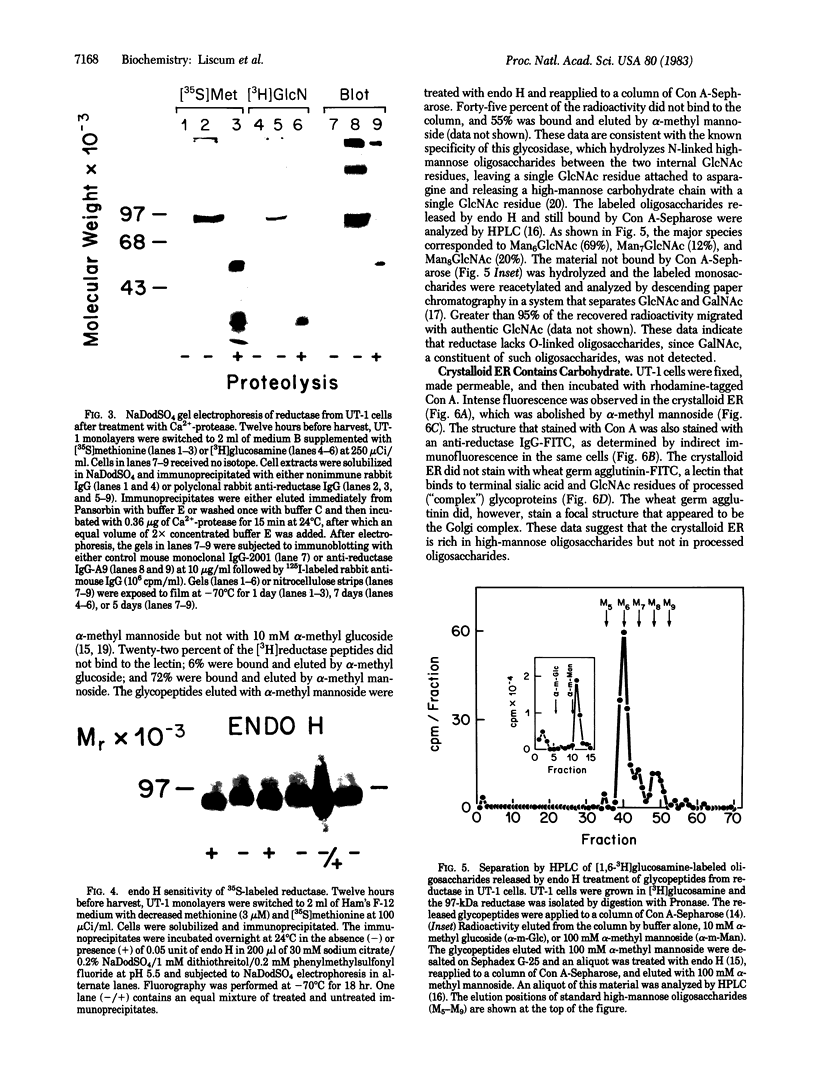
Abstract
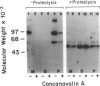
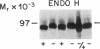
3-Hydroxy-3-methylglutaryl-CoA reductase (EC 1.1.1.34) is an abundant protein of the crystalloid endoplasmic reticulum of UT-1 cells, a line of cultured hamster cells that over-produces the reductase as a result of gene amplification. In the current studies, we show that reductase in UT-1 cells is a glycoprotein. The solubilized enzyme (Mr = 97,000) from UT-1 cells, Chinese hamster ovary cells, and rat liver was adsorbed quantitatively and specifically to concanavalin A-Sepharose. UT-1 cells incorporated [1,6-3H]glucosamine into the reductase; after release with endo-N-acetylglucosaminidase H most of the radioactivity was found in N-linked "high-mannose" chains, including Man6(GlcNAc)2, Man7(GlcNAc)2, and Man8(GlcNAc)2. The carbohydrate of the reductase was localized to a 30- to 35-kilodalton fragment that was separable proteolytically from a cytoplasmic 53-kilodalton fragment that contained the active site of the enzyme. We conclude that 3-hydroxy-3-methylglutaryl-CoA reductase is a transmembrane glycoprotein with an active site facing the cytoplasm and a carbohydrate-bearing site oriented toward the lumen of the endoplasmic reticulum.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anderson R. G., Orci L., Brown M. S., Garcia-Segura L. M., Goldstein J. L. Ultrastructural analysis of crystalloid endoplasmic reticulum in UT-1 cells and its disappearance in response to cholesterol. J Cell Sci. 1983 Sep;63:1–20. doi: 10.1242/jcs.63.1.1. [DOI] [PubMed] [Google Scholar]
- Baenziger J. U., Fiete D. Structural determinants of concanavalin A specificity for oligosaccharides. J Biol Chem. 1979 Apr 10;254(7):2400–2407. [PubMed] [Google Scholar]
- Beg Z. H., Brewer H. B., Jr Regulation of liver 3-hydroxy-3-methylglutaryl-CoA reductase. Curr Top Cell Regul. 1981;20:139–184. doi: 10.1016/b978-0-12-152820-1.50008-0. [DOI] [PubMed] [Google Scholar]
- Brown M. S., Goldstein J. L. Multivalent feedback regulation of HMG CoA reductase, a control mechanism coordinating isoprenoid synthesis and cell growth. J Lipid Res. 1980 Jul;21(5):505–517. [PubMed] [Google Scholar]
- CARDINI C. E., LELOIR L. F. Enzymatic formation of acetylgalactosamine. J Biol Chem. 1957 Mar;225(1):317–324. [PubMed] [Google Scholar]
- Chin D. J., Luskey K. L., Anderson R. G., Faust J. R., Goldstein J. L., Brown M. S. Appearance of crystalloid endoplasmic reticulum in compactin-resistant Chinese hamster cells with a 500-fold increase in 3-hydroxy-3-methylglutaryl-coenzyme A reductase. Proc Natl Acad Sci U S A. 1982 Feb;79(4):1185–1189. doi: 10.1073/pnas.79.4.1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin D. J., Luskey K. L., Faust J. R., MacDonald R. J., Brown M. S., Goldstein J. L. Molecular cloning of 3-hydroxy-3-methylglutaryl coenzyme a reductase and evidence for regulation of its mRNA. Proc Natl Acad Sci U S A. 1982 Dec;79(24):7704–7708. doi: 10.1073/pnas.79.24.7704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cummings R. D., Kornfeld S. Fractionation of asparagine-linked oligosaccharides by serial lectin-Agarose affinity chromatography. A rapid, sensitive, and specific technique. J Biol Chem. 1982 Oct 10;257(19):11235–11240. [PubMed] [Google Scholar]
