Abstract
Previously, we reported that in clam oocytes, cytoplasmic polyadenylation element-binding protein (CPEB) co-immunoprecipitates with p47, a member of the highly conserved RCK family of RNA helicases which includes Drosophila Me31B and Saccharomyces cerevisiae Dhh1. Xp54, the Xenopus homologue, with helicase activity, is a component of stored mRNP. In tethered function assays in Xenopus oocytes, we showed that MS2–Xp54 represses the translation of non-adenylated firefly luciferase mRNAs and that mutations in two core helicase motifs, DEAD and HRIGR, surprisingly, activated translation. Here we show that wild-type MS2–Xp54 tethered to the reporter mRNA 3′-untranslated region (UTR) represses translation in both oocytes and eggs in an RNA-dependent complex with endogenous Xp54. Injection of mutant helicases or adenylated reporter mRNA abrogates this association. Thus Xp54 oligomerization is a hallmark of translational repression. Xp54 complexes, which also contain CPEB and eIF4E in oocytes, change during meiotic maturation. In eggs, CPEB is degraded and, while eIF4E still interacts with Xp54, this interaction becomes RNA dependent. Supporting evidence for RNA-mediated oligomerization of endogenous Xp54, and RNA-independent association with CPEB and eIF4E in oocytes was obtained by gel filtration. Altogether, our data are consistent with a model in which the active form of the Xp54 RNA helicase is an oligomer in vivo which, when tethered, via either MS2 or CPEB to the 3′UTR, represses mRNA translation, possibly by sequestering eIF4E from the translational machinery.
INTRODUCTION
During meiotic maturation and early embryogenesis, a period of development that occurs in the absence of transcription, specific maternal mRNAs which are translationally dormant in the oocyte undergo a dramatic activation. Their products, including c-mos, cyclins, wee-1 and ribonucleotide reductase, allow the completion of meiosis and entry into mitotic cleavages. These mRNAs are distinguished by the presence of one or more cytoplasmic polyadenylation elements (CPE), typically U4–6A1–2U, near to the ubiquitous nuclear polyadenylation signal AAUAAA (1,2). These 3′-untranslated region (UTR) elements mediate repression in the oocyte, and polyadenylation and translational activation in the egg, in clams, frogs and mice (3–5). Increases in poly(A) length may stimulate translation by stabilizing the interactions between initiation factors (eIF4G–eIF4E) and poly(A)-binding protein (PABP), which circularize eukaryotic mRNAs by their association with the 5′ cap structure and the 3′ poly(A) tail (6,7). ‘Housekeeping’ mRNAs, lacking 3′UTR CPEs, are deadenylated during meiotic maturation, and released from polysomes (2); the lack of decapping activity (8) may explain why non-adenylated mRNAs are uniquely stable in early development, until the mid-blastula transition (9,10).
CPEs mediate their function by their interaction with CPEB (CPE-binding protein), first cloned and characterized in Xenopus (11). The CPEB family includes clam p82 (12), Drosophila Orb (13), Caenorhabditis elegans CPB-1–4 (14), Aplysia CPEB (15) as well as the more closely related zebrafish, mouse and human homologues (16–18). CPEB is a critical regulator of gene expression in early development. Several partners of CPEB have been proposed to mediate its role in translational repression, including maskin (19), Pumilio (20) and the DEAD-box helicase Xp54 (21). In stage VI oocytes, Xenopus CPEB represses translation of CPE-containing mRNAs by preventing eIF4E–eIF4G interactions through its bridging partner maskin, which contains a peptide sequence that resembles one conserved among eIF4E-binding proteins (19). Maskin levels are regulated during oogenesis, with significant amounts only detectable during late stage V/VI, suggesting that repression earlier in oogenesis relies on other mechanisms (22). During meiotic maturation, the inhibitory maskin–eIF4E link is severed (19,23). A conserved partner of CPEB is the RNA-binding protein Pumilio/FBF, a member of the PUF family (24). In C.elegans, FBF binds CPEB and NANOS-3 to control the sperm–oocyte switch (14,25), while in Xenopus oocytes, Pumilio in association with CPEB enhances translational repression of cyclin B1 mRNA (26). Early in meiosis, vertebrate CPEBs are phosphorylated by Eg2/aurora kinase (27) which recruits to CPE-containing mRNAs the cytoplasmic forms of cleavage and polyadenylation specificity factors and thus promotes polyadenylation (28,29). Subsequent phosphorylation by cdc2 mediates CPEB degradation (30–32).
We identified a CPEB co-immunoprecipitating protein as the DEAD-box RNA helicase, p47, in Spisula oocytes (21). Clam p47 is a member of a helicase SF2/DDX6 subfamily, highly conserved from trypanosomes to man (21), with homologues in Xenopus [Xp54 (33)], Drosophila [Me31B (34)], C.elegans [Cgh-1 (35)] and Saccharomyces cerevisisae [Dhh1 (36,37)]. Members of the DEAD-box superfamily are involved in a variety of cellular processes, including splicing, ribosome biogenesis, RNA transport, degradation and translation, although their precise contribution to most of these processes is not known. They are characterized by nine conserved domains, including the eponymous DEAD motif and the newly identifed Q motif, with varying roles in catalysis and substrate binding. Helicases use ATP hydrolysis to unwind short RNA duplexes and may also influence rearrangements of large RNA structures or protein–RNA interactions. Their ATPase activity is stimulated or dependent on RNA binding, but it is unclear to what extent helicases recognize specific RNA sequences [reviewed in Fuller-Pace (38), Lüking et al. (39), Tanner et al. (40) and Linder (41)].
Xenopus p54, an integral component of stored mRNP in oocytes, is present at constant levels throughout oogenesis, possesses bona fida ATP-dependent duplex unwinding activity and is a shuttling protein implicated in the nuclear assembly of stored mRNP particles (32,33,42). To test its function in translation, we tethered Xp54 via 3′UTR MS2-binding sites to reporter firefly luciferase mRNA, microinjected into oocytes (21). Xp54 repressed translation 3- to 5-fold and, strikingly, mutations in the DEAD (ATP hydrolysis) and HRIGR (ATP-binding) helicase motifs activated translation 3- to 4-fold, relative to MS2. We showed that these effects were at the level of translation, rather than RNA stability, and that they required MS2-binding sites (21). These data taken together imply that the helicase activity of Xp54 is required for repression (21).
Interestingly, the Drosophila homologue Me31B mediates the translational silencing of oskar and BicD mRNAs in oocytes (34), while yeast Dhh1 enhances decapping and interacts with both the decapping and deadenylase complexes (36,37) in discrete cytoplasmic processing bodies (43,44). Cgh-1 of C.elegans is localized to P granules and other mRNPs (35) while Me31B is concentrated in the sponge bodies in germline cells during oogenesis (34) and Xp54 is present in the Balbioni body in early Xenopus oocytes (42). Thus RCK helicases are present in distinct cytoplasmic RNP particles in many cell types. It is also interesting to note that the Xenopus and human proteins can complement dhh1Δ yeast cells (44,45). Collectively, these data highlight the conserved and critical role of this RNA helicase in downregulating mRNA expression, and hint at a possible link between decapping and masking, possibly at the level of eIF4E, the cap-binding protein and the 5′ cap structure.
Here, we report that Xp54 and Xenopus CPEB interact in vivo and that wild-type tethered MS2–Xp54 represses translation while mutant helicases (DQAD and HRIGQ forms) activate translation, in both oocytes and eggs. Wild-type, but not mutant MS2–Xp54, oligomerizes, in an RNA-dependent manner. In cells injected with adenylated reporter mRNA, constitutively activated, wild-type MS2–Xp54 does not associate with endogenous Xp54. Thus oligomerization of tethered Xp54 is a hallmark of translational repression. Supporting evidence for RNA-mediated oligomerization of native Xp54, and association with CPEB, was obtained by gel filtration. We also find that the interaction between Xp54 and eIF4E changes during meiotic maturation and discuss a model for the role of Xp54 in translational repression.
