Serial norovirus infections with multiple genotypes were found among a Peruvian birth cohort early in infancy. Protection against the subsequent infection was genotype specific, suggesting that norovirus vaccines may need to target multiple genotypes.
Keywords: norovirus, infant diarrhea, gastroenteritis, birth cohort, natural infection
Abstract
Background. Human noroviruses are among the most common enteropathogens globally, and are a leading cause of infant diarrhea in developing countries. However, data measuring the impact of norovirus at the community level are sparse.
Methods. We followed a birth cohort of children to estimate norovirus infection and diarrhea incidence in a Peruvian community. Stool samples from diarrheal episodes and randomly selected nondiarrheal samples were tested by polymerase chain reaction for norovirus genogroup and genotype. Excretion duration and rotavirus coinfection were evaluated in a subset of episodes.
Results. Two hundred twenty and 189 children were followed to 1 and 2 years of age, respectively. By 1 year, 80% (95% confidence interval [CI], 75%–85%) experienced at least 1 norovirus infection and by 2 years, 71% (95% CI, 65%–77%) had at least 1 episode of norovirus-associated diarrhea. Genogroup II (GII) infections were 3 times more frequent than genogroup 1 (GI) infections. Eighteen genotypes were found; GII genotype 4 accounted for 41%. Median excretion duration was 34.5 days for GII vs 8.5 days for GI infection (P = .0006). Repeat infections by the same genogroup were common, but repeat infections by the same genotype were rare. Mean length-for-age z score at 12 months was lower among children with prior norovirus infection compared to uninfected children (coefficient: −0.33 [95% CI, −.65 to −.01]; P = .04); the effect persisted at 24 months.
Conclusions. Norovirus infection occurs early in life and children experience serial infections with multiple genotypes, suggesting genotype-specific immunity. An effective vaccine would have a substantial impact on morbidity, but may need to target multiple genotypes.
(See the Editorial Commentary by Lopman and Kang on pages 492–4.)
Noroviruses, RNA viruses with >36 recognized genotypes [1, 2], are the leading cause of gastroenteritis outbreaks [3, 4]. Noroviruses have recently been recognized as the second most common etiology of infant diarrhea and hospitalization for diarrhea, and cause an estimated 200 000 child deaths annually in developing countries [5]. With rotavirus vaccine introduction, the relative importance of norovirus is increasing [6, 7]. However, the need for reverse transcription polymerase chain reaction (RT-PCR) has impeded studies in resource-limited settings. The impact of norovirus at the community level is poorly understood, because few population- or clinic-based studies have been conducted [8–12]. In this study, the first community-based birth cohort to examine norovirus epidemiology, we followed Peruvian infants to investigate norovirus incidence, determinants of norovirus diarrhea, excretion duration, and evidence for protection from subsequent infection.
METHODS
Ethics Statement
The study was approved by the institutional review boards of Asociación Benéfica PRISMA, Universidad Peruana Cayetano Heredia (UPCH), Johns Hopkins University, the Centers for Disease Control and Prevention (CDC), and the European Union. Each woman provided written informed consent for her infant.
Recruitment and Data Collection
Participants were recruited in Las Pampas de San Juan de Miraflores, a shantytown with approximately 50 000 inhabitants in southern Lima. Pregnant women and those with newborns younger than 3 months were randomly selected from a complete community census. Exclusion criteria were hospitalization for >1 month at birth, any congenital defect, twin birth, and birth weight <1500 g. None of the participants received rotavirus immunization; all were beyond the eligible age when it was introduced in Lima. From June 2007 to April 2011, field workers visited each household twice weekly to compile a daily record of fever, vomiting, frequency of liquid or semiliquid stools, and breastfeeding status (categorized as exclusive breastfeeding, formula only, mix of formula and breast milk, or weaned). Length and weight were measured weekly until 2 months and then monthly [13]. Stool samples were collected weekly, regardless of symptoms. One specimen was requested during each diarrheal episode. Samples were transported to UPCH within 12 hours for storage at −50°C.
Sample Selection
Norovirus testing was conducted in all specimens from diarrheal episodes and a random sample of nondiarrheal stools. One nondiarrheal specimen per child per month was tested in the first year of life. Due to resource limitations, in the second year of life, testing was conducted in 2 nondiarrheal samples frequency-matched by month with the corresponding diarrheal stools. Real-time RT-PCR for rotavirus [14] was performed in an age-stratified random sample from each of the following groups: norovirus-positive with and without diarrhea; norovirus-negative with and without diarrhea. To evaluate excretion duration, we selected an age-stratified random sample of 46 norovirus-positive index specimens and tested weekly specimens collected before and after the index specimens. These samples were selected blindly and not selected by genotype. We defined the end of an infection episode when 2 sequential specimens were negative. The duration of excretion were defined as the dates between the first and last positive specimens.
Definitions
A day of diarrhea was defined by the presence of ≥3 liquid or semiliquid stools in 24 hours. For infants younger than 2 months, the definition was based on the mother's or caretaker's assessment that the child had diarrhea [15]. An episode ended when the child had 2 consecutive days without diarrhea. Weaning was defined as breastfeeding interruption for 7 days or more.
Norovirus was diagnosed by positive real-time RT-PCR results. Infection with a different genotype at any time was considered a new infection. The same genogroup or genotype detected after ≥30 days with 2 intervening negative samples was considered a new infection. If only 1 negative sample existed, a 60-day interval was required to define a new infection. “Norovirus-associated diarrhea” was defined based on positive results by PCR in a stool taken during or within 7 days of the beginning or end of the diarrheal episode. Diarrhea onset date was defined as the start date for symptomatic infection. An “asymptomatic” infection occurred when no symptoms were reported within 7 days of the positive specimen that defined the beginning of the infection episode. If positive results were found in a nondiarrheal specimen but diarrhea occurred within 30 days or vomiting within 7 days without norovirus testing, the episode was coded as “undefined.”
