Abstract
Instability of repetitive DNA sequences within the genome is associated with a number of human diseases. The expansion of trinucleotide repeats is recognized as a major cause of neurological and neuromuscular diseases, and progress in understanding the mutations over the last 20 years has been substantial. Here we provide a brief summary of progress with an emphasis on technical advances at different stages.
Keywords: Trinucleotide repeat, Genomic instability, Triplet repeat expansion, Huntington’s disease, Fragile X syndrome, Myotonic dystrophy, Spinocerebellar ataxia, Neurodegeneration
1 Introduction
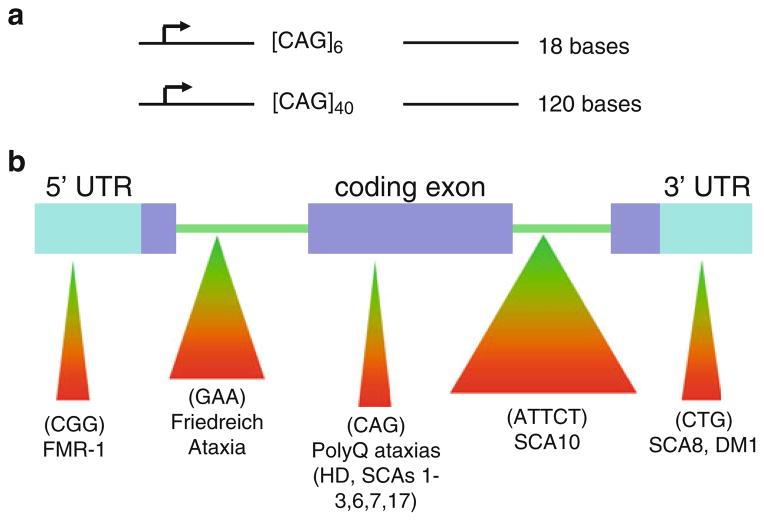
The unstable and dynamic transmission of simple repetitive elements in DNA is a new type of mutation, which has changed the face of genetics [1]. The mutation, referred to as “trinucleotide repeat (TNR) expansion,” occurs when the number of triplets present in a mutated gene is greater than the number found in a normal gene [1–3]. Additionally, the number of triplets in the disease gene continues to increase as the disease gene is inherited (Fig. 1a). As the TNR number grows, the growing triplet tract alters gene expression and/or function of the gene product; TNRs residing in a coding sequence of a gene typically produce a faulty protein, while expansion of a TNR in a noncoding gene region suppresses protein expression, alters its splicing, or may influence aspects of antisense regulation (Fig. 1b) [1–3]. For example, in Huntington’s disease (HD), unaffected individuals may have roughly 6–29 CAG triplets in both alleles; yet, in HD patients, the disease allele may contain 36 to hundreds of CAG triplets. As the TNR repeat number grows, the growing polyglutamine tract produces an HD gene product (called huntingtin) with increasingly aberrant properties that causes death of brain cells controlling movement [1–3].
Fig. 1.
Trinucleotide repeat expansion disorders caused by triplet repeats in coding and noncoding gene regions. (a) Inheritance of disease genes and parent–child transmission causes rapid expansion of repeat regions. (b) Triplet repeats residing in coding and noncoding sequences of a gene have significant impact on human health and underlie many severe neurological disorders
The triplet repeat tract is one of many microsatellites that are defined as simple sequences 1–6 nucleotides that are repeated multiple times [4]. Microsatellites constitute 30 % of the human genome in most species [5, 6], but the triplet repeat has taken on special significance due to its highly unstable nature. Alterations in the lengths of repetitive DNA over evolutionary time scales create diversity in the species [7, 8]. However, the alteration in TNR length observed in human expansion diseases is rapid and surprising. Mammals have developed systems for resisting rapid changes that could be deleterious [1]. Consequently, the major concept of mutation was the Mendelian transmission of a stable nucleotide sequence change, most often as a missense change [9], or a stable translocation in DNA [7] that altered gene expression [10, 11]. Yet, these simple triplets beyond a critical threshold length override genomic safeguards and expand during most parent–child transmissions and during the lifetime of an organism. The list of diseases associated with insertion and deletions of microsatellite sequences has grown over the years, and the tracts in these disease genes do not exclusively comprise three nucleotide repetitive units [2, 3]. However, the consequences of triplet instability on human health are profound: the unstable maintenance of simple repeats at or near genes leads to the mutation underlying dozens of severe neuromuscular and neurodegenerative disorders (Figs. 1 and 2) [2, 3].
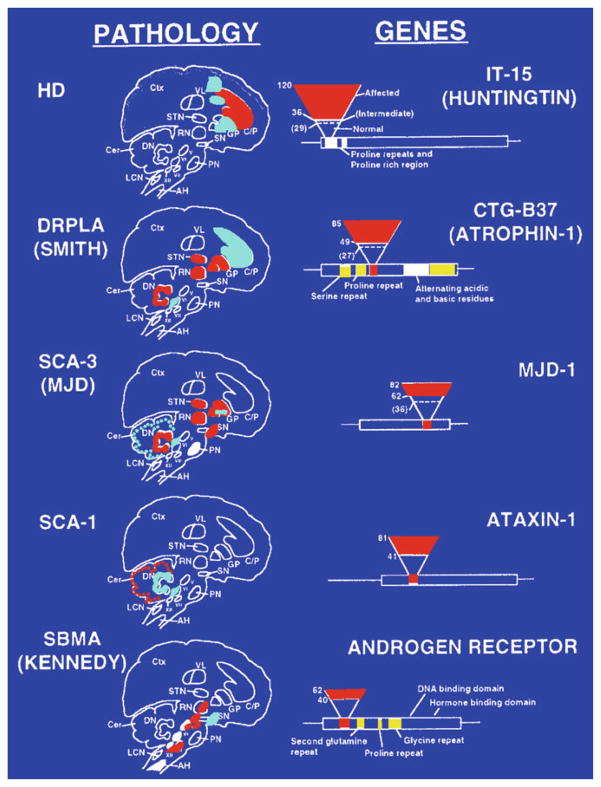
Fig. 2.
