A, comparative analysis of PrP migration in GSS Y145Stop (lanes 2 and 3), F198S (lanes 4 and 5), and “anchorless PrP” Tg mice (lanes 6 and 7), after PK treatment under standard (−) and inactivating (+) conditions (panel a); immunoblot with 3F4 shows that the 8-kDa PrP internal fragment in PrP-CAA, GSS and “anchorless PrP” mice co-migrates with the synthetic 82–146 PrP peptide (odd lanes, arrows); conversely, immunoblot with 6G11 failed to detect internal PrP fragments (panel b). B, 1D- and 2D-PAGE analyses of PK-treated BHs from “anchorless PrP” Tg mice, show that the 8-kDa band, generated in Tg mice under inactivating conditions, migrates as a single basic spot (panels a and b, arrow); the detection of multiple 8-kDa spots in Y145stop (panels c and d) and F198S BHs (panels e and f) is indicative of N- and C-terminal ragged ends within PrP residues 146 to 153. Noteworthy, the ladder pattern of Y145Stop and F198S under SDS-PAGE separation (panels c and e), is simplified under 2D-PAGE, because of the solubilization of 8-kDa aggregates by 8 m urea.
Additional evidence that the 8-kDa PrP fragment detected in “anchorless PrP” Tg mice shared its electrophoretic properties with the 82–146 synthetic peptide and with the internal fragment generated by PK digestion of Y145stop and F189S mutations, was provided by results obtained following 2D-PAGE separation (Fig. 6B, panels a–f). However, whereas the 8-kDa fragment migrated as a single spot in “anchorless PrP” Tg mice, in Y145Stop and F198S mutations the internal PK-resistant fragments separated as multiple spots, within an 8.4–9.6 pI zone, as an effect of ragged N and C termini.
DISCUSSION
Over the last few years, the identification and characterization of GPI-anchored and GPI-anchorless forms of PrPSc in human and animal prion diseases has been a matter of extensive investigations, aimed at addressing how the diverging physical properties of prions may influence propagation, pathogenesis, and neuropathology of prion disorders. In vitro studies have previously shown that anchorless PrP undergoes an aberrant metabolism, which includes defective glycosylation, altered transport to the secretory pathway, and lack of recycling through to the plasma membrane (13, 14). These features are in keeping with the present evidence that endogenous proteolysis of brain tissues from RML-infected “anchorless PrP” Tg mice generate minimal amounts of the C2 fragment (mostly derived from PrPSc) and almost undetectable quantities of the C1 fragment (generated by α-cleavage of PrPC). This is at variance with “anchored PrP” C57BL mice infected with RML that showed well-detectable levels of both fragments. In addition to different levels of PrP species recovered in untreated brain homogenates, under exogenous proteolytic treatment, “anchorless PrP” Tg mice lacked C-terminal truncated fragments, indicating that their generation occurs as a result of an aberrant cleavage of anchored PrPSc. Further differences between “anchorless PrP” Tg mice and “anchored PrP” C57BL mice were observed after PK digestion under inactivating conditions, due to the pruning of anchorless prions, but not anchored prions, at the N and C termini, as a likely effect of conformational changes exposing the anchorless C terminus to proteolytic cleavage. Although the relative amount of internal PrPSc fragments was small, these results suggest that PrP species lacking the amino and carboxyl termini represent a molecular fingerprint of anchorless prions. Intriguingly, a 10-kDa internal fragment has been detected in brain tissues of uninfected Tg mice overexpressing anchorless PrP, and in Tg mice co-expressing anchored and anchorless PrP (12). Noteworthy, the aforementioned transgenic mice develop a spontaneous neurologic illness characterized by large PrP amyloid deposits composed by a 10-kDa PK-resistant fragment. Among human prion diseases, anchorless C-terminally truncated PrPSc fragments are a feature of PrP-CAA and GSS stop codon mutations Y145X, Q160X, Y163X, Y226X, and Q227X (22–25). In addition, in a number of GSS cases associated with point mutations, such as F198S, impaired GPI anchoring has been suggested to cause C-terminal truncation of PrPSc (12). PrP-CAA and/or parenchymal amyloid PrPSc deposition, generating a 7–11 kDa PK-resistant PrP internal fragment, characterize the above mutations at neuropathology, in the absence of spongiosis. Taken together, experimental and human conditions characterized by lack of the GPI anchor or altered GPI anchoring, share common pathogenic mechanisms leading to accumulation of an internal PrP fragment under amyloid plaques.