Real-time RT-PCR and Genotyping
Aliquots of 0.1 g (formed) or 0.1 mL (watery) stools were resuspended to 1 mL and RNA was extracted using silica particles with guanidinium thiocyanate [16]. A segment of the ORF1–ORF2 junction was amplified by TaqMan real-time RT-PCR, using published primers and probes for genogroups I (GI) and II (GII) [17]. The detection limit was determined by standard curves using 10-fold serial dilutions (106 to 100) of GII.4 and GI.3 RNA transcripts provided by the National Calicivirus Laboratory, CDC. The detection limit was 10 copies corresponding to cycle threshold (Ct) 37 for GI and 38 for GII.
Real-time PCR–positive samples were genotyped by conventional PCR targeting the capsid N/S domain (primers G1SKF/G1SKR for GI, G2SKF/G2SKR for GII) [18]. DNA sequences were assembled with ChromasPro (Technelysium Pty Ltd, Tewantin, Australia), aligned with ClustalX (http://www.clustal.org). Genotypes and GII.4 variants were identified using NoroNet (http://www.noronet.nl) [19].
Statistical Analysis
Cumulative incidence rates were estimated by Kaplan-Meier survival analyses. Analyses of asymptomatic infection focused on infants up to 12 months due to the smaller number of nondiarrheal stools tested in the second year of life. To evaluate protection associated with previous norovirus infections, we used Cox proportional hazard models with the Breslow method for handling ties [20, 21]. We used logistic regression with generalized estimating equations to test associations with diarrhea during norovirus infection. Variables with P < .1 in univariable analyses were tested by stepwise selection in multivariable models. We also performed a cross-sectional analysis to estimate the unadjusted attributable fraction comparing the prevalence of norovirus in diarrhea and nondiarrhea samples. (the proportion of diarrhea theoretically eliminated if norovirus were eliminated) [22]. Episode duration, maximum daily number of diarrheal stools, days of vomiting, and reported fever were compared by genogroup and GII genotype using the χ2 test or Mann-Whitney U test. The same variables were compared for the first and second infection within the same child. To evaluate associations between norovirus infection and growth, multiple linear regression models were generated with the outcome length-for-age and weight-for-age z scores (LAZ and WAZ, respectively) [13]. Analyses adjusted for breastfeeding prevalence included data only from children with ≥6 months of follow-up. To test if the observed frequencies of genotypes in repeated infections were significantly different than expected by chance, we generated a simulation of 1 million children with a mean of 3 norovirus infections randomly distributed according to the genotype prevalence found in the study population. The genotype frequencies expected by chance were compared with the observed frequencies by a 2-sample proportion test for a binomial distribution. Analyses were performed using Stata software, version 12 (StataCorp, College Station, Texas).
RESULTS
A total of 291 children participated, yielding 4978 child-months of follow-up. Median age at recruitment was 19 days (range, 0–97 days). The number of children followed to 6 months, 1 year, and 2 years of age was 256, 220, and 189, respectively. Of study households, 89% had continuous indoor running water and 91% had an indoor toilet. Background characteristics were comparable for the 189 children who completed follow-up and 102 children who withdrew before the study ended (Supplementary Table 1). Norovirus testing was performed in 1495 diarrheal and 3690 randomly selected nondiarrheal stools. An additional 789 stools included in the random selection were found to have been collected within 7 days of a diarrheal episode; data from these specimens were included in norovirus diarrhea incidence analyses, but not in calculation of the attributable fraction (see below).
Norovirus Incidence
In total, 607 norovirus infections were identified (140 [23.1%] GI, 460 [75.8%] GII, and 7 [1.1%] GI/GII mixed infections). Two hundred seventy-five (45.3%; 60 GI, 211 GII, 4 GI/GII mixed) infections were associated with diarrhea.
In the first year of life, 321 infections occurred during 2694 child-months of follow-up. The incidence rate was 8.6 (95% confidence interval [CI], 7.1–10.4) and 15.7 (95% CI, 13.6–18.1) per 100 child-months for ages 0–5 months and 6–11 months, respectively. Eighty percent of children had at least 1 infection and 38% had at least 2 infections by 1 year of age (Figure 1A). Of 208 first infections in the first year of life, 46% (95) were asymptomatic, 33% (69) symptomatic, and 21% (44) undefined. A significantly higher percentage of infections was asymptomatic among infants younger than 6 months (55% [66/119]) compared to those aged 6–11 months (36% [73/202]) (adjusted odds ratio [AOR], 2.2 [95% CI, 1.3–3.8]; P < .001). Exclusive breastfeeding during age 3–5 months was protective (AOR, 0.5 [95% CI, .3–.9]; P = .03). There was no significant difference in diarrhea risk between GI and GII infections (AOR, 1.6 [95% CI, .8–3.0]).