Pathophysiology of trinucleotide repeat disorders and effect of repeat number. Affected brain regions and regions of neuronal loss in neurological disorders associated with TNR expansion are shown on the left. Red regions indicate the major affected areas and areas of neuronal loss. In HD, patients with 36–120 CAG repeats are affected by the disease and show neuronal loss in regions of the brain that control movement. Schematic representations of affected genes are shown on the left. Repeat regions within each gene are indicated by the small bar in the coding region of the gene. The inverted triangle represents an increasing number of repeats. The base of the triangle represents unaffected individuals; dotted lines indicate unaffected carriers for disease, and the red part of the triangle indicates affected individuals. C/P caudate/putamen, CTX cortex, GP globus pallidus, STN subthalamic nucleus, VL ventrolateral thalamic nucleus, SN substantia nigra
Advances in understanding the expansion diseases, at each step of the discovery process, have relied on the development of new methods and approaches. In the first edition of Trinucleotide repeat protocols [12], methods for generating transgenic animals and cellular models for disease, as well as methods for quantifying triplet repeat lengths in cells, humans, and in animals were highlighted. The second edition of Trinucleotide repeat protocols builds on and extends these themes to include more sophisticated TNR detection methods, new animal models for quantifying expansion, and new experimental approaches that address both TNR disease pathogenesis and therapeutics.
2 Early Clinical Detection of TNRs and Historical Perspective
Approaches to evaluating the changing length of mutagenic TNR tracts and its effects on pathophysiology have coevolved over the years. Intergenerational trinucleotide expansion explained a long observed clinical feature of genetic disease called “anticipation,” in which disease onset inversely correlates with the length of the repeating segment, and the disease phenotype of those affected worsens from generation to generation (Fig. 3) [13, 14]. Indeed, the link between the TNR and the onset of disease took nearly 200 years to recognize [13]. Anticipation in genetic disorders was documented as early as the mid-nineteenth century, yet most geneticists had dismissed anticipation as an ascertainment bias, i.e., the relationship between the diagnosis and symptom onset was not recorded accurately [14]. However, the discovery of CGG expansion in the 5′ untranslated region of the fragile X mental retardation 1 (FMR1) gene in fragile X syndrome (FXS) (1991) [15], CAG expansion in the coding sequence of X-linked spinal and bulbar muscular atrophy (SMBA) (1991) [16], CTG expansion in the 3′ untranslated region of myotonic dystrophy protein kinase (DMPK) in myotonic dystrophy type 1 (DM1) (1992) [17], and CAG expansion in the exon 1 coding sequence of Huntington’s disease (HD) (1993) [18] provided evidence that expansion of triplet tracts was the underlying mutation in disease. Furthermore, the anticipation phenotype might arise from a changing number of repeats in the dynamic expansion. The results led to speculation that expansion was likely to be the underlying cause of additional diseases that shared an anticipation phenotype, a prediction that has been corroborated [13].
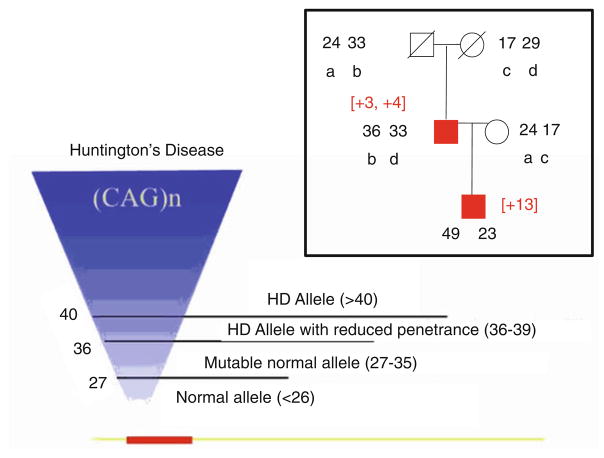
Fig. 3.
Effects of CAG expansion in the huntingtin gene. A representative two-generation pedigree of an HD family. Squares are males; circles are females. Red boxes indicate affected individuals. Open circles are unrelated spouses. Black numbers represent CAG repeat number in each allele of the affected family members. Small black letters indicate the alleles present in father-to-son transmission. The number in brackets represents the size of the CAG expansion during inheritance. The relationship between HD and CAG repeat number (left). In this schematic representation of the HD gene, the open bar represents the coding region of the Huntington’s gene (called huntingtin); the small red bar indicates the position of the CAG repeat stretch located within the N-terminal portion of the coding sequence. The inverted triangle represents an increasing number of CAG repeats. The base of triangle represents unaffected individuals with 6–26 CAG repeats; lines indicate unaffected carriers for disease with 27–35 CAG repeats; and the top part of the triangle indicates affected individuals with 36–120 CAG repeats
Over the next decade, relating the length and dynamics of the triplet repeat mutation to its effects on the disease phenotype took center stage. Early focus was placed on documenting the TNR dynamics during parent–child transmission (Fig. 4) [19–21]. Indeed, the correlation between the length of the parental disease tract and the onset of disease in the progeny is so strong that repeat length is often used clinically to diagnose the disease, to predict disease onset, and to provide insight into aspects of disease outcome (Fig. 4) [19–21]. Repeat expansions are difficult to measure, and although approaches are now well established, developing methods for accurate detection took years. Polymerase chain reaction (PCR) gives rise to multiple shadow bands that are generated during sizing of the repeats [22] and rendered tract sizing difficult to measure accurately. Thus, it was difficult to consider questions such as whether disease was dominated by smaller numbers of short expansions or a small number of long expansions. The need to more stringently link repeat length and pathophysiology prompted development of small pool PCR (SP-PCR) [23, 24], which provided a method to precisely quantify the frequency of the repeat changes. The method has been valuable in assessing the variability of expansion in blood, sperm, and in somatic cells from human patients and mouse samples [12, 23, 24]. Long CGG tracts, even today, are measured by Southern blot, since many polymerases have difficulty in traversing heavily CG-rich tracts if they are long [12]. Together, these methods provide up to 99 % accuracy needed for clinical analysis.
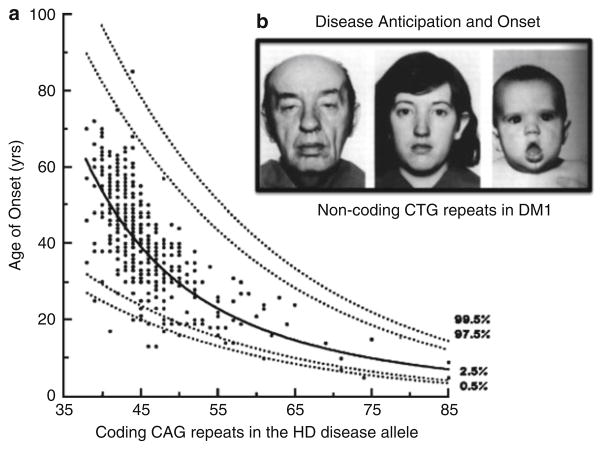
Fig. 4.