Recently, Notari et al. reported on the presence of anchorless PrPSc in different sCJD subtypes, hence widening the spectrum of molecular PrP quasispecies detected in this human disorder (16). Anchorless PrP forms were tentatively identified based on the immunodecoration of PK-resistant species migrating 2 kDa faster than the unglycosylated PrP27–30, under SDS-PAGE separation. These findings are potentially relevant and raise concern on the potential spreading of anchorless PrP species to body fluids and peripheral tissues of sCJD patients, in view of the propensity of anchorless prions to accumulate in non-neural tissues.
The contribution of the anchor to the molecular mass of PrP has been previously calculated to account for 2–3 kDa and 4 kDa, under experimental conditions using, respectively, hydrofluoric acid and proaerolysin (16, 26). These data are in keeping with our findings showing a 2.5 kDa difference between “anchored PrP” C57BL and “anchorless PrP” Tg mice in the orthogonal migration of either native or PK-resistant PrPSc isoforms. Moreover, our 2D-PAGE immunoblot study discloses a basic shift of 1 pH unit for anchorless species, hence providing an additional parameter for separating anchored and anchorless prions.
Having determined the molecular coordinates of anchorless PrP in Tg mice we focused on the identification of possible anchorless PrPSc forms in sCJD, taking also advantages of a panel of synthetic PrP peptides, as indicators of migration under two-dimensional separation. Based on previous studies, major PK-cleavage sites of PrPSc occur at G78-G82 for type-1 PrPSc, or ∼10 amino acids upstream the synthetic 90–230 PrP peptide migrating at ∼16 kDa, and G92-S97 for type-2 PrPSc, a few amino acids downstream the above peptide (19). Therefore, the observed migration of the PrP27–30 unglycosylated fragment at 19.5 kDa for type-1 PrPSc and 17 for type-2 PrPSc, is in keeping with their expected molecular masses, taken also in due account the 2.5 kDa contribution of the GPI-anchor moieties. Moreover, the horizontal migration of the synthetic 90–230 PrP milestone at pI 7.9, provided an additional watershed for the detection of anchorless PrP species, knowing that the GPI anchor cause an acidic shift of 1 pH unit. Accordingly, the migration of the core fragment at pI 7.0 in sCJD cases with type-1 PrPSc, and in a 6.0–7.0 pI range in MM2 sCJD, rule out the presence of anchorless PrP species in these sCJD subtypes. At variance, in MV2 and VV2 sCJD and in vCJD, the detection of a few spots with a focusing mobility toward basic pI zones, might apparently suggest the presence of anchorless PrP species. However, basic spots did not show changes in their molecular masses, as compared with acidic spots, therefore suggesting that these isoforms may represent molecular PrP species with modified anchor composition. The latter hypothesis is further supported by the immunodetection of basic spots with mAb 3E2 in a pattern similar to mAb 3F4. These results rule out the possibility of PrPSc truncation at the C terminus, which is expected to compromise antibody recognition. Within this context, the finding of Notari et al. that putative anchorless forms were virtually undetectable by 2301 antiserum is not supportive for their presence, being instead a further proof that the extreme C-terminal sequence of PrP molecule is less accessible to mAb recognition following SDS-PAGE separation, unless exposed to highly denaturing conditions (27, 28). This is also the case of our diverging results obtained with mAb 3E2, which efficiently recognized PrP following 2D-PAGE separation, while leaving PrP unstained in SDS-PAGE immunoblots, thus replicating previous results with mAb SP-214 (18).
