Abstract
Viruses employ a variety of strategies to usurp and control cellular activities through the orchestrated recruitment of macromolecules to specific cytoplasmic or nuclear compartments. Formation of such specialized virus-induced cellular microenvironments, which have been termed viroplasms, virus factories, or virus replication centers, complexes, or compartments, depends on molecular interactions between viral and cellular factors that participate in viral genome expression and replication and are in some cases associated with sites of virion assembly. These virus-induced compartments function not only to recruit and concentrate factors required for essential steps of the viral replication cycle but also to control the cellular mechanisms of antiviral defense. In this review, we summarize characteristic features of viral replication compartments from different virus families and discuss similarities in the viral and cellular activities that are associated with their assembly and the functions they facilitate for viral replication.
INTRODUCTION
Viral genomes are replicated, expressed, and assembled in association with intracellular structures that are formed or reorganized by viral and cellular macromolecules. These complex molecular assemblies occupy either cytoplasmic or nuclear sites, where the viral genome and viral and cellular proteins accumulate. The molecules and activities that are associated with these compartments are diverse but, invariably, include components that direct viral genome replication and expression. Formation of compartments where viral genomes are replicated and assembled has been described for negative-sense RNA (−), double-stranded RNA (dsRNA), and positive-sense RNA (+) viruses. In the case of positive-strand RNA genomes of a variety of virus families, RNA replication is associated with extensive reorganization and generation of single or double membranous structures coopted by membrane-associated viral replicase complexes (RCs). RCs formed by RNA (+) viruses have been the subject of intense investigation, and it is for these virus families that more information is available and has been included in several excellent reviews (1–5). RNA and DNA viruses that replicate in the cytoplasm form viral factories or viroplasms that require relocalization of organelles, reorganization of the endoplasmic reticulum (ER), Golgi apparatus, endosome, lysosome, mitochondria, or other cellular membranes, and changes in the distribution and dynamics of the cytoskeleton (1–5). In the case of DNA viruses that replicate in the nucleus, formation of RCs does not seem to require membranous structures but, rather, a reorganization of nuclear components that accompanies formation of these compartments. This regorganization includes the redistribution of chromatin and components of nuclear domains, such as promyelocytic leukemia protein (PML) nuclear bodies (PML-NB), Cajal bodies (CB), interchromatin granules (IG), or the nucleolus.
Although assembly of RCs and their components varies between different virus families, several fundamental similarities exist between RNA viruses, and some parallels can be made with DNA viruses. These similarities might originate from the requirement to control common cellular factors that participate in viral genome replication or transcription and the ability to coopt cellular biosynthetic pathways, as well as the evasion of common mechanisms of the cellular antiviral response. In general terms, RCs concentrate and compartmentalize viral and cellular molecules that are required for DNA or RNA synthesis and, as a consequence of their assembly and architecture, provide a scaffold that maximizes the efficiency of viral replication and simultaneously conceals the viral genomes and their products from detection by defense mechanisms (Table 1). Interestingly, many cellular factors that regulate pathways that are central to normal cell metabolism are also associated with RCs and, although they may play indirect roles, understanding their impact on virus replication should shed light onto cellular mechanisms that are at the basis of virus-cell interactions.
TABLE 1.
Features of RCs induced by different virus families included in this reviewa
| Virus family | Virus genus and species | Structure in close proximity to RCs | Proteins associated with RCs |
Altered cellular activities | |
|---|---|---|---|---|---|
| Viral proteins | Cellular proteins | ||||
| Poxviridae | VV | MTOC (12) | RNA polymerase, transcription factor VITF-3, E3 dsRNA binding protein (15, 18) | Nuclear transcription factors (G3BP, Caprin-1), translation initiation factors eIF4G and eIF4E (15) | Transport of viral core via microtubules to MTOC; vimentin rearrangement; recruitment of ribosomes, chaperones, and mitochondria to RCs; rearrangement of ER membranes (12, 15) |
| Asfarviridae | ASFV | MTOC (36, 46) | DNA polymerase, topoisomerase, helicase, ligase, DNA binding protein (42, 43) | Translation initiation factors eIF4G and eIF4E (45, 54) | Phosphorylation of lamins A/C and disruption of lamins A/C network; vimentin rearrangement; recruitment of ribosomes, chaperones, ubiquitin, proteasomes, and mitochondria to RCs; reorganization of microtubulues; redistribution of ER membranes, loss of trans-Golgi network, reorganization of cytoskeleton (36, 44–47, 49–52) |
| Herpesviridae | HSV-1 | PML-NBs (67–72) | Immediate-early regulatory proteins ICP0, ICP4, and ICP27, ssDNA binding protein ICP8, replication initiation protein UL9, heterotrimeric helicase/primase complex, DNA polymerase, polymerase accessory protein U42, structural proteins (72, 77, 82, 85, 90, 92, 93) | PML, Sp100, Daxx, SUMO E3 ligase PIAS2β, RNA polymerase II, DNA polymerase α, DNA polymerase γ, topoisomerase II, DDR proteins, replication protein A (73, 74, 82, 83, 93, 96) | Disruption of PML-NBs; proteasomal degradation of PML and Sp100; recruitment of PQC system to VICE domain; degradation of DDR proteins; exploitation of cellular DDR (74–77, 86–89, 103, 104) |
| Herpesviridae | HCMV | PML-NBs (111, 114, 120) | IE1, IE2, UL112-113 proteins, DNA polymerase UL54, polymerase-associated processivity factor UL44, ssDNA binding protein UL57, heterotrimeric helicase UL105/primase UL70 complex, terminase subunit pUL56 (123, 128–131) | Proteasomes, replication protein A, p53, RNA polymerase II (122, 127, 139) | Disruption of PML-NBs; recruitment of proteasomes to periphery of RCs; accumulation of p53 in RCs (109, 112, 123, 136, 139) |
| Herpes-viridae | KSHV | PML-NBs (147, 151) | LANA2, ssDNA binding protein Orf6, polymerase processivity factor Orf 59, polymerase Orf9, primase/helicase tripartite subcomplex (147) | SUMO-2/3 modification of PML, disruption of PML-NBs (147, 151) | |
| Herpesviridae | HVS | PML-NBs (148) | Degradation of PML-NB component Sp100 (148) | ||
| Herpesviridae | EBV | PML-NBs (152) | BZLF, BNRF1, DNA polymerase processivity factor BMRF1 (152, 153) | PCNA, replication protein C (154, 156) | Disruption of PML-NBs by dispersal of PML-NB components; induction of ATM-mediated DDR (155, 156) |
| Herpesviridae | VZV | PML-NBs (158) | Disruption of PML-NBs (158, 159) | ||
| Adenoviridae | Adenovirus | PML-NBs (162, 163) | E1A, E4orf3, E1B-55K, E4orf6, ssDNA binding protein DBP, protein primer terminal protein, E2B DNA polymerase (162, 180, 181, 185, 186) | Nuclear factor 1; cellular proteins involved in DNA replication, transcription, and splicing factors (188, 189, 191) | Disruption of PML-NBs, redistribution of PML-NB components in track-like structures; targeting MRN complex into tracks and inhibition of DDR; exploitation of Cajal body and interchromatin granule components (162–172, 181, 183, 184) |
| Parvoviridae | AAV | PML-NBs (in Ad-infected cells) (221) | AAV Rep proteins, Ad DBP (221) | Replication protein A, replication protein C, DNA polymerase δ/PCNA, DDR proteins (222, 223) | Usurpation of Ad RCs (221) |
| Parvoviridae | Parvovirus | Distinct from any prominent nuclear structure (238) | Nonstructural proteins NS1 and NS2, capsid proteins VP1 and VP2 (238, 239) | DNA polymerase δ/PCNA, DNA polymerase α, replication factor A, replication factor C, DDR sensor and signaling proteins (240–242) | DDR (242, 246) |
| Polyomaviridae | BKV, JCV, SV40 | PML-NBs (248, 249, 251, 252) | Large TAg (251, 253) | DDR proteins, DNA polymerase α/primase, DNA polymerase δ/PCNA, replication protein A, replication protein C, topoisomerases I and II (249, 251) | Disruption of PML-NBs in BK virus infection (249) |
| Papillomaviridae | HPV | PML-NBs (268, 279, 281, 283, 289) | DNA helicase E1, regulatory protein E2, capsid proteins L1 and L2 (273, 276, 277) | DNA polymerase α/primase, DNA polymerase δ/PCNA, replication protein A, topoisomerases I and II, DDR proteins (274, 275, 278, 279) | Exploitation of SUMOylation system; reorganization of PML-NBs; redistribution of DDR proteins (284, 287) |
The reference(s) supporting the reported information in each cell is provided in parentheses for each virus species.
CYTOPLASMIC DNA VIRUS FACTORIES
Nucleocytoplasmic large DNA viruses.
The nucleocytoplasmic large DNA viruses (NCLDV) encompass seven families of large eukaryotic DNA viruses that infect a wide range of hosts, from algae to insects and mammals: Ascoviridae, Asfarviridae, Iridoviridae, Mimiviridae, Phycodnaviridae, and Poxviridae, as well as a recently proposed family that includes Lausannevirus and Marseillevirus (6), all of which possess a large genome ranging from 150 to 1.2 Mb. Besides sharing several core genes, they all form complex cytoplasmic RCs, called viral factories, that are involved in viral replication and assembly and therefore display a dynamic architecture through the viral replication cycle. However, some NCLDV members replicate exclusively in the cytoplasm, whereas others replicate in a time-dependent manner in both the nucleus and cytoplasm (7, 8).
RCs of poxviruses and African swine fever virus (ASFV) have been studied in greater detail and have been included in recent reviews (4, 9). Like aggresomes, which are formed as a cellular response to protein aggregation (10), NCLDV viral factories assemble at the microtubule-organizing center (MTOC), depend on microtubule-associated dynein motor dynamics, redistribute vimentin filaments into a cage-like structure, and recruit mitochondria, chaperones, ubiquitin, and proteasomes (4, 11) (Fig. 1).
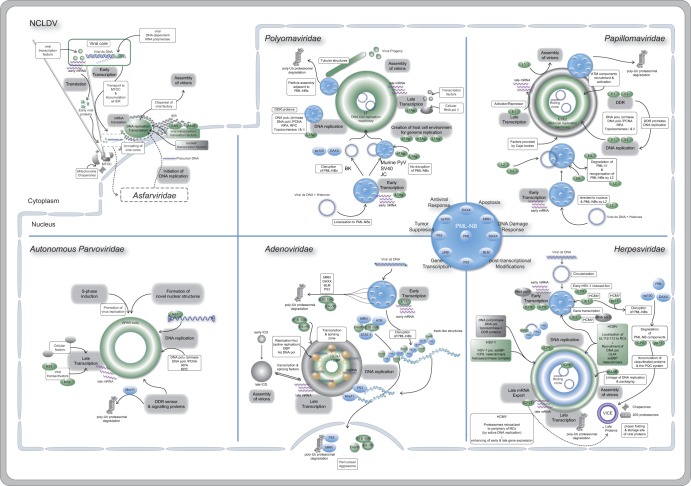
FIG 1.
Scheme of DNA virus replication compartments, indicating localization within the cytoplasm or nucleus. In particular, the virus families Polyomaviridae, Papillomaviridae, Adenoviridae, and Herpesviridae as well as NCLDV and autonomous parvoviruses are shown. Viral factors and proteins are indicated in green and cellular ones are gray, with PML-NBs, which play a prominent role during replication of Polyomaviridae, Papillomaviridae, Adenoviridae, and Herpesviridae, shown in blue.
Poxviridae.
Poxviridae possess the unusual characteristic of replicating exclusively in the cytoplasm. Upon membrane fusion and release into the cytoplasm, the poxvirus core is transported via microtubules (MT) toward the MTOC. Such transport is associated with vimentin rearrangement and recruitment of chaperones and mitochondria to perinuclear sites, where replication compartments are finally formed (12). Early gene transcription, however, is already initiated within the virus core by the viral DNA-dependent RNA polymerase (Pol) and viral transcription factors packaged into the virus particles together with the genome. Thus, a set of about 100 early mRNAs is transcribed and subsequently translocated into the cytoplasm where, while still associated with MT, they are engaged by ribosomes for translation. Meanwhile, viral cores accumulate close to the rough endoplasmic reticulum (rER). In the case of vaccinia virus (VV), the prototype and most-studied member of Poxviridae, early proteins are required to uncoat the viral core and to release the viral DNA, as soon as the viral core reaches the ER (13). The disassembly steps for genome uncoating depend on ubiquitin-mediated proteasomal degradation of previously ubiquitinated capsid proteins, and a Cullin3-based ubiquitin ligase is required for genome replication (14). The genome released from the core associates with a set of viral proteins that participate in DNA replication and organization of the RC precursor (reviewed in reference 12), as DNA replication proceeds, intermediate and late mRNA, as well as ribosomes and translation factors, accumulate in expanding DNA factories (15), which are later released from the rER when virion assembly starts (16, 17).
Poxviruses encode over 130 proteins, including factors involved in transcription (18), encompassing more than 20 viral proteins involved in RNA synthesis (reviewed in reference 19), as well as DNA replication (reviewed in reference 20). Although these include a nearly complete repertoire of viral proteins responsible for viral gene expression and DNA replication, the extensive rearrangement of ER membranes is likely to recruit and control many cellular proteins. Indeed, a recent RNA interference screen identified 188 cellular factors required for VV infection, including both nuclear and cytoplasmic functions (14). During intermediate and late times of infection, cellular protein synthesis is downregulated (21–23) and cellular mRNA is degraded by a viral decapping enzyme (24, 25). Furthermore, Poxviridae have been suggested to promote selective and augmented translation of viral mRNAs in in vitro studies (26). The highly complex ER-surrounded RCs have been compared to mininuclei, where viral DNA as well as transcription and translation factors are enclosed inside a membranous compartment. However, although linked transcription and translation has recently been demonstrated (15), compartmentalization of the virus factories leads to separation of gene expression and genome replication, which occur in distinguishable subcompartments within or on the surface of virus factories. Katsafanas and Moss showed that viral RNA transcription occurs within the factory, as both the viral G8R RNA and poly(U) RNA localized to distinct compartments that those authors called cavities or tunnels of the virus factories. Moreover, poly(U) labeling showed reduced cytoplasmic staining compared to uninfected cells, consistent with cellular mRNA degradation during intermediate times of infection (15). The viral E3 dsRNA binding protein that counteracts activation of the dsRNA-sensing kinase (protein kinase R [PKR]) was also observed in RCs, as was the viral intermediate transcription factor 3. Nuclear factors required for transcription of the viral genome are also recruited to these sites (18), as are the cellular G3BP and Caprin-1, both known to be required for viral intermediate transcription in vitro (15). The cellular translation initiation factors eukaryotic initiation factor 4G (eIF4G) and eIF4E, as well as ribosomes, are also present in viral RCs; however, a large pool of ribosomes remains in the cytoplasm, suggesting that preferential translation of viral mRNA could be achieved by sequestering initiation factors. Translation of viral mRNA occurs within viral RCs, as β-galactosidase expressed from a recombinant VV was confined to these compartments (15). This compartmentalization of translation factors on the one hand would be relevant for efficient expression of viral genes and on the other hand for simultaneous suppression of cellular protein synthesis.
During the late phase of infection, when virus assembly is initiated, the replication sites are released from the rER and crescent-shaped membranes associate with viral DNA to initiate a complex assembly process (reviewed in reference 27).
Asfarviridae.
Like Poxviridae, ASFV, the only member of the Asfarviridae family at this time, replicates in virus factories that are distributed in perinuclear cytoplasmic sites. However, in contrast to poxviruses, ASFV not only recruits nuclear factors to these factories but also initiates genome replication within the cell nucleus (28–31). It is believed that short precursor DNA fragments are subsequently exported from the nucleus to the cytoplasmic RCs, where they are used as primers for full-length genome replication (30–32). Interestingly, cytoplasmic DNA synthesis is independent of the nucleus (33). Given its large genome, it is thought that the virus encodes proteins that actively transport the viral genome into the nucleus as well as factors that direct the precursor DNA exit to the cytoplasm. While two viral structural proteins, p37 and p14, might be involved in nuclear entry (34), the viral matrix protein p37, which exhibits nucleocytoplasmic shuttling activities, might trigger nuclear egress and transport of viral precursor DNA to RCs (34, 35).