- Edwards P. A., Lemongello D., Kane J., Shechter I., Fogelman A. M. Properties of purified rat hepatic 3-hydroxy-3-methylglutaryl coenzyme A reductase and regulation of enzyme activity. J Biol Chem. 1980 Apr 25;255(8):3715–3725. [PubMed] [Google Scholar]
- Faust J. R., Luskey K. L., Chin D. J., Goldstein J. L., Brown M. S. Regulation of synthesis and degradation of 3-hydroxy-3-methylglutaryl-coenzyme A reductase by low density lipoprotein and 25-hydroxycholesterol in UT-1 cells. Proc Natl Acad Sci U S A. 1982 Sep;79(17):5205–5209. doi: 10.1073/pnas.79.17.5205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanover J. A., Lennarz W. J. Transmembrane assembly of membrane and secretory glycoproteins. Arch Biochem Biophys. 1981 Oct 1;211(1):1–19. doi: 10.1016/0003-9861(81)90423-9. [DOI] [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- Liscum L., Luskey K. L., Chin D. J., Ho Y. K., Goldstein J. L., Brown M. S. Regulation of 3-hydroxy-3-methylglutaryl coenzyme A reductase and its mRNA in rat liver as studied with a monoclonal antibody and a cDNA probe. J Biol Chem. 1983 Jul 10;258(13):8450–8455. [PubMed] [Google Scholar]
- Luskey K. L., Faust J. R., Chin D. J., Brown M. S., Goldstein J. L. Amplification of the gene for 3-hydroxy-3-methylglutaryl coenzyme A reductase, but not for the 53-kDa protein, in UT-1 cells. J Biol Chem. 1983 Jul 10;258(13):8462–8469. [PubMed] [Google Scholar]
- Okada Y., Frey A. B., Guenthner T. M., Oesch F., Sabatini D. D., Kreibich G. Studies on the biosynthesis of microsomal membrane proteins. Site of synthesis and mode of insertion of cytochrome b5, cytochrome b5 reductase, cytochrome P-450 reductase and epoxide hydrolase. Eur J Biochem. 1982 Feb;122(2):393–402. doi: 10.1111/j.1432-1033.1982.tb05894.x. [DOI] [PubMed] [Google Scholar]
- Rodriguez Boulan E., Sabatini D. D., Pereyra B. N., Kreibich G. Spatial orientation of glycoproteins in membranes of rat liver rough microsomes. II. Transmembrane disposition and characterization of glycoproteins. J Cell Biol. 1978 Sep;78(3):894–909. doi: 10.1083/jcb.78.3.894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodwell V. W., Nordstrom J. L., Mitschelen J. J. Regulation of HMG-CoA reductase. Adv Lipid Res. 1976;14:1–74. doi: 10.1016/b978-0-12-024914-5.50008-5. [DOI] [PubMed] [Google Scholar]
- Varki A., Kornfeld S. The spectrum of anionic oligosaccharides released by endo-beta-N-acetylglucosaminidase H from glycoproteins. Structural studies and interactions with the phosphomannosyl receptor. J Biol Chem. 1983 Mar 10;258(5):2808–2818. [PubMed] [Google Scholar]
- Volpe J. J., Goldberg R. I. Effect of tunicamycin on 3-hydroxy-3-methylglutaryl coenzyme A reductase in C-6 glial cells. J Biol Chem. 1983 Aug 10;258(15):9220–9226. [PubMed] [Google Scholar]