Significance
Exactly how memory cells are selected into a recall response to acute viral infection (stochastic or deterministic) remains unresolved. This paper demonstrates definitively that selection of virus-specific CD8+ T cells from memory occurs via active selection of particular T-cell clones after secondary virus infection and that this selection appears to be based on the avidity of the T-cell receptor (TCR) for the virus-derived peptide (p) + major histocompatibility complex class I molecule. We also show that despite clear clonal preferences, there is no global narrowing of epitope-specific TCR diversity in the recall response. We propose that the immune system has evolved such a strategy to optimize cytotoxic T-lymphocyte responses while safeguarding TCR repertoire diversity.
Keywords: recall CD8+ T-cell response, clonal selection, antiviral immunity, memory CD8+ T cells, influenza virus
Abstract
The recall of memory CD8+ cytotoxic T lymphocytes (CTLs), elicited by prior virus infection or vaccination, is critical for immune protection. The extent to which this arises as a consequence of stochastic clonal expansion vs. active selection of particular clones remains unclear. Using a parallel adoptive transfer protocol in combination with single cell analysis to define the complementarity determining region (CDR) 3α and CDR3β regions of individual T-cell receptor (TCR) heterodimers, we characterized the antigen-driven recall of the same memory CTL population in three individual recipients. This high-resolution analysis showed reproducible enrichment (or diminution) of particular TCR clonotypes across all challenged animals. These changes in clonal composition were TCRα− and β chain–dependent and were directly related to the avidity of the TCR for the virus-derived peptide (p) + major histocompatibility complex class I molecule. Despite this shift in clonotype representation indicative of differential selection, there was no evidence of overall repertoire narrowing, suggesting a strategy to optimize CTL responses while safeguarding TCR diversity.
Virus-specific CD8+ cytotoxic T lymphocytes (CTLs) are key for effective pathogen clearance. To exert their antiviral effects, naïve CD8+ CTL precursors (CTLps) must first be activated through the specific recognition of virus-derived peptides (p) in the context of major histocompatibility complex class I molecules (MHCI) expressed on the surface of dendritic cells. Ligation of these pMHCI epitopes is mediated via specific T-cell receptor (TCR) αβ heterodimers, leading to the recruitment, proliferation, and activation of antigen-specific CTLs. Subsequent to virus clearance, CTL populations contract to form a stable pool of resting memory cells, typically at around 5–10% of their acute phase numbers (1, 2). On secondary virus encounter, this memory pool of CD8+ T cells is able to expand rapidly, providing potent immune protection (2).
An understanding of CD8+ T-cell recruitment/expansion into the recall response has significant implications for effective vaccine strategies. If recruitment and expansion occur stochastically, a memory population in which highly effective clones predominate is desirable. Alternatively, if selection is deterministic, the basic requirement would be the presence of high-quality clones in the memory population for selective expansion by antigen-driven mechanisms. Moreover, different mechanisms of memory recruitment have different long-term implications for the clonal diversity of epitope-specific populations, especially with repeated virus exposure. Repeated selection of particular clones inherent in deterministic clonal selection has the potential consequence of narrowing the CTL repertoire, whereas stochastic recruitment/expansion seems more likely to maintain CTL diversity, shown to be beneficial for virus control (3–7). Despite the implications, there remains conjecture about how epitope-specific CD8+ T cells are recruited from the memory pool. Studies have indicated a focusing of the recall response relative to the memory pool, arising from only a subpopulation of cells being recruited or expanded (8), whereas others have indicated that primary and secondary responses are either highly similar or randomly different, prompting the conclusion that selection from the memory pool likely occurs as a result of stochastic T-cell selection (9–13).
Using secondary virus challenge of influenza A virus–primed B6 mice as a model of acute localized infection, we investigated the recruitment/expansion of memory CD8+ T-cell clones specific for the immunodominant DbNP366 epitope. We used a parallel adoptive transfer method, in which a population of memory CD8+ T cells from one individual is rechallenged with virus in three independent recipients, to definitively ascertain whether changes in clonal prevalence between memory and recall populations occur in a stochastic or deterministic fashion. Using high-resolution single cell TCRβ or αβ analysis, our data show evidence of active selection for particular clones following secondary infection, without a reduction in overall TCR diversity. Further, the selection for particular T-cell clones appears to be based on the avidity of the TCR–pMHCI interaction. Thus, it seems that the recall response may be optimized without diminishing the breadth of TCR use that may be critical for effective virus control.
Results
The CD8+ T-cell response to influenza virus in B6 mice is characterized by an immunodominant response to the DbNP366 epitope in both primary and recall responses (1). Within the dominant TCR Vβ (TRBV) 13-1+ subset of the DbNP366-specific response, the immune repertoire is markedly hierarchical and public, with clones (defined by TCR complementarity determining region (CDR) 3β sequences) comprising up to 80% of the TRBV13-1+ response and frequently found in other individuals (13, 14). Consequently, the DbNP366-specific TRBV13-1+ CD8+ T-cell population represents an ideal endogenous polyclonal repertoire for the identification of dominant clones and alterations in clone frequency.
Reproducible Changes in TCRβ Clonotype Frequency in the Recall Response.
