Abstract
MicroRNA-126 (miR-126) is a microRNA predominately expressed by endothelial cells and controls angiogenesis. We found miR-126 was required for the innate response to pathogen-associated nucleic acids, and that miR-126-deficient mice had increased susceptibility to pseudotyped-HIV infection. miRNA profiling and deep-sequencing indicated that miR-126 was highly and specifically expressed by plasmacytoid dendritic cells (pDCs). miR-126 controlled the survival and function of pDCs, and regulated expression ofinnate response genes, including Tlr7, Tlr9 and Nfkb1, as well as Kdr, which encodes VEGF-receptor 2 (VEGFR2). Deletion of Kdr in DCs resulted in reduced type I interferon production, supporting a role for VEGFR2 in miR-126 regulation of pDCs. These studies identify the miR-126–VEGFR2 axis as an important regulator of the innate response that operates through multiscale control of pDCs.
Introduction
Mammals have evolved an interconnected molecular and cellular network of nucleic acid sensors to rapidly respond to incoming viruses and bacteria1,2. Sentinel cells of the hematopoietic system, predominately macrophages and dendritic cells (DCs), are distributed throughout the body and express Toll-like receptors (TLRs) that recognize the nucleic acids carried by pathogens2,3. These sensors represent the first line of defense against pathogen invaders.
TLR7 or TLR9 are triggered by their pathogen-associated ligands, single-stranded RNA (ssRNA) and unmethylated CpG DNA (CpG), respectively, initiating a signaling cascade in which the adapter protein MyD88 forms a complex with the kinases IRAK1 and IRAK4 and the E3 ubiquitin ligase TRAF6 (ref. 4). This leads to the activation of transcription factors, such as interferon regulatory factors (IRF)-3, 7 and 8, STAT3 and STAT5, and NF-κB, which modulate specific genes that drive the maturation of the TLR-expressing cell, and the production of type I interferons (IFN-α/β) and other pro-inflammatory molecules2,5. This response slows the spread of infection and recruits the adaptive immune system.
Recently, microRNAs (miRNAs) have emerged as important contributors to the antiviral response6. miRNAs are short, non-coding RNAs that incorporate into Argonaute (Ago) proteins7. miRNAs guide Ago to transcripts with complementary sequences, and, once bound, Ago can prevent expression of the transcript either by blocking translation of the mRNA or by promoting the mRNA’s decay7. miRNA profiling has identified a number of miRNAs that are induced by TLR stimulation, such as miR-21, miR-146 and miR-155, whose expression is upregulated in macrophages and DCs following exposure to CpG and other TLR ligands8-10.
Although several of the TLR-responsive miRNAs have been shown to propagate or dampen the innate response after initial triggering9,11-13, it is not well known whether any miRNAs are involved in preconditioning the responsiveness of the host to pathogen infection in steady-state, prior to infection. Here, we set out to investigate this issue. Unexpectedly, we found that miR-126, a miRNA that has mostly been associated with the regulation of vasculogenesis and angiogenesis14-16, controlled the IFN-α/β response to pathogen-associated nucleic acids through regulation of plasmacytoid dendritic cell (pDC) homeostasis and function. We further show that miR-126 operated, in part, by regulating another proangiogenic gene, Kdr, in pDCs. This work identifies the existence of an endothelial cell pathway operating in pDCs to control the innate response, and assigns an important new function to a clinically targeted receptor and miRNA.
Results
miR-126 is required for the innate response to nucleic acids
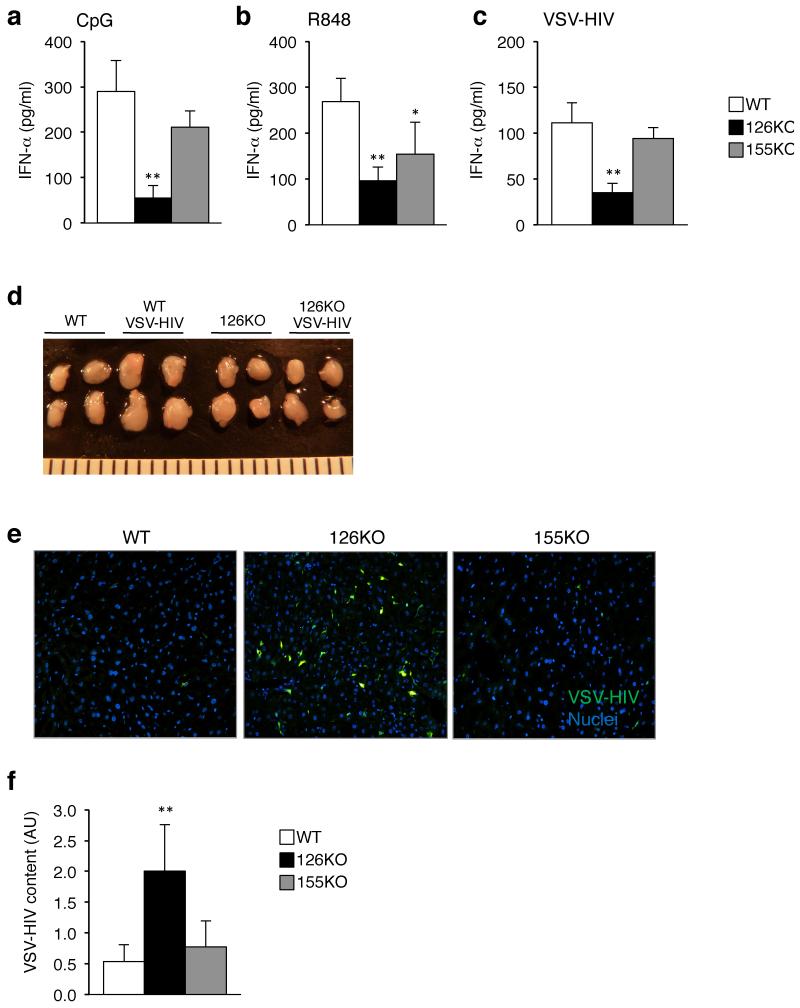
In an effort to understand the relevance of miR-155 in the early interferon response, we injected Mir155−/− mice with the TLR9 agonist CpG-A, and measured serum concentrations of IFN-α after 3 h. As a comparison, we injected CpG-A into wild-type mice, and mice deficient in miR-126, a miRNA that is predominately expressed in endothelial cells and helps to control angiogenesis in the embryo15,16. In both wild-type and Mir155−/− mice, there was a similar increase in circulating IFN-α following CpG-A injection (Fig. 1a). Unexpectedly though, Mir126−/− mice produced less IFN-α than wild-type or Mir155−/− mice. Even at a higher dose of CpG, and examined over time, there was a significant difference in IFN-α production between Mir126−/− mice and littermate controls (Supplementary Fig. 1a). Moreover, interleukin 6 (IL-6) production was also much lower in the absence of miR-126 (Supplementary Fig. 1b), indicating that loss of miR-126 impairs the innate response to unmethylated DNA. To determine whether Mir126−/− mice were impaired in their response to other TLR stimuli, we injected them, along with the Mir155−/− and wild-type mice, with the TLR7 agonist R848, and measured IFN-α 2 h later. Similar to what we observed with the TLR9 agonist, Mir126−/− mice treated with the TLR7 agonist had significantly less circulating IFN-α compared to wild-type mice (Fig. 1b).
Figure 1. The IFN-α/β response is impaired in mice deficient in miR-126.
(a) Measurement of serum IFN-α concentrations in response to TLR9 stimulation. CpG-A 2216 (2 μg) + DOTAP was injected into Mir155−/−, Mir126−/− and wild-type (WT) mice. Serum was collected 3 h, and IFN-α was measured by ELISA. Results are the mean ± s.d. (n = 3-5). (b) Measurement of serum IFN-α concentrations in response to TLR7 stimulation. R848 (2 μg) was injected into Mir155−/−, Mir126−/− and WT mice and 2 h later serum was collected for quantitation of IFN-α concentrations. Results are the mean ± s.d. (n = 3-5). (c) Measurement of serum IFN-α concentrations in response to HIV injection. VSV-pseudotyped, non-replicating HIV (VSV-HIV) was injected into Mir155−/−, Mir126−/− and WT mice and blood was collected 3 h later for quantitation of IFN-α. All results are shown as mean ± s.d. (n = 3-5). (d) Images of the brachial and inguinal lymph nodes taken from mice 3 h after injection of VSV-HIV. (e) Fluorescent microscopy images of the liver taken from mice 5 days after injection of VSV-HIV (encoding GFP). Representative image of 4 sections per mouse (n = 3-7). GFP (green), DAPI stained nuclei (blue). GFP+ cells are mostly macrophages and DCs. The liver was analyzed because it is the main site of infection following intravenous injection. (f) Quantitation of integrated HIV genomes in the liver 5 days after virus injection. Data are the mean ± s.d.; arbitrary units (AU) after normalization for 3-5 mice per group. AU was calculated by comparing CT values to a standard curve made from the HIV genome. *P < 0.05 vs WT, **P < 0.01 vs WT. Data is representative of at least 2 independent experiments.
The initial sensing of HIV is mediated by TLR7 and TLR9 (ref. 17,18), which triggers release of IFN-α/β that block HIV infection of cells19. To determine if miR-126 is also required for sensing HIV, we injected Mir126−/− mice with a VSV-pseudotyped HIV (VSV-HIV). As expected, in wild-type mice circulating concentrations of IFN-α rapidly rose following injection, but in the Mir126−/− mice circulating IFN-α concentrations were barely above background (Fig. 1c). In wild-type mice there was an enlargement of the lymph nodes (Fig. 1d), and an overall increase in cellularity, including the number of neutrophils, monocytes and B cells (data not shown). In the Mir126−/− mice, VSV-HIV did not cause lymph node enlargement or a change in cellularity, providing additional evidence that Mir126−/− mice do not respond normally to infection.
