Abstract
Purpose of review
The aim of this review is to describe disease mechanisms by which chromosome 9 open reading frame 72 (C9ORF72) repeat expansions could lead to amyotrophic lateral sclerosis (ALS) and frontotemporal dementia (FTD), and to discuss these diseases in relation to other non-coding repeat expansion disorders.
Recent findings
ALS and FTD are complex neurodegenerative disorders with a considerable clinical and pathological overlap, and this overlap is further substantiated by the recent discovery of C9ORF72 repeat expansions. These repeat expansions are currently the most important genetic cause of familial ALS and FTD, accounting for approximately 34.2% and 25.9% of the cases. Clinical phenotypes associated with these repeat expansions are highly variable, and combinations with mutations in other ALS- and/or FTD-associated genes may contribute to this pleiotropy. It is challenging, however, to diagnose patients with C9ORF72 expansions, not only because of large repeat sizes, but also due to somatic heterogeneity. Most other non-coding repeat expansion disorders share an RNA gain-of-function disease mechanism, a mechanism that could underlie the development of ALS and/or FTD as well.
Summary
The discovery of C9ORF72 repeat expansions provides novel insights into the pathogenesis of ALS and FTD, and highlights the importance of non-coding repeat expansions and RNA toxicity in neurodegenerative diseases.
Keywords: Amyotrophic lateral sclerosis, frontotemporal dementia, non-coding repeat expansion disorders, C9ORF72, genetics
Introduction
Amyotrophic lateral sclerosis (ALS) and frontotemporal dementia (FTD) are fatal neurodegenerative disorders for which no effective treatments are available. ALS is the most frequent motor neuron disease, resulting in progressive weakness and death from respiratory failure, typically within three years of symptom onset [1*]. FTD is the second most common cause of early-onset dementia, and is characterized by behavior and personality changes and/or language dysfunction, due to degeneration of the frontal and temporal cortex [2–5]. Most FTD patients die 5–10 years after symptom onset.
Clinicopathological studies have long supported the concept that ALS and FTD may represent a disease continuum with a shared underlying pathogenesis [6*,7*,8]. They often co-occur in a family, and the prevalence of frontal lobe impairment in ALS populations may approach 50% [9–11]. Similarly, as many as half of FTD patients develop clinical symptoms of motor neuron dysfunction [12]. Especially important was the identification of the transactive response DNA-binding protein 43 (TDP-43) in 2006, as the major inclusion protein in the vast majority of ALS patients and in the most common pathological subtype of FTD, now referred to as frontotemporal lobar degeneration with TDP-43 pathology (FTLD-TDP) [13,14].
Both diseases are etiologically complex with genetic and presumably environmental factors contributing to its onset [15**,16]. A positive family history has been reported in ~10% of ALS patients, and up to 50% of FTD patients [2,17]. Nonetheless, no genes were identified that sufficiently explained the growing class of families in which affected members developed either ALS or FTD or both (ALS-FTD). Last year, however, two independent studies identified hexanucleotide repeat expansions in the chromosome 9 open reading frame 72 gene (C9ORF72) [18**,19**]. This important discovery raised new hope with clinicians and researchers for the development of treatments, and provided novel avenues for studying and understanding the disease pathogenesis of ALS and FTD.
In this review, we underscore the importance of the C9ORF72 mutations in ALS and FTD, and review current hypotheses related to its disease mechanism(s) and associated phenotypes. We also discuss this novel mutation in relation to other repeat expansion disorders, especially myotonic dystrophy type 1 (DM1).
Identification of repeat expansions in C9ORF72 and possible disease mechanism(s)
Previously, genetic studies convincingly linked ALS and FTD to a region on chromosome 9p21 [20–24,25*,26,27]. Although these studies were able to minimize the region, the genetic defect remained elusive until last year, when two independent groups described a GGGGCC hexanucleotide repeat in a non-coding region of C9ORF72 [18**,19**]. One of these groups focused on family VSM-20, a large ALS-FTD family, and used primers flanking the repeat region to amplify the region and to determine the size of the repeat in affected and unaffected family members [18**]. Intriguingly, their results appeared to indicate that all affected individuals were homozygous for the repeat, while none of their affected children seemed to have inherited their alleles. Their findings underlined the need for an alternative methodology, since a pathogenic repeat expansion may not be amplified by a conventional PCR, and therefore, they developed a repeat-primed PCR assay, resulting in the identification of the pathogenic repeat expansion. The other group employed next-generation sequencing to study a Welsh ALS-FTD family [19**]. Their efforts revealed a drop off in the sequence coverage in the region of the repeat, emphasizing the polymorphic nature of this region, which eventually led to the detection of the pathogenic repeat expansion.
The GGGGCC hexanucleotide repeat is located between two five prime non-coding exons of C9ORF72, which encodes a completely uncharacterized protein with unknown function. Two different isoforms of the protein are predicted to be generated from a total of three or more different C9ORF72 transcripts. Unexpectedly, several groups showed reduced levels of at least one C9ORF72 transcript in expanded-repeat carriers, suggesting a possible loss-of-function disease mechanism [18**,19**,28*]. The accumulation of transcripts containing the GGGGCC repeat as nuclear RNA foci in the frontal cortex and spinal cord of C9ORF72 mutation carriers, however, has also been demonstrated, favoring a toxic RNA gain-of-function disease mechanism in line with most other non-coding expansion disorders [18**].
Frequency of C9ORF72 repeat expansions
Until the discovery of the pathogenic repeat expansion in C9ORF72 only 20–30% of the familial ALS cases could be explained by mutations in the superoxide dismutase-1 gene (SOD1), and the genes encoding TDP-43 (TARDBP) and fused in sarcoma (FUS), while mutations in the microtubule-associated protein tau gene (MAPT), progranulin gene (GRN), and, less commonly, the valosin containing protein gene (VCP) and the charged multivesicular body protein 2b gene (CHMP2B) were responsible for 20–30% of the FTD cases [15**,29**,30*,31*]. Mutations in these genes were also present in 1–5% of the sporadic ALS and FTD cases.
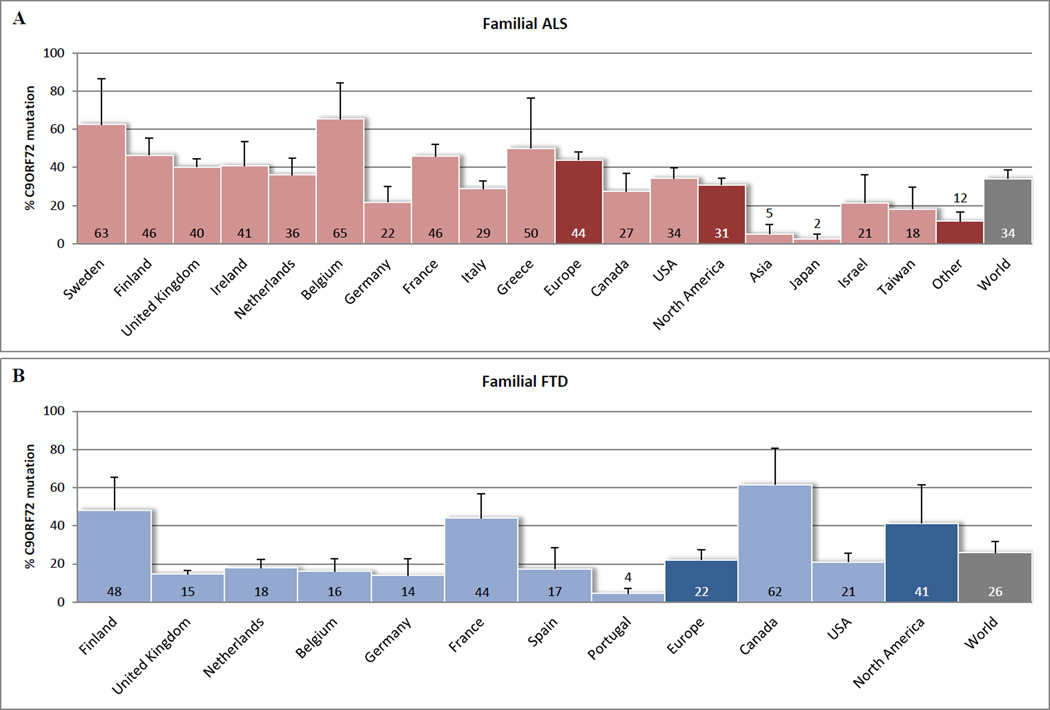
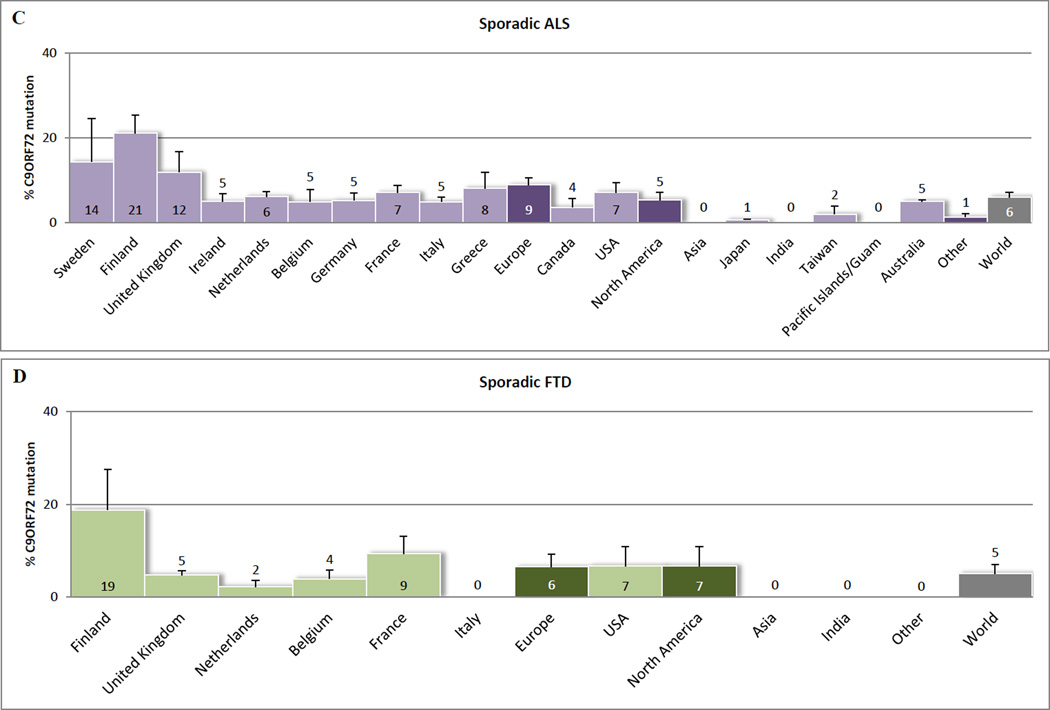
To date, less than a year after the discovery, more than thirty articles have described frequencies of C9ORF72 repeat expansions in ALS and FTD populations: from the United States of America to Europe, Australia and Asia [18**,19**,28*,29**,32**–41*,42**–46*,47**–49*,50**–52*,53**–56*,57**]. In Figure 1, we have provided a graphic representation of these frequencies. Even though heterogeneity between populations and study design hampers this comparison (Supplementary Table 1) our representation suggests that C9ORF72 mutations account for 34.2% (standard error [SE] 4.5) of the familial ALS cases, 5.9% (SE 1.3) of the sporadic ALS cases, 25.9% (SE 5.9) of the familial FTD cases, 5.1% (SE 2.0) of the sporadic FTD cases, and 0.17% (SE 0.07) of the control subjects. Hence, these frequencies underline that C9ORF72 repeat expansions are currently the major genetic cause of ALS and/or FTD worldwide.