MATERIALS AND METHODS
Cloning of Xenopus CPEB
The CPEB expression plasmid was constructed by inserting the open reading frame (ORF) of CPEB, as an NdeI–XhoI fragment, into pET21b (Novagen). The cDNA was produced by PCR from a Xenopus cDNA library (46) using oligodeoxynucleotides 5′ CCA GGA GCC TGC CAT ATG GCC TTC CCA CTG 3′ and 5′ GTT GTT CCA ATG CTC GAG GCT GGA GTC ACG 3′; NdeI and XhoI sites are underlined. The amino acid sequence of the bacterially expressed protein differed from that of the endogenous protein by the addition of a C-terminal extension with the sequence LEHHHHHH. Haemagglutinin (HA)-tagged CPEB was produced by insertion of the CPEB ORF as a EcoRI–NotI fragment into a construct containing a 3× HA tag, β-globin 5′ and 3′UTRs and a 3′ A23C30 tail. The cDNA was produced by PCR from pET21b-CPEB using oligodeoxynucleotides 5′ GAG GAA TTC ATA TGG CCT TCC CAC TGA AAG 3′ and 5′ GTG GGC GGC CGC TTA GCT GGA GTC ACG ACT 3′ (sites underlined) and cloned into an EcoRI–NotI-digested vector derived from pS664TEN (a gift from S. Morley); the vector backbone was substituted as an HindIII–PstI fragment into pGEM2 (Promega) with a deleted EcoRI site, retaining the β-globin UTRs and the A23 tail, but allowing subsequent digestion with SmaI and transcription with T7 RNA polymerase. Fused in-frame at the N-terminus are three copies of an influenza HA epitope tag.
Xenopus oocyte preparation
Ovarian lobes were removed from Xenopus laevis females. Stage VI oocytes were isolated and stored at all times in modified Barth’s solution [8.8 mM NaCl, 1 mM KCl, 330 µM Ca(NO3)2, 410 µM CaCl2, 820 µM MgSO4, 2.4 mM NaHCO3, 10 mM HEPES-NaOH pH 7.4]. Oocytes were microinjected as required. Maturation was performed, if necessary, by treatment of oocytes with 10 µg/ml progesterone at 20°C for 16 h. Extracts were made either crude in minimal modified Barth’s solution or diluted in an appropriate buffer, and clarified twice by centrifugation at 9300 g for 10 min at 4°C. When 35S labelling was required, 5–10 oocytes were transferred, 3 h post-injection, into 50 µl of modified Barth’s solution containing [35S]promix (Amersham) at 1 µCi/µl and incubated for 16 h at 20°C. After removing excess label, oocytes were washed in modified Barth’s solution and processed as normal.
Tethering
The MSP vector and the Luc-MS2 reporter were supplied by N.K. Gray (47). cDNAs encoding MS2–Xp54 and its mutated forms have been described previously (21): plasmids encoding fusion proteins were linearized with HindIII and Luc-MS2 with BglII prior to transcription with T7 RNA polymerase. Luc-MS2 reporter constructs requiring a poly(A) tail were modified using Escherichia coli poly(A) polymerase from a poly(A) tailing kit (Ambion), following the manufacturer’s instructions to add a tail of about 150 A residues.
Oocyte micromanipulation was performed essentially as described above. A 50 nl aliquot (at ∼500 ng/µl) of a solution of mRNAs encoding fusion proteins was injected into stage VI oocytes. After 3 h, oocytes were treated with progesterone and/or [35S]promix, as required and, after a further 3 h, injected with 10 nl of a solution containing Luc-MS2 reporter mRNA (10 ng/µl) and Renilla luciferase control mRNA (0.35 ng/µl). Incubation was continued overnight before harvesting.
Three to five pools each containing 3–5 oocytes were assayed per experimental point. Oocytes/eggs were homogenized in lysis buffer (50 µl/oocyte; Promega) and 10 µl samples assayed for firefly and Renilla luciferases using a Dual-Luciferase assay system (Promega). Relative light determinations were measured in a TD20/20 Turner Designs luminometer.
Immunoprecipitation
Immunoprecipitation experiments were performed from oocytes injected with MS2 fusion proteins, using mouse monoclonal anti-MS2 antibodies (a generous gift of Mike Kiledjian; Nelson Biological Laboratories, New Jersey). Where oocytes were co-injected with HA-tagged CPEB, 50 nl of an RNA mixture containing 500 ng/µl MS2–Xp54 (or MS2) and 500 ng/µl HA-CPEB was injected and oocytes incubated for 16 h. Oocytes co-injected with reporter constructs were manipulated as described above. S10 lysates were prepared from 20 oocytes/eggs in 1 ml of NET buffer [50 mM Tris–HCl pH 7.5, 150 mM NaCl, 0.5% (v/v) NP-40, 1 mM EDTA pH 8.0, 0.25% (w/v) gelatin, 0.02% (w/v) NaN3] + 2% (w/v) bovine serum albumin (BSA). Lysates were incubated, with gentle mixing, with 0.1 µl of anti-MS2 antibody for 2 h at 4°C, prior to addition of 10 µl of protein G–Sepharose beads for 2 h at 4°C. Beads were washed in 3 × 1 ml of NET buffer and bound proteins eluted in 20 µl of protein sample buffer. A 10–20 µl aliquot of each sample was separated by SDS–PAGE and proteins detected by silver staining, autoradiography or western blotting. For immunoprecipitations involving RNase A-treated extracts, Xenopus extracts were supplemented with RNase A to 20 pg/µl extract and incubated at 20°C for 10 min, prior to clarification by centrifugation at 9300 g for 10 min at 4°C. This treatment was sufficient to degrade mRNA, as judged by in vitro translation of phenol-extracted RNA. Immunoprecipitation reactions for the isolation of proteins for peptide sequencing were performed essentially as described above by scaling up reaction volumes 25-fold; volumes of protein G–Sepharose, however, were scaled up 10-fold.
Mass spectrometry analysis
Proteins within the gel-excised bands were first reduced, carboxyamido methylated, and then digested to peptides using trypsin on a MassPrepStation (Micromass, Manchester, UK). The resulting peptides were subjected to LC–MS/MS. The liquid chromatographic separation was achieved with a PepMap C18, 180 µm i.d., 15 cm column (LC Packings, Amsterdam). The mass spectrometer was a QTof (Micromass). Fragmentation data were used to search the National Center for Biotechnology Information database using the MASCOT search engine (http://www.matrixscience.com). Probability-based MASCOT scores were used to evaluate identifications. Only matches with P < 0.05 for random occurrence were considered significant (further explanation of MASCOT scores can be found at http://www.matrixscience.com).
Preparation of anti-Xp54 antibody
Anti-Xp54 antibody was obtained from rabbits immunized with acrylamide gel fragments containing recombinant Xp54 (CovalAb). The Xp54 expression plasmid was constructed by inserting the complete ORF of Xp54 as an NheI–SalI fragment into NheI–XhoI-digested pGIT [the 5′UTR and 3′UTR-A33C18 of Xenopus globin were introduced into the XbaI and BlpI sites of pET21d (Novagen), in order to create a single vector suitable for protein expression in bacteria and in vitro synthesis of RNA for microinjections into Xenopus oocytes]. The cDNA was produced by PCR from Xp54 clone B2 (33), using oligodeoxynucleotides 5′ CGC GCT AGC ATG AGC ACC GCC AGA A 3′ and 5′ ACG GTC GAC TTA AGG TTT GTC TTC C 3′. A culture of transformed BL21 star cells was grown to an A600 of approximately 0.3 and induced for 4 h at 37°C by the addition of 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG). Cells were harvested by centrifugation at 5000 g for 10 min and resuspended in 0.01–0.02 volumes of NTA buffer [300 mM NaCl, 1% (v/v) Triton X-100, 50 mM sodium phosphate buffer, pH 7.8]. Cells were lysed using a French pressure cell, and cell debris, including the insoluble Xp54 helicase, pelleted at 8000 g for 20 min. Pellets were resuspended in NTA buffer and aliquots run on SDS–polyacrylamide preparative gels. Coomassie-stained Xp54-containing gel slices were excised and used for immunization.