In wild-type and Mir155−/− mice injected with VSV-HIV there were few infected cells, as indicated by the lack of cells expressing green florescent protein (GFP), which is encoded by the virus, and measurement of viral integration (Fig. 1e,f). In Mir126−/− mice, there was widespread infection throughout the tissue indicating that IFN-α/β production was sufficiently diminished by the loss of miR-126 to result in increased cell infection. This data provides clear evidence that Mir126−/− mice are functionally impaired in their response to TLR7 and TLR9 stimulation, and reveal that miR-126 is required for a normal IFN-α/β response to pathogen-associated nucleic acids.
pDCs express high concentrations of miR-126
miR-126 expression was reported to be predominately endothelial cell specific16, however transcriptional analysis across the hematopoietic system, and in fibroblasts, epithelial cells, and endothelial cells, indicated that TLR7 and TLR9 is only expressed by macrophages, B cells and DCs, and not by endothelial cells or other non-hematopoietic cells (Supplementary Fig. 2a). Thus, it was unclear why there was an impaired TLR7 and TLR9 response in the Mir126−/− mice, and so we decided to look at whether any of the TLR expressing cells also expressed miR-126.
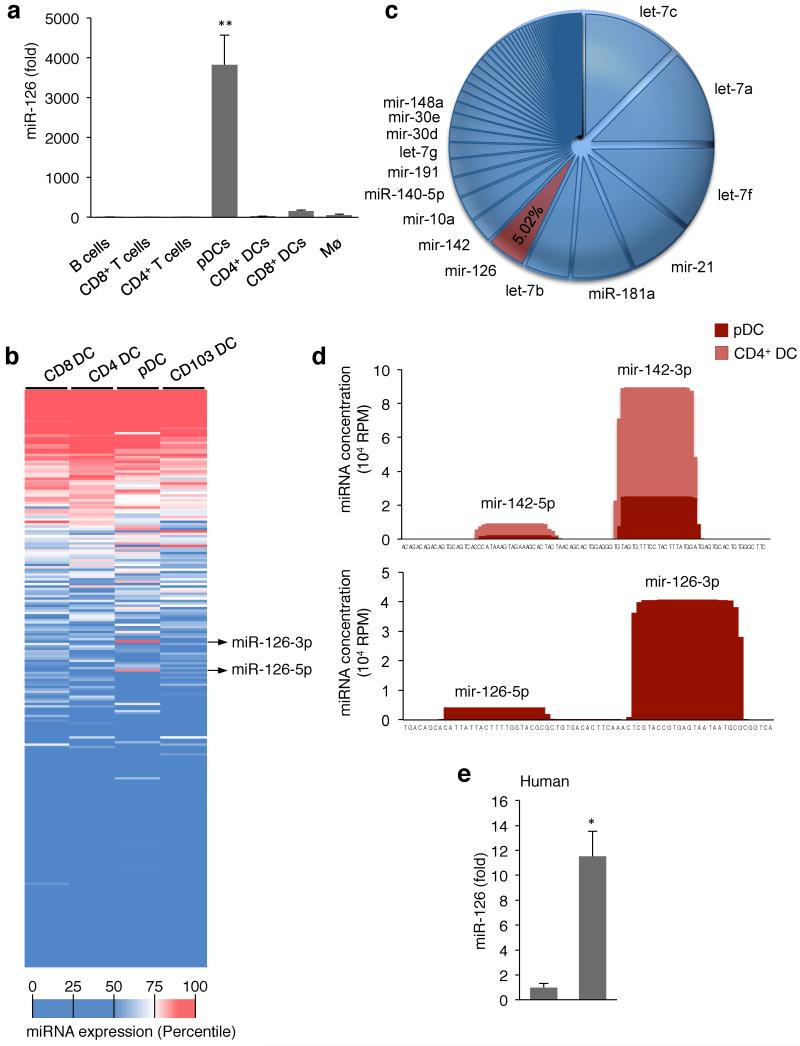
We isolated B cells, macrophages, CD4+ and CD8+ T cells, conventional CD4+ and CD8+ DCs, and pDCs from the spleens of mice. miR-126 was highly and specifically expressed only in pDCs among the hematopoietic cells analyzed (Fig. 2a). Expanded analysis of 365 different miRNAs across DC subsets revealed that while there were other differentially expressed miRNAs between DCs, miR-126 was the only miRNA that was uniquely expressed in one subset, the pDCs (Fig. 2b, Supplementary Fig. 2b). To confirm these findings, we isolated CD4+ DCs and pDCs, and deep-sequenced the small RNA fraction of the cells. In support of the qPCR analysis, miR-126 was detected in pDCs but not CD4+ DCs. miR-126 was one of the most abundant miRNA in pDCs with more than 40,000 reads per million or 5% of the total miRNA pool of the pDCs (Fig. 2c,d).
Figure 2. miR-126 is highly and specifically expressed by plasmacytoid DCs.
(a) Measurement of miR-126 expression in TLR7 and TLR9 expressing immune cells. Splenic B cells, CD8+ T cells, CD4+ T cells, macrophages, CD4+ DCs, CD8+ DCs and plasmacytoid DCs (pDCs) were sorted by flow cytometry. Small RNA was extracted and miR-126-3p, miR-21 and miR-16 were measured by quantitative PCR (qPCR). miR-126 abundance was normalized to miR-16 and miR-21, and calibrated across samples relative to expression in B cells (lowest value). Results are shown as mean ± s.d. (n = 4). **P < 0.01. (b) Profile of 365 miRNAs in mouse DCs. CD4+ DCs, CD8+ DCs and pDCs from the spleen and CD103+ DCs from the lungs were sorted by flow cytometry, small RNA was extracted, and the expression of 365 miRNAs was measured by qPCR (n = 3-5 replicates per cell type). The data was normalized using array means, and plotted in decreasing order according to the miRNA expression average across the groups. The color scale denotes the relative abundance of each miRNA. (c,d) miR-126 abundance in pDCs and CD4+ DCs. Small RNA was extracted from sorted splenic CD4+ DCs and pDCs and subjected to deep-sequencing. The relative frequency of each miRNA in pDCs is shown (c), as well as the read density across the miR-126 gene for pDCs and CD4+ DCs, and across the Mir142 gene, as a comparison. (e) miR-126 expression in human DCs. Human pDCs and conventional DCs (cDCs) were isolated from blood, small RNA was extracted, and qPCR analysis was performed to measure expression of miR-126 and miR-21. miR-126 abundance was normalized to miR-21 and calibrated relative to that expressed in cDCs. Results are shown as the mean ± s.d. (n = 3). *P <0.05 vs cDCs.
To determine if human pDCs also expressed miR-126, we isolated pDCs and cDCs from human peripheral blood by flow cytometry. Total RNA was extracted from the cells, and we measured expression of miR-126, along with miR-16, miR-21, and miR-142-3p. Whereas the other three miRNAs were similarly expressed between pDCs and cDCs, miR-126 expression was 12-fold higher in pDCs compared to cDCs from human peripheral blood (Fig. 2e).
pDCs are the first and strongest producers of IFN-α during viral infection due to their steady-state expression of TLR7, TLR9 and abundant IRF7 (ref. 20). Since pDCs were the only TLR7 and TLR9 expressing cells in which miR-126 was detected and pDCs are major IFN-α/β producing cells21, these results suggest that the diminished IFN-α/β response to ssRNA and unmethylated DNA in Mir126−/− mice may be due to a defect in pDCs.
miR-126 controls pDC homeostasis
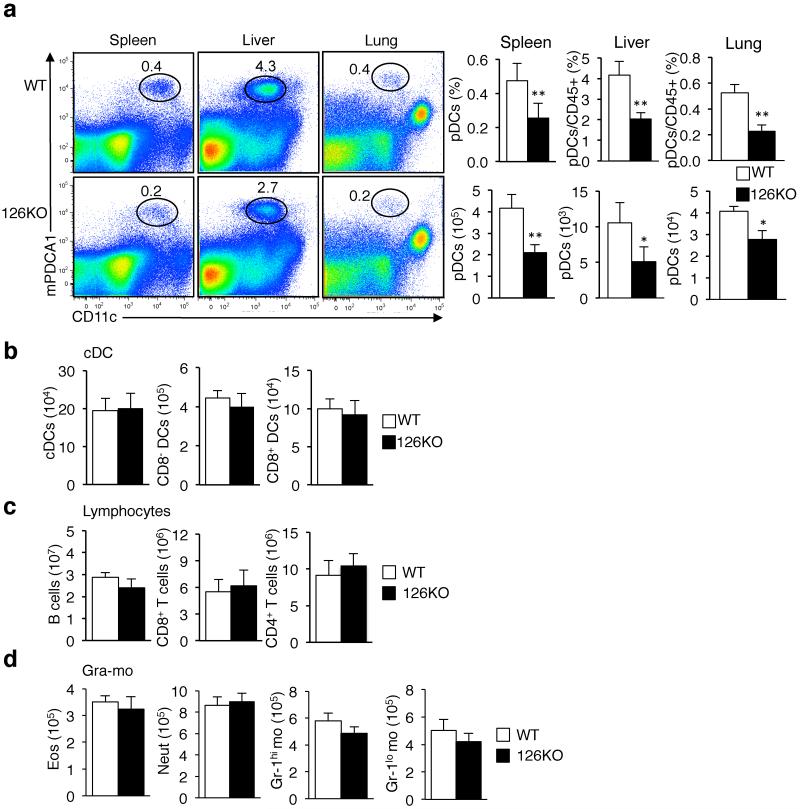
To investigate how the loss of miR-126 may have affected the innate response to TLR stimulation, we examined the frequency of TLR9 expressing cell types, as well as other immune cells, in Mir126−/− mice and littermate controls. In each of the tissues examined a clear pattern emerged in which the frequency and absolute number of pDCs was decreased by 50% or more in the Mir126−/− mice compared to the controls (Fig. 3a). In contrast, there was no change in the frequency of other DC subsets (CD4+ and CD8+ DCs), B cells, T cells, granulocytes, or monocytes (Fig. 3b-d and Supplementary Fig. 3). These findings suggest the impaired response to TLR7 and TLR9 stimulation in Mir126−/− mice is due to a specific deficiency in pDCs, and not because of a general immune cell deficiency, and reveal that miR-126 plays an important role in pDC homeostasis.