Figure 1.
C9ORF72 mutation frequencies worldwide. Percentages reported in (A) familial ALS patients, (B) familial FTD patients, (C) sporadic ALS patients, and (D) sporadic FTD patients. Cohorts of less than ten subjects were not included. Only reported subjects with more than 29 repeat expansions were considered to have a C9ORF72 mutation. In general, error bars represent 95% confidence intervals, as calculated with the Wald method. If multiple studies were conducted for one country, then error bars represent standard errors. For these countries the average of all conducted studies was calculated, applying equal weight to all of them. The same method was used to calculate an average for all reported countries in Europe, countries in North America, other countries (Other), and an overall average (World). Graphpad Prism version 5.04 (http://www.graphpad.com) was used to perform these analyses. When mutation frequencies of other genes were reported, they were incorporated, to give a better impression of the actual mutation percentage in the general ALS/FTD population. If authors stated that the same (sub)group was used in multiple studies, then this (sub)group was only included in one of the studies. Majounie et al. [47**], Chio et al. [49*], and Renton et al. [19**], however, did include the same subset of 29 familial ALS samples without specification, and therefore, this relatively small amount of samples could not be excluded. More details about the studies included in our comparison can be found in Supplementary Table 1.
Oligogenic etiology
Interestingly, more than twenty patients have been described that harbor mutations in C9ORF72 in combination with mutations in other ALS- and/or FTD-associated genes [29**,33*,36*,42**,43*,51*,58*,59*]. In these patients, mutations were also detected in SOD1, TARDBP, FUS, angiogenin (ANG), optineurin (OPTN), ubiquilin-2 (UBQLN2), vesicle-associated membrane protein B (VAPB), D-amino-acid oxidase (DAO), peripherin (PRPH), GRN and presenilin-2 (PSEN2). It is important to note, however, that more than half of these additional mutations have also been reported in control subjects, and that their effects, as predicted by in silico programs, remain unclear (Table 1). Although it could therefore be argued that they merely represent benign polymorphisms, a recent ALS study has demonstrated that the frequency of multiple mutations is higher than expected on the basis of chance [29**]. This could indicate that these mutations act as disease modifiers, which could contribute to the pleiotropy that is encountered in patients with C9ORF72 mutations.
Table 1.
Patients reported with C9ORF72 repeat expansions in combination with mutations in other genes.
| Gene 1 | Gene 2 | Mutation 2 | ALS | FTD | Gender | Age at Onset (y) |
Site of Onset |
Duration (m) |
Reported in controls |
Prediction PolyPhen-2 |
Prediction PMut | Additional information |
|
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Ratti 2012 [33*] | C9ORF72 | TARDBP | p.A382T | FALS | N/A | M | 47 | Spinal | N/A | Yes | Benign | Neutral | [60] |
| C9ORF72 | PRPH | p.R133P | SALS | N/A | M | 70 | Spinal | 24 | No | Probably damaging | Pathological | [61*] | |
| Lattante 2012 [36*] | C9ORF72 | ANG | p.I46V | SALS | N/A | F | 38 | Spinal | > 38 | Yes | Benign | Neutral | [36*] |
| Van Blitterswijk 2012 [29**] | C9ORF72 | TARDBP | p.N352S | FALS | No | M | 42 | Cervical | > 91 | No | Benign | Pathological | [29**][62*] |
| C9ORF72 | TARDBP | p.N352S | FALS | No | F | 47 | Cervical | > 15 | No | Benign | Pathological | [29**][62*] | |
| C9ORF72 | SOD1 | p.D90A | FALS | No | F | 51 | Cervical | 77 | Yes | Benign | Pathological | [29**][62*] | |
| C9ORF72 | FUS | p.Q210H | FALS | No | M | 58 | Lumbosacral | 25 | Yes | Probably damaging * | Neutral | [29**][62*] | |
| Van Blitterswijk 2012b [58*] | C9ORF72 | VAPB | p.V234I | FALS | No | F | 65 | Lumbosacral | 34 | No | Benign * | Neutral | [58*][62*] |
| Millecamps 2012 [42**] | C9ORF72 | ANG | p.K41I (p.K17I) | FALS | No | F | 47 | Lumbosacral | 27 | Yes | Benign | Pathological | [63] |
| C9ORF72 | DAO | p.R38H | FALS | N/A | F | 42 | Lumbosacral | 7 | Yes | Benign | Pathological | [64] | |
| C9ORF72 | OPTN | p.D128EfsX22 | FALS | N/A | M | 46 | N/A | 21 | Yes | N/A | N/A | [65*] | |
| C9ORF72 | UBQLN2 | p.G502_I504del | FALS | No | F | 52 | Bulbar | 27 | Yes | N/A | N/A | [66*] | |
| C9ORF72 | SOD1 | p.D110Y | FALS | N/A | N/A | 59 | Cervical | 42 | No | Benign | Neutral | [63] | |
| C9ORF72 | FUS | p.R521C | FALS | N/A | N/A | 40 | Bulbar | 14 | No | Benign | Pathological | [63] | |
| Ferrari 2012 [43*] | C9ORF72 | GRN | p.Y294C | No | FTD, familial | M | 53 | Behavioral-variant | > 96 | No | Probably damaging | Pathological | [43*] |
| C9ORF72 | PSEN2 | p.I146V | No | FTD, familial | M | 59 | Behavioral-variant | > 108 | No | Benign | Neutral | [43*] | |
| Cooper-Knock 2012 [51*] | C9ORF72 | TARDBP | p.A321V | FALS | No | F | 37 | Cervical | 58 | No | Benign | Pathological | [67] |
| C9ORF72 | FUS | p.G174del | FALS | No | F | 62 | Bulbar | 24 | Yes | N/A | N/A | [68] | |
| C9ORF72 | OPTN | p.E322K | FALS | No | F | 50 | Bulbar | 29 | Yes | N/A | N/A | [51*] | |
| Chio 2012b [59*] | C9ORF72 | TARDBP | p.A382T | FALS | FTD | M | 43 | Bulbar | 34 | Yes | Benign | Neutral | [59*] |
| C9ORF72 | TARDBP | p.A382T | FALS | No | M | 35 | Cervical | > 42 | Yes | Benign | Neutral | [59*] | |
| Total | 90% familial | 21% FTD | 47% M | 50 y | 25% bulbar | 42 m | 57% yes |
Abbreviations: ALS = amyotrophic lateral sclerosis, FALS = familial ALS, SALS = sporadic ALS, FTD = frontotemporal dementia, N/A = not available, M = male, F = female, y = years, and m = months.
Predictions were performed on 07-18-2012 by PolyPhen-2 version 2.2 (http://genetics.bwh.harvard.edu/pph2/) and PMut (http://mmb2.pcb.ub.es:8080/PMut/).
Multiple sequence alignments used by the previous version of PolyPhen-2 resulted in ‘unknown’ for p.Q210H in FUS and ‘possibly damaging’ for p.V234I in VAPB.
Clinicopathological phenotypes associated with C9ORF72 repeat expansions
Clinical data of patients with C9ORF72 mutations demonstrates that approximately 55.8% (SE 2.4) is male; the mean age at onset is 56.1 years (SE 0.9), and the mean disease duration is 49.9 months (SE 4.9) (Table 2 and Supplementary Table 2) [18**,28*,36*–38*,42**,44*–47**,49*,50**–52*,53**–56*,57**,69*,70*]. Importantly, age at onset and disease duration are highly variable, even within a single family. Based on the current literature, the age at onset ranges from 27 to 83 years [46*,47**,56*], and the disease duration varies between 3 and 264 months [47**,52*,54*]. Nevertheless, there seems to be a tendency towards a younger age at onset and shorter disease duration in patients with C9ORF72 repeat expansions, as compared to patients without them [28*,45*,46*,49*,51*,56*].
Table 2.
Clinical characteristics of ALS and FTD patients with C9ORF72 mutations.
| Cohort | % Male (SE) | % Bulbar (SE) | Mean Age at Onset (SE) | Median Age at Onset (SE) | Mean Duration (SE) | Median Duration (SE) | % Dementia (SE) | % Behavioral (SE) | % FTD-MND (SE) |
|---|---|---|---|---|---|---|---|---|---|
| ALSa | 55.31 (3.21) | 34.58 (2.55) | 56.28 (1.20) | 56.50 (0.93) | 35.43 (1.80) | 31.40 (2.56) | 26.95 (4.94) | ||
| FTDb | 56.91 (3.29) | 85.08 (6.12) | 55.75 (0.80) | 54.10 (2.10) | 75.33 (6.35) | 80.95 (5.08) | 30.57 (3.43) | ||
| Total | 55.84 (2.37) | 44.20 (4.99) | 56.13 (0.88) | 55.90 (0.88) | 49.94 (4.87) |
Abbreviation: SE = standard error.