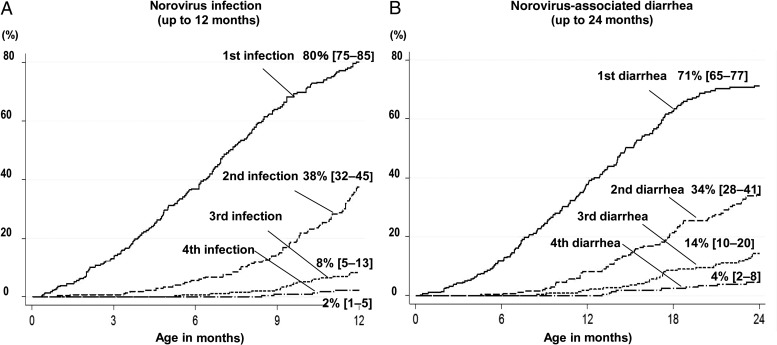
Figure 1.
Cumulative incidence of first and subsequent norovirus-associated infection and diarrhea in a birth cohort of 291 children. A, Survival curves show cumulative incidence of the first through fourth norovirus infections during the first year of life. B, Curves show the cumulative incidence of the first through fourth episodes of norovirus diarrhea for children aged 0–2 years. Percentages show the cumulative incidence, and 95% confidence intervals (shown in brackets) are based on Kaplan-Meier survival analysis.
Among infants aged 0–5, 6–11, and 12–24 months, norovirus-associated diarrhea incidence was 2.4 (95% CI, 1.7–3.4), 6.6 (95% CI, 2.1–8.0), and 7.0 (95% CI, 6.1–8.4) per 100 child-months, respectively. At least 1 episode of norovirus-associated diarrhea occurred in 38% and 71% of children by age 1 and 2 years, respectively (Figure 1B).
The cross-sectional analysis of stool prevalence included data from 1495 diarrheal and 3690 nondiarrheal stools. Norovirus occurred in 22.8% (95% CI, 20.7%–25.0% [341/1495]) of diarrheal stools and 13.3% (95% CI, 12.2%–14.4% [491/3690]) of nondiarrheal stools. The association of norovirus with diarrhea was stronger in the second (odds ratio [OR], 2.8 [95% CI, 2.2–3.5]) than the first year of life (0–5 months: OR, 1.6 [95% CI, .9–1.8]; 6–11 months: OR, 1.6 [95% CI, 1.2–2.1]). The norovirus attributable fraction was 7.8% in the first year and 23.1% in the second year of life (Figure 2). Ct values were lower in diarrheal than nondiarrheal samples (GI: median, 28.2 vs 31.0 [P < .066]; GII: median, 26.4 vs 30.1 [P = .0001], respectively; Supplementary Figure 1).
Figure 2.
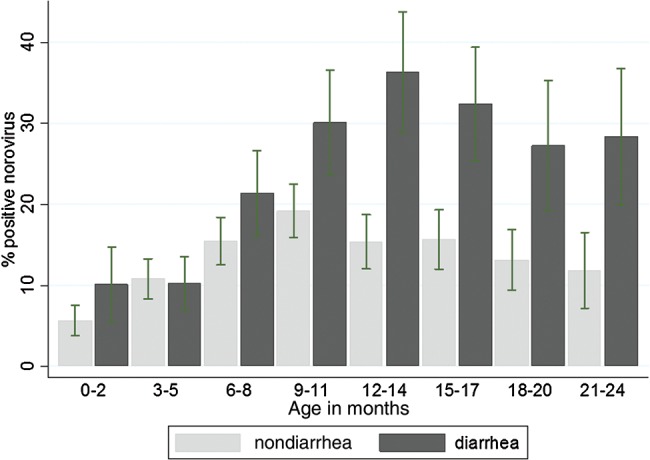
Prevalence of norovirus in diarrheal and nondiarrheal stool samples by age. Prevalences are based on cross-sectional analysis of norovirus polymerase chain reaction in 1495 specimens collected during diarrheal episodes and 3690 randomly selected nondiarrheal specimens; 789 specimens collected within 7 days of diarrhea were excluded.
Rotavirus Coinfection
Rotavirus occurred in 3.7% (2/54) and 7.4% (4/54) of asymptomatic and diarrheal norovirus-positive specimens, and 17.2% (10/58) and 11.6% (8/69) of asymptomatic and diarrheal norovirus-negative specimens, respectively. Among the 4 diarrheal episodes with both pathogens, rotavirus was the more likely etiology in 1 (18.1 vs GI 27.8 for rotavirus vs norovirus) and norovirus in 2 episodes based on relative Ct values (37.8 vs GII 25.9 and 39.0 vs GI 22.4); the fourth episode had relatively high Ct values in both (37.7 vs GII 36.4) [23].
Protection Associated With Previous Infection
One or more previous norovirus infections significantly decreased the risk of subsequent norovirus infection compared to no previous infection (Table 1). GII infection conferred significant protection against subsequent GII infection and diarrhea, but no such effect was seen for GI infection against later GI infection or diarrhea.
Table 1.