Figure 3. Loss of miR-126 impairs plasmacytoid DC homeostasis.
Eight-week old Mir126−/− mice and WT littermates were analyzed to determine the frequency and absolute number of pDCs (CD11cint, B220+, PDCA1+ cells) (a); splenic CD4+ DCs and CD8+ DCs (b); B cells and CD4+ and CD8+ T cells (c); eosinophils (Eos), neutrophils (Neut) and monocytes (mo) (d). Representative flow cytometry plots are shown. All graphs present the mean ± s.d. of the frequency and number of cells (n = 5-6). Data is representative of at least 5 independent analyses of mouse cohorts over a 18 month period.
miR-126 regulates pDC survival
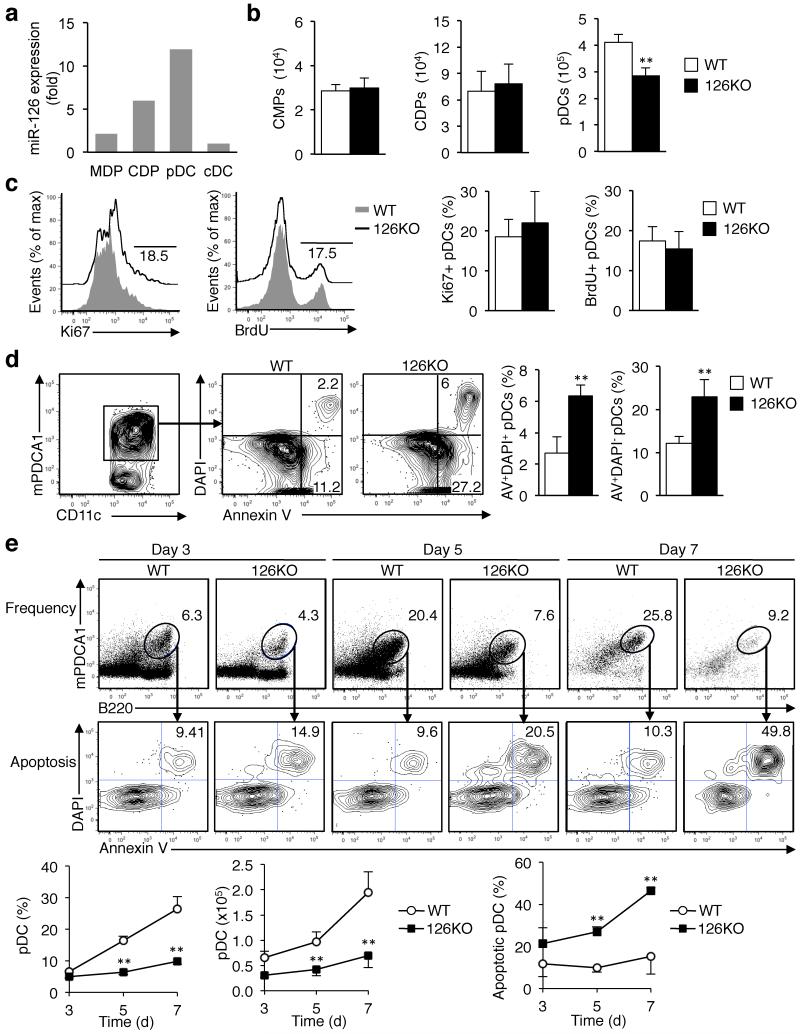
pDCs develop in the bone marrow (BM) from myeloid progenitors common to all DCs, and are released as mature pDCs in to the blood20,22. Within the BM, common myeloid progenitors (CMPs) can differentiate to common DC precursors (CDPs), which can then become pDCs or pre-conventional DCs (cDCs)22. To understand at which stage of pDC development miR-126 may be critical, we isolated CMPs, CDPs, pDCs and pre-conventional cDCs from the BM, and measured miR-126 expression. miR-126 expression increased during differentiation with maximum amounts found in pDCs, and virtually no miR-126 in cDCs (Fig. 4a). Flow cytometry analysis indicated that the frequency and number of CMPs and CDPs was normal in Mir126−/− mice, but the number of pDCs is decreased by 30% in the BM (Fig. 4b).
Figure 4. miR-126 controls plasmacytoid DC survival.
(a) Measurement of miR-126 during DC differentiation. CMPs, CDPs, pre-conventional DCs, and pDCs were isolated from the bone marrow (BM) and sorted by flow cytometry, and miR-126 was measured by qPCR. The data was normalized on miR-16 and miR-21 and calibrated relative to that expressed by cDCs. (b) CMPs, CDPs, and pDCs were quantified in the BM of Mir126−/− mice and WT littermates by flow cytometry. Results are shown as the mean ± s.d. (n = 4-6). (c) Measurement of pDC proliferation was performed by Ki67 staining or BrdU incorporation analysis of pDCs from Mir126−/− mice or WT littermates. Graphs present the mean ± s.d. (n = 3-4). Representative histograms are shown. Results of one out of three independent experiments are shown. (d) Apoptosis of pDCs was analyzed by quantification of Annexin V+ and DAPI+ pDCs in the BM of Mir126−/− mice and WT littermates. Results are mean ± s.d. (n = 4). (e) The number and frequency of pDCs, and their apoptotic status, in Flt3L cultures was determined by flow cytometry analysis of CD11c+ B220+ PDCA1+ cells stained with Annexin V and DAPI. Representative flow cytometry plots of the cultures at the indicated time points during differentiation are shown. The graphs present the mean ± s.d. (n = 4). *P < 0.05 vs WT, **P < 0.01 vs WT. Results of one out of two independent experiments are shown.
Since we could detect pDCs in the Mir126−/− mice, we speculated that the impaired homeostasis was due to a defect in pDC replication or survival. To assess proliferation, we quantified the number of Ki67+ and BrdU-positive pDCs in the BM where pDCs develop. This revealed no difference in the proliferation rate between Mir126−/− pDCs and wild-type pDCs (Fig. 4c), suggesting that loss of miR-126 did not decrease the number of pDCs by reducing their capacity to proliferate. We next measured apoptosis of pDCs by Annexin V (AV) and DAPI staining by flow cytometry. The ratio of both AV+DAPI+ and pre-apoptotic AV+DAPI- cells was more than 2-fold higher in Mir126−/− pDCs, indicating that increased apoptosis was responsible for the reduced number of pDCs in these mice (Fig. 4d).
To further understand how pDC homeostasis is impaired in the absence of miR-126, we took advantage of an in vitro model of DC development, which uses Flt3 ligand (Flt3L) to drive DC progenitors to differentiate into DC subsets, including pDCs, which have similar molecular and phenotypic profiles to in vivo DCs22. BM was collected from Mir126−/− mice and littermate controls, cultured with Flt3L, and DC differentiation was followed over time. After 3 days, there were similar numbers of pDCs between wild-type and Mir126−/− cultures, as measured by the frequency of CD11c+B220+mPDCA1+ cells. However, by day 5, there was a 40% reduction in Mir126−/− pDCs compared to wild-type pDCs, and by day 7 the number of Mir126−/− pDCs was 60% lower than wild-type pDCs (Fig. 4e). There were no statistical differences in the numbers of DC progenitors or cDCs between the wild-type and Mir126−/− cultures (data not shown).
Proliferation during in vitro development was assessed by dye eFluor670 dilution and staining of pDCs, and no difference in Mir126−/− pDC proliferation was found in vitro (Supplementary Fig. 4). However, the percentage of pDCs undergoing apoptosis in the Flt3L cultures, as measured by Annexin V and DAPI staining, was significantly higher in the Mir126−/− pDCs compared to wild-type pDCs (Fig. 4e). These results recapitulate our in vivo findings showing that the reduced numbers of pDCs in Mir126−/− mice is due to an increase in the rate of pDC apoptosis, and indicate that miR-126 controls pDC survival.
miR-126 is necessary for normal pDC function
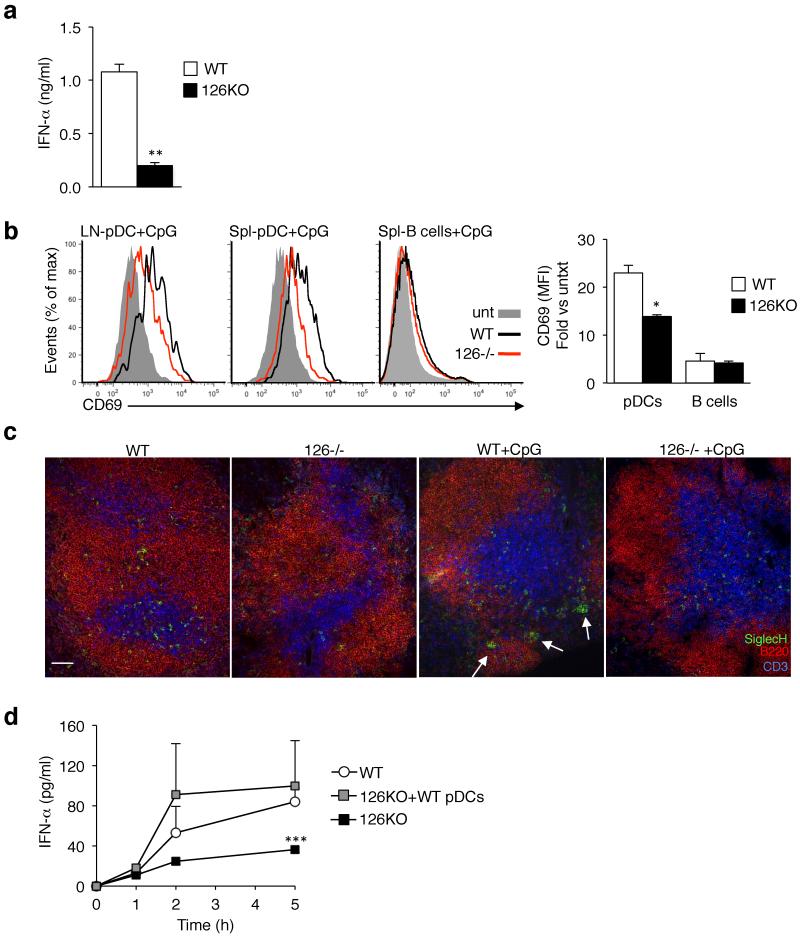
Although there was a major decrease in the number of Mir126−/− pDCs, there were still cells in the Mir126−/− mice and cultures that expressed pDC markers. To determine if the pDCs that develop without miR-126 were functional, we isolated pDCs from wild-type and Mir126−/− mice, and stimulated matched numbers of cells with CpG-A ex vivo. Whereas wild-type pDCs produced more than 1,000 pg/ml of IFN-α, Mir126−/− pDCs produced less than 200 pg/ml (Fig. 5a).