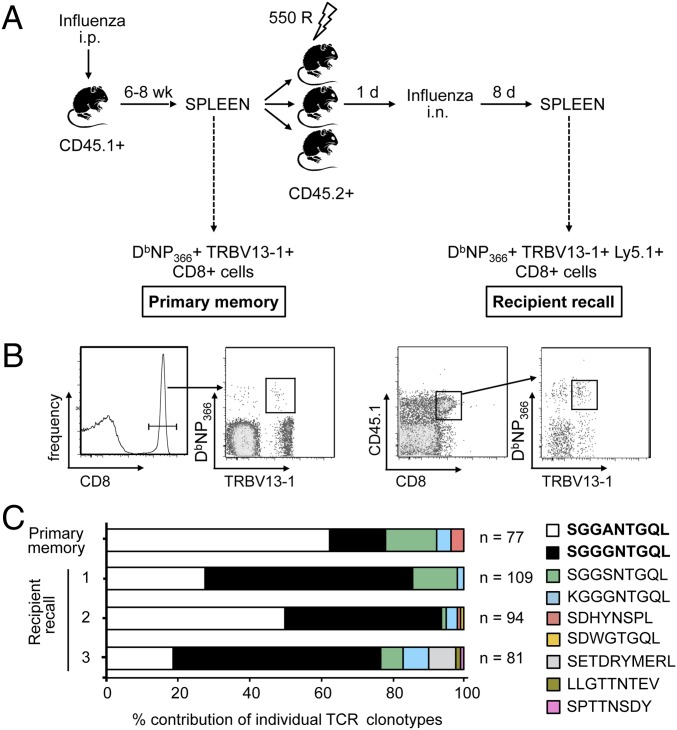
To measure changes in CTL clone representation from primary memory to acute recall and to determine whether such alterations were a result of stochastic or deterministic mechanisms, a parallel adoptive transfer approach was used. A small sample of splenocytes from a mouse primed with PR8 influenza virus 6–8 wk earlier was characterized with respect to DbNP366-specific TRBV13-1+ CD8+ TCRβ clonotype use (donor memory), whereas the majority of the population was split three ways and transferred into sublethally irradiated recipient mice (Fig. 1A). Recipient mice were then challenged intranasally (i.n.) 24 h later with the serologically distinct (H3N2) HKx31 influenza virus that shares the identical PR8 nucleoprotein (NP). Profiles of TCRβ use were then characterized for individual CD45.1+ CD8+ TRBV13-1+ DbNP366-specific CTLs recovered from the spleen at the acute phase (day 8) of the CTL recall response (Fig. 1B).
Fig. 1.
Consistent selection of TCRβ clonotypes following secondary virus infection. Individual memory phase (6–8 wk after infection) DbNP366+ TRBV13-1+ CD45.1+ CD8+ T cells were sorted from a sample of spleen. Remaining splenocytes (2–3 × 107 cells per mouse) were transferred into three sublethally irradiated (550 rad) recipient mice (CD45.2+). Recipient mice were infected (i.n.) 1 d later with ×31 influenza virus and, on day 8, individual DbNP366+ TRBV13-1+ CD45.1+ CD8+ T cells were sorted (A). Representative flow cytometry plots of sorted memory DbNP366+ TRBV13-1+ CD8+ T cells or acute recall DbNP366+ TRBV13-1+ CD45.1+ CD8+ T cells (B). TRBV13-1–specific nested single-cell RT-PCR was performed on sorted cells to determine CDR3β use. Plots represent the percentage contribution of individual TCRβ clonotypes to the total sequenced TCRβ population, shown for one representative experiment of a total of six (C). n, number of sequences.
The transition in DbNP366-specific clonal prevalence from the resting memory to the d8 recall response is shown for the first experiment in Fig. 1C. A noticeable shift in TRBV13-1+ CD8+ DbNP366-specific clonotype representation was observed between the transferred, memory population, and the responders recalled following secondary challenge (Fig. 1C). In particular, we observed an increase in prevalence of one TCRβ clonotype (SGGGNTGQL) in the recall response (Fig. 1C, black bars, and Table S1). Intriguingly, this was remarkably consistent for all three recipients, with the same T-cell clonotype being preferentially expanded despite its relatively low frequency in the adoptively transferred memory population (P = 4.21E−18, comparing memory to recipients). This pattern was highly reproducible, with six separate experiments yielding similar results (Table S1). In addition, the selective increase in clonotype prevalence was compensated by a reduction in the frequency of one or two other prominent T-cell clonotypes, a pattern that was observed for all three of the mice analyzed in this initial experiment (Fig. 1C, white bars, and Table S1). Overall, the consistency with which clonotypes were enriched or diminished in three independent recall responses strongly suggests a deterministic bias in the recall of CTL-mediated immunity.
Although the TCRβ clonotypes that consistently changed in prevalence following virus challenge were not identical for all six experiments, in three of six experiments, the SGGGNTGQL CDR3β clonotype was reproducibly increased in prevalence following virus challenge, whereas CTLs expressing SGGANTGQL decreased in four of six experiments (Table S1). Despite this trend, TCRβ clonotypes identified as notably increased or decreased in prevalence in one, or a few, experiments were also found to remain stable in some instances. This variability could reflect differential TCRα pairing, although it is also possible that the context of the repertoire (i.e., the suitability of a clone in the context of all other clones) determines which T cells are selectively expanded in the recall response.
T-Cell Selection into the Recall Response Is Based on the TCRαβ Phenotype.
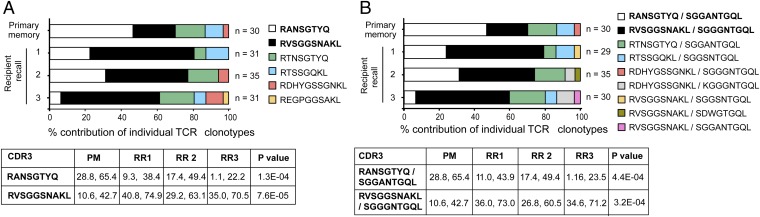
Given that the TCR CDR3β analysis indicated preferential recruitment/expansion of particular clones in the recall response, we questioned the extent to which the TCRα chain contributed to this selection. Using a multiplex RT-PCR approach (15) allowing simultaneous characterization of TCRα and β transcripts from a single lymphocyte, we found that donor memory and recipient recall responses (CD8+TRBV13-1+) showed reproducible enrichment of a TCRα (RVSGGSNAKL) in each of the three mice following secondary virus challenge, relative to donor memory (P = 7.6E−05; Fig. 2A). Moreover, a different TCRα (RANSGTYQ), present at a relatively high frequency in the donor, decreased in prevalence in all three recipients (Fig. 2A; P = 1.3E−04).
Fig. 2.