For ALS and FTD cohorts the average of studies reporting clinical characteristics was calculated, applying equal weight to all of them. Cohorts of less than five subjects were not included. Only subjects with more than 29 repeat expansions were considered to have a C9ORF72 mutation.
Approximately 29.3% (SE 3.3) of the patients with C9ORF72 mutations displays symptoms of both ALS and FTD (Table 2). Of the patients with ALS, 44.2% (SE 5.0) presents with a bulbar onset of symptoms, which is higher than the expected frequency of 19–30% [72]. In 81.0% (SE 5.0) of the FTD patients with C9ORF72 mutations, the behavioral variant is detected, while the expected frequency is ~50% [31*].
Additional symptoms have also been described in patients with C9ORF72 mutations, including signs of parkinsonism and psychotic phenomena [46*,50**,51*,54*,55*,69*,73,74*–77*]. Furthermore, in patients with clinical diagnoses of Alzheimer’s disease (AD), Parkinson disease (PD), corticobasal syndrome (CBS), and olivopontocerebellar degeneration (OPCD) C9ORF72 mutations have been detected as well, but they appear to be rare, and may be due to clinical misdiagnoses (<3%) [75*,78*–80*,81]. All these findings highlight the substantial clinical heterogeneity that is detected in patients with C9ORF72 mutations, both between and in families [82**].
Apart from clinical studies, neuropathological investigations have shown that repeat expansions in C9ORF72 are characterized by TDP-43 pathology in various neuroanatomical regions, and ubiquitin-positive but TDP-43-negative neuronal cytoplasmic inclusions in the cerebellar granular layer, hippocampal pyramidal neurons and other neuroanatomical sites, which are unique to C9ORF72 repeat expansions carriers [18**,28*,45*,50**,51*,52*,54*–57**,69*,70*,83*]. Several research groups are now focusing on identifying the nature of the ubiquitinated protein in these TDP-43 negative inclusions, as it may shed light on the disease mechanism(s) associated with C9ORF72 expansions.
Critical issues associated with size and sequence composition of C9ORF72 repeats
In the general population, the vast majority of the C9ORF72 alleles contain two to thirty GGGGCC hexanuceotide repeats [18**,19**]. Affected individuals with C9ORF72 mutations harbor one normal allele and one expanded allele with hundreds to thousands of these repeats [18**,19**]. Consequently, a cut-off of thirty repeats is commonly used to differentiate between pathogenic and non-pathogenic repeat sizes [19**]. It is important to realize, however, that repeat sizes of thirty or more are also present in approximately 0.17% of the control subjects (Supplementary Table 1) [18**,19**,28*,29**,33*,35*,38*,42**–44*,46*,47**,49*,51*,53**,54*,57**], and therefore, a cut-off of thirty repeats should be used with caution.
A second problem relates to the repeat-primed PCR method that is commonly used to screen ALS and FTD patients. Even though this method is fast and cost effective, it does not provide an accurate estimate of the number of repeats. Therefore, Southern blot analysis should be used to estimate the number of repeats in expanded repeat carriers, however, its application appears to be challenging, and currently only a handful of samples has been tested [18**,39*]. Moreover, neither repeat-primed PCR methods nor Southern blot analysis are able to reveal the actual DNA composition of the expanded repeat, and thus, new protocols and methods are needed to guarantee more reliable diagnostic testing.
Finally, somatic heterogeneity is likely to be common in repeat expansion carriers, resulting in varying repeat sizes in different tissues from a single patient. As a result, repeat sizes determined using DNA extracted from whole blood may not adequately reflect the C9ORF72 repeat sizes in a patient’s brain or spinal cord tissue, and may hamper genotype-phenotype correlations.
Can we learn from other repeat expansion disorders?
Pathogenic repeat expansions have already been identified in at least 24 other neurological disorders [84–86**] (Table 3). In coding regions, repeat expansions can cause long stretches of amino acids, for instance, of polyglutamine or polyalanine. These stretches can disrupt the normal function of encoded proteins and result in toxic aggregate formation [104,105**]. The underlying mechanism of non-coding repeat expansions disorders, on the other hand, most commonly involves an RNA gain-of-function, independently of the encoded proteins [106]. This mechanism has been thoroughly studied for DM1, which is caused by more than fifty CTG repeats in the three prime untranslated region of the dystrophia myotonica-protein kinase gene (DMPK) [107,108]. These CTG repeat expansions result in flawed RNA transcripts that prevent translation into proteins, and can cause nuclear retention in RNA foci [109,110]. It is thought that these RNA foci will alter the function of one or more RNA-binding proteins, such as muscleblind-like 1, resulting in downstream changes in gene expression and/or alternative splicing of a range of transcripts [111–113].
Table 3.
Neurological disorders caused by expanded repeats.
| Disease | Repeat Unit | Repeat Locus |
Repeat Location |
Affected Gene |
Disease Causing Repeat Length |
Mechanisms of Pathogenesis |
|---|---|---|---|---|---|---|
| Myotonic Dystrophy type 1 (DM1) | CTG | 19q13 | 3’ UTR | DMPK | 50–6500 | Altered RNA function |
| Myotonic Dystrophy type 2 (DM2) | CCTG | 3q21 | Intron | CNBP | 75–11,000 | Altered RNA function |
| Spinocerebellar ataxia 1 (SCA1) | CAG | 6p23 | Coding | ATXN1 | > 44 | Polyglutamine gain-of-function |
| Spinocerebellar ataxia 2 (SCA2) | CAG | 12q24 | Coding | ATXN2 | > 32 | Polyglutamine gain-of-function |
| Spinocerebellar ataxia 3 (SCA3) | CAG | 14q24-q32 | Coding | ATXN3 | > 52 | Polyglutamine gain-of-function |
| Spinocerebellar ataxia 6 (SCA6) | CAG | 19p13 | Coding | CACNA1A | 20–33 | Polyglutamine gain-of-function |
| Spinocerebellar ataxia 7 (SCA7) | CAG | 3q21 | Coding | ATXN7 | 37–460 | Polyglutamine gain-of-function |
| Spinocerebellar ataxia 8 (SCA8) | CTG/CAG | 13q21 | 3’ UTR | ATXN8 | 80–1300 | Polyglutamine gain-of-function |
| Spinocerebellar ataxia 10 (SCA10) | ATTCT | 22q13 | Intron | ATXN10 | 800–4500 | Altered RNA function |
| Spinocerebellar ataxia 12 (SCA12) | CAG | 5q31-q33 | 5’ UTR | PPP2R2B | 55–78 | Unknown |
| Spinocerebellar ataxia 17 (SCA17) | CAG | 6q27 | Coding | TBP | 49–66 | Polyglutamine gain-of-function |
| Spinocerebellar ataxia 31 (SCA 31) | TGGAA | 16q21-q22 | Intron | TK2-BEAN | 2.5- to 3.8-kb | RNA gain-of-function |
| Spinocerebellar ataxia 36 (SCA 36) | GGCCTG | 20p13 | Intron | NOP56 | 1500–2500 | RNA gain-of-function |
| Fragile X mental retardation 1 (FMR1) | CGG | Xq27 | 5’ UTR | FMR1 | > 200 | Altered RNA function |
| Fragile X-associated tremor ataxia syndrome (FXTAS) | CGG | Xq27 | 5’ UTR | FMR1 | 55–200 | RNA gain-of-function |
| Fragile X mental retardation 2 (FMR2) | CCG | Xq28 | 5’ UTR | FMR2 | 200–900 | Loss of protein function |
| Huntington’s disease (HD) | CAG | 4p16 | Coding | HTT | > 35 | Polyglutamine gain-of-function |
| Huntington’s disease-like 2 (HDL2) | CTG | 16q24 | 3’ UTR | JPH3 | > 41 | Altered RNA function |
| Friedreich’s Ataxia (FRDA) | GAA | 9q13 | Intron | FXN | 66–1700 | Loss of protein function |
| Epilepsy progressive myoclonia (EPM1) | CCCCGCGCGCGCGCG | 21q22 | Promoter | CSTB | 30–75 | Loss of protein function |
| Oculopharyngeal muscular dystrophy (OPMD) | GCG | 14q11 | Coding | PABPN1 | 11–17 | Polyalanine gain-of-function |
| Spinal and bulbar muscular atrophy (SBMA) | CAG | Xq12 | Coding | AR | > 37 | Polyglutamine gain-of-function |
| X-linked mental retardation | GCG | Xp21 | Coding | ARX | 17–23 | Loss of protein function |
| Dentatorubral-pallidoluysian atrophy (DRPLA) | CAG | 12p13 | Coding | ATN1 | 48–93 | Polyglutamine gain-of-function |
| ALS and/or FTD | GGGGCC | 9p21 | Intron | C9ORF72 | Up to thousands | RNA gain-of-function? |
RNA foci have also been detected in a growing number of other non-coding repeat expansion disorders, including myotonic dystrophy type 2 (DM2), Fragile X-associated tremor ataxia syndrome (FXTAS), Huntington’s disease-like 2 (HDL2), spinocerebellar ataxia type 36 (SCA36), spinocerebellar ataxia type 31 (SCA31), spinocerebellar ataxia type 8 (SCA8), spinocerebellar ataxia type 10 (SCA10) [85,86**,112,114–116], and now C9ORF72-associated ALS and FTD [18**,19**]. These findings support a common RNA gain-of-function mechanism in many non-coding repeat expansion disorders [104,105**,117], and further support the importance of these foci in C9ORF72-positive patients [18**]. To better understand the role of RNA misprocessing in C9ORF72 mutation carriers, total RNA sequencing studies in affected tissue should now be performed to identify the specific downstream targets affected in these patients.
Previous studies performed on DM1 can also help us learn about their diagnostic challenges and therapeutic strategies for C9ORF72 repeat expansions. Patients with DM1 can harbor several thousands of repeats, and the number of repeats shows a high degree of instability [118]. This instability appears to predispose towards further expansion, and could be associated with the progressive nature of the disease [119]. It could also account for the differences in DM1 alleles that are detected both between and within tissues of the same patient [120–126]. Furthermore, it explains why children may inherit repeat lengths that are considerably longer than those of their parents [127], and why an earlier age at onset or increased severity are reported in successive generations (genetic anticipation) [108,121,128–133]. The degree of instability associated with C9ORF72 repeat expansions still needs to be determined; however, some studies have suggested that families with C9ORF72 mutations can display anticipation as well [18**,19**,28*,40*,43*,49*,50**,51*,56*,69*,74*,77*].