Figure 5. pDCs are functionally impaired in the absence of miR-126.
(a) Measurement of IFN-α from freshly isolated pDCs stimulated with CpG-A. pDCs were isolated from the spleen and lymph nodes of Mir126−/− mice and littermate controls (WT) by bead selection. 2.5 × 104 cells were seeded and stimulated with ODN 2216 (5 μM) in a 96-round bottom well plate, and IFN-α was measured in the supernatant after 12 h by ELISA. The graph presents the mean ± s.d. (n = 4). **P < 0.01 vs WT. (b) In vivo activation of pDCs following TLR9 stimulation was assessed by measuring surface CD69 on pDCs in the spleen (Spl) and lymph nodes (LN) of Mir126−/− mice and WT littermates following injection of ODN 2216 (1.5 μg). CD69 expression on B cells from the same mice is shown for comparison. Representative histograms are shown. Graphs present the mean ± s.d. (n = 4) of the mean fluorescence intensity (MFI) of CD69. *P<0.05 vs WT. Results of one out of two independent experiments are shown. (c) Florescence microscopy analysis of pDC biodistribution in vivo. Splenic frozen tissue sections from the mice treated in (b), then stained for SiglecH (green), B220 (red) and CD3 (blue) to label pDCs, B cells and T cells, respectively. Arrows point to pDC clusters that form after activation. Scale bar is 50 μm. Representative images are shown from n = 4 animals. (d) Measurement of serum IFN-α concentrations in Mir126−/− mice following adoptive transfer of WT pDCs. Mir126−/− mice or Mir126−/− mice in which we transferred WT pDCs were injected with VSV-HIV. Serum was collected at the indicated times, and IFN-α was measured by ELISA. Results are the mean ± s.d. (n = 3-4).
Next, we assessed pDC maturation in vivo by injecting the Mir126−/− mice or littermate controls with CpG-A. After 3 h, we examined the expression of CD69, an activation marker, and found that Mir126−/− pDCs were less activated than wild-type pDCs (Fig. 5b). Since pDCs are known to migrate after activation23,241, we looked at their biodistribution in the spleen after injection of CpG. In steady-state, both wild-type and Mir126−/− pDCs were localized in the T cell zone, albeit at a reduced frequency in the Mir126−/− mice (Fig. 5c). At 3 h after injection of CpG, wild-type pDCs migrated to the outer marginal zone and formed clusters, as previously reported24. Instead, in Mir126−/− mice, the pDCs remained in the T cell zone after CpG injection, which further suggests that their function is impaired. These results demonstrate that, in addition to controlling pDC survival, miR-126 is necessary for normal pDC function.
Wild-type pDCs restore IFN-α in Mir126−/− mice
The reduced numbers and function of Mir126−/− pDCs suggested that this was the reason for the impaired innate response of Mir126−/− mice. To test this hypothesis, we isolated pDCs from wild-type mice, and transferred them to Mir126−/− mice. In parallel, we injected the mice with VSV-HIV, and followed the IFN-α response. In Mir126−/− mice there was a small increase in serum IFN-α over time (up to 36 pg/ml), but in Mir126−/− mice that received wild-type pDCs, IFN-α concentrations were more than 100 pg/ml by 5 h, similar to wild-type mice (Fig. 5d). Taken together, our findings indicate that the diminished innate response in Mir126−/− mice is due to miR-126 control of pDC homeostasis and function.
Mir126−/− pDCs exhibit a reduced innate response profile
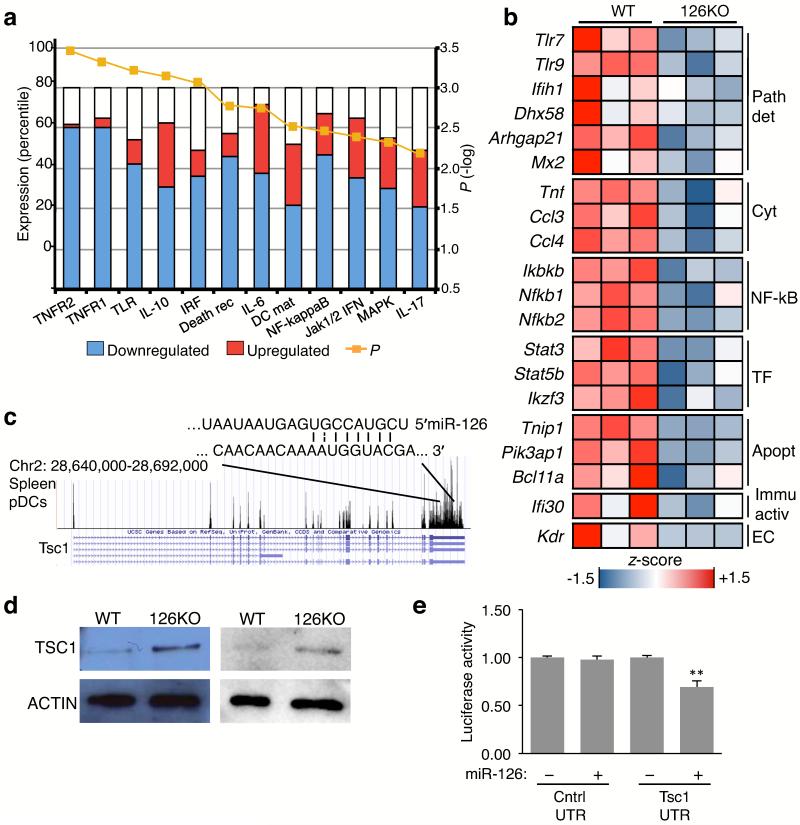
To understand why activation was impaired in miR-126-deficient pDCs, we sorted pDCs from Mir126−/− mice and littermate controls, and performed transcriptional profiling. The expression of 253 genes were altered by loss of miR-126, with most of the genes clustering in specific innate immune response pathways, including the TNF receptor pathway, the NF-κB pathway, and the TLR-induced pathway (Fig. 6a). Both Tlr7 and Tlr9 expression were significantly lower in Mir126−/− pDCs (Fig. 6b), along with other key genes encoding viral detectors, such as the helicase Dhx58 (Lpg2)25, Arhgap21 (ref. 3), and Ifih1 and Mx2, which are expressed by pDCs even in steady-state26.
Figure 6. miR-126 regulates key innate response genes in pDCs and directly targets the mTOR pathway.
(a) pDCs were isolated from the spleen of Mir126−/− mice and WT littermates, sorted by flow cytometry, and gene expression was measured by microarray. Analysis of molecular pathways altered in Mir126−/− pDCs was performed by Ingenuity® by using Fisher’s exact test, Benjamini-Hochberg correction for multiple testing. Yellow line is the −log(P-value). (b) Heat-map representation of genes that were significantly changed in Mir126−/− pDCs compared to WT pDCs, and known to be involved in the innate response. Each column is an individual mouse. Data is color coded to reflect gene expression z-scores. Genes are grouped as: Pat det (pathogen detection related genes), Cyt (cytokines), NF-κB (NF-ΚB family genes), TF (transcription factors), Apopt (apoptosis related genes), Immu active (immune activation related genes), EC (endothelial cell-specific genes). (c) RNA-seq analysis of Tsc1 mRNA expression in flow-sorted WT splenic pDCs. The miR-126 recognition site in the Tsc1 3′UTR is shown. (d) TSC1 protein expressed in splenic pDCs of Mir126−/− and WT littermates, isolated by flow cytometry and measured by immunoblot. Two independent biological replicates are shown. (e) Analysis of miR-126 regulation of Tsc1 by 3′UTR assay. Plasmids encoding luciferase upstream of the Tsc1 3′UTR sequence, or the 3′UTR of a gene with no putative miR-126 binding site were co-transfected into HEK293T cells with either a control plasmid or a plasmid expressing miR-126. Results are shown as mean ± s.d. (n = 3). *P < 0.05 vs control plasmid.
Ccl4 and Ccl3 are expressed by pDCs27, however Mir126−/− pDCs expressed less of both chemokines. Tnf was another cytokine gene whose expression was less in Mir126−/− pDCs. The expression of Nfkb1 and Nfkb2, encoding subunits of NF-κB, were reduced in Mir126−/− pDCs. Moreover, the key transcription factors involved in TLR signaling and cytokine production, encoded by Stat3 and Stat5b28, were similarly decreased. These studies indicate that miR-126 is necessary for pDCs to establish a normal molecular configuration for responding to pathogen-associated nucleic acids, and provide mechanistic insight into why Mir126−/− pDCs have an impaired response to ssRNA and CpG DNA.
miR-126 suppresses translation of Tsc1
We performed in silico analysis to cross identify genes that contain a miR-126 target site and that participate in an innate response or survival pathway (Supplementary Table 1). Of note, the Tsc1 transcript, which encodes a negative regulator of mammalian target of rapamycin (mTOR), was identified as a target for miR-126 and expressed by pDCs (Fig. 6c). Since the mTOR pathway regulates pDC survival29 and cooperate with TLR signaling in pDCs30, we decided to investigate if miR-126 directly regulates TSC1.