Consistent selection of TCRα and TCRαβ clonotypes following secondary virus infection. The adoptive transfer and infection strategy outlined in Fig. 1A was used to simultaneously determine TRB13-1 and TRAV use for the experiments outlined in Fig. 1C. Plots represent the percentage contribution of individual TCRα clonotypes to the total sequenced TCRα population, irrespective of CDR3β use (A). Plots represent the percentage contribution of individual TCRαβ clonotypes to the total sequenced TCRαβ population (B). n, number of sequences. Tables contain the 95% CI for each designated clonotype contribution, and P values test the hypothesis that a given sequence in donor and recipient populations comes from the same underlying distribution.
Given that these separate analyses of CDR3α and CDR3β prominence in the recall response both show reproducible and similar patterns of clonotype enrichment or loss (Figs. 1C and 2A), it is likely that these predominantly represent CTLs expressing a single TCRαβ. As observed for the analysis of TCRα and β chains alone, characterization of the combined TCRαβ signatures showed a consistent (between the three recipients) increase (P = 3.24E−04) in the frequency of one TCRαβ (RVSGGSNAKL/SGGGNTGQL)-defined clonotype (Fig. 2B), together with a corresponding and reproducible decrease (P = 4.4E−04) in the prevalence of another (RANSGTYQ/SGGANTGQL), relative to donor memory. The patterns of clonotype selection were remarkably similar, irrespective of the chain being analyzed. Interestingly, some TCRβ or TCRα clonotypes identified as being selectively enriched or diminished in the recall response were identified in partnership with other TCRα or β chains, respectively, with no notable alteration in frequency (Fig. 2B). Collectively, these data indicate that enrichment of a particular clonotype in the recall response represents active selection via the combined TCRαβ heterodimer.
Homeostatic Proliferation Does Not Notably Alter Clonal Prevalence.
To exclude the possibility that the alterations in clonotype prevalence were due to homeostatic proliferation in the ∼24 h after memory cell transfer before infection, splenocytes from a long-term influenza virus–primed animal were adoptively transferred into three irradiated recipients (Fig. 1A), and the splenic DbNP366+ TRBV13-1+ CD45.1+ CD8+ T cells were analyzed 24 h later (Fig. S1A). Comparison of TCR CDR3β use between donor memory and pooled recipient recall sets showed that all populations were dominated by the SGGSNTGQL TCRβ clonotype, with a prevalence of 68.2% and 73.3%, respectively (Fig. S1B). Furthermore, the relative contribution of this CDR3β signature in the recipients showed no substantial or consistent (across three recipients) difference from the donor population (P = 0.67). Although there was some variability between mice and a few low-frequency clonotypes were observed only in individuals, this was likely due to the low numbers of DbNP366-specific cells that could be retrieved from recipients in the absence of a secondary virus challenge.
Collectively, these data indicate that the distribution of CTL clonotypes is not substantially or reproducibly impacted by homeostatic proliferation during the 24 h after memory cell transfer.
Route of Influenza Inoculation in Recipient Mice Does Not Drive Changes in Clonal Prevalence.
The experiments described in Fig. 1 were performed using the well-characterized i.p. prime (nonreplicative infection) and i.n. challenge (replicative infection) protocol (12, 13, 16). To ensure that the different route of virus exposure was not responsible for the TCR selection observed, we performed the same transfer experiment (Fig. 1A) by both priming and challenging mice with virus via the i.p. route (Fig. S2A). Single cell analysis of CDR3β clonotype use showed that, similar to previous experiments, the prevalence of the familiar (Figs. 1 and 2 and Table S1) SGGSNTGQL increased from 22.2% in donor memory to an average of 93.3% in the pooled recipients (P = 5.27E−17). Thus, the differential selection of TCR clonotypes from the memory pool into the recall response is not due to route of virus inoculation.
Clonotype Prevalence in the Recall Response Reflects TCR/pMHCI Avidity.
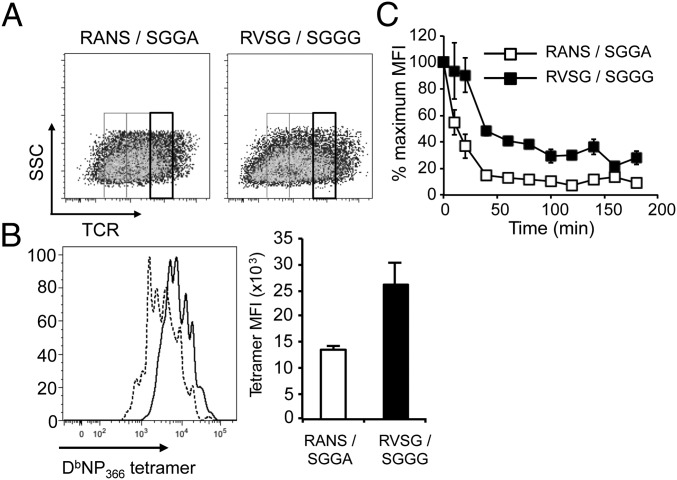
To address whether the change in size for particular CTL clones in the recall response was based on differential TCR binding characteristics, we transiently transfected 293T cells with retroviral constructs encoding the designated TCRαβ heterodimers along with a fluorescent reporter gene (GFP or AmCyan). These cells were stained for TCRβ and divided according to TCR expression levels (Fig. 3A). Analysis of each of these populations (TCRlo, int, hi) showed that the TCR that was found to be enriched in the in vivo recall responses (RVSGGSNAKL/SGGGNTGQL) bound the DbNP366 tetramer more effectively than was the case for the diminished TCR (RANSGTYQ/SGGANTGQL) (Fig. 3B). Further, the enriched TCR had a substantially slower off-rate of pMHC binding (Fig. 3C). Collectively, considering the absence of accessory molecules on 293T cells, these data indicate that the prevalence of CTL clones in the recall response correlates directly with the intrinsic affinity of the TCR.
Fig. 3.