Purification of anti-CPEB peptide antibody
Rabbit anti-peptide Xenopus CPEB antibody was described previously (32). His-tagged CPEB was expressed in BL21 star cells, as described above for Xp54, and 10 µg purified from the post-lysis insoluble fraction by SDS–PAGE and western blotting. The membrane was stained briefly with Ponceau S [0.2% (w/v) dye in 0.3% (w/v) trichloroacetic acid] prior to excision of the CPEB-containing band. Membrane fragments were washed in 3 × 500 µl of phosphate-buffered saline (PBS), prior to incubation in 5% (w/v) BSA in PBS at 4°C for 1 h, with agitation. A 200 µl aliquot of antiserum was incubated with the blocked membrane for 16 h at 4°C, depleted serum removed and membranes washed in 3 × 1 ml of PBS. Bound antibody was eluted in 100 µl of 0.1 M glycine, pH 2.5 for 10 min at 20°C, neutralized with 10 µl of 1 M Tris, pH 8.0, and diluted to 200 µl in PBS.
Immunoblotting
Protein samples were separated by SDS–PAGE on 15% (w/v) acrylamide gels and transferred onto an Immobilon P membrane (Millipore) using a semidry blotting apparatus. Rabbit anti-Xp54 antibody was used at 1:10 000, affinity-purified rabbit anti-CPEB antibody at 1:10 000 and eIF4E antibody (gift of Dr Simon Morley) at 1:30 000. Western blots were subsequently detected by enhanced chemiluminescence (ECL).
FPLC gel filtration
A 24 ml Superose 6 HR 10/30 column was washed with ddH2O at 0.2 ml/min for 70 min, followed by 0.5 ml/min for 60 min, and equilibrated in eluent buffer (34.2 mM Na2HPO4, 15.8 mM NaH2PO4, 150 mM NaCl). The void volume was determined by running 1 mg/ml dextran blue and the column calibrated with thyroglobulin (669 kDa), ferritin (440 kDa), catalase (232 kDa), aldolase (158 kDa), ovalbumin (43 kDa), chymotrypsinogen A (25 kDa) and RNase A (13 kDa) (all Amersham Biosciences) in eluent buffer at a flow rate of 0.5 ml/min. Elution volumes for each protein were used to produce a calibration curve from which further approximate molecular weights were determined. A 200 µl aliquot of stage V/VI S10 extract was fractionated through the column at a flow rate of 0.5 ml/min, and 0.3 ml fractions collected. Alternate fractions were separated by SDS–PAGE and visualized by western blotting. Where required, lysate was treated with RNase A, 167 µg/ml for 10 min at 20°C prior to loading on the column.
RESULTS
Xenopus CPEB and Xp54 interact in vivo
We previously showed that clam p47 helicase and clam p82/CPEB co-precipitate in oocyte lysates, using both anti-CPEB and anti-helicase antibodies (21). Here we examined whether the Xenopus forms of the RNA helicase and CPEB interact in oocytes. As Xp54 and XCPEB proteins have very similar sizes on SDS–PAGE, and these are close to the size of the heavy chain of IgG, and both available antibodies were raised in rabbits, we could not examine such an interaction with the endogenous proteins. Thus, oocytes were co-injected with HA-CPEB and MS2–Xp54 mRNA or control MS2 mRNA and labelled with [35S]promix. Oocytes were harvested after 22 h and lysates used in immunoprecipitation experiments with MS2 antibodies and protein G–Sepharose. In vitro translated proteins, labelled in the rabbit reticulocyte lysate cell-free system, were analysed alongside the precipitated proteins to show protein size (Fig. 1). The results indicate that HA-CPEB co-precipitates with MS2–Xp54, but not with MS2 control protein. Furthermore, experiments reported below show that MS2–Xp54 associates with endogenous CPEB, and that the native proteins co-purify in gel filtration (see Figs 5 and 6). We conclude that as in Spisula, Xenopus CPEB and Xp54 interact in vivo.
Figure 1.
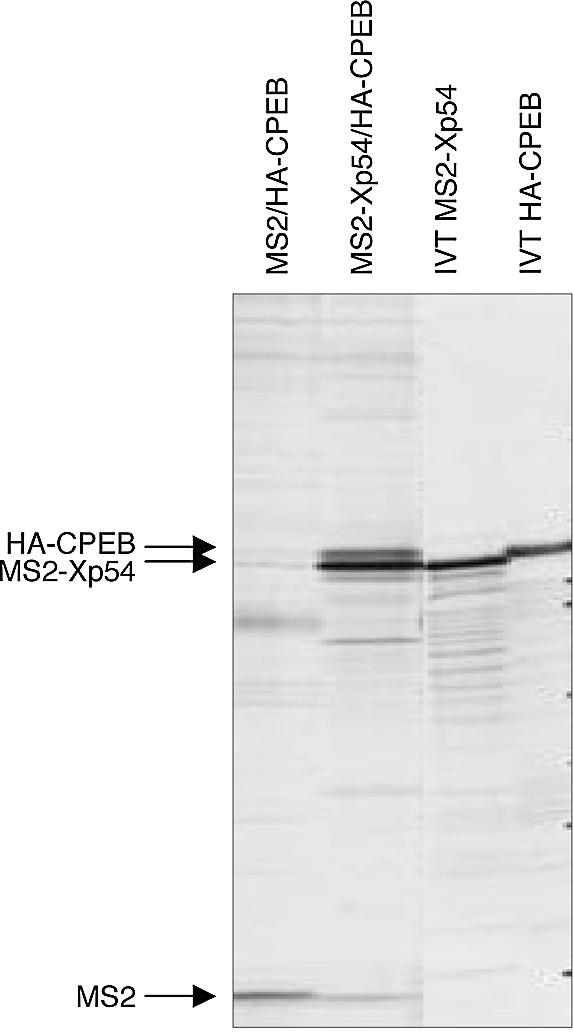
Xenopus CPEB and Xp54 interact in vivo. Oocytes were co-injected with HA-CPEB and MS2–Xp54 or MS2 control mRNAs as indicated and labelled with [35S]promix. Oocytes were harvested after 22 h and lysates used in immunoprecipitation experiments with anti-MS2 antibodies and protein G–Sepharose. In vitro translated (IVT) MS2–Xp54 and HA-CPEB are shown for size comparison.
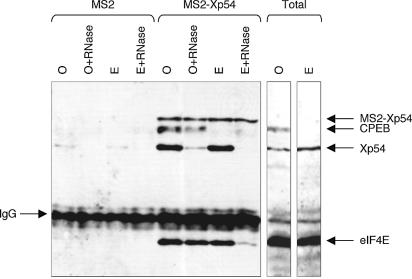
Figure 5.
Changes in interactions between MS2–Xp54 and endogenous Xp54 (p50), CPEB and eIF4E during meiotic maturation. Xenopus stage VI oocytes were injected with mRNA encoding MS2 or MS2–Xp54 proteins, and some oocytes were matured with progesterone. After a further 18 h, groups of 10 cells were used to make S10 lysates. Lysates, untreated or treated with RNase, were used in immunoprecipitation experiments with anti-MS2 antibody and subjected to SDS–PAGE and western blot analysis with Xp54, CPEB and eIF4E antibodies. The IgG band is indicated. Also shown are lanes of total oocyte and egg proteins (one cell equivalent). A representative experiment is shown.
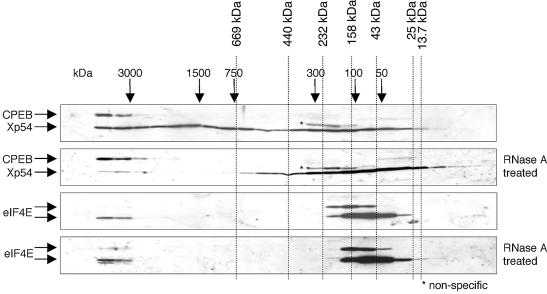
Figure 6.