We sorted pDCs by flow cytometry from the spleens of Mir126−/− mice and littermate controls and measured TSC1 protein by immunoblot. More TSC1 was present in Mir126−/− pDCs compared to wild-type pDCs (Fig. 6d), suggesting that miR-126 decreases the amount of TSC1 protein expressed in wild-type pDCs. To confirm the transcript is directly regulated by miR-126, the 3′ untranslated region (UTR) of Tsc1 was cloned downstream of luciferase in a reporter plasmid. The plasmid was transfected into 293T cells (which do not express miR-126) along with either a control plasmid or a plasmid expressing miR-126. In the cells overexpressing miR-126, there was a 30% reduction in luciferase expression demonstrating that miR-126 directly regulates TSC1 protein abundance (Fig. 6e). This result demonstrates that miR-126 controls a negative regulator of the mTOR pathway, and helps to explain the impaired survival and TLR response of pDCs in the absence of miR-126.
Deletion of Kdr partially phenocopies loss of miR-126
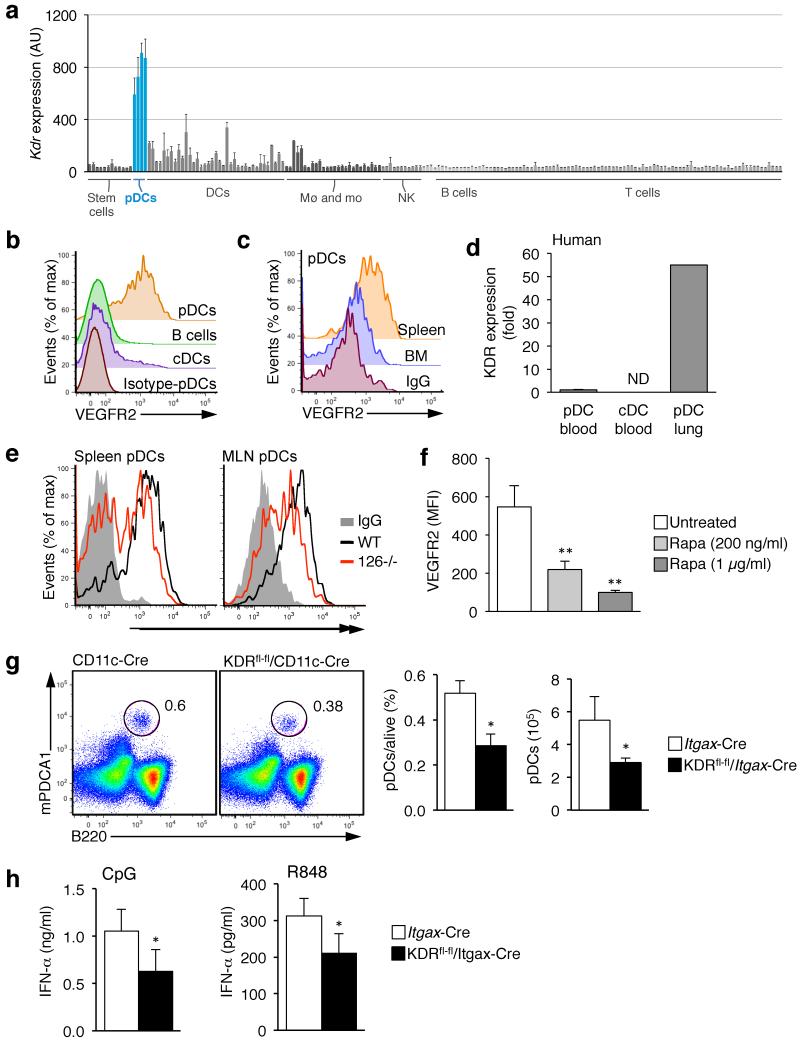
Interestingly, one of the genes whose expression was downregulated in Mir126−/− pDCs was Kdr, which encodes VEGFR2 (Fig. 6b). VEGFR2 is a receptor tyrosine kinase that is the main receptor for VEGF-A31. Kdr is a haploinsufficient gene, which is essential for vasculogenesis during fetal development, as well as in angiogenesis during wound healing and tumor growth32. VEGFR2 reporter mice revealed that VEGFR2 is only expressed by endothelial cells in steady-state33. However, examination of Immgen data, which contains the transcriptional profiles of 234 immune cell types26, revealed that Kdr is also highly expressed on pDCs, while its expression was low or undetectable on all other hematopoietic cell types (Fig. 7a).
Figure 7. VEGFR2 is important for pDC function and its expression is regulated by miR-126.
(a) VEGFR2 (Kdr) expression pattern across the murine immune system as determined by the Immgen Consortium. Expression of each gene was measured by Affymetrix gene array on 234 different populations of immune cells that were flow sorted to high purity. Shown is the mean ± s.d. (n ≥ 3). (b) VEGFR2 surface expression was determined on conventional DCs (cDC), B cells and pDCs from the spleen by flow cytometry. (c) The expression of VEGFR2 on pDCs from the spleen and bone marrow (BM). (d) KDR expression in human DCs. Human pDCs and conventional DCs (cDCs) were isolated from blood, and pDCs were isolated from the lungs of a human donor. qPCR analysis was performed to measure expression of KDR. (e) VEGFR2 surface expression was measured on pDCs taken from the spleen and mesenteric lymph nodes (MLN) of Mir126−/− mice and controls. Shown is a representative histogram (n = 5) of one representative experiment out of two. (f) VEGFR2 surface expression was measured on pDCs after treatment with the mTOR inhibitor Rapamycin. Splenocytes were cultured from wild-type mice with the indicated dose of rapamycin for 16 h, and VEGFR2 was measured on pDCs (PDCA+SiglecH+B220+CD11cint) by flow cytometry. Shown is the mean ± s.d. (n = 3). (g) Analysis of pDC frequency in the spleen of 2-month old Itgax-Cre Kdrfl/fl mice and WT littermates (Kdrfl/fl and Itgax-Cre). pDCs were defined as CD11cint B220+ SiglecH+ PDCA1+ cells. Shown is a representative flow cytometry plot (top) and a graph of the mean ± s.d. (n ≥ 3). (h) Measurement of serum IFN-α concentrations in Itgax-Cre Kdrfl/fl mice and WT littermates after injection of CpG-A or R848. Shown is the mean ± s.d. (n =3-8). Results of one of two independent experiments are shown.
To confirm this finding, we examined the expression of VEGFR2 on the surface of pDCs by flow cytometry. Abundant VEGFR2 was detected on B220+PDCA1+ cells, but not B cells or cDCs (Fig. 7b and Supplementary Fig. 5a). We confirmed that VEGFR2 was also expressed by human pDCs, but not cDCs, by isolating pDCs and cDCs from human blood and lung, and measuring VEGFR2 gene expression by qPCR. Though in humans, as well as mice, VEGFR2 was only detected on pDCs from the tissue, and not those isolated from blood (in human) or the BM (in mouse), suggesting that VEGFR2 expression may only be upregulated in pDCs once they enter a tissue (Fig. 7c,d).
We measured VEGFR2 expression on Mir126−/− pDCs in the spleen and lymph nodes by flow cytometry. Consistent with reduced Kdr mRNA (Fig. 6b), less VEGFR2 was expressed in pDCs in the absence of miR-126 (Fig. 7e). Since we had identified miR-126 as directly regulating TSC1 we wondered whether the mTOR pathway may control VEGFR2 expression in pDCs. To test this, we cultured total splenocytes from wild-type mice in the presence or absence of rapamycin, then measured VEGFR2 expression. Rapamycin significantly downregulated VEGFR2 expression in a dose-dependent manner (Fig. 7f). A similar downregulation of VEGFR2 occurred in pDCs in vivo when we treated mice with rapamycin (data not shown). These results indicate that miR-126 positively regulates VEGFR2 expression in pDCs, and suggests that miR-126 controls the mTOR pathway.
In endothelial cells, signaling through VEGFR2 promotes cell survival32. Thus, we hypothesized that VEGFR2 may have a similar role in pDCs. To test our hypothesis, we crossed Itgax-Cre mice, which express Cre in DCs34, with Kdrfl/fl mice, to generate mice in which the Kdr gene is deleted in DCs (Itgax-CreKdrfl/fl, Supplementary Fig. 5b,c). The frequency of pDCs in the spleen of littermate controls was ~0.5% (Fig. 7g). Instead, in Itgax-CreKdrfl/fl mice, the frequency of pDCs was 0.3%, a 40% reduction, which is comparable to what we observed in Mir126−/− mice. Moreover, consistent with the fact that we do not detect VEGFR2 on BM pDCs, the number of pDCs in the BM was not changed in Itgax-CreKdrfl/fl mice (Supplementary Fig. 5d).
Finally, we examined the IFN-α response in the DC-specific VEGFR2-deficient mice. Itgax-CreKdrfl/fl mice or littermate controls were injected with R848 or CpG-A, and serum concentrations of IFN-α were measured. For both ligands there was a significant reduction in IFN-α in Itgax-CreKdrfl/fl mice compared to wild-type mice (Fig. 7h), which partially recapitulates the impaired IFN-α/β response observed in Mir126−/− mice. These studies provide the first evidence that VEGF signaling is important for pDC homeostasis and function, and strongly suggest that miR-126 regulates pDCs, at least in part, through its control of VEGFR2 expression (Supplementary Fig. 6).