For diagnostic purposes, DM1 flow charts have been developed. Firstly, a conventional PCR has to be performed to determine whether an individual has two alleles with a low number of repeats. If only one allele size is detected, then additional testing is necessary. Alleles up to ~100 CTG repeats can be identified with a repeat-primed PCR, both robustly and reliably [118,134,135**]. Because of extinction of the signal in higher size regions, however, no trustworthy information about the precise length of the expanded repeats can be obtained. Moreover, rare interruptions in the CTG repeats may result in a failure to detect expansions [135**]. Alleles containing 100 CTG repeats and over, can be assessed with Southern blot analysis. This method is the gold standard, but it is time-consuming and requires a large amount of genomic DNA [118,135**]. Southern blot analysis often results in a diffused band, consistent with somatic heterogeneity and correlating with the age of the patient [136]. Therefore, diagnostic DM1 tests usually report a size range (5–35, 36–50, 50–150 and >150), instead of an exact repeat length, and are based on comparisons with molecular weight standards and/or characterized control samples [135**]. If more is known about the C9ORF72 repeat sizes in control subjects and ALS and/or FTD patients, it would be possible to develop a similar flow chart and size range for diagnostic C9ORF72 testing.
Lastly, promising therapeutic strategies are currently being developed for DM1, including antisense oligonucleotides that hybridize to cellular mRNAs, inhibit gene expression, and could target mRNA for degradation, which could halt progression or reverse damage induced by toxic RNA [137**]. It is the hope that a similar approach may one day be used in FTD and ALS patients with C9ORF72 repeat expansions.
What is the role of other repeat expansions in FTD and ALS?
Previously, other repeat expansions have already been implicated in ALS and/or FTD, these include repeat expansions in the ataxin-2 gene (ATXN2) and in the non-imprinted Prader-Willi/Angelman syndrome region protein 1 gene (NIPA1) [138*–140*,141]. In addition, a rare intronic GGCCTG repeat expansion in the NOP56 ribonucleoprotein homolog gene (NOP56) has recently been identified in patients with spinocerebellar ataxia and motor neuron involvement [86**].
These expansions, however, may only represent the tip of the iceberg, and the actual contribution of repeat expansions to the etiology of ALS and/or FTD may be much higher. For example, the C9ORF72 mutation is only one out of 109 repeats in the human genome with at least three GGGGCC repeat units, several of which are in non-coding regions, and expansions of either one of these repeats could be implicated in disease. Unfortunately, until novel methods are developed that are designed to systematically screen for repeat expansions on a genome-wide level, the actual contribution of repeat expansions to the pathogenesis of ALS and/or FTD remains undetermined.
Conclusions and future directions
Despite the excitement, it is important to acknowledge that it is still early days, and several key questions related to the C9ORF72 mutation presently remain unanswered: Are the RNA foci observed in C9ORF72 repeat carriers toxic and which downstream targets are affected? Which protein accumulates in the ubiquitinated TDP-43-negative inclusions and do they have a role in disease pathogenesis? What determines whether someone develops ALS, FTD or both? Which role does the repeat length play in determining disease onset and presentation, and is there a minimal number of repeats needed for pathogenicity?
In this review, we have provided an up-to-date overview of the current C9ORF72 literature, and we have discussed this mutation in relation to other non-coding repeat expansion disorders, which emphasizes the crucial role of repeat expansions, and RNA toxicity, in a broad range of neurodegenerative disorders.
Supplementary Material
Key points.
C9ORF72 repeat expansions are currently the major genetic cause of ALS and/or FTD worldwide, accounting for approximately 34.2% of the familial ALS cases, and 25.9% of the familial FTD cases.
Mutations in other ALS- and/or FTD-associated genes may act as disease modifiers, which could contribute to the pleiotropy that is encountered in patients with C9ORF72 mutations.
Somatic heterogeneity is likely to be common in repeat expansion carriers, and as a result, repeat sizes determined using DNA extracted from whole blood may not adequately reflect the C9ORF72 repeat sizes in a patient’s brain or spinal cord tissue.
C9ORF72 expansions are probably pathogenic due to a toxic RNA gain-of-function mechanism in line with most other non-coding repeat expansion disorders.
The actual contribution of repeat expansions to the etiology of ALS and/or FTD may be much higher; non-coding expansions that have been identified thus far may only represent the tip of the iceberg.
Acknowledgements
The research in the authors’ laboratory is funded by NIH grants P50 AG016574, R01 NS065782 and R01 AG026251, the ALS Therapy Alliance, and the Consortium for Frontotemporal dementia (CFR).
Footnotes
Disclosure statement: No actual or potential conflicts of interest have been reported.
References
Papers of particular interest, published within the annual period of review, have been highlighted as:
* of special interest
** of outstanding interest
- 1. Kiernan MC, Vucic S, Cheah BC, et al. Amyotrophic lateral sclerosis. Lancet. 2011;377:942–955. doi: 10.1016/S0140-6736(10)61156-7. This review summerizes the etiology of amyotrophic lateral sclerosis, its clinical characteristics, its diagnosis, its prognosis, and its management.
- 2.Graff-Radford NR, Woodruff BK. Frontotemporal dementia. Semin Neurol. 2007;27:48–57. doi: 10.1055/s-2006-956755. [DOI] [PubMed] [Google Scholar]
- 3.Rascovsky K, Hodges JR, Knopman D, et al. Sensitivity of revised diagnostic criteria for the behavioural variant of frontotemporal dementia. Brain. 2011;134(Pt 9):2456–2477. doi: 10.1093/brain/awr179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gorno-Tempini ML, Hillis AE, Weintraub S, et al. Classification of primary progressive aphasia and its variants. Neurology. 2011;76:1006–1014. doi: 10.1212/WNL.0b013e31821103e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.The Lund and Manchester Groups. Clinical and neuropathological criteria for frontotemporal dementia. J Neurol Neurosurg Psychiatry. 1994;57:416–418. doi: 10.1136/jnnp.57.4.416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ferrari R, Kapogiannis D, Huey ED, Momeni P. FTD and ALS: a tale of two diseases. Curr Alzheimer Res. 2011;8:273–294. doi: 10.2174/156720511795563700. Reviews the clinical, pathological and genetic features of frontotemporal dementia and amyotrophic lateral sclerosis, underlining the overlap between these two diseases.
- 7. Fecto F, Siddique T. Making connections: pathology and genetics link amyotrophic lateral sclerosis with frontotemporal lobe dementia. J Mol Neurosci. 2011;45:663–675. doi: 10.1007/s12031-011-9637-9. Commonalities between amyotrophic lateral sclerosis and frontotemporal dementia are reviewed.
- 8.Lillo P, Hodges JR. Frontotemporal dementia and motor neurone disease: overlapping clinic-pathological disorders. J Clin Neurosci. 2009;16:1131–1135. doi: 10.1016/j.jocn.2009.03.005. [DOI] [PubMed] [Google Scholar]
- 9.Giordana MT, Ferrero P, Grifoni S, et al. Dementia and cognitive impairment in amyotrophic lateral sclerosis: a review. Neurol Sci. 2011;32:9–16. doi: 10.1007/s10072-010-0439-6. [DOI] [PubMed] [Google Scholar]
- 10.Lomen-Hoerth C, Murphy J, Langmore S, et al. Are amyotrophic lateral sclerosis patients cognitively normal? Neurology. 2003;60:1094–1097. doi: 10.1212/01.wnl.0000055861.95202.8d. [DOI] [PubMed] [Google Scholar]
- 11.Phukan J, Pender NP, Hardiman O. Cognitive impairment in amyotrophic lateral sclerosis. Lancet Neurol. 2007;6:994–1003. doi: 10.1016/S1474-4422(07)70265-X. [DOI] [PubMed] [Google Scholar]
- 12.Lomen-Hoerth C, Anderson T, Miller B. The overlap of amyotrophic lateral sclerosis and frontotemporal dementia. Neurology. 2002;59:1077–1079. doi: 10.1212/wnl.59.7.1077. [DOI] [PubMed] [Google Scholar]
- 13.Neumann M, Sampathu DM, Kwong LK, et al. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science. 2006;314:130–133. doi: 10.1126/science.1134108. [DOI] [PubMed] [Google Scholar]
- 14.Mackenzie IR, Neumann M, Bigio EH, et al. Nomenclature for neuropathologic subtypes of frontotemporal lobar degeneration: consensus recommendations. Acta Neuropathol. 2009;117:15–18. doi: 10.1007/s00401-008-0460-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Andersen PM, Al-Chalabi A. Clinical genetics of amyotrophic lateral sclerosis: what do we really know? Nat Rev Neurol. 2011;7:603–615. doi: 10.1038/nrneurol.2011.150. This article discusses how familial amyotrophic lateral sclerosis is frequently underdiagnosed and how apparently sporadic amyotrophic lateral sclerosis may represent familial amyotrophic lateral sclerosis with reduced disease penetrance. Furthermore, it hypothesizes that most amyotrophic lateral sclerosis cases could be attributed to oligogenic inheritance.
- 16.Paulson HL, Igo I. Genetics of dementia. Semin Neurol. 2011;31:449–460. doi: 10.1055/s-0031-1299784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gros-Louis F, Gaspar C, Rouleau GA. Genetics of familial and sporadic amyotrophic lateral sclerosis. Biochim Biophys Acta. 2006;1762:956–972. doi: 10.1016/j.bbadis.2006.01.004. [DOI] [PubMed] [Google Scholar]
- 18. DeJesus-Hernandez M, Mackenzie IR, Boeve BF, et al. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron. 2011;72:245–256. doi: 10.1016/j.neuron.2011.09.011. Reports the identification of non-coding C9ORF72 repeat expansions in patients with frontotemporal dementia and amyotrophic lateral sclerosis.
- 19. Renton AE, Majounie E, Waite A, et al. A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron. 2011;72:257–268. doi: 10.1016/j.neuron.2011.09.010. In this paper, the hexanucleotide repeat expansion in C9ORF72 is reported in patients with amyotrophic lateral sclerosis and frontotemporal dementia.