Effect of repeat number on age of onset and anticipation. (a) Age of onset is inversely correlated with repeat number. Increasing CAG repeats in the HD disease gene decreases the age of onset of symptoms of the disease. Early onset/juvenile HD symptoms can be seen at repeat numbers above 60. (b) Anticipation phenotype in a three-generation family showing the increased severity in successive generations. Grandfather (left), symptoms of myotonia since age 50, but no significant disability. Mother (middle), myotonia since late teens. Son (right), congenital myotonic dystrophy
3 Genetic Modifiers of Disease: Somatic Expansion and Chromatin
The ability to link clinical age of onset with a precise length of the trinucleotide tract brought to light a second key feature of expansion disease (Fig. 4); onset is highly variable [2, 19–21]. For example, in HD, the inherited repeat length can predict the average age of onset (Fig. 4), but onset in an individual patient with a given repeat length can vary as much as fourfold (onset from 18 to 80 years of age) from the predicted average (Fig. 4). The high degree of variability has suggested the existence of genetic modifiers, and the hunt for these factors has captured significant attention [25–29]. However, to date, the most important modifier influencing disease onset appears to be the variability of CAG tract length itself [30, 31].
During the last decade, somatic variations in repeat copy number have been measured in most human triplet diseases [32–39], and in most mouse models for triplet diseases, including FXS [40, 41], DM-1 [42, 43], SCA1 [44], and HD [45–48], among many others. But do somatic variations in repeat length influence the onset of disease? In many cases somatic expansions are comparable in size to those that are inherited. In HD, for example, pathophysiology is so dependent on the size of the inherited repeat that growth by a single CAG repeat can reduce the age of onset of an affected patient by 5 years. Heterogeneous and prominent expansions of up to thousands occur in the striatum, the most vulnerable region of the brain [30, 32]. Thus, some of the CAG repeat expansions in the HD brain far exceed those that are inherited. By logical extension, then, the somatic growth of the repeat in human brain cells would be predicted to cause a more severe disease phenotype with age since disease proteins of longer polyglutamine tracts are progressively produced. In animals, somatic expansion of the CAG tract continues progressively throughout the lifetime of animals as they develop toxic phenotypes.
Whether somatic expansion influences the onset of disease is, however, difficult to rigorously determine in humans, since their brains cannot be evaluated until death. Nonetheless, recent evidence in human postmortem HD brain suggests that the somatic repeat length matters [48]. Somatic instability in the cortex region of the brain from a cohort of HD individuals exhibited phenotypic extremes of young and old disease onset as predicted by the length of a reference allele measured in the cerebellum [48]. After accounting for reference repeat length, larger repeat length gains associated with earlier disease onset. Many variables need to be sorted out before firm conclusions can be drawn. Humans vary in their diets, lifestyles, and how they deal with stress. Moreover, the effects of the inherited repeat and the somatic alleles on disease onset cannot be cleanly separated in postmortem brains. However, the human data are consistent with the hypothesis that somatic HD CAG repeat length expansions in target tissues contribute to the HD pathogenic process.
Far less is known concerning a second genetic modifier, the effects of chromatin packaging on triplet repeat instability [49]. Expansion must operate within the context of chromatin, and there is growing interest in exploring how chromatin structure and epigenetic modifications influence expansion [49]. For example, a study of the human Ataxin-7 locus in transgenic mice has established a link between binding of CCCTC binding factor (CTCF, a regulatory protein implicated in DNA conformation and genomic imprinting) and regulation of repeat instability [50]. Mutation of the CCCTC sequence in the Ataxin-7 locus, a CTCF binding site next to an expanded repeat sequence, enhanced instability [50]. Currently, the links among epigenetic changes and expansion remain enigmatic, but the influence of genome locus, posttranslational modification of histones, and DNA methylation on TNR expansion will be key issues to explore.
4 The Genetic Mechanism of Expansion
Linking the length and dynamics of the triplet repeat mutation lies at the heart of triplet repeat disorders, and interest in the genetic mechanisms for the expansion exploded concomitantly with discovery of the disease genes. Structural and molecular analyses between 1995 and 2005 were key in solidifying the hypothesis that stable secondary structures were the intermediates of expansion [2, 3, 49]. The frequency of expansion depends on structure-forming sequences and increases with the length of CGG [51], CAG [52], CTG [53], and GAA [54] tracts. Visualizing a hairpin structure in vivo is not possible at current resolution limits. However, the model is consistent with most genetic data, and structure-forming heteroduplex loops comprising triplet repeats are refractory to repair during meiosis in yeast, while other repeats and random sequence loops are removed [55].
Expansions occur in both dividing and nondividing cells [2, 3, 49] implying that the mutation could occur under conditions of proliferation or repair. A case for DNA repair as an expansion mechanism arose from a seminal work in 1999 indicating that MutS-homologue 2 (MSH2), part of the mismatch repair recognition complex, caused rather than corrected expansion [56, 57]. In mouse models of HD [58] or DM1 [59], loss of the MSH2–MSH3 complex or its binding partner, PMS2 [60], suppresses expansion. MSH2–MSH6 had small, tissue-specific effects [58, 59]. Since then, mouse models harboring triplet repeat tracts of disease length have been crossed with animals lacking key DNA damage recognition proteins for the major pathways of DNA repair [2]. Loss of XPC (nucleotide excision repair) [61], Cockayne’s syndrome B (CSB) (transcription coupled repair) [62], Rad52 (homologous recombination) [42], Rad54 (homologous recombination) [42], DNA-PK Catalytic subunit (nonhomologous end joining) [42], or Ku (nonhomologous end joining) [42] has little effect on expansion. However, recent findings imply that expansion occurs in the process of removing oxidative DNA damage that accumulates with age. Furthermore, in these cases, expansion is driven by the action of 7,8-dihyro-8-oxoguanine DNA glycosylase (OGG1) [63–65], a member of the base excision repair pathway. The results in mice, however, have not always agreed with those of cell lines or flies. Indeed, in mice, loss of CSB has little effect on expansion. However, in Drosophila melanogaster, transcriptional induction of a transgene (under the control of regulatory protein GAl4) that contains exon 10 of ataxin 3 (ATXN3, also known as SCA3) results in expansions and contractions of the TNR tract within this exon, and loss of XPG in this line reduced instability [66]. In human cell lines, contraction of long CAG repeats within an intron of an active hypox-anthine guanine phosphoribosyl transferase (HPRT) minigene [67] is reduced by small interfering RNA knockdown of ERCC1, XPG, and CSB. Thus, whatever the mechanism, the looped DNA remains uncorrected. The resulting insertion and deletion mutations, in this case, will be “site-specific” in that they are limited to particular locations where the repair-resistant lesions reside. The properties of trinucleotide expansion characterize this type of mutation.