Discussion
These studies reveal that miR-126 regulates the innate response to pathogen-associated nucleic acids. This finding was unanticipated since miR-126 has mostly been associated with control of angiogenesis14-16. Our initial observation was that Mir126−/− mice produced significantly lower amounts of IFN-α in response to ssRNA and CpG DNA, as well as HIV, and this increases host permissiveness to virus infection. We can conclude that the defect is due to miR-126 control of pDC homeostasis and function because pDCs are the only TLR7 and TLR9 expressing cells that express miR-126, and the frequency of pDCs is reduced in the absence of miR-126. Moreover, we specifically show that miR-126-deficient pDCs have a reduced activation capacity.
pDCs were only fully discovered just over a decade ago23,35,36, and the molecular underpinnings controlling their homeostasis and function are still mostly unknown20. Our transcriptional profiling indicated that in the absence of miR-126 pDCs exhibited less expression of genes that are crucial for the innate recognition and response process. Expression of Tlr7 and Tlr9 was decreased, as were genes encoding key transcription factors and signaling factors involved in the expression of IFN-α/β and other pro-inflammatory cytokines, such as Nfkb1, Stat3 and Arhgap21 (refs. 3,37). Though the differences in expression of some of these genes were modest, the coordinated reduction of so many factors that participate in the same pathways is what likely results in the major impairment in IFN-α production from Mir126−/− pDCs. Since the expression of these genes is reduced in non-activated pDCs, our results indicate that miR-126 functions in steady-state to control the readiness of pDCs to sense and respond to pathogens. In addition to controlling the IFN-α/β response through regulation of key innate pathways, miR-126 also functions by regulating pDC homeostasis. Although it appears that pDC differentiation is initiated even in the absence of miR-126, we found miR-126 controls pDC frequency by controlling pDC survival.
The identification of a gene that controls pDC survival is an important finding since very little is known about what controls the lifespan of a pDC in steady-state20. Only recently was it shown that the PI(3)K-mTOR pathway has a role in human pDC survival in vitro29. However, since this pathway is a commonly used survival pathway38, the discovery of upstream factors, particularly ones that are unique to pDCs amongst hematopoietic cells, is relevant since it provides a potential target for controlling pDC survival and function.
Our data indicate that miR-126 can directly suppress TSC1 abundance. Because TSC1 is a negative regulator of mTOR38, miR-126-mediated suppression of TSC1 would allow enhanced signaling through this pathway and increase pDC survival. In addition to the role of mTOR in cell survival, it also cooperates with components of the TLR9 pathway to induce IFN-α/β expression30,39. Thus, miR-126-mediated suppression of TSC1 would also serve to enhance the TLR9 response, and further explain why there is an impaired IFN-α/β response in the absence of miR-126.
We also show that pDCs express abundant VEGFR2, and that VEGFR2 signaling contributes to pDC homeostasis and function. VEGFR2 expression is less in the absence of miR-126, which indicates that miR-126 positively regulates VEGFR2 expression in pDCs. We found that mTOR positively regulates VEGFR2 expression. Thus, it is likely that miR-126 suppression of TSC1, which increases mTOR activity, is how miR-126 controls VEGFR2 abundance. Since VEGFR2 signaling also negatively regulates TSC1, this pathway represents a feed forward mechanism to enhance mTOR activity, and increase pDC survival and IFN-α/β production.
VEGFR2 is essential for vascularization, and many tumor cell types produce VEGF-A, the ligand for VEGFR2, to promote angiogenesis. This scenario may explain why large numbers of pDCs are found in some solid tumors40. There are a number of drugs that are designed to block VEGFR2 signaling, such as bevacizumab (Avastin), which are used for cancer therapy as well as the treatment of macular degeneration41. Our results suggest that it will be important to examine the effect of these drugs on innate immunity as they may have unintended consequences on pDCs.
This work uncovers a new function for the miR-126-VEGFR2 pathway, and a new regulator of the innate response to pathogen-associated RNA and DNA. Since pDCs play an important role in autoimmunity42-46, it will be worth examining whether dysregulated expression of miR-126 is associated with the etiology of any of these conditions. Already there has been a report that miR-126 can be found in the blood of patients with systemic lupus erythematosus (SLE)47, but more detailed analysis will be necessary. Given the fact that both the VEGF pathway, and now miRNAs, are druggable targets41,48, our findings have potential implications for the treatment of cancer and autoimmune disease.
Methods
Mice
Mir126−/− mice15 and Kdrfl/fl mice were generated previously33. Mir155−/− mice49 and CD11c-Cre (Itgax-Cre)34 mice were purchased from Jackson Laboratories. CD45.1 mice were purchased from the National Cancer Institute. For the activation of TLRs, mice were treated with the TLR9 agonist CpG-A (ODN 2216, Invivogen), TLR7 agonist R848 (Invivogen), TLR3 agonist polyI:C (Invivogen) and VSV-pseudotyped non-replicating HIV (VSV-HIV)50. Mice received 1.5 μg of CpG-A (ODN 2216) + DOTAP, 2 μg of R848, and 6 × 108 or 2 × 108 transducing units (TU) of VSV-HIV via tail vein. Blood was collected from tail at the indicated time points for serum collection or, alternatively, mice were sacrificed at 3 h or 5 d later, respectively, for blood and tissue collection. IFN-α and IL-6 were determined by ELISA (Thermo Fisher and BD Biosciences). All animal procedures were performed according to protocols approved by the Mount Sinai School of Medicine Institutional Animal Care and Use Committee.
Immunostaining
Spleens were harvested and frozen directly in OCT. 8-μm spleen sections were fixed for 15 min with cold acetone prior to staining. Staining was performed at 4°C for 2 h with anti-SiglecH (clone 440c, Sigma), Alexa Fluor 488-conjugated anti-CD3 (clone 17A2, Biolegend) and Cyanin5-conjugated anti-B220 (clone RA3-6B2, eBioscience). Immunodetection of SiglecH was performed using Alexa Fluor 568-conjugated goat anti-rat IgG (Invitrogen). Sections were blocked with 5% rat serum in PBS before staining with rat CD3 and B220 antibodies. Images were obtained with an upright wide-field microscope (Axioplan2; Zeiss) equipped with a 20× lens, and analyzed with ImageJ software. Livers were fixed in 4% paraformaldehyde, equilibrated in 20% sucrose and embedded in OCT prior to sectioning. 5-μm cryostat sections were stained with rat rabbit anti-GFP-Alexa488 (Invitrogen) and DAPI (Vector Laboratories) for nuclei labeling. Images were captured with a Coolsnap HQ camera and Eclipse E800 (Nikon) microscope.
HIV content in tissue
The HIV content was determined in the liver 5 days after VSV-HIV injection by quantitative PCR as previously described50.
Flow cytometry analysis and mouse cell purification
Spleen, lungs and liver were digested in HBSS (Hank’s buffer) containing 10% FBS and 0.2 mg/ml collagenase IV (Sigma-Aldrich) for 30 min. After filtration through a 70-μm cell strainer (BD Biosciences), red blood cells (RBC) were lysed with RBC lysis buffer (eBioscience) for 3 minutes. The hematopoietic cell fraction of the liver was purified by gradient centrifugation in Percoll as previously described50. BM single cells suspension was obtained by flushing tibiae and femurs. RBC lysis was similarly performed.
Samples were stained with: SiglecH (eBio440c)-APC, mPDCA1 (eBio129c)-PE and (eBio927)-APC, B220 (RA3-6B2)-APC-eFluor780 and FITC, CD11c (N418)-APC-Alexa780 and PE-Cy7, CD19 (eio1D3)-PercPCy5.5 and PECy7, F4/80 (BM-8)-PercPCy5.5 and -FITC, CD11b (M1/70)-PercP-Cy5.5, CD45.2 (104)-APC-Alexa780 and PerCP-Cy5.5, CD45.1 (A20)-eFluor450, MHCII IA/IE (M5/114.15.2)-eFluor450, CD69 (H1.2F3)-PE, CD40 (1C10)-PE, CD80 (16-10A1)-PE, CD86 (GL1)-PE, CD8 (53-6.7)-PerCPCy5.5 and FITC, CD4 (RM4-5)-APC and eFluro450, CD135 (A2F10)-PE, CD117 (2B8)-PECy7, CD115 (AFS98)-APC, Gr-1 (RB6-8C5)-PE, CD16/32 (93)-PerCP-Cy5.5, Sca1 (D7)-FITC, CD127 (A7R34)-PECy7, CD309 (VEGFR2) (Avas12a1)-PE and biotin, Streptavidin-PE from eBioscience and SiglecF (cloneE50-2440)-AlexaFluor647 from BD Pharmingen. DAPI was used to stain dead cells.
For in vivo proliferation of pDCs, BM cells were stained for pDC markers (CD11cint B220+ PDCA1+) and permeabilized by FoxP3 fixation/permeabilization kit (eBiosciences) and stained with Ki67 (SolA15)-PE (eBiosciences). Alternatively, mice received 1 mg of BrdU by i.p. injection and sacrificed for the collection of BM cells 24 h later. BrdU staining kit for Flow Cytometry (eBiosciences) was used following manufacturer’s instructions. Apoptosis of BM pDCs in vivo was measured by AnnexinV-FITC staining (BD Pharmingen). LSR-Fortessa and LSR-II (BD) were used to acquire the samples and FlowJo® was used to analyze the data.
For purification of pDCs, single-cell suspensions were pre-enriched by negative selection with Plasmacytoid DC Isolation kit II from Miltenyi Biotec. For CMP and CDP isolation, lineage negative cells were pre-enriched with Lineage negative selection kit II from Miltenyi Biotec.
pDC adoptive transfer
pDCs were isolated from spleens and lymph nodes obtained from WT CD45.1 mice by mechanical disruption to obtain single cell suspension and using magnetic beads and columns (Plasmacytoid DC isolation and LS columns, Miltenyi) following manufacturer’s instructions. For high purity, cells were passed through 2 consecutive columns. Purity was checked by B220, PDCA1 and SiglecH staining prior to injection and purity of >94% was confirmed. Mir126−/− mice received 2.5 × 106 pDCs via the tail vein injection. VSV- HIV (6 × 106 TU/mouse) was injected into WT, Mir126−/− or Mir126−/− mice that had received WT pDCs. Blood was collected from the tail at the indicated time points.