Altered clonal prevalence correlates with TCR avidity. 293T cells were transfected with MSCV vectors encoding the CD3γδε and ζ chains (GFP) and either of the designated DbNP366-specific TCRs, identified by expression of GFP or AmCyan. After 48 h, cells were stained with anti–TCRβ-APC and DbNP366-PE tetramer. GFP+ (RVSG/SGGG) or GFP+AmCyan+ (RANS/SGGA) cells were gated for uniformly high TCR expression (A). A representative and mean level of tetramer binding (MFI) is shown for triplicate samples of TCRhi cells (B), and the rate of tetramer dissociation in the presence of an anti-H2Db antibody was determined on triplicate samples of transfected cells (C).
Partial Reversion of Clonal Prevalence from Recall to Secondary Memory.
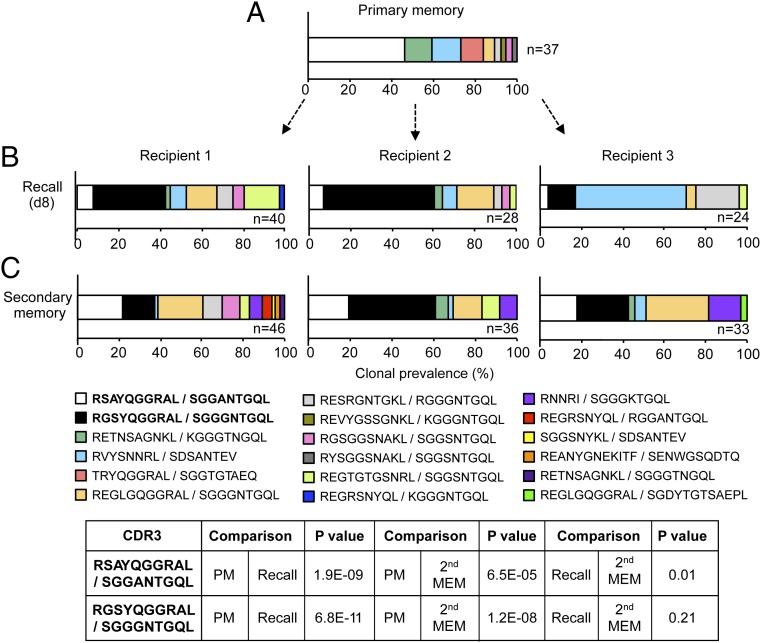
We next investigated the fate of these reproducibly enriched or diminished clones following contraction into secondary memory. To address this, the adoptive transfer system described above was modified to analyze both the acute recall and secondary memory profiles. The three recipients were bled (day 8) for analysis of the acute response, and memory phase TCR use was analyzed later (days 60–80) from the spleen. As observed previously in the acute recall repertoire, there was a significant diminution in prevalence of the SGGANTGQL CDR3β clonotype from 45.9% in donor memory to an average of 6.3% in the recipients (P = 1.9E−09), and a significant increase (P = 6.8E−11) in the abundance of a SGGGNTGQL-expressing TCR clone from 0% in donor memory to an average of 33.7% in the recipients (Fig. 4, compare A and B). Interestingly, these TCRβ chains paired with different TCRα chains to those previously observed (Fig. 2), reflecting a greater diversity of TCRα, relative to TCRβ, use. Furthermore, the TCR that was enriched in the recall response (RGSYQGGRAL/SGGANTGQL) bound significantly more tetramer than the diminished TCR (RSAYQGGRAL/SGGANTGQL) after retroviral expression in 293T cells (as previously described), indicative of higher TCR affinity/avidity (Fig. S3A), although the rate of tetramer dissociation appeared similar for the two TCRs (Fig. S3B).
Fig. 4.
Reversion of TCRβ clonal prevalence in secondary memory. The strategy outlined in Fig. 1A was modified such that blood was analyzed 8 d after secondary infection, and splenocytes were analyzed 60 d after infection (second memory). Plots represent the percentage contribution of individual TCRαβ clonotypes to the total sequenced TCRαβ population for primary memory (A), acute recall (B), and secondary memory (C) populations. n, number of paired CDR3αβ amino acid sequences obtained. P values test the hypothesis that a given sequence in donor and recipient populations comes from the same underlying distribution. Shown is one of two representative experiments.
Analysis of the secondary memory pool showed that the RSAYQGGRAL/SGGANTGQL clone regained prevalence (average of 19.8%) relative to that found for the acute recall response (P = 0.01), whereas the RGSYQGGRAL/SGGGNTGQL clone lost prevalence (average of 27.04%), although the latter was not significant (Fig. 4, compare B and C). Thus, based on the profiles for these two clonotypes, the secondary memory repertoire appears, in part, to revert toward the primary memory distribution, suggesting that preferential expansion is matched by enhanced contraction of these clones in secondary memory. This finding was further demonstrated by calculation of the prevalence of the RSAYQGGRAL/SGGANTGQL (diminished) and RGSYQGGRAL/SGGGNTGQL (enriched) clones in the total CD8+ T-cell pool in acute recall compared with secondary memory. The enriched clone showed a notably reduced prevalence (2.4-fold) in secondary memory, whereas the diminished clone showed a slightly elevated prevalence (1.7-fold). Given that all clones would be contracting to some extent and none would undergo a net expansion from acute recall to secondary memory, these data therefore indicate that the RGSYQGGRAL/SGGGNTGQL clone undergoes more pronounced contraction than the more stable RSAYQGGRAL/SGGANTGQL clone. This experiment was repeated with similar results (Fig. S4).
CTL Clonal Diversity and Distribution Across the Stages of the Antiviral Response.