Overall, our present data do not support recent findings purporting to show the presence of pools of anchored and anchorless PrPSc in sCJD with type-1 and type-2A PrPSc. Regarding the detection of anchorless prions in MV2 sCJD subtype, immunoblots with mAb 12B2, which has a high affinity for type-1 PrPSc, clearly showed that the two-dimensional separation of type-1 isoforms at 19.5 kDa and type-2 PrPSc spots at 17 kDa within a 6.0–7.0 pI range, were consistent with an electrophoretic migration highly suggestive for anchored PrP species. These data are in accordance with recent mass-spectrometry-based analysis of anchorless PrP, showing that PK treatment of anchorless PrPSc generates several N-terminal ragged ends, leaving unaffected the C terminus (29). Finally, the lack of generation of N- and C-terminally truncated PrPSc fragments in BHs from all sCJD subtypes and vCJD, after PK digestion under inactivating conditions, is an additional argument against the presence of anchorless prions in these human disorders.
It should be noted that the PrP region encompassing residues 89–140 is an integral part of the minimal sequence able to sustain prion replication in cell culture and transgenic mice, and that it plays a dual role in the PrPC to PrPSc conversion and in propagating strain properties (30). Although this study provides additional evidence that the absence of GPI anchor might favor the formation GSS-associated PrPSc internal fragments, the exact mechanism responsible for generation of amyloidogenic PrP quasispecies remains still unknown.
Acknowledgments
We thank Dr. Bruce Chesebro, Laboratory of Persistent Viral Diseases, Rocky Mountain Laboratories, National Institute of Allergy and Infectious Diseases, Hamilton, MT 59840 for providing scrapie-infected “anchorless PrP” transgenic and wild type mice, and for critical reading of the manuscript.
This work was supported in part by RF2009-1474758 (to G. Z.) and by “Disabilità cognitiva e comportamentale nelle demenze e nelle psicosi” from Fondazione Cariverona (to S. M.).
- TSEs
- transmissible spongiform encephalopathies
- BH
- brain homogenates
- sCJD
- sporadic Creutzfedlt-Jakob disease
- GSS
- Gerstmann-Sträussler-Scheinker syndrome
- PRNP
- human prion protein gene
- PrPC
- cellular prion protein
- PrPTSE
- disease-associated PrP
- GPI
- glycosylphosphatidylinositol moiety
- GPI-Tg mice
- GPI negative transgenic mice
- PK
- proteinase- K.
REFERENCES
- 1. Prusiner S. B. (1998) Prions. Proc. Natl. Acad. Sci. U.S.A. 95, 13363–13383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Caughey B., Baron G. S., Chesebro B., Jeffrey M. (2009) Getting a grip on prions: oligomers, amyloids, and pathological membrane interactions. Annu. Rev. Biochem. 78, 177–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Stahl N., Baldwin M. A., Burlingame A. L., Prusiner S. B. (1990) Identification of glycoinositol phospholipid linked and truncated forms of the scrapie prion protein. Biochemistry 29, 8879–8884 [DOI] [PubMed] [Google Scholar]
- 4. Stahl N., Borchelt D. R., Prusiner S. B. (1990) Differential release of cellular and scrapie prion proteins from cellular membranes by phosphatidylinositol-specific phospholipase C. Biochemistry 29, 5405–5412 [DOI] [PubMed] [Google Scholar]
- 5. Chen S. G., Teplow D. B., Parchi P., Teller J. K., Gambetti P., Autilio-Gambetti L. (1995) Truncated forms of the human prion protein in normal brain and in prion diseases. J. Biol. Chem. 270, 19173–19180 [DOI] [PubMed] [Google Scholar]