5 Protein and RNA-Mediated Toxicity of Triplet Repeat Expansion Disorders
Quantifying the effects of expansion on the pathology of triplet expansion disease in humans has been the focus of clinical and basic research for nearly 20 years. Most early work in model systems focused on the toxicity induced by the expanded gene products of repeats as dictating DNA structural abnormalities or toxicity from mutant protein expression. However, more recently, RNA toxicity has come to the forefront as an additional mechanism. Over the past 10 years, a growing list of neurological disorders have been identified that share an RNA-dependent mechanism of toxicity. For example, myotonic dystrophy type 1 (DM1) is caused by a CTG expansion that is transcribed but not translated [68]. CCTG expansion in an intron causes myotonic dystrophy type 2 (DM2) [69]. These findings highlighted a new type of mechanism in which microsatellite expansion mutations cause disease through an RNA gain-of-function mechanism. The growing number of dominant noncoding expansion disorders includes fragile X tremor ataxia syndrome (FXTAS), spinocerebellar ataxia type 8 (SCA8), SCA10, SCA12, and Huntington’s disease-like 2 (HDL2) [70]. Pathogenic molecular events initiated by expanded RNA products are often mediated through aberrant effects comprising RNA-binding proteins that interact with the repeats. Expanded RNA products are known to aggregate into nuclear RNA foci [71]. These are dynamic, protein-containing structures that are hallmarks of myotonic dystrophy (DM) and may play important roles in other RNA-toxic disorders [72]. The mechanisms of toxic RNA-mediated pathogenesis are as yet poorly understood. However, the effects of RNA toxicity are likely to be as complex as protein-induced toxicity and are likely to include transcriptional alterations, sequestration of mRNA-associated protein complexes, aberrant mRNA splicing and processing, abnormal signaling cascades, and failure of protein quality control pathways [73, 74].
The largest group of triplet repeat diseases remains those in which the expansion resides in the gene coding sequences and comprise expression of expanded polyglutamine tracts. Why expanded proteins result in systemic toxicity or why an expanded gene product preferentially kills neurons in select brain regions is poorly understood. However, the expanded polyglutamine proteins have effects on diverse biological transactions including vesicle trafficking, lipid metabolism, mitochondrial (MT) dysfunction [75], autophagy [76–78], and transcription [76, 79]. Whether all of these defects have equivalent impacts on toxicity is unknown. However, three common processes have received significant attention: autophagy, trafficking, and MT dysfunction, all of which are intertwined with lipid homeostasis. There is a general consensus that mhtt influences membrane trafficking, lysosomal biogenesis, and autophagy, the “waste management” system of the cell [80]. Autophagy depends on membrane fusion events of endosomes, peroxisomes, and dysfunctional MT with the lysosomes for export out of the cell. Defects in autophagic responses have been noted in HD [76], SMBA [77], and PD [78, 79]. Similarly, mitochondrial defects are measurable by changes in membrane potential, low threshold for mitochondrial transition pore opening, and low ATP production. Indeed, HD, Alzheimer’s (AD), and Parkinson’s (PD) share the property that MT struggle to meet the energy demands of the cell [75]. Imbalance and/or alterations in lipid metabolism are likely to affect organelle integrity and may connect or influence all of these phenotypes.
6 Therapeutics
Efforts to screen for compounds that reduce protein-dependent toxicity and oxidative DNA damage have been intense, but have not yet yielded therapeutics that effectively offset the onset of disease [81–83]. Although the search for small molecules continues, novel therapeutic strategies towards triplet repeat disease have advanced on three fronts: DNA, RNA, and protein. If somatic expansion is a factor in onset and progression of disease, then blocking somatic expansion should be beneficial. Thus, therapeutic approaches to reduce expansion have garnered intense interest, although effective molecular inhibitors of DNA expansion have yet to be identified. DNA repair proteins promote expansion, and inhibiting them is a possible strategy to regulate triplet repeat length. However, DNA repair plays essential roles in mutation avoidance at other sites. Thus, a challenge in this effort will be to identify molecules that inhibit expansion without altering the normal level of DNA repair.
Perhaps the most rapidly evolving technologies in the triplet field are therapeutics targeting the aberrant protein products or RNA using siRNA [84, 85]. siRNA “knockdown” of the mutant huntingtin protein indicates that silencing the mutant HD transgene significantly inhibits neurodegeneration, improves motor control, and extends survival of HD mice [86, 87]. The product of the normal gene is required for cell survival and development. Therefore, a major challenge in any siRNA approach is to inhibit expression of the mutant gene while allowing expression of its normal counterpart. With future improvement of mutant allele selectivity (preserving the expression of the neuroprotective wild-type allele), target specificity, efficacy, and safety, as well as optimization of delivery methods, the efficacy of small noncoding RNA-based therapeutic applications is likely to improve [88].
Additional strategies against RNA-induced toxicity include the use of antisense RNA. CAG repeat antisense oligonucleotides are effective in DM1 models by altering the protein interactions or metabolism of the toxic RNA [89]. The CAG repeat antisense oligonucleotides are designed to bind to the CUG repeat RNA and to block toxic RNA–protein interactions. Release of the expanded CUG transcripts from nuclear foci presumably facilitates their transport to the cytoplasm, where they undergo rapid decay. As an added benefit, somatic instability of CTG repeats in the DMPK gene is suppressed after treatment with antisense oligonucleotides [89]. These results raise the possibility that early intervention with antisense oligonucleotides can reduce RNA or protein toxicity and may have the additional benefit of stabilizing CTG:CAG repeats at subpathogenic lengths.