- 20.Boxer AL, Mackenzie IR, Boeve BF, et al. Clinical, neuroimaging and neuropathological features of a new chromosome 9p-linked FTD-ALS family. J Neurol Neurosurg Psychiatry. 2011;82:196–203. doi: 10.1136/jnnp.2009.204081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gijselinck I, Engelborghs S, Maes G, et al. Identification of 2 Loci at chromosomes 9 and 14 in a multiplex family with frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Arch Neurol. 2010;67:606–616. doi: 10.1001/archneurol.2010.82. [DOI] [PubMed] [Google Scholar]
- 22.Le Ber I, Camuzat A, Berger E, et al. Chromosome 9p-linked families with frontotemporal dementia associated with motor neuron disease. Neurology. 2009;72:1669–1676. doi: 10.1212/WNL.0b013e3181a55f1c. [DOI] [PubMed] [Google Scholar]
- 23.Luty AA, Kwok JB, Thompson EM, et al. Pedigree with frontotemporal lobar degeneration--motor neuron disease and Tar DNA binding protein-43 positive neuropathology: genetic linkage to chromosome 9. BMC Neurol. 2008;8:32. doi: 10.1186/1471-2377-8-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Morita M, Al-Chalabi A, Andersen PM, et al. A locus on chromosome 9p confers susceptibility to ALS and frontotemporal dementia. Neurology. 2006;66:839–844. doi: 10.1212/01.wnl.0000200048.53766.b4. [DOI] [PubMed] [Google Scholar]
- 25. Pearson JP, Williams NM, Majounie E, et al. Familial frontotemporal dementia with amyotrophic lateral sclerosis and a shared haplotype on chromosome 9p. J Neurol. 2011;258:647–655. doi: 10.1007/s00415-010-5815-x. The clinical phenotype and pathology of a family with frontotemporal dementia and amyotrophic lateral sclerosis linked to 9p21 is described.
- 26.Valdmanis PN, Dupre N, Bouchard JP, et al. Three families with amyotrophic lateral sclerosis and frontotemporal dementia with evidence of linkage to chromosome 9p. Arch Neurol. 2007;64:240–245. doi: 10.1001/archneur.64.2.240. [DOI] [PubMed] [Google Scholar]
- 27.Vance C, Al-Chalabi A, Ruddy D, et al. Familial amyotrophic lateral sclerosis with frontotemporal dementia is linked to a locus on chromosome 9p13.2-21.3. Brain. 2006;129:868–876. doi: 10.1093/brain/awl030. [DOI] [PubMed] [Google Scholar]
- 28. Gijselinck I, Van Langenhove T, van der Zee J, et al. A C9orf72 promoter repeat expansion in a Flanders-Belgian cohort with disorders of the frontotemporal lobar degeneration-amyotrophic lateral sclerosis spectrum: a gene identification study. Lancet Neurol. 2012;11:54–65. doi: 10.1016/S1474-4422(11)70261-7. This study examines Belgian patients with frontotemporal dementia and amyotrophic lateral sclerosis, and confirms the presence of C9ORF72 repeat expansions by using association and segregation studies, genomic sequencing, repeat genotyping and expression studies.
- 29. van Blitterswijk M, van Es MA, Hennekam EA, et al. Evidence for an oligogenic basis of amyotrophic lateral sclerosis. Hum Mol Genet. 2012;21:3776–3784. doi: 10.1093/hmg/dds199. Reports familial amyotrophic lateral sclerosis patients with mutations in multiple genes that are associated with amyotrophic lateral sclerosis, providing evidence for an oligogenic etiology of familial amyotrophic lateral sclerosis.
- 30. Rohrer JD, Warren JD. Phenotypic signatures of genetic frontotemporal dementia. Curr Opin Neurol. 2011;24:542–549. doi: 10.1097/WCO.0b013e32834cd442. In this review, clinical and neuroanatomical phenotypes associated with autosomal-dominant frontotemporal dementia are considered.
- 31. Seelaar H, Rohrer JD, Pijnenburg YA, et al. Clinical, genetic and pathological heterogeneity of frontotemporal dementia: a review. J Neurol Neurosurg Psychiatry. 2011;82:476–486. doi: 10.1136/jnnp.2010.212225. This study summarizes clinical, neuropsychological, imaging, genetic, and pathological features of frontotemporal dementia.
- 32. Conte A, Lattante S, Luigetti M, et al. Classification of familial amyotrophic lateral sclerosis by family history: effects on frequency of genes mutation. J Neurol Neurosurg Psychiatry. 2012 doi: 10.1136/jnnp-2012-302897. Paper that proposes a classification for familial amyotrophic lateral sclerosis based on family history.
- 33. Ratti A, Corrado L, Castellotti B, et al. C9ORF72 repeat expansion in a large Italian ALS cohort: evidence of a founder effect. Neurobiol Aging. 2012;33:2528.e7–2528.e14. doi: 10.1016/j.neurobiolaging.2012.06.008. Repeat expansions in C9ORF72 are assessed in amyotrophic lateral sclerosis patients of Italian descent.
- 34. Chester C, de Carvalho M, Miltenberger G, et al. Rapidly progressive frontotemporal dementia and bulbar amyotrophic lateral sclerosis in Portuguese patients with C9orf72 mutation. Amyotroph Lateral Scler. 2012 doi: 10.3109/17482968.2012.690418. Portuguese patients with familial frontotemporal dementia and familial amyotrophic lateral sclerosis are tested for repeat expansions in C9ORF72.
- 35. Ogaki K, Li Y, Atsuta N, et al. Analysis of C9orf72 repeat expansion in 563 Japanese patients with amyotrophic lateral sclerosis. Neurobiol Aging. 2012;33:2527.e11–2527.e16. doi: 10.1016/j.neurobiolaging.2012.05.011. Japanese patients with familial and sporadic amyotrophic lateral sclerosis are screened for C9ORF72 mutations, which demonstrates that these mutations are rare.
- 36. Lattante S, Conte A, Zollino M, et al. Contribution of major amyotrophic lateral sclerosis genes to the etiology of sporadic disease. Neurology. 2012;79:66–72. doi: 10.1212/WNL.0b013e31825dceca. The frequency of mutations in major genes associated with amyotrophic lateral sclerosis is investigated in familial and sporadic amyotrophic lateral sclerosis patients of Italian descent.
- 37. Smith BN, Newhouse S, Shatunov A, et al. The C9ORF72 expansion mutation is a common cause of ALS+/−FTD in Europe and has a single founder. Eur J Hum Genet. 2012 doi: 10.1038/ejhg.2012.98. European cohorts of patients with amyotrophic lateral sclerosis and/or frontotemporal dementia are screened for mutations in C9ORF72, demonstrating a common founder.
- 38. Tsai CP, Soong BW, Tu PH, et al. A hexanucleotide repeat expansion in C9ORF72 causes familial and sporadic ALS in Taiwan. Neurobiol Aging. 2012;33:2232, e2211–e2238. doi: 10.1016/j.neurobiolaging.2012.05.002. In a Taiwanese cohort of familial and sporadic amyotrophic lateral sclerosis patients repeat expansions in C9ORF72 are detected.
- 39. Ishiura H, Takahashi Y, Mitsui J, et al. C9ORF72 Repeat Expansion in Amyotrophic Lateral Sclerosis in the Kii Peninsula of JapanC9ORF72 Repeat Expansion in ALS. Arch Neurol. 2012 doi: 10.1001/archneurol.2012.1219. This study investigates C9ORF72 repeat expansions in Japanese familial and sporadic amyotrophic lateral sclerosis patients, and shows that they are uncommon.
- 40. Pamphlett R, Cheong PL, Trent RJ, Yu B. Transmission of C9orf72 hexanucleotide repeat expansions in sporadic amyotrophic lateral sclerosis: an Australian trio study. Neuroreport. 2012;23:556–559. doi: 10.1097/WNR.0b013e3283544718. In an Australian trio study, no evidence of repeat instability, causing repeat expansions in C9ORF72, is identified in sporadic amyotrophic lateral sclerosis patients.
- 41. Dols-Icardo O, Suarez-Calvet M, Hernandez I, et al. Expansion mutation in C9ORF72 does not influence plasma progranulin levels in frontotemporal dementia. Neurobiol Aging. 2012;33:1851, e1817–e1859. doi: 10.1016/j.neurobiolaging.2012.03.005. This study demonstrates that plasma progranulin levels are not a reliable biomarker to detect C9ORF72 mutation carriers.
- 42. Millecamps S, Boillee S, Le Ber I, et al. Phenotype difference between ALS patients with expanded repeats in C9ORF72 and patients with mutations in other ALS-related genes. J Med Genet. 2012;49:258–263. doi: 10.1136/jmedgenet-2011-100699. Genotype-phenotype analyses are performed for familial and sporadic amyotrophic lateral sclerosis patients, comparing patients with and without C9ORF72 repeat expansions.
- 43. Ferrari R, Mok K, Moreno JH, et al. Screening for C9ORF72 repeat expansion in FTLD. Neurobiol Aging. 2012;33:1850. doi: 10.1016/j.neurobiolaging.2012.02.017. e1851-1850 e1811. In a cohort from the United States of America, the prevalence of C9ORF72 mutations is assessed in familial and sporadic patients with frontotemporal dementia.
- 44. Mok KY, Koutsis G, Schottlaender LV, et al. High frequency of the expanded C9ORF72 hexanucleotide repeat in familial and sporadic Greek ALS patients. Neurobiol Aging. 2012;33:1851, e1851–e1855. doi: 10.1016/j.neurobiolaging.2012.02.021. C9ORF72 repeat expansions are frequently detected in familial and sporadic amyotrophic lateral sclerosis patients from Greece.
- 45. Brettschneider J, Van Deerlin VM, Robinson JL, et al. Pattern of ubiquilin pathology in ALS and FTLD indicates presence of C9ORF72 hexanucleotide expansion. Acta Neuropathol. 2012;123:825–839. doi: 10.1007/s00401-012-0970-z. Patients from the United States of America with amyotrophic lateral sclerosis and frontotemporal dementia (both familial and sporadic) are screened for C9ORF72 hexanucleotide repeat expansions, and their clinical phenotypes and neuropathology are compared.