- 6. Notari S., Qing L., Pocchiari M., Dagdanova A., Hatcher K., Dogterom A., Groisman J. F., Lumholtz I. B., Puopolo M., Lasmezas C., Chen S. G., Kong Q., Gambetti P. (2012) Assessing prion infectivity of human urine in sporadic Creutzfeldt-Jakob disease. Emerg. Infect. Dis. 18, 21–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Castagna A., Campostrini N., Farinazzo A., Zanusso G., Monaco S., Righetti P. G. (2002) Comparative two-dimensional mapping of prion protein isoforms in human cerebrospinal fluid and central nervous system. Electrophoresis 23, 339–346 [DOI] [PubMed] [Google Scholar]
- 8. Dagdanova A., Ilchenko S., Notari S., Yang Q., Obrenovich M. E., Hatcher K., McAnulty P., Huang L., Zou W., Kong Q., Gambetti P., Chen S. G. (2010) Characterization of the prion protein in human urine. J. Biol. Chem. 285, 30489–30495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Stahl N., Borchelt D. R., Hsiao K., Prusiner S. B. (1987) Scrapie prion protein contains a phosphatidylinositol glycolipid. Cell 51, 229–240 [DOI] [PubMed] [Google Scholar]
- 10. Nishina K. A., Supattapone S. (2007) Immunodetection of glycophosphatidylinositol-anchored proteins following treatment with phospholipase C. Anal. Biochem. 363, 318–320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Campana V., Caputo A., Sarnataro D., Paladino S., Tivodar S., Zurzolo C. (2007) Characterization of the properties and trafficking of an anchorless form of the prion protein. J. Biol. Chem. 282, 22747–22756 [DOI] [PubMed] [Google Scholar]
- 12. Stöhr J., Watts J. C., Legname G., Oehler A., Lemus A., Nguyen H. O., Sussman J., Wille H., DeArmond S. J., Prusiner S. B., Giles K. (2011) Spontaneous generation of anchorless prions in transgenic mice. Proc. Natl. Acad. Sci. U.S.A. 108, 21223–21228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chesebro B., Trifilo M., Race R., Meade-White K., Teng C., LaCasse R., Raymond L., Favara C., Baron G., Priola S., Caughey B., Masliah E., Oldstone M. (2005) Anchorless prion protein results in infectious amyloid disease without clinical scrapie. Science 308, 1435–1439 [DOI] [PubMed] [Google Scholar]
- 14. Horiuchi M., Caughey B. (1999) Specific binding of normal prion protein to the scrapie form via a localized domain initiates its conversion to the protease-resistant state. EMBO J. 18, 3193–3203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Johnson R. T. (2005) Prion diseases. The Lancet Neurology 4, 635–642 [DOI] [PubMed] [Google Scholar]
- 16. Notari S., Strammiello R., Capellari S., Giese A., Cescatti M., Grassi J., Ghetti B., Langeveld J. P., Zou W. Q., Gambetti P., Kretzschmar H. A., Parchi P. (2008) Characterization of truncated forms of abnormal prion protein in Creutzfeldt-Jakob disease. J. Biol. Chem. 283, 30557–30565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zerr I., Kallenberg K., Summers D. M., Romero C., Taratuto A., Heinemann U., Breithaupt M., Varges D., Meissner B., Ladogana A., Schuur M., Haik S., Collins S. J., Jansen G. H., Stokin G. B., Pimentel J., Hewer E., Collie D., Smith P., Roberts H., Brandel J. P., van Duijn C., Pocchiari M., Begue C., Cras P., Will R. G., Sanchez-Juan P. (2009) Updated clinical diagnostic criteria for sporadic Creutzfeldt-Jakob disease. Brain 132, 2659–2668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zanusso G., Farinazzo A., Prelli F., Fiorini M., Gelati M., Ferrari S., Righetti P. G., Rizzuto N., Frangione B., Monaco S. (2004) Identification of distinct N-terminal truncated forms of prion protein in different Creutzfeldt-Jakob disease subtypes. J. Biol. Chem. 279, 38936–38942 [DOI] [PubMed] [Google Scholar]