With the advent of inducible pluripotent stem cells (iPS), stem cell replacement therapy is a promising avenue [90, 91]. Stem cells are derived from fetal tissue but have the potential to develop into normal brain cells if surgically implanted into the brain [92], and surgical strategies for transplantation of stem cells to replace lost neurons in the brain are being explored [90–92]. Induction of pluripotent stem cells (iPS) by addition of transcription factor “cocktails” is being tested to directly reprogram human somatic cells for replacement therapy [93, 94]. Thus, iPS cells provide hope that the severity of pathophysiology might be reduced or its onset delayed by replacing dying neurons with renewable cells. These procedures are experimental, and it is not yet clear if they will be effective. However, embryonic grafts have improved motor functions in both animals and in humans [90, 93–95].
7 Conclusions
Our understanding of the pathogenic mechanisms for trinucleotide repeat (TNR) expansion has advanced substantially in recent years. While many aspects of toxicity and the mutational mechanisms remain enigmatic, new models for DNA, RNA, and protein provide increasing ability to inhibit decline for these deadly diseases. Although no effective therapy is yet around the corner, hope for intervention is on the horizon.
Acknowledgments
I would like to thank Christie A. Canaria, Virginia Platt (NIH/NIA T32-AG00266), Do Yup Lee, Nelson Chan, Ella Xun, James Lim, and support from the National Institutes of Health Grants NS069177, NS40738, NS062384, and NS060115.
References
- 1.Hegde MV, Saraph AA. Unstable genes unstable mind: beyond the central dogma of molecular biology. Med Hypotheses. 2011;77:165–170. doi: 10.1016/j.mehy.2011.03.051. [DOI] [PubMed] [Google Scholar]
- 2.McMurray CT. Mechanisms of trinucleotide repeat instability during human development. Nat Rev Genet. 2010;11:786–799. doi: 10.1038/nrg2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.La Spada AR, Taylor JP. Repeat expansion disease: progress and puzzles in disease pathogenesis. Nat Rev Genet. 2010;11:247–258. doi: 10.1038/nrg2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Toth G, Gaspari Z, Jurka J. Microsatellites in different eukaryotic genomes: survey and analysis. Genome Res. 2000;10:967–981. doi: 10.1101/gr.10.7.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jurka J, Pethiyagoda C. Simple repetitive DNA sequences from primates: compilation and analysis. J Mol Evol. 1995;40:120–126. doi: 10.1007/BF00167107. [DOI] [PubMed] [Google Scholar]
- 6.Beckman JS, Weber JL. Survey of human and rat microsatellites. Genomics. 1992;12:627–631. doi: 10.1016/0888-7543(92)90285-z. [DOI] [PubMed] [Google Scholar]
- 7.Jeffreys AJ, Holloway JK, Kauppi L, et al. Meiotic recombination hot spots and human DNA diversity. Philos Trans R Soc Lond B Biol Sci. 2004;359:141–152. doi: 10.1098/rstb.2003.1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kashi Y, King DG. Simple sequence repeats as advantageous mutators in evolution. Trends Genet. 2006;22:253–259. doi: 10.1016/j.tig.2006.03.005. [DOI] [PubMed] [Google Scholar]
- 9.Strachan T, Read AP. Human molecular genetics. Vol. 2. Wiley-Liss; New York: 1999. [Google Scholar]
- 10.Djian P, Hancock JM, Chana HS. Codon repeats in genes associated with human diseases: fewer repeats in the genes of nonhuman primates and nucleotide substitutions concentrated at the sites of reiteration. Proc Natl Acad Sci U S A. 1996;93:417–421. doi: 10.1073/pnas.93.1.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yant SR, Wu X, Huang Y, et al. High-resolution genome-wide mapping of transposon integration in mammals. Mol Cell Biol. 2005;25:2085–2094. doi: 10.1128/MCB.25.6.2085-2094.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kohwi Y. Methods in molecular biology. Vol. 277. Humana; Totowa, NJ: 2004. Trinucleotide repeat protocols. [Google Scholar]
- 13.Friedman JE. Anticipation in hereditary disease: the history of a biomedical concept. Hum Genet. 2011;130:705–714. doi: 10.1007/s00439-011-1022-9. [DOI] [PubMed] [Google Scholar]
- 14.Maltecca F, Filla A, Castaldo I, et al. Intergenerational instability and marked anticipation in SCA-17. Neurology. 2003;61:1441–1443. doi: 10.1212/01.wnl.0000094123.09098.a0. [DOI] [PubMed] [Google Scholar]
- 15.Verkerk AJ, Pieretti M, Sutcliffe JS, et al. Identification of a gene (FMR-1) containing a CGG repeat coincident with a breakpoint cluster region exhibiting length variation in fragile X syndrome. Cell. 1991;65:905–914. doi: 10.1016/0092-8674(91)90397-h. [DOI] [PubMed] [Google Scholar]
- 16.La Spada AR, Wilson EM, Lubahn DB, et al. Androgen receptor gene mutations in X-linked spinal and bulbar muscular atrophy. Nature. 1991;352:77–79. doi: 10.1038/352077a0. [DOI] [PubMed] [Google Scholar]
- 17.Mahadevan M, Tsilfidis C, Sabourin L, et al. Myotonic dystrophy mutation: an unstable CTG repeat in the 3′ untranslated region of the gene. Science. 1992;255:1253–1255. doi: 10.1126/science.1546325. [DOI] [PubMed] [Google Scholar]
- 18.The Huntington’s Disease Collaborative Research Group. A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington’s disease chromosomes. Cell. 1993;72:971–983. doi: 10.1016/0092-8674(93)90585-e. [DOI] [PubMed] [Google Scholar]
- 19.Duyao M, Ambrose C, Myers R, et al. Trinucleotide repeat length instability and age of onset in Huntington’s disease. Nat Genet. 1993;4:387–392. doi: 10.1038/ng0893-387. [DOI] [PubMed] [Google Scholar]
- 20.Andresen JM, Gayan J, Djousse L, et al. The relationship between CAG repeat length and age of onset differs for Huntington’s disease patients with juvenile onset or adult onset. Ann Hum Genet. 2007;71:295–301. doi: 10.1111/j.1469-1809.2006.00335.x. [DOI] [PubMed] [Google Scholar]