- 46. Sabatelli M, Conforti FL, Zollino M, et al. C9ORF72 hexanucleotide repeat expansions in the Italian sporadic ALS population. Neurobiol Aging. 2012;33:1848, e1815–e1820. doi: 10.1016/j.neurobiolaging.2012.02.011. A large cohort of sporadic amyotrophic lateral sclerosis patients of Italian descent is tested for C9ORF72 mutations.
- 47. Majounie E, Renton AE, Mok K, et al. Frequency of the C9orf72 hexanucleotide repeat expansion in patients with amyotrophic lateral sclerosis and frontotemporal dementia: a cross-sectional study. Lancet Neurol. 2012;11:323–330. doi: 10.1016/S1474-4422(12)70043-1. Patients from 17 regions worldwide are screened for repeat expansions in C9ORF72; expansions are relatively common in patients from Finland, and are rare in patients from Asia and India.
- 48. Khan BK, Yokoyama JS, Takada LT, et al. Atypical, slowly progressive behavioural variant frontotemporal dementia associated with C9ORF72 hexanucleotide expansion. J Neurol Neurosurg Psychiatry. 2012;83:358–364. doi: 10.1136/jnnp-2011-301883. Two patients are described with the slowly progressive behavioral variant of frontotemporal dementia and C9ORF72 mutations.
- 49. Chio A, Borghero G, Restagno G, et al. Clinical characteristics of patients with familial amyotrophic lateral sclerosis carrying the pathogenic GGGGCC hexanucleotide repeat expansion of C9ORF72. Brain. 2012;135:784–793. doi: 10.1093/brain/awr366. C9ORF72 repeat expansions are determined in familial amyotrophic lateral sclerosis patients from Italy and Germany; their clinical characteristics are described as well.
- 50. Boeve BF, Boylan KB, Graff-Radford NR, et al. Characterization of frontotemporal dementia and/or amyotrophic lateral sclerosis associated with the GGGGCC repeat expansion in C9ORF72. Brain. 2012;135:765–783. doi: 10.1093/brain/aws004. Clinical phenotypes, neuropsychological tests, neuroimaging results, and neuropathology are reported of C9ORF72 patients from the United States of America with amyotrophic lateral sclerosis and frontotemporal dementia (both familial and sporadic).
- 51. Cooper-Knock J, Hewitt C, Highley JR, et al. Clinico-pathological features in amyotrophic lateral sclerosis with expansions in C9ORF72. Brain. 2012;135:751–764. doi: 10.1093/brain/awr365. This study reports the clinicopathological features of familial and sporadic amyotrophic lateral sclerosis patients with repeat expansions in C9ORF72 from the United Kingdom.
- 52. Mahoney CJ, Beck J, Rohrer JD, et al. Frontotemporal dementia with the C9ORF72 hexanucleotide repeat expansion: clinical, neuroanatomical and neuropathological features. Brain. 2012;135:736–750. doi: 10.1093/brain/awr361. Patients with frontotemporal dementia from the United Kingdom, both familial and sporadic, are screened for C9ORF72 mutations, and their clinical, neuroimaging, and histopathological analyses are presented.
- 53. Byrne S, Elamin M, Bede P, et al. Cognitive and clinical characteristics of patients with amyotrophic lateral sclerosis carrying a C9orf72 repeat expansion: a population-based cohort study. Lancet Neurol. 2012;11:232–240. doi: 10.1016/S1474-4422(12)70014-5. Familial and sporadic amyotrophic lateral sclerosis patients of Irish descent are tested for mutations in C9ORF72, and their cognitive and clinical characteristics are reported.
- 54. Simon-Sanchez J, Dopper EG, Cohn-Hokke PE, et al. The clinical and pathological phenotype of C9ORF72 hexanucleotide repeat expansions. Brain. 2012;135:723–735. doi: 10.1093/brain/awr353. Clinical and pathological features are described for Dutch frontotemporal dementia patients with C9ORF72 repeat expansions, both familial and sporadic.
- 55. Snowden JS, Rollinson S, Thompson JC, et al. Distinct clinical and pathological characteristics of frontotemporal dementia associated with C9ORF72 mutations. Brain. 2012;135:693–708. doi: 10.1093/brain/awr355. This paper highlights the clinical and pathological characteristics of frontotemporal dementia patients from the United Kingdom with C9ORF72 repeat expansions.
- 56. Stewart H, Rutherford NJ, Briemberg H, et al. Clinical and pathological features of amyotrophic lateral sclerosis caused by mutation in the C9ORF72 gene on chromosome 9p. Acta Neuropathol. 2012;123:409–417. doi: 10.1007/s00401-011-0937-5. In this article, C9ORF72 patients with familial and sporadic amyotrophic lateral sclerosis from Canada are discussed, describing their clinical and pathological characteristics.
- 57. Al-Sarraj S, King A, Troakes C, et al. p62 positive, TDP-43 negative, neuronal cytoplasmic and intranuclear inclusions in the cerebellum and hippocampus define the pathology of C9orf72-linked FTLD and MND/ALS. Acta Neuropathol. 2011;122:691–702. doi: 10.1007/s00401-011-0911-2. In patients with familial and sporadic frontotemporal dementia and amyotrophic lateral sclerosis, from the United Kingdom, the presence of C9ORF72 mutations is determined and their neuropathological characteristics are reported.
- 58.van Blitterswijk M, van Es MA, Koppers M, et al. VAPB and C9orf72 mutations in 1 familial amyotrophic lateral sclerosis patient. Neurobiol Aging. 2012 doi: 10.1016/j.neurobiolaging.2012.07.004. [DOI] [PubMed] [Google Scholar]
- 59. Chio A, Restagno G, Brunetti M, et al. ALS/FTD phenotype in two Sardinian families carrying both C9ORF72 and TARDBP mutations. J Neurol Neurosurg Psychiatry. 2012;83:730–733. doi: 10.1136/jnnp-2012-302219. This article presents Italian families with mutations in both C9ORF72 and TARDBP.
- 60.Corrado L, Ratti A, Gellera C, et al. High frequency of TARDBP gene mutations in Italian patients with amyotrophic lateral sclerosis. Hum Mutat. 2009;30:688–694. doi: 10.1002/humu.20950. [DOI] [PubMed] [Google Scholar]
- 61. Corrado L, Carlomagno Y, Falasco L, et al. A novel peripherin gene (PRPH) mutation identified in one sporadic amyotrophic lateral sclerosis patient. Neurobiol Aging. 2011;32:552, e551–e556. doi: 10.1016/j.neurobiolaging.2010.02.011. The novel peripherin mutation turned out to be present in a patient with a C9ORF72 repeat expansion.
- 62.van Rheenen W, van Blitterswijk M, Huisman MH, et al. Hexanucleotide repeat expansions in C9ORF72 in the spectrum of motor neuron diseases. Neurology. 2012 doi: 10.1212/WNL.0b013e3182661d14. [DOI] [PubMed] [Google Scholar]
- 63.Millecamps S, Salachas F, Cazeneuve C, et al. SOD1, ANG, VAPB, TARDBP, and FUS mutations in familial amyotrophic lateral sclerosis: genotype-phenotype correlations. J Med Genet. 2010;47:554–560. doi: 10.1136/jmg.2010.077180. [DOI] [PubMed] [Google Scholar]
- 64.Millecamps S, Da Barroca S, Cazeneuve C, et al. Questioning on the role of D amino acid oxidase in familial amyotrophic lateral sclerosis. Proc Natl Acad Sci U S A. 2010;107:E107. doi: 10.1073/pnas.1006190107. author reply E108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Millecamps S, Boillee S, Chabrol E, et al. Screening of OPTN in French familial amyotrophic lateral sclerosis. Neurobiol Aging. 2011;32:557, e511–e553. doi: 10.1016/j.neurobiolaging.2010.11.005. This study shows that variants in optineurin are rare in French patients with familial amyotrophic lateral sclerosis.
- 66. Millecamps S, Corcia P, Cazeneuve C, et al. Mutations in UBQLN2 are rare in French amyotrophic lateral sclerosis. Neurobiol Aging. 2012;33:839, e831–e833. doi: 10.1016/j.neurobiolaging.2011.11.010. One deletion in UBQLN2 is identified in a French patient with amyotrophic lateral sclerosis, however, this mutation turned out to be present in a patient with a C9ORF72 mutation.
- 67.Kirby J, Goodall EF, Smith W, et al. Broad clinical phenotypes associated with TAR-DNA binding protein (TARDBP) mutations in amyotrophic lateral sclerosis. Neurogenetics. 2010;11:217–225. doi: 10.1007/s10048-009-0218-9. [DOI] [PubMed] [Google Scholar]
- 68.Hewitt C, Kirby J, Highley JR, et al. Novel FUS/TLS mutations and pathology in familial and sporadic amyotrophic lateral sclerosis. Arch Neurol. 2010;67:455–461. doi: 10.1001/archneurol.2010.52. [DOI] [PubMed] [Google Scholar]
- 69. Hsiung GY, DeJesus-Hernandez M, Feldman HH, et al. Clinical and pathological features of familial frontotemporal dementia caused by C9ORF72 mutation on chromosome 9p. Brain. 2012;135:709–722. doi: 10.1093/brain/awr354. This paper contains a detailed analysis of the clinical and neuropathological features of Canadian families with frontotemporal dementia due to C9ORF72 mutations.
- 70. Murray ME, DeJesus-Hernandez M, Rutherford NJ, et al. Clinical and neuropathologic heterogeneity of c9FTD/ALS associated with hexanucleotide repeat expansion in C9ORF72. Acta Neuropathol. 2011;122:673–690. doi: 10.1007/s00401-011-0907-y. The neuropathology and clinical features are investigated of American patients with frontotemporal dementia and amyotrophic lateral sclerosis that harbor C9ORF72 repeat expansions.
- 71. Brettschneider J, Toledo JB, Van Deerlin VM, et al. Microglial activation correlates with disease progression and upper motor neuron clinical symptoms in amyotrophic lateral sclerosis. PLoS One. 2012;7:e39216. doi: 10.1371/journal.pone.0039216. This study displays that microglial pathology is more extensive in the medulla and motor cortex of patients with C9ORF72 repeat expansions.