Human DC purification
Human lung tissues were obtained from consenting patients undergoing lung resection surgery at The Mount Sinai Medical Center (New York, NY). Tissues were taken from noninvolved lung areas in patients with pulmonary resection surgeries for either pulmonary metastatic disease or primary non-small-cell lung cancer. All protocols were approved by IRB (08-1236-0001–08-1236-0005) at MSSM. Care was taken to obtain lung tissue as distant as possible from any primary lesions and anatomic abnormalities and in a way to proportionally represent subpleural (distal) and deeper (proximal) anatomic regions of lung. Human lung tissue was processed analogously to the mice protocol.
PBMCs were isolated from leukapheresis using Ficoll-Paque Plus density gradient centrifugation (StemCell Technologies). The following antibodies for human antigens were used: CD11c (B-ly6)-Pacific Blue from BD, (HLA-DR (LN3)-PE was from eBioscience and CD303 (BDCA2)-APC was from Miltenyi Biotec.
miRNA Quantification
The small RNA fraction of flow cytometry–sorted cells was extracted using the miRNAesy kit (Qiagen) according to the manufacturer’s instructions. For analysis of miRNA expression, reverse transcription (RT) was carried out on 10 ng of small RNA using the Reverse Transcription miRNA Taqman Kit (Applied Biosystems) specific for the miRNA (hsa-miR-126-3p, hsa-miR-16, and hsa-miR-21). For each cDNA sample, real-time quantitative PCR (qPCR) was performed using miRNA-specific primers/probe on an ABI Prism 7900HT Real-Time PCR System (Applied Biosystems).
For small RNA deep-sequencing (miR-seq), small RNA libraries were prepared using the TruSeq Small RNA Sample Preparation Kit (Illumina) according to the manufacturer’s instructions. The small RNA library quality was assessed on the Agilent 2100 Bioanalyzer (Agilent), and the quantity was determined using a PicroGreen assay. Each small RNA library was sequenced on an Illumina Genome Analyzer II Platform (Illumina). The sequences were mapped using the small RNA dashboard software.
mRNA expression analysis
Transcriptional profiling of Mir126−/− and control pDCs was performed similar to our previously described protocol26. Briefly, spleens were collected from mice and made into single cell suspensions. pDCs were enriched using the negative selection kit and magnetic columns (Plasmacytoid DC isolation, Miltenyi). Enriched fraction was stained with CD11c, B220, CD19, PDCA1, SiglecH, CD19, CD11b and sorted by flow cytometry in Influx Cell Sorter (BD). RNA was extracted with Trizol (Qiagen) and amplified and hybridized on the Affymetrix Mouse Gene 1.0 ST array according to the manufacturer’s instructions. RNA processing and microarray analysis with the Affymetrix MoGene 1.0 ST array was prepared according to standard operating procedures of the ImmGen Project (Harvard University, Boston, MA) and data were normalized as previously described26.
Gene expression was further analyzed by quantitative-PCR (qPCR). Total RNA was extracted from flow-sorted pDCs by using Tripure Isolation Reagent (Roche Applied Science,) and Glycogen-blue (Ambion, Life Technologies) according to the manufacturers’ instructions. For qPCR, 0.2-1 μg total RNA was reverse-transcribed for 1 h at 37 °C using RNA-to-cDNA kit (Applied Biosystems). qPCR was performed using the SYBR green qPCR master mix 2x (Fermentas, Thermo Scientific) and the following primers: Mouse Actin Fw: 5′-CTAAGGCCAACCGTGAAAAG-3′, Mouse Actin Rev: 5′-ACCAGAGGCATACAGGGACA-3′; Mouse KDR exon3 Fw: 5′-ATGCTCAGCGTGATTCTGAGG-3′, Mouse KDR exon3 Rv: 5′-ACAGTGGAGGCTATGTCGACG-3′, Human Kdr Fw: 5′-GAACATTTGGGAAATCTCTTGC-3′, Human Kdr Rev: 5′-CGGAAGAACAATGTAGTCTTTGC-3′, Human Actin Fw: 5′-GCACTCTTCCAGCCTTCC-3′, Human Actin Rev: 5′-AGAAAGGGTGTAACGCAACTAAG-3′.
Immunoblot analysis
For analysis of protein expression in pDCs, spleens and lymph nodes were collected from 2–3 Mir126−/− mice or age matched littermate controls per replicate, single cell suspensions were made, and pDCs were sorted by flow cytometry as described above to high purity. The pDCs were homogenized in protein lysis buffer after being washed with PBS. Protein lysates (~10 μg) were separated by 6% and 10% SDS-PAGE, transferred to nitrocellulose membranes, and probed with primary antibodies against TSC1 (Cell Signaling) and β-actin (Abcam). Detection was performed using horseradish peroxidase-labeled anti-goat immunoglobulin G or horseradish peroxidase-labeled anti-rabbit immunoglobulin G (DakoCytomation) and ECL Plus Western Blotting Detection Reagent (GE Healthcare Life Sciences).
Cell culture
Spleen and lymph node pDCs were freshly isolated as above by magnetic negative bead-selection and checked for purity (>85%). 2.5 ×104 cells were seeded in round bottom 96-well plates in complete RPMI 1640 media and treated with 5 μM of CpG-A (ODN 2216) for 12 h. Bone marrow cells were obtained by flushing tibiae and femurs with media, lysing RBCs, and plating the remaining cells in RPMI 1640 media supplemented with 150 ng/ml of Flt3L (Preprotech). Flt3L-supplemented media was added at day 3 of culture. CpG-A (ODN 1585), or R848 were added at the indicated concentration at day 6 and cells and media were collected for analysis after overnight incubation. IFN-α concentrations were determined in supernatant by ELISA (Thermo Scientific). When relevant, the values were adjusted for cell number per well. Proliferation of pDCs in Flt3L cultures was determined by dilution of eFluor670 dye. Apoptosis was quantified by Annexin V-FITC staining.
Luciferase assays
Predicted binding sequences for miR-126 were identified by TargetScan6.1. To study the regulation of the putative target Tsc1 by miR-126, 1.5 × 105 HEK293Tcells were cotransfected using Lipofectamin 2000 (Invitrogen) with vectors encoding luciferase with either the 3′UTR Tsc1 or a mock 3′UTR (Arntl) and a plasmid driving expression miR-126 or a control plasmid. For normalization, a plasmid encoding Renilla was included in the transfection in each condition. Luciferase activity was detected with the Dual luciferase reporter kit (Promega) 48 h after transfection.
Statistical analysis
Differences between groups were compared by Student’s t-test. A P value < 0.05 was considered statistically significant. Pathway analysis was performed by Ingenuity® by using Fisher’s exact test, Benjamini-Hochberg correction for multiple testing.
Supplementary Material
Acknowledgments
We would like to thank C.J. Kuo (Stanford University) for the Mir126−/− mice. We would also like to thank P. Sathe for helpful discussions and J. Ochando for reading the manuscript and the Flow Cytometry Core and the Mouse Genetics and Mouse Targeting facility for technical assistance. B.D.B. is supported by NIH grants DP2DK083052 and 1R01AI104848. M.M. is supported by NIH grants CA154947A, AI10008, and AI089987. J.A. is supported by a Beatriu de Pinós postdoctoral fellowship, the Robin Neustein Award, and a Juvenile Diabetes Research Foundation (JDRF) postdoctoral fellowship.
Footnotes
Accession codes.
Microarray data have been deposited in the Gene Expression Omnibus (GSE51255)
Author contributions
J.A. designed and performed research and analyzed data. A.R., N.T., H.S., M.L., D.H., C.B., L.A.G., A.B., performed research. M.M. designed the project and analyzed data. B.D.B. designed and coordinated the project and analyzed data.
Competing financial interests
The authors declare no competing financial interests.