- 21.Filla A, De Michele G, Cavalcanti F, et al. The relationship between trinucleotide (GAA) repeat length and clinical features in Friedreich ataxia. Am J Hum Genet. 1996;59:554–560. [PMC free article] [PubMed] [Google Scholar]
- 22.Bovo D, Rugge M, Shiao YH. Origin of spurious multiple bands in the amplification of microsatellite sequences. Mol Pathol. 1999;52:50–51. doi: 10.1136/mp.52.1.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leeflang EP, Zhang L, Tavare S, et al. Single sperm analysis of the trinucleotide repeats in the Huntington’s disease gene: quantification of the mutation frequency spectrum. Hum Mol Genet. 1995;4:1519–1526. doi: 10.1093/hmg/4.9.1519. [DOI] [PubMed] [Google Scholar]
- 24.Monckton DG, Wong LJ, Ashizawa T, et al. Somatic mosaicism, germline expansions, germline reversions and intergenerational reductions in myotonic dystrophy males: small pool PCR analyses. Hum Mol Genet. 1995;4:1–8. doi: 10.1093/hmg/4.1.1. [DOI] [PubMed] [Google Scholar]
- 25.Gayan J, Brocklebank D, Andresen JM, et al. Genomewide linkage scan reveals novel loci modifying age of onset of Huntington’s disease in the Venezuelan HD kindreds. Genet Epidemiol. 2008;32:445–453. doi: 10.1002/gepi.20317. [DOI] [PubMed] [Google Scholar]
- 26.Arning L, Monte D, Hansen W, et al. ASK1 and MAP2K6 as modifiers of age at onset in Huntington’s disease. J Mol Med (Berl) 2008;86:485–490. doi: 10.1007/s00109-007-0299-6. [DOI] [PubMed] [Google Scholar]
- 27.Djousse L, Knowlton B, Hayden MR, et al. Evidence for a modifier of onset age in Huntington disease linked to the HD gene in 4p16. Neurogenetics. 2004;5:109–114. doi: 10.1007/s10048-004-0175-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee JM, Gillis T, Mysore JS, et al. Common SNP-based haplotype analysis of the 4p16.3 Huntington disease gene region. Am J Hum Genet. 2012;90:434–444. doi: 10.1016/j.ajhg.2012.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li JL, Hayden MR, Warby SC, et al. Genome-wide significance for a modifier of age at neurological onset in Huntington’s disease at 6q23-24: the HD MAPS study. BMC Med Genet. 2006;7:71. doi: 10.1186/1471-2350-7-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Veitch NJ, Ennis M, McAbney JP, et al. Inherited CAG.CTG allele length is a major modifier of somatic mutation length variability in Huntington disease. DNA Repair (Amst) 2007;6:789–796. doi: 10.1016/j.dnarep.2007.01.002. [DOI] [PubMed] [Google Scholar]
- 31.Lee JM, Ramos EM, Lee JH, et al. CAG repeat expansion in Huntington disease determines age at onset in a fully dominant fashion. Neurology. 2012;78:690–695. doi: 10.1212/WNL.0b013e318249f683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shelbourne PF, Keller-McGandy C, Bi WL, et al. Triplet repeat mutation length gains correlate with cell-type specific vulnerability in Huntington disease brain. Hum Mol Genet. 2007;16:1133–1142. doi: 10.1093/hmg/ddm054. [DOI] [PubMed] [Google Scholar]
- 33.De Rooij KE, De Koning Gans PA, Roos RA, et al. Somatic expansion of the (CAG)n repeat in Huntington disease brains. Hum Genet. 1995;95:270–274. doi: 10.1007/BF00225192. [DOI] [PubMed] [Google Scholar]
- 34.Telenius H, Kremer B, Goldberg YP, et al. Somatic and gonadal mosaicism of the Huntington disease gene CAG repeat in brain and sperm. Nat Genet. 1994;6:409–414. doi: 10.1038/ng0494-409. [DOI] [PubMed] [Google Scholar]
- 35.Nolin SL, Ding XH, Houck GE, et al. Fragile X full mutation alleles composed of few alleles: implications for CGG repeat expansion. Am J Med Genet A. 2008;146A:60–65. doi: 10.1002/ajmg.a.32087. [DOI] [PubMed] [Google Scholar]
- 36.Koefoed P, Hasholt L, Fenger K, et al. Mitotic and meiotic instability of the CAG trinucleotide repeat in spinocerebellar ataxia type 1. Hum Genet. 1998;103:564–569. doi: 10.1007/s004390050870. [DOI] [PubMed] [Google Scholar]
- 37.Goellner GM, Tester D, Thibodeau S, et al. Different mechanisms underlie DNA instability in Huntington disease and colorectal cancer. Am J Hum Genet. 1997;60:879–890. [PMC free article] [PubMed] [Google Scholar]
- 38.Norremolle A, Sorensen SA, Fenger K, et al. Correlation between magnitude of CAG repeat length alterations and length of the paternal repeat in paternally inherited Huntington’s disease. Clin Genet. 1995;47:113–117. doi: 10.1111/j.1399-0004.1995.tb03941.x. [DOI] [PubMed] [Google Scholar]
- 39.Martorell L, Gamez J, Cayuela ML, et al. Germline mutational dynamics in myotonic dystrophy type 1 males: allele length and age effects. Neurology. 2004;62:269–274. doi: 10.1212/wnl.62.2.269. [DOI] [PubMed] [Google Scholar]
- 40.Qin M, Entezam A, Usdin K, et al. A mouse model of the fragile X premutation: effects on behavior, dendrite morphology, and regional rates of cerebral protein synthesis. Neurobiol Dis. 2011;42:85–98. doi: 10.1016/j.nbd.2011.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Entezam A, Biacsi R, Orrison B, et al. Regional FMRP deficits and large repeat expansions into the full mutation range in a new Fragile X premutation mouse model. Gene. 2007;395:125–134. doi: 10.1016/j.gene.2007.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Savouret C, Brisson E, Essers J, et al. CTG repeat instability and size variation timing in DNA repair-deficient mice. EMBO J. 2003;22:2264–2273. doi: 10.1093/emboj/cdg202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fortune MT, Vassilopoulos C, Coolbaugh MI, et al. Dramatic, expansion-biased, age-dependent, tissue-specific somatic mosaicism in a transgenic mouse model of triplet repeat instability. Hum Mol Genet. 2000;9:439–445. doi: 10.1093/hmg/9.3.439. [DOI] [PubMed] [Google Scholar]
- 44.Sato T, Oyake M, Nakamura K, et al. Transgenic mice harboring a full-length human mutant DRPLA gene exhibit age-dependent intergenerational and somatic instabilities of CAG repeats comparable with those in DRPLA patients. Hum Mol Genet. 1999;8:99–106. doi: 10.1093/hmg/8.1.99. [DOI] [PubMed] [Google Scholar]