- 72.Huisman MH, de Jong SW, van Doormaal PT, et al. Population based epidemiology of amyotrophic lateral sclerosis using capture-recapture methodology. J Neurol Neurosurg Psychiatry. 2011;82:1165–1170. doi: 10.1136/jnnp.2011.244939. [DOI] [PubMed] [Google Scholar]
- 73.Floris G, Borghero G, Cannas A, et al. Frontotemporal dementia with psychosis, parkinsonism, visuo-spatial dysfunction, upper motor neuron involvement associated to expansion of C9ORF72: a peculiar phenotype? J Neurol. 2012;259:1749–1751. doi: 10.1007/s00415-012-6444-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Savica R, Adeli A, Vemuri P, et al. Characterization of a Family With c9FTD/ALS Associated With the GGGGCC Repeat Expansion in C9ORF72. Arch Neurol. 2012 doi: 10.1001/archneurol.2012.772. Presents the clinical, neuropsychological, and neuroimaging findings of a C9ORF72 family with frontotemporal dementia and amyotrophic lateral sclerosis.
- 75. Lindquist S, Duno M, Batbayli M, et al. Corticobasal and ataxia syndromes widen the spectrum of C9ORF72 hexanucleotide expansion disease. Clin Genet. 2012 doi: 10.1111/j.1399-0004.2012.01903.x. Repeat expansions in C9ORF72 are assessed in patients with early onset autosomal dominant dementia, broadening the spectrum associated with these expansions.
- 76. Luigetti M, Quaranta D, Conte A, et al. Frontotemporal dementia, Parkinsonism and lower motor neuron involvement in a patient with C9ORF72 expansion. Amyotroph Lateral Scler. 2012 doi: 10.3109/17482968.2012.692383. A patient with a C9ORF72 mutation is reported with a complex phenotype consisting of frontotemporal dementia, parkinsonism, and amyotrophic lateral sclerosis.
- 77. Arighi A, Fumagalli GG, Jacini F, et al. Early Onset Behavioral Variant Frontotemporal Dementia due to the C9ORF72 Hexanucleotide Repeat Expansion: Psychiatric Clinical Presentations. J Alzheimers Dis. 2012;31:447–452. doi: 10.3233/JAD-2012-120523. This article emphasizes that patients with mutations in C9ORF72 can present with atypical features, including psychiatric symptoms.
- 78. Rollinson S, Halliwell N, Young K, et al. Analysis of the hexanucleotide repeat in C9ORF72 in Alzheimer's disease. Neurobiol Aging. 2012;33:1846, e1845–e1846. doi: 10.1016/j.neurobiolaging.2012.01.109. No C9ORF72 repeat expansions are detected in patients with Alzheimer's disease.
- 79. Wallon D, Rovelet-Lecrux A, Deramecourt V, et al. Definite Behavioral Variant of Frontotemporal Dementia with C9ORF72 Expansions Despite Positive Alzheimer's Disease Cerebrospinal Fluid Biomarkers. J Alzheimers Dis. 2012 doi: 10.3233/JAD-2012-120877. In less than 3% of the patients with Alzheimer's disease mutations in C9ORF72 are detected.
- 80. Majounie E, Abramzon Y, Renton AE, et al. Repeat expansion in C9ORF72 in Alzheimer's disease. N Engl J Med. 2012;366:283–284. doi: 10.1056/NEJMc1113592. Repeat expansions in C9ORF72 are absent in a cohort of Parkinson's disease patients.
- 81.Majounie E, Abramzon Y, Renton AE, et al. Large C9orf72 repeat expansions are not a common cause of Parkinson's disease. Neurobiol Aging. 2012;33:2527.e1–2527.e2. doi: 10.1016/j.neurobiolaging.2012.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Rademakers R, Neumann M, Mackenzie IR. Advances in understanding the molecular basis of frontotemporal dementia. Nat Rev Neurol. 2012;8:423–434. doi: 10.1038/nrneurol.2012.117. This review focuses on the molecular basis of frontotemporal dementia, describing the role of tau, progranulin, TDP-43, FET proteins, and C9ORF72 repeat expansions.
- 83. Troakes C, Maekawa S, Wijesekera L, et al. An MND/ALS phenotype associated with C9orf72 repeat expansion: Abundant p62-positive, TDP-43-negative inclusions in cerebral cortex, hippocampus and cerebellum but without associated cognitive decline. Neuropathology. 2011 doi: 10.1111/j.1440-1789.2011.01286.x. The distinctive cortical and cerebellar pathology of C9ORF72 mutations is presented.
- 84.La Spada AR, Taylor JP. Repeat expansion disease: progress and puzzles in disease pathogenesis. Nat Rev Genet. 2010;11:247–258. doi: 10.1038/nrg2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sato N, Amino T, Kobayashi K, et al. Spinocerebellar ataxia type 31 is associated with "inserted" penta-nucleotide repeats containing (TGGAA)n. Am J Hum Genet. 2009;85:544–557. doi: 10.1016/j.ajhg.2009.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Kobayashi H, Abe K, Matsuura T, et al. Expansion of intronic GGCCTG hexanucleotide repeat in NOP56 causes SCA36, a type of spinocerebellar ataxia accompanied by motor neuron involvement. Am J Hum Genet. 2011;89:121–130. doi: 10.1016/j.ajhg.2011.05.015. This study identifies an intronic repeat in NOP56, which causes an RNA gain-of-function, and subsequently, spinocerebellar ataxia type 36.
- 87.Todd PK, Paulson HL. RNA-mediated neurodegeneration in repeat expansion disorders. Ann Neurol. 2010;67:291–300. doi: 10.1002/ana.21948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Paulson HL. The spinocerebellar ataxias. J Neuroophthalmol. 2009;29:227–237. doi: 10.1097/WNO0b013e3181b416de. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Durr A. Autosomal dominant cerebellar ataxias: polyglutamine expansions and beyond. Lancet Neurol. 2010;9:885–894. doi: 10.1016/S1474-4422(10)70183-6. [DOI] [PubMed] [Google Scholar]
- 90. Verbeek DS, van de Warrenburg BP. Genetics of the dominant ataxias. Semin Neurol. 2011;31:461–469. doi: 10.1055/s-0031-1299785. In this article the dominantly inherited spinocerebellar ataxias are reviewed.
- 91. Seidel K, Siswanto S, Brunt ER, et al. Brain pathology of spinocerebellar ataxias. Acta Neuropathol. 2012;124:1–21. doi: 10.1007/s00401-012-1000-x. Review that reports the genetic background, clinical features, and neuropathological characteristics of patients with spinocerebellar ataxias.
- 92. Whaley NR, Fujioka S, Wszolek ZK. Autosomal dominant cerebellar ataxia type I: a review of the phenotypic and genotypic characteristics. Orphanet J Rare Dis. 2011;6:33. doi: 10.1186/1750-1172-6-33. This paper reviews the characteristics of spinocerebellar ataxia subtypes.
- 93.Finsterer J. Bulbar and spinal muscular atrophy (Kennedy's disease): a review. Eur J Neurol. 2009;16:556–561. doi: 10.1111/j.1468-1331.2009.02591.x. [DOI] [PubMed] [Google Scholar]
- 94.Brais B. Oculopharyngeal muscular dystrophy: a late-onset polyalanine disease. Cytogenet Genome Res. 2003;100:252–260. doi: 10.1159/000072861. [DOI] [PubMed] [Google Scholar]
- 95.Gusella JF, MacDonald ME, Ambrose CM, Duyao MP. Molecular genetics of Huntington's disease. Arch Neurol. 1993;50:1157–1163. doi: 10.1001/archneur.1993.00540110037003. [DOI] [PubMed] [Google Scholar]
- 96.Li LB, Bonini NM. Roles of trinucleotide-repeat RNA in neurological disease and degeneration. Trends Neurosci. 2010;33:292–298. doi: 10.1016/j.tins.2010.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Palazzolo I, Gliozzi A, Rusmini P, et al. The role of the polyglutamine tract in androgen receptor. J Steroid Biochem Mol Biol. 2008;108:245–253. doi: 10.1016/j.jsbmb.2007.09.016. [DOI] [PubMed] [Google Scholar]
- 98.Ishikawa K, Durr A, Klopstock T, et al. Pentanucleotide repeats at the spinocerebellar ataxia type 31 (SCA31) locus in Caucasians. Neurology. 2011;77:1853–1855. doi: 10.1212/WNL.0b013e3182377e3a. [DOI] [PubMed] [Google Scholar]
- 99.Duyao M, Ambrose C, Myers R, et al. Trinucleotide repeat length instability and age of onset in Huntington's disease. Nat Genet. 1993;4:387–392. doi: 10.1038/ng0893-387. [DOI] [PubMed] [Google Scholar]
- 100.Takano H, Cancel G, Ikeuchi T, et al. Close associations between prevalences of dominantly inherited spinocerebellar ataxias with CAG-repeat expansions and frequencies of large normal CAG alleles in Japanese and Caucasian populations. Am J Hum Genet. 1998;63:1060–1066. doi: 10.1086/302067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Majounie E, Wardle M, Muzaimi M, et al. Case control analysis of repeat expansion size in ataxia. Neurosci Lett. 2007;429:28–32. doi: 10.1016/j.neulet.2007.09.055. [DOI] [PubMed] [Google Scholar]
- 102.Cossee M, Faivre L, Philippe C, et al. ARX polyalanine expansions are highly implicated in familial cases of mental retardation with infantile epilepsy and/or hand dystonia. Am J Med Genet A. 2011;155A:98–105. doi: 10.1002/ajmg.a.33785. [DOI] [PubMed] [Google Scholar]
- 103.Gecz J, Cloosterman D, Partington M. ARX: a gene for all seasons. Curr Opin Genet Dev. 2006;16:308–316. doi: 10.1016/j.gde.2006.04.003. [DOI] [PubMed] [Google Scholar]
- 104.Orr HT, Zoghbi HY. Trinucleotide repeat disorders. Annu Rev Neurosci. 2007;30:575–621. doi: 10.1146/annurev.neuro.29.051605.113042. [DOI] [PubMed] [Google Scholar]
- 105. Klein AF, Gasnier E, Furling D. Gain of RNA function in pathological cases: Focus on myotonic dystrophy. Biochimie. 2011;93:2006–2012. doi: 10.1016/j.biochi.2011.06.028. Review that investigates an RNA gain-of-function mechanism for non-coding repeat expansion disorders.