Changes in Hazard Ratios of Sequential Norovirus Infection by Number of Previous Infections
| No. of Previous Infections | No. of Sequential Episodes | Incidence per 100 Child-moa | Hazard Ratio (95% CI) for Subsequent Eventb |
|
|---|---|---|---|---|
| Adjusted | P Value | |||
| Previous norovirus infection | ||||
| 0 | 197 | 12.46 | Ref | |
| 1 | 86 | 12.04 | 0.74 (.57–.95) | .020 |
| ≥2 | 27 | 11.07 | 0.58 (.38–.90) | .014 |
| Norovirus diarrhea | ||||
| 0 | 66 | 4.17 | Ref | |
| 1 | 38 | 5.32 | 0.79 (.53–1.18) | .256 |
| ≥2 | 9 | 3.69 | 0.44 (.18–1.10) | .079 |
| Asymptomatic infection | ||||
| 0 | 87 | 5.50 | Ref | |
| 1 | 33 | 4.62 | 0.76 (.50–1.15) | .135 |
| ≥2 | 11 | 4.51 | 0.70 (.38–1.27) | .120 |
| Symptom undefined | ||||
| 0 | 44 | 2.78 | Ref | |
| 1 | 15 | 2.10 | 0.54 (.30–.97) | .040 |
| ≥2 | 7 | 2.87 | 0.62 (.31–1.24) | .173 |
| Sequential infection by the same genogroup | ||||
| Previous GII infection | ||||
| 0 | 184 | 10.97 | Ref | |
| 1 | 60 | 8.30 | 0.55 (.41–.74) | <.001 |
| ≥2 | 6 | 4.32 | 0.23 (.11–.48) | <.001 |
| GII diarrhea | ||||
| 0 | 67 | 3.99 | Ref | |
| 1 | 26 | 3.60 | 0.55 (.34–.87) | .011 |
| ≥2 | 2 | 1.44 | 0.18 (.05–.68) | .012 |
| Previous GI infection | ||||
| 0 | 56 | 2.42 | Ref | |
| ≥1 | 7 | 3.07 | 0.93 (.40–2.18) | .629 |
| GI diarrhea | ||||
| 0 | 17 | 0.74 | Ref | |
| ≥1 | 3 | 1.31 | 1.06 (.21–5.26) | .944 |
| Background characteristics | ||||
| Age group | ||||
| 0–5 mo | 108 | 8.61 | Ref | |
| 6–11 mo | 202 | 15.72 | 2.12 (1.65–2.73) | <.001 |
| Days with exclusive breastfeeding during age 3–5 mo, % | ||||
| <50% | 140 | 13.47 | Ref. | |
| ≥50% | 170 | 11.33 | 0.82 (.66–1.02) | .069 |
| Potable water available 24 h/day in household | ||||
| Yes | 262 | 11.54 | Ref. | |
| No | 48 | 17.80 | 1.59 (1.15–2.20) | .005 |
| Season | ||||
| Winter (June–August) | 45 | 6.66 | Ref | |
| Fall (March–May) | 67 | 10.58 | 1.51 (1.03–2.21) | .020 |
| Summer (December–February) | 86 | 15.09 | 2.18 (1.54–3.09) | <.001 |
| Spring (September–November) | 112 | 16.95 | 2.51 (1.78–3.53) | <.001 |
Abbreviations: CI, confidence interval; GI, genogroup I; GII, genogroup II.
a Follow-up time was 1581 child-months for the group with no previous infections, 714 child-months for those with 1 previous infection, and 244 child-months for those with ≥2 previous infections. Thirty-five children with <6 months of follow-up were excluded from analysis; these children had 11 norovirus infections. Therefore, 310 infections were included in this analysis.
b Analyses were adjusted for age (0–5 months and 6–11 months), percentage of days with exclusive breastfeeding during 3–5 months of age, and having running water in household for 24 hours/day. Analyses of background characteristics were also adjusted for norovirus infection status. Sex, birth weight, mother's education, mother's age, household income, ability to store water, and having toilet with drainage were not significantly associated with norovirus infection or diarrhea.
c P values in bold are significant at <.05 level.
Genotypes
Of 769 real-time RT-PCR positive specimens tested, 573 (75%) were amplified by conventional PCR and 535 had readable sequences. In total, 18 genotypes were identified from 347 infection episodes (Supplementary Table 2). The most frequent genotype, GII.4, occurred in 141 (40.6%) typed infection episodes (77 diarrheal, 39 asymptomatic, and 25 undefined). GII.4 was more frequent in symptomatic (49/90 [54%]) than asymptomatic (36/90 [40%]) infections in the first year of life (P = .0523). Four GII.4 variants (2006b [77]; 2007 [52]; 2008 [11]; and 2010 [1]) were detected.
Overall, 81 children had 1, 58 had 2, 52 had 3, and 53 had 4–8 norovirus infections detected. Repeat infection by the same genogroup was common (34 GI, 231 GII). However, of the 151 sets of repeat infections with genotype data, only 9 (6.0%) were of the same genotype (1 GI.5, 7 GII.4, and 1 GII.6). Among 7 sets with repeat GII.4 infection, 2 were by the same variant (one each for 2006b and 2007), and 5 by different variants (three 2006b–2007, two 2006b–2008). Thus, of 151 repeat infections, 147 (97%) were by a different genotype or GII.4 variant. The observed frequency of repeat infection was significantly lower than expected by chance for GII.6, GII.4 2006b, and GII.4 2007 (P = .0063, P < .0001, and P = .0072, respectively), but not for GI.5, GII.4 2008, and GII.4 2010.