Size and protein ligands of Xp54 complexes assessed by gel filtration of Xenopus oocyte lysate. A 24 ml Superose 6 HR 10/30 column was equilibrated in eluent buffer (34.2 mM Na2HPO4, 15.8 mM NaH2PO4, 150 mM NaCl) prior to calibration with thyroglobulin (669 kDa), ferritin (440 kDa), catalase (232 kDa), aldolase (158 kDa), ovalbumin (43 kDa), chymotrypsinogen A (25 kDa) and RNase A (13 kDa). Actual elution volumes for each protein are marked by dotted lines and used to produce a calibration curve from which further approximate molecular weights were determined (marked by arrows). A 200 µl aliquot of stage V/VI S10 extract was fractionated through the column at a flow rate of 0.5 ml/min and 0.3 ml fractions collected. Alternate fractions were separated by SDS–PAGE and visualized by western blotting using antibodies as indicated. [Two proteins, of 22 and 27 kDa, react with the eIF4E antibody in Xenopus oocytes; both of these appear to be bona fide cap-binding proteins as they are selectively retained on a cap–Sepharose column (not shown).] A non-specific protein is shown by an asterisk. Where indicated, lysate was treated with RNase A, 167 µg/ml, 10 min, 20°C prior to loading on the column. This treatment did not significantly affect the fractionation behaviour of the Coomassie blue-stainable proteins (not shown).
Tethered Xp54 represses translation in oocytes and in eggs
To examine the role of Xp54 in translation, we used the tethered function assay (48). As Xp54 associates with CPEB, a specific RNA-binding protein that binds 3′UTR CPEs, Xp54 was fused with MS2 coat protein and tethered to the 3′UTR of a reporter RNA via MS2 coat protein-binding sites. First, mRNA encoding MS2–Xp54 was microinjected into oocytes, some of which were matured with progesterone (see Materials and Methods). After several hours of incubation to allow the synthesis of fusion protein, firefly luciferase mRNA containing MS2-binding sites in its 3′UTR was injected, along with an internal control Renilla luciferase mRNA lacking any regulatory elements in its 3′UTR (Fig. 2). Variants of Xp54, containing single amino acid changes (DEAD→DQAD) and (HRIGR→HRIGQ) in the conserved helicase motifs II and VI, which reduce or abolish ATPase hydrolysis and ATP-binding, respectively [reviewed in Linder (41)], were also examined (Fig. 2). We monitored the effects of MS2–Xp54, relative to MS2 alone, on the ratio of firefly to Renilla luciferase activities. As the levels of MS2, and all Xp54 fusion proteins were approximately equal (see Figs 3B, 4 and 5), and firefly reporter RNA levels were not affected by ectopic Xp54 expression [not shown, see Minshall et al. (21)], the effects were exerted at the level of translation. We estimate that ∼1–5 ng of Xp54 is expressed ectopically in each oocyte in our conditions, in contrast to the 50–100 ng levels of the endogenous helicase (not shown); precluding any possible effects of wild-type or dominant-negative forms of the helicase on total protein synthesis.
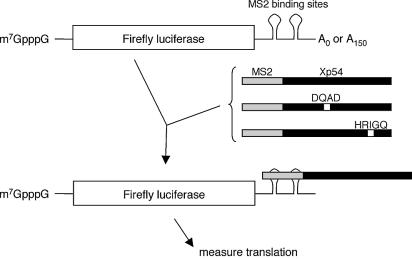
Figure 2.
Tethering strategy. The tethered function approach for assaying translational effects of Xp54 on firefly luciferase mRNA. Xenopus oocytes are first injected with mRNAs encoding MS2 fusion proteins as indicated. Following 6 h incubation to allow expression, firefly luciferase mRNA with 3′UTR MS2-binding sites, in a non-adenylated or adenylated form, is co-injected with a control Renilla firefly luciferase mRNA that lacks 3′UTR regulatory sequences. Activities of both luciferases are then assayed after a further 18 h incubation.
Figure 3.
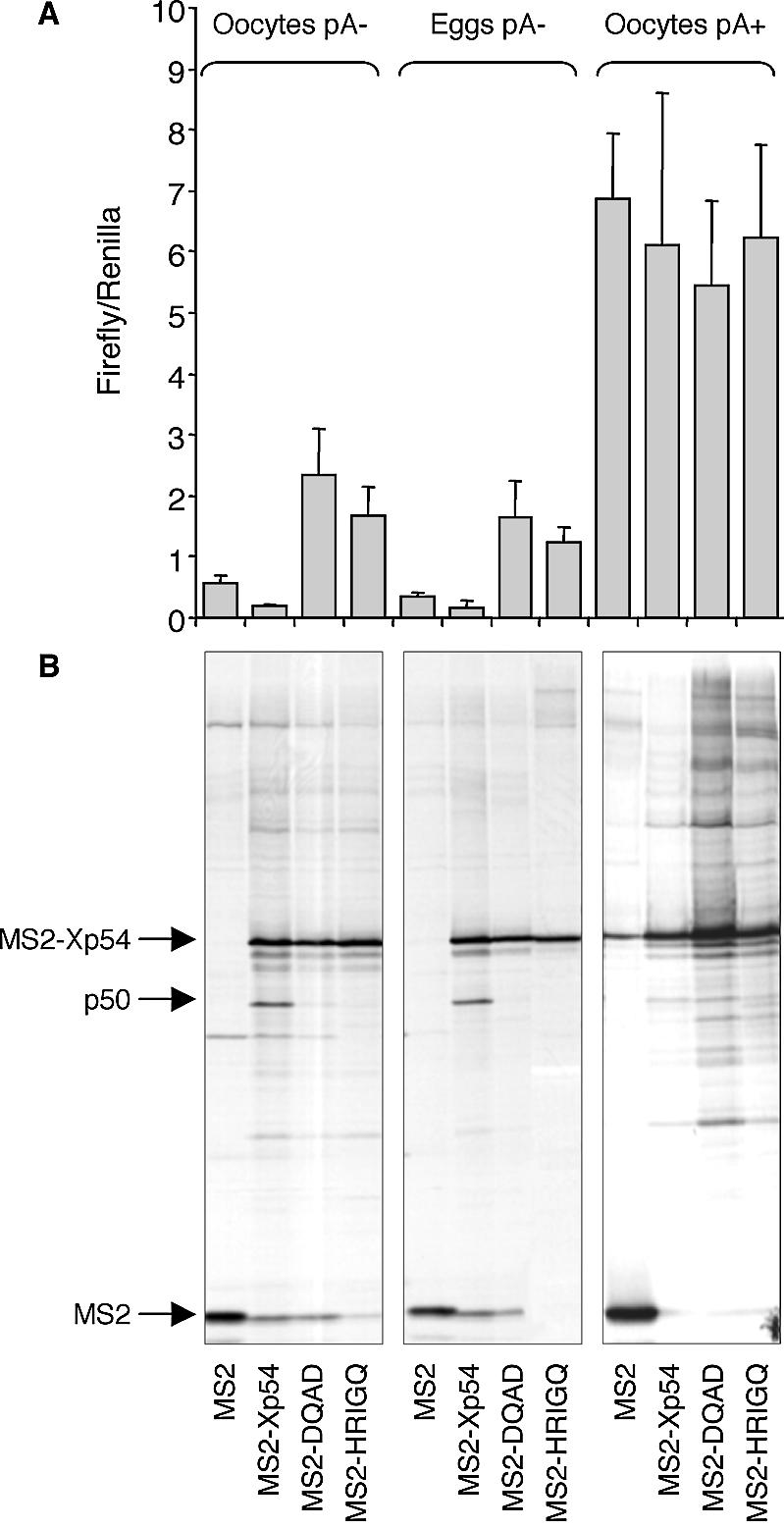
(A) Tethered wild-type Xp54 represses non-adenylated firefly reporter mRNA in both oocytes and eggs, while mutant helicase forms activate translation, but poly(A)+ reporter mRNA, containing a tail of approximately 150 A residues, is activated to similar levels regardless of the tethered protein. Xenopus stage VI oocytes were first injected with mRNAs encoding MS2–protein fusions as indicated. Following a 3 h incubation, experiments requiring maturation were treated with progesterone and after a further 3 h all oocytes were co-injected with two luciferase mRNAs, encoding control Renilla luciferase that lacks 3′UTR regulatory sequences, and firefly luciferase mRNA containing MS2-binding sites. After a further 18 h, five pools each containing five oocytes were assayed per experimental point from which mean values and standard deviations were determined. (B) A 50 kDa protein co-immunoprecipitates with MS2–Xp54 in oocytes and eggs, in a tethered assay, but not with its mutated forms DQAD (abrogates ATPase activity) and HRIGQ (abrogates ATP binding), when the reporter RNA is non-adenylated, nor with MS2–Xp54 in oocytes when the reporter mRNA is polyadenylated. Oocytes labelled with [35S]promix were used in immunoprecipitation experiments with anti-MS2 antibodies. Immunoprecipitated proteins were subjected to gel electrophoresis and autoradiography. A representative experiment is shown.