References
- 1.McCartney SA, Colonna M. Viral sensors: diversity in pathogen recognition. Immunol. Rev. 2009;227:87–94. doi: 10.1111/j.1600-065X.2008.00726.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kawai T, Akira S. Toll-like receptors and their crosstalk with other innate receptors in infection and immunity. Immunity. 2011;34:637–650. doi: 10.1016/j.immuni.2011.05.006. [DOI] [PubMed] [Google Scholar]
- 3.Chevrier N, et al. Systematic discovery of TLR signaling components delineates viral-sensing circuits. Cell. 2011;147:853–867. doi: 10.1016/j.cell.2011.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yang K, et al. Human TLR-7-, -8-, and -9-mediated induction of IFN-alpha/beta and -lambda Is IRAK-4 dependent and redundant for protective immunity to viruses. Immunity. 2005;23:465–478. doi: 10.1016/j.immuni.2005.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kumar H, Kawai T, Akira S. Toll-like receptors and innate immunity. Biochem. Biophys. Res. Commun. 2009;388:621–625. doi: 10.1016/j.bbrc.2009.08.062. [DOI] [PubMed] [Google Scholar]
- 6.Baltimore D, et al. MicroRNAs: new regulators of immune cell development and function. Nat. Immunol. 2008;9:839–845. doi: 10.1038/ni.f.209. [DOI] [PubMed] [Google Scholar]
- 7.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Taganov KD, Boldin MP, Chang K-J, Baltimore D. NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc. Natl. Acad. Sci. U.S.A. 2006;103:12481–12486. doi: 10.1073/pnas.0605298103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.O’Connell RM, et al. MicroRNA-155 is induced during the macrophage inflammatory response. Proc. Natl. Acad. Sci. U.S.A. 2007;104:1604–1609. doi: 10.1073/pnas.0610731104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brown BD, et al. Endogenous microRNA can be broadly exploited to regulate transgene expression according to tissue, lineage and differentiation state. Nat. Biotechnol. 2007;25:1457–1467. doi: 10.1038/nbt1372. [DOI] [PubMed] [Google Scholar]
- 11.O’Connell RM, Chaudhuri AA, Rao DS, Baltimore D. Inositol phosphatase SHIP1 is a primary target of miR-155. Proc. Natl. Acad. Sci. U.S.A. 2009;106:7113–7118. doi: 10.1073/pnas.0902636106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhou H, et al. miR-155 and its star-form partner miR-155* cooperatively regulate type I interferon production by human plasmacytoid dendritic cells. Blood. 2010;116:5885–5894. doi: 10.1182/blood-2010-04-280156. [DOI] [PubMed] [Google Scholar]
- 13.O’Neill LA, Sheedy FJ, McCoy CE. MicroRNAs: the fine-tuners of Toll-like receptor signalling. Nat. Rev. Immunol. 2011;11:163–175. doi: 10.1038/nri2957. [DOI] [PubMed] [Google Scholar]
- 14.Fish JE, et al. miR-126 regulates angiogenic signaling and vascular integrity. Dev. Cell. 2008;15:272–284. doi: 10.1016/j.devcel.2008.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kuhnert F, et al. Attribution of vascular phenotypes of the murine Egfl7 locus to the microRNA miR-126. Development. 2008;135:3989–3993. doi: 10.1242/dev.029736. [DOI] [PubMed] [Google Scholar]
- 16.Wang S, et al. The endothelial-specific microRNA miR-126 governs vascular integrity and angiogenesis. Dev. Cell. 2008;15:261–271. doi: 10.1016/j.devcel.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brown BD, et al. In vivo administration of lentiviral vectors triggers a type I interferon response that restricts hepatocyte gene transfer and promotes vector clearance. Blood. 2007;109:2797–2805. doi: 10.1182/blood-2006-10-049312. [DOI] [PubMed] [Google Scholar]
- 18.Beignon A-S, et al. Endocytosis of HIV-1 activates plasmacytoid dendritic cells via Toll-like receptor-viral RNA interactions. J. Clin. Invest. 2005;115:3265–3275. doi: 10.1172/JCI26032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Meylan PR, Guatelli JC, Munis JR, Richman DD, Kornbluth RS. Mechanisms for the inhibition of HIV replication by interferons-alpha, -beta, and -gamma in primary human macrophages. Virology. 1993;193:138–148. doi: 10.1006/viro.1993.1110. [DOI] [PubMed] [Google Scholar]
- 20.Reizis B, Bunin A, Ghosh HS, Lewis KL, Sisirak V. Plasmacytoid dendritic cells: recent progress and open questions. Annu. Rev. Immunol. 2011;29:163–183. doi: 10.1146/annurev-immunol-031210-101345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Colonna M, Trinchieri G, Liu Y-J. Plasmacytoid dendritic cells in immunity. Nat. Immunol. 2004;5:1219–1226. doi: 10.1038/ni1141. [DOI] [PubMed] [Google Scholar]
- 22.Naik SH, et al. Development of plasmacytoid and conventional dendritic cell subtypes from single precursor cells derived in vitro and in vivo. Nat. Immunol. 2007;8:1217–1226. doi: 10.1038/ni1522. [DOI] [PubMed] [Google Scholar]
- 23.Cella M, et al. Plasmacytoid monocytes migrate to inflamed lymph nodes and produce large amounts of type I interferon. Nat. Med. 1999;5:919–923. doi: 10.1038/11360. [DOI] [PubMed] [Google Scholar]
- 24.Asselin-Paturel C, et al. Type I interferon dependence of plasmacytoid dendritic cell activation and migration. J. Exp. Med. 2005;201:1157–1167. doi: 10.1084/jem.20041930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vitour D, Meurs EF. Regulation of interferon production by RIG-I and LGP2: a lesson in self-control. Sci. STKE. 2007;2007:pe20. doi: 10.1126/stke.3842007pe20. [DOI] [PubMed] [Google Scholar]
- 26.Miller JC, et al. Deciphering the transcriptional network of the dendritic cell lineage. Nat. Immunol. 2012;13:888–899. doi: 10.1038/ni.2370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Krug A, et al. IFN-producing cells respond to CXCR3 ligands in the presence of CXCL12 and secrete inflammatory chemokines upon activation. J. Immunol. 2002;169:6079–6083. doi: 10.4049/jimmunol.169.11.6079. [DOI] [PubMed] [Google Scholar]
- 28.Li HS, et al. The signal transducers STAT5 and STAT3 control expression of Id2 and E2-2 during dendritic cell development. Blood. 2012;120:4363–4373. doi: 10.1182/blood-2012-07-441311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van de Laar L, et al. PI3K-PKB hyperactivation augments human plasmacytoid dendritic cell development and function. Blood. 2012;120:4982–4991. doi: 10.1182/blood-2012-02-413229. [DOI] [PubMed] [Google Scholar]
- 30.Cao W, et al. Toll-like receptor-mediated induction of type I interferon in plasmacytoid dendritic cells requires the rapamycin-sensitive PI(3)K-mTOR-p70S6K pathway. Nat. Immunol. 2008;9:1157–1164. doi: 10.1038/ni.1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Quinn TP, Peters KG, De Vries C, Ferrara N, Williams LT. Fetal liver kinase 1 is a receptor for vascular endothelial growth factor and is selectively expressed in vascular endothelium. Proc. Natl. Acad. Sci. U.S.A. 1993;90:7533–7537. doi: 10.1073/pnas.90.16.7533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Carmeliet P, Jain RK. Molecular mechanisms and clinical applications of angiogenesis. Nature. 2011;473:298–307. doi: 10.1038/nature10144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ding B-S, et al. Inductive angiocrine signals from sinusoidal endothelium are required for liver regeneration. Nature. 2010;468:310–315. doi: 10.1038/nature09493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ghosh HS, Cisse B, Bunin A, Lewis KL, Reizis B. Continuous expression of the transcription factor e2-2 maintains the cell fate of mature plasmacytoid dendritic cells. Immunity. 2010;33:905–916. doi: 10.1016/j.immuni.2010.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Grouard G, et al. The enigmatic plasmacytoid T cells develop into dendritic cells with interleukin (IL)-3 and CD40-ligand. J. Exp. Med. 1997;185:1101–1111. doi: 10.1084/jem.185.6.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Siegal FP, et al. The nature of the principal type 1 interferon-producing cells in human blood. Science. 1999;284:1835–1837. doi: 10.1126/science.284.5421.1835. [DOI] [PubMed] [Google Scholar]
- 37.Stepkowski SM, Chen W, Ross JA, Nagy ZS, Kirken RA. STAT3: an important regulator of multiple cytokine functions. Transplantation. 2008;85:1372–1377. doi: 10.1097/TP.0b013e3181739d25. [DOI] [PubMed] [Google Scholar]
- 38.Plas DR, Thomas G. Tubers and tumors: rapamycin therapy for benign and malignant tumors. Curr. Opin. Cell Biol. 2009;21:230–236. doi: 10.1016/j.ceb.2008.12.013. [DOI] [PubMed] [Google Scholar]
- 39.Guiducci C, et al. PI3K is critical for the nuclear translocation of IRF-7 and type I IFN production by human plasmacytoid predendritic cells in response to TLR activation. Journal of Experimental Medicine. 2008;205:315–322. doi: 10.1084/jem.20070763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Conrad C, et al. Plasmacytoid dendritic cells promote immunosuppression in ovarian cancer via ICOS costimulation of Foxp3(+) T-regulatory cells. Cancer Res. 2012;72:5240–5249. doi: 10.1158/0008-5472.CAN-12-2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ferrara N, Mass RD, Campa C, Kim R. Targeting VEGF-A to treat cancer and age-related macular degeneration. Annu. Rev. Med. 2007;58:491–504. doi: 10.1146/annurev.med.58.061705.145635. [DOI] [PubMed] [Google Scholar]
- 42.Nestle FO, et al. Plasmacytoid predendritic cells initiate psoriasis through interferon-alpha production. J. Exp. Med. 2005;202:135–143. doi: 10.1084/jem.20050500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lande R, et al. Plasmacytoid dendritic cells sense self-DNA coupled with antimicrobial peptide. Nature. 2007;449:564–569. doi: 10.1038/nature06116. [DOI] [PubMed] [Google Scholar]
- 44.Guiducci C, et al. Autoimmune skin inflammation is dependent on plasmacytoid dendritic cell activation by nucleic acids via TLR7 and TLR9. Journal of Experimental Medicine. 2010;207:2931–2942. doi: 10.1084/jem.20101048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Diana J, et al. Crosstalk between neutrophils, B-1a cells and plasmacytoid dendritic cells initiates autoimmune diabetes. Nat. Med. 2013;19:65–73. doi: 10.1038/nm.3042. [DOI] [PubMed] [Google Scholar]
- 46.Banchereau J, Pascual V. Type I interferon in systemic lupus erythematosus and other autoimmune diseases. Immunity. 2006;25:383–392. doi: 10.1016/j.immuni.2006.08.010. [DOI] [PubMed] [Google Scholar]
- 47.Wang H, Peng W, Ouyang X, Li W, Dai Y. Circulating microRNAs as candidate biomarkers in patients with systemic lupus erythematosus. Transl Res. 2012;160:198–206. doi: 10.1016/j.trsl.2012.04.002. [DOI] [PubMed] [Google Scholar]
- 48.Janssen HLA, et al. Treatment of HCV Infection by Targeting MicroRNA. N. Engl. J. Med. 2013 doi: 10.1056/NEJMoa1209026. doi:10.1056/NEJMoa1209026. [DOI] [PubMed] [Google Scholar]
- 49.O’Connell RM, et al. MicroRNA-155 promotes autoimmune inflammation by enhancing inflammatory T cell development. Immunity. 2010;33:607–619. doi: 10.1016/j.immuni.2010.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Agudo J, et al. A TLR and non-TLR mediated innate response to lentiviruses restricts hepatocyte entry and can be ameliorated by pharmacological blockade. Mol. Ther. 2012;20:2257–2267. doi: 10.1038/mt.2012.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.