Excretion Duration
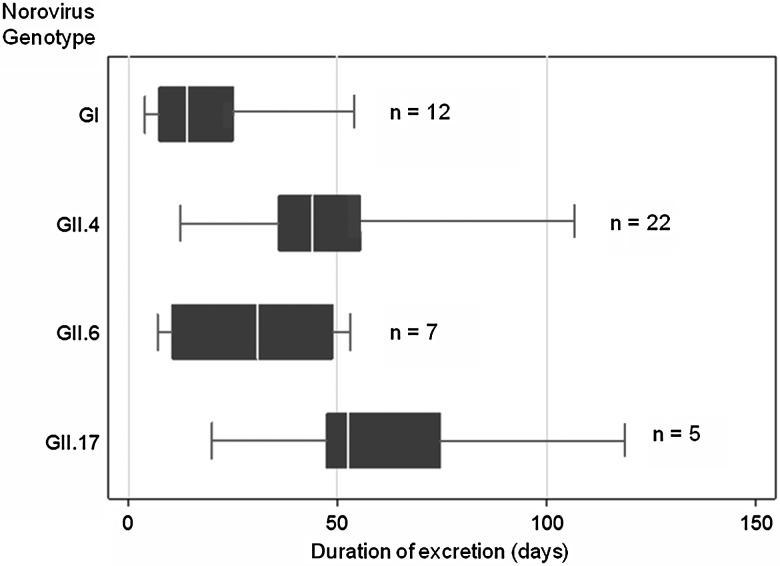
Length of viral shedding was comparable in diarrheal (n = 24; median, 31.5 days) and asymptomatic episodes (n = 14; median, 30 days; P = .83; 8 undefined episodes excluded). Median GII excretion duration was 34.5 days (n = 34; interquartile range [IQR], 28–47; max = 98), with 74% (25/34) lasting >30 days and 18% (6/34) >60 days (Figure 3). GI excretion was of shorter duration (n = 12; median, 8.5 days; IQR, 3.5–19.5; max = 49) than GII (P = .0006). Seven infections yielded positive results only in the first weekly stool (4 GI, 3 GII). In 35 of the 39 remaining episodes, the lowest Ct values were observed in the first or second week (8 GI, 27 GII). The lowest Ct occurred later than the second week of excretion in only 4 (9%) episodes (all GII).
Figure 3.
Duration of norovirus shedding by real-time reverse transcription polymerase chain reaction in 46 randomly selected infection episodes. The boxes represent 25th percentile, median, and 75th percentile, and the whiskers show the minimum and maximum duration of shedding in days. Abbreviations: GI, genogroup I; GII, genogroup II.
Symptoms by Genogroup
Of 275 norovirus-associated diarrhea episodes, 54 were GI and 209 GII only; 4 mixed infections and 8 with incomplete clinical information were excluded. GII episodes were more likely than GI episodes to be associated with fever (26% vs 13%, P = .04), ≥3 days of vomiting (9% vs 0%, P = .04), and longer duration of symptoms (median, 2 days [range, 1–10] vs 3 days [range, 1–17]; P = .06). The median daily number of diarrheal stools was 4 for both GI and GII (P = .79). GII.4 episodes were more likely to have longer duration of episodes compared to other GII genotypes (median, 3 days [range, 1–17] vs 2 days [range, 1–10]; P = .047). One study child was admitted to hospital for severe diarrhea due to norovirus GII.6. Among 163 children with repeated norovirus infection, 71 had repeated norovirus diarrhea and 54 had repeated GII diarrhea. There was no decrease in likelihood of fever, vomiting, maximum number of diarrheal stools, or symptom duration in second episodes compared with first episodes for all norovirus or only GII episodes.
Norovirus and Linear Growth
Mean LAZ and WAZ at 12 months were significantly lower in children with norovirus infection during the first year of life compared to those not infected, in models adjusted for birth weight, breastfeeding status, and household income (coefficient: −0.33 [95% CI, −.65 to −.01], P = .04 and −0.55 [95% CI, −.87 to −.23], P < .01, respectively; Tables 2 and 3). Having indoor connection of the water for 24 hours daily, having a toilet, ability to store the water or not, and mother's education level were not associated with LAZ or WAZ. Decrease in LAZ and WAZ at 12 months was more significant by GII infection than by GI. The effect of norovirus and GII infection on LAZ and WAZ persisted at 24 months (coefficient: −0.43 [95% CI, −.79 to −.07], P = .02 and –0.40 [95% CI, −.71 to −.08], P = .01, respectively).
Table 2.
Effect of Norovirus Infection on Linear Growth Based on Generalized Linear Regression Models for Length-for-Age z Score at 12 Months of Age
| Variable | LAZ Coefficient | 95% CI | P Value | |
|---|---|---|---|---|
| No. of NoV infectionsa | None | Ref. | Ref. | |
| ≥1 | −0.33 | (−.65 to −.01) | .04 | |
| No. of GI infectionsa | None | Ref. | Ref. | |
| ≥1 | −0.06 | (−.36 to −.24) | .71 | |
| No. of GII infectionsa | None | Ref. | Ref. | |
| ≥1 | −0.28 | (−.57 to −.01) | .06 | |
| Diarrheal episodes during first year of lifea | ≤4 | Ref. | Ref. | |
| >4 | 0.05 | (−.22 to .31) | .73 | |
| Days of diarrhea during first year of lifea | ≤11 | Ref. | Ref. | |
| >11 | 0.05 | (−.31 to .22) | .74 | |
| Birth weight, gb | ≤3300 | Ref. | Ref. | |
| >3300 | 0.65 | (.39 to −.90) | <.001 | |
| Days of exclusive breastfeeding at age 3–5 mo, %b | <50 | Ref. | Ref. | |
| ≥50 | −0.28 | (−.54 to −.02) | .04 | |
| Monthly income, USDb | 51–150 | Ref. | Ref. | |
| ≥151 | 0.42 | (.10 to .74) | .01 |
Outcome expressed as adjusted LAZ coefficients.
Abbreviations: CI, confidence interval; GI, genogroup I; GII, genogroup II; LAZ, length-for-age z score; NoV, norovirus; USD, US dollars.
a Coefficients were adjusted for exclusive breastfeeding, birth weight, and income.
b Coefficients were adjusted for norovirus infection in addition to the 3 variables above. Of 220 children who completed 1 year of follow-up, 4 were excluded due to incomplete data on household income or birth weight.
c P values in bold are significant at <.05 level.