Figure 4.
The interaction between MS2–Xp54 and p50 is RNA dependent. A 50 kDa protein co-immunoprecipitates with MS2–Xp54 in an RNA-dependent manner. Xenopus stage VI oocytes were injected with mRNAs encoding MS2–protein fusions as indicated. Oocytes were labelled with [35S]promix, and lysates were subjected to treatment with RNase A as required (20 ng/ml lysate for 10 min at 20°C) and used in immunoprecipitation experiments with anti-MS2 antibodies. Immunoprecipitated proteins were subjected to gel electrophoresis and autoradiography.
First, we observed that Xp54 tethered to the 3′UTR represses translation of firefly luciferase reporter RNA in both oocytes and eggs to very similar extents (approximately 3–4 times) relative to MS2 (Fig. 3A). In contrast, mutant helicases activated translation, in both cell types, by ∼3- to 5-fold, relative to MS2 (Fig. 3A). Thus the effects of ectopic Xp54, all variants, on reporter mRNA were the same irrespective of cell type, implying that the helicase is not functionally modified during maturation. During meiotic maturation, some maternal mRNAs are rapidly mobilized into polysomes by a process involving cytoplasmic polyadenylation (see Introduction). In this experiment, the firefly reporter RNA was not adenylated prior to injection and, due to lack of CPE and hexanucleotide elements in its 3′UTR, was not adenylated during maturation. When firefly reporter mRNA was adenylated prior to microinjection (by an estimated 150 nt, see Materials and Methods), its translation was stimulated ∼10-fold in oocytes (Fig. 3A), as expected from previous studies showing the stimulatory effect of a poly(A) tail. This stimulation of translation was not affected by co-expression of either wild-type or mutant helicases [Fig. 3A and Minshall et al. (21)]. Thus Xp54, while able to repress translation in eggs when the tethering mRNA is non-adenylated, does not repress the translation of polyadenylated RNA.
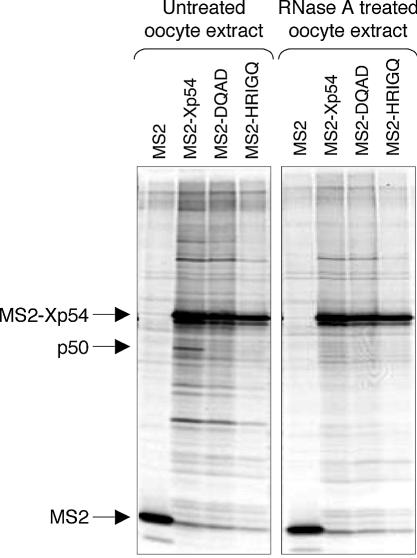
Xp54 represses translation in association with p50
Next, we examined the possible co-purification of endogenous proteins with wild-type and mutant (DQAD/HRIGQ) Xp54 proteins fused with MS2. Oocytes or eggs, co-injected with firefly luciferase reporter mRNA and MS2–Xp54 mRNAs or MS2 control mRNA, were incubated in Barth’s solution containing [35S]promix, to allow sensitive detection of protein ligands. Cells were harvested after 22 h and lysates used in immunoprecipitation experiments with anti-MS2 antibodies and protein G–Sepharose. Several proteins co-precipitated with Xp54 proteins specifically, as they were absent from the MS2 control samples (first lane in each panel). Strikingly, one protein, of ∼50 kDa, consistently co-precipitated with wild-type MS2–Xp54, but not with either of the two mutant helicases (Fig. 3B). Moreover, MS2–Xp54 does not co-purify with endogenous p50 in cells injected with the polyadenylated reporter RNA (Fig. 3B). Altogether, the data shown in Figure 3 indicate that recruitment of newly synthesized p50 by MS2–Xp54 correlates with the ability of the tethered helicase to repress translation. In parallel experiments in which the lysates were treated with RNase A prior to immunoprecipitation, we noted that the association of p50 with MS2–Xp54 was RNA dependent (Fig. 4).
Identification of p50 as Xp54
Large-scale immunoprecipitations were performed using lysates from unlabelled oocytes microinjected with MS2–Xp54 mRNA. The protein G–Sepharose MS2 antibody-bound proteins were stained with Coomassie colloidal blue, and the band migrating at ∼50 kDa was submitted for mass spectrometer peptide sequencing following trypsin digestion (see Materials and Methods). The highest probability-based Mowse score, 236, in a MASCOT search of NCBInr (MATRIX Science; http://www.matrixscience.com), was shown by vertebrate p54 helicase proteins with eight matching peptides (GNEFEDYCLK, SGAYLIPLLER, VMATTGGTNLR, LDDTVHVVIATPGR, GVTQYYAYV TER, VFHDFR, NLVCTDLFTR and LAETYLHR).
To confirm that the 50 kDa polypeptide interacting with the wild-type helicase was Xp54, control MS2 and MS2–Xp54 mRNAs were injected into oocytes, some of which were matured into eggs, and the MS2 immunoprecipitation reactions were analysed by western blot with Xp54 antibody. As shown in Figure 5, this antibody recognizes the MS2–Xp54 helicase and a protein of ∼50 kDa which co-precipitates with MS2–Xp54, in an RNA-dependent manner, in both oocytes and eggs. (A very low level of non-specific binding of Xp54 to MS2, also RNase-sensitive, was also noted.) On the basis of both the protein sequence and the western data, we conclude that Xp54 interacts with itself, as a dimer or higher oligomer, via RNA. From the higher ECL signal ratio of endogenous Xp54 to MS2–Xp54 (Fig. 5), it seems likely that at least a portion of the immunoprecipitated Xp54 is in an oligomeric form. Since using antibodies in immunoprecipitation and westerns assays may not yield quantitative data, we have not sought to determine the precise oligomerization number.
Changes in Xp54–CPEB–eIF4E interactions during meiotic maturation
To confirm and extend the nature of the interactions seen previously between Xp54 and CPEB-tagged proteins in oocytes (Fig. 1), the MS2 western blot was overprobed with CPEB antibody (Fig. 5). Endogenous CPEB interacts specifically with MS2–Xp54 in oocytes (Fig. 5), but, in contrast to the MS2–Xp54:Xp54 interaction, which is sensitive to RNase, the co-precipitation of CPEB with MS2–Xp54 does not require RNA (Fig. 5; see Discussion). In eggs, due to CPEB degradation during meiotic maturation (30–32), the total levels of CPEB are very much reduced and CPEB is not detectable in MS2–Xp54 immunoprecipitates (Fig. 5).
To examine possible interactions between Xp54 and components of the translational machinery, the MS2 western blot was also overprobed with eIF4E antibody (Fig. 5). Interestingly, the cap-binding initiation factor interacts with MS2–Xp54 in an RNA-independent manner in oocytes. More unexpectedly, the interaction between Xp54 and eIF4E changes from being independent of RNA in oocytes to being mediated by RNA in eggs (Fig. 5). Altogether, the MS2–Xp54 pull-down experiment confirms the identification of p50 as Xp54, as well as the interaction between Xp54 and CPEB. Moreover, evidence was obtained of dynamic changes in Xp54–CPEB–eIF4E interactions during meiotic maturation.
Xp54 is present in large and intermediate sized complexes
To extend our tethering and pull-down experiments, we sought an independent way to assess the protein ligands of endogenous Xp54. Lysates, prepared from stage V/VI oocytes, either in an untreated form or after treatment with RNase A, were analysed by gel filtration on a Superose 6 HR 10/30 column by FPLC (Fig. 6). Previous workers had shown that in stage V and VI oocytes, Xp54 is entirely cytoplasmic (42). Column fractions were analysed by western blotting using Xp54, CPEB and eIF4E antibodies. In untreated lysates, Xp54 fractionated with a broad profile throughout the column, in fractions ranging from ∼50 kDa to >2 MDa, representing monomer/dimer and larger complexes. Most of the large and intermediate sized complexes are sensitive to RNase, since in the nuclease-treated lysate the majority of Xp54 is found in fractions that correspond approximately to a monomer species. A minor fraction of Xp54, in the large complexes, is resistant to RNase (Fig. 6). In contrast, CPEB is found only in large, RNase-resistant complexes, in association with an Xp54 monomer. These large complexes also contain a fraction of eIF4E. These results support those obtained in tethering and immunoprecipitation studies. First, it appears that both native and tagged Xp54 is found in RNA-dependent dimer (and higher oligomer) forms, and secondly, both associate, directly or indirectly, with CPEB in oocytes in very large complexes which contain additional protein partners, including eIF4E.