- 19. Zanusso G., Righetti P. G., Ferrari S., Terrin L., Farinazzo A., Cardone F., Pocchiari M., Rizzuto N., Monaco S. (2002) Two-dimensional mapping of three phenotype-associated isoforms of the prion protein in sporadic Creutzfeldt-Jakob disease. Electrophoresis 23, 347–355 [DOI] [PubMed] [Google Scholar]
- 20. Parchi P., Castellani R., Capellari S., Ghetti B., Young K., Chen S. G., Farlow M., Dickson D. W., Sima A. A., Trojanowski J. Q., Petersen R. B., Gambetti P. (1996) Molecular basis of phenotypic variability in sporadic Creutzfeldt-Jakob disease. Ann. Neurol. 39, 767–778 [DOI] [PubMed] [Google Scholar]
- 21. Parchi P., Zou W., Wang W., Brown P., Capellari S., Ghetti B., Kopp N., Schulz-Schaeffer W. J., Kretzschmar H. A., Head M. W., Ironside J. W., Gambetti P., Chen S. G. (2000) Genetic influence on the structural variations of the abnormal prion protein. Proc. Natl. Acad. Sci. U.S.A. 97, 10168–10172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ghetti B., Piccardo P., Spillantini M. G., Ichimiya Y., Porro M., Perini F., Kitamoto T., Tateishi J., Seiler C., Frangione B., Bugiani O., Giaccone G., Prelli F., Goedert M., Dlouhy S. R., Tagliavini F. (1996) Vascular variant of prion protein cerebral amyloidosis with tau-positive neurofibrillary tangles: the phenotype of the stop codon 145 mutation in PRNP. Proc. Natl. Acad. Sci. U.S.A. 93, 744–748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Jansen C., Parchi P., Capellari S., Vermeij A. J., Corrado P., Baas F., Strammiello R., van Gool W. A., van Swieten J. C., Rozemuller A. J. (2010) Prion protein amyloidosis with divergent phenotype associated with two novel nonsense mutations in PRNP. Acta Neuropathol. 119, 189–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ghetti B., Dlouhy S. R., Giaccone G., Bugiani O., Frangione B., Farlow M. R., Tagliavini F. (1995) Gerstmann-Sträussler-Scheinker disease and the Indiana kindred. Brain Pathol. 5, 61–75 [DOI] [PubMed] [Google Scholar]
- 25. Jayadev S., Nochlin D., Poorkaj P., Steinbart E. J., Mastrianni J. A., Montine T. J., Ghetti B., Schellenberg G. D., Bird T. D., Leverenz J. B. (2011) Familial prion disease with Alzheimer disease-like tau pathology and clinical phenotype. Ann. Neurol. 69, 712–720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zou W. Q., Capellari S., Parchi P., Sy M. S., Gambetti P., Chen S. G. (2003) Identification of novel proteinase K-resistant C-terminal fragments of PrP in Creutzfeldt-Jakob disease. J. Biol. Chem. 278, 40429–40436 [DOI] [PubMed] [Google Scholar]
- 27. Williamson R. A., Peretz D., Pinilla C., Ball H., Bastidas R. B., Rozenshteyn R., Houghten R. A., Prusiner S. B., Burton D. R. (1998) Mapping the prion protein using recombinant antibodies. J. Virol. 72, 9413–9418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kim C. L., Umetani A., Matsui T., Ishiguro N., Shinagawa M., Horiuchi M. (2004) Antigenic characterization of an abnormal isoform of prion protein using a new diverse panel of monoclonal antibodies. Virology 320, 40–51 [DOI] [PubMed] [Google Scholar]
- 29. Vázquez-Fernández E., Alonso J., Pastrana M. A., Ramos A., Stitz L., Vidal E., Dynin I., Petsch B., Silva C. J., Requena J. R. (2012) Structural organization of mammalian prions as probed by limited proteolysis. PLoS One 7, e50111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Helmus J. J., Surewicz K., Nadaud P. S., Surewicz W. K., Jaroniec C. P. (2008) Molecular conformation and dynamics of the Y145Stop variant of human prion protein in amyloid fibrils. Proc. Natl. Acad. Sci. U.S.A. 105, 6284–6289 [DOI] [PMC free article] [PubMed] [Google Scholar]