Table 3.
Effect of Norovirus Infection on Linear Growth Based on Generalized Linear Regression Models for Weight-for-Age z Score at 12 Months of Age
| Variables | WAZ Coefficient | 95% CI | P Value | |
|---|---|---|---|---|
| No. of NoV infectionsa | None | Ref. | Ref. | |
| ≥1 | −0.55 | (−.87 to −.23) | .001 | |
| No. of GI infectionsa | None | Ref. | Ref. | |
| ≥1 | −0.16 | (−.47 to .15) | .30 | |
| No. of GII infectionsa | None | Ref. | Ref. | |
| ≥1 | −0.42 | (−.72 to −.12) | .01 | |
| Diarrheal episodes during first year of lifea | ≤4 | Ref. | Ref. | |
| >4 | −0.05 | (−.32 to .22) | .70 | |
| Days of diarrhea during first year of lifea | ≤11 | Ref. | Ref. | |
| >11 | −0.02 | (−.28 to .25) | .90 | |
| Birth weight, gb | ≤3300 | Ref. | Ref. | |
| >3300 | 0.64 | (.38 to .90) | <.001 | |
| Days of exclusive breastfeeding at age 3–5 mo, %b | <50 | Ref. | Ref. | |
| ≥50 | −0.36 | (−.62 to −.10) | .01 | |
| Monthly income, USDb | 51–150 | Ref. | Ref. | |
| ≥151 | 0.27 | (−.05 to .59) | .10 |
Outcome expressed as adjusted WAZ coefficients.
Abbreviations: CI, confidence interval; GI, genogroup I; GII, genogroup II; NoV, norovirus; USD, US dollars; WAZ, weight-for age z score.
a Coefficients were adjusted for exclusive breastfeeding, birth weight, and income.
b Coefficients were adjusted for norovirus infection in addition to the 3 variables above. Of 220 children who completed 1 year of follow-up, 4 were excluded due to incomplete data on household income or birth weight.
c P values in bold are significant at <.05 level.
DISCUSSION
Our data represent the first population-based norovirus birth cohort and underscore the importance of this virus as a cause of endemic diarrhea in young children [5, 24–27]. In this periurban shantytown, norovirus infected 80% of infants by age 12 months and children had up to 8 infection episodes by age 2 years. The majority of infections during the first 6 months were asymptomatic, possibly reflecting the effect of breastfeeding and transferred maternal antibodies. Infection was more strongly associated with diarrhea in the second year of life than the first. Genogroup II infections were more frequent, more likely to cause fever and vomiting, and had longer excretion periods than GI. As in previous studies, we found that gastroenteritis was associated with lower Ct values, implying higher viral loads, than asymptomatic infection [28].
The prolonged shedding shown in this study and others [8, 29, 30] was remarkable. In our data, 74% of GII infections lasted >1 month. Because of the long excretion period, we included stringent requirements to define the end of infection episodes and associations with diarrhea. In addition, only a proportion of weekly surveillance stools were tested. Our incidence estimates are therefore conservative. Nevertheless, we found that nearly all children had been infected and 71% had norovirus diarrhea at least once by age 2 years. The low infective dose, prolonged excretion, resistance to disinfection, and multiple routes of transmission [3, 4] combine to make norovirus infection nearly universal early in life. Children with norovirus in their first year of life were significantly shorter for their age than those without infection, and at 2 years, this linear growth deficit persisted. Prolonged infection, even when asymptomatic, may cause subclinical chronic intestinal inflammation decreasing nutrient absorption and growth [31]. Among those tested, 7% and 4% of norovirus-positive diarrheal and nondiarrheal stools were also positive for rotavirus. Limited testing for other enteric pathogens means that some study children with norovirus likely had undetected copathogens contributing to their symptoms. Some analyses were limited by the 20% gap between norovirus episodes identified by real-time RT-PCR and those genotyped by the conventional PCR assay. The lower rate of amplification may have resulted from the longer product targeted by the C capsid PCR, primer-template mismatch, and/or low viral loads in some specimens [32, 33]. Our typing method did not include polymerase region type or evaluation of multiple genotypes [34], which may have resulted in underreporting of genotype diversity. Nevertheless, our data confirm the utility of the C capsid PCR to type the majority of noroviruses [19].
Our data demonstrated a high degree of norovirus genetic diversity. Repeat infections by the same genogroup were common, but 97% of repeat infections were by a different genotype or GII.4 variant. These data suggest that children may develop genotype-specific immunity with only a modest level of cross-protection even within the genogroup. This observation will need to be taken into account in development of vaccines, which may need to include multiple genotypes to achieve clinical protection. Furthermore, the observation of repeat infections by different GII.4 variants suggest that molecular monitoring may be necessary to ensure that the most relevant variants are included in vaccines over time.
In summary, this study demonstrates a high incidence of multiple norovirus infections with remarkable genetic diversity and prolonged excretion in infants living in a periurban community. Norovirus is a frequent cause of diarrhea severe enough for hospitalization [25], with an estimated 200 000 child deaths each year from the virus [5]. Clearly, development of an effective norovirus vaccine represents the next issue of importance to control diarrheal deaths and hospitalization. A vaccine based on the Norwalk virus-like particle demonstrated protection against homotypic experimental challenge; other approaches are under development. However, our study demonstrates that children can mount an effective immune response to norovirus but this response is restricted by viral genotype. Thus, our data indicate that vaccines may need to include multiple genotypes and GII.4 variants.