DISCUSSION
Oligomerization of Xp54 and other RNA/DNA helicases
Using the tethered function approach and gel filtration analysis, we have shown that the DEAD-box helicase Xp54 oligomerizes in an RNA-dependent manner, and that this self-association correlates with the ability of the helicase to repress translation of reporter RNA (Figs 3, 4 and 6). Self-association is not observed when the tethered Xp54 is inactivated by mutations in the DEAD and HRIGR motifs, nor when the reporter RNA is pre-adenylated and not subject to Xp54 control (Fig. 3). Moreover, our gel filtration data support the existence of a substantial fraction of Xp54 helicase in RNase-sensitive oligomeric forms. [Interestingly, similar biochemical fractionation studies also indicate that Dhh1 is present in very large complexes in yeast cells (44,49)]. Thus, our data imply that Xp54 helicase is active as an oligomer during the translational repression in vivo. One possible explanation of these results is that the inactivating mutations abrogate RNA binding, resulting in loss of oligomerization. The DEAD-box (motif II), the Walker B motif of ATP-binding proteins, contains the highly conserved glutamic residue required for ATP hydrolysis. DQAD or DAAD mutants of eIF4A, Dbp5 and Ded1 bind ATP and RNA with wild-type (or near wild-type) affinities, but lack ATPase activity (50–52), while the principal role of motif VI (HRIGR) is now understood to be to bind ATP [reviewed in Linder (41)]. Based on these examples, it appears unlikely that both the Xp54 mutations resulted in the loss of RNA binding; however, until we can produce significant amounts of soluble proteins, and identify the RNA target(s), we cannot be entirely certain of this conclusion.
Xp54 joins a growing list of helicases that function as dimer/oligomers. A well-characterized example is the hepatitis C NS3 protein, the non-structural viral protein with protease, NTPase and RNA/DNA helicase activities. The crystal structure report of Cho et al. (53) suggests that NS3 helicase, a member of the DEXH family of helicases, forms a dimer, with a cleft between the components of the dimer through which a single-stranded nucleotide can pass as the helicase unwinds the RNA. Dimerization or oligomerization of NS3 helicase requires the presence of single-stranded DNA and is necessary for optimal helicase activity (54,55). A dimer of E.coli UvrD, a DNA helicase, is also the active form of the helicase in vitro. Two UvrD monomers must bind the DNA substrate, and both must be able to hydrolyse ATP, in order to form a functional unwinding complex (56). The highly related DEAD box RNA helicases p68 and p72, involved in both transcription and mRNA processing, exist as homodimers and, more abundantly, as heterodimers, as well as monomers, in cells (57). Mutation of DEAD to NEAD, rendering them ATPase and helicase inactive, weakens their interactions and prevents association with fibrillarin in the yeast two-hybrid system. Dimerization is mediated by a large part of the conserved core of the helicases, but in this case does not require RNA (57). While exact details vary, in the case of those helicases that are known to dimerize, directly or indirectly via nucleic acid substrate, self-association is required for helicase function.
CPEB–Xp54 association
We also showed that Xp54 interacts with CPEB, in both immunoprecipitation and gel filtration analyses, thus extending our previous report that Spisula p82/CPEB binds the p47 homologue in oocytes (21). This interaction may also be conserved in Drosophila. Thus Me31B forms a complex with Exuperantia (Exu), a protein involved in RNA localization, and Ypsilon Schachtel (Yps), the Y-box FRGY2 homologue, in an RNase-sensitive manner (34). Exu binds directly to Yps in oocytes (58). Furthermore, yps interacts genetically with orb, and, Orb is physically associated with both the Exu and Yps proteins, in an RNase-sensitive manner (59). While these findings, taken altogether, are suggestive of a conserved CPEB/RCK association, it remains to be established whether Orb and Me31B themselves interact, and whether such an interaction is mediated by RNA. Previously we reported that in clam oocytes, p82/CPEB bound p47 helicase via RNA (21). This conclusion was based on the observation that RNase treatment of lysates prior to immunoprecipitation with p82 antibodies prevented co-immunoprecipitation of p47. We also showed that RNase treatment of lysates prior to immunoprecipitation with p47 antibodies only partially reduced the levels of co-immunoprecipitating p82. Then, we ascribed these differences to the varying abilities of the antibodies to bind RNP complexes and protect their contents from nuclease (21). However, the data shown here, using essentially the same protocols but in Xenopus oocytes, clearly show that CPEB and Xp54 associate in the absence of RNA (Fig. 5). We believe the two sets of data can be reconciled in light of the observation that Xp54 oligomerizes in an RNA-dependent manner (Figs 4 and 5). Thus, most of the Xp54 polypeptides are lost upon nuclease treatment from CPEB precipitates, and the one attached monomer may fall below the level of detection. In the reverse reaction, all helicase polypetides are recovered, with the associated CPEB. Nevertheless, it seems likely that CPEB and Xp54 do not interact directly, but do so via another protein(s), as we have been unable to detect binding beween the two proteins synthesized in the reticulocyte lysate (data not shown). We are currently determining the domains in each protein that are required for their association in oocytes.
Role of Xp54 in translational repression
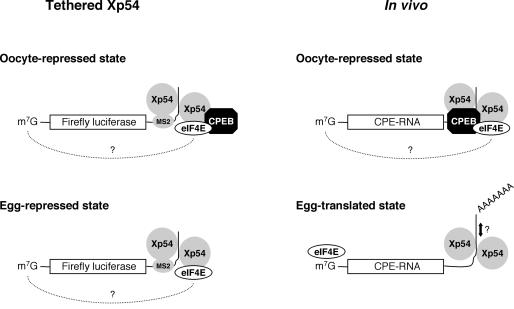
On the basis of our data, we propose the following model for Xp54 function in repression (Fig. 7). When tethered to the 3′UTR of non-adenylated mRNA, Xp54 oligomerizes and represses translation. Tethering to this location may be artificially assisted as with the MS2 system, or provided by CPEB which specifically binds the U-rich CPEs in translationally regulated RNAs. Since tethered, oligomeric Xp54 is able to inhibit translation in both oocytes (MS2 and CPEB as tethers) and eggs (MS2 tether), i.e. in the presence or absence of CPEB, we conclude that an Xp54–CPEB interaction is not required for repression. Tethered Xp54 which represses translation in oocytes interacts with eIF4E, possibly via CPEB and maskin (19), and may sequester it from the translational machinery. In progesterone-matured eggs, in the absence of CPEB, Xp54 remains capable of RNA-dependent self-association; however, it is presumably no longer anchored to the CPE motifs owing to the lack of CPEB. This release may render it capable of reorganizing protein complexes and allowing polyadenylation in vivo. When reporter RNA is injected in a polyadenylated form, the 3′ extension appears to prevent recruitment of additional Xp54 molecules, implying its function in the change between repressed and activated states. The interaction with eIF4E also changes dramatically, from being RNA independent in oocytes to requiring RNA in eggs, and may reflect a functional relocation from 3′UTR-based protein ligands to the 5′ cap structure. Interestingly, a dynamic eIF4E–cap interaction may also underlie the decapping enhancer function of Dhh1 in yeast (36).
Figure 7.
Model of Xp54 function in translation. Oligomerization of Xp54 and tethering to RNA is required for repression. Tethering is achieved via an MS2 fusion on Luc-MS2 RNA in both oocytes and eggs. CPEB is not necessary for repression in this tethering system. For endogenous messages, Xp54 dimerizes and is anchored to the RNA by CPEB in oocytes. In eggs, CPEB is degraded, thus releasing Xp54 from its CPE anchor. Xp54 can then act to remodel mRNP complexes, releasing eIF4E to bind the 5′ cap and allowing polyadenylation leading to translational activation. The dotted line linking the 3′ and 5′ ends of mRNA indicates a possible circular form of repressed mRNA, in which Xp54 sequesters eIF4E from proper interactions with eIF4G and eIF4A.