- 106.Wheeler TM, Thornton CA. Myotonic dystrophy: RNA-mediated muscle disease. Curr Opin Neurol. 2007;20:572–576. doi: 10.1097/WCO.0b013e3282ef6064. [DOI] [PubMed] [Google Scholar]
- 107.Brook JD, McCurrach ME, Harley HG, et al. Molecular basis of myotonic dystrophy: expansion of a trinucleotide (CTG) repeat at the 3' end of a transcript encoding a protein kinase family member. Cell. 1992;69:385. doi: 10.1016/0092-8674(92)90418-c. [DOI] [PubMed] [Google Scholar]
- 108.Mahadevan M, Tsilfidis C, Sabourin L, et al. Myotonic dystrophy mutation: an unstable CTG repeat in the 3' untranslated region of the gene. Science. 1992;255:1253–1255. doi: 10.1126/science.1546325. [DOI] [PubMed] [Google Scholar]
- 109.Taneja KL, McCurrach M, Schalling M, et al. Foci of trinucleotide repeat transcripts in nuclei of myotonic dystrophy cells and tissues. J Cell Biol. 1995;128:995–1002. doi: 10.1083/jcb.128.6.995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Davis BM, McCurrach ME, Taneja KL, et al. Expansion of a CUG trinucleotide repeat in the 3' untranslated region of myotonic dystrophy protein kinase transcripts results in nuclear retention of transcripts. Proc Natl Acad Sci U S A. 1997;94:7388–7393. doi: 10.1073/pnas.94.14.7388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Miller JW, Urbinati CR, Teng-Umnuay P, et al. Recruitment of human muscleblind proteins to (CUG)(n) expansions associated with myotonic dystrophy. EMBO J. 2000;19:4439–4448. doi: 10.1093/emboj/19.17.4439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.White MC, Gao R, Xu W, et al. Inactivation of hnRNP K by expanded intronic AUUCU repeat induces apoptosis via translocation of PKCdelta to mitochondria in spinocerebellar ataxia 10. PLoS Genet. 2010;6:e1000984. doi: 10.1371/journal.pgen.1000984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Wheeler TM, Lueck JD, Swanson MS, et al. Correction of ClC-1 splicing eliminates chloride channelopathy and myotonia in mouse models of myotonic dystrophy. J Clin Invest. 2007;117:3952–3957. doi: 10.1172/JCI33355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Sellier C, Rau F, Liu Y, et al. Sam68 sequestration and partial loss of function are associated with splicing alterations in FXTAS patients. EMBO J. 2010;29:1248–1261. doi: 10.1038/emboj.2010.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Rudnicki DD, Holmes SE, Lin MW, et al. Huntington's disease--like 2 is associated with CUG repeat-containing RNA foci. Ann Neurol. 2007;61:272–282. doi: 10.1002/ana.21081. [DOI] [PubMed] [Google Scholar]
- 116.Daughters RS, Tuttle DL, Gao W, et al. RNA gain-of-function in spinocerebellar ataxia type 8. PLoS Genet. 2009;5:e1000600. doi: 10.1371/journal.pgen.1000600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Ranum LP, Cooper TA. RNA-mediated neuromuscular disorders. Annu Rev Neurosci. 2006;29:259–277. doi: 10.1146/annurev.neuro.29.051605.113014. [DOI] [PubMed] [Google Scholar]
- 118.Radvansky J, Kadasi L. The expanding world of myotonic dystrophies: how can they be detected? Genet Test Mol Biomarkers. 2010;14:733–741. doi: 10.1089/gtmb.2010.0073. [DOI] [PubMed] [Google Scholar]
- 119.Martorell L, Monckton DG, Gamez J, et al. Progression of somatic CTG repeat length heterogeneity in the blood cells of myotonic dystrophy patients. Hum Mol Genet. 1998;7:307–312. doi: 10.1093/hmg/7.2.307. [DOI] [PubMed] [Google Scholar]
- 120.Anvret M, Ahlberg G, Grandell U, et al. Larger expansions of the CTG repeat in muscle compared to lymphocytes from patients with myotonic dystrophy. Hum Mol Genet. 1993;2:1397–1400. doi: 10.1093/hmg/2.9.1397. [DOI] [PubMed] [Google Scholar]
- 121.Lavedan C, Hofmann-Radvanyi H, Shelbourne P, et al. Myotonic dystrophy: size- and sex-dependent dynamics of CTG meiotic instability, and somatic mosaicism. Am J Hum Genet. 1993;52:875–883. [PMC free article] [PubMed] [Google Scholar]
- 122.Jansen G, Willems P, Coerwinkel M, et al. Gonosomal mosaicism in myotonic dystrophy patients: involvement of mitotic events in (CTG)n repeat variation and selection against extreme expansion in sperm. Am J Hum Genet. 1994;54:575–585. [PMC free article] [PubMed] [Google Scholar]
- 123.Thornton CA, Johnson K, Moxley RT., 3rd Myotonic dystrophy patients have larger CTG expansions in skeletal muscle than in leukocytes. Ann Neurol. 1994;35:104–107. doi: 10.1002/ana.410350116. [DOI] [PubMed] [Google Scholar]
- 124.Monckton DG, Wong LJ, Ashizawa T, Caskey CT. Somatic mosaicism, germline expansions, germline reversions and intergenerational reductions in myotonic dystrophy males: small pool PCR analyses. Hum Mol Genet. 1995;4:1–8. doi: 10.1093/hmg/4.1.1. [DOI] [PubMed] [Google Scholar]
- 125.Zatz M, Passos-Bueno MR, Cerqueira A, et al. Analysis of the CTG repeat in skeletal muscle of young and adult myotonic dystrophy patients: when does the expansion occur? Hum Mol Genet. 1995;4:401–406. doi: 10.1093/hmg/4.3.401. [DOI] [PubMed] [Google Scholar]
- 126.Kinoshita M, Takahashi R, Hasegawa T, et al. (CTG)n expansions in various tissues from a myotonic dystrophy patient. Muscle Nerve. 1996;19:240–242. doi: 10.1002/(SICI)1097-4598(199602)19:2<240::AID-MUS21>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 127.Turner C, Hilton-Jones D. The myotonic dystrophies: diagnosis and management. J Neurol Neurosurg Psychiatry. 2010;81:358–367. doi: 10.1136/jnnp.2008.158261. [DOI] [PubMed] [Google Scholar]
- 128.McComas AJ, Sica RE, Toyonaga K. Incidence, severity, and time-course of motoneurone dysfunction in myotonic dystrophy: their significance for an understanding of anticipation. J Neurol Neurosurg Psychiatry. 1978;41:882–893. doi: 10.1136/jnnp.41.10.882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Harley HG, Rundle SA, Reardon W, et al. Unstable DNA sequence in myotonic dystrophy. Lancet. 1992;339:1125–1128. doi: 10.1016/0140-6736(92)90729-m. [DOI] [PubMed] [Google Scholar]
- 130.Martorell L, Monckton DG, Sanchez A, et al. Frequency and stability of the myotonic dystrophy type 1 premutation. Neurology. 2001;56:328–335. doi: 10.1212/wnl.56.3.328. [DOI] [PubMed] [Google Scholar]
- 131.Korneluk RG, Narang MA. Anticipating anticipation. Nat Genet. 1997;15:119–120. doi: 10.1038/ng0297-119. [DOI] [PubMed] [Google Scholar]
- 132.Zheng CJ, Byers B, Moolgavkar SH. Allelic instability in mitosis: a unified model for dominant disorders. Proc Natl Acad Sci U S A. 1993;90:10178–10182. doi: 10.1073/pnas.90.21.10178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Harper PS, Harley HG, Reardon W, Shaw DJ. Anticipation in myotonic dystrophy: new light on an old problem. Am J Hum Genet. 1992;51:10–16. [PMC free article] [PubMed] [Google Scholar]
- 134.Warner JP, Barron LH, Goudie D, et al. A general method for the detection of large CAG repeat expansions by fluorescent PCR. J Med Genet. 1996;33:1022–1026. doi: 10.1136/jmg.33.12.1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Kamsteeg EJ, Kress W, Catalli C, et al. Best practice guidelines and recommendations on the molecular diagnosis of myotonic dystrophy types 1 and 2. Eur J Hum Genet. 2012 doi: 10.1038/ejhg.2012.108. In this review guidelines for the diagnosis of myotonic dystrophy type 1 and type 2 are discussed.
- 136.Wong LJ, Ashizawa T, Monckton DG, et al. Somatic heterogeneity of the CTG repeat in myotonic dystrophy is age and size dependent. Am J Hum Genet. 1995;56:114–122. [PMC free article] [PubMed] [Google Scholar]
- 137. Foff EP, Mahadevan MS. Therapeutics development in myotonic dystrophy type 1. Muscle Nerve. 2011;44:160–169. doi: 10.1002/mus.22090. This article reviews how understanding the RNA toxicity in myotonic dystrophy has created novel therapeutic strategies.
- 138. Blauw HM, van Rheenen W, Koppers M, et al. NIPA1 polyalanine repeat expansions are associated with amyotrophic lateral sclerosis. Hum Mol Genet. 2012;21:2497–2502. doi: 10.1093/hmg/dds064. Study that demonstrates that repeat expansions in NIPA1 are associated with amyotrophic lateral sclerosis.
- 139. Ross OA, Rutherford NJ, Baker M, et al. Ataxin-2 repeat-length variation and neurodegeneration. Hum Mol Genet. 2011;20:3207–3212. doi: 10.1093/hmg/ddr227. Ataxin-2 glutamine repeats are assessed in the spectrum of neurodegenerative diseases, and confirm its role as risk factor for ALS, possibly for other neurodegenerative diseases as well.
- 140. Van Damme P, Veldink JH, van Blitterswijk M, et al. Expanded ATXN2 CAG repeat size in ALS identifies genetic overlap between ALS and SCA2. Neurology. 2011;76:2066–2072. doi: 10.1212/WNL.0b013e31821f445b. This paper reveals a genetic overlap between amyotrophic lateral sclerosis and spinocerebellar ataxia type 2, while both diseases are associated with ataxin-2 repeat sizes.
- 141.Elden AC, Kim HJ, Hart MP, et al. Ataxin-2 intermediate-length polyglutamine expansions are associated with increased risk for ALS. Nature. 2010;466:1069–1075. doi: 10.1038/nature09320. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.