Supplementary Data
Supplementary materials are available at Clinical Infectious Diseases online (http://cid.oxfordjournals.org/). Supplementary materials consist of data provided by the author that are published to benefit the reader. The posted materials are not copyedited. The contents of all supplementary data are the sole responsibility of the authors. Questions or messages regarding errors should be addressed to the author.
Notes
Acknowledgments. We are grateful to the National Calicivirus laboratory for providing RNA transcripts. We thank the community of San Juan de Miraflores for their long-term collaboration. In addition, we acknowledge M. De la Cruz, O. Navarro, J. Castro, R. Cabrera, P. Maguiña, R. Jimenez, G. Vivanco, J.B. Phu, and D. Sara for their technical support.
Author contributions. M. S., R. H. G., V. C., M. K., J. C., W. C., and R. E. B. conceived and planned the study, S. A., S. E., and D. V. developed and processed the laboratory assays. M. S., R. H. G., L. C., and V. C. oversaw field activities. M. S., M. Z., S. L., and C. B. assisted in analysis of data. M. S., D. V., R. H. G., R. E. B., J. C., and C. B. contributed to writing the manuscript.
Disclaimer. The findings and conclusions in this report are those of the author(s) and do not necessarily represent the official position of the Centers for Disease Control and Prevention (CDC). Mention of company names or products does not constitute endorsement by the CDC.
Financial support. This work was supported by the Sixth Framework Programme of the European Union, Project CONTENT INCO-CT-2006-032136, Population Health Metrics Research Consortium Project, and the CDC.
Potential conflicts of interest. All authors: No reported conflicts.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.Kageyama T, Shinohara M, Uchida K, et al. Coexistence of multiple genotypes, including newly identified genotypes, in outbreaks of gastroenteritis due to norovirus in Japan. J Clin Microbiol. 2004;42:2988–95. doi: 10.1128/JCM.42.7.2988-2995.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hall AJ, Vinjé J, Lopman B, et al. Updated norovirus outbreak management and disease prevention guidelines. MMWR Morb Mortal Wkly Rep. 2011;60(RR03):1–15. [PubMed] [Google Scholar]
- 3.Glass RI, Parashar UD, Estes MK. Norovirus gastroenteritis. N Engl J Med. 2009;361:1776–85. doi: 10.1056/NEJMra0804575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lopman B, Gastanaduy P, Park GW, Hall AJ, Parashar UD, Vinje J. Environmental transmission of norovirus gastroenteritis. Curr Opin Virol. 2012;2:96–102. doi: 10.1016/j.coviro.2011.11.005. [DOI] [PubMed] [Google Scholar]
- 5.Patel MM, Widdowson MA, Glass RI, Akazawa K, Vinje J, Parashar UD. Systematic literature review of role of noroviruses in sporadic gastroenteritis. Emerg Infect Dis. 2008;14:1224–31. doi: 10.3201/eid1408.071114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hemming M, Rasanen S, Huhti L, Paloniemi M, Salminen M, Vesikari T. Major reduction of rotavirus, but not norovirus, gastroenteritis in children seen in hospital after the introduction of RotaTeq vaccine into the National Immunization Programme in Finland. Eur J Pediatr. 2013;172:739–46. doi: 10.1007/s00431-013-1945-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Payne DC, Vinje J, Szilagyi PG, et al. Norovirus and medically attended gastroenteritis in U.S. children. N Engl J Med. 2013;368:1121–30. doi: 10.1056/NEJMsa1206589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.O'Ryan ML, Lucero Y, Prado V, et al. Symptomatic and asymptomatic rotavirus and norovirus infections during infancy in a Chilean birth cohort. Pediatr Infect Dis J. 2009;28:879–84. doi: 10.1097/INF.0b013e3181a4bb60. [DOI] [PubMed] [Google Scholar]
- 9.Phillips G, Tam CC, Conti S, et al. Community incidence of norovirus-associated infectious intestinal disease in England: improved estimates using viral load for norovirus diagnosis. Am J Epidemiol. 2010;171:1014–22. doi: 10.1093/aje/kwq021. [DOI] [PubMed] [Google Scholar]
- 10.Karsten C, Baumgarte S, Friedrich AW, et al. Incidence and risk factors for community-acquired acute gastroenteritis in north-west Germany in 2004. Eur J Clin Microbiol Infect Dis. 2009;28:935–43. doi: 10.1007/s10096-009-0729-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hall AJ, Rosenthal M, Gregoricus N, et al. Incidence of acute gastroenteritis and role of norovirus, Georgia, USA, 2004–2005. Emerg Infect Dis. 2011;17:1381–8. doi: 10.3201/eid1708.101533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rockx B, De Wit M, Vennema H, et al. Natural history of human calicivirus infection: a prospective cohort study. Clin Infect Dis. 2002;35:246–53. doi: 10.1086/341408. [DOI] [PubMed] [Google Scholar]
- 13.de Onis M, Onyango A, Borghi E, Siyam A, Blossner M, Lutter C. Worldwide implementation of the WHO Child Growth Standards. Public Health Nutr. 2012;15:1603–10. doi: 10.1017/S136898001200105X. [DOI] [PubMed] [Google Scholar]