We also showed that mutant tethered Xp54 helicases that fail to oligomerize do not repress translation but in fact strongly activate translation [see also Minshall et al. (21)]. We are currently investigating the basis of this stimulation; it is possible that a further unidentified component of the mRNP complex is responsible for this dramatic activation. We speculate that the oligomeric form of wild-type Xp54 prevents this protein from binding the complex or from carrying out its activation role.
Acknowledgments
ACKNOWLEDGEMENTS
We gratefully acknowledge the Cambridge Centre for Proteomics, in particular its director Dr Kathryn Lilley, for help with mass spectrometry and peptide sequencing. We also thank the following for gifts of clones and antibodies: Dr John Sommerville for the B2 Xp54 clone, Dr Mike Kiledjian for MS2 antibody, and Dr Simon Morley for eIF4E antibody. This work was funded by the Wellcome Trust.
REFERENCES
- 1.Richter J.D. (2000) In Sonenberg,N., Hershey,J. and Mathews,M. (eds), Translational Control of Gene Expression. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 785–806. [Google Scholar]
- 2.Wickens M., Goodwin,E., Kimble,J., Strickland,S. and Hentze,M. (2000) In Sonenberg,N., Hershey,J. and Mathews,M. (eds), Translational Control of Gene Expression. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 295–370. [Google Scholar]
- 3.Minshall N., Walker,J., Dale,M. and Standart,N. (1999) Dual roles of p82, the clam CPEB homolog, in cytoplasmic polyadenylation and translational masking. RNA, 5, 27–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.deMoor C. and Richter,J.D. (1999) Cytoplasmic polyadenylation elements mediate masking and unmasking of cyclin B1 mRNA. EMBO J., 18, 2294–2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tay J., Hodgman,R. and Richter,J. (2000) The control of cyclin B1 mRNA translation during mouse oocyte maturation. Dev. Biol., 221, 1–9. [DOI] [PubMed] [Google Scholar]
- 6.Sachs A. (2000) In Sonenberg,N., Hershey,J. and Mathews,M. (eds), Translational Control of Gene Expression. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 7.Wakiyama M., Imataka,H. and Sonenberg,N. (2000) Interaction of eIF4G with poly(A)-binding protein stimulates translation and is critical for Xenopus oocyte maturation. Curr. Biol., 10, 1147–1150. [DOI] [PubMed] [Google Scholar]
- 8.Zhang S., Williams,C.J., Wormington,M., Stevens,A. and Peltz,S.W. (1999) Monitoring mRNA decapping activity. Methods, 17, 46–51. [DOI] [PubMed] [Google Scholar]
- 9.Audic Y.F.O. and Osborne,H.B. (1997) Postfertilisation deadenylation of mRNAs in Xenopus laevis embryos is sufficient to cause their degradation at the blastula stage. Mol. Cell. Biol., 17, 209–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Voeltz G.K. and Steitz,J.A. (1998) AUUUA sequences direct mRNA deadenylation uncoupled from decay during Xenopus early development. Mol. Cell. Biol., 18, 7537–7545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hake L.E., Mendez,R. and Richter,J.D. (1998) Specificity of RNA binding by CPEB: requirement for RNA recognition motifs and a novel zinc finger. Mol. Cell. Biol., 18, 685–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Walker J., Minshall,C., Hake,L., Richter,J. and Standart,N. (1999) The clam 3′UTR masking element-binding protein p82 is a member of the CPEB family. RNA, 5, 14–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lantz V., Ambrosio,L. and Schedl,P. (1992) The Drosophila orb gene is predicted to encode sex-specific germline RNA-binding proteins and has localised transcripts in ovaries and early emryos. Development, 115, 75–88. [DOI] [PubMed] [Google Scholar]
- 14.Luitjens C., Gallegos,M., Kraemer,B., Kimble,J. and Wickens,M. (2000) CPEB proteins control two key steps in spermatogenesis in C.elegans. Genes Dev., 14, 2596–2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu J. and Schwartz,J.H. (2002) The cytoplasmic polyadenylation element binding protein and polyadenylation of messenger RNA in Aplysia neurons. Brain Res., 959, 68–76. [DOI] [PubMed] [Google Scholar]
- 16.Gebauer F. and Richter,J. (1996) Mouse cytoplasmic polyadenylylation element binding protein: an evolutionarily conserved protein that interacts with the cytoplasmic polyadenylylation elements of c-mos mRNA. Proc. Natl Acad. Sci. USA, 93, 14602–14607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bally-Cuif L., Schatz,W.J. and Ho,R.K. (1998) Characterisation of the zebrafish Orb/CPEB-related RNA-binding protein and localization of maternal components in the zebrafish oocyte. Mech. Dev., 77, 31–47. [DOI] [PubMed] [Google Scholar]
- 18.Welk J.F., Charlesworth,A., Smith,G.D. and MacNicol,A.M. (2001) Identification and characterisation of the gene encoding human cytoplasmic polyadenylation element binding protein. Gene, 263, 113–120. [DOI] [PubMed] [Google Scholar]
- 19.Stebbins-Boaz B., Cao,Q., de Moor,C.H., Mendez,R. and Richter,J.D. (1999) Maskin is a CPEB-associated factor that transiently interacts with eIF-4E. Mol. Cell, 4, 1017–1027. [DOI] [PubMed] [Google Scholar]
- 20.Nakahata S., Katsu,Y., Mita,K., Inoue,K., Nagahama,Y. and Yamashita,M. (2001) Biochemical identification of Xenopus Pumilio as a sequence-specific cyclin B1 mRNA-binding protein that physically interacts with a nanos homolog (Xcat-2) and a cytoplasmic polyadenylation element-binding protein (CPEB). J. Biol. Chem., 276, 20945–20953. [DOI] [PubMed] [Google Scholar]
- 21.Minshall N., Thom,G. and Standart,N. (2001) A conserved role of a DEAD box helicase in mRNA masking. RNA, 7, 1728–1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Groisman I., Huang,Y.-S., Mendez,R., Cao,Q., Therkauf,W. and Richter,J. (2000) CPEB, maskin and cyclin B1 mRNA at the mitotic apparatus: implications for local translational control of cell division. Cell, 103, 435–447. [DOI] [PubMed] [Google Scholar]
- 23.Cao Q. and Richter,J.D. (2002) Dissolution of the maskin–eIF4E complex by cytoplasmic polyadenylation and poly(A)-binding protein controls cyclin B1 mRNA translation and oocyte maturation. EMBO J., 21, 3852–3862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wickens M., Bernstein,D.S., Kimble,J. and Parker,R. (2002) A PUF family portrait: 3′UTR regulation as a way of life. Trends Genet., 18, 150–158. [DOI] [PubMed] [Google Scholar]
- 25.Crittenden S.L., Bernstein,D.S., Bachorik,J.L., Thompson,B.E., Gallegos,M., Petcherski,A.G., Moulder,G., Barstead,R., Wickens,M. and Kimble,J. (2002) A conserved RNA-binding protein controls germline stem cells in Caenorhabditis elegans. Nature, 417, 660–663. [DOI] [PubMed] [Google Scholar]
- 26.Nakahata S., Kotani,T., Mita,K., Kawasaki,T., Katsu,Y., Nagahama,Y. and Yamashita,M. (2003) Involvement of Xenopus Pumilio in the translational regulation that is specific to cyclin B1 mRNA during oocyte maturation. Mech. Dev., 120, 865–880. [DOI] [PubMed] [Google Scholar]
- 27.Mendez R., Hake,L.E., Andresson,T., Littlepage,L.E., Ruderman,J.V. and Richter,J.D. (2000) Phosphorylation of CPE binding factor by Eg2 regulates translation of c-mos mRNA. Nature, 404, 302–307. [DOI] [PubMed] [Google Scholar]