Upon viral entry and virion release from the endosome-lysosome system into the cytoplasm, the ASFV core is transported to the MTOC (36). Meanwhile, transcription of early genes and posttranscriptional modification of the mRNA occur inside the virion; this is ensured by packaged proteins such as the viral RNA polymerase and, hence, independent of cellular enzymes (37–41). Late gene expression, however, depends on viral DNA replication and viral early proteins. For DNA synthesis, the ASFV genome encodes a set of proteins responsible for cytoplasmic DNA replication, including DNA polymerase, topoisomerase, helicase, ligase, and DNA binding proteins (42, 43).
ASFV induces major changes in the organization of the infected cell. During the early phase, lamins A/C are phosphorylated and the nuclear membrane network next to sites where newly synthesized viral DNA accumulates is disrupted (44). Components of the nucleolus (B23) and splicing speckles (SC35) are also redistributed within the nucleus, and RNA polymerase II dephosphorylation and degradation are induced and contribute to disrupt cellular transcription (44). Furthermore, ASFV induces shutoff of host protein synthesis by recruiting eIFs and ribosomes to RCs (45). At later time points, when viral DNA accumulates in cytoplasmic factories adjacent to the outer nuclear membrane, the lamin A/C network is further disrupted and sequestered to nuclear and cytoplasmic foci. The latter are recruited to the RCs along with other nuclear membrane components (44). Similar to other viruses, cytoplasmic ASFV factories have been compared with aggresomes, since emerging viral factories form at the MTOC and become surrounded by vimentin and recruit cellular chaperones, ubiquitin, proteasomes, and mitochondria. Furthermore, similar to aggresomes, ASFV factories require microtubules and dynein motor proteins for their formation (36, 46).
Early ASFV replication sites appear as punctuate foci of extranuclear viral DNA, while structural proteins are spread throughout the cytoplasm. Later on, these proteins are actively transported to the MTOC via microtubules, where they fuse with ASFV factories (47, 48). Interestingly, the reorganization of microtubules seems to be critical for DNA replication, as well as late gene transcription (46, 47, 49), presumably by stabilizing viral RCs and concentrating cellular and viral proteins required for replication at the MTOC (46, 47, 49). Further reorganization of cellular components is induced as replication progresses and involves the relocation of mitochondria and chaperones as well as the redistribution of ER membranes to RCs, the loss of the trans-Golgi network, and extensive reorganization of the cytoskeleton (47, 50–52). ASFV encodes a viral homologue of cellular ubiquitin-conjugating enzymes (UBCs), suggesting that ASFV might manipulate ubiquitin-mediated host cell responses or modifications of viral or host proteins (53).
Subdivision of poxviral factories allows the separation of viral gene expression and DNA replication (15). Similarly, during ASFV infection, host initiation factors are recruited to viral RCs, whereas viral RNA and host ribosomes are localized to the factories' peripheries (54).
The ASFV replication cycle primarily takes place in these virus factories, which are first observed 6 to 8 h postinfection (p.i.). During ongoing infection, amorphic and circular membranous material as well as increasing numbers of immature and mature viral particles accumulate in ASFV factories 12 to 24 h p.i. and follow a complex morphogenetic process analogous to that for poxviruses (reviewed in reference 55).
Similar to poxviruses and ASFV, other members of the remaining NCLDV families also form cytoplasmic factories that share many of the features described above; however, the genomes of the iridoviruses (56) and phycodnaviruses (57) are initially shuttled into the nucleus, where replication begins, whereas genome replication of mimiviruses seems to take place entirely in the cytoplasm (6).
NUCLEAR DNA VIRUS REPLICATION CENTERS
Like the NCLDV, DNA viruses that replicate in the nucleus use aggresome-like structures as sites for assembly of nuclear factories. However, the ultrastructure and detailed architecture of the nuclear replication and assembly sites has not been studied in as much detail as cytoplasmic factories. The accumulated evidence and recent biochemical studies have found multiple interactions between viral genomes and RCs with nuclear components and domains, and the architecture of viral replication compartments has been revealed by a variety of fluorescence and electron microscopy studies.
Upon internalization of virus particles, viral cores are transported to the MTOC by cellular dynein/dynactin motors, and viral genomes are delivered into the nucleus by disassembly of the viral capsid at the nuclear pore (reviewed in references 5, 58, 59, and 60).
Inside the nucleus, viral genomes colonize specific nuclear sites by utilizing cellular components to ensure efficient viral gene expression and replication. These so-called replication centers or compartments (RCs) often form adjacent to PML-NBs, nuclear structures that have been implicated in DNA repair, transcriptional regulation, cell senescence, apoptosis, and the interferon-induced antiviral state. The major structural component of PML-NBs is the PML protein, also known as TRIM19, a member of the tripartite motif (TRIM) family that functions as the essential organizer of the PML-NB and regulates protein transit by SUMO-mediated protein interactions (reviewed in reference 61). In most cases, components of the PML-NB are targeted to the viral genomes as part of a cellular defense mechanism. Subsequently, cellular factors relocalized to these sites are either utilized by the virus for its replication (DNA Pol, RNA Pol, transcription factors, posttranscriptional processing, and mRNA export) and/or inhibited by the virus to prevent/control the cellular antiviral defense (PML, p53, ATM, ATR, Daxx, STAT, interferon regulatory factors) (62–64) (Fig. 1; Table 1).
The association of PML-NBs with nuclear aggresomes provides a link for RCs to the storage as well as removal of (misfolded) proteins (63, 65). In contrast to cytoplasmic replicating viruses, which assemble progeny particles in close association with cellular membranes, virions of DNA viruses replicating in the nucleus are often assembled in close proximity to viral transcription and DNA replication centers devoid of a surrounding membrane (1, 5).
Herpesviridae.
Upon infection with different members of the Herpesviridae family, similar replication centers are formed (Fig. 1). Proteins of the PML-NB are recruited to the viral genome, and viral RCs containing the parental genome form adjacent to PML-NBs. However, RCs are established by different mechanisms, as described here, in particular, for herpes simplex virus 1 (HSV-1) and human cytomegalovirus (HCMV).
HSV-1 (alphaherpesvirus).
Shortly after the HSV-1 genome has entered the nucleus, the host cell antiviral defense is activated and components of PML-NBs are delivered to sites associated with the parental genome (66–71). The recruitment of PML, Sp100, and Daxx depends on their SUMO interaction motifs. Furthermore, SUMOylated proteins as well as the cellular SUMO E3 ligase PIAS2β are found in these HSV-1-induced foci. Thus, it has recently been proposed that the cellular defense against foreign DNA is regulated by the SUMO pathway (72). The viral regulatory protein ICP0 colocalizes with these foci and induces the proteasome-mediated degradation of PML and Sp100 by acting as an E3 ubiquitin ligase (73–77), thus leading to the disruption of PML-NBs. Interestingly, ICP0 specifically targets SUMO-1-modified forms of the PML-NB proteins for proteasomal degradation (78–80). Ongoing replication requires the recruitment of different viral and cellular proteins to the virus-induced foci, which mature to viral replication centers (81–83). As the name implies, these compartments provide an optimal environment for viral gene expression, DNA synthesis, and assembly of progeny nucleocapsids (84, 85). Additionally, aggresome-like structures are formed adjacent to RCs early during infection. These so-called virus-induced chaperone-enriched (VICE) domains contain molecular chaperones, especially the Hsp70 chaperone family member Hsc70, and 20S proteasomes that are sequestered from a diffuse nuclear distribution, as well as ubiquinated proteins (86–89). Both molecular chaperones as well as the ubiquitin proteasome system are key components of cellular protein quality control (PQC). Initially, ubiquitylation of cellular proteins (see below) during viral infection leads to the formation of the VICE domains, an additional nuclear aggresome-like structure (86). Hence, the cellular PQC system recruited to these structures can be exploited for different processes during HSV-1 replication. Thus, molecular chaperones allow assembly and proper folding of viral multimeric protein complexes, including the RC scaffold, the helicase/primase complex, and the viral capsids. VICE domains additionally serve as storage sites for structural proteins of the viral capsid that have been made in excess compared to those packaged into capsids. Thus, it has been suggested that VICE domains are assemblons that form late during infection (90, 91). Viral gene expression in nuclear RCs starts immediately after entry of the viral genome into the nucleus. The linear DNA is circularized, and immediate-early (IE), early (E), and late (L) viral gene products are sequentially expressed. Active herpesviral gene expression in RCs requires the translocation of viral ICP0, ICP4, and ICP27 proteins as well as RNA polymerase II to these sites (92, 93). Both ICP0 and ICP4 stimulate transcription of E and L genes, whereby ICP4 functions as a major transcriptional activator and is required for the progression beyond the immediate-early phase. In contrast, ICP27 mediates export of the majority of herpesviral mRNAs, interacting with the cellular export receptors Tap/Nxf and Aly/Ref (94). The accumulation of viral proteins and viral template DNA as well as the translocation of cellular proteins to the RCs allow efficient gene expression as well as viral genome replication. Among the herpesviral proteins associated with RCs are the single-stranded DNA (ssDNA) binding protein ICP8, heterotrimeric helicase/primase complex, and the HSV-1 polymerase. Furthermore, cellular DNA polymerases α and γ and topoisomerase II, as well as DNA repair and recombination proteins, are recruited to the RCs (82, 83, 95, 96).
Progeny virion assembly, including capsid formation and DNA packaging, is a tightly regulated process that takes place in the nuclear RCs (97, 98). Typically, this process involves proteins forming an internal scaffold where the capsid shell is assembled and the genome is subsequently incorporated (99–101). Thereafter, the assembled nucleocapsid exits the nucleus by a unique budding mechanism through the nuclear membrane. Finally, the virion is transported via secretory transport vesicles to the plasma membrane and is released upon membrane fusion (102).
Common features of DNA viruses are the formation of E3 ubiquitin ligases, as described above for HSV-1 ICP0, as well as the deregulation of the cellular DNA damage response (DDR). Most interestingly, both the degradation of cellular proteins as well as the modulation of the DDR are linked to a considerable extent. Already in the early phase of HSV-1 infection, proteins involved in homologous recombination (HR) (83) as well as nonhomologous end joining (NHEJ) of DNA (95, 103) are recruited to viral RCs. Interestingly, in contrast to adenoviruses, which inhibit this intrinsic antiviral mechanism (see below), the cellular DDR appears to be beneficial for HSV-1. Nevertheless, HSV-1 also manipulates the DDR by inactivating some of its components and utilizing others. ICP0 interferes with both DDR pathways by inducing the degradation of different target proteins, among these, the catalytic subunit of the DNA-dependent protein kinase DNA-PK (104, 105), which is involved in sensing of NHEJ, as well as the E3 ligases RNF8 and RNF168 (106, 107), which promote ATM tethering to sites of DNA damage, i.e., the sensing mechanism of HR. In contrast, the Mre11-Rad50-Nbs1 (MRN) complex as well as ATM kinase activation, both of which are involved in NHEJ, seem to be beneficial for HSV-1 replication (103). Interestingly, although phosphorylated replication protein A (RPA) is excluded from RCs and sequestered to adjacent VICE domains, thereby preventing normal ATR signaling (108), ATR and ATRIP localize in RCs and have been shown to be conducive to HSV-1 replication (73).
HCMV (betaherpesvirus).
Replication centers of human cytomegaloviruses form adjacent to PML-NBs. Different studies have suggested that the recruitment of PML proteins to the viral nucleocapsid shortly after its delivery into the nucleus is part of the antiviral host defense. However, this is subsequently hampered by different HCMV proteins. The cellular transcriptional repressor Daxx inhibits HCMV gene expression from the major IE promoter. This is circumvented by the viral tegument protein pp71 (109–111). Interestingly, pp71 stimulates proteasomal degradation of Daxx by a mechanism that is independent of a functional ubiquitin pathway (112). In addition, pp71 was found to trigger Daxx SUMOylation, although so far no function for this modification has been described (113). Furthermore, the viral transactivator IE1 colocalizes with PML-NBs and disrupts these structures (114) by interfering with SUMO modification of PML (115). Thus, both pp71 and IE1 cooperate in the disruption of cellular PML-NBs and antagonize host defense mechanisms. In addition, studies on these HCMV proteins revealed a link to the cellular SUMOylation pathway, as has been observed for other DNA viruses. Consistent with these effects on cellular PML-NBs, downregulation of Daxx and/or PML as well as of the PML-NB component Sp100 was found to enhance viral replication (116–119). Moreover, the HCMV kinase pUL97 seems to be an additional viral protein that interferes with PML function. pUL97 inhibits the complex formation between PML and the retinoblastoma protein (Rb) by hyperphosphorylating Rb (120, 121). Interestingly, the kinase activity of pUL97 also inhibits the formation of nuclear aggresomes (121). In contrast to the viral tegument protein pp65, which induces inappropriate aggregation and sequestration of viral tegument and capsid proteins, pUL97 prevents this undesirable effect. Thus, pUL97 is proposed to play an important role in progeny virus assembly.
Similar to HSV-1, HCMV expresses its genes in a temporally regulated cascade, with the replication cycle being divided into IE, E, and L phases. The immediate-early proteins IE1 and IE2 stimulate viral E and L gene expression, with IE2 the driving force of HCMV replication in general. As would be expected, RNA Pol II and splicing factors colocalize with IE genes in RCs (122). While IE2 colocalizes with the viral RCs by interacting with the viral genome, IE1 localization to PML-NBs has been reported to depend on interaction with PML (123).
Recently, it was reported that efficient viral replication requires active cellular proteasomes (124–126). HCMV infection specifically leads to enhanced proteasomal pathway activity. Furthermore, active viral DNA replication induces the recruitment of proteasomes to the periphery of viral RCs, where active RNA transcription takes place. This is consistent with the observation that the cellular proteasomal pathway is exploited to enhance early and late gene expression (127).
Already early during HCMV infection, the UL112-113 proteins are localized together with IE2 to prereplication foci and are found in RCs throughout the viral life cycle (128–130). UL112-113 encodes four phosphoproteins, which are supposed to recruit proteins involved in DNA replication, to viral RCs (128, 131). These viral replication core proteins include the DNA polymerase UL54, the polymerase-associated processivity factor UL44, and the ssDNA binding protein UL57, as well as a heterotrimer consisting of the DNA helicase UL105, the primase UL70, and the primase-associated factor (132, 133). Upon entry into the nucleus, the HCMV genome is circularized, and a rolling circle mechanism of replication produces concatemers late in infection (136, 135). The HCMV terminase subunit pUL56 potentially links DNA replication with packaging. Interestingly, this protein is involved in cleavage and packaging of progeny viral genomes (136) and also localizes to viral RCs (137).
As described for HSV-1, DNA viruses often interfere with the cellular DDR machinery. HCMV infection induces the accumulation of the tumor suppressor p53 (138–140). Simultaneously, the virus compromises the ability of p53 to transactivate cellular downstream targets (138, 140–142). Thus, HCMV may exploit p53 functions to fully activate the infected cells, in parallel inhibiting its ability to regulate DDR as well as apoptosis. Interestingly, p53 as well as RPA are both relocalized to viral RCs (143). It could therefore be that both cellular proteins are utilized to help in viral DNA damage control and/or to promote efficient viral DNA synthesis, as has been reported for other DNA viruses (144–146).
Other herpesviruses.
Interestingly, apart from HSV-1 and HCMV, various herpesviruses counteract the antiviral properties of PML-NBs and lead to the disruption of these bodies and/or relocalization of specific PML-NB components. Thus, members of the gammaherpesvirus family, like Kaposi's sarcoma-associated herpesvirus (KSHV) (147), herpesvirus saimiri (HVS) (148), and Epstein-Barr virus (EBV) (152), express proteins with properties similar to HSV-1 ICP0. Furthermore, viral replication compartments develop in association with PML-NB remnants (147, 148, 152).
For KSHV, it has been shown that shortly after infection, PML-NB components localize to viral genomes and prereplication compartments (147). Interestingly, PML is SUMO-2 modified by LANA2, the latency-associated nuclear antigen 2. Since SUMO-2/3 modification of PML has been described to induce PML degradation via ubiquitination by the cellular RNF4 ubiquitin ligase (150), this modification most likely leads to proteasome-mediated degradation of PML and hence to PML-NB disruption. Thereby, LANA2 seems to interfere with PML-mediated transcriptional repression (151). DNA replication during the lytic cycle of KSHV infection requires six viral proteins, including the ssDNA binding protein Orf6, the polymerase processivity factor Orf59, and the polymerase Orf9, as well as a primase/helicase tripartite subcomplex. Interestingly, transient cotransfection of these factors results in the formation of nuclear structures resembling replication compartments. Furthermore, these pseudo-RCs are surrounded by PML-NBs (147).
As mentioned above, HVS also antagonizes the PML-NB-mediated, intrinsic host cell defense. Specifically, the HVS tegument protein Orf3 induces degradation of the PML-NB component Sp100, whereas the distribution and/or functionality of PML and Daxx remains unchanged. Although PML restricts HVS replication, it might play an important role for the establishment of latent infection (148).
Herpesviruses can cause both latent and lytic infection of the host cell. During EBV latency, PML-NBs remain intact, and replication of the virus is not associated with these nuclear structures. However, the switch from latent to lytic EBV replication immediately disrupts PML-NBs by dispersing Sp100, Daxx, and NDP55. In contrast, PML is dispersed only after the onset of lytic replication (149). For the disruption of PML-NBs, the EBV immediate-early protein BZLF alone is sufficient. Interestingly, BZLF competes with PML for free SUMO-1, which leads to a decrease of SUMO-1-modified PML (152). Furthermore, EBV expresses a tegument protein that specifically targets the intrinsic host cell defense, thereby activating viral early gene transcription. This protein, BNRF1, disrupts the cellular Daxx-ATRX chromatin remodeling complex, which seems to be required for the switch from latent to lytic infection by alteration of viral chromatin (153). Further proteins localizing to replication compartments and interacting with the EBV DNA polymerase processivity factor BMRF1 include the proliferating cell nuclear antigen (PCNA) and replication factor C (RFC), as well as several proteins of the cellular mismatch repair system. Thereby, EBV might prevent recombination events between homologous stretches in its genome sequences (154). Interestingly, active EBV DNA replication induces an ATM-mediated DDR (155), which is modified by the viral BGLF4 kinase. Ultimately, this results in an optimal environment for viral genome amplification, while host cell DNA synthesis is blocked (156, 157).
Similar to HSV-1, the varicella-zoster virus (VZV) protein Orf61, a homologue of HSV-1 ICP0, disrupts cellular PML-NBs. The SUMO-interacting motifs (SIM) present in Orf61 are critical for both the interaction and dispersal of these nuclear structures (158). A three-dimensional (3D) reconstruction using serial section array scanning electron microscopy confirmed that PML-NBs are disassembled by the interaction of Orf61 SIM with PML and revealed that VZV capsids become entrapped in large PML cages (159).
Taken together, replication of herpesviruses occurs adjacent to cellular PML nuclear bodies, which are known to negatively restrict viral replication. Interestingly, herpesviruses evolved various mechanisms to counteract the PML-NB function. Therefore, it is most likely that different herpesviruses target similar cellular pathways, involving DDR, chromatin remodelling, cell cycle control, and transcription machinery, to foster their replication. So far, these mechanisms have been best studied for HSV-1 and HCMV; however, recent results revealed the first detailed insights into replication of other members of the Herpesviridae.
Adenoviridae.
Upon infection of the cell, adenoviral (Ad) particles are transported toward the MTOC and disassembled at the nuclear pore. Immediately after the viral genome is released into the nucleus, it is targeted by the host cell repression machinery. PML-NB components, like Daxx, thereby mediate transcriptional repression of the Ad genome, specifically, of the immediate-early E1A promoter (reviewed in references 64 and 68). Interestingly, this is counteracted by the capsid protein VI, which enters the nucleus in association with the Ad genome (160). In the course of an adenoviral infection, Daxx is targeted for proteasomal degradation by the early E1B-55K protein (161).
Another early protein, E4orf3, associates with PML nuclear bodies and induces their disruption from punctuate foci into track-like structures (162, 163). Specifically, PML, Sp100, Daxx, SUMO-1, and TIF1α are relocated into these tracks by E4orf3 (163–166), which might resemble nuclear aggresomes (167). These PML-containing structures are thereafter localized adjacent to viral RCs. Thus, the antiviral response is inhibited, whereas specific components of these nuclear bodies could be exploited by the virus for efficient replication (162, 163).
Apart from the interferon-dependent antiviral defense mediated by PML-NB components, E4orf3 targets the Mre11-Rad50-Nbs1 (MRN) DNA damage-sensing complex into tracks (168). Ultimately, the MRN complex is redistributed to cytoplasmic aggresomes by E4orf3 (169). Furthermore, Mre11 and other cellular proteins are ubiquitinated by E1B-55K and E4orf6, which assemble a Cullin-dependent E3 ubiquitin ligase complex and thus target proteins for proteasomal degradation (reviewed in reference 170). Besides Mre11, other proteins of the cellular DNA damage response, including the DNA ligase IV and Bloom helicase (BLM), are conducted to E1B-55K/E4orf6 complex-dependent degradation to prevent concatenation of viral dsDNA genomes mediated by this pathway (168, 171, 172).
Another target protein of the E3 ubiquitin ligase complex is the tumor suppressor protein p53. During the early phase of expression, p53 is induced by E1A (see below). Since p53 transactivates several downstream proteins, p53 induction by E1A is potently proapoptotic (173). Besides host cell proteins like the tumor suppressor p53, E1A also activates the transcription of viral early genes. Both are mediated by the exclusive binding of E1A to the retinoblastoma tumor suppressor (pRB) (174–177). This leads to dissociation of pRB from the E2F transcription factor and subsequently to the constitutive activation of the E2F-responsive cellular and the viral E2 early promoter (178, 179). Thus, E1A economically induces cell cycle progression as well as expression of viral proteins required for viral DNA replication.
The transition to the late phase of Ad infection is marked by the onset of genome replication and subsequent expression of genes from the major late transcription unit (MLTU). This switch coincides with formation of the viral RCs (180) and further reorganization of nuclear bodies (181–185). Adenoviral DNA replication is initiated by a unique priming mechanism that involves a viral protein primer, the terminal protein (TP), and is accomplished by the viral E2B DNA polymerase. Additionally, viral and cellular proteins involved in host cell DNA replication are recruited to sites of viral DNA synthesis (186, 187). Single-stranded DNA products are associated with the ssDNA binding protein (DBP) and form nuclear inclusions that are variable in size and appear as crescents or spheres (180, 185). Surrounding these sites of ssDNA accumulation is the peripheral replicative zone, where both replication and transcription take place. Punctate sites of active replication are arranged as a ring surrounding ssDNA. The replicated dsDNA, however, is displaced from these replication foci to the periphery, where it serves as a template for the transcription of viral late genes (185). Active transcription leads to the formation of a ring-shaped transcription and splicing zone surrounding ssDNA and replication foci (188). This zone contains dsDNA and nascent viral RNA, as well as transcription and splicing factors. These factors, which play an important role in enhancing Ad gene expression, are recruited from CB, which progressively disassemble during the late phase, and replication centers coalesce as they occupy regions of interchromatin granules (181, 183, 185, 189, 191).
Transcription and splicing, on the other hand, are tightly linked to the preferential export of Ad transcripts in the late phase of infection. Ongoing Ad replication leads to the concentration of splicing factors and viral RNA in interchromatin granules, which become progressively larger (182, 190). This might enhance processing or trafficking, as well as transport of RNAs. The preferential export of Ad late transcripts is mediated by the early Ad proteins E1B-55K and E4orf6. Interestingly, both proteins are present in viral RCs (191). The underlying mechanism, however, is still unclear, although recent results point to different scenarios. First, the activity of the E3 ubiquitin ligase complex is necessary for efficient export (189, 192). Since during Ad replication the transport of bulk cellular mRNA is simultaneously blocked (193, 194), it is likely that one or more proteins implicated in their export are targeted for proteasomal degradation, which might, in turn, promote preferential export of viral transcripts. Second, further studies suggested that the cellular export receptor of bulk cellular mRNA, TAP/NXF1, participates in the export of Ad late transcripts (195). An interesting hypothesis is that cellular export factors are relocalized to viral transcription and replication centers and thus promote efficient export of newly transcribed mRNAs out of the nucleus. Simultaneously, these proteins would be depleted in the rest of the nuclear compartment, which could also account for the blockage of cellular mRNA transport. However, this only applies to the late phase of infection, when RCs are formed. During the early phase Ad exploits the CRM1-dependent export pathway for its transcripts (196).
Similar to the Herpesviridae, Ad replication centers form adjacent to PML-NBs and are therefore likely to both counteract PML-dependent antiviral activities and benefit from components and the scaffold from the PML-NB. Interestingly, the E1B-55K and E4orf3 proteins, are known to participate in SUMOylation of cellular substrates (197, 198), and both associate with PML-NBs (199).
Parvoviridae.
Members of the family Parvoviridae differ in some interesting aspects from other viruses that replicate in vertebrates. In contrast to DNA tumor viruses, parvoviruses are not capable of inducing S-phase entry of the host cell; rather, the virus remains inactive unless the host cell itself enters the S phase. Furthermore, parvoviruses that infect vertebrates can be distinguished into two groups: autonomously replicating viruses and viruses that depend on coinfection of a helper virus and therefore are classified as dependoviruses (200).
Adeno-associated viruses.
Members belonging to the Dependovirus genus are the so-called adeno-associated viruses (AAV), which were first described as requiring adenovirus coinfection (201, 202). More recently, HSV-1 and -2, CMV, and pseudorabies virus have been identified as helper viruses (203–206). In the absence of a helper virus, the AAV genome integrates into the host cell genome (207, 208). In the event of helper virus infection of such latently infected cells, however, AAV gene expression is reactivated and the replication cycle progresses (209).
AAV contain two open reading frames that encode four nonstructural (Rep) and three structural (Cap) proteins (210–212), as well as a recently identified protein involved in capsid assembly, the assembly-associated protein AAP (213). Rep78 and Rep68 are known to play a key role during AAV DNA amplification (211, 212), whereas Rep52 and Rep40 are assumed to be involved in DNA packaging (214, 215).
Furthermore, AAV utilizes helper viral as well as cellular proteins for its own replication. So far, adenoviruses have been most extensively studied in the context of AAV helper virus, and thus will be mainly discussed here. Various Ad early gene products involved in regulating different processes during Ad infection are required for complete Ad helper virus function (208). Thus, Ad E1A enhances transcription of both Ad early genes as well as AAV Rep and Cap genes (216), and it activates the cellular gene expression required for S-phase entry and synthesis of DNA replication proteins (208). Additionally, the Ad-dependent E3 ubiquitin ligase regulates AAV gene expression, most likely by facilitating mRNA transport and promoting AAV DNA replication (217). The adenoviral DNA binding protein DBP stimulates AAV gene expression and DNA elongation, as well as AAV particle formation (218, 219). Thus, DBP is the only Ad protein, which has been proposed to be directly involved in AAV genome amplification (219), in contrast to the Ad protein primer and DNA polymerase, which are both dispensable for AAV replication.
Intriguingly, AAV further usurps Ad replication compartments for its own DNA replication in PML-NB-associated RCs (220). Therefore, AAV genome and Rep proteins are relocalized to Ad RCs (221). To ensure AAV DNA amplification, apart from Ad DBP, cellular components are also required in these nuclear structures, including RPA, the replication factor C (RFC), PCNA, and DNA polymerase δ (222, 223). Thus, an adenovirus coinfection not only provides AAV with essential Ad proteins and factors but also reprograms the host cell accordingly. However, assembly of AAV progeny virions is not conducted in the RCs but instead takes place in the nucleolus (213, 224).
Interestingly, host cell functions can compensate for a helper virus infection under specific conditions that induce a cellular stress response (225, 226). In agreement, the celluar DNA damage response is also induced during Ad coinfection. Recently, two independent studies found that DDR proteins become activated and in part relocalized to RCs, e.g., DNA-PK, phosphorylated Nbs1, Ku70/Ku86, and/or ATM (227, 228). Inhibition experiments have suggested that there is a distinct, yet unclear, requirement for cellular DDR proteins to foster or inhibit AAV replication (227).
Autonomous parvoviruses.
In contrast to the dependoviruses, autonomous parvoviruses are strictly dependent on host cell functions, especially on progression of the cell into the S phase (229, 230). The genome of autonomous parvoviruses comprises two transcription units, encoding the nonstructural proteins NS1 and NS2 and the capsid proteins VP1 and VP2 (231). NS1 is a multifunctional phosphoprotein that acts as a transcriptional activator of viral promoters as well as initiator and helicase in parvoviral DNA replication (232–237). Furthermore, NS1 colocalizes with replicating viral DNA in virus-induced nuclear foci (238, 239).
These compartments represent parvovirus replication centers termed autonomous PV-associated replication (APAR) bodies. Besides NS1 and parvovirus DNA, host cell replication proteins that facilitate viral genome amplification, like PCNA, RPA, and RFC, as well as DNA polymerase α and δ, accumulate in these bodies (238–241). Interestingly, the parvovirus RCs are distinct from any prominent nuclear structure, such as PML-NBs, nucleoli, coiled bodies, and/or speckled domains (238). Thus, parvovirus induces the formation of novel structures in the host cell nucleus and simultaneously recruits required cellular proteins to these compartments. Accumulation of viral DNA and capsid components leads to the expansion of these RCs, as they ultimately occupy most of the host cell nucleus (242). To date, controversial data have been collected on whether capsid assembly occurs in the nuclear (224, 243, 244) or cytoplasmic (245) compartment. It seems likely that the capsid protein VP2 triggers cytoplasmic assembly of VP1/VP2 oligomers and, additionally, mediates nuclear import of these complexes by its nuclear import signal. Sequentially, viral capsids might be assembled in the nuclear compartment (246).
Similar to AAV, minute virus of mice (MVM), an autonomous parvovirus, has recently been described to induce a robust DNA damage response and to exploit this cellular pathway for its own benefit. DDR sensor proteins (Nbs1, Mre11, ATM, DNA-PKs, and RPA) and signaling proteins (γH2AX, Ku70, and Ku86) accumulate in APAR bodies in response to active viral DNA replication. The virus-regulated modifications of cellular DDR include proteasome-dependent degradation of Mre11 and, seemingly, utilization of ATM, since inhibitors of this kinase restrict MVM replication. In contrast to AAV, DDR signaling in MVM-infected cells is mediated by this ATM kinase (247).
Polyomaviridae.
Binding of cellular transcription factors to the polyomaviral genome mediates its import into the nucleus. Subsequently, the genome is delivered to specific nuclear sites, namely, PML-NB (248).
During progression of the viral life cycle, various viral and cellular proteins colocalize with the polyomaviral (PyV) genome at PML-NBs (Fig. 1). These factors include proteins involved in gene expression and DNA replication as well as viral capsid proteins. Altogether, this suggests that these nuclear sites may be important for PyV replication. Intriguingly, studies on various PyV types have drawn different scenarios. Thus, the absence of PML, the main structural component of PML-NBs, either has no effect on or even enhances viral replication (249, 250). Specifically, BK viruses (BKV) seem to counteract PML-NB functions by dramatic reorganization of these structures and dispersal of two of its major components: Sp100 and Daxx (249). In contrast, although PML restricts JC virus replication, this virus does not directly modulate PML-NBs (250). Similarly, simian virus 40 (SV40) neither relocalizes PML-NB components nor disrupts these structures. Interestingly, transfection of the viral large T antigen TAg alone already induces its localization to PML-NBs. Thus, the deposition of viral RCs at PML-NBs seems not only to be a passive effect mediated by a PML-dependent antiviral defense mechanism, but also PyV seems to actively induce RCs at these sites (249, 250). The most detailed study on PyV factories was performed recently using a murine polyomavirus, and the investigators showed that although PyV RCs are located adjacent to PML-NBs, the establishment of these centers as well as effective virus growth did not depend on the PML protein (251).
In general, PyV DNA replication has been localized adjacent to PML-NBs (248, 249, 252, 253). Interestingly, SV40 gene expression also takes place in PyV RCs at PML-NBs. However, the spatial restriction to PML-NBs only accounts for DNA amplification, while transcription seems to be an indirect consequence following genome delivery to these nuclear sites (254).
Concomitant with the onset of viral DNA synthesis, late gene transcription is initiated. Following transcription of late genes, the structural proteins VP1, VP2, and VP3 are transported into the nucleus via their nuclear localization signal (NLS) (255, 256, 257). However, capsid proteins are not imported as monomers but as capsid subunits, especially as VP1 pentamers and VP2/3 complexes (258). Since large TAg and VP1 have been found to colocalize adjacent to PML-NBs, a spatial coupling of DNA replication and capsid assembly has been proposed (248, 252, 253). Intriguingly, Garcea and coworkers established a new and unanticipated mechanism of virion assembly (251) that is contrary to the assembly process of larger DNA viruses, including herpes- and adenoviruses, namely, the encapsidation of the viral genome into preformed capsids (98, 259). Already by 1980, polymerization of capsid subunits onto the viral genome had been proposed (260). However, VP1 pentamers seem to first assemble tubular structures, in which the viral genome is integrated, and finally icosahedral virion particles are formed by a budding mechanism (251).
Similar to parvoviruses and papillomaviruses (see below), polyomaviruses actively recruit cellular DDR proteins to the RCs and exploit their functions (261–264). Specifically, PyV recruit proteins involved in homologous recombination, such as ATM kinase and the MRN complex (261, 263), and activate these pathways (265). Both relocalization and activation are essential for PyV growth (261–265). Interestingly, the multifunctional large TAg induces replication foci containing PML and relocalizes DDR proteins to these structures, although this has been shown to be PML independent (251, 263). Similar to adenoviruses, Mre11 is recruited to SV40 replication sites and is later degraded (263).
Papillomaviridae.
Compared to Herpesviridae and Adenoviridae, little is known about productive replication of papillomaviruses, as studying these viruses has been hindered by the absence of an appropriate cell culture system.
Papillomaviruses specifically infect squamous epithelial cells. The productive replication of papillomaviruses can be divided into an early and late phase, with the late phase, including viral DNA synthesis, production of capsid proteins, and virion assembly, being exclusively restricted to differentiated epithelial cells. It is likely that, upon infection, genome transport toward the nucleus is regulated by the L2 capsid protein via interaction with microtubules (266). Since cell division is required for nuclear genome translocation and expression (267), the viral genome may enter the cell nucleus only during mitotic membrane breakdown.
Comparable to other nuclear DNA viruses, the papillomavirus genome localizes to PML-NBs (Fig. 1). Intriguingly, these nuclear structures foster papillomavirus transcription (268), in contrast to most DNA viruses, which developed mechanisms to antagonize the restrictive function of PML-NBs.
In general, viral DNA replication is achieved by two early viral proteins, E1 and E2 (269–272). In contrast to E2, which is only required for initiation, E1 possesses ATPase and DNA helicase activities, thereby also triggering elongation of DNA synthesis (273, 274). Except for E1 and E2, the residual replication initiation machinery, including DNA polymerase α/primase, DNA polymerase δ/PCNA, RPA, and topoisomerases I and II, is provided by the host cell (275, 276).
Interestingly, like other DNA viruses, papillomavirus also seems to exploit the cellular SUMOylation system. Thus, E1 interacts with the cellular SUMO-conjugating enzyme Ubc9, seemingly triggering SUMO-1 modification of this viral protein. It has been shown that these functions are essential for the intranuclear accumulation of the protein as well as the efficient origin-dependent replication of the viral genome (277, 278).
Furthermore, papillomavirus DNA replication is tightly linked with the cellular DNA damage response. E1 and E2 both activate and relocalize the cellular DDR to viral RCs. The targeting of the cellular DDR leads to growth suppression of the host cell. While E2 recruits ATM DDR components to the viral RCs, E1 specifically activates this cellular ATM DDR pathway (279). Both growth arrest and ATM activation depend on the ATPase activity of E1 and its binding to the viral origin of replication. Interestingly, the cellular DDR is activated to facilitate viral DNA amplification, thus stimulating papillomavirus replication (280).
Prior to initial gene expression, the viral genome is localized to PML-NBs by the viral capsid protein L2, supporting viral early gene transcription (268). The presence of the HPV genome in basal cells leads to an increased total number of PML-NBs as well as elevated levels of posttranslationally modified PML protein (281). The HPV E6 oncoprotein has been described to colocalize with PML-NBs (282) and to induce proteasomal degradation of PML-IV, seemingly overcoming restrictive functions of these nuclear bodies (283). Interestingly, these cellular structures are also associated with papillomaviral RCs (269, 284–288). It has been proposed that L2 localizes to PML-NBs, where it associates with E2 and recruits the viral genome to sites of assembly. Subsequently, L1 could be recruited to RCs for progeny virion assembly (268, 284). Although, different studies observed no requirement of PML-NBs for papillomavirus DNA replication (281, 288), it is evident that these nuclear structures become reorganized by L2 (289) and are target sites for virion assembly.
CONCLUSIONS
As obligate intracellular parasites, viruses have coevolved with the host cell. Consequently, the changes in cellular activities and architecture that are induced in a virus-infected cell are likely to reflect a “tug-of-war” between the opposing cellular defenses and the mechanisms viruses employ to divert or utilize the cellular machinery and metabolic pathways. Assembly of virus inclusions or aggregates is an essential step for productive replication of RNA and DNA viruses from a wide variety of different families, and although these structures display different subcellular localizations, architectures, and compositions, they seem to share several fundamental similarities: (i) virus factories function as scaffolds that anchor viral genomes, concentrate viral and cellular macromolecules, and facilitate the assembly of replication complexes. RNA and DNA viruses that replicate in the cytoplasm make use of cellular membranes as scaffolds, whereas nuclear DNA viruses mostly rely on the nature and organization of nuclear domains, in particular PML-NB, for assembly of replication compartments that employ protein scaffolds. (ii) Although it is often not clear how viral genomes are directed to specific subcellular locations, the assembly of replication compartments is accompanied by the reorganization of the cytoskeleton and associated motor proteins. Often viral genomes are directed to the MTOC, where perinuclear aggresomes assemble, as has been found for togaviruses, flaviviruses, bunyaviruses, coronaviruses, and arteriviruses, as well as for several of the NCLDV. Interestingly, the genomes of DNA viruses are targeted to or by PML-NBs and also make use of nuclear and in some cases cytoplasmic aggresomes. This suggests that a cellular response dedicated to counter invasion of foreign/misfolded proteins may be responsible for targeting viral genomes and structural proteins, thus initiating an aggresome response that is coopted by viruses to generate assembly sites. Viruses from many different families either employ or modulate the ubiquitin-proteasome pathway for degradation of cellular substrates, making aggresomes appropriate sites where chaperones, ubiquitin, and 26S proteasomes are concentrated. (iii) Formation of cytoplasmic viral factories functions to conceal the viral genome and transcripts from an arsenal of cellular defense mechanisms that include the cytoplasmic dsRNA-sensing kinase (PKR), as well as the Toll-like receptors (TLRs) and cytoplasmic retinoic acid-inducible gene I (RIG-I) helicase receptors, which trigger the production of type I interferon. In the nucleus, PML-NBs are primary sites of the interferon-dependent as well as the intrinsic response to infection (64, 67, 290). Many proteins encoded by DNA and RNA viruses colocalize and perturb PML-NBs, suggesting that, as in the case of cytoplasmic aggresomes, alteration of PML-NBs may be a viral strategy to evade a cellular defense mechanism. Cellular activities that are regulated by PML-NBs include DNA damage repair, apoptosis, the ubiquitin pathway, and gene expression (136). Interestingly, as described in the previous sections, DNA viruses that replicate in the nucleus modulate, relocate, or induce the degradation of cellular factors implicated in each of these activities.
Progress in understanding the molecular mechanisms that underlie the formation of replication centers formed by DNA viruses is likely to continue to advance rapidly in the near future. For example, three-dimensional analyses of infected-cells via electron tomography will help in the study of their architecture, while biochemical studies should help determine their protein and nucleic acid compositions, as well as the interactions that are established between viral and cellular components.
ACKNOWLEDGMENTS
R.A.G. and T.D. received support from the Research Group Linkage Program of the Alexander von Humboldt Foundation. The Heinrich Pette Institute is supported by the Freie und Hansestadt Hamburg and the Bundesministerium für Gesundheit. R.A.G. received grants from CONACyT-SEP (CB-2011-01-168497) and Promep-SEP.
Footnotes
Published ahead of print 20 November 2013
REFERENCES
- 1.Netherton C, Moffat K, Brooks E, Wileman T. 2007. A guide to viral inclusions, membrane rearrangements, factories, and viroplasm produced during virus replication. Adv. Virus Res. 70:101–182. 10.1016/S0065-3527(07)70004-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Paul D, Bartenschlager R. 2013. Architecture and biogenesis of plus-strand RNA virus replication factories. World J. Virol. 2:32–48. 10.5501/wjv.v2.j2.32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dreux M, Chisari FV. 2010. Viruses and the autophagy machinery. Cell Cycle 9:1295–1307. 10.4161/cc.9.7.11109 [DOI] [PubMed] [Google Scholar]
- 4.Netherton CL, Wileman T. 2011. Virus factories, double membrane vesicles and viroplasm generated in animal cells. Curr. Opin. Virol. 1:381–387. 10.1016/j.coviro.2011.09.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Novoa RR, Calderita G, Arranz R, Fontana J, Granzow H, Risco C. 2005. Virus factories: associations of cell organelles for viral replication and morphogenesis. Biol. Cell 97:147–172. 10.1042/BC20040058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Colson P, De Lamballerie X, Yutin N, Asgari S, Bigot Y, Bideshi DK, Cheng X-W, Federici BA, Van Etten JL, Koonin EV, La Scola B, Raoult D. 2013. “Megavirales,” a proposed new order for eukaryotic nucleocytoplasmic large DNA viruses. Arch. Virol. 158:2517–2521. 10.1007/s00705-013-1768-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Iyer LM, Aravind L, Koonin EV. 2001. Common origin of four diverse families of large eukaryotic DNA viruses. J. Virol. 75:11720–11734. 10.1128/JVI.75.23.11720-11734.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Iyer LM, Balaji S, Koonin EV, Aravind L. 2006. Evolutionary genomics of nucleo-cytoplasmic large DNA viruses. Virus Res. 117:156–184. 10.1016/j.virusres.2006.01.009 [DOI] [PubMed] [Google Scholar]
- 9.Netherton CL, Wileman TE. 2013. African swine fever virus organelle rearrangements. Virus Res. 173:76–86. 10.1016/j.virusres.2012.12.014 [DOI] [PubMed] [Google Scholar]
- 10.Kopito RR. 2000. Aggresomes, inclusion bodies and protein aggregation. Trends Cell Biol. 10:524–530. 10.1016/S0962-8924(00)01852-3 [DOI] [PubMed] [Google Scholar]
- 11.Wileman T. 2007. Aggresomes and pericentriolar sites of virus assembly: cellular defense or viral design? Annu. Rev. Microbiol. 61:149–167. 10.1146/annurev.micro.57.030502.090836 [DOI] [PubMed] [Google Scholar]
- 12.Schramm B, Locker JK. 2005. Cytoplasmic organization of POXvirus DNA replication. Traffic 6:839–846. 10.1111/j.1600-0854.2005.00324.x [DOI] [PubMed] [Google Scholar]
- 13.Mallardo M, Leithe E, Schleich S, Roos N, Doglio L, Krijnse-Locker J. 2002. Relationship between vaccinia virus intracellular cores, early mRNAs, and DNA replication sites. J. Virol. 76:5167–5183. 10.1128/JVI.76.10.5167-5183.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mercer J, Snijder B, Sacher R, Burkard C, Bleck CKE, Stahlberg H, Pelkmans L, Helenius A. 2012. RNAi screening reveals proteasome- and Cullin3-dependent stages in vaccinia virus infection. Cell Rep. 2:1036–1047. 10.1016/j.celrep.2012.09.003 [DOI] [PubMed] [Google Scholar]
- 15.Katsafanas GC, Moss B. 2007. Colocalization of transcription and translation within cytoplasmic poxvirus factories coordinates viral expression and subjugates host functions. Cell Host Microbe 2:221–228. 10.1016/j.chom.2007.08.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tolonen N, Doglio L, Schleich S, Krijnse-Locker J. 2001. Vaccinia virus DNA replication occurs in endoplasmic reticulum-enclosed cytoplasmic mini-nuclei. Mol. Biol. Cell 12:2031–2046. 10.1019/mbc.12.7.2031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Doglio L, De Marco A, Schleich S, Roos N, Krijnse-Locker J. 2002. The vaccinia virus E8R gene product: a viral membrane protein that is made early in infection and packaged into the virions' core. J. Virol. 76:9773–9786. 10.1128/JVI.76.19.9773-9786.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Broyles SS. 2003. Vaccinia virus transcription. J. Gen. Virol. 84:2293–2303. 10.1099/vir.0.18942-0 [DOI] [PubMed] [Google Scholar]
- 19.Moss B. 2001. Poxviridae: the viruses and their replication, p 2849–2883 In] ?>Knipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA, Roizman B, Straus SE. (ed), Fields virology, 4th ed. Lippincot Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 20.Beaud G. 1995. Vaccinia virus DNA replication: a short review. Biochimie 77:774–779. 10.1016/0300-9084(96)88195-8 [DOI] [PubMed] [Google Scholar]
- 21.Brum LM, Lopez MC, Varela JC, Baker HV, Moyer RW. 2003. Microarray analysis of A549 cells infected with rabbitpox virus (RPV): a comparison of wild-type RPV and RPV deleted for the host range gene, SPI-1. Virology 315:322–334. 10.1016/S0042-6822(03)00532-4 [DOI] [PubMed] [Google Scholar]
- 22.Guerra S, López-Fernández LA, Pascual-Montano A, Muñoz M, Harshman K, Esteban M. 2003. Cellular gene expression survey of vaccinia virus infection of human HeLa cells. J. Virol. 77:6493–6506. 10.1128/JVI.77.11.6493-6506.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rice AP, Roberts BE. 1983. Vaccinia virus induces cellular mRNA degradation. J. Virol. 47:529–539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Parrish S, Moss B. 2007. Characterization of a second vaccinia virus mRNA-decapping enzyme conserved in poxviruses. J. Virol. 81:12973–12978. 10.1128/JVI.01668-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shors T, Keck JG, Moss B. 1999. Down regulation of gene expression by the vaccinia virus D10 protein. J. Virol. 73:791–796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bablanian R, Goswami SK, Esteban M, Banerjee AK, Merrick WC. 1991. Mechanism of selective translation of vaccinia virus mRNAs: differential role of poly(A) and initiation factors in the translation of viral and cellular mRNAs. J. Virol. 65:4449–4460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Condit RC, Moussatche N, Traktman P. 2006. In a nutshell: structure and assembly of the vaccinia virion. Adv. Virus Res. 66:31–124. 10.1016/S0065-3527(06)66002-8 [DOI] [PubMed] [Google Scholar]
- 28.Ballester M, Galindo-Cardiel I, Gallardo C, Argilaguet JM, Segalés J, Rodriguez JM, Rodriguez F. 2010. Intranuclear detection of African swine fever virus DNA in several cell types from formalin-fixed and paraffin-embedded tissues using a new in situ hybridisation protocol. J. Virol. Methods 168:38–43. 10.1016/j.viromet.2010.04.013 [DOI] [PubMed] [Google Scholar]
- 29.Brookes SM, Dixon LK, Parkhouse RM. 1996. Assembly of African swine fever virus: quantitative ultrastructural analysis in vitro and in vivo. Virology 224:84–92. 10.1006/viro.1996.0509 [DOI] [PubMed] [Google Scholar]
- 30.Garcia-Beato R, Salas ML, Vinuela E, Salas J. 1992. Role of the host cell nucleus in the replication of African swine fever virus DNA. Virology 188:637–649. 10.1016/0042-6822(92)90518-T [DOI] [PubMed] [Google Scholar]
- 31.Rojo G, Garcia-Beato R, Vinuela E, Salas ML, Salas J. 1999. Replication of African swine fever virus DNA in infected cells. Virology 257:524–536. 10.1006/viro.1999.9704 [DOI] [PubMed] [Google Scholar]
- 32.Ortín J, Enjuanes L, Vinuela E. 1979. Cross-links in African swine fever virus DNA. J. Virol. 31:579–583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Caeiro F, Meireles M, Ribeiro G, Costa JV. 1990. In vitro DNA replication by cytoplasmic extracts from cells infected with African swine fever virus. Virology 179:87–94. 10.1016/0042-6822(90)90277-X [DOI] [PubMed] [Google Scholar]
- 34.Eulalio A, Nunes-Correia I, Carvalho AL, Faro C, Citovsky V, Simoes S, Pedroso de Lima MC. 2004. Two African swine fever virus proteins derived from a common precursor exhibit different nucleocytoplasmic transport activities. J. Virol. 78:9731–9739. 10.1128/JVI.78.18.9731-9739.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Eulalio A, Nunes-Correia I, Salas J, Salas ML, Simoes S, Pedroso de Lima MC. 2007. African swine fever virus p37 structural protein is localized in nuclear foci containing the viral DNA at early post-infection times. Virus Res. 130:18–27. 10.1016/j.virusres.2007.05.009 [DOI] [PubMed] [Google Scholar]
- 36.Stefanovic S, Windsor M, Nagata K-I, Inagaki M, Wileman T. 2005. Vimentin rearrangement during African swine fever virus infection involves retrograde transport along microtubules and phosphorylation of vimentin by calcium calmodulin kinase II. J. Virol. 79:11766–11775. 10.1128/JVI.79.18.11766-11775.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kuznar J, Salas ML, Vinuela E. 1980. DNA-dependent RNA polymerase in African swine fever virus. Virology 101:169–175. 10.1016/0042-6822(80)90493-6 [DOI] [PubMed] [Google Scholar]
- 38.Salas ML, Kuznar J, Vinuela E. 1981. Polyadenylation, methylation, and capping of the RNA synthesized in vitro by African swine fever virus. Virology 113:484–491. 10.1016/0042-6822(81)90176-8 [DOI] [PubMed] [Google Scholar]
- 39.Carvalho ZG, Rodrigues-Pousada C. 1986. African swine fever virus gene expression in infected Vero cells. J. Gen. Virol. 67:1343–1350 [DOI] [PubMed] [Google Scholar]
- 40.Salas ML, Rey-Campos J, Almendral JM, Talavera A, Vinuela E. 1986. Transcription and translation maps of African swine fever virus. Virology 152:228–240. 10.1016/0042-6822(86)90387-9 [DOI] [PubMed] [Google Scholar]
- 41.Santaren JF, Vinuela E. 1986. African swine fever virus-induced polypeptides in Vero cells. Virus Res. 5:391–405. 10.1016/0168-1702(86)90031-6 [DOI] [PubMed] [Google Scholar]
- 42.Esteves A, Ribeiro G, Costa JV. 1987. DNA-binding proteins specified by African swine fever virus. Virology 161:403–409. 10.1016/0042-6822(87)90133-4 [DOI] [PubMed] [Google Scholar]
- 43.Yáñez RJ, Rodriguez JM, Nogal ML, Yuste L, Enríquez C, Rodriguez JF, Vinuela E. 1995. Analysis of the complete nucleotide sequence of African swine fever virus. Virology 208:249–278. 10.1006/viro.1995.1149 [DOI] [PubMed] [Google Scholar]
- 44.Ballester M, Rodríguez-Cariño C, Pérez M, Gallardo C, Rodríguez JM, Salas ML, Rodriguez F. 2011. Disruption of nuclear organization during the initial phase of African swine fever virus infection. J. Virol. 85:8263–8269. 10.1128/JVI.00704-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sánchez EG, Quintas A, Nogal M, Castelló A, Revilla Y. 2013. African swine fever virus controls the host transcription and cellular machinery of protein synthesis. Virus Res. 173:58–75. 10.1016/j.virusres.2012.10.025 [DOI] [PubMed] [Google Scholar]
- 46.Heath CM, Windsor M, Wileman T. 2001. Aggresomes resemble sites specialized for virus assembly. J. Cell Biol. 153:449–455. 10.1083/jcb.153.3.449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Carvalho ZG, De Matos AP, Rodrigues-Pousada C. 1988. Association of African swine fever virus with the cytoskeleton. Virus Res. 11:175–192. 10.1016/0168-1702(88)90042-1 [DOI] [PubMed] [Google Scholar]
- 48.De Matos AP, Carvalho ZG. 1993. African swine fever virus interaction with microtubules. Biol. Cell 78:229–234. 10.1016/0248-4900(93)90134-Z [DOI] [PubMed] [Google Scholar]
- 49.Alonso C, Miskin J, Hernaez B, Fernandez-Zapatero P, Soto L, Canto C, Rodriguez-Crespo I, Dixon L, Escribano JM. 2001. African swine fever virus protein p54 interacts with the microtubular motor complex through direct binding to light-chain dynein. J. Virol. 75:9819–9827. 10.1128/JVI.75.20.9819-9827.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rojo G, Chamorro M, Salas ML, Vinuela E, Cuezva JM, Salas J. 1998. Migration of mitochondria to viral assembly sites in African swine fever virus-infected cells. J. Virol. 72:7583–7588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Breese SSJ, DeBoer CJ. 1966. Electron microscope observations of African swine fever virus in tissue culture cells. Virology 28:420–428. 10.1016/0042-6822(66)90054-7 [DOI] [PubMed] [Google Scholar]
- 52.McCrossan M, Windsor M, Ponnambalam S, Armstrong J, Wileman T. 2001. The trans Golgi network is lost from cells infected with African swine fever virus. J. Virol. 75:11755–11765. 10.1128/JVI.75.23.11755-11765.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hingamp PM, Arnold JE, Mayer RJ, Dixon LK. 1992. A ubiquitin conjugating enzyme encoded by African swine fever virus. EMBO J. 11:361–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Castelló A, Quintas A, Sánchez EG, Sabina P, Nogal M, Carrasco L, Revilla Y. 2009. Regulation of host translational machinery by African swine fever virus. PLoS Pathog. 5(8):e1000562. 10.1371/journal.ppat.1000562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Salas ML, Andrés G. 2013. African swine fever virus morphogenesis. Virus Res. 173:29–41. 10.1016/j.virusres.2012.09.016 [DOI] [PubMed] [Google Scholar]
- 56.Williams T, Barbosa-Solomieu V, Chinchar VG. 2005. A decade of advances in iridovirus research. Adv. Virus Res. 65:173–248. 10.1016/S0065-3527(05)65006-3 [DOI] [PubMed] [Google Scholar]
- 57.Yamada T, Onimatsu H, Van Etten JL. 2006. Chlorella viruses. Adv. Virus Res. 66:293–336. 10.1016/S0065-3527(06)66006-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jiang M, Imperiale MJ. 2012. Design stars: how small DNA viruses remodel the host nucleus. Future Virol. 7:445–459. 10.2217/fvl.12.38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Whittaker GR, Kann M, Helenius A. 2000. Viral entry into the nucleus. Annu. Rev. Cell Dev. Biol. 16:627–651. 10.1146/annurev.cellbio.16.1.627 [DOI] [PubMed] [Google Scholar]
- 60.Smith AE, Helenius A. 2004. How viruses enter animal cells. Science 304:237–242. 10.1126/science.1094823 [DOI] [PubMed] [Google Scholar]
- 61.Carracedo A, Ito K, Pandolfi PP. 2011. The nuclear bodies inside out: PML conquers the cytoplasm. Curr. Opin. Cell Biol. 23:360–366. 10.1016/j.ceb.2011.03.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Everett RD. 2001. DNA viruses and viral proteins that interact with PML nuclear bodies. Oncogene 20:7266–7273. 10.1038/sj.onc.1204759 [DOI] [PubMed] [Google Scholar]
- 63.Everett RD. 2006. Interactions between DNA viruses, ND10 and the DNA damage response. Cell. Microbiol. 8:365–374. 10.1111/j.1462-5822.2005.00677.x [DOI] [PubMed] [Google Scholar]
- 64.Geoffroy M-C, Chelbi-Alix MK. 2011. Role of promyelocytic leukemia protein in host antiviral defense. J. Interferon Cytokine Res. 31:145–158. 10.1089/jir.2010.0111 [DOI] [PubMed] [Google Scholar]
- 65.Fu L, Gao Y-S, Tousson A, Shah A, Chen T-LL, Vertel BM, Sztul E. 2005. Nuclear aggresomes form by fusion of PML-associated aggregates. Mol. Biol. Cell 16:4905–4917. 10.1091/mbc.E05-01-0019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Saffert RT, Kalejta RF. 2008. Promyelocytic leukemia-nuclear body proteins: herpesvirus enemies, accomplices, or both? Future Virol. 3:265–277. 10.2217/17460794.3.3.265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tavalai N, Stamminger T. 2008. New insights into the role of the subnuclear structure ND10 for viral infection. Biochim. Biophys. Acta 1783:2207–2221 [DOI] [PubMed] [Google Scholar]
- 68.Everett RD, Chelbi-Alix MK. 2007. PML and PML nuclear bodies: implications in antiviral defence. Biochimie 89:819–830. 10.1016/j.biochi.2007.01.004 [DOI] [PubMed] [Google Scholar]
- 69.Maul GG, Ishov AM, Everett RD. 1996. Nuclear domain 10 as preexisting potential replication start sites of herpes simplex virus type-1. Virology 217:67–75. 10.1006/viro.1996.0094 [DOI] [PubMed] [Google Scholar]
- 70.Everett RD, Murray J. 2005. ND10 components relocate to sites associated with herpes simplex virus type 1 nucleoprotein complexes during virus infection. J. Virol. 79:5078–5089. 10.1128/JVI.79.8.5078-5089.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Everett RD, Sourvinos G, Leiper C, Clements JB, Orr A. 2004. Formation of nuclear foci of the herpes simplex virus type 1 regulatory protein ICP4 at early times of infection: localization, dynamics, recruitment of ICP27, and evidence for the de novo induction of ND10-like complexes. J. Virol. 78:1903–1917. 10.1128/JVI.78.4.1903-1917.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cuchet-Lourenço D, Boutell C, Lukashchuk V, Grant K, Sykes A, Murray J, Orr A, Everett RD. 2011. SUMO pathway dependent recruitment of cellular repressors to herpes simplex virus type 1 genomes. PLoS Pathog 7(7):e1002123. 10.1371/journal.ppat.1002123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mohni KN, Livingston CM, Cortez D, Weller SK. 2010. ATR and ATRIP are recruited to herpes simplex virus type 1 replication compartments even though ATR signaling is disabled. J. Virol. 84:12152–12164. 10.1128/JVI.01643-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Maul GG, Everett RD. 1994. The nuclear location of PML, a cellular member of the C3HC4 zinc-binding domain protein family, is rearranged during herpes simplex virus infection by the C3HC4 viral protein ICP0. J. Gen. Virol. 75:1223–1233 [DOI] [PubMed] [Google Scholar]
- 75.Maul GG, Guldner HH, Spivack JG. 1993. Modification of discrete nuclear domains induced by herpes simplex virus type 1 immediate early gene 1 product (ICP0). J. Gen. Virol. 74:2679–2690 [DOI] [PubMed] [Google Scholar]
- 76.Boutell C, Sadis S, Everett RD. 2002. Herpes simplex virus type 1 immediate-early protein ICP0 and is isolated RING finger domain act as ubiquitin E3 ligases in vitro. J. Virol. 76:841–850. 10.1128/JVI.76.2.841-850.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Everett RD, Maul GG. 1994. HSV-1 IE protein Vmw110 causes redistribution of PML. EMBO J. 13:5062–5069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chelbi-Alix MK, de The H. 1999. Herpes virus induced proteasome-dependent degradation of the nuclear bodies-associated PML and Sp100 proteins. Oncogene 18:935–941. 10.1038/sj.onc.1202366 [DOI] [PubMed] [Google Scholar]
- 79.Parkinson J, Everett RD. 2000. Alphaherpesvirus proteins related to herpes simplex virus type 1 ICP0 affect cellular structures and proteins. J. Virol. 74:10006–10017. 10.1128/JVI.74.21.10006-10017.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Everett RD, Freemont P, Saitoh H, Dasso M, Orr A, Kathoria M, Parkinson J. 1998. The disruption of ND10 during herpes simplex virus infection correlates with the Vmw110- and proteasome-dependent loss of several PML isoforms. J. Virol. 72:6581–6591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Burkham J, Coen DM, Weller SK. 1998. ND10 protein PML is recruited to herpes simplex virus type 1 prereplicative sites and replication compartments in the presence of viral DNA polymerase. J. Virol. 72:10100–10107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Carrington-Lawrence SD, Weller SK. 2003. Recruitment of polymerase to herpes simplex virus type 1 replication foci in cells expressing mutant primase (UL52) proteins. J. Virol. 77:4237–4247. 10.1128/JVI.77.7.4237-4247.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wilkinson DE, Weller SK. 2004. Recruitment of cellular recombination and repair proteins to sites of herpes simplex virus type 1 DNA replication is dependent on the composition of viral proteins within prereplicative sites and correlates with the induction of the DNA damage response. J. Virol. 78:4783–4796. 10.1128/JVI.78.9.4783-4796.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lamberti C, Weller SK. 1996. The herpes simplex virus type 1 UL6 protein is essential for cleavage and packaging but not for genomic inversion. Virology 226:403–407. 10.1006/viro.1996.0668 [DOI] [PubMed] [Google Scholar]
- 85.Quinlan MP, Chen LB, Knipe DM. 1984. The intranuclear location of a herpes simplex virus DNA-binding protein is determined by the status of viral DNA replication. Cell 36:857–868. 10.1016/0092-8674(84)90035-7 [DOI] [PubMed] [Google Scholar]
- 86.Burch AD, Weller SK. 2004. Nuclear sequestration of cellular chaperone and proteasomal machinery during herpes simplex virus type 1 infection. J. Virol. 78:7175–7185. 10.1128/JVI.78.13.7175-7185.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Burch AD, Weller SK. 2005. Herpes simplex virus type 1 DNA polymerase requires the mammalian chaperone hsp90 for proper localization to the nucleus. J. Virol. 79:10740–10749. 10.1128/JVI.79.16.10740-10749.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Li L, Johnson LA, Dai-Ju JQ, Sandri-Goldin RM. 2008. Hsc70 focus formation at the periphery of HSV-1 transcription sites requires ICP27. PLoS One 3(1):e1491. 10.1371/journal.pone.0001491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Livingston CM, DeLuca NA, Wilkinson DE, Weller SK. 2008. Oligomerization of ICP4 and rearrangement of heat shock proteins may be important for herpes simplex virus type 1 prereplicative site formation. J. Virol. 82:6324–6336. 10.1128/JVI.00455-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ward PL, Ogle WO, Roizman B. 1996. Assemblons: nuclear structures defined by aggregation of immature capsids and some tegument proteins of herpes simplex virus 1. J. Virol. 70:4623–4631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hutchinson I, Whiteley A, Browne H, Elliott G. 2002. Sequential localization of two herpes simplex virus tegument proteins to punctate nuclear dots adjacent to ICP0 domains. J. Virol. 76:10365–10373. 10.1128/JVI.76.20.10365-10373.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Everett RD, Sourvinos G, Orr A. 2003. Recruitment of herpes simplex virus type 1 transcriptional regulatory protein ICP4 into foci juxtaposed to ND10 in live, infected cells. J. Virol. 77:3680–3689. 10.1128/JVI.77.6.3680-3689.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Jenkins HL, Spencer CA. 2001. RNA polymerase II holoenzyme modifications accompany transcription reprogramming in herpes simplex virus type 1-infected cells. J. Virol. 75:9872–9884. 10.1128/JVI.75.20.9872-9884.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Tian X, Devi-Rao G, Golovanov AP, Sandri-Goldin RM. 2013. The interaction of the cellular export adaptor protein Aly/REF with ICP27 contributes to the efficiency of herpes simplex virus 1 mRNA export. J. Virol. 87:7210–7217. 10.1128/JVI.00738-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Taylor TJ, Knipe DM. 2004. Proteomics of herpes simplex virus replication compartments: association of cellular DNA replication, repair, recombination, and chromatin remodeling proteins with ICP8. J. Virol. 78:5856–5866. 10.1128/JVI.78.11.5856-5866.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ebert SN, Subramanian D, Shtrom SS, Chung IK, Parris DS, Muller MT. 1994. Association between the p170 form of human topoisomerase II and progeny viral DNA in cells infected with herpes simplex virus type 1. J. Virol. 68:1010–1020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Bowman BR, Welschhans RL, Jayaram H, Stow ND, Preston VG, Quiocho FA. 2006. Structural characterization of the UL25 DNA-packaging protein from herpes simplex virus type 1. J. Virol. 80:2309–2317. 10.1128/JVI.80.5.2309-2317.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Brown JC, Newcomb WW. 2011. Herpesvirus capsid assembly: insights from structural analysis. Curr. Opin. Virol. 1:142–149. 10.1016/j.coviro.2011.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Newcomb WW, Homa FL, Thomsen DR, Booy FP, Trus BL, Steven AC, Spencer JV, Brown JC. 1996. Assembly of the herpes simplex virus capsid: characterization of intermediates observed during cell-free capsid formation. J. Mol. Biol. 263:432–446 [DOI] [PubMed] [Google Scholar]
- 100.Newcomb WW, Homa FL, Brown JC. 2005. Involvement of the portal at an early step in herpes simplex virus capsid assembly. J. Virol. 79:10540–10546. 10.1128/JVI.79.16.10540-10546.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Yang K, Homa F, Baines JD. 2007. Putative terminase subunits of herpes simplex virus 1 form a complex in the cytoplasm and interact with portal protein in the nucleus. J. Virol. 81:6419–6433. 10.1128/JVI.00047-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Arvin A, Campadelli-Fiume G, Mocarski E, Moore PS, Roizman B, Whitley R, Yamanishi K. 2007. The egress of alphaherpesviruses from the cell. Cambridge University Press, Cambridge, United Kingdom: [PubMed] [Google Scholar]
- 103.Lilley CE, Carson CT, Muotri AR, Gage FH, Weitzman MD. 2005. DNA repair proteins affect the lifecycle of herpes simplex virus 1. Proc. Natl. Acad. Sci. U. S. A. 102:5844–5849. 10.1073/pnas.0501916102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Lees-Miller SP, Long MC, Kilvert MA, Lam V, Rice SA, Spencer CA. 1996. Attenuation of DNA-dependent protein kinase activity and its catalytic subunit by the herpes simplex virus type 1 transactivator ICP0. J. Virol. 70:7471–7477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Parkinson J, Lees-Miller SP, Everett RD. 1999. Herpes simplex virus type 1 immediate-early protein vmw110 induces the proteasome-dependent degradation of the catalytic subunit of DNA-dependent protein kinase. J. Virol. 73:650–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Lilley CE, Chaurushiya MS, Boutell C, Landry S, Suh J, Panier S, Everett RD, Stewart GS, Durocher D, Weitzman MD. 2010. A viral E3 ligase targets RNF8 and RNF168 to control histone ubiquitination and DNA damage responses. EMBO J. 29:943–955. 10.1038/emboj.2009.400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Chaurushiya MS, Lilley CE, Aslanian A, Meisenhelder J, Scott DC, Landry S, Ticau S, Boutell C, Yates JR, Schulman BA, Hunter T, Weitzman MD. 2012. Viral E3 ubiquitin ligase-mediated degradation of a cellular E3: viral mimicry of a cellular phosphorylation mark targets the RNF8 FHA domain. Mol. Cell 46:79–90. 10.1016/j.molcel.2012.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Wilkinson DE, Weller SK. 2006. Herpes simplex virus type I disrupts the ATR-dependent DNA-damage response during lytic infection. J. Cell Sci. 119:2695–2703. 10.1242/jcs.02981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Marshall KR, Rowley KV, Rinaldi A, Nicholson IP, Ishov AM, Maul GG, Preston CM. 2002. Activity and intracellular localization of the human cytomegalovirus protein pp71. J. Gen. Virol. 83:1601–1612 [DOI] [PubMed] [Google Scholar]
- 110.Cantrell SR, Bresnahan WA. 2005. Interaction between the human cytomegalovirus UL82 gene product (pp71) and hDaxx regulates immediate-early gene expression and viral replication. J. Virol. 79:7792–7802. 10.1128/JVI.79.12.7792-7802.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Ishov AM, Vladimirova OV, Maul GG. 2002. Daxx-mediated accumulation of human cytomegalovirus tegument protein pp71 at ND10 facilitates initiation of viral infection at these nuclear domains. J. Virol. 76:7705–7712. 10.1128/JVI.76.15.7705-7712.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Hwang J, Kalejta RF. 2007. Proteasome-dependent, ubiquitin-independent degradation of Daxx by the viral pp71 protein in human cytomegalovirus-infected cells. Virology 367:334–338. 10.1016/j.virol.2007.05.037 [DOI] [PubMed] [Google Scholar]
- 113.Hwang J, Kalejta RF. 2009. Human cytomegalovirus protein pp71 induces Daxx SUMOylation. J. Virol. 83:6591–6598. 10.1128/JVI.02639-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Korioth F, Maul GG, Plachter B, Stamminger T, Frey J. 1996. The nuclear domain 10 (ND10) is disrupted by the human cytomegalovirus gene product IE1. Exp. Cell Res. 229:155–158. 10.1006/excr.1996.0353 [DOI] [PubMed] [Google Scholar]
- 115.Lee H-R, Kim D-J, Lee J-M, Choi CY, Ahn B-Y, Hayward GS, Ahn J-H. 2004. Ability of the human cytomegalovirus IE1 protein to modulate sumoylation of PML correlates with its functional activities in transcriptional regulation and infectivity in cultured fibroblast cells. J. Virol. 78:6527–6542. 10.1128/JVI.78.12.6527-6542.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Preston CM, Nicholl MJ. 2006. Role of the cellular protein hDaxx in human cytomegalovirus immediate-early gene expression. J. Gen. Virol. 87:1113–1121. 10.1099/vir.0.81566-0 [DOI] [PubMed] [Google Scholar]
- 117.Tavalai N, Papior P, Rechter S, Leis M, Stamminger T. 2006. Evidence for a role of the cellular ND10 protein PML in mediating intrinsic immunity against human cytomegalovirus infections. J. Virol. 80:8006–8018. 10.1128/JVI.00743-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Tavalai N, Papior P, Rechter S, Stamminger T. 2008. Nuclear domain 10 components promyelocytic leukemia protein and hDaxx independently contribute to an intrinsic antiviral defense against human cytomegalovirus infection. J. Virol. 82:126–137. 10.1128/JVI.01685-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Adler M, Tavalai N, Müller R, Stamminger T. 2011. Human cytomegalovirus immediate-early gene expression is restricted by the nuclear domain 10 component Sp100. J. Gen. Virol. 92:1532–1538. 10.1099/vir.0.030981-0 [DOI] [PubMed] [Google Scholar]
- 120.Alcalay M, Tomassoni L, Colombo E, Stoldt S, Grignani F, Fagioli M, Szekely L, Helin K, Pelicci PG. 1998. The promyelocytic leukemia gene product (PML) forms stable complexes with the retinoblastoma protein. Mol. Cell. Biol. 18:1084–1093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Prichard MN, Sztul E, Daily SL, Perry AL, Frederick SL, Gill RB, Hartline CB, Streblow DN, Varnum SM, Smith RD, Kern ER. 2008. Human cytomegalovirus UL97 kinase activity is required for the hyperphosphorylation of retinoblastoma protein and inhibits the formation of nuclear aggresomes. J. Virol. 82:5054–5067. 10.1128/JVI.02174-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Snaar SP, Vincent M, Dirks RW. 1999. RNA polymerase II localizes at sites of human cytomegalovirus immediate-early RNA synthesis and processing. J. Histochem. Cytochem. 47:245–254. 10.1177/002215549904700213 [DOI] [PubMed] [Google Scholar]
- 123.Sourvinos G, Tavalai N, Berndt A, Spandidos DA, Stamminger T. 2007. Recruitment of human cytomegalovirus immediate-early 2 protein onto parental viral genomes in association with ND10 in live-infected cells. J. Virol. 81:10123–10136. 10.1128/JVI.01009-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Kaspari M, Tavalai N, Stamminger T, Zimmermann A, Schilf R, Bogner E. 2008. Proteasome inhibitor MG132 blocks viral DNA replication and assembly of human cytomegalovirus. FEBS Lett. 582:666–672. 10.1016/j.febslet.2008.01.040 [DOI] [PubMed] [Google Scholar]
- 125.Prosch S, Priemer C, Hoflich C, Liebenthaf C, Babel N, Kruger DH, Volk HD. 2003. Proteasome inhibitors: a novel tool to suppress human cytomegalovirus replication and virus-induced immune modulation. Antivir. Ther. 8:555–567 [PubMed] [Google Scholar]
- 126.Sadanari H, Tanaka J, Li Z, Yamada R, Matsubara K, Murayama T. 2009. Proteasome inhibitor differentially regulates expression of the major immediate early genes of human cytomegalovirus in human central nervous system-derived cell lines. Virus Res. 142:68–77. 10.1016/j.virusres.2009.01.010 [DOI] [PubMed] [Google Scholar]
- 127.Tran K, Mahr JA, Spector DH. 2010. Proteasome subunits relocalize during human cytomegalovirus infection, and proteasome activity is necessary for efficient viral gene transcription. J. Virol. 84:3079–3093. 10.1128/JVI.02236-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Ahn JH, Jang WJ, Hayward GS. 1999. The human cytomegalovirus IE2 and UL112-113 proteins accumulate in viral DNA replication compartments that initiate from the periphery of promyelocytic leukemia protein-associated nuclear bodies (PODs or ND10). J. Virol. 73:10458–10471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Penfold ME, Mocarski ES. 1997. Formation of cytomegalovirus DNA replication compartments defined by localization of viral proteins and DNA synthesis. Virology 239:46–61. 10.1006/viro.1997.8848 [DOI] [PubMed] [Google Scholar]
- 130.Yamamoto T, Suzuki S, Radsak K, Hirai K. 1998. The UL112/113 gene products of human cytomegalovirus which colocalize with viral DNA in infected cell nuclei are related to efficient viral DNA replication. Virus Res. 56:107–114. 10.1016/S0168-1702(98)00032-X [DOI] [PubMed] [Google Scholar]
- 131.Kim Y-E, Ahn J-H. 2010. Role of the specific interaction of UL112-113 p84 with UL44 DNA polymerase processivity factor in promoting DNA replication of human cytomegalovirus. J. Virol. 84:8409–8421. 10.1128/JVI.00189-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Chee MS, Bankier AT, Beck S, Bohni R, Brown CM, Cerny R, Horsnell T, Hutchison CA, Kouzarides T, Martignetti JA. 1990. Analysis of the protein-coding content of the sequence of human cytomegalovirus strain AD169. Curr. Top. Microbiol. Immunol. 154:125–169 [DOI] [PubMed] [Google Scholar]
- 133.Davison AJ, Dolan A, Akter P, Addison C, Dargan DJ, Alcendor DJ, McGeoch DJ, Hayward GS. 2003. The human cytomegalovirus genome revisited: comparison with the chimpanzee cytomegalovirus genome. J. Gen. Virol. 84:17–28. 10.1099/vir.0.18606-0 [DOI] [PubMed] [Google Scholar]
- 134.LaFemina RL, Hayward GS. 1983. Replicative forms of human cytomegalovirus DNA with joined termini are found in permissively infected human cells but not in non-permissive Balb/c-3T3 mouse cells. J. Gen. Virol. 64:373–389 [DOI] [PubMed] [Google Scholar]
- 135.McVoy MA, Nixon DE, Adler SP. 1997. Circularization and cleavage of guinea pig cytomegalovirus genomes. J. Virol. 71:4209–4217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Bogner E, Radsak K, Stinski MF. 1998. The gene product of human cytomegalovirus open reading frame UL56 binds the pac motif and has specific nuclease activity. J. Virol. 72:2259–2264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Giesen K, Radsak K, Bogner E. 2000. Targeting of the gene product encoded by ORF UL56 of human cytomegalovirus into viral replication centers. FEBS Lett. 471:215–218. 10.1016/S0014-5793(00)01407-1 [DOI] [PubMed] [Google Scholar]
- 138.Jault FM, Jault JM, Ruchti F, Fortunato EA, Clark C, Corbeil J, Richman D, Spector DH. 1995. Cytomegalovirus infection induces high levels of cyclins, phosphorylated Rb, and p53, leading to cell cycle arrest. J. Virol. 69:6697–6704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Muganda P, Mendoza O, Hernandez J, Qian Q. 1994. Human cytomegalovirus elevates levels of the cellular protein p53 in infected fibroblasts. J. Virol. 68:8028–8034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Speir E, Modali R, Huang ES, Leon MB, Shawl F, Finkel T, Epstein SE. 1994. Potential role of human cytomegalovirus and p53 interaction in coronary restenosis. Science 265:391–394. 10.1126/science.8023160 [DOI] [PubMed] [Google Scholar]
- 141.Bresnahan WA, Boldogh I, Thompson EA, Albrecht T. 1996. Human cytomegalovirus inhibits cellular DNA synthesis and arrests productively infected cells in late G1. Virology 224:150–160. 10.1006/viro.1996.0516 [DOI] [PubMed] [Google Scholar]
- 142.Muralidhar S, Doniger J, Mendelson E, Araujo JC, Kashanchi F, Azumi N, Brady JN, Rosenthal LJ. 1996. Human cytomegalovirus mtrII oncoprotein binds to p53 and down-regulates p53-activated transcription. J. Virol. 70:8691–8700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Fortunato EA, Spector DH. 1998. p53 and RPA are sequestered in viral replication centers in the nuclei of cells infected with human cytomegalovirus. J. Virol. 72:2033–2039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Li R, Botchan MR. 1993. The acidic transcriptional activation domains of VP16 and p53 bind the cellular replication protein A and stimulate in vitro BPV-1 DNA replication. Cell 73:1207–1221. 10.1016/0092-8674(93)90649-B [DOI] [PubMed] [Google Scholar]
- 145.Prelich G, Kostura M, Marshak DR, Mathews MB, Stillman B. 1987. The cell-cycle regulated proliferating cell nuclear antigen is required for SV40 DNA replication in vitro. Nature 326:471–475. 10.1038/326471a0 [DOI] [PubMed] [Google Scholar]
- 146.Wold MS, Weinberg DH, Virshup DM, Li JJ, Kelly TJ. 1989. Identification of cellular proteins required for simian virus 40 DNA replication. J. Biol. Chem. 264:2801–2809 [PubMed] [Google Scholar]
- 147.Full F, Reuter N, Zielke K, Stamminger T, Ensser A. 2012. Herpesvirus saimiri antagonizes nuclear domain 10-instituted intrinsic immunity via an ORF3-mediated selective degradation of cellular protein Sp100. J. Virol. 86:3541–3553. 10.1128/JVI.06992-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Wu FY, Ahn JH, Alcendor DJ, Jang WJ, Xiao J, Hayward SD, Hayward GS. 2001. Origin-independent assembly of Kaposi's sarcoma-associated herpesvirus DNA replication compartments in transient cotransfection assays and association with the ORF-K8 protein and cellular PML. J. Virol. 75:1487–1506. 10.1128/JVI.75.3.1487-1506.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Bell P, Lieberman PM, Maul GG. 2000. Lytic but not latent replication of Epstein-Barr virus is associated with PML and induces sequential release of nuclear domain 10 proteins. J. Virol. 74:11800–11810. 10.1128/JVI.74.24.11800-11810.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Lallemand-Breitenbach V, Jeanne M, Benhenda S, Nasr R, Lei M, Peres L, Zhou J, Zhu J, Raught B, de Thé H. 2008. Arsenic degrades PML or PML-RARα through a SUMO-triggered RNF4/ubiquitin-mediated pathway. Nat. Cell Biol. 10:547–555. 10.1038/ncb1717 [DOI] [PubMed] [Google Scholar]
- 151.Marcos-Villar L, Lopitz-Otsoa F, Gallego P, Muñoz-Fontela C, González-Santamaría J, Campagna M, Shou-Jiang G, Rodriguez MS, Rivas C. 2009. Kaposi's sarcoma-associated herpesvirus protein LANA2 disrupts PML oncogenic domains and inhibits PML-mediated transcriptional repression of the survivin gene. J. Virol. 83:8849–8858. 10.1128/JVI.00339-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Adamson AL, Kenney S. 2001. Epstein-Barr virus immediate-early protein BZLF1 is SUMO-1 modified and disrupts promyelocytic leukemia bodies. J. Virol. 75:2388–2399. 10.1128/JVI.75.5.2388-2399.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Tsai K, Thikmyanova N, Wojcechowskyj JA, Delecluse H-J, Lieberman PM. 2011. EBV tegument protein BNRF1 disrupts DAXX-ATRX to activate viral early gene transcription. PLoS Pathog. 7(11):e1002376. 10.1371/journal.ppat.1002376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Daikoku T, Kudoh A, Sugaya Y, Iwahori S, Shirata N, Isomura H, Tsurumi T. 2006. Postreplicative mismatch repair factors are recruited to Epstein-Barr virus replication compartments. J. Biol. Chem. 281:11422–11430. 10.1074/jbc.M510314200 [DOI] [PubMed] [Google Scholar]
- 155.Kudoh A, Fujita M, Zhang L, Shirata N, Daikoku T, Sugaya Y, Isomura H, Nishiyama Y, Tsurumi T. 2005. Epstein-Barr virus lytic replication elicits ATM checkpoint signal transduction while providing an S-phase-like cellular environment. J. Biol. Chem. 280:8156–8163. 10.1074/jbc.M411405200 [DOI] [PubMed] [Google Scholar]
- 156.Kudoh A, Iwahori S, Sato Y, Nakayama S, Isomura H, Murata T, Tsurumi T. 2009. Homologous recombinational repair factors are recruited and loaded onto the viral DNA genome in Epstein-Barr virus replication compartments. J. Virol. 83:6641–6651. 10.1128/JVI.00049-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Kudoh A, Daikoku T, Ishimi Y, Kawaguchi Y, Shirata N, Iwahori S, Isomura H, Tsurumi T. 2006. Phosphorylation of MCM4 at sites inactivating DNA helicase activity of the MCM4-MCM6-MCM7 complex during Epstein-Barr virus productive replication. J. Virol. 80:10064–10072. 10.1128/JVI.00678-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Wang L, Oliver SL, Sommer M, Rajamani J, Reichelt M, Arvin AM. 2011. Disruption of PML nuclear bodies is mediated by ORF61 SUMO-interacting motifs and required for varicella-zoster virus pathogenesis in skin. PLoS Pathog. 7(8):e1002157. 10.1371/journal.ppat.1002157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Reichelt M, Joubert L, Perrino J, Koh AL, Phanwar I, Arvin AM. 2012. 3D reconstruction of VZV infected cell nuclei and PML nuclear cages by serial section array scanning electron microscopy and electron tomography. PLoS Pathog. 8(6):e1002740. 10.1371/journal.ppat.1002740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Schreiner S, Martinez R, Groitl P, Rayne F, Vaillant R, Wimmer P, Bossis G, Sternsdorf T, Marcinowski L, Ruzsics Z, Dobner T, Wodrich H. 2012. Transcriptional activation of the adenoviral genome is mediated by capsid protein VI. PLoS Pathog. 8(2):e1002549. 10.1371/journal.ppat.1002549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Schreiner S, Wimmer P, Sirma H, Everett RD, Blanchette P, Groitl P, Dobner T. 2010. Proteasome-dependent degradation of Daxx by the viral E1B-55K protein in human adenovirus-infected cells. J. Virol. 84:7029–7038. 10.1128/JVI.00074-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Carvalho T, Seeler JS, Ohman K, Jordan P, Pettersson U, Akusjarvi G, Carmo-Fonseca M, Dejean A. 1995. Targeting of adenovirus E1A and E4-ORF3 proteins to nuclear matrix-associated PML bodies. J. Cell Biol. 131:45–56. 10.1083/jcb.131.1.45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Doucas V, Ishov AM, Romo A, Juguilon H, Weitzman MD, Evans RM, Maul GG. 1996. Adenovirus replication is coupled with the dynamic properties of the PML nuclear structure. Genes Dev. 10:196–207. 10.1101/gad.10.2.196 [DOI] [PubMed] [Google Scholar]
- 164.Stracker TH, Lee DV, Carson CT, Araujo FD, Ornelles DA, Weitzman MD. 2005. Serotype-specific reorganization of the Mre11 complex by adenoviral E4orf3 proteins. J. Virol. 79:6664–6673. 10.1128/JVI.79.11.6664-6673.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Ullman AJ, Hearing P. 2008. Cellular proteins PML and Daxx mediate an innate antiviral defense antagonized by the adenovirus E4 ORF3 protein. J. Virol. 82:7325–7335. 10.1128/JVI.00723-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Yondola MA, Hearing P. 2007. The adenovirus E4 ORF3 protein binds and reorganizes the TRIM family member transcriptional intermediary factor 1 alpha. J. Virol. 81:4264–4271. 10.1128/JVI.02629-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Evans JD, Hearing P. 2005. Relocalization of the Mre11-Rad50-Nbs1 complex by the adenovirus E4 ORF3 protein is required for viral replication. J. Virol. 79:6207–6215. 10.1128/JVI.79.10.6207-6215.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Stracker TH, Carson CT, Weitzman MD. 2002. Adenovirus oncoproteins inactivate the Mre11-Rad50-NBS1 DNA repair complex. Nature 418:348–352. 10.1038/nature00863 [DOI] [PubMed] [Google Scholar]
- 169.Araujo FD, Stracker TH, Carson CT, Lee DV, Weitzman MD. 2005. Adenovirus type 5 E4orf3 protein targets the Mre11 complex to cytoplasmic aggresomes. J. Virol. 79:11382–11391. 10.1128/JVI.79.17.11382-11391.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Schreiner S, Wimmer P, Dobner T. 2012. Adenovirus degradation of cellular proteins. Future Microbiol. 7:211–225. 10.2217/fmb.11.153 [DOI] [PubMed] [Google Scholar]
- 171.Baker A, Rohleder KJ, Hanakahi LA, Ketner G. 2007. Adenovirus E4 34k and E1b 55K oncoproteins target host DNA ligase IV for proteasomal degradation. J. Virol. 81:7034–7040. 10.1128/JVI.00029-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Orazio NI, Naeger CM, Karlseder J, Weitzman MD. 2011. The adenovirus E1b55K/E4orf6 complex induces degradation of the Bloom helicase during infection. J. Virol. 85:1887–1892. 10.1128/JVI.02134-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Debbas M, White E. 1993. Wild-type p53 mediates apoptosis by E1A, which is inhibited by E1B. Genes Dev. 7:546–554. 10.1101/gad.7.4.546 [DOI] [PubMed] [Google Scholar]
- 174.Buchkovich K, Dyson N, Whyte P, Harlow E. 1990. Cellular proteins that are targets for transformation by DNA tumour viruses. Ciba Found. Symp. 150:262–271 [DOI] [PubMed] [Google Scholar]
- 175.Mittnacht S, Lees JA, Desai D, Harlow E, Morgan DO, Weinberg RA. 1994. Distinct sub-populations of the retinoblastoma protein show a distinct pattern of phosphorylation. EMBO J. 13:118–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Giordano A, McCall C, Whyte P, Franza BR. 1991. Human cyclin A and the retinoblastoma protein interact with similar but distinguishable sequences in the adenovirus E1A gene product. Oncogene 6:481–485 [PubMed] [Google Scholar]
- 177.Dyson N, Buchkovich K, Whyte P, Harlow E. 1989. Cellular proteins that are targetted by DNA tumor viruses for transformation. Int. Symp. Princess Takamatsu Cancer Res. Fund. 20:191–198 [PubMed] [Google Scholar]
- 178.Bagchi S, Raychaudhuri P, Nevins JR. 1990. Adenovirus E1A proteins can dissociate heteromeric complexes involving the E2F transcription factor: a novel mechanism for E1A trans-activation. Cell 62:659–669 [DOI] [PubMed] [Google Scholar]
- 179.Cress WD, Nevins JR. 1996. A role for a bent DNA structure in E2F-mediated transcription activation. Mol. Cell. Biol. 16:2119–2127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Puvion-Dutilleul F, Puvion E. 1990. Replicating single-stranded adenovirus type 5 DNA molecules accumulate within well-delimited intranuclear areas of lytically infected HeLa cells. Eur. J. Cell Biol. 52:379–388 [PubMed] [Google Scholar]
- 181.James NJ, Howell GJ, Walker JH, Blair GE. 2010. The role of Cajal bodies in the expression of late phase adenovirus proteins. Virology 399:299–311. 10.1016/j.virol.2010.01.013 [DOI] [PubMed] [Google Scholar]
- 182.Bridge E, Pettersson U. 1996. Nuclear organization of adenovirus RNA biogenesis. Exp. Cell Res. 229:233–239. 10.1006/excr.1996.0365 [DOI] [PubMed] [Google Scholar]
- 183.Bridge E, Xia DX, Carmo-Fonseca M, Cardinali B, Lamond AI, Pettersson U. 1995. Dynamic organization of splicing factors in adenovirus-infected cells. J. Virol. 69:281–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Gama-Carvalho M, Condado I, Carmo-Fonseca M. 2003. Regulation of adenovirus alternative RNA splicing correlates with a reorganization of splicing factors in the nucleus. Exp. Cell Res. 289:77–85. 10.1016/S0014-4827(03)00251-9 [DOI] [PubMed] [Google Scholar]
- 185.Pombo A, Ferreira J, Bridge E, Carmo-Fonseca M. 1994. Adenovirus replication and transcription sites are spatially separated in the nucleus of infected cells. EMBO J. 13:5075–5085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Murti KG, Davis DS, Kitchingman GR. 1990. Localization of adenovirus-encoded DNA replication proteins in the nucleus by immunogold electron microscopy. J. Gen. Virol. 71:2847–2857 [DOI] [PubMed] [Google Scholar]
- 187.Bosher J, Dawson A, Hay RT. 1992. Nuclear factor I is specifically targeted to discrete subnuclear sites in adenovirus type 2-infected cells. J. Virol. 66:3140–3150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Puvion-Dutilleul F, Bachellerie JP, Visa N, Puvion E. 1994. Rearrangements of intranuclear structures involved in RNA processing in response to adenovirus infection. J. Cell Sci. 107:1457–1468 [DOI] [PubMed] [Google Scholar]
- 189.Woo JL, Berk AJ. 2007. Adenovirus ubiquitin-protein ligase stimulates viral late mRNA nuclear export. J. Virol. 81:575–587. 10.1128/JVI.01725-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Aspegren A, Rabino C, Bridge E. 1998. Organization of splicing factors in adenovirus-infected cells reflects changes in gene expression during the early to late phase transition. Exp. Cell Res. 245:203–213. 10.1006/excr.1998.4264 [DOI] [PubMed] [Google Scholar]
- 191.Ornelles DA, Shenk T. 1991. Localization of the adenovirus early region 1B 55-kilodalton protein during lytic infection: association with nuclear viral inclusions requires the early region 4 34-kilodalton protein. J. Virol. 65:424–429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Blanchette P, Kindsmüller K, Groitl P, Dallaire F, Speiseder T, Branton PE, Dobner T. 2008. Control of mRNA export by adenovirus E4orf6 and E1B55K proteins during productive infection requires E4orf6 ubiquitin ligase activity. J. Virol. 82:2642–2651. 10.1128/JVI.02309-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Babiss LE, Ginsberg HS, Darnell JE. 1985. Adenovirus E1B proteins are required for accumulation of late viral mRNA and for effects on cellular mRNA translation and transport. Mol. Cell. Biol. 5:2552–2558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Flint SJ, Gonzalez RA. 2003. Regulation of mRNA production by the adenoviral E1B 55-kDa and E4 Orf6 proteins. Curr. Top. Microbiol. Immunol. 272:287–330 [DOI] [PubMed] [Google Scholar]
- 195.Yatherajam G, Huang W, Flint SJ. 2011. Export of adenoviral late mRNA from the nucleus requires the Nxf1/Tap export receptor. J. Virol. 85:1429–1438. 10.1128/JVI.02108-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Schmid M, Gonzalez RA, Dobner T. 2012. CRM1-dependent transport supports cytoplasmic accumulation of adenoviral early transcripts. J. Virol. 86:2282–2292. 10.1128/JVI.06275-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Sohn S-Y, Hearing P. 2012. Adenovirus regulates sumoylation of Mre11-Rad50-Nbs1 components through a paralog-specific mechanism. J. Virol. 86:9656–9665. 10.1128/JVI.01273-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Pennella MA, Liu Y, Woo JL, Kim CA, Berk AJ. 2010. Adenovirus E1B 55-kilodalton protein is a p53-SUMO1 E3 ligase that represses p53 and stimulates its nuclear export through interactions with promyelocytic leukemia nuclear bodies. J. Virol. 84:12210–12225. 10.1128/JVI.01442-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Wimmer P, Schreiner S, Everett RD, Sirma H, Groitl P, Dobner T. 2010. SUMO modification of E1B-55K oncoprotein regulates isoform-specific binding to the tumour suppressor protein PML. Oncogene 29:5511–5522. 10.1038/onc.2010.284 [DOI] [PubMed] [Google Scholar]
- 200.Berns KI. 1990. Parvovirus replication. Microbiol. Mol. Biol. Rev. 54:316–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Atchison RW, Casto BC, Hammon WM. 1965. Adenovirus-associated defective virus particles. Science 149:754–756. 10.1126/science.149.3685.754 [DOI] [PubMed] [Google Scholar]
- 202.Hoggan MD, Blacklow N R, Rowe WP. 1966. Studies of small DNA viruses found in various adenovirus preparations: physical, biological, and immunological characteristics. Proc. Natl. Acad. Sci. U. S. A. 55:1467–1474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Atchison RW. 1970. The role of herpesviruses in adenovirus-associated virus replication in vitro. Virology 42:155–162. 10.1016/0042-6822(70)90248-5 [DOI] [PubMed] [Google Scholar]
- 204.Buller RM, Janik JE, Sebring ED, Rose JA. 1981. Herpes simplex virus types 1 and 2 completely help adenovirus-associated virus replication. J. Virol. 40:241–247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.McPherson RA, Rosenthal LJ, Rose JA. 1985. Human cytomegalovirus completely helps adeno-associated virus replication. Virology 147:217–222 [DOI] [PubMed] [Google Scholar]
- 206.Bauer HJ, Monreal G. 1986. Herpesviruses provide helper functions for avian adeno-associated parvovirus. J. Gen. Virol. 67:181–185 [DOI] [PubMed] [Google Scholar]
- 207.Carter BJ. 1992. Adeno-associated virus vectors. Curr. Opin. Biotechnol. 3:533–539 [DOI] [PubMed] [Google Scholar]
- 208.Muzyczka N. 1992. Use of adeno-associated virus as a general transduction vector for mammalian cells. Curr. Top. Microbiol. Immunol. 158:97–129 [DOI] [PubMed] [Google Scholar]
- 209.Leonard CJ, Berns KI. 1994. Adeno-associated virus type 2: a latent life cycle. Prog. Nucleic Acids Res. Mol. Biol. 48:29–52 [DOI] [PubMed] [Google Scholar]
- 210.Srivastava A, Lusby EW, Berns KI. 1983. Nucleotide sequence and organization of the adeno-associated virus 2 genome. 45:555–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Tratschin JD, Miller IL, Carter BJ. 1984. Genetic analysis of adeno-associated virus: properties of deletion mutants constructed in vitro and evidence for an adeno-associated virus replication function. J. Virol. 51:611–619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Hermonat PL, Labow MA, Wright R, Berns KI, Muzyczka N. 1984. Genetics of adeno-associated virus: isolation and preliminary characterization of adeno-associated virus type 2 mutants. J. Virol. 51:329–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Sonntag F, Schmidt K, Kleinschmidt JA. 2010. A viral assembly factor promotes AAV2 capsid formation in the nucleolus. Proc. Natl. Acad. Sci. U. S. A. 107:10220–10225. 10.1073/pnas.1001673107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Chejanovsky N, Carter BJ. 1989. Replication of a human parvovirus nonsense mutant in mammalian cells containing an inducible amber suppressor. Virology 171:239–247. 10.1016/0042-6822(89)90531-X [DOI] [PubMed] [Google Scholar]
- 215.Hölscher C, Kleinschmidt JA, Bürkle A. 1995. High-level expression of adeno-associated virus (AAV) Rep78 or Rep68 protein is sufficient for infectious-particle formation by a rep-negative AAV mutant. J. Virol. 69:6880–6885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Chang LS, Shi Y, Shenk T. 1989. Adeno-associated virus P5 promoter contains an adenovirus E1A-inducible element and a binding site for the major late transcription factor. 63:3479–3488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Samulski RJ, Shenk T. 1988. Adenovirus E1B 55-Mr polypeptide facilitates timely cytoplasmic accumulation of adeno-associated virus mRNAs. J. Virol. 62:206–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Chang LS, Shenk T. 1990. The adenovirus DNA-binding protein stimulates the rate of transcription directed by adenovirus and adeno-associated virus promoters. J. Virol. 64:2103–2109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Jay FT, Laughlin CA, Carter BJ. 1981. Eukaryotic translational control: adeno-associated virus protein synthesis is affected by a mutation in the adenovirus DNA-binding protein. Proc. Natl. Acad. Sci. U. S. A. 78:2927–2931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Fraefel C, Bittermann AG, Büeler H, Heid I, Bächi T, Ackermann M. 2004. Spatial and temporal organization of adeno-associated virus DNA replication in live cells. J. Virol. 78:389–398. 10.1128/JVI.78.1.389-398.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Weitzman MD, Fisher KJ, Wilson JM. 1996. Recruitment of wild-type and recombinant adeno-associated virus into adenovirus replication centers. J. Virol. 70:1845–1854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Nash K, Chen W, McDonald WF, Zhou X, Muzyczka N. 2007. Purification of host cell enzymes involved in adeno-associated virus DNA replication. J. Virol. 81:5777–5787. 10.1128/JVI.02651-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Ni TH, McDonald WF, Zolotukhin I, Melendy T, Waga S, Stillman B, Muzyczka N. 1998. Cellular proteins required for adeno-associated virus DNA replication in the absence of adenovirus coinfection. J. Virol. 72:2777–2787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Wistuba A, Kern A, Weger S, Grimm D, Kleinschmidt JA. 1997. Subcellular compartmentalization of adeno-associated virus type 2 assembly. J. Virol. 71:1341–1352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Schlehofer JR, Ehrbar M, zur Hausen H. 1986. Vaccinia virus, herpes simplex virus, and carcinogens induce DNA amplification in a human cell line and support replication of a helpervirus dependent parvovirus. Virology 152:110–117. 10.1016/0042-6822(86)90376-4 [DOI] [PubMed] [Google Scholar]
- 226.Yalkinoglu AO, Heilbronn R, Bürkle A, Schlehofer JR, zur Hausen H. 1988. DNA amplification of adeno-associated virus as a response to cellular genotoxic stress. Cancer Res. 48:3123–3129 [PubMed] [Google Scholar]
- 227.Collaco RF, Bevington JM, Bhrigu V, Kalman-Maltese V, Trempe JP. 2009. Adeno-associated virus and adenovirus coinfection induces a cellular DNA damage and repair response via redundant phosphatidylinositol 3-like kinase pathways. Virology 392:24–33. 10.1016/j.virol.2009.06.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Schwartz RA, Carson CT, Schuberth C, Weitzman MD. 2009. Adeno-associated virus replication induces a DNA damage response coordinated by DNA-dependent protein kinase. J. Virol. 83:6269–6278. 10.1128/JVI.00318-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Tattersall P. 1972. Replication of the parvovirus MVM. I. Dependence of virus multiplication and plaque formation on cell growth. J. Virol. 10:586–590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Wolter S, Richards R, Armentrout RW. 1980. Cell cycle-dependent replication of the DNA of minute virus of mice, a parvovirus. Biochim. Biophys. Acta 607:420–431 [DOI] [PubMed] [Google Scholar]
- 231.Parrish CR. 1991. Mapping specific functions in the capsid structure of canine parvovirus and feline panleukopenia virus using infectious plasmid clones. 183:195–205 [DOI] [PubMed] [Google Scholar]
- 232.Jindal HK, Yong CB, Wilson GM, Tam P, Astell CR. 1994. Mutations in the NTP-binding motif of minute virus of mice (MVM) NS-1 protein uncouple ATPase and DNA helicase functions. J. Biol. Chem. 269:3283–3289 [PubMed] [Google Scholar]
- 233.Christensen J, Cotmore SF, Tattersall P. 1995. Minute virus of mice transcriptional activator protein NS1 binds directly to the transactivation region of the viral P38 promoter in a strictly ATP-dependent manner. J. Virol. 69:5422–5430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Rhode SL, Paradiso PR. 1989. Parvovirus replication in normal and transformed human cells correlates with the nuclear translocation of the early protein NS1. J. Virol. 63:349–355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Doerig C, Hirt B, Beard P, Antonietti JP. 1988. Minute virus of mice non-structural protein NS-1 is necessary and sufficient for trans-activation of the viral P39 promoter. J. Gen. Virol. 69:2563–2573 [DOI] [PubMed] [Google Scholar]
- 236.Lorson C, Burger LR, Mouw M, Pintel DJ. 1996. Efficient transactivation of the minute virus of mice P38 promoter requires upstream binding of NS1. J. Virol. 70:834–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Rhode SL. 1985. trans-Activation of parvovirus P38 promoter by the 76K noncapsid protein. J. Virol. 55:886–889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Cziepluch C, Lampel S, Grewenig A, Grund C, Lichter P, Rommelaere J. 2000. H-1 parvovirus-associated replication bodies: a distinct virus-induced nuclear structure. J. Virol. 74:4807–4815. 10.1128/JVI.74.10.4807-4815.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Young PJ, Jensen KT, Burger LR, Pintel DJ, Lorson CL. 2002. Minute virus of mice NS1 interacts with the SMN protein, and they colocalize in novel nuclear bodies induced by parvovirus infection. J. Virol. 76:3892–3904. 10.1128/JVI.76.8.3892-3904.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Bashir T, Rommelaere J, Cziepluch C. 2001. In vivo accumulation of cyclin A and cellular replication factors in autonomous parvovirus minute virus of mice-associated replication bodies. J. Virol. 75:4394–4398. 10.1128/JVI.75.9.4394-4398.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Christensen J, Tattersall P. 2002. Parvovirus initiator protein NS1 and RPA coordinate replication fork progression in a reconstituted DNA replication system. J. Virol. 76:6518–6531. 10.1128/JVI.76.13.6518-6531.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Ihalainen TO, Niskanen EA, Jylhävä J, Paloheimo O, Dross N, Smolander H, Langowski J, Timonen J, Vihinen-Ranta M. 2009. Parvovirus induced alterations in nuclear architecture and dynamics. PLoS One 4(6):e5948. 10.1371/journal.pone.0005948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Hoque M, Ishizu K, Matsumoto A, Han SI, Arisaka F, Takayama M, Suzuki K, Kato K, Kanda T, Watanabe H, Handa H. 1999. Nuclear transport of the major capsid protein is essential for adeno-associated virus capsid formation. J. Virol. 73:7912–7915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Lombardo E, Ramirez JC, Agbandje-McKenna M, Almendral JM. 2000. A beta-stranded motif drives capsid protein oligomers of the parvovirus minute virus of mice into the nucleus for viral assembly. J. Virol. 74:3804–3814. 10.1128/JVI.74.8.3804-3814.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Yuan W, Parrish CR. 2001. Canine parvovirus capsid assembly and differences in mammalian and insect cells. Virology 279:546–557. 10.1006/viro.2000.0734 [DOI] [PubMed] [Google Scholar]
- 246.Lombardo E, Ramírez JC, Garcia J, Almendral JM. 2002. Complementary roles of multiple nuclear targeting signals in the capsid proteins of the parvovirus minute virus of mice during assembly and onset of infection. J. Virol. 76:7049–7059. 10.1128/JVI.76.14.7049-7059.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Adeyemi RO, Landry S, Davis ME, Weitzman MD, Pintel DJ. 2010. Parvovirus minute virus of mice induces a DNA damage response that facilitates viral replication. PLoS Pathog. 6(10):e1001141. 10.1371/journal.ppat.1001141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Ishov AM, Maul GG. 1996. The periphery of nuclear domain 10 (ND10) as site of DNA virus deposition. J. Cell Biol. 134:815–826. 10.1083/jcb.134.4.815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Jiang M, Entezami P, Gamez M, Stamminger T, Imperiale MJ. 2011. Functional reorganization of promyelocytic leukemia nuclear bodies during BK virus infection. mBio 2(1):e00281–10. 10.1028/mBio.00281-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Gasparovic ML, Maginnis MS, O'Hara BA, Dugan AS, Atwood WJ. 2009. Modulation of PML protein expression regulates JCV infection. Virology 390:279–288. 10.1016/j.virol.2009.05.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Erickson KD, Bouchet-Marquis C, Heiser K, Szomolanyi-Tsuda E, Mishra R, Lamothe B, Hoenger A, Garcea RL. 2012. Virion assembly factories in the nucleus of polyomavirus-infected cells. PLoS Pathog. 8(4):e1002630. 10.1371/journal.ppat.1002630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Jul-Larsen A, Visted T, Karlsen BO, Rinaldo CH, Bjerkvig R, Lønning PE, Bøe SO. 2004. PML-nuclear bodies accumulate DNA in response to polyomavirus BK and simian virus 40 replication. Exp. Cell Res. 298:58–73. 10.1016/j.yexcr.2004.03.045 [DOI] [PubMed] [Google Scholar]
- 253.Shishido-Hara Y, Ichinose S, Higuchi K, Hara Y, Yasui K. 2004. Major and minor capsid proteins of human polyomavirus JC cooperatively accumulate to nuclear domain 10 for assembly into virions. J. Virol. 78:9890–9903. 10.1128/JVI.78.18.9890-9903.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Tang Q, Bell P, Tegtmeyer P, Maul GG. 2000. Replication but not transcription of simian virus 40 DNA is dependent on nuclear domain 10. J. Virol. 74:9694–9700. 10.1128/JVI.74.20.9694-9700.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Gharakhanian E, Takahashi J, Kasamatsu H. 1987. The carboxyl 35 amino acids of SV40 Vp3 are essential for its nuclear accumulation. Virology 157:440–448 [DOI] [PubMed] [Google Scholar]
- 256.Gharakhanian E, Kasamatsu H. 1990. Two independent signals, a nuclear localization signal and a Vp1-interactive signal, reside within the carboxy-35 amino acids of SV40 Vp3. Virology 178:62–71. 10.1016/0042-6822(90)90379-6 [DOI] [PubMed] [Google Scholar]
- 257.Ishii N, Minami N, Chen EY, Medina AL, Chico MM, Kasamatsu H. 1996. Analysis of a nuclear localization signal of simian virus 40 major capsid protein Vp1. J. Virol. 70:1317–1322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Lin W, Hata T, Kasamatsu H. 1984. Subcellular distribution of viral structural proteins during simian virus 40 infection. J. Virol. 50:363–371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Ostapchuk P, Hearing P. 2005. Control of adenovirus packaging. J. Cell Biochem. 96:25–35. 10.1002/jcb.20523 [DOI] [PubMed] [Google Scholar]
- 260.Garber EA, Seidman MM, Levine AJ. 1980. Intracellular SV40 nucleoprotein complexes: synthesis to encapsidation. Virology 107:389–401. 10.1016/0042-6822(80)90306-2 [DOI] [PubMed] [Google Scholar]
- 261.Dahl J, You J, Benjamin TL. 2005. Induction and utilization of an ATM signaling pathway by polyomavirus. J. Virol. 79:13007–13017. 10.1128/JVI.79.20.13007-13017.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Shi Y, Dodson GE, Shaikh S, Rundell K, Tibbetts RS. 2005. Ataxia-telangiectasia-mutated (ATM) is a T-antigen kinase that controls SV40 viral replication in vivo. J. Biol. Chem. 280:40195–40200. 10.1074/jbc.C500400200 [DOI] [PubMed] [Google Scholar]
- 263.Zhao X, Madden-Fuentes RJ, Lou BX, Pipas JM, Gerhardt J, Rigell CJ, Fanning E. 2008. Ataxia telangiectasia-mutated damage-signaling kinase- and proteasome-dependent destruction of Mre11-Rad50-Nbs1 subunits in simian virus 40-infected primate cells. J. Virol. 82:5316–5328. 10.1128/JVI.02677-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Hein J, Boichuk S, Wu J, Cheng Y, Freire R, Jat PS, Roberts TM, Gjoerup OV. 2009. Simian virus 40 large T antigen disrupts genome integrity and activates a DNA damage response via Bub1 binding. J. Virol. 83:117–127. 10.1128/JVI.01515-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Boichuk S, Hu L, Hein J, Gjoerup OV. 2010. Multiple DNA damage signaling and repair pathways deregulated by simian virus 40 large T antigen. J. Virol. 84:8007–8020. 10.1128/JVI.00334-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Florin L, Becker KA, Lambert C, Nowak T, Sapp C, Strand D, Streeck RE, Sapp M. 2006. Identification of a dynein interacting domain in the papillomavirus minor capsid protein L2. J. Virol. 80:6691–6696. 10.1128/JVI.00057-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Darshan MS, Lucchi J, Harding E, Moroianu J. 2004. The L2 minor capsid protein of human papillomavirus type 16 interacts with a network of nuclear import receptors. J. Virol. 78:12179–12188. 10.1128/JVI.78.22.12179-12188.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Day PM, Baker CC, Lowy DR, Schiller JT. 2004. Establishment of papillomavirus infection is enhanced by promyelocytic leukemia protein (PML) expression. Proc. Natl. Acad. Sci. U. S. A. 101:14252–14257. 10.1073/pnas.0404229101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Ustav M, Stenlund A. 1991. Transient replication of BPV-1 requires two viral polypeptides encoded by the E1 and E2 open reading frames. EMBO J. 10:449–457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Chiang CM, Ustav M, Stenlund A, Ho TF, Broker TR, Chow LT. 1992. Viral E1 and E2 proteins support replication of homologous and heterologous papillomaviral origins. Proc. Natl. Acad. Sci. U. S. A. 89:5799–5803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Sverdrup F, Khan SA. 1994. Replication of human papillomavirus (HPV) DNAs supported by the HPV type 18 E1 and E2 proteins. J. Virol. 68:505–509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Kadaja M, Silla T, Ustav E, Ustav M. 2009. Papillomavirus DNA replication: from initiation to genomic instability. Virology 384:360–368. 10.1016/j.virol.2008.11.032 [DOI] [PubMed] [Google Scholar]
- 273.Sedman J, Stenlund A. 1998. The papillomavirus E1 protein forms a DNA-dependent hexameric complex with ATPase and DNA helicase activities. J. Virol. 72:6893–6897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Liu JS, Kuo SR, Broker TR, Chow LT. 1995. The functions of human papillomavirus type 11 E1, E2, and E2C proteins in cell-free DNA replication. J. Biol. Chem. 270:27283–27291 [DOI] [PubMed] [Google Scholar]
- 275.Chow LT, Broker TR. 1994. Papillomavirus DNA replication. Intervirology 37:150–158 [DOI] [PubMed] [Google Scholar]
- 276.Park P, Copeland W, Yang L, Wang T, Botchan MR, Mohr IJ. 1994. The cellular DNA polymerase alpha-primase is required for papillomavirus DNA replication and associates with the viral E1 helicase. Proc. Natl. Acad. Sci. U. S. A. 91:8700–8704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Rangasamy D, Wilson VG. 2000. Bovine papillomavirus E1 protein is sumoylated by the host cell Ubc9 protein. J. Biol. Chem. 275:30487–30495. 10.1074/jbc.M003898200 [DOI] [PubMed] [Google Scholar]
- 278.Yasugi T, Vidal M, Sakai H, Howley PM, Benson JD. 1997. Two classes of human papillomavirus type 16 E1 mutants suggest pleiotropic conformational constraints affecting E1 multimerization, E2 interaction, and interaction with cellular proteins. J. Virol. 71:5942–5951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Kadaja M, Isok-Paas H, Laos T, Ustav E, Ustav M. 2009. Mechanism of genomic instability in cells infected with the high-risk human papillomaviruses. PLoS Pathog. 5(4):e1000397. 10.1371/journal.ppat.1000397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Sakakibara N, Mitra R, McBride AA. 2011. The papillomavirus E1 helicase activates a cellular DNA damage response in viral replication foci. J. Virol. 85:8981–8995. 10.1128/JVI.00541-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Nakahara T, Lambert PF. 2007. Induction of promyelocytic leukemia (PML) oncogenic domains (PODs) by papillomavirus. Virology 366:316–329. 10.1016/j.virol.2007.04.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Guccione E, Massimi P, Bernat A, Banks L. 2002. Comparative analysis of the intracellular location of the high- and low-risk human papillomavirus oncoproteins. Virology 293:20–25. 10.1006/viro.2001.1290 [DOI] [PubMed] [Google Scholar]
- 283.Guccione E, Lethbridge KJ, Killick N, Leppard KN, Banks L. 2004. HPV E6 proteins interact with specific PML isoforms and allow distinctions to be made between different POD structures. Oncogene 23:4662–4672. 10.1038/sj.onc.1207631 [DOI] [PubMed] [Google Scholar]
- 284.Day PM, Roden RB, Lowy DR, Schiller JT. 1998. The papillomavirus minor capsid protein, L2, induces localization of the major capsid protein, L1, and the viral transcription/replication protein, E2, to PML oncogenic domains. J. Virol. 72:142–150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Swindle CS, Zou N, Van Tine BA, Shaw GM, Engler JA, Chow LT. 1999. Human papillomavirus DNA replication compartments in a transient DNA replication system. J. Virol. 73:1001–1009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Heino P, Zhou J, Lambert PF. 2000. Interaction of the papillomavirus transcription/replication factor, E2, and the viral capsid protein, L2. Virology 276:304–314. 10.1006/viro.2000.0342 [DOI] [PubMed] [Google Scholar]
- 287.Florin L, Sapp C, Streeck RE, Sapp M. 2002. Assembly and translocation of papillomavirus capsid proteins. J. Virol. 76:10009–10014. 10.1128/JVI.76.19.10009-10014.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Becker KA, Florin L, Sapp C, Maul GG, Sapp M. 2004. Nuclear localization but not PML protein is required for incorporation of the papillomavirus minor capsid protein L2 into virus-like particles. J. Virol. 78:1121–1128. 10.1128/JVI.78.3.1121-1128.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Florin L, Schäfer F, Sotlar K, Streeck RE, Sapp M. 2002. Reorganization of nuclear domain 10 induced by papillomavirus capsid protein l2. Virology 295:97–107. 10.1006/viro.2002.1360 [DOI] [PubMed] [Google Scholar]
- 290.Moshe A, Gorovits R. 2012. Virus-induced aggregates in infected cells. Viruses 4:2218–2232. 10.3390/v4102218 [DOI] [PMC free article] [PubMed] [Google Scholar]