- 14.Freeman MM, Kerin T, Hull J, McCaustland K, Gentsch J. Enhancement of detection and quantification of rotavirus in stool using a modified real-time RT-PCR assay. J Med Virol. 2008;80:1489–96. doi: 10.1002/jmv.21228. [DOI] [PubMed] [Google Scholar]
- 15.Mazumder S, Taneja S, Bhandari N, et al. Effectiveness of zinc supplementation plus oral rehydration salts for diarrhoea in infants aged less than 6 months in Haryana state, India. Bull World Health Organ. 2010;88:754–60. doi: 10.2471/BLT.10.075986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Apaza S, Espetia S, Gilman RH, et al. Detection and genogrouping of noroviruses from children's stools by TaqMan One-Step RT-PCR. J Vis Exp. 2012;65:e3232. doi: 10.3791/3232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kageyama T, Kojima S, Shinohara M, et al. Broadly reactive and highly sensitive assay for Norwalk-like viruses based on real-time quantitative reverse transcription-PCR. J Clin Microbiol. 2003;41:1548–57. doi: 10.1128/JCM.41.4.1548-1557.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kojima S, Kageyama T, Fukushi S, et al. Genogroup-specific PCR primers for detection of Norwalk-like viruses. J Virol Methods. 2002;100:107–14. doi: 10.1016/s0166-0934(01)00404-9. [DOI] [PubMed] [Google Scholar]
- 19.Kroneman A, Vennema H, Deforche K, et al. An automated genotyping tool for enteroviruses and noroviruses. J Clin Virol. 2011;51:121–5. doi: 10.1016/j.jcv.2011.03.006. [DOI] [PubMed] [Google Scholar]
- 20.Gladstone BP, Ramani S, Mukhopadhya I, et al. Protective effect of natural rotavirus infection in an Indian birth cohort. N Engl J Med. 2011;365:337–46. doi: 10.1056/NEJMoa1006261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Velazquez FR, Matson DO, Calva JJ, et al. Rotavirus infections in infants as protection against subsequent infections. N Engl J Med. 1996;335:1022–8. doi: 10.1056/NEJM199610033351404. [DOI] [PubMed] [Google Scholar]
- 22.Blackwelder WC, Biswas K, Wu Y, et al. Statistical methods in the Global Enteric Multicenter Study (GEMS) Clin Infect Dis. 2012;55(suppl 4):S246–53. doi: 10.1093/cid/cis788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Phillips G, Lopman B, Tam CC, Iturriza-Gomara M, Brown D, Gray J. Diagnosing rotavirus A associated IID: using ELISA to identify a cut-off for real time RT-PCR. J Clin Virol. 2009;44:242–5. doi: 10.1016/j.jcv.2008.12.001. [DOI] [PubMed] [Google Scholar]
- 24.Monica B, Ramani S, Banerjee I, et al. Human caliciviruses in symptomatic and asymptomatic infections in children in Vellore, South India. J Med Virol. 2007;79:544–51. doi: 10.1002/jmv.20862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Parashar UD, Li JF, Cama R, et al. Human caliciviruses as a cause of severe gastroenteritis in Peruvian children. J Infect Dis. 2004;190:1088–92. doi: 10.1086/423324. [DOI] [PubMed] [Google Scholar]
- 26.Yori PP, Schwab K, Gilman RH, et al. Norovirus highly prevalent cause of endemic acute diarrhea in children in the peruvian Amazon. Pediatr Infect Dis J. 2009;28:844–7. doi: 10.1097/INF.0b013e3181a24730. [DOI] [PubMed] [Google Scholar]
- 27.Zhang S, Chen TH, Wang J, et al. Symptomatic and asymptomatic infections of rotavirus, norovirus, and adenovirus among hospitalized children in Xi'an, China. J Med Virol. 2011;83:1476–84. doi: 10.1002/jmv.22108. [DOI] [PubMed] [Google Scholar]
- 28.Phillips G, Lopman B, Tam CC, Iturriza-Gomara M, Brown D, Gray J. Diagnosing norovirus-associated infectious intestinal disease using viral load. BMC Infect Dis. 2009;9:63. doi: 10.1186/1471-2334-9-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kirkwood CD, Streitberg R. Calicivirus shedding in children after recovery from diarrhoeal disease. J Clin Virol. 2008;43:346–8. doi: 10.1016/j.jcv.2008.08.001. [DOI] [PubMed] [Google Scholar]
- 30.Siebenga JJ, Beersma MF, Vennema H, van Biezen P, Hartwig NJ, Koopmans M. High prevalence of prolonged norovirus shedding and illness among hospitalized patients: a model for in vivo molecular evolution. J Infect Dis. 2008;198:994–1001. doi: 10.1086/591627. [DOI] [PubMed] [Google Scholar]
- 31.Humphrey JH. Child undernutrition, tropical enteropathy, toilets and handwashing. Lancet. 2009;374:1032–35. doi: 10.1016/S0140-6736(09)60950-8. [DOI] [PubMed] [Google Scholar]
- 32.Pang X, Lee B, Chui L, Preiksaitis JK, Monroe SS. Evaluation and validation of real-time reverse transcription-PCR assay using the LightCycler system for detection and quantitation of norovirus. J Clin Microbiol. 2004;42:4679–85. doi: 10.1128/JCM.42.10.4679-4685.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.George S, Menon VK, Ramani S, Kang G. Comparison of primers for the detection of genogroup II noroviruses in India. Indian J Med Microbiol. 2012;30:24–9. doi: 10.4103/0255-0857.93016. [DOI] [PubMed] [Google Scholar]
- 34.Bull RA, Eden JS, Luciani F, McElroy K, Rawlinson WD, White PA. Contribution of intra- and interhost dynamics to norovirus evolution. J Virol. 2012;86:3219–29. doi: 10.1128/JVI.06712-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.