- 45.Kennedy L, Shelbourne PF. Dramatic mutation instability in HD mouse striatum: does polyglutamine load contribute to cell-specific vulnerability in Huntington’s disease? Hum Mol Genet. 2000;9:2539–2544. doi: 10.1093/hmg/9.17.2539. [DOI] [PubMed] [Google Scholar]
- 46.Kaytor MD, Burright EN, Duvick LA, et al. Increased trinucleotide repeat instability with advanced maternal age. Hum Mol Genet. 1997;6:2135–2139. doi: 10.1093/hmg/6.12.2135. [DOI] [PubMed] [Google Scholar]
- 47.Kennedy L, Evans E, Chen CM, et al. Dramatic tissue-specific mutation length increases are an early molecular event in Huntington disease pathogenesis. Hum Mol Genet. 2003;12:3359–3367. doi: 10.1093/hmg/ddg352. [DOI] [PubMed] [Google Scholar]
- 48.Swami M, Hendricks AE, Gillis T, et al. Somatic expansion of the Huntington’s disease CAG repeat in the brain is associated with an earlier age of disease onset. Hum Mol Genet. 2009;18:3039–3047. doi: 10.1093/hmg/ddp242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dion V, Wilson JH. Instability and chromatin structure of expanded trinucleotide repeats. Trends Genet. 2009;25:288–297. doi: 10.1016/j.tig.2009.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Libby RT, Hagerman KA, Pineda VV, et al. CTCF cis-regulates trinucleotide repeat instability in an epigenetic manner: a novel basis for mutational hot spot determination. PLoS Genet. 2008;4:e1000257. doi: 10.1371/journal.pgen.1000257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fry M, Loeb LA. The fragile X syndrome d(CGG)n nucleotide repeats form a stable tetrahelical structure. Proc Natl Acad Sci U S A. 1994;91:4950–4954. doi: 10.1073/pnas.91.11.4950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pearson CE, Tam M, Wang YH, et al. Slipped-strand DNAs formed by long (CAG)*(CTG) repeats: slipped-out repeats and slip-out junctions. Nucleic Acids Res. 2002;30:4534–4547. doi: 10.1093/nar/gkf572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pearson CE, Wang YH, Griffith JD, et al. Structural analysis of slipped-strand DNA (S-DNA) formed in (CTG)n. (CAG)n repeats from the myotonic dystrophy locus. Nucleic Acids Res. 1998;26:816–823. doi: 10.1093/nar/26.3.816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gacy AM, Goellner GM, Spiro C, et al. GAA instability in Friedreich’s Ataxia shares a common, DNA-directed and intraallelic mechanism with other trinucleotide diseases. Mol Cell. 1998;1:583–593. doi: 10.1016/s1097-2765(00)80058-1. [DOI] [PubMed] [Google Scholar]
- 55.Moore H, Greenwell PW, Liu CP, et al. Triplet repeats form secondary structures that escape DNA repair in yeast. Proc Natl Acad Sci U S A. 1999;96:1504–1509. doi: 10.1073/pnas.96.4.1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Manley K, Shirley TL, Flaherty L, et al. Msh2 deficiency prevents in vivo somatic instability of the CAG repeat in Huntington disease transgenic mice. Nat Genet. 1999;23:471–473. doi: 10.1038/70598. [DOI] [PubMed] [Google Scholar]
- 57.McMurray CT. Hijacking of the mismatch repair system to cause CAG expansion and cell death in neurodegenerative disease. DNA Repair (Amst) 2008;7:1121–1134. doi: 10.1016/j.dnarep.2008.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kovtun IV, McMurray CT. Trinucleotide expansion in haploid germ cells by gap repair. Nat Genet. 2001;27:407–411. doi: 10.1038/86906. [DOI] [PubMed] [Google Scholar]
- 59.van den Broek WJ, Nelen MR, Wansink DG, et al. Somatic expansion behaviour of the (CTG)n repeat in myotonic dystrophy knock-in mice is differentially affected by Msh3 and Msh6 mismatch-repair proteins. Hum Mol Genet. 2002;11:191–198. doi: 10.1093/hmg/11.2.191. [DOI] [PubMed] [Google Scholar]
- 60.Gomes-Pereira M, Fortune MT, Ingram L, et al. Pms2 is a genetic enhancer of trinucleotide CAG.CTG repeat somatic mosaicism: implications for the mechanism of triplet repeat expansion. Hum Mol Genet. 2004;13:1815–1825. doi: 10.1093/hmg/ddh186. [DOI] [PubMed] [Google Scholar]
- 61.Dragileva E, Hendricks A, Teed A, et al. Intergenerational and striatal CAG repeat instability in Huntington’s disease knock-in mice involve different DNA repair genes. Neurobiol Dis. 2009;33:37–47. doi: 10.1016/j.nbd.2008.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lin Y, Wilson JH. Transcription-induced CAG repeat contraction in human cells is mediated in part by transcription-coupled nucleotide excision repair. Mol Cell Biol. 2007;27:6209–6217. doi: 10.1128/MCB.00739-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kovtun IV, Liu Y, Bjoras M, et al. OGG1 initiates age-dependent CAG trinucleotide expansion in somatic cells. Nature. 2007;447:447–452. doi: 10.1038/nature05778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Goula AV, Berquist BR, Wilson DM, 3rd, et al. Stoichiometry of base excision repair proteins correlates with increased somatic CAG instability in striatum over cerebellum in Huntington’s disease transgenic mice. PLoS Genet. 2009;5:e1000749. doi: 10.1371/journal.pgen.1000749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jarem DA, Wilson NR, Schermerhorn KM, et al. Incidence and persistence of 8-oxo-7,8-dihydroguanine within a hairpin intermediate exacerbates a toxic oxidation cycle associated with trinucleotide repeat expansion. DNA Repair (Amst) 2011;10:887–896. doi: 10.1016/j.dnarep.2011.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jung J, Bonini N. CREB-binding protein modulates repeat instability in a Drosophila model for polyQ disease. Science. 2007;315:1857–1859. doi: 10.1126/science.1139517. [DOI] [PubMed] [Google Scholar]
- 67.Lin Y, Dion V, Wilson JH. Transcription promotes contraction of CAG repeat tracts in human cells. Nat Struct Mol Biol. 2006;13:179–180. doi: 10.1038/nsmb1042. [DOI] [PubMed] [Google Scholar]
- 68.Larkin K, Fardaei M. Myotonic dystrophy—a multigene disorder. Brain Res Bull. 2001;56:389–395. doi: 10.1016/s0361-9230(01)00656-6. [DOI] [PubMed] [Google Scholar]
- 69.Schneider-Gold C, Timchenko LT. CCUG repeats reduce the rate of global protein synthesis in myotonic dystrophy type 2. Rev Neurosci. 2010;21:19–28. doi: 10.1515/revneuro.2010.21.1.19. [DOI] [PubMed] [Google Scholar]
- 70.Dick KA, Margolis JM, Day JW, et al. Dominant non-coding repeat expansions in human disease. Genome Dyn. 2006;1:67–83. doi: 10.1159/000092501. [DOI] [PubMed] [Google Scholar]
- 71.Echeverria GV, Cooper TA. RNA-binding proteins in microsatellite expansion disorders: mediators of RNA toxicity. Brain Res. 2012;1462:100–111. doi: 10.1016/j.brainres.2012.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wojciechowska M, Krzyzosiak WJ. Cellular toxicity of expanded RNA repeats: focus on RNA foci. Hum Mol Genet. 2011;20:3811–3821. doi: 10.1093/hmg/ddr299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tan H, Xu Z, Jin P. Role of noncoding RNAs in trinucleotide repeat neurodegenerative disorders. Exp Neurol. 2012;235(2):469–475. doi: 10.1016/j.expneurol.2012.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Todd PK, Paulson HL. RNA-mediated neurodegeneration in repeat expansion disorders. Ann Neurol. 2010;67:291–300. doi: 10.1002/ana.21948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lin MT, Beal MF. Mitochondrial dysfunction and oxidative stress in neurodegenerative diseases. Nature. 2006;443:787–795. doi: 10.1038/nature05292. [DOI] [PubMed] [Google Scholar]
- 76.Vidal RL, Figueroa A, Court FA, et al. Targeting the UPR transcription factor XBP1 protects against Huntington’s disease through the regulation of FoxO1 and autophagy. Hum Mol Genet. 2012;21(10):2245–2262. doi: 10.1093/hmg/dds040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yu Z, Wang AM, Adachi H, et al. Macroautophagy is regulated by the UPR-mediator CHOP and accentuates the phenotype of SBMA mice. PLoS Genet. 2011;7:e1002321. doi: 10.1371/journal.pgen.1002321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Jimenez-Sanchez M, Thompson F, Zavodsky E, et al. Autophagy and polyglutamine diseases. Prog Neurobiol. 2011;97(2):67–82. doi: 10.1016/j.pneurobio.2011.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Schapira AH, Jenner P. Etiology and pathogenesis of Parkinson’s disease. Mov Disord. 2011;26:1049–1055. doi: 10.1002/mds.23732. [DOI] [PubMed] [Google Scholar]
- 80.Ravikumar B, Duden R, Rubinsztein DC. Aggregate-prone proteins with polyglutamine and polyalanine expansions are degraded by autophagy. Hum Mol Genet. 2002;11:1107–1117. doi: 10.1093/hmg/11.9.1107. [DOI] [PubMed] [Google Scholar]
- 81.Lopez Castel A, Cleary JD, Pearson CE. Repeat instability as the basis for human diseases and as a potential target for therapy. Nat Rev Mol Cell Biol. 2010;11:165–170. doi: 10.1038/nrm2854. [DOI] [PubMed] [Google Scholar]
- 82.Ranganathan S, Fischbeck KH. Therapeutic approaches to spinal and bulbar muscular atrophy. Trends Pharmacol Sci. 2010;31:523–527. doi: 10.1016/j.tips.2010.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Schulz JB, Boesch S, Burk K, et al. Diagnosis and treatment of Friedreich ataxia: a European perspective. Nat Rev Neurol. 2009;5:222–234. doi: 10.1038/nrneurol.2009.26. [DOI] [PubMed] [Google Scholar]
- 84.Sah DW, Aronin N. Oligonucleotide therapeutic approaches for Huntington disease. J Clin Invest. 2011;121:500–507. doi: 10.1172/JCI45130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Fiszer A, Olejniczak M, Switonski PM, et al. An evaluation of oligonucleotide-based therapeutic strategies for polyQ diseases. BMC Mol Biol. 2012;13:6. doi: 10.1186/1471-2199-13-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Boudreau RL, McBride JL, Martins I, et al. Nonallele-specific silencing of mutant and wild-type huntingtin demonstrates therapeutic efficacy in Huntington’s disease mice. Mol Ther. 2009;17:1053–1063. doi: 10.1038/mt.2009.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Harper SQ, Staber PD, He X, et al. RNA interference improves motor and neuropathological abnormalities in a Huntington’s disease mouse model. Proc Natl Acad Sci USA. 2005;102:5820–5825. doi: 10.1073/pnas.0501507102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zhang Y, Friedlander RM. Using non-coding small RNAs to develop therapies for Huntington’s disease. Gene Ther. 2011;18:1139–1149. doi: 10.1038/gt.2011.170. [DOI] [PubMed] [Google Scholar]
- 89.Nakamori M, Gourdon G, Thornton CA. Stabilization of expanded (CTG)*(CAG) repeats by antisense oligonucleotides. Mol Ther. 2011;19:2222–2227. doi: 10.1038/mt.2011.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Benraiss A, Goldman SA. Cellular therapy and induced neuronal replacement for Huntington’s disease. Neurotherapeutics. 2011;8:577–590. doi: 10.1007/s13311-011-0075-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lindvall O, Bjorklund A. Cell therapeutics in Parkinson’s disease. Neurotherapeutics. 2011;8:539–548. doi: 10.1007/s13311-011-0069-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Daadi MM. Novel paths towards neural cellular products for neurological disorders. Regen Med. 2011;6:25–30. doi: 10.2217/rme.11.85. [DOI] [PubMed] [Google Scholar]
- 93.Park IH, Zhao R, West JA, et al. Reprogramming of human somatic cells to pluripotency with defined factors. Nature. 2008;451:141–146. doi: 10.1038/nature06534. [DOI] [PubMed] [Google Scholar]
- 94.Takahashi K, Tanabe K, Ohnuki M, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 95.Chen SJ, Chang CM, Tsai SK, et al. Functional improvement of focal cerebral ischemia injury by subdural transplantation of induced pluripotent stem cells with fibrin glue. Stem Cells Dev. 2010;19:1757–1767. doi: 10.1089/scd.2009.0452. [DOI] [PubMed] [Google Scholar]