- 28.Mendez R., Murthy,K.G.K., Ryan,K., Manley,J.L. and Richter,J.D. (2000) Phosphorylation of CPEB by Eg2 mediates the recruitment of CPSF into an active cytoplasmic polyadenylation complex. Mol. Cell, 6, 1253–1259. [DOI] [PubMed] [Google Scholar]
- 29.Dickson K.S., Thompson,S.R., Gray,N.K. and Wickens,M. (2001) Poly(A) polymerase and the regulation of cytoplasmic polyadenylation. J. Biol. Chem., 276, 41810–41816. [DOI] [PubMed] [Google Scholar]
- 30.Reverte C.G., Ahearn,M.D. and Hake,L.E. (2001) CPEB degradation during Xenopus oocyte maturation requires a PEST domain and the 26S proteasome. Dev. Biol., 231, 447–458. [DOI] [PubMed] [Google Scholar]
- 31.Mendez R., Barnard,D. and Richter,J.D. (2002) Differential mRNA translation and meiotic progression require cdc2-mediated CPEB destruction. EMBO J., 21, 1833–1844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thom G., Minshall,N., Git,A., Argasinska,J. and Standart,N. (2003) Role of cdc2 kinase phosphorylation and conserved N-terminal proteolysis motifs in cytoplasmic polyadenylation-element-binding protein (CPEB) complex dissociation and degradation. Biochem. J., 370, 91–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ladomery M., Wade,E. and Sommerville,J. (1997) Xp54, the Xenopus homologue of human RNA helicase p54, is an integral component of stored mRNP particles in oocytes. Nucleic Acids Res., 25, 965–973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nakamura A., Amikura,R., Hanyu,K. and Kobayashi,S. (2001) Me31B silences translation of oocyte-localising RNAs through the formation of cytoplasmic RNP complex during Drosophila oogenesis. Development, 128, 3233–3242. [DOI] [PubMed] [Google Scholar]
- 35.Navarro R.E., Shim,E.Y., Kohara,Y., Singson,A. and Blackwell,T.K. (2001) cgh-1 a conserved predicted helicase required for gametogenesis and protection from physiological germline apoptosis in C.elegans.Development, 128, 3221–3232. [DOI] [PubMed] [Google Scholar]
- 36.Coller J.M., Tucker,M., Sheth,U. and Parker,R. (2001) The DEAD box helicase, Dhh1p, functions in mRNA decapping and interacts with both the decapping and deadenylase complexes. RNA, 7, 1717–1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fischer N. and Weis,K. (2002) The DEAD box protein Dhh1 stimulates the decapping enzyme Dcp1. EMBO J., 21, 2788–2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fuller-Pace F.V. (1994) RNA helicases: modulators of RNA structure. Trends Cell Biol., 4, 271–274. [DOI] [PubMed] [Google Scholar]
- 39.Lüking A., Stahl,U. and Schimdt,U. (1998) The protein family of RNA helicases. Crit. Rev. Biochem. Mol. Biol., 33, 259–296. [DOI] [PubMed] [Google Scholar]
- 40.Tanner N.K., Cordin,O., Banroques,J., Doere,M. and Linder,P. (2003) The Q motif: a newly identified motif in DEAD box helicases may regulate ATP binding and hydrolysis. Mol. Cell, 11, 127–138. [DOI] [PubMed] [Google Scholar]
- 41.Linder P. (2003) Yeast RNA helicases of the DEAD-box family involved in translation initiation. Biol. Cell, 95, 157–167. [DOI] [PubMed] [Google Scholar]
- 42.Smillie D.A. and Sommerville,J. (2002) RNA helicase p54 (DDX6) is a shuttling protein involved in nuclear assembly of stored mRNP particles. J. Cell Sci., 115, 395–407. [DOI] [PubMed] [Google Scholar]
- 43.Sheth U. and Parker,R. (2003) Decapping and decay of messenger RNA occur in cytoplasmic processing bodies. Science, 300, 805–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tseng-Rogenski S.S.-I., Chong,J.-L., Thomas,C.B., Enomoto,S., Berman,J. and Chang,T.-H. (2003) Functional conservation of Dhh1p, a cytoplasmic DExD/H-box protein present in large complexes. Nucleic Acids Res., 31, 4995–5002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Westmoreland T.J., Olson,J.A., Saito,B.S., Huper,G., Marks,J.R. and Bennett,C.B. (2003) Dhh1 regulates the G1/S-checkpoint following DNA damage or BRCA1 expression in yeast. J. Surg. Res., 113, 62–73. [DOI] [PubMed] [Google Scholar]
- 46.Lemaire P., Garrett,N. and Gurdon,J. (1995) Expression cloning of Siamois, a Xenopus homeobox gene expressed in dorsal-vegetal cells of blastulae and able to induce a complete secondary axis. Cell, 81, 85–94. [DOI] [PubMed] [Google Scholar]
- 47.Gray N., Coller,J., Dickson,K. and Wickens,M. (2000) Multiple portions of poly(A)-binding protein stimulate translation in vivo. EMBO J., 19, 4723–4733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Coller J. and Wickens,M. (2002) Tethered function assays using 3′ untranslated regions. Methods, 26, 142–150. [DOI] [PubMed] [Google Scholar]
- 49.Maillet L. and Collart,M.A. (2002) Interaction between Not1p, a component of the Ccr4–not complex, a global regulator of transcription and Dhh1p, a putative RNA helicase. J. Biol. Chem., 277, 2835–2842. [DOI] [PubMed] [Google Scholar]
- 50.Pause A. and Sonenberg,N. (1992) Mutational analysis of a DEAD box RNA helicase: the mammalian translation initiation factor eIF-4A. EMBO J., 11, 2643–2654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Iost I., Dreyfus,M. and Linder,P. (1999) Ded1p, a DEAD-box protein required for translation initiation in Saccharomyces cerevisiae, is an RNA helicase. J. Biol. Chem., 274, 17677–17683. [DOI] [PubMed] [Google Scholar]
- 52.Schmitt C., von Kobbe,C., Bachi,A., Pante,N., Rodrigues,J.P., Wilm,M., Seraphin,B., Carmo-Fonseca,M. and Izaurralde,E. (1999) Dbp5, a DEAD-box protein required for mRNA export, is recruited to the cytoplasmic fibrils of nuclear pore complex via a conserved interaction with CAN/Nup159p. EMBO J., 18, 4332–4347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cho H.-S., Ha,N.-C., Kang,L.-W., Chung,K.M., Back,S.H., Jang,S.K. and Oh,B.-H. (1998) Crystal structure of RNA helicase from genotype 1b hepatitis C virus. J. Biol. Chem., 273, 15045–15052. [DOI] [PubMed] [Google Scholar]
- 54.Levin M.K. and Patel,S.S. (1999) The helicase from hepatitis C virus is active as an oligomer. J. Biol. Chem., 274, 31839–31846. [DOI] [PubMed] [Google Scholar]
- 55.Khu Y.-L., Koh,E., Lim,S.P., Tan,Y.H., Brenner,S., Lim,S.G., Hong,W.J. and Goh,P.-Y. (2001) Mutations that affect dimer formation and helicase activity of the hepatitis C virus helicase. J. Virol., 75, 205–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Maluf N.K., Fischer,C.J. and Lohman,T.M. (2003) A dimer of Escherichia coli UvrD is the active form of the helicase in vitro. J. Mol. Biol., 325, 913–935. [DOI] [PubMed] [Google Scholar]
- 57.Ogilvie V.C., Wilson,B.J., Nicol,S.M., Morrice,N.A., Saunders,L.R., Barber,G.N. and Fuller-Pace,F.V. (2003) The highly related DEAD box RNA helicases p68 and p72 exist as heterodimers in cells. Nucleic Acids Res., 31, 1470–1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wilhelm J.E., Mansfield,J., Hom-Booher,N., Wang,S., Turck,C.W., Hazelrigg,T. and Vale,R.D. (2000) Isolation of a ribonucleoprotein complex involved in mRNA localization in Drosophila oocytes. J. Cell Biol., 148, 427–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mansfield J.H., Wilhelm,J.E. and Hazelrigg,T. (2002) Ypsilon Schachtel, a Drosophila Y-box protein, acts antagonistically to Orb in the oskar mRNA localization and translation pathway. Development, 129, 197–209. [DOI] [PubMed] [Google Scholar]