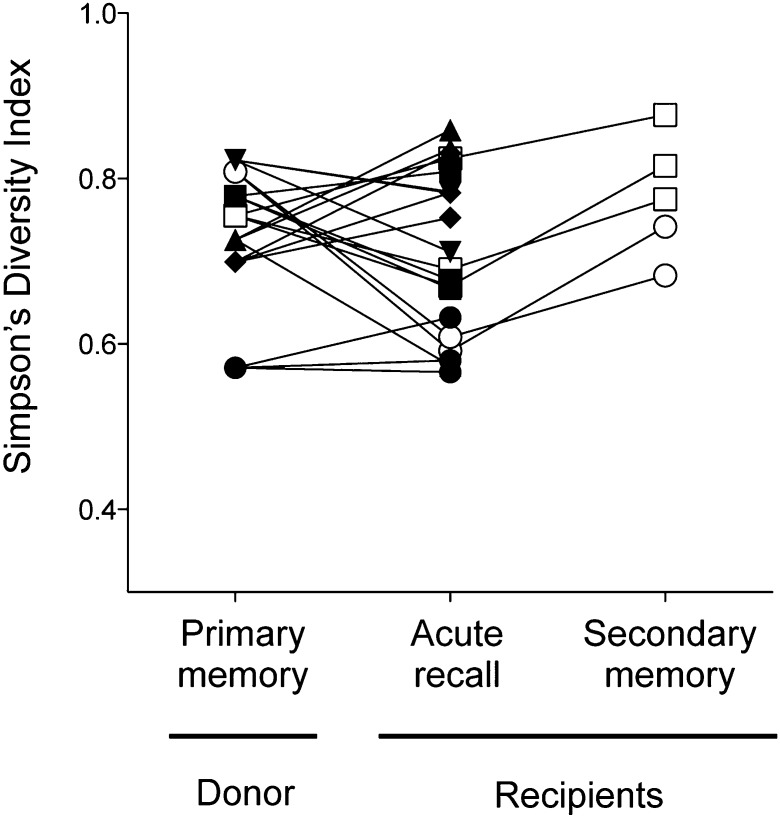
Prior analyses of CTL responses demonstrated a focusing (or narrowing) of TCR β repertoire use with successive antigen encounters (17–19). Given that we observed reproducible shifts in clonal prevalence from memory to the recall response that (in secondary memory) appeared to trend back toward that observed following initial priming, we investigated the extent to which such changes in the repertoire impacted its overall diversity. Fig. 5 shows Simpson’s diversity index (SDI; a function of clone number and distribution) values (20–22) for the TCRβ repertoires analyzed at the indicated stages, with related repertoires connected. Of the 20 repertoires analyzed in the acute recall response (those shown in Table S1 and three recipients from experiment in Fig. 4), only 11 showed a narrowing in diversity, with 2 remaining unchanged, and in 7 populations TCRβ diversity increased relative to the donor memory population (Fig. 5). Overall, there was no statistically significant change in clonal diversity (P = 0.45). Similar results were obtained when analysis was restricted to evenness of clonal distribution using the Gini coefficient (21, 23) (Fig. S5). Intriguingly, in all five of the repertoires that were analyzed in secondary memory, there was a shift toward increased diversity (and increased evenness), although the sample size was too low to provide statistical significance at a P < 0.05 level with a two-sided nonparametric test. Nevertheless, the data suggest that, where selective clonal expansion in the recall response results in narrowing of TCR diversity, that diversity is regained following contraction into secondary memory. In summary, we find no evidence of a global narrowing of the virus-specific CTL repertoire subsequent to secondary virus infection, either acutely or in the secondary memory phase.
Fig. 5.
No evidence of global narrowing of TCR repertoire diversity in acute recall or secondary memory. TCRβ clonotypic diversity within primary memory, recall, and secondary memory populations was measured using Simpson’s diversity index. Symbols represent populations derived from the same experiment. Lines connect related populations.
Discussion
Previous studies have variously indicated that T-cell recruitment/expansion from memory pools is deterministic, with a focusing of TCR use for both CD8+ and CD4+ T-cell recall responses (8, 17), corresponding to an increase in repertoire affinity (8, 14, 17–19, 24), or stochastic, with recall CD8+ T-cell populations showing either similar TCR use to the memory populations from which they were derived (9–11, 13) or seemingly random clonotypic expansions (12, 25). Here, we used a parallel adoptive transfer strategy to determine the reproducibility of a particular recall response generated from the same memory population in different recipients. We observed clear differences between the donor memory and recipient recall populations that were reproducible between all three independently generated recall responses, typically involving one or two clonotypes with either substantially increased or decreased prevalence. Overall, these data clearly demonstrate active repertoire selection in an acute antiviral CTL response and show that this selection occurs at the level of individual T-cell clones, is dependent on both TCRα and β chains, and correlates with TCR avidity for the immunogenic pMHCI.
The extent to which clonal selection modifies the global repertoire appears to be variable, with TCR diversity and distribution increased or decreased in different experiments. It is possible that this reflects the different extents to which preferred clones are expanded and nonpreferred clones are diminished. Moreover, clones were observed whose prevalence altered inconsistently in the three recipients. These data suggest that the preference for particular clonotypes in the recall response is not necessarily as extreme as has been previously demonstrated for other infections such as Listeria (8). It supports a model whereby all clones above a certain affinity threshold can contribute to the response (26, 27), although preferential expansion of particular clonotypes allows for potentially higher functioning T cells to assume dominance (28, 29). Collectively, this has the biological advantage of a recall response with enhanced function while maintaining TCR diversity, shown to be beneficial for virus control (4, 6) and for retaining the plasticity to respond to viral escape mutants (3, 5). Intriguingly, for those repertoires analyzed here where clonal diversity appeared to narrow slightly in recall, diversity was restored after contraction into secondary memory. These data suggest that not only do CD8+ T cells expressing a relatively high avidity TCR receive a (qualitatively or quantitatively) distinct signal to those expressing low avidity TCRs, resulting in their increased expansion, it appears that such clones retain a memory of their increased division, resulting in their increased contraction after viral clearance. This intrinsic register of cell division was first proposed by Sun et al. (30) when the enhanced CD4+ and CD8+ T-cell expansion observed in cells expressing a chimeric IL-7 receptor was matched by their contraction, such that memory numbers were unchanged. Other studies have also observed a link between T-cell division and death (31).
Interestingly, in six independent experiments analyzing TCRβ clonotype use within the dominant TRBV13-1+ DbNP366-specific populations, there was a trend toward (SGGGNTGQL) and against (SGGANTGQL) particular TCRβ clonotypes. We have previously shown lower TCR avidity in DbNP366-specific CD8+ T cells expressing, almost exclusively, the TCRβ SGGANTGQL clonotype (32). Several antiviral CTL responses in humans are characterized by the existence of heavily biased TCR use both within and between individuals (33). The dominance of these clones has been linked to the nature of the structural fit between the TCR and the pMHC (34, 35). Indeed, it has been suggested that the prolonged antigen presentation associated with chronic CMV and EBV infections, drives the preferential selection of high avidity CTL clones (36). Our data demonstrate that the avidity of the pMHC–TCR interaction is also a key determinant of clonal prevalence in the recall response to acute infections caused by readily eliminated viruses. In addition, it indicates that avidity maturation observed in T-cell responses from memory to recall (14, 18, 24) is due, at least in part, to the preferential expansion of clones expressing high avidity TCRs rather than simply reflecting an optimization of signaling via the altered organization, activation, and/or expression of TCR or signaling molecules (35, 37–39).
The vast majority of T-cell repertoire analyses to date have focused on the TCRβ chain, primarily due to a lack of reagents for specific TCRα chains. Using a strategy of unparalleled resolution to characterize the precise TCRβ and α chain use (at the level of CDR3 sequence) (15) from individual TRBV13-1+ DbNP366-specific cells, we identified the role of TCRα and β chain pairing in conferring optimal DbNP366 specificity. Although specific TCRα and TCRβ clonotypes were able to pair with a number of different TCRβ or α clonotypes, respectively, selection for particular T cells in the recall response was based on both the TCRα and β chains. This selection is in accord with other studies that have demonstrated an equivalent role for the TCRα and β chains in conferring specificity and in mediating contact with peptide (40, 41).
In summary, we demonstrated that the expansion of influenza virus–specific CD8+ T cells in the recall response is deterministic and appears to be based on TCR binding characteristics. Surprisingly, such deliberate repertoire selection can occur at the level of specific clones defined by unique CDR3 amino acid sequences. Despite this shift in clonotype representation, there was no evidence for global narrowing of the repertoire, with any narrowing observed in acute recall repertoires reversed in secondary memory. These data reveal an effective strategy for both safeguarding the flexibility of the repertoire and ensuring an effective response.
Materials and Methods
Mice and Virus Infections.
Female CD45.1+ C57BL/6J (B6; H-2b) and CD45.2+ B6 mice were bred and housed in specific pathogen-free conditions in the animal facility of the Department of Microbiology and Immunology, University of Melbourne (Parkville, VIC, Australia). Donor mice were inoculated i.p. 6–8 wk previously with 1.5 × 107 plaque forming units (PFUs) of the A/Puerto Rico/8/34 (PR8, H1N1) influenza virus. Recipient mice were sublethally irradiated with 550 rad before adoptive transfer and, following transfer, were either inoculated i.p. with 1.5 × 107 PFU of the PR8 influenza virus or infected i.n. with 1 × 104 PFU of the A/Hong Kong/X31 (X31, H3N2) influenza virus. All experimental procedures were reviewed and approved by the University of Melbourne Animal Experimentation Ethics Committee.
Adoptive Transfer.
Splenocytes (typically 2–3 × 107 cells per mouse) were transferred via i.v. injection into sublethally irradiated CD45.2+ B6 recipient mice. Recipient mice were infected i.n. with 1 × 104 PFUs or i.p. with 1.5 × 107 PFU of ×31 influenza virus 24 h after cell transfer and killed 8 d thereafter for analysis of the acute recall response from splenocytes. In some instances, recipient mice were killed 24 h after cell transfer for splenic repertoire analysis.
Isolation of Individual CD8+ T Cells by Sorting.
Enriched splenocytes or peripheral blood lymphocytes (PBLs; SI Materials and Methods) were stained with phycoerythrin (PE) or allophycocyanin (APC)-conjugated tetrameric complexes of the influenza virus H-2Db MHC class I glycoprotein and NP366–374 (ASNENMETM) peptide for 1 h at room temperature. Cells were then stained with various combinations of conjugated antibodies specific for Vβ8.3, CD8α (53-6.7), and CD45.1-PE (A20) (BD Pharmingen and BioLegend). Individual DbNP366+ TRBV13-1(Vβ8.3)+ CD8+ T cells or DbNP366+ TRBV13-1+ CD45.1+ CD8+ T cells were sorted using a BD FACSAria (BD Biosciences).
RT-PCR and Sequencing.
Single-cell RT and PCR amplification of TRBV13-1+ TCR CDR3β regions was performed as previously described (12). Multiplex single-cell RT and PCR amplification of TCR CDR3α and CDR3β regions was performed for amplification of cells expressing TRBV13-1 and unknown TCR Vα (TRAV), using TRBV13-1–specific oligonucleotides and a panel of TRAV specific oligonucleotides, as previously described (15). PCR products were purified with ExoSAP-IT reagent (USB Corporation) and sequenced with corresponding TRAC or TRBC internal primers (12, 15).
Statistics.
Statistics were performed as described in SI Materials and Methods.
Transfection of 293T Cells and Analysis of TCR Binding Characteristics.
293T cells were transfected with a mouse stem cell virus (MSCV)-based retroviral vector containing αβTCR sequence + internal ribosome entry site (IRES)-GFP (pMIG) or IRES-AmCyan (pMIC), as well as pMIG encoding the CD3γδε and ζ subunits, according to ref. 42. Transfected 293T cells were labeled with DbNP366-PE tetramer 48 h later, followed either by conjugated antibodies specific for CD8α and TCRβ or incubated for designated times at 37 °C with anti-H-2Db antibody (28-14-8; BD PharMingen) (50 μg/mL) and then with CD8α Ab (43).
Supplementary Material
Acknowledgments
This work was supported by National Health and Medical Research Council (NHMRC) Project Grants AI628316 and AI1046333 (to N.L.L.G.), National Institute of Health Grants AI091938 (to P.G.T. and A.H.) and AI107625 (to P.G.T.), a Sylvia and Charles Viertel Senior Medical Research fellowship (to N.L.L.G.), an NHMRC Biomedical Postgraduate Scholarship ID520643 (to T.C.), and an Australian Research Council Future Fellowship (to S.J.T.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1323736111/-/DCSupplemental.
References
- 1.Belz GT, Xie W, Altman JD, Doherty PC. A previously unrecognized H-2D(b)-restricted peptide prominent in the primary influenza A virus-specific CD8(+) T-cell response is much less apparent following secondary challenge. J Virol. 2000;74(8):3486–3493. doi: 10.1128/jvi.74.8.3486-3493.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Williams MA, Bevan MJ. Effector and memory CTL differentiation. Annu Rev Immunol. 2007;25:171–192. doi: 10.1146/annurev.immunol.25.022106.141548. [DOI] [PubMed] [Google Scholar]
- 3.Cornberg M, et al. Narrowed TCR repertoire and viral escape as a consequence of heterologous immunity. J Clin Invest. 2006;116(5):1443–1456. doi: 10.1172/JCI27804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Messaoudi I, Guevara Patiño JA, Dyall R, LeMaoult J, Nikolich-Zugich J. Direct link between mhc polymorphism, T cell avidity, and diversity in immune defense. Science. 2002;298(5599):1797–1800. doi: 10.1126/science.1076064. [DOI] [PubMed] [Google Scholar]
- 5.Price DA, et al. T cell receptor recognition motifs govern immune escape patterns in acute SIV infection. Immunity. 2004;21(6):793–803. doi: 10.1016/j.immuni.2004.10.010. [DOI] [PubMed] [Google Scholar]
- 6.Wang GC, Dash P, McCullers JA, Doherty PC, Thomas PG. T cell receptor αβ diversity inversely correlates with pathogen-specific antibody levels in human cytomegalovirus infection. Sci Transl Med. 2012;4(128):28ra42. doi: 10.1126/scitranslmed.3003647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Luo W, et al. Limited T cell receptor repertoire diversity in tuberculosis patients correlates with clinical severity. PLoS ONE. 2012;7(10):e48117. doi: 10.1371/journal.pone.0048117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Busch DH, Pilip I, Pamer EG. Evolution of a complex T cell receptor repertoire during primary and recall bacterial infection. J Exp Med. 1998;188(1):61–70. doi: 10.1084/jem.188.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blattman JN, Sourdive DJ, Murali-Krishna K, Ahmed R, Altman JD. Evolution of the T cell repertoire during primary, memory, and recall responses to viral infection. J Immunol. 2000;165(11):6081–6090. doi: 10.4049/jimmunol.165.11.6081. [DOI] [PubMed] [Google Scholar]
- 10.Maryanski JL, Jongeneel CV, Bucher P, Casanova JL, Walker PR. Single-cell PCR analysis of TCR repertoires selected by antigen in vivo: A high magnitude CD8 response is comprised of very few clones. Immunity. 1996;4(1):47–55. doi: 10.1016/s1074-7613(00)80297-6. [DOI] [PubMed] [Google Scholar]
- 11.Sourdive DJ, et al. Conserved T cell receptor repertoire in primary and memory CD8 T cell responses to an acute viral infection. J Exp Med. 1998;188(1):71–82. doi: 10.1084/jem.188.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Turner SJ, Diaz G, Cross R, Doherty PC. Analysis of clonotype distribution and persistence for an influenza virus-specific CD8+ T cell response. Immunity. 2003;18(4):549–559. doi: 10.1016/s1074-7613(03)00087-6. [DOI] [PubMed] [Google Scholar]
- 13.Kedzierska K, Turner SJ, Doherty PC. Conserved T cell receptor usage in primary and recall responses to an immunodominant influenza virus nucleoprotein epitope. Proc Natl Acad Sci USA. 2004;101(14):4942–4947. doi: 10.1073/pnas.0401279101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhong W, Reinherz EL. In vivo selection of a TCR Vbeta repertoire directed against an immunodominant influenza virus CTL epitope. Int Immunol. 2004;16(11):1549–1559. doi: 10.1093/intimm/dxh156. [DOI] [PubMed] [Google Scholar]
- 15.Dash P, et al. Paired analysis of TCRα and TCRβ chains at the single-cell level in mice. J Clin Invest. 2011;121(1):288–295. doi: 10.1172/JCI44752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.La Gruta NL, et al. Epitope-specific TCRbeta repertoire diversity imparts no functional advantage on the CD8+ T cell response to cognate viral peptides. Proc Natl Acad Sci USA. 2008;105(6):2034–2039. doi: 10.1073/pnas.0711682102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McHeyzer-Williams MG, Davis MM. Antigen-specific development of primary and memory T cells in vivo. Science. 1995;268(5207):106–111. doi: 10.1126/science.7535476. [DOI] [PubMed] [Google Scholar]
- 18.Busch DH, Pamer EG. T cell affinity maturation by selective expansion during infection. J Exp Med. 1999;189(4):701–710. doi: 10.1084/jem.189.4.701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kedl RM, Schaefer BC, Kappler JW, Marrack P. T cells down-modulate peptide-MHC complexes on APCs in vivo. Nat Immunol. 2002;3(1):27–32. doi: 10.1038/ni742. [DOI] [PubMed] [Google Scholar]
- 20.Magurran AE. Measuring Biological Diversity. Blackwell Publishing, Malden, MA; 2004. [Google Scholar]
- 21.Thomas PG, Handel A, Doherty PC, La Gruta NL. Ecological analysis of antigen-specific CTL repertoires defines the relationship between naive and immune T-cell populations. Proc Natl Acad Sci USA. 2013;110(5):1839–1844. doi: 10.1073/pnas.1222149110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Venturi V, Kedzierska K, Turner SJ, Doherty PC, Davenport MP. Methods for comparing the diversity of samples of the T cell receptor repertoire. J Immunol Methods. 2007;321(1-2):182–195. doi: 10.1016/j.jim.2007.01.019. [DOI] [PubMed] [Google Scholar]
- 23.Sadras V, Bongiovanni R. Use of Lorenz curves and Gini coefficients to assess yield inequality within paddocks. Field Crops Res. 2004;90(2-3):303–310. [Google Scholar]
- 24.Savage PA, Boniface JJ, Davis MM. A kinetic basis for T cell receptor repertoire selection during an immune response. Immunity. 1999;10(4):485–492. doi: 10.1016/s1074-7613(00)80048-5. [DOI] [PubMed] [Google Scholar]
- 25.Lin MY, Welsh RM. Stability and diversity of T cell receptor repertoire usage during lymphocytic choriomeningitis virus infection of mice. J Exp Med. 1998;188(11):1993–2005. doi: 10.1084/jem.188.11.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Malherbe L, Hausl C, Teyton L, McHeyzer-Williams MG. Clonal selection of helper T cells is determined by an affinity threshold with no further skewing of TCR binding properties. Immunity. 2004;21(5):669–679. doi: 10.1016/j.immuni.2004.09.008. [DOI] [PubMed] [Google Scholar]
- 27.Schmid DA, et al. Evidence for a TCR affinity threshold delimiting maximal CD8 T cell function. J Immunol. 2010;184(9):4936–4946. doi: 10.4049/jimmunol.1000173. [DOI] [PubMed] [Google Scholar]
- 28.Zehn D, Lee SY, Bevan MJ. Complete but curtailed T-cell response to very low-affinity antigen. Nature. 2009;458(7235):211–214. doi: 10.1038/nature07657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hommel M, Hodgkin PD. TCR affinity promotes CD8+ T cell expansion by regulating survival. J Immunol. 2007;179(4):2250–2260. doi: 10.4049/jimmunol.179.4.2250. [DOI] [PubMed] [Google Scholar]
- 30.Sun JC, Lehar SM, Bevan MJ. Augmented IL-7 signaling during viral infection drives greater expansion of effector T cells but does not enhance memory. J Immunol. 2006;177(7):4458–4463. doi: 10.4049/jimmunol.177.7.4458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nolz JC, Rai D, Badovinac VP, Harty JT. Division-linked generation of death-intermediates regulates the numerical stability of memory CD8 T cells. Proc Natl Acad Sci USA. 2012;109(16):6199–6204. doi: 10.1073/pnas.1118868109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kedzierska K, et al. Terminal deoxynucleotidyltransferase is required for the establishment of private virus-specific CD8+ TCR repertoires and facilitates optimal CTL responses. J Immunol. 2008;181(4):2556–2562. doi: 10.4049/jimmunol.181.4.2556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gras S, Kjer-Nielsen L, Burrows SR, McCluskey J, Rossjohn J. T-cell receptor bias and immunity. Curr Opin Immunol. 2008;20(1):119–125. doi: 10.1016/j.coi.2007.12.001. [DOI] [PubMed] [Google Scholar]
- 34.Kjer-Nielsen L, et al. A structural basis for the selection of dominant alphabeta T cell receptors in antiviral immunity. Immunity. 2003;18(1):53–64. doi: 10.1016/s1074-7613(02)00513-7. [DOI] [PubMed] [Google Scholar]
- 35.Stewart-Jones GB, McMichael AJ, Bell JI, Stuart DI, Jones EY. A structural basis for immunodominant human T cell receptor recognition. Nat Immunol. 2003;4(7):657–663. doi: 10.1038/ni942. [DOI] [PubMed] [Google Scholar]
- 36.Price DA, et al. Avidity for antigen shapes clonal dominance in CD8+ T cell populations specific for persistent DNA viruses. J Exp Med. 2005;202(10):1349–1361. doi: 10.1084/jem.20051357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Slifka MK, Whitton JL. Functional avidity maturation of CD8(+) T cells without selection of higher affinity TCR. Nat Immunol. 2001;2(8):711–717. doi: 10.1038/90650. [DOI] [PubMed] [Google Scholar]
- 38.Cawthon AG, Alexander-Miller MA. Optimal colocalization of TCR and CD8 as a novel mechanism for the control of functional avidity. J Immunol. 2002;169(7):3492–3498. doi: 10.4049/jimmunol.169.7.3492. [DOI] [PubMed] [Google Scholar]
- 39.Kersh EN, et al. TCR signal transduction in antigen-specific memory CD8 T cells. J Immunol. 2003;170(11):5455–5463. doi: 10.4049/jimmunol.170.11.5455. [DOI] [PubMed] [Google Scholar]
- 40.Jorgensen JL, Esser U, Fazekas de St Groth B, Reay PA, Davis MM. Mapping T-cell receptor-peptide contacts by variant peptide immunization of single-chain transgenics. Nature. 1992;355(6357):224–230. doi: 10.1038/355224a0. [DOI] [PubMed] [Google Scholar]
- 41.Valkenburg SA, et al. Fixing an irrelevant TCR alpha chain reveals the importance of TCR beta diversity for optimal TCR alpha beta pairing and function of virus-specific CD8+ T cells. Eur J Immunol. 2010;40(9):2470–2481. doi: 10.1002/eji.201040473. [DOI] [PubMed] [Google Scholar]
- 42.Szymczak AL, et al. Correction of multi-gene deficiency in vivo using a single ‘self-cleaving’ 2A peptide-based retroviral vector. Nat Biotechnol. 2004;22(5):589–594. doi: 10.1038/nbt957. [DOI] [PubMed] [Google Scholar]
- 43.La Gruta NL, Doherty PC, Turner SJ. A correlation between function and selected measures of T cell avidity in influenza virus-specific CD8+ T cell responses. Eur J Immunol. 2006;36(11):2951–2959. doi: 10.1002/eji